Monogalactosyldiacylglycerol is required for membrane-associated processes of the protochlorophyllide synthesis pathway and the formation of protochlorophyllide-enzyme complexes in Arabidopsis etioplasts.
Abstract
Cotyledon cells of dark-germinated angiosperms develop etioplasts that are plastids containing unique internal membranes called prolamellar bodies (PLBs). Protochlorophyllide (Pchlide), a precursor of chlorophyll, accumulates in PLBs and forms a ternary complex with NADPH and light-dependent NADPH:protochlorophyllide oxidoreductase (LPOR), which allows for the rapid formation of chlorophyll after illumination while avoiding photodamage. PLBs are 3D lattice structures formed by the lipid bilayer rich in monogalactosyldiacylglycerol (MGDG). Although MGDG was found to be required for the formation and function of the thylakoid membrane in chloroplasts in various plants, the roles of MGDG in PLB formation and etioplast development are largely unknown. To analyze the roles of MGDG in etioplast development, we suppressed MGD1 encoding the major isoform of MGDG synthase by using a dexamethasone-inducible artificial microRNA in etiolated Arabidopsis (Arabidopsis thaliana) seedlings. Strong MGD1 suppression caused a 36% loss of MGDG in etiolated seedlings, together with a 41% decrease in total Pchlide content. The loss of MGDG perturbed etioplast membrane structures and impaired the formation of the photoactive Pchlide-LPOR-NADPH complex and its oligomerization, without affecting LPOR accumulation. The MGD1 suppression also impaired the formation of Pchlide from protoporphyrin IX via multiple enzymatic reactions in etioplast membranes, which suggests that MGDG is required for the membrane-associated processes in the Pchlide biosynthesis pathway. Suppressing MGD1 at several germination stages revealed that MGDG biosynthesis at an early germination stage is particularly important for Pchlide accumulation. MGDG biosynthesis may provide a lipid matrix for Pchlide biosynthesis and the formation of Pchlide-LPOR complexes as an initial step of etioplast development.
Angiosperms germinating in darkness develop etioplasts instead of chloroplasts in cotyledon cells (Solymosi and Schoefs, 2010). Etioplasts have unique internal membrane structures called prolamellar bodies (PLBs), 3D lattice structures of membrane tubules, in addition to lamellar prothylakoid (PT) membranes (Gunning, 1965; Kowalewska et al., 2016). Etioplasts do not contain chlorophyll (Chl) but rather accumulate small amounts of a Chl precursor, protochlorophyllide (Pchlide; Masuda, 2008). The accumulation of Pchlide in the dark may be an advantage for forming Chl rapidly after light exposure. However, as for Chl and other Chl intermediates, Pchlide is a photosensitizer, and excess accumulation of Pchlide causes photodamage and cell death with light irradiation (Triantaphylidès and Havaux, 2009). Thus, the amount of Pchlide in the dark is strictly controlled by multiple regulatory mechanisms of the biosynthetic pathway (Tanaka et al., 2011). In addition, plants develop a photoprotective system in etioplasts to avoid oxidative damage from photoreactive Pchlide (Solymosi and Schoefs, 2010).
In PLBs, a major portion of Pchlide forms the ternary complex with light-dependent NADPH:protochlorophyllide oxidoreductase (LPOR) and NADPH, which further aggregates into large oligomers (Schoefs and Franck, 2003). LPOR is the membrane-associated enzyme that reduces Pchlide to chlorophyllide (Chlide), the immediate precursor of Chl, by using NADPH and light energy (Heyes and Hunter, 2005). The photoactive form of Pchlide bound by LPOR at the active site is converted instantaneously to Chlide in the light without generating singlet oxygen, whereas nonphotoactive Pchlide, which is not bound to the LPOR active site, easily generates singlet oxygen and causes photobleaching with light (op den Camp et al., 2003). After the photoconversion of Pchlide, LPOR also functions in preventing photodamage from Chlide by forming Chlide-LPOR-NADPH ternary complexes (Solymosi and Schoefs, 2010). Thus, the formation of pigment-LPOR complexes in PLBs is essential for the rapid and safe conversion of etioplasts to chloroplasts during the dark-to-light transition.
The tetrapyrrole biosynthetic pathway in plants has been well characterized as described in comprehensive reviews (Beale, 1999; Moulin and Smith, 2005; Masuda and Fujita, 2008; Tanaka et al., 2011; Brzezowski et al., 2015) and is briefly summarized as follows. The biosynthesis of all tetrapyrroles including Pchlide starts from the formation of 5-aminolevulinic acid (ALA) in plastids. ALA biosynthesis is the rate-limiting process of the tetrapyrrole biosynthesis pathway, with glutamyl-tRNA reductase (GluTR) subjected to strict transcriptional and posttranslational regulation as a key enzyme of this step. Subsequently, a cascade of enzymatic reactions takes place to form protoporphyrin IX (Proto IX), the last common precursor shared by the Chl and heme biosynthesis pathways. Insertion of Mg2+ into Proto IX by Mg-chelatase (MgCh) yields Mg-Proto IX for Chl biosynthesis, whereas insertion of Fe2+ by ferrochelatase results in heme b in one step. In the Chl biosynthesis pathway, S-adenosyl-l-methionine:Mg-Proto IX methyltransferase (MgMT) esterifies Mg-Proto IX into Mg-Proto IX monomethylester (Mg-Proto IX ME), which is further metabolized to Pchlide by Mg-Proto IX ME cyclase (MgCY). Then, Pchlide is converted to Chlide by Pchlide reductase. Angiosperms possess only LPOR, which absolutely requires light for catalysis, whereas cyanobacteria, algae, and most other plants have the dark-operative (light-independent) type of Pchlide reductase in addition to LPOR. Therefore, in angiosperms, the Chl biosynthesis pathway is paused at Pchlide in the dark; only after light exposure does LPOR reduce Pchlide to Chlide. In addition, 3,8-divinyl Pchlide a 8-vinyl reductase, which reduces the 8-vinyl group on the B pyrrole ring, is involved in Chlide formation. Chlide is subsequently esterified with a phytol chain by Chl synthase to result in Chl.
PLB is a lipid bilayer membrane structure rich in glycerolipids relative to proteins and pigments (Selstam and Sandelius, 1984; Williams et al., 1998). The lipid composition of the PLB membrane is similar to that of the thylakoid membrane in chloroplasts, with two galactolipids, monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG), accounting for ∼50% and ∼30% of total lipids, respectively, in both membranes (Selstam and Sandelius, 1984; Dorne et al., 1990). These galactolipids also are predominant lipid constituents of the PTs and the envelopes of etioplasts. In plants, MGDG is synthesized in plastid envelops in a one-step reaction by MGDG synthase, which transfers the Gal moiety from UDP-Gal to diacylglycerol (Benning and Ohta, 2005). DGDG is synthesized from MGDG, so MGDG biosynthesis also is essential for DGDG biosynthesis. Three isoforms of MGDG synthase, namely MGD1, MGD2, and MGD3, have been identified in Arabidopsis (Arabidopsis thaliana); inner envelope-localized MGD1 is responsible for most of the MGDG biosynthesis for thylakoid biogenesis, whereas outer envelope-localized MGD2 and MGD3 mainly provide MGDG for DGDG biosynthesis specifically under phosphate-starved conditions (Kobayashi et al., 2009b).
The requirement of MGDG biosynthesis for chloroplast development has been demonstrated by analyses of MGD1 mutants. Partial deficiency of MGDG by knockdown mutations in MGD1 decreased the amount of the thylakoid membrane, Chl content, and photosynthetic activity in Arabidopsis (Jarvis et al., 2000; Fujii et al., 2014) and tobacco (Nicotiana tabacum; Wu et al., 2013). A knockout mutation of MGD1 (mgd1-2) in Arabidopsis, which resulted in severe loss of both galactolipids, strongly impaired thylakoid membrane development and completely abolished photosynthetic activity (Kobayashi et al., 2007, 2013). MGDG biosynthesis also is required for the coordinated expression of nucleus- and plastid-encoded photosynthesis-associated genes responsible for chloroplast development (Kobayashi et al., 2013; Fujii et al., 2014). Thus, MGDG biosynthesis is one of the determinant steps of chloroplast biogenesis in plants.
Considering the abundance of galactolipids in PLBs, MGDG biosynthesis also may play a crucial role in etioplast development in the dark. In fact, MGDG is involved in making PLB-like cubic structures in vitro (Brentel et al., 1985), and the interaction between LPOR and MGDG has been hypothesized to contribute to PLB formation (Klement et al., 1999; Engdahl et al., 2001; Selstam et al., 2002). Very recently, Gabruk et al. (2017) demonstrated that MGDG promotes the oligomerization of the Pchlide-LPOR complexes in vitro. However, mutant analyses so far have not provided conclusive information on the role of MGDG in PLB formation and etioplast development. Double knockout mutations of MGD2 and MGD3 did not affect galactolipid content in etiolated seedlings, which suggests that the remaining MGD1 is the main isoform responsible for galactolipid biosynthesis in etioplasts as in chloroplasts (Kobayashi et al., 2009a). However, Jarvis et al. (2000) reported that the MGD1 knockdown mutant (mgd1-1), with 42% reduced MGDG content in light-grown leaves, showed no noticeable defects in etioplast development. Meanwhile, the knockout mgd1-2 mutant cannot be used for the analysis of etioplast development because it does not develop cotyledons, owing to severe inhibition of embryogenesis (Kobayashi et al., 2007).
Recently, we generated Arabidopsis transgenic lines carrying a dexamethasone (DEX)-inducible artificial microRNA targeting MGD1 (amiR-MGD1; Fujii et al., 2014). In the previous study, these lines showed up to 75% reduction in MGD1 expression in a DEX-dependent manner, which resulted in up to 90% reduction in MGDG content in light-grown seedlings as compared with the DEX-free (−DEX) control. In amiR-MGD1 lines, we could eliminate the effect of the MGD1 deficiency on embryo development by harvesting seeds under −DEX conditions. Taking advantage of the inducible knockdown system of amiR-MGD1 lines, we investigated the roles of MGD1 in galactolipid biosynthesis, Pchlide biosynthesis, the formation of photoactive Pchlide-LPOR-NADPH complexes, and the development of PLBs during etiolated seedling growth.
RESULTS
Screening of Phenotypically Homogenous Lines of amiR-MGD1
We previously reported that the T3 generation of amiR-MGD1 lines grown under DEX-treated (+DEX) conditions showed various color phenotypes from green to white in cotyledons even within a single homozygous line, presumably due to fluctuations of the suppression levels of MGD1 expression (Fujii et al., 2014). Because the heterogeneity in a single amiR-MGD1 line prevented detailed analyses of etiolated seedlings, we attempted to isolate phenotypically homogenous amiR-MGD1 lines. Screening of T4 generations of the amiR-MGD1 line 4 (L4; Fujii et al., 2014) under +DEX conditions identified several lines showing homogenous cotyledon phenotypes (Supplemental Fig. S1A). In L4-01, L4-03, L4-04, L4-07, L4-09, and L4-11 lines, all seedlings had albino cotyledons under +DEX conditions and Chl content decreased to 10% of the −DEX control (Supplemental Fig. S1B). By contrast, in L4-02, L4-05, L4-06, L4-08, and L4-10, all seedlings had green cotyledons regardless of DEX treatment. In these green lines, Chl content was decreased only slightly with DEX treatment (Supplemental Fig. S1B). Phenotypically homogenous lines also were obtained from the amiR-MGD1 L2 line (Supplemental Fig. S1C), so these phenomena are not specific to the L4 line.
To examine the relationship between cotyledon phenotypes and suppression levels of MGD1 expression, we determined MGD1 mRNA levels in 5-d-old seedlings of homogenous L4 lines (Supplemental Fig. S1D). In DEX-induced albino lines (L4-01, L4-03, and L4-04), MGD1 mRNA levels were decreased to 20% or less, whereas those in green lines (L4-02, L4-05, and L4-06) ranged from ∼35% to ∼50% as compared with the −DEX control. These data are consistent with a previous report showing that reducing MGD1 expression to ∼25% but not to ∼40% of wild-type levels caused an albino cotyledon phenotype (Fujii et al., 2014). We conclude that L4-01, L4-03, L4-04, L4-07, L4-09, and L4-11 were homogenous lines that strongly suppressed MGD1 expression in a DEX-dependent manner. Because these six lines, originally derived from a single T2 plant, showed almost the same phenotype in MGD1 expression, Chl content, and seedling growth under +DEX conditions, we used a mixture of these lines, called L4w, for further analyses. We also used a mixture of L4-02, L4-05, L4-06, L4-08, and L4-10, called L4g, showing the homogenous green cotyledon phenotype under +DEX conditions. Likewise, we used L2-01, called L2w, and a mixture of L2-03 and L2-04, called L2g, for homogenous L2 lines with white and green cotyledons, respectively, under +DEX conditions.
MGD1 Suppression Decreases MGDG Content in Etiolated Seedlings
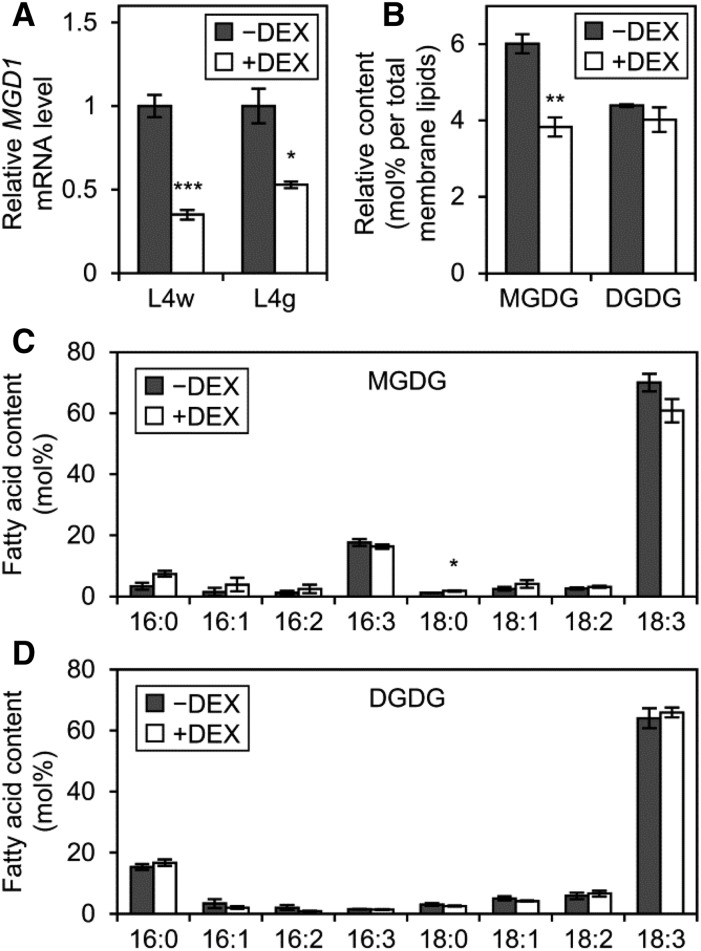
To reveal the roles of MGD1 during etioplast development, we investigated etiolated amiR-MGD1 seedlings under −DEX and +DEX conditions. In 4-d-old L4w and L4g seedlings grown under +DEX conditions in the dark, MGD1 mRNA levels were decreased to 35% and 53%, respectively, of the −DEX control (Fig. 1A). This result confirms the stronger MGD1 suppression in L4w than L4g even in etiolated seedlings. A similar result was observed in L2 seedlings (Supplemental Fig. S2A). To assess whether the MGD1 suppression affected galactolipid biosynthesis in etiolated seedlings, we analyzed galactolipid content in etiolated L4w seedlings (Fig. 1B). In L4w seedlings, the proportion of MGDG in total membrane lipids was decreased from 6 mol % under −DEX conditions to 3.8 mol % under +DEX conditions. By contrast, the proportion of DGDG did not differ noticeably between +DEX and −DEX seedlings, so the MGDG-to-DGDG ratio decreased from 1.37 in the −DEX control to 0.96 in +DEX seedlings. For both MGDG (Fig. 1C) and DGDG (Fig. 1D), fatty acid compositions were not greatly altered by DEX treatment. The DEX-dependent decrease in relative MGDG content without altered DGDG content in etiolated L4w seedlings allowed us to investigate the specific effects of the MGDG deficiency in etioplast development.
Figure 1.
Effect of MGD1 suppression on galactolipid biosynthesis in etiolated seedlings. A, Quantitative reverse transcription-PCR analysis of MGD1 mRNA levels in 4-d-old etiolated seedlings of amiR-MGD1 under +DEX and −DEX conditions. Data are presented as fold difference from the −DEX control after normalizing to the control gene ACTIN8. Data are means ± se from 13 (L4w) or three (L4g) independent experiments. B, Accumulation of MGDG and DGDG in 4-d-old etiolated seedlings of amiR-MGD1 L4w. C and D, Fatty acid composition of MGDG (C) and DGDG (D) in 4-d-old etiolated seedlings of amiR-MGD1 L4w. In B to D, data are means ± se from three independent experiments. Asterisks indicate significant differences from the −DEX control (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, Student’s t test).
MGD1 Suppression Impairs Pchlide Accumulation in Etiolated Seedlings
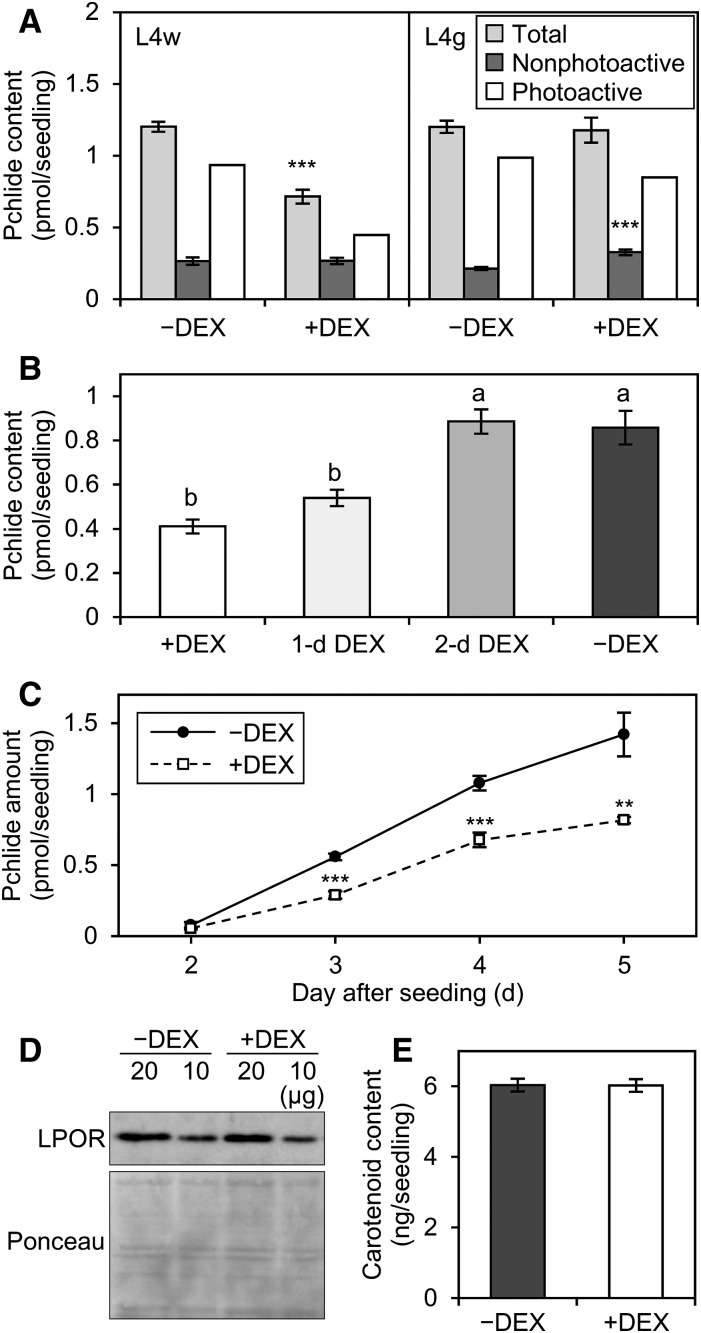
To reveal the contribution of MGDG to Pchlide accumulation, we quantified the total amount of Pchlide in amiR-MGD1 seedlings (Fig. 2A). DEX treatment from the beginning of germination decreased total Pchlide content in etiolated L4w seedlings to 59% of the −DEX control without affecting the size of cotyledons (Supplemental Fig. S3). By contrast, total Pchlide content in L4g seedlings was not changed by DEX treatment (Fig. 2A). Similar data were obtained from L2 lines (Supplemental Fig. S2, B and C). Thus, similar to Chl content, Pchlide content was affected only when MGD1 expression was strongly suppressed. The data also show that DEX treatment itself had no effect on Pchlide accumulation. To examine when MGDG biosynthesis was required for Pchlide accumulation, we delayed the start time of DEX treatment after seeding and determined Pchlide content in etiolated 4-d-old L4w seedlings (Fig. 2B). DEX treatment from 1 d after seeding decreased Pchlide accumulation in L4w seedlings but to a slightly smaller extent than with DEX treatment from the beginning. By contrast, DEX treatment from 2 d after seeding did not decrease Pchlide content. These data suggest that the MGD1 expression at the very early stage of germination is particularly important for Pchlide accumulation.
Figure 2.
Contribution of MGDG to the accumulation of Pchlide and carotenoids in etiolated seedlings. A, Pchlide content in 4-d-old etiolated seedlings of amiR-MGD1 grown under −DEX and +DEX conditions. Total and nonphotoactive Pchlide were extracted before and after flash treatment, respectively. Data are means ± se from 12 (L4w) or 15 (L4g) independent experiments. The amount of photoactive Pchlide was estimated by subtracting the amount of nonphotoactive Pchlide from total Pchlide. B, Pchlide content in 4-d-old etiolated L4w seedlings treated with DEX at different times after seeding. L4w plants were treated with DEX from the beginning of seeding (+DEX), 1 d after seeding (1-d DEX), or 2 d after seeding (2-d DEX). Seedlings grown in the absence of DEX were analyzed as the untreated control (−DEX). Data are means ± se from seven to 12 independent experiments. Different letters indicate significant differences (P < 0.05, Tukey-Kramer multiple comparison test). C, Pchlide accumulation in etiolated amiR-MGD1 L4w seedlings grown for 2 to 5 d under −DEX and +DEX conditions. Data are means ± se from six to 12 independent experiments. D, Immunoblot analysis of total LPOR proteins (∼37 kD) in 4-d-old etiolated seedlings of amiR-MGD1 L4w. As a loading control, Ponceau-stained proteins between ∼25 and ∼50 kD blotted onto a membrane are shown. Representative data from three biologically independent experiments are shown. E, Total carotenoid content in 4-d-old etiolated seedlings of amiR-MGD1 L4w. Data are means ± se from eight independent experiments. In A, C, and E, asterisks indicate significant differences from the −DEX control (**, P < 0.01; and ***, P < 0.001, Student’s t test).
Pchlide in etiolated seedlings is distinguished as photoactive and nonphotoactive forms; only photoactive Pchlide can be converted immediately into Chlide by a short light treatment (Schoefs, 2001). To assess which type of Pchlide was decreased by MGDG deficiency in etiolated L4w seedlings, we determined Pchlide content after a 0.7-ms light flash, which represented the amount of nonphotoactive Pchlide (Fig. 2A). The amount of nonphotoactive Pchlide was not changed by DEX treatment, so the proportion of photoactive to total Pchlide was decreased from 78% in −DEX seedlings to 63% in +DEX seedlings. A similar result in L2w (Supplemental Fig. S2B) supports that MGD1 suppression mainly decreases photoactive Pchlide content in the dark. Meanwhile, etiolated L4g seedlings showed a slight increase in nonphotoactive Pchlide content (Fig. 2A).
We also performed a time-course analysis of Pchlide accumulation during etioplast development in L4w seedlings (Fig. 2C). In 2-d-old etiolated seedlings, Pchlide content was very low under both +DEX and −DEX conditions. However, in −DEX seedlings, Pchlide content was increased sharply until 5 d after seeding. Although Pchlide content also was increased gradually in +DEX seedlings, the rate was much lower than in the −DEX control. Because photoactive Pchlide is associated with LPOR proteins, we examined total LPOR content in 4-d-old etiolated L4w seedlings by using polyclonal anti-LPOR antibodies that recognize all Arabidopsis LPOR isoforms (Rowe and Griffiths, 1995; Masuda et al., 2003). Immunoblot analysis showed that the MGD1 suppression by DEX treatment did not noticeably change LPOR levels in etiolated seedlings (Fig. 2D).
According to Moro et al. (2004), the inhibition of carotenoid biosynthesis in etiolated seedlings impairs the accumulation of photoactive Pchlide and the formation of PLBs without altering LPOR protein levels. To investigate whether MGDG deficiency affects carotenoid biosynthesis in etiolated seedlings, we measured total carotenoid content in 4-d-old etiolated L4w seedlings (Fig. 2E). Carotenoid content was not affected by DEX treatment in etiolated seedlings. In addition, absorption spectra between ∼420 and ∼490 nm, mostly derived from a composite of carotenoids in etiolated seedlings (Böddi et al., 1989), were similar in +DEX and −DEX seedlings (Supplemental Fig. S4), so the carotenoid composition also may be unchanged by the MGD1 suppression.
Although our data from the amiR-MGD1 lines suggest that MGD1 is required for the accumulation of photoactive Pchlide, Aronsson et al. (2008) previously reported that the ratio of photoactive to nonphotoactive Pchlide was increased in the etiolated mgd1-1 mutant. Thus, we investigated the amount of total and nonphotoactive Pchlide in etiolated seedlings of mgd1-1 and corresponding wild-type (Columbia) in the same experimental condition as amiR-MGD1 (Supplemental Fig. S2D). In our experiments, the mgd1-1 mutation decreased total Pchlide content in etiolated seedlings without affecting the ratio of photoactive to nonphotoactive Pchlide. The discrepancy between our results and those by Aronsson et al. (2008) might be due to some differences in experimental conditions; for example, Aronsson et al. (2008) grew seedlings on soil for 5 d, whereas we grew them on agar-solidified medium containing 1% Suc for 4 d.
MGD1 Suppression Impairs the Formation of the Photoactive Pchlide-LPOR-NADPH Complex and Its Oligomerization
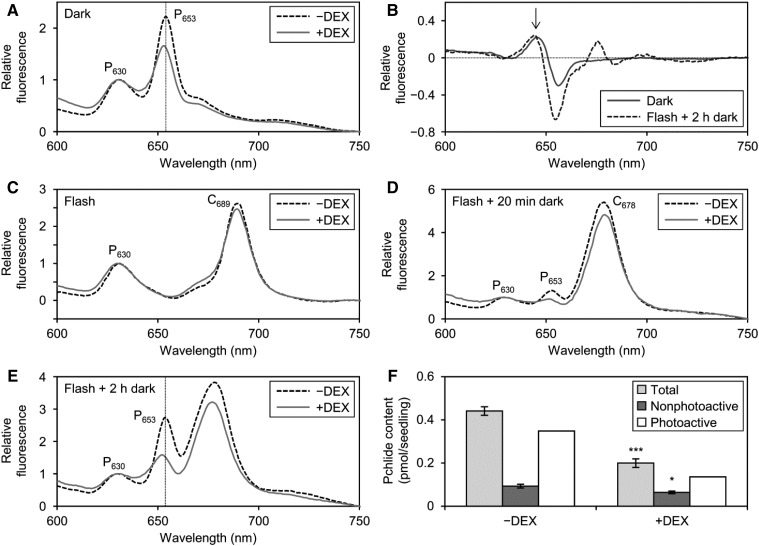
To evaluate the role of MGDG in the formation of the Pchlide-LPOR-NADPH ternary complex in PLBs, we measured the fluorescence spectra of Pchlide at 77K in etiolated amiR-MGD1 L4w seedlings (Fig. 3A). Photoactive and nonphotoactive forms of Pchlide can be optically distinguished with their fluorescence peaks at about 655 and 633 nm, respectively, under 77K (Schoefs, 2001; Solymosi et al., 2007). Furthermore, photoactive Pchlide includes two forms, one emitting fluorescence at ∼645 nm at 77K in the dimeric ternary complex, which is minor in typical etioplasts in several plants, and the other, the major form in PLBs, emitting fluorescence at ∼657 nm in oligomeric aggregates of the complex (Böddi et al., 1989; Schoefs, 2001). In our experimental conditions, we observed two fluorescence bands peaking at about 630 nm (P630) and 653 nm (P653) in the −DEX control. In +DEX seedlings, the peak position of P653 but not P630 was slightly blue shifted as compared with that in the −DEX control (Fig. 3A; Table I). A difference spectrum between +DEX and −DEX samples revealed increased fluorescence at ∼645 nm with decreased fluorescence at ∼655 nm in +DEX seedlings relative to the −DEX control (Fig. 3B). These data suggest an increase in the dimeric ternary complex and a decrease in large aggregates with DEX treatment. The blue shift of P653 was not found in etiolated seedlings of +DEX L4g (Table I; Supplemental Fig. S5A) and mgd1-1 (Supplemental Fig. S5, B and C), confirming that the spectral change observed in L4w was caused by strong MGD1 suppression.
Figure 3.
Role of MGDG in the formation of the Pchlide-LPOR-NADPH complex and processes after illumination. A, C, D, and E, In situ 77K fluorescence spectra under 440-nm excitation in etiolated cotyledons of amiR-MGD1 L4w grown for 4 d under +DEX and −DEX conditions. Samples were frozen in liquid nitrogen without flash treatment (A), immediately after a 0.7-ms flash (C), and after additional dark incubation for 20 min (D) or 2 h (E) following flash treatment. Representative data from three or more biologically independent experiments are shown. Vertical dotted lines in A and E represent the peak wavelength of fluorescence from photoactive Pchlide (P653) in −DEX seedlings. B, +DEX minus −DEX difference spectra in continuous dark (Dark) or 2 h of dark after flash irradiation (Flash + 2 h dark). Data are means of eight (Dark) or three (Flash + 2 h dark) independent experiments. The arrow indicates the fluorescence peak from the dimeric Pchlide-LPOR-NADPH complex at ∼645 nm. F, Pchlide content in 4-d-old etiolated amiR-MGD1 L4w seedlings dark incubated for 20 min after flash treatment. Data are means ± se from five to seven independent experiments. The amount of photoactive Pchlide was estimated by subtracting the amount of nonphotoactive Pchlide from total Pchlide. Asterisks indicate significant differences from each form of Pchlide of the −DEX control (*, P < 0.05 and ***, P < 0.001, Student’s t test).
Table I. Peak positions of Pchlide fluorescence bands in 4-d-old etiolated cotyledons of amiR-MGD1 L4w and L4g under 77K.
Fluorescence data were obtained every 0.2 nm. Data are means ± se from three to six independent experiments. Asterisks indicate significant differences from the −DEX control (*, P < 0.05 and **, P < 0.01, Student’s t test).
| Lines | Conditions | Fluorescence Bands | DEX Treatment | Peak Wavelength |
|---|---|---|---|---|
| nm | ||||
| L4w | Dark (before flash) | P630 | −DEX | 630.3 ± 0.1 |
| +DEX | 630.0 ± 0.2 | |||
| P653 | −DEX | 653.2 ± 0.1 | ||
| +DEX | 652.6 ± 0.1** | |||
| L4g | Dark (before flash) | P630 | −DEX | 629.8 ± 0.2 |
| +DEX | 630.1 ± 0.1 | |||
| P653 | −DEX | 653.1 ± 0.0 | ||
| +DEX | 653.1 ± 0.1 | |||
| L4w | Flash + 2 h of dark | P653 | −DEX | 653.3 ± 0.4 |
| +DEX | 652.1 ± 0.2* |
To examine whether the MGD1 suppression affected the rapid photoconversion of Pchilde, we treated etiolated L4w seedlings with a 0.7-ms light flash before 77K fluorescence measurement (Fig. 3C). In both −DEX and +DEX seedlings, P653 was no longer observed after flash irradiation, whereas P630 remained, which substantiates that these two bands were attributed to photoactive and nonphotoactive Pchlide, respectively. In addition, a prominent band peaking at 689 nm (C689), which originates from the Chlide-LPOR-NADP+ complex (Schoefs, 2001; Solymosi et al., 2007), emerged after flash irradiation of both −DEX and +DEX seedlings. Thus, the Pchlide-LPOR-NADPH complex was converted efficiently to the Chlide-LPOR-NADP+ complex with flash irradiation even when MGDG biosynthesis was suppressed.
After illumination, the fluorescence maximum around 690 nm shifted gradually to ∼680 nm in the dark, presumably as a result of disaggregation or the rearrangement of large oligomers of Chlide-LPOR complexes (Shibata, 1957; Smeller et al., 2003; Solymosi et al., 2007). During this process, called the Shibata shift, major spectral changes are completed within 20 min after irradiation, followed by small shifts continuing until 2 or 3 h. To investigate the contribution of MGDG to subsequent processes after Pchlide photoconversion, we incubated etiolated L4w seedlings in the dark for 20 min (Fig. 3D) or 2 h (Fig. 3E) after flash irradiation and measured fluorescence spectra at 77K. In both −DEX and +DEX seedlings, C689 was shifted completely to a peak at 678 nm (C678) during the first 20 min after flash irradiation. After dark incubation for 2 h, the peak wavelength of C678 was shifted slightly to 677 nm, regardless of DEX treatment. These data suggest that the MGD1 suppression did not remarkably affect the Shibata shift in etiolated seedlings.
Meanwhile, DEX treatment largely affected the regeneration of photoactive Pchlide during dark incubation after illumination. In −DEX seedlings, the P653 emission from photoactive Pchlide emerged at 20 min after dark incubation and became more prominent after 2 h (Fig. 3, D and E). The P653 emission also was increased in +DEX seedlings during dark incubation, but to a lesser extent than in the −DEX control. Moreover, the peak wavelength of P653 after 2 h of dark incubation was slightly blue shifted in +DEX seedlings, as was observed before flash irradiation (Table I). Consistent with this result, a band peaking at around 645 nm, which would originate from the dimer of the Pchlide-LPOR-NADPH ternary complex, was again observed in a difference spectrum between +DEX and −DEX samples (Fig. 3B). To evaluate the regeneration activity of photoactive Pchlide after flash irradiation, we determined the Pchlide content in L4w seedlings incubated in the dark for 20 min after flash irradiation (Fig. 3F). In +DEX seedlings, total Pchlide content was 55% lower than in the −DEX control, mainly due to a reduction in photoactive Pchlide content. These data suggest that MGDG deficiency impairs the regeneration of the photoactive Pchlide-LPOR-NADPH complex after flash irradiation.
MGD1 Suppression Affects Membrane Organization in Etioplasts
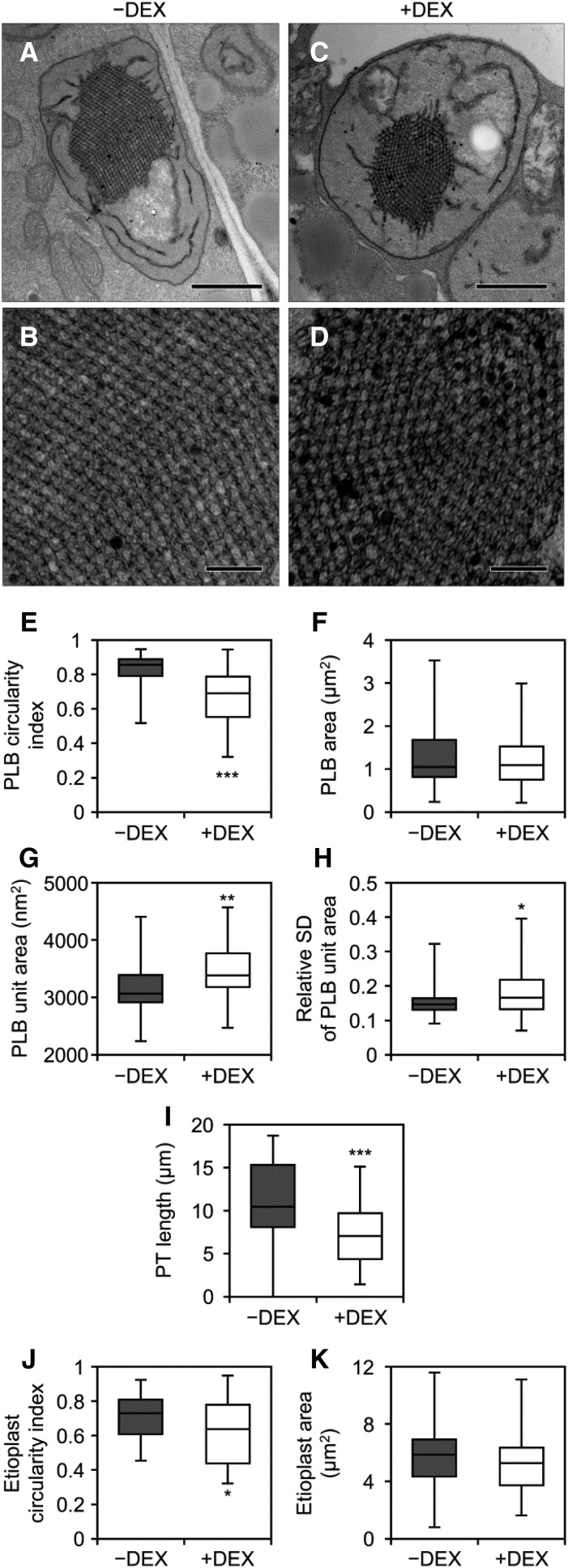
To assess the role of MGDG in PLB formation, we observed the ultrastructure of etioplasts in 4-d-old L4w seedlings by transmission electron microscopy (Fig. 4; Supplemental Fig. S6). In the −DEX control, etioplasts had the regular lattice membrane structure connecting to short lamellae (Fig. 4, A and B; Supplemental Fig. S6, A–H). Although DEX treatment did not abolish the development of etioplasts (Fig. 4, C and D; Supplemental Fig. S6, I–S), some etioplasts in +DEX seedlings had irregularly shaped PLBs. Quantitative analysis revealed that the circularity but not the total size of PLBs was decreased in +DEX seedlings compared with the −DEX control (Fig. 4, E and F). In addition, the unit size of the PLB lattice was increased by DEX treatment (Fig. 4G), with relative sd values of the unit area in a PLB, used as an index of irregularity of the lattice crystalline structure, also increased in +DEX seedlings (Fig. 4H; Supplemental Fig. S7). Besides the morphological changes in PLBs, the length of PTs was shorter in +DEX seedlings than in the −DEX control (Fig. 4I). Moreover, the shape of etioplasts was disordered by MGD1 suppression, as represented by a decreased circularity of etioplasts in +DEX seedlings (Fig. 4J). Intrusion of cytosolic regions into etioplasts was more frequently observed in +DEX seedlings (13 of 46 etioplasts; Supplemental Fig. S6, K and P–S, red arrowheads) than in the control (1 of 46 etioplasts), although the size of the etioplast was not largely different in both conditions (Fig. 4K).
Figure 4.
Ultrastructure of etioplasts in cotyledon cells of 4-d-old etiolated amiR-MGD1 L4w seedlings. A and C, Images of whole etioplasts in cotyledons grown under −DEX (A) and +DEX (C). Bars = 1 μm. B and D, Magnified images of PLB lattices in A (B) and C (D). Bars = 200 nm. For more images, see Supplemental Figure S6. E to K, Quantitative data of circularity index (E) and area (F) of PLBs, area of a single PLB unit (G), relative sd value (sd/average) of the unit area in a PLB (H), length of PTs (I), and circularity index (J) and area (K) of etioplasts. The horizontal line in each box represents the median value of the distribution. The top and bottom of each box represent the upper and lower quartiles, respectively. The whiskers represent the range. Data were obtained from 46 different etioplasts. In H, the relative sd value was calculated from 20 units of the PLB in each etioplast. The distribution of the PLB unit area in each etioplast is shown in Supplemental Figure S7. Asterisks indicate significant differences from the −DEX control (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, Welch’s t test).
MGD1 Suppression Impairs Membrane-Associated Processes of the Pchlide Biosynthesis Pathway
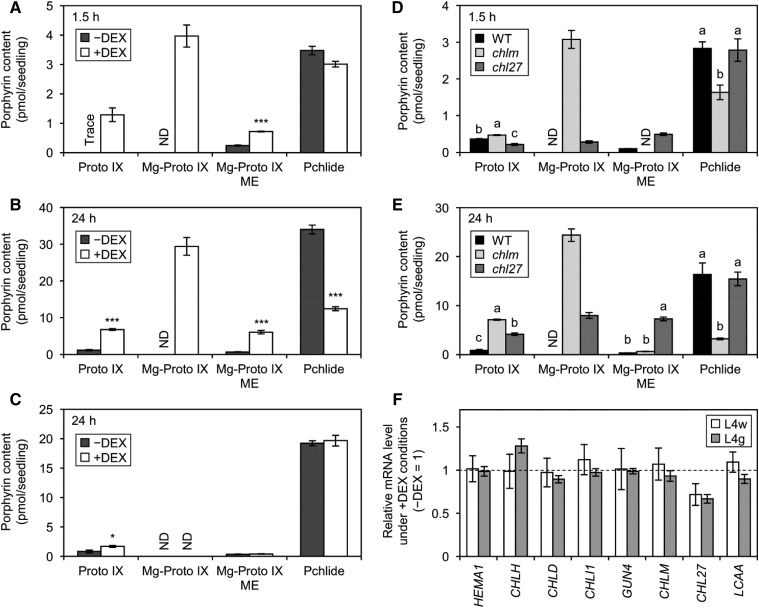
The decreased content of total Pchlide in +DEX L4w seedlings suggested impaired Pchlide biosynthesis by MGDG deficiency. To evaluate the effect of MGDG deficiency on the Pchlide biosynthesis pathway, we measured porphyrin intermediates in amiR-MGD1 etiolated seedlings. Porphyrin pigments in the Chl biosynthesis pathway, such as Proto IX, Mg-Proto IX, Mg-Proto IX ME, and Pchlide, can be distinguished by their own unique fluorescence characteristics, whereas ALA and other nonporphyrin intermediates do not emit fluorescence. Porphyrin pigments extracted from 4-d-old etiolated seedlings were separated and detected by HPLC with a fluorescence detector (Supplemental Fig. S8A). In both −DEX and +DEX seedlings, no porphyrin pigments besides Pchlide were detected. Because porphyrin metabolism in the dark is strictly regulated particularly at the ALA biosynthesis step (Brzezowski et al., 2015), it is not surprising that toxic porphyrin intermediates were not detected in amiR-MGD1 even under +DEX conditions.
To bypass the rate-limiting step of ALA biosynthesis, we fed 10 mm ALA to dark-grown amiR-MGD1 L4w seedlings for 1.5 h (Fig. 5A; Supplemental Fig. S8A) and 24 h (Fig. 5B). This method is commonly used to measure the activity of porphyrin metabolism in planta (Terry and Kendrick, 1999; Tottey et al., 2003). In −DEX seedlings, Pchlide content was increased 3- and 30-fold with ALA feeding for 1.5 and 24 h, respectively (Fig. 5, A and B; compare with Fig. 2A). Small amounts of Proto IX and Mg-Proto IX ME also were accumulated with ALA feeding, but Mg-Proto IX was undetectable even after ALA feeding for 24 h. In +DEX seedlings, Pchlide accumulated more slowly than in the −DEX control; the content after 24 h of ALA feeding was only 30% of the −DEX control level (Fig. 5B). Instead, +DEX seedlings accumulated a substantial amount of Mg-Proto IX, which was not detected in the −DEX control, in addition to larger amounts of Proto IX and Mg-Proto IX ME than in −DEX seedlings. Excess accumulation of porphyrin intermediates was already observed after 1.5 h of ALA feeding (Fig. 5A) and enhanced after 24 h (Fig. 5B). We also measured porphyrin levels in amiR-MGD1 L4g seedlings treated with ALA for 24 h (Fig. 5C). DEX treatment to L4g etiolated seedlings neither decreased Pchlide content nor enhanced the accumulation of other intermediates, so changes in the porphyrin profile in L4w were not due to side effects of DEX treatment but resulted from strong MGD1 suppression and consequent MGDG deficiency. In addition, there were no notable differences in porphyrin accumulation between etiolated mgd1-1 and wild-type seedlings with ALA feeding for 24 h (Supplemental Fig. S8C).
Figure 5.
Effect of MGD1 suppression on Pchlide biosynthesis in the dark. A to C, Accumulation of porphyrin pigments in etiolated amiR-MGD1 L4w seedlings fed ALA for 1.5 h (A) and 24 h (B) and L4g seedlings fed ALA for 24 h (C). Seedlings were grown in the dark under +DEX or −DEX conditions for 4 d before ALA feeding. D and E, Accumulation of porphyrin pigments in 4-d-old etiolated wild-type (WT), chlm, and chl27 seedlings fed ALA for 1.5 h (D) and 24 h (E). In A to E, data are means ± se from three to six independent experiments. ND, Not detected; Trace, trace amount. F, Quantitative reverse transcription-PCR analysis of the mRNA expression of genes involved in Pchlide biosynthesis in L4w and L4g seedlings grown in the dark for 4 d. mRNA levels in +DEX seedlings are presented as fold differences from the −DEX controls (broken line) after normalizing to the control gene ACTIN8. Data are means ± se from 10 (L4w) or three (L4g) independent experiments. In A, B, C, and F, asterisks indicate significant differences from the −DEX control (*, P < 0.05 and ***, P < 0.001, Student’s t test). In D and E, different letters indicate significant differences (P < 0.05, Tukey-Kramer multiple comparison test).
To address which processes of porphyrin biosynthesis are particularly affected by MGDG deficiency in etioplasts, we compared porphyrin profiles in +DEX L4w seedlings with those in Chl biosynthesis mutants after ALA feeding (Fig. 5, D and E; Supplemental Fig. S8A). chlm (Mochizuki et al., 2008) and chl27/crd1 (Ankele et al., 2007; Mochizuki et al., 2008) are T-DNA insertion knockdown mutants of genes for MgMT and MgCY, respectively. Reflecting the role of MgMT in the conversion of Mg-Proto IX to Mg-Proto IX ME, Mg-Proto IX was accumulated prominently in the chlm mutant with ALA feeding for 1.5 h and increased further with 24 h of feeding. In chlm, Proto IX content also was higher than that in wild-type seedlings, particularly with 24 h of ALA feeding. Meanwhile, Mg-Proto IX ME, the product of the MgMT reaction, was undetectable in chlm with 1.5 h of ALA feeding and remained at low levels with 24 h of feeding. In contrast to the high accumulation of Mg-Proto IX and Proto IX, Pchlide formation was strongly inhibited in this mutant. In the chl27 mutant, which is partially deficient in MgCY activity converting Mg-Proto IX ME to Pchlide, Mg-Proto IX ME was strongly accumulated along with Proto IX and Mg-Proto IX with ALA feeding. However, the chl27 mutation did not notably affect the Pchlide accumulation with ALA feeding. These data reveal that +DEX amiR-MGD1 L4w seedlings showed a porphyrin profile similar to that of chlm, but unlike chlm, L4w also highly accumulated Proto IX and Mg-Proto IX ME even after short ALA feeding (Fig. 5A).
To reveal the mechanism for how MGD1 suppression affects the Pchlide biosynthesis pathway, we investigated mRNA levels of genes involved in Pchlide biosynthesis in amiR-MGD1 etiolated seedlings (Fig. 5F). HEMA1 encodes the major isoform of GluTR. CHLH, CHLD, and CHLI1 encode H, D, and the major isoform of I subunits of MgCh, respectively, whereas GUN4 encodes the GUN4 protein, which is required for MgCh activity. CHLM is a single gene for MgMT. CHL27 and LOW CHLOROPHYLL ACCUMULATION A (LCAA) encode two membrane-bound subunits constituting MgCY (Tanaka et al., 2011; Albus et al., 2012). In etiolated L4w seedlings, steady-state mRNA levels of these Pchlide synthesis genes were unchanged by DEX treatment. However, the mRNA level of CHL27 was decreased slightly in +DEX L4w, but a similar decrease also was observed in +DEX L4g seedlings. Because +DEX L4g seedlings showed normal porphyrin metabolism (Fig. 5C), the decreased CHL27 expression does not likely affect the MgCY activity.
MGD1 Suppression Is Unlikely to Affect the Expression of Photosynthesis-Associated Genes in Etiolated Seedlings
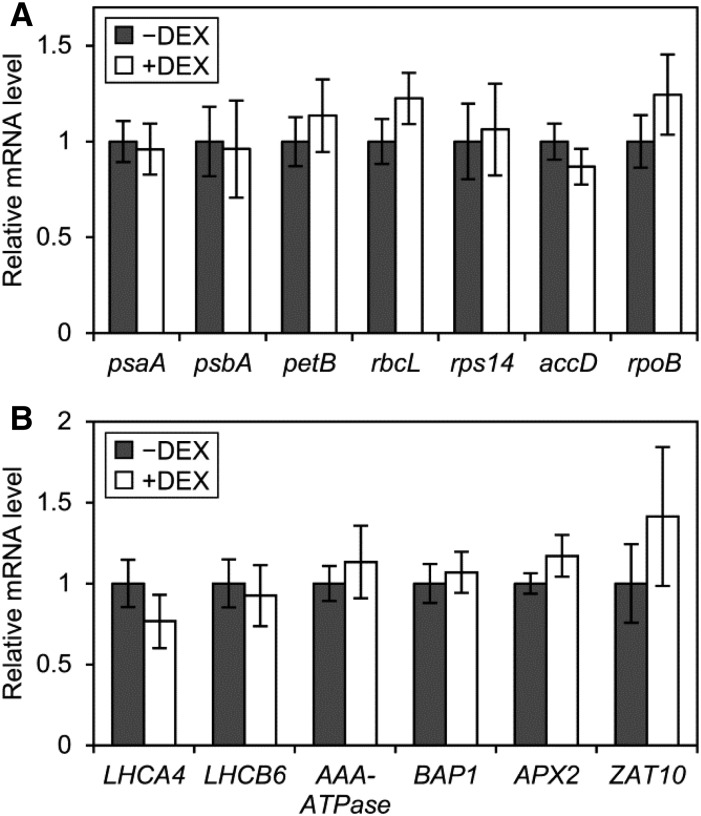
We previously revealed, in the mgd1-2 mutant, that galactolipid biosynthesis and subsequent thylakoid development are crucial for the expression of photosynthesis-associated genes encoded in the nucleus and plastids in light-grown Arabidopsis seedlings (Kobayashi et al., 2013). Indeed, MGD1 suppression in amiR-MGD1 during an early stage of chloroplast development down-regulated photosynthesis-associated genes (Fujii et al., 2014). We investigated whether the decreased MGDG content in etioplasts also affects the mRNA expression of plastid-encoded photosynthesis-associated genes in etiolated amiR-MGD1 L4w seedlings (Fig. 6A). The genes psaA and psbA encode PsaA and D1 proteins in reaction centers of PSI and PSII, respectively, and petB encodes the cytochrome b6 subunit. The genes rbcL and rps14 are for the large subunit of Rubisco and the 14S subunit of plastidic 30S ribosome, respectively. The genes accD and rpoB encode the βCT subunit of acetyl-CoA carboxylase and the β-subunit of plastid-encoded RNA polymerase, respectively. The mRNA levels of these genes were unchanged with DEX treatment in etiolated L4w seedlings, which suggests that partial MGDG deficiency does not affect plastid-encoded gene expression in etioplasts. We also examined the mRNA levels of two photosynthesis-associated nuclear genes, LHCA4 and LHCB6, encoding the light-harvesting complex (LHC) I subunit 4 and the LHCII subunit 6, respectively, and found no altered expression of these genes with MGD1 suppression in etiolated seedlings (Fig. 6B).
Figure 6.
Quantitative reverse transcription-PCR analysis of mRNA levels of photosynthesis-associated and reactive oxygen species-responsive genes in amiR-MGD1 L4w etiolated seedlings grown for 4 d under −DEX and +DEX conditions. A, Genes encoded in the plastid genome. B, Photosynthesis-associated and reactive oxygen species-responsive genes encoded in the nucleus. In A and B, mRNA levels are presented as fold difference from the −DEX control after normalizing to the control gene ACTIN8. Data are means ± se from 10 independent experiments. None of the genes showed significant differences between +DEX and −DEX seedlings (P > 0.05, Student’s t test).
Impaired porphyrin metabolism often causes the production of reactive oxygen species and oxidative damage under light (Triantaphylidès and Havaux, 2009). Moreover, a Chl metabolite, pheophorbide a, is reported to induce cell death with increased hydrogen peroxide even in the dark (Hirashima et al., 2009). AAA-ATPase and BON ASSOCIATED PROTEIN1 (BAP1) are singlet oxygen-induced genes (Simková et al., 2012), whereas ASCORBATE PEROXIDASE2 (APX2) and ZAT ZINK FINGER PROTEIN10 (ZAT10) are induced by superoxide and altered plastid redox state (Pogson et al., 2008). mRNA levels of these four nucleus-encoded genes were not changed by MGD1 suppression (Fig. 6B), so the partial MGDG deficiency in the dark does not globally affect gene expression in plastids and the nucleus.
DISCUSSION
Identification of Homogenous Albino Lines of amiR-MGD1 Transgenic Arabidopsis
In this study, we identified homogenous T4 amiR-MGD1 lines that develop albino cotyledons in the presence of DEX under light (Supplemental Fig. S1). MGD1 mRNA levels in these lines were reduced to less than 20% of the −DEX control with DEX treatment. We also obtained homogenous green amiR-MGD1 lines, showing MGD1 mRNA levels reduced by DEX treatment to 35% to 50% of the −DEX control but Chl content decreased only slightly. These results are consistent with a previous observation in T3 amiR-MGD1 lines that strong suppression of MGD1 expression to less than 30% of wild-type levels caused a white cotyledon phenotype, whereas milder MGD1 suppression (∼50% of wild-type levels) resulted in a wild-type-like green cotyledon phenotype (Fujii et al., 2014). Similar results in two independent amiR-MGD1 lines (L2 and L4; Supplemental Fig. S1) suggest that these phenomena are not due to the positional effect of the transgene. Under our growth conditions, mgd1-1 seedlings grown in the light showed a phenotype similar to that of +DEX green seedlings of amiR-MGD1 lines: Chl content was decreased only slightly in mgd1-1, whereas the MGD1 mRNA level was reduced to 38% of the wild-type level (Supplemental Fig. S9). These data suggest the existence of a threshold level of MGD1 expression at ∼35% of the wild-type level to maintain regular chloroplast development in cotyledons (Supplemental Fig. S10). Galactolipid biosynthesis would be one of the determinant processes in chloroplast development (Kobayashi, 2016), and decreased MGD1 expression below the threshold may cause severe deficiency of MGDG and subsequent discontinuation of chloroplast development at early stages in cotyledons. We note that the homogeneity of the T4 amiR-MGD1 lines is not genetically fixed across generations. L4-01 plants, one of the DEX-dependent albino T4 lines, generated T5 lines with various cotyledon color phenotypes under +DEX conditions (Supplemental Fig. S11). The suppression levels of MGD1 expression in amiR-MGD1 lines may be prone to fluctuation in response to the growth conditions of parent plants, although the underlying mechanism remains to be elucidated.
MGD1 Is Responsible for MGDG Biosynthesis in Etioplasts
Our analyses with homogenous amiR-MGD1 L4w plants revealed that DEX treatment suppressed the MGD1 expression in etiolated seedlings to 35% of the −DEX control, which resulted in decreased MGDG content to 64% of the control level (Fig. 1). By contrast, loss of function of both MGD2 and MGD3 did not affect galactolipid content in etiolated seedlings (Kobayashi et al., 2009a). Therefore, MGD1 plays a central role in MGDG biosynthesis during etioplast development. Although MGDG is used as a substrate for DGDG biosynthesis, MGD1 suppression did not change DGDG content, so the MGDG-DGDG ratio was reduced in +DEX etiolated L4w seedlings (Fig. 1B). This result is consistent with previous reports of light-grown seedlings showing that partial decreases in MGD1 activity primarily result in a loss of MGDG without affecting DGDG biosynthesis (Jarvis et al., 2000; Wu et al., 2013; Fujii et al., 2014). Although we cannot exclude that MGD2 and MGD3, localized to the outer envelope membrane of plastids and using diacylglycerol pools different from the MGD1 pathway localized to the inner envelope (Kobayashi et al., 2009b), specifically function to produce DGDG in etioplasts, unchanged fatty acid compositions in both galactolipids by DEX treatment (Fig. 1, C and D) imply a negligible contribution of the MGD2/MGD3-mediated galactolipid biosynthesis pathway.
MGDG Is Required for the Pchlide Biosynthesis Pathway in Etioplasts
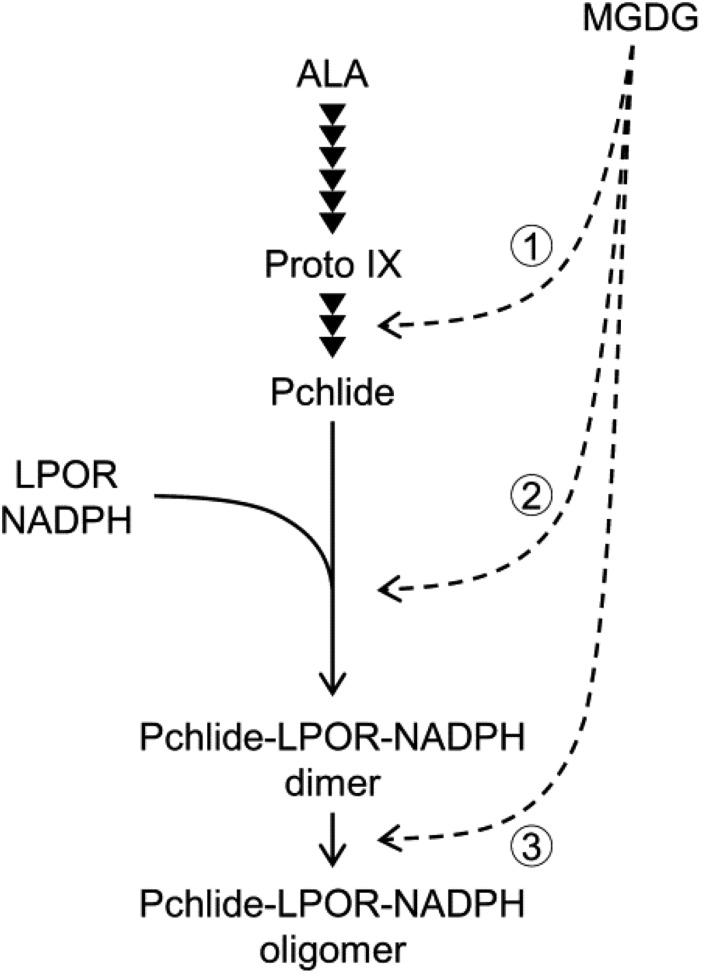
Strong suppression of MGD1 expression decreased the total amount of Pchlide in etiolated L2w and L4w seedlings (Fig. 2A; Supplemental Fig. S2B). Moreover, the rate of Pchlide accumulation was remarkably lower in +DEX L4w seedlings than in the −DEX control (Fig. 2C), which suggests that Pchlide biosynthesis is strongly retarded by MGDG deficiency. ALA feeding experiments revealed that Mg-Proto IX metabolism in the Pchlide biosynthesis pathway was particularly impaired by strong MGD1 suppression (Fig. 5, B and C). The porphyrin profile in +DEX L4w seedlings after ALA feeding (Fig. 5, A and B) was similar to that in the ALA-fed chlm mutant (Fig. 5, D and E), so the conversion of Mg-Proto IX to Mg-Proto IX ME by CHLM-encoded MgMT may be strongly impaired by MGDG deficiency. However, unlike the chlm mutant, +DEX L4w seedlings also rapidly accumulated Proto IX and Mg-Proto IX ME after ALA feeding. Thus, MGDG deficiency also may impair the metabolisms of Proto IX by MgCh and Mg-Proto IX ME by MgCY. Despite strong impairments of the Pchlide biosynthesis pathway, particularly Mg-Proto IX metabolism, no porphyrin intermediates were accumulated in +DEX etiolated L4w seedlings in the absence of ALA (Supplemental Fig. S8A). The deficiency of MgCh, MgMT, and MgCY activities by genetic manipulation is reported to suppress ALA biosynthesis (Papenbrock et al., 2000; Alawady and Grimm, 2005; Peter et al., 2010; Schlicke et al., 2014), possibly via increased metabolic flow from Proto IX to heme and consequent feedback inhibition of GluTR activity by heme (Vothknecht et al., 1996; Terry and Kendrick, 1999; Goslings et al., 2004), which prevents the accumulation of porphyrin intermediates in mutant plants. Therefore, impaired porphyrin metabolism by MGDG deficiency likely down-regulates ALA biosynthesis in a feedback manner, which may further reduce Pchlide accumulation in +DEX L4w seedlings in the dark. Meanwhile, the biosynthetic pathway from ALA to Proto IX may not be affected by MGDG deficiency, because the sum of Proto IX, Mg-Proto IX, Mg-Proto IX ME, and Pchlide content after ALA feeding was not obviously reduced in +DEX seedlings, or seemed even higher, as compared with that in the −DEX control (Fig. 5, A and B). Therefore, MGDG is required for efficient porphyrin metabolism from Proto IX to Pchlide (Fig. 7, arrow 1) and somehow affects ALA biosynthesis but is less important for the pathway from ALA to Proto IX.
Figure 7.
Roles of MGDG in Pchlide biosynthesis and the formation of photoactive Pchlide-LPOR-NADPH complexes during etioplast development. Arrowheads indicate enzymatic steps in the Pchlide biosynthesis pathway from ALA. Most of the Pchlide synthesized in etioplasts forms the photoactive ternary complex with LPOR and NADPH, and the photoactive complex exists as the dimer or further aggregates into oligomeric complexes. MGDG is required for the Pchlide biosynthesis pathway from Proto IX to Pchlide (arrow 1), the formation of the photoactive Pchlide-LPOR-NADPH ternary complex (arrow 2), and the oligomerization of the ternary complex (arrow 3).
Quantitative reverse transcription-PCR analysis suggested that impaired Pchlide biosynthesis in +DEX L4w seedlings was not attributed to the transcriptional modification of genes involved in this pathway (Fig. 5F). Considering that the Chl biosynthesis pathway downstream from Proto IX formation takes place in plastid membranes rich in MGDG, several possibilities for impaired porphyrin metabolisms by MGD1 suppression can be considered. A 36% loss of MGDG in +DEX L4w seedlings slightly but significantly changed membrane structures in etioplasts, although it did not severely perturb etioplast development (Fig. 4; Supplemental Fig. S6). Thus, a disordered local lipid environment by MGDG deficiency rather than inhibited etioplast biogenesis may affect the Pchlide biosynthesis pathway. Because MgCh, MgMT, and MgCY are bound to plastid membranes (Masuda and Fujita, 2008), altered lipid compositions with reduced MGDG content, which would change the fluidity and/or local structures of the membrane (Demé et al., 2014), may affect the functional status of these enzymes. Moreover, enzymes in the Chl biosynthesis pathway, including MgCh, MgMT, and MgCY, may form heterocomplexes to channel Chl intermediates efficiently (Tanaka and Tanaka, 2007; Wang and Grimm, 2015). Thus, MGDG deficiency may perturb the localization of MgCh, MgMT, or MgCY to the proper sites of membranes in etioplasts or may impair the formation of multiple complexes and channeling of Chl intermediates, which results in the accumulation of the intermediates. Recently, Kopečná et al. (2015) showed that a deficiency of phosphatidylglycerol, a major phospholipid in the thylakoid membrane of chloroplasts and cyanobacteria, in Synechocystis PCC 6803 inhibits the Chl biosynthesis pathway particularly at the conversion of Mg-Proto IX ME to Pchlide. The authors hypothesized that phosphatidylglycerol is an essential component in a membrane microdomain where Chl biosynthesis may take place along with the synthesis of PSI proteins. Although the exact site of the last several steps of Pchlide biosynthesis in plant etioplasts remains unclear, MGDG also may function to provide a specific lipid environment for the Pchlide biosynthesis pathway. Another possibility is that MGDG acts as an essential cofactor or activator of enzymes in the Pchlide biosynthesis pathway. However, Synechocystis MgCh subunits expressed in Escherichia coli (Jensen et al., 1996) and tobacco subunits expressed in yeast (Papenbrock et al., 1997) could reconstitute the chelatase activity in vitro. Recombinant Arabidopsis MgMT expressed in E. coli also exerted its catalytic activity in vitro (Block et al., 2002). E. coli and yeast do not contain galactolipids, so MGDG is not essential for MgCh and MgMT activity in vitro, although we do not exclude that MGDG functions to enhance the activity of these enzymes.
MGDG Facilitates the Formation of the Photoactive Pchlide-LPOR-NADPH Complex and Its Oligomerization
A photoconversion assay of Pchlide revealed that the strong MGD1 suppression particularly impaired the accumulation of photoactive Pchlide (Fig. 2A; Supplemental Fig. S2B). Consistent with this finding, the regeneration of photoactive Pchlide after flash irradiation was retarded in MGD1-suppressed seedlings (Fig. 3F). By contrast, the total amount of LPOR proteins was not changed with MGD1 suppression (Fig. 2D). Thus, the decreased photoactive Pchlide in +DEX L4w seedlings is not due to a deficiency of LPOR proteins. These results suggest that MGDG deficiency perturbs the formation of the photoactive Pchlide-LPOR-NADPH ternary complex. The impaired complex formation together with reduced Pchlide biosynthesis resulted in a preferential decrease in photoactive Pchlide content in +DEX L4w and L2w seedlings, with nonphotoactive Pchlide levels virtually unchanged owing to the increased ratio of the nonphotoactive form to the photoactive form (Fig. 2A; Supplemental Fig. S2B). The amount of photoactive Pchlide also was decreased slightly in +DEX L4g seedlings, while the total Pchlide level was unchanged (Fig. 2A). Thus, the weak MGD1 suppression in L4g seedlings may partially impair the formation of the photoactive Pchlide complex without affecting total Pchlide biosynthesis activity. Meanwhile, in mgd1-1 seedlings, which showed no retarded porphyrin metabolism after ALA feeding (Supplemental Fig. S8C), amounts of both Pchlide forms were decreased similarly, resulting in a significant decrease in the total Pchlide content (Supplemental Fig. S2D). Unlike in amiR-MGD1 lines, MGD1 expression in mgd1-1 is suppressed continuously by a T-DNA insertion in the MGD1 promoter region. The different profiles of MGD1 suppression between mgd1-1 and amiMGD1 lines may partially change the profiles of Pchlide accumulation during etioplast development.
In addition to the impaired formation of the photoactive Pchlide complex, strong MGD1 suppression caused a blue shift of the fluorescence band emitted from photoactive Pchlide, with increased fluorescence around 645 nm (Fig. 3, A, B, and E; Table I). Fluorescence bands peaking at ∼645 and ∼655 nm may be derived from the dimer of the Pchlide-LPOR-NADPH ternary complex and its large aggregates, respectively (Schoefs, 2001). Increased fluorescence at 645 nm with decreased fluorescence at 655 nm in +DEX seedlings relative to the −DEX control (Fig. 3B) suggest that the MGDG deficiency impairs the formation of large aggregates of the Pchlide-LPOR-NADPH ternary complex from the dimer. Consistently, in vitro experiments showed that MGDG strongly enhances the oligomerization of the Pchlide-LPOR complexes, presumably by interacting with LPOR (Gabruk et al., 2017). Therefore, an MGDG-rich lipid environment in etioplasts may contribute to forming the photoactive ternary complex efficiently and developing it into large aggregates (Fig. 7, arrows 2 and 3). Meanwhile, all photoactive Pchlide in large aggregates or the dimeric complex was instantaneously photoconverted to Chlide by flash irradiation even in MGD1-suppressed seedlings, which indicates that the high ratio of MGDG in membranes is not essential for the catalytic activity of the ternary complex. The Shibata shift after flash irradiation also was unaffected by MGDG deficiency, so MGDG may not play a crucial role in the disaggregation of large aggregates of Chlide-LPOR complexes.
MGDG Contributes to the Formation of the Membrane Structure in Etioplasts
A 36% reduction in the relative MGDG content in etiolated L4w seedlings disordered the entire shape and the lattice structure of PLBs in etioplasts (Fig. 4, E, G, and H). Because MGDG in PLBs has been suggested to facilitate the formation of the cubic phase structure (Brentel et al., 1985), the high presence of MGDG may be required for the structural organization of the PLB membrane in addition to or through the formation of the photoactive Pchlide-LPOR-NADPH complex and its oligomerization. Carotenoids, the major lipophilic pigments in etioplasts, also are deeply involved in the formation and maintenance of PLB structures, as demonstrated by the disrupted PLB structure in etiolated seedlings with disordered carotenoid biosynthesis (Park et al., 2002; Moro et al., 2004). However, MGD1 suppression altered neither total carotenoid content nor carotenoid composition in etiolated L4w seedlings (Fig. 2E; Supplemental Fig. S4), so carotenoid functions would not be associated with the distorted PLB structures by MGDG deficiency.
Comprehensive studies with LPOR-overexpressing and LPOR-deficient etiolated seedlings indicated that the total amount of LPOR proteins is well associated with the size of PLBs (Sperling et al., 1998; Franck et al., 2000; Masuda et al., 2003). In fact, in etiolated +DEX L4w seedlings, which showed no significant reduction in LPOR levels (Fig. 2D), the size of PLBs was similar to that of the −DEX control despite decreased MGDG content (Fig. 4F). By contrast, the size of PTs was reduced by the MGD1 suppression (Fig. 4I). The maintained PLB size in MGDG-deficient L4w seedlings by LPORs may result in the decreased formation of PTs due to a deficiency of membrane lipids, although we cannot exclude the possibility that MGDG plays a specific role in PT development. In addition, irregularly shaped etioplasts and the intrusion of cytosolic regions into etioplasts were observed frequently in +DEX L4w cotyledons (Fig. 4J; Supplemental Fig. S6). Because MGDG accounts for 47% of the total membrane lipids in the etioplast envelope (Selstam and Sandelius, 1984), MGDG also may be important for maintaining the structure of the etioplast envelope.
MGDG Biosynthesis at an Early Germination Stage Is a Prerequisite for Pchlide Accumulation
Although the DEX treatment from the beginning of or 1 d after seeding decreased Pchlide content in etiolated L4w seedlings, that from 2 d after seeding no longer inhibited Pchlide accumulation (Fig. 2B). Considering that Pchlide begins to accumulate from 2 d after seeding (Fig. 2C), the MGD1 suppression after this time would be less effective, presumably because the MGD1 protein or galactolipid is already synthesized to some extent at that stage. In fact, public transcriptome data show that MGD1 is expressed in dry seeds and during the early germination stage (Winter et al., 2007; Bassel et al., 2008). Moreover, lipid analysis in cucumber (Cucumis sativus) indicates that galactolipid biosynthesis starts early after germination in the dark (Ohta et al., 1995). Thus, the MGD1 expression and subsequent galactolipid biosynthesis likely occur before Pchlide biosynthesis, which may be essential for effective Pchlide biosynthesis and the formation of the Pchlide-LPOR-NADPH ternary complex. A similar result was obtained in +DEX amiR-MGD1 seedlings grown under continuous light (Fujii et al., 2014): although MGD1 suppression initiated within 3 d after seeding severely impaired Chl accumulation, suppression after 3 d did not inhibit the greening of cotyledons. MGDG biosynthesis may be a prerequisite as one of the initial processes of etioplast and chloroplast biogenesis, because galactolipid-rich lipid bilayers provide a matrix for various processes on the membrane, such as Pchlide biosynthesis and the formation of Pchlide-LPOR complexes during etioplast development and Chl accumulation and the formation of photosynthetic Chl-protein complexes during chloroplast development.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Light Treatment
The amiR-MGD1 transgenic lines were the Landsberg erecta ecotype (Fujii et al., 2014), and the mgd1-1 (Jarvis et al., 2000), chlm (SALK_110265; Mochizuki et al., 2008), and chl27/crd1 (SALK_009052; Ankele et al., 2007; Mochizuki et al., 2008) mutants were the Columbia ecotype of Arabidopsis (Arabidopsis thaliana). Seeds were surface sterilized and then cold treated in water at 4°C for 4 d in the dark before seeding. Plants were grown on Murashige and Skoog medium (adjusted to pH 5.7 with KOH) containing 1% (w/v) Suc solidified with 0.8% (w/v) agar except for the experiments in Figure 2B, in which plants were grown in liquid medium with gentle rotation. All plants were grown at 23°C in a growth chamber. For light-grown seedlings, plants were illuminated with continuous white light (∼30 μmol photos m−2 s−1). For etiolated seedlings, cold-treated seeds were illuminated with room light for ∼3 h at room temperature to synchronize germination and then germinated in darkness. Unless stated otherwise, light-grown and etiolated seedlings were grown for 5 and 4 d, respectively. For DEX treatment, DEX (Wako) was added to a final concentration of 10 μm in the medium from a 50 mm stock in dimethyl sulfoxide. Etiolated seedlings were sampled under dim green light unless stated otherwise. To obtain seeds, parental plants were grown on watered soil in the absence of DEX at ∼23°C under continuous white light (∼30 μmol photos m−2 s−1).
The photoconversion of photoactive Pchlide into Chlide involved a single flash of white light for 0.7 ms (Power ratio 1/2) from PZ42X electronic flash equipment (Sunpak).
Quantitative Reverse Transcription-PCR Analysis
Total RNA extraction, genomic DNA digestion, reverse transcription, cDNA amplification, and normalization of transcript abundance were performed as described (Fujii et al., 2014). The gene-specific primers used in cDNA amplification are listed in Supplemental Table S1.
Immunoblot Analysis
Total proteins were extracted and solubilized from seedlings crushed into powder in liquid nitrogen by adding sample buffer and incubating at 95°C for 5 min. Protein content was determined by using the RC DC Protein Assay (Bio-Rad) with bovine serum albumin as a standard. Ten and 20 μg of total proteins were subjected to SDS-PAGE on a gel containing 12.5% (w/v) polyacrylamide for separation, then electrotransferred to nitrocellulose membranes (Amersham Protran Premium 0.2 NC; GE Healthcare). Protein bands reacting with a primary antibody against total LPOR protein (Rowe and Griffiths, 1995; Masuda et al., 2003) were secondarily labeled with goat anti-rabbit IgG secondary antibody conjugated with horseradish peroxidase (Thermo Scientific). The secondary antibody was detected using a chemiluminescence reagent (Pierce Western Blotting Substrate Plus; Thermo Scientific) and an imager (ImageQuant LAS 4000 mini; GE Healthcare). For the loading control, proteins blotted on the membranes were stained with 0.1% (w/v) Ponceau S in 5% (v/v) acetic acid solution.
Lipid Analysis
Extraction of total lipids, separation by thin-layer chromatography, and visualization of lipids were performed as described (Kobayashi et al., 2006; Fujii et al., 2014). MGDG, DGDG, and a mixture of other glycerolipids were isolated from silica gel plates. Fatty acids in each lipid fraction were methyl esterified by incubation in 1 m HCl in methanol at 85°C for 1.5 h and quantified by gas chromatography (GC-17A; Shimadzu) with myristic acid as an internal standard.
Determination of Chl, Pchlide, and Carotenoids
Pigments were extracted by incubating intact seedlings in 1 mL of 80% (v/v) acetone at 4°C in the dark for 3 d (for Chl) or overnight (for Pchlide and carotenoids). Chl content was determined spectrophotometrically by measuring the absorbance of the extract at 663 and 645 nm with an Ultrospec 2100 pro (GE Healthcare) or a V-730 BIO (JASCO) spectrophotometer as described (Melis et al., 1987). In 20 etiolated Arabidopsis seedlings used for carotenoid determination, Chl and Pchlide were spectrophotometrically undetectable; thus, carotenoid content was determined by simply measuring the absorbance of the extract at 470 nm with the following formula: 1,000 × A470/198 (μg carotenoids mL−1; Lichtenthaler, 1987).
Pchlide content was determined by measuring fluorescence emission at 634 nm under 433-nm excitation with an RT-5300PC spectrofluorometer (Shimadzu) by using a Pchlide standard of known concentration. The concentration of the Pchlide standard, which was extracted from etiolated cucumber (Cucumis sativus) cotyledons, was determined by measuring the absorbance of the extract at 663, 645, and 626 nm (Anderson and Boardman, 1964). Many etiolated angiosperms including Arabidopsis accumulate both monovinyl-Pchlide and divinyl-Pchlide (Tanaka et al., 2011). Because the Mr values of monovinyl- and divinyl-Pchlide, 613 and 611, respectively, were very close, we used the Mr of monovinyl-Pchlide, the major form in mature etiolated Arabidopsis seedlings (Nagata et al., 2007), for calculation. To determine nonphotoactive Pchlide content, intact seedlings were irradiated with a single flash of light before extraction.
ALA Feeding and HPLC Analysis of Porphyrin Pigments
For porphyrin determination, intact seedlings were incubated in the dark in a solution containing 10 mm ALA, 10 mm MES-KOH (pH 5.7), and 5 mm MgCl2, with or without 10 μm DEX, with gentle rotation at 23°C in a growth chamber. Pigments were extracted by incubating intact seedlings in 100 μL of N,N-dimethylformamide at 4°C in the dark overnight. HPLC analysis was performed basically as described (Zapata et al., 2000) with some modifications. Pigments in 10 μL of extract were separated by using an HPLC system consisting of an L-2130 pump (Hitachi), a Rheodyne 7725i injector with a 20-μL sample loop (IDEX Health and Science), a COSMOSIL 5C18-MS-II guard column (Nakalai Tesque), and a reverse-phase C8 column (Symmetry C8 column, 100 Å, 3.5 µm, 4.6 × 150 mm; Waters) and detected by using an RF-550 spectrofluorometric detector (Shimadzu). The mobile phase consisted of two solvents: A (50% [v/v] methanol, 25% [v/v] acetonitrile, and 25% [v/v] 0.25 m pyridine in ultrapure water [adjusted to pH 5 with acetic acid]) and B (20% [v/v] methanol, 60% [v/v] acetonitrile, and 20% [v/v] acetone). Pigments were eluted with a linear gradient from 100% A to 70% A plus 30% B over 8 min and to 2% A plus 98% B over 0.5 min for Proto IX detection; 100% A to 76% A plus 24% B over 6.4 min and to 2% A plus 98% B over 0.6 min for Mg-Proto IX (ME) detection; and 100% A to 79% A plus 21% B over 5.6 min and to 2% A plus 98% B over 0.4 min for Pchlide detection, followed by isocratic elution with 2% A and 98% B for 5 min for all cases (Supplemental Fig. S8B). The flow rate was 1.2 mL min−1. Pigments were detected by measuring fluorescence emission at 634 nm under 400-nm excitation (Proto IX), at 595 nm under 420-nm excitation (Mg-Proto IX [ME]), and at 634 nm under 440-nm excitation (Pchlide; Supplemental Fig. S8A). Pigments were identified and quantified by comparing retention times and absorption spectra of standard pigments of Proto IX, Mg-Proto IX (Frontier Science), and Pchlide (from cucumber as described above). The concentration of standard Proto IX and Mg-Proto IX was determined by absorption at 404 and 417 nm by using the V-730 BIO spectrophotometer (JASCO) and calculated with extinction coefficients of 1.08244 and 1.659 × 105 m−1 cm−1, respectively (Kopetz et al., 2004). For Mg-Proto IX ME quantification, standard curves of Mg-Proto IX were used because they have the same spectral property.
In Situ Fluorescence Spectroscopy
Fluorescence emission spectra were obtained directly from excised cotyledons placed between two thin acrylic resin plates by using an RF-5300PC spectrofluorometer (Shimadzu) under 440-nm excitation at 77K in liquid nitrogen. Slit widths for excitation and emission were 3 and 5 nm, respectively. Fluorescence data were obtained every 1 nm (Fig. 3; Supplemental Fig. S5, A and B) and 0.2 nm wavelength (Table I; Supplemental Fig. S5C). Obtained spectra were normalized at the maxima between 620 and 640 nm as 1 and the fluorescence at 750 nm as 0. For measurement before and after photoconversion, cotyledons placed between plates were frozen before or immediately after flash treatment. For measurement of the Shibata shift and regeneration of photoactive Pchlide, intact seedlings on the agar-solidified medium were flash irradiated and incubated at 23°C in darkness for 20 min or 2 h before spectrum measurements.
Cotyledon Size Measurement
Etiolated cotyledons were observed by using an MZ16 FA stereomicroscope (Leica) with a VB-7010 CCD camera (KEYENCE). Cotyledons were excised from the seedlings and stuck on adhesive tape to observe the front view. The area of cotyledons was determined by using ImageJ software (https://imagej.nih.gov/ij/).
Transmission Electron Microscopy Analysis
Samples were fixed with 4% glutaraldehyde and 4% paraformaldehyde in a 50 mm sodium cacodylate buffer, pH 7, at 4°C for 2 h and washed with the same buffer at 4°C overnight. Then they were postfixed with 2% OsO4 in a 50 mm sodium cacodylate buffer at 4°C for 2 h. The fixed samples were run through an alcohol series and embedded in Spurr resin. Ultrathin sections (80 nm thick) were cut with a diamond knife on an ULTRACUT E ultramicrotome (Leica) and transferred to formvar-coated grids. They were double stained with 1% (v/v) uranyl acetate for 20 min and with lead citrate solution for 10 min. After washing with distilled water, the samples were observed with a JEM-1400 transmission electron microscope (JEOL).
Quantitative analysis of etioplast ultrastructures was performed with ImageJ software. Etioplasts with no clear PLBs or two or more PLBs were eliminated from the analysis. The circularity index of PLBs and etioplasts was calculated as follows: 4 × π × area/(perimeter2). The unit of PLBs was defined as a low-electron-density region surrounded by a high-density membrane area in the lattice of PLBs.
Accession Numbers
Sequence data of the genes investigated in this article can be found in The Arabidopsis Information Resource under the following accession numbers: ACT8 (AT1G49240), MGD1 (AT4G31780), HEMA1 (AT1G58290), CHLH (AT5G13630), CHLD (AT1G08520), CHLI1 (AT4G18480), GUN4 (AT3G59400), CHLM (AT4G25080), CHL27 (AT3G56940), LCAA (AT5G58250), psaA (ATCG00350), psbA (ATCG00020), petB (ATCG00720), rbcL (ATCG00490), rps14 (ATCG00330), accD (ATCG00500), rpoB (ATCG00190), LHCA4 (AT3G47470), LHCB6 (AT1G15820), AAA-ATPase (AT3G28580), BAP1 (AT3G61190), APX2 (AT3G09640), and ZAT10 (AT1G27730).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. T4 generation of amiR-MGD1 transgenic lines.
Supplemental Figure S2. MGD1 expression and Pchlide accumulation in etiolated seedlings of amiR-MGD1 L2 lines and mgd1-1.
Supplemental Figure S3. Size of cotyledons in 4-d-old etiolated seedlings of amiR-MGD1 L4w.
Supplemental Figure S4. Absorbance spectra of pigments extracted from 4-d-old etiolated seedlings of amiR-MGD1 L4w grown under −DEX and +DEX conditions.
Supplemental Figure S5. In situ 77K Pchlide fluorescence spectra in etiolated cotyledons of amiR-MGD1 L4w and mgd1-1.
Supplemental Figure S6. Ultrastructure of etioplasts in cotyledon cells of 4-d-old etiolated seedlings of amiR-MGD1 L4w.
Supplemental Figure S7. Distribution of the PLB unit area in each etioplast of amiR-MGD1 L4w seedlings.
Supplemental Figure S8. HPLC analysis of 4-d-old etiolated seedlings.
Supplemental Figure S9. MGD1 expression and Chl accumulation in mgd1-1 seedlings grown under the light.
Supplemental Figure S10. Correlation between MGD1 mRNA level and Chl content.
Supplemental Figure S11. T5 generation of amiR-MGD1 transgenic lines.
Supplemental Table S1. Oligonucleoide primers used for quantitative reverse transcription-PCR analysis.
Acknowledgments
We thank Paul Jarvis (Department of Plant Sciences, University of Oxford) for supplying the mgd1-1 mutant, Nobuyoshi Mochizuki (Department of Botany, Graduate School of Science, Kyoto University) for the chlm and chl27 mutants, and Megumi Kobayashi (Department of Chemical and Biological Sciences, Faculty of Science, Japan Women’s University) for technical assistance in transmission electron microscopy analysis.
Glossary
- PLB
prolamellar body
- PT
prothylakoid
- Chl
chlorophyll
- Pchlide
protochlorophyllide
- Chlide
chlorophyllide
- ALA
5-aminolevulinic acid
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- DEX
dexamethasone
Footnotes
This work was supported by the Japan Society for the Promotion of Science (KAKENHI grant no. 16J10176 to S.F., grant no. 26711016 to K.K., grant no. 16K07393 to T.M., and grant no. 26440170 to N.N.).
References
- Alawady AE, Grimm B (2005) Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J 41: 282–290 [DOI] [PubMed] [Google Scholar]
- Albus CA, Salinas A, Czarnecki O, Kahlau S, Rothbart M, Thiele W, Lein W, Bock R, Grimm B, Schöttler MA (2012) LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol 160: 1923–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Boardman NK (1964) Studies on the greening of dark-grown bean plants. Aust J Biol Sci 17: 93–101 [Google Scholar]
- Ankele E, Kindgren P, Pesquet E, Strand A (2007) In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 19: 1964–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H, Schöttler MA, Kelly AA, Sundqvist C, Dörmann P, Karim S, Jarvis P (2008) Monogalactosyldiacylglycerol deficiency in Arabidopsis affects pigment composition in the prolamellar body and impairs thylakoid membrane energization and photoprotection in leaves. Plant Physiol 148: 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Fung P, Chow TF, Foong JA, Provart NJ, Cutler SR (2008) Elucidating the germination transcriptional program using small molecules. Plant Physiol 147: 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale SI. (1999) Enzymes of chlorophyll biosynthesis. Photosynth Res 60: 43–73 [Google Scholar]
- Benning C, Ohta H (2005) Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem 280: 2397–2400 [DOI] [PubMed] [Google Scholar]
- Block MA, Tewari AK, Albrieux C, Maréchal E, Joyard J (2002) The plant S-adenosyl-L-methionine:Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. Eur J Biochem 269: 240–248 [DOI] [PubMed] [Google Scholar]
- Böddi B, Lindsten A, Ryberg M, Sundqvist C (1989) On the aggregational states of protochlorophyllide and its protein complexes in wheat etioplasts. Physiol Plant 76: 135–143 [Google Scholar]
- Brentel I, Selstam E, Lindblom G (1985) Phase equilibria of mixtures of plant galactolipids: the formation of a bicontinuous cubic phase. Biochim Biophys Acta 812: 816–826 [Google Scholar]
- Brzezowski P, Richter AS, Grimm B (2015) Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim Biophys Acta 1847: 968–985 [DOI] [PubMed] [Google Scholar]
- Demé B, Cataye C, Block MA, Maréchal E, Jouhet J (2014) Contribution of galactoglycerolipids to the 3-dimensional architecture of thylakoids. FASEB J 28: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Dorne AJ, Joyard J, Douce R (1990) Do thylakoids really contain phosphatidylcholine? Proc Natl Acad Sci USA 87: 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engdahl S, Aronsson H, Sundqvist C, Timko MP, Dahlin C (2001) Association of the NADPH:protochlorophyllide oxidoreductase (POR) with isolated etioplast inner membranes from wheat. Plant J 27: 297–304 [DOI] [PubMed] [Google Scholar]
- Franck F, Sperling U, Frick G, Pochert B, van Cleve B, Apel K, Armstrong GA (2000) Regulation of etioplast pigment-protein complexes, inner membrane architecture, and protochlorophyllide a chemical heterogeneity by light-dependent NADPH:protochlorophyllide oxidoreductases A and B. Plant Physiol 124: 1678–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Kobayashi K, Nakamura Y, Wada H (2014) Inducible knockdown of MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE1 reveals roles of galactolipids in organelle differentiation in Arabidopsis cotyledons. Plant Physiol 166: 1436–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabruk M, Mysliwa-Kurdziel B, Kruk J (2017) MGDG, PG and SQDG regulate the activity of light-dependent protochlorophyllide oxidoreductase. Biochem J 474: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Goslings D, Meskauskiene R, Kim C, Lee KP, Nater M, Apel K (2004) Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J 40: 957–967 [DOI] [PubMed] [Google Scholar]
- Gunning BES. (1965) The greening process in plastids. 1. The structure of the prolamellar body. Protoplasma 60: 111–130 [Google Scholar]
- Heyes DJ, Hunter CN (2005) Making light work of enzyme catalysis: protochlorophyllide oxidoreductase. Trends Biochem Sci 30: 642–649 [DOI] [PubMed] [Google Scholar]
- Hirashima M, Tanaka R, Tanaka A (2009) Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol 50: 719–729 [DOI] [PubMed] [Google Scholar]
- Jarvis P, Dörmann P, Peto CA, Lutes J, Benning C, Chory J (2000) Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc Natl Acad Sci USA 97: 8175–8179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Gibson LCD, Henningsen KW, Hunter CN (1996) Expression of the chlI, chlD, and chlH genes from the cyanobacterium Synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J Biol Chem 271: 16662–16667 [DOI] [PubMed] [Google Scholar]
- Klement H, Helfrich M, Oster U, Schoch S, Rüdiger W (1999) Pigment-free NADPH:protochlorophyllide oxidoreductase from Avena sativa L: purification and substrate specificity. Eur J Biochem 265: 862–874 [DOI] [PubMed] [Google Scholar]
- Kobayashi K. (2016) Role of membrane glycerolipids in photosynthesis, thylakoid biogenesis and chloroplast development. J Plant Res 129: 565–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H (2009a) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H (2007) Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci USA 104: 17216–17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Masuda T, Takamiya K, Ohta H (2006) Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J 47: 238–248 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Nakamura Y, Ohta H (2009b) Type A and type B monogalactosyldiacylglycerol synthases are spatially and functionally separated in the plastids of higher plants. Plant Physiol Biochem 47: 518–525 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Narise T, Sonoike K, Hashimoto H, Sato N, Kondo M, Nishimura M, Sato M, Toyooka K, Sugimoto K, et al. (2013) Role of galactolipid biosynthesis in coordinated development of photosynthetic complexes and thylakoid membranes during chloroplast biogenesis in Arabidopsis. Plant J 73: 250–261 [DOI] [PubMed] [Google Scholar]
- Kopečná J, Pilný J, Krynická V, Tomčala A, Kis M, Gombos Z, Komenda J, Sobotka R (2015) Lack of phosphatidylglycerol inhibits chlorophyll biosynthesis at multiple sites and limits chlorophyllide reutilization in Synechocystis sp. strain PCC 6803. Plant Physiol 169: 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopetz KJ, Kolossov VL, Rebeiz CA (2004) Chloroplast Biogenesis 89: development of analytical tools for probing the biosynthetic topography of photosynthetic membranes by determination of resonance excitation energy transfer distances separating metabolic tetrapyrrole donors from chlorophyll a acceptors. Anal Biochem 329: 207–219 [DOI] [PubMed] [Google Scholar]
- Kowalewska ŁM, Mazur R, Suski S, Garstka M, Mostowska A (2016) Three-dimensional visualization of the tubular-lamellar transformation of the internal plastid membrane network during runner bean chloroplast biogenesis. Plant Cell 28: 875–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Masuda T. (2008) Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth Res 96: 121–143 [DOI] [PubMed] [Google Scholar]
- Masuda T, Fujita Y (2008) Regulation and evolution of chlorophyll metabolism. Photochem Photobiol Sci 7: 1131–1149 [DOI] [PubMed] [Google Scholar]
- Masuda T, Fusada N, Oosawa N, Takamatsu K, Yamamoto YY, Ohto M, Nakamura K, Goto K, Shibata D, Shirano Y, et al. (2003) Functional analysis of isoforms of NADPH:protochlorophyllide oxidoreductase (POR), PORB and PORC, in Arabidopsis thaliana. Plant Cell Physiol 44: 963–974 [DOI] [PubMed] [Google Scholar]
- Melis A, Spangfort M, Andersson B (1987) Light-absorption and electron-transport balance between photosystem II and photosystem I in spinach chloroplasts. Photochem Photobiol 45: 129–136 [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A (2008) The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA 105: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro I, Dalla Vecchia F, La Rocca N, Navari-Izzo F, Quartacci MF, Di Baccio D, Rüdiger W, Rascio N (2004) Impaired carotenogenesis can affect organization and functionality of etioplast membranes. Physiol Plant 122: 123–132 [Google Scholar]
- Moulin M, Smith AG (2005) Regulation of tetrapyrrole biosynthesis in higher plants. Biochem Soc Trans 33: 737–742 [DOI] [PubMed] [Google Scholar]
- Nagata N, Tanaka R, Tanaka A (2007) The major route for chlorophyll synthesis includes [3,8-divinyl]-chlorophyllide a reduction in Arabidopsis thaliana. Plant Cell Physiol 48: 1803–1808 [DOI] [PubMed] [Google Scholar]
- Ohta H, Shimojima M, Ookata K, Masuda T, Shioi Y, Takamiya K (1995) A close relationship between increases in galactosyltransferase activity and the accumulation of galactolipids during plastid development in cucumber seedlings. Plant Cell Physiol 36: 1115–1120 [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J, Gräfe S, Kruse E, Hänel F, Grimm B (1997) Mg-chelatase of tobacco: identification of a Chl D cDNA sequence encoding a third subunit, analysis of the interaction of the three subunits with the yeast two-hybrid system, and reconstitution of the enzyme activity by co-expression of recombinant CHL D, CHL H and CHL I. Plant J 12: 981–990 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Mock HPP, Tanaka R, Kruse E, Grimm B (2000) Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol 122: 1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter E, Rothbart M, Oelze ML, Shalygo N, Dietz KJ, Grimm B (2010) Mg protoporphyrin monomethylester cyclase deficiency and effects on tetrapyrrole metabolism in different light conditions. Plant Cell Physiol 51: 1229–1241 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Rowe JD, Griffiths WT (1995) Protochlorophyllide reductase in photosynthetic prokaryotes and its role in chlorophyll synthesis. Biochem J 311: 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicke H, Hartwig AS, Firtzlaff V, Richter AS, Glässer C, Maier K, Finkemeier I, Grimm B (2014) Induced deactivation of genes encoding chlorophyll biosynthesis enzymes disentangles tetrapyrrole-mediated retrograde signaling. Mol Plant 7: 1211–1227 [DOI] [PubMed] [Google Scholar]
- Schoefs B. (2001) The protochlorophyllide-chlorophyllide cycle. Photosynth Res 70: 257–271 [DOI] [PubMed] [Google Scholar]
- Schoefs B, Franck F (2003) Protochlorophyllide reduction: mechanisms and evolutions. Photochem Photobiol 78: 543–557 [DOI] [PubMed] [Google Scholar]
- Selstam E, Sandelius AS (1984) A comparison between prolamellar bodies and prothylakoid membranes of etioplasts of dark-grown wheat concerning lipid and polypeptide composition. Plant Physiol 76: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selstam E, Schelin J, Brain T, Williams WP (2002) The effects of low pH on the properties of protochlorophyllide oxidoreductase and the organization of prolamellar bodies of maize (Zea mays). Eur J Biochem 269: 2336–2346 [DOI] [PubMed] [Google Scholar]
- Shibata K. (1957) Spectroscopic studies on chlorophyll formation in intact leaves. J Biochem 44: 147–173 [Google Scholar]
- Simková K, Moreau F, Pawlak P, Vriet C, Baruah A, Alexandre C, Hennig L, Apel K, Laloi C (2012) Integration of stress-related and reactive oxygen species-mediated signals by topoisomerase VI in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 16360–16365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeller L, Solymosi K, Fidy J, Böddi B (2003) Activation parameters of the blue shift (Shibata shift) subsequent to protochlorophyllide phototransformation. Biochim Biophys Acta 1651: 130–138 [DOI] [PubMed] [Google Scholar]
- Solymosi K, Schoefs B (2010) Etioplast and etio-chloroplast formation under natural conditions: the dark side of chlorophyll biosynthesis in angiosperms. Photosynth Res 105: 143–166 [DOI] [PubMed] [Google Scholar]
- Solymosi K, Smeller L, Ryberg M, Sundqvist C, Fidy J, Böddi B (2007) Molecular rearrangement in POR macrodomains as a reason for the blue shift of chlorophyllide fluorescence observed after phototransformation. Biochim Biophys Acta 1768: 1650–1658 [DOI] [PubMed] [Google Scholar]
- Sperling U, Franck F, van Cleve B, Frick G, Apel K, Armstrong GA (1998) Etioplast differentiation in Arabidopsis: both PORA and PORB restore the prolamellar body and photoactive protochlorophyllide-F655 to the cop1 photomorphogenic mutant. Plant Cell 10: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T (2011) Tetrapyrrole metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0145, doi/10.1199/tab.0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Terry MJ, Kendrick RE (1999) Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol 119: 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Scheller HV, Merchant S, Jensen PE (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA 100: 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C, Havaux M (2009) Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci 14: 219–228 [DOI] [PubMed] [Google Scholar]
- Vothknecht UC, Kannangara CG, von Wettstein D (1996) Expression of catalytically active barley glutamyl tRNAGlu reductase in Escherichia coli as a fusion protein with glutathione S-transferase. Proc Natl Acad Sci USA 93: 9287–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Grimm B (2015) Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth Res 126: 189–202 [DOI] [PubMed] [Google Scholar]
- Williams WP, Selstam E, Brain T (1998) X-ray diffraction studies of the structural organisation of prolamellar bodies isolated from Zea mays. FEBS Lett 422: 252–254 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Ping W, Wu H, Li M, Gu D, Xu Y (2013) Monogalactosyldiacylglycerol deficiency in tobacco inhibits the cytochrome b6f-mediated intersystem electron transport process and affects the photostability of the photosystem II apparatus. Biochim Biophys Acta 1827: 709–722 [DOI] [PubMed] [Google Scholar]
- Zapata M, Rodríguez F, Garrido JL (2000) Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar Ecol Prog Ser 195: 29–45 [Google Scholar]