Novel missense alleles of the Arabidopsis PEX1 ATPase reveal essential peroxisomal roles that impact embryogenesis and plant growth.
Abstract
A variety of metabolic pathways are sequestered in peroxisomes, conserved organelles that are essential for human and plant survival. Peroxin (PEX) proteins generate and maintain peroxisomes. The PEX1 ATPase facilitates recycling of the peroxisome matrix protein receptor PEX5 and is the most commonly affected peroxin in human peroxisome biogenesis disorders. Here, we describe the isolation and characterization of, to our knowledge, the first Arabidopsis (Arabidopsis thaliana) pex1 missense alleles: pex1-2 and pex1-3. pex1-2 displayed peroxisome-related defects accompanied by reduced PEX1 and PEX6 levels. These pex1-2 defects were exacerbated by growth at high temperature and ameliorated by growth at low temperature or by PEX6 overexpression, suggesting that PEX1 enhances PEX6 stability and vice versa. pex1-3 conferred embryo lethality when homozygous, confirming that PEX1, like several other Arabidopsis peroxins, is essential for embryogenesis. pex1-3 displayed symptoms of peroxisome dysfunction when heterozygous; this semidominance is consistent with PEX1 forming a heterooligomer with PEX6 that is poisoned by pex1-3 subunits. Blocking autophagy partially rescued PEX1/pex1-3 defects, including the restoration of normal peroxisome size, suggesting that increasing peroxisome abundance can compensate for the deficiencies caused by pex1-3 and that the enlarged peroxisomes visible in PEX1/pex1-3 may represent autophagy intermediates. Overexpressing PEX1 in wild-type plants impaired growth, suggesting that excessive PEX1 can be detrimental. Our genetic, molecular, and physiological data support the heterohexamer model of PEX1-PEX6 function in plants.
Critical steps of vital metabolic pathways are housed in single lipid bilayer-bound organelles known as peroxisomes. For example, plant peroxisomes house enzymes for reactive oxygen species detoxification, photorespiration, the glyoxylate cycle, and β-oxidation (for review, see Hu et al., 2012; Reumann and Bartel, 2016). Proteins necessary for peroxisome biogenesis and maintenance are termed peroxins (PEX proteins), and most known peroxins facilitate protein import into the peroxisome matrix from their site of synthesis in the cytosol. Matrix proteins generally have a peroxisomal targeting signal (PTS1 or PTS2; Gould et al., 1989; Swinkels et al., 1991), which is recognized in the cytosol by receptors PEX5 (McCollum et al., 1993) or PEX7 (Marzioch et al., 1994), respectively. The binding of PEX7 to PEX5 in plants (Nito et al., 2002) and mammals (Braverman et al., 1998) couples the import of PTS2 and PTS1 proteins (Hayashi et al., 2005; Woodward and Bartel, 2005; Khan and Zolman, 2010; Ramón and Bartel, 2010). The actual transport step is not fully elucidated, but in yeast, PEX5 docks at the PEX14 peroxisomal membrane protein (Hayashi et al., 2000; Stanley and Wilmanns, 2006), where PEX5 membrane insertion allows cargo delivery into the peroxisomal matrix (Meinecke et al., 2010). Once in the peroxisome, the N-terminal region containing the PTS2 signal is cleaved in plants by the protease DEG15 (Helm et al., 2007). After cargo delivery, yeast PEX5 is monoubiquitinated or diubiquitinated (Kragt et al., 2005; Williams et al., 2007) to signal for retrotranslocation to the cytosol by PEX1 and PEX6 (Platta et al., 2005), allowing PEX5 reuse (Dammai and Subramani, 2001). Alternatively, polyubiquitinated PEX5 can be degraded by the proteasome (for review, see Platta and Erdmann, 2007). Although PEX5 ubiquitination has not been demonstrated directly in plants, analyses of PEX5 levels and localization in Arabidopsis (Arabidopsis thaliana) mutants defective in the peroxisomal ubiquitination machinery implicate similar pathways for recycling or degrading plant PEX5 (Mano et al., 2006; Ratzel et al., 2011; Kao et al., 2016).
PEX1 and PEX6 are the only members of the AAA family (for ATPases associated with diverse cellular activities; for review, see Patel and Latterich, 1998) implicated in peroxisome biogenesis and are closely related to p97, which functions in endoplasmic reticulum-associated protein degradation to retrotranslocate endoplasmic reticulum proteins to the cytosol (Ye et al., 2001). PEX1 and PEX6 have two ATPase domains, named D1 and D2 (Fig. 1D). Each ATPase domain usually contains Walker A and Walker B motifs needed for nucleotide binding and hydrolysis, respectively (for review, see Grimm et al., 2012) and an SRH domain needed for ATPase activity (Karata et al., 1999). In humans, mutations in PEX1 are the most common cause of peroxisome biogenesis disorders, a spectrum of generally fatal conditions in which peroxisomes are not formed or inefficiently import matrix proteins (for review, see Waterham and Ebberink, 2012). Similarly, yeast null pex1 mutants lack functional peroxisomes (Erdmann et al., 1989), and defects in the yeast PEX1 D2 lead to empty vesicles rather than functional peroxisomes (Birschmann et al., 2005).
Figure 1.
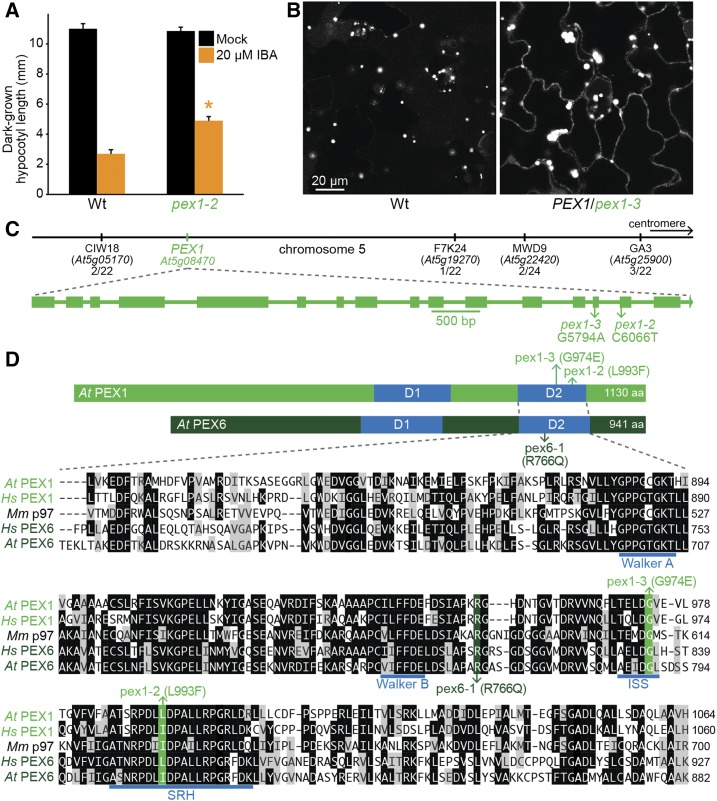
Identification of two mutations in PEX1 that modify the predicted second AAA ATPase domain of PEX1. A, Hypocotyl lengths of 5-d-old dark-grown seedlings (n = 20). Error bars show se, the asterisk indicates a statistically significant difference through Student’s t test (P < 0.001), and data are representative of three replicates. Wt, Wild type. B, Confocal micrographs of representative cotyledon epidermal cells of 8-d-old seedlings expressing peroxisome-targeted fluorescence (GFP-PTS1). Bar = 20 µm. C, The chromosome map indicates the positions of markers used to map pex1-3 and the ratios of recombinants to the number of chromosomes assessed. The gene diagram, with exons as rectangles and introns as lines, indicates the positions of pex1 mutations. D, Diagram of PEX1 and PEX6 proteins indicates the position of the AAA domains (D1 and D2) and the pex1 and pex6-1 mutations. The Arabidopsis (At) second AAA ATPase domain of PEX1 was aligned to that of human (Hs) PEX1, mouse (Mm) p97, human PEX6, and Arabidopsis PEX6 using ClustalW on Lasergene MegAlign (DNAStar). Residues are shaded when identical (black) or chemically similar (gray) in at least three sequences. The locations of the pex1-2, pex1-3, and pex6-1 lesions (green), the second region of homology (SRH; Santos, 2006), the intersubunit signaling (ISS) motif (Augustin et al., 2009), and the Walker A and B motifs needed for ATPase activity (blue) are indicated.
In yeast, PEX1 and PEX6 form a heterohexamer (Saffian et al., 2012) composed of three units of each peroxin arranged in a ring of alternating subunits with a central pore (Gardner et al., 2015) through which the complex might pull PEX5 (Ciniawsky et al., 2015). The PEX1-PEX6 interaction in yeast and humans requires the PEX1 D2 and D1 regions (Birschmann et al., 2005; Tamura et al., 2006). In addition to supporting heterohexamer assembly, ATP hydrolysis elicits conformational changes in the PEX1-PEX6 complex that facilitate the retrotranslocation of ubiquitinated PEX5 in yeast (for review, see Platta et al., 2016) and humans (Tamura et al., 2006). PEX5 retrotranslocation requires ATP hydrolysis by both PEX1 and PEX6 D2 regions (Platta et al., 2005; Tamura et al., 2006). Defects in PEX1 or PEX6 can lead to inefficient PEX5 retrotranslocation from the peroxisomal membrane accompanied by PEX5 polyubiquitination and degradation (for review, see Platta et al., 2016). Saccharomyces cerevisiae pex1 mutants accumulate polyubiquitinated PEX5 (Kragt et al., 2005), and pex1 mutants in human cells (Dodt and Gould, 1996) and Pichia pastoris (Collins et al., 2000) have reduced PEX5 levels.
The PEX6 N-terminal region recruits the PEX1-PEX6 heterohexamer to the peroxisome by interacting with the cytosolic domain of a tail-anchored peroxisomal membrane protein: PEX15 in yeast (Birschmann et al., 2003), PEX26 in human (Matsumoto et al., 2003), and PEX26, also known as ABERRANT PEROXISOME MORPHOLOGY9 or DAYU (Goto et al., 2011; Li et al., 2014), in Arabidopsis. Human PEX26 also binds to PEX14 and dissociates upon PEX1-PEX6 ATPase activity (Tamura et al., 2014). Because PEX14 recruits PEX5 to the peroxisome, this finding is consistent with the hypothesis that PEX26 positions PEX1-PEX6 in proximity to PEX14-bound PEX5 to facilitate PEX5 retrotranslocation to the cytosol.
Peroxisomes perform vital functions in plants. Arabidopsis is an oilseed plant that catabolizes triacylglycerol stored in the seed to support early seedling growth. Because peroxisomes are the sole site of fatty acid β-oxidation in plants (for review, see Graham, 2008), pex mutants can display reduced growth that is ameliorated by supplementation with an alternative fixed carbon source such as Suc (for review, see Bartel et al., 2014). Because peroxisomes also house the β-oxidation of the auxin precursor indole-butyric acid (IBA) to the active auxin indole-3-acetic acid (Zolman et al., 2000; Strader et al., 2010), screening for IBA resistance can yield mutants defective in peroxisome biogenesis (for review, see Bartel et al., 2014). Such mutants often are also resistant to the synthetic IBA analog 2,4-dichlorophenoxybutyric acid (2,4-DB), which is similarly β-oxidized to the auxinic herbicide 2,4-dichlorophenoxyacetic acid (Wain and Wightman, 1954; Hayashi et al., 1998). At a cellular level, defects in peroxisomal size, positioning, and matrix protein import can be visualized by tagging GFP with a PTS, which also has formed the basis of screens for mutants with peroxisome defects (Mano et al., 2006; Burkhart et al., 2013; Rinaldi et al., 2016). Because PTS2 cleavage occurs after import, mutations in peroxin genes often impair PTS2 processing (for review, see Bartel et al., 2014). The PTS2-processing protease DEG15 is a PTS1 protein (Helm et al., 2007), and so PTS2-processing defects can reflect PTS1 and/or PTS2 import defects.
As in yeast and mammals, Arabidopsis PEX6 binds PEX26, and PEX1 localization to peroxisomes requires both PEX6 and PEX26 (Goto et al., 2011). Arabidopsis pex6-1 carries a missense mutation in the D2 and has reduced PEX5 levels (Zolman and Bartel, 2004) due to proteasomal PEX5 degradation (Kao and Bartel, 2015), and PEX5 overexpression partially rescues pex6-1 defects (Zolman and Bartel, 2004; Burkhart et al., 2013), suggesting that reduced PEX5 function contributes to the defects of some plant pex mutants. PEX1 expression increases under starvation and senescence conditions (Charlton et al., 2005) and following wounding or hydrogen peroxide treatment (Lopez-Huertas et al., 2000), suggesting a role for PEX1 in oxidative stress responses. An RNA interference (RNAi) knockdown line targeting PEX1 shows peroxisome-related physiological defects (Nito et al., 2007). A reverse-genetic screen for EMBRYO-DEFECTIVE (EMB) genes identified two insertional mutations in PEX1 (emb2817-1 and emb2817-2) that confer embryos arresting at the preglobular stage (http://www.seedgenes.org/; Muralla et al., 2011).
Although mutants in almost all predicted single-gene peroxins have been isolated in forward-genetic screens for peroxisomal defects in Arabidopsis (for review, see Bartel et al., 2014), viable pex1 mutants have not been reported in plants. Here, we describe two Arabidopsis mutants in PEX1, pex1-2 and pex1-3, that display distinct peroxisome-related defects and harbor missense mutations in the region encoding the PEX1 D2. We found that PEX1 contributed to PEX6 accumulation and peroxisomal matrix protein import and that excessive PEX1 caused growth defects in the wild type. The pex1-3 mutation could not be recovered as a homozygote, confirming that PEX1 is necessary for embryogenesis, and was semidominant, consistent with the heterohexamer model of PEX1-PEX6 function.
RESULTS
pex1 Mutants Were Isolated from Screens for Peroxisomal Defects
Two pex1 mutants emerged from different screens for Arabidopsis seedlings with peroxisomal defects. We isolated pex1-2 from a screen for IBA resistance in dark-grown seedlings (Strader et al., 2011; Kao et al., 2016). Dark-grown pex1-2 seedlings displayed longer hypocotyls than the wild type on inhibitory concentrations of IBA (Fig. 1A). Whole-genome sequencing of pooled backcrossed lines revealed several mutations, including pex1-2 (Supplemental Fig. S1A; Supplemental Data Set S1). This C6606-to-T mutation changed Leu-993 to Phe in the D2 SRH (Fig. 1, C and D). The analogous residue is conserved as an Ile or Leu in human PEX1, Arabidopsis and human PEX6, and mouse p97 (Fig. 1D).
We isolated pex1-3 in a screen for seedlings displaying a pattern of peroxisome-targeted GFP (GFP-PTS1) distribution that differed from that of the wild type (Rinaldi et al., 2016). The pex1-3 mutant displayed larger GFP-PTS1 puncta than the wild type combined with GFP-PTS1 mislocalized to the cytosol (Fig. 1B). Recombination mapping suggested a causal heterozygous mutation at the top of chromosome 5 (Fig. 1C). Whole-genome sequencing of two backcrossed lines revealed a heterozygous pex1-3 mutation (Supplemental Fig. S1B; Supplemental Data Set S1) that changed Gly-974 to Glu. Gly-974 is in the D2 intersubunit signaling motif (Augustin et al., 2009) and is conserved in other AAA proteins, including human PEX1, Arabidopsis and human PEX6, and mouse p97 (Fig. 1D).
pex1-3 Is Semidominant and Confers Embryo Lethality When Homozygous
The peroxisomal defects of pex1-3 (Fig. 1B) segregated in every generation, and we were unable to obtain a homogenous population. This persistent heterogeneity suggested a semidominant causal mutation that conferred lethality when homozygous. Indeed, our recombination mapping (Fig. 1C) and whole-genome sequencing (Supplemental Fig. S1B) both revealed heterozygosity at the top of chromosome 5, consistent with a lethal mutation in this region.
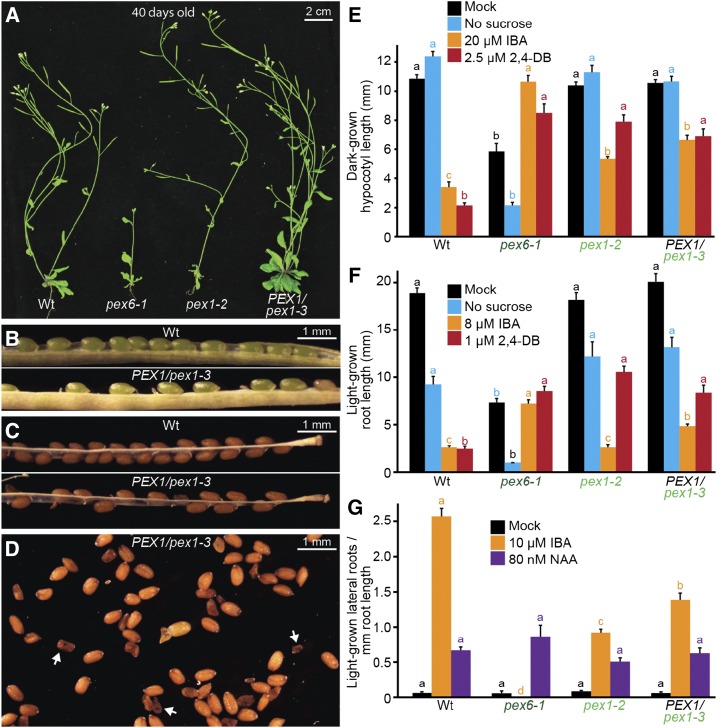
Adult PEX1/pex1-3 plants resembled wild-type plants in overall morphology (Fig. 2A). However, when we examined seed development in PEX1/pex1-3 plants, we found empty or missing seeds in green seedpods (Fig. 2B) and shriveled seeds among plump healthy seeds in mature seedpods (Fig. 2, C and D), consistent with the possibility that pex1-3/pex1-3 homozygous embryos arrest and die during development. Moreover, 1,188 individual progeny of PEX1/pex1-3 heterozygotes were genotyped in the course of this research: 431 were wild type, 757 were PEX1/pex1-3 heterozygotes, and none were pex1-3/pex1-3 homozygotes. This 1:1.9:0 ratio resembles the 1:2:0 ratio expected for Mendelian segregation of a homozygous-lethal mutation. Therefore, we characterized the effects of the pex1-3 mutation as a heterozygote.
Figure 2.
pex1-3 is a semidominant mutation that is lethal when homozygous, and pex1 mutants display IBA and 2,4-DB resistance in hypocotyl elongation, 2,4-DB resistance in root elongation, and IBA resistance in lateral root promotion. A, Photographs of 40-d-old plants. Bar = 2 cm. B, Green seedpods from a PEX1/pex1-3 parent contained aborted seeds. Bar = 1 mm. C, Mature seedpods contained missing and shriveled seeds. Bar = 1 mm. D, Mature seeds from a PEX1/pex1-3 parent included shriveled seeds (e.g. white arrows). Bar = 1 mm. E, Hypocotyl lengths of 5-d-old dark-grown seedlings (n ≥ 19). F, Root lengths of 8-d-old light-grown seedlings (n ≥ 13). G, Seedlings were grown in the absence of hormone for 4 d and then transferred to either medium without hormone or supplemented with the indicated hormone for an additional 4 d. The numbers of lateral roots and root lengths were measured, and the ratio is shown (n ≥ 12). Error bars in E to G show se, and data are representative of three replicates. Significant differences (one-way ANOVA, P < 0.001) for each growth condition are marked with different letters above the bars. NAA, 1-Naphthaleneacetic acid; Wt, wild type.
Mutations in PEX1 Lead to Peroxisomal Defects
We compared our pex1 mutants to pex6-1, a mutant defective in the PEX1-interacting ATPase (Zolman and Bartel, 2004; Goto et al., 2011), using various assays that monitor peroxisome function. Like PEX1/pex1-3, adult pex1-2 plants generally resembled wild-type plants (Fig. 2A), whereas pex6-1 is a pale-green dwarf plant (Fig. 2A; Zolman and Bartel, 2004). pex1-2 and PEX1/pex1-3 were partially resistant to the inhibitory effects of 2,4-DB on hypocotyl and root elongation (Fig. 2, E and F), somewhat resistant to the inhibitory effects of IBA on hypocotyl elongation in the dark (Fig. 2E), but less noticeably resistant to the inhibitory effects of IBA on root elongation in the light (Fig. 2F). In contrast, pex6-1 roots and hypocotyls are clearly resistant to both precursors (Fig. 2, E and F; Zolman and Bartel, 2004; Burkhart et al., 2013). Similarly, pex1-2 and PEX1/pex1-3 displayed only partial resistance to the promotion of lateral roots by IBA (Fig. 2G), compared with the complete resistance of pex6-1 in this assay (Zolman and Bartel, 2004). This IBA resistance was not due to a general auxin-response defect, as the mutants produced lateral roots like the wild type in response to the auxin analog 1-naphthaleneacetic acid (Fig. 2G), which does not require β-oxidation for activity.
pex1 mutants did not display consistent growth defects attributable to reduced fatty acid mobilization. Unlike pex6-1, pex1-2 and PEX1/pex1-3 resembled the wild type when grown on medium without Suc (Fig. 2, E and F). Together, both physiological assays (Fig. 2, E–G) and adult morphology (Fig. 2A) indicated that peroxisome function was less disrupted in pex1-2 and PEX1/pex1-3 than in the pex6-1 mutant.
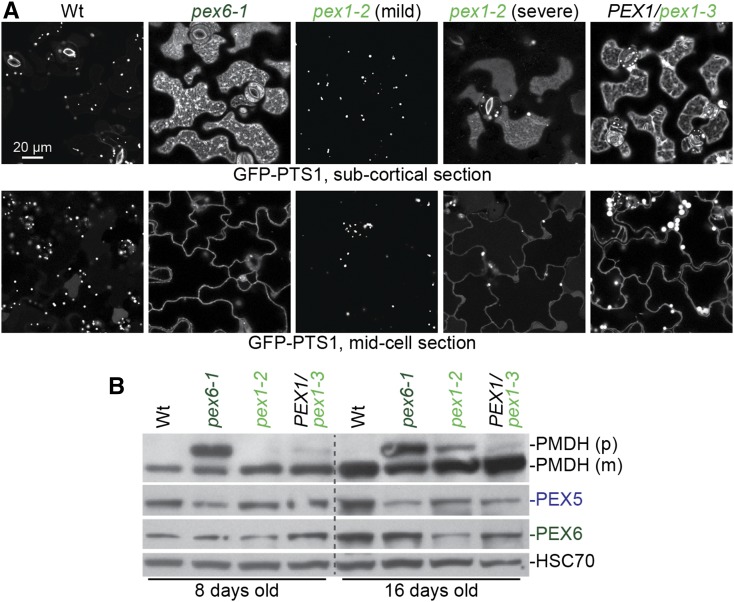
Along with physiological defects, both pex1 mutants displayed defects in matrix protein import. pex6-1 shows substantial GFP-PTS1 mislocalization to the cytosol and few GFP-PTS1 puncta (Fig. 3A; Burkhart et al., 2014), indicating a strong defect in peroxisomal matrix protein import. Similarly, PEX1/pex1-3 seedlings showed both cytosolic GFP-PTS1 localization and GFP-PTS1 puncta, some of which were larger than wild-type puncta (Figs. 1B and 3A). In contrast, pex1-2 was variable in this assay: some 8-d-old seedlings appeared similar to wild-type seedlings, whereas others showed GFP-PTS1 mislocalization to the cytosol along with GFP-PTS1 puncta (Fig. 3A).
Figure 3.
pex1 mutants display peroxisomal matrix protein import defects. A, Confocal micrographs of cotyledon epidermal cells of 8-d-old seedlings expressing peroxisome-targeted fluorescence (GFP-PTS1). Representative confocal micrographs demonstrating the range of phenotypes observed in pex1-2 are shown, from no apparent GFP-PTS1 mislocalization (mild) to substantial GFP-PTS1 mislocalization (severe). Images were taken at two planes, a subcortical section showing diffuse cytosolic fluorescence (top row) and a midcell section where cytosolic fluorescence outlines the cell (bottom row). Data are representative of three replicates. Bar = 20 µm. B, Seedlings were grown in the absence of hormone for 4 d and then transferred to medium supplemented with 10 µm IBA for an additional 4 d. Seedlings were then used for immunoblotting (8-d-old samples) or moved to medium without hormone for an additional 8 d prior to immunoblotting (16-d-old samples). For PEX1/pex1-3 lanes, progeny of a PEX1/pex1-3 individual were plated, and only individuals that displayed IBA resistance in lateral root production were collected. Seedling extracts were prepared for immunoblot analysis and serially probed with antibodies recognizing the indicated proteins. Peroxisomal malate dehydrogenase (PMDH) is translated as a precursor (p) with a PTS2 that is cleaved inside the peroxisome to yield mature (m) protein. HSC70 is a loading control. Data are representative of three replicates. Wt, Wild type.
In a second assay to monitor matrix protein import, we examined PTS2 processing in the mutants. Whereas pex6-1 had a notable PTS2-processing defect (Fig. 3B; Zolman et al., 2005), PEX1/pex1-3 presented only a slight PTS2-processing defect in young seedlings (Fig. 3B), and pex1-2 displayed a slight PTS2-processing defect in older plants (Fig. 3B).
PEX1 Overexpression Complements pex1 Mutants
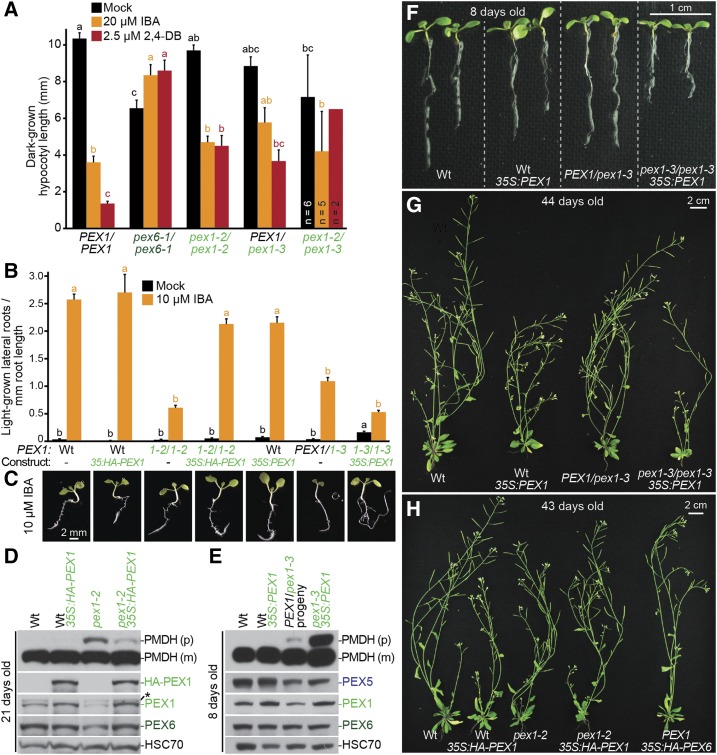
To confirm that the mutations we identified in PEX1 were responsible for the observed phenotypes, we performed a complementation assay by crossing pex1-2/pex1-2 and PEX1/pex1-3. The pex1-2/pex1-3 transheterozygote was underrepresented in the resultant F1: 37 individuals were pex1-2/pex1-3, whereas 79 were PEX1/pex1-2. This 1:2 ratio deviated from the expected 1:1 ratio, suggesting that some pex1-2/pex1-3 individuals died during embryogenesis. Furthermore, of the 37 pex1-2/pex1-3 transheterozygous individuals recovered, 24 barely germinated and did not develop into seedlings. Among the 13 transheterozygous pex1-2/pex1-3 seedlings that emerged from the seed coat, the two on a plate containing 2,4-DB had elongated hypocotyls (Fig. 4A), also suggesting failure to complement. The pex1-2/pex1-3 transheterozygotes that developed into seedlings did not survive to adulthood. This failure of pex1-2 to complement the lethality of pex1-3 suggested that both identified pex1 mutations were causal.
Figure 4.
PEX1 overexpression rescues IBA resistance in lateral root production and PTS2-processing defects in pex1-2 and lethality in pex1-3. A, pex1-2 and pex1-3 fail to complement each other. Hypocotyl lengths of 5-d-old dark-grown seedlings were measured. Error bars show se. For most lines, n ≥ 13; for the pex1-2/pex1-3 transheterozygote, most seedlings did not develop, and only seedlings that germinated and emerged from the seed coat were measured (n is indicated in each bar). Significant differences (one-way ANOVA, P < 0.001) for each growth condition are marked with different letters above the bars. pex1-2/pex1-3 was excluded from the 2,4-DB ANOVA for significance because of the low sample number. B, Seedlings were grown in the absence of hormone for 4 d and then transferred to either medium without hormone or supplemented with IBA for an additional 4 d. The numbers of lateral roots and root lengths were measured, and the ratio is shown. Error bars show se (n ≥ 14). Data are representative of three replicates. C, Micrographs of representative 8-d-old IBA-treated seedlings from B. Bar = 2 mm. D, Twenty-one-day-old plants were prepared for immunoblot analysis and serially probed with antibodies recognizing the indicated proteins. PMDH is translated as a precursor (p) with a PTS2 that is cleaved inside the peroxisome to yield mature (m) protein. HSC70 is a loading control. The asterisk indicates a cross-reacting band. E, Eight-day-old seedlings were prepared for immunoblot analysis and serially probed with antibodies recognizing the indicated proteins. Progeny of a PEX1/pex1-3 individual were pooled for immunoblotting. PMDH is translated as a precursor with a PTS2 that is cleaved inside the peroxisome to yield mature protein. HSC70 is a loading control. F, Photograph of 8-d-old seedlings grown in the light on agar-based medium. Bar = 1 cm. G, Photograph of 44-d-old soil-grown plants. Bar = 2 cm. H, Photograph of 43-d-old soil-grown plants. Bar = 2 cm. Wt, Wild type.
We also introduced constructs that used the cauliflower mosaic virus 35S promoter to express untagged or HA-tagged PEX1 cDNAs into the pex1 mutants and tested for complementation. HA-PEX1 expression in pex1-2 restored IBA sensitivity in lateral root promotion (Fig. 4, B and C) and improved PTS2 processing (Fig. 4D), again indicating that the pex1-2 mutation was causal. Moreover, we were able to obtain homozygous pex1-3/pex1-3 lines overexpressing PEX1. These lines produced viable offspring, indicating that the pex1-3 mutation was responsible for the lethality. pex1-3 35S:PEX1 homozygotes were smaller than the wild type as seedlings (Fig. 4F) and adults (Fig. 4G) and still displayed peroxisomal defects, including IBA resistance in lateral root production (Fig. 4, B and C) and incomplete PTS2 processing (Fig. 4E). These defects may persist because levels of wild-type PEX1 are insufficient to counteract the defective pex1-3 protein that remains in these plants or because PEX1 is overexpressed to detrimental levels.
We transformed wild-type plants with constructs overexpressing PEX1 or PEX6 in parallel. Transformants for 35S:HA-PEX6 were easy to obtain and resembled the wild type in growth (Fig. 4H), whereas transformants for 35S:HA-PEX1 were fewer than 35S:HA-PEX6 transformants and smaller than the wild type, suggesting that PEX1 overexpression is deleterious. To directly compare the consequences of PEX1 overexpression in the wild type and pex1-2, we crossed a pex1-2 line rescued with 35S:HA-PEX1 (Fig. 4, B–D) to the wild type and isolated the transgene in a wild-type PEX1 background. Interestingly, pex1-2 carrying 35S:HA-PEX1 was morphologically more similar to the wild type than the wild type was to 35S:HA-PEX1 (Fig. 4H). Thus, the HA-PEX1 levels produced by this transformation event are sufficient to restore PEX1 function in pex1-2 but appear to cause growth defects in a wild-type background, suggesting that excessive PEX1, or PEX1 expressed ectopically in tissues normally lacking PEX1, is deleterious. Consistent with this hypothesis, we found that PEX1 and HA-PEX1 levels were increased only modestly compared with endogenous PEX1 levels (Fig. 4, D and E) both in the wild type and in lines that restore viability to pex1-3 and IBA responsiveness and PTS2 processing to pex1-2.
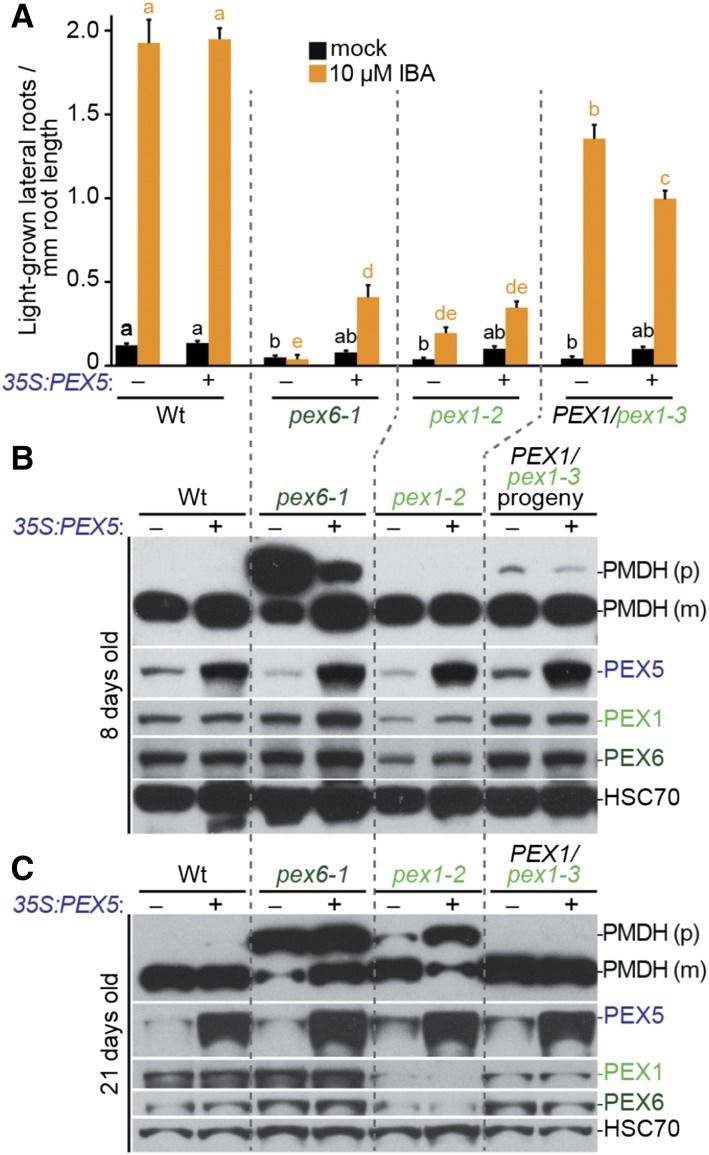
PEX5 Overexpression Fails to Rescue pex1 Mutants
PEX5 overexpression partially rescues pex6-1 defects in growth, Suc dependence, and PTS2 processing (Zolman and Bartel, 2004; Burkhart et al., 2013), presumably because PEX5 is excessively organelle associated (Ratzel et al., 2011) and degraded by the proteasome in pex6-1 (Kao and Bartel, 2015). Unlike in pex6-1, PEX5 levels generally resembled wild-type levels in PEX1/pex1-3 (Figs. 3B, 4E, and 5, B and C), and we did not find consistently low PEX5 levels in pex1-2 (Figs. 3B and 5, B and C). Also unlike pex6-1 seedlings, PEX5 was distributed in organellar and cytosolic fractions similarly to the wild type in extracts from pex1-2 and PEX1/pex1-3 progeny (Supplemental Fig. S2). Furthermore, overexpressing PEX5 in pex1-2 or PEX1/pex1-3 did not improve IBA responsiveness (Fig. 5A) or PTS2 processing (Fig. 5, B and C). In fact, PEX5 overexpression worsened the PTS2-processing defect of pex1-2 (Fig. 5C).
Figure 5.
PEX5 overexpression did not rescue pex1 mutants. A, Seedlings were grown in the absence of hormone for 4 d and then transferred to either medium without hormone or supplemented with IBA for an additional 4 d. The numbers of lateral roots and root lengths were measured, and the ratio is shown. Error bars show se (n ≥ 20). Significant differences (one-way ANOVA, P < 0.001) for each growth condition are marked with different letters above the bars. B and C, Eight-day-old seedlings (B) or 21-d-old plants (C) were prepared for immunoblot analysis and serially probed with antibodies recognizing the indicated proteins. PMDH is translated as a precursor (p) with a PTS2 that is cleaved inside the peroxisome to yield mature (m) protein. HSC70 is a loading control. Wt, Wild type.
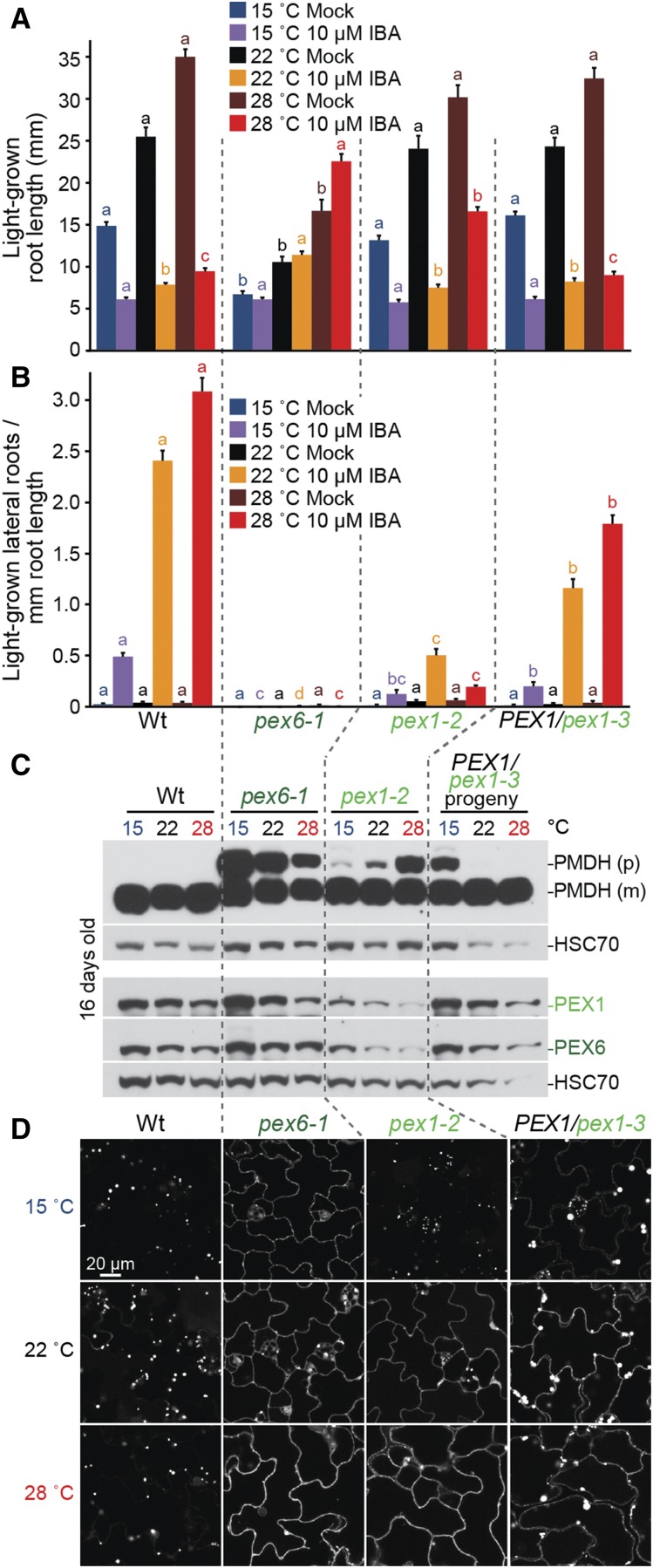
pex1-2 Protein Accumulation and pex1-2 Peroxisomal Function Are Reduced at Elevated Growth Temperature
We noticed that older pex1-2 seedlings accumulated less PEX1 protein than the wild type or the PEX1/pex1-3 heterozygote (Fig. 4D). To further investigate the connection between PEX1 protein levels and peroxisomal defects, we examined peroxisome-related phenotypes following growth at temperatures higher (28°C) or lower (15°C) than our standard growth temperature (22°C), anticipating that higher temperatures might exacerbate pex1-2 defects if they were caused by pex1-2 protein misfolding and consequent degradation.
Growth at mildly elevated temperature increases wild-type hypocotyl elongation (Gray et al., 1998), root elongation (Rogg et al., 2001), and lateral root production (Wang et al., 2016). We found that growth at 28°C increased wild-type root elongation (Fig. 6A) and IBA-responsive lateral root production (Fig. 6B). pex6-1 was resitant to IBA (Fig. 6, A and B) and displayed GFP-PTS1 import defects (Fig. 6D) and impaired PTS2 processing (Fig. 6C) at all growth temperatures assayed, although PTS2-processing defects were more apparent at lower temperatures (Fig. 6C), perhaps due to deleterious accumulation of PEX5 in the peroxisomal membrane (Kao and Bartel, 2015). PEX1/pex1-3 generally responded to temperature similarly to the wild type (Fig. 6, A and B), and cytosolic mislocalization of GFP-PTS1 in PEX1/pex1-3 seedlings was apparent at all three growth temperatures (Fig. 6D), suggesting that the defects in this mutant are not related to PEX1-PEX6 protein levels. In fact, lower temperature appeared to worsen rather than ameliorate the PTS2-processing defect in PEX1/pex1-3 (Fig. 6C).
Figure 6.
pex1-2 accumulation is temperature dependent. A and B, Seedlings were grown in the absence of hormone for 4 d at 22°C and then transferred to either medium without hormone or supplemented with the indicated hormone for an additional 4 d at 15°C, 22°C, or 28°C. The numbers of lateral roots and root lengths were measured, and root lengths (A) and the density of lateral roots (B) are shown (n ≥ 16). Error bars show se, and data are representative of three replicates. Significant differences (one-way ANOVA, P < 0.001) for each growth condition are marked with different letters above the bars. C, Seedlings grown for 8 d at 22°C and 8 d at the indicated temperatures were prepared for immunoblot analysis and serially probed with antibodies recognizing the indicated proteins. PMDH and thiolase are translated as precursors (p); the PTS2 region is cleaved inside the peroxisome to yield mature (m) proteins. HSC70 is a loading control. D, Confocal micrographs of cotyledon epidermal cells of seedlings grown for 4 d at 22°C and 4 d at the indicated temperatures expressing peroxisome-targeted fluorescence (GFP-PTS1). Confocal micrographs demonstrating the most defective GFP-PTS1 import observed in pex1-2 (severe) are shown. Data are representative of three replicates. Wt, Wild type. Bar = 20 µm.
In contrast to PEX1/pex1-3, high temperature exacerbated pex1-2 IBA resistance in hypocotyl elongation (Fig. 6A) and lateral root production (Fig. 6B). Moreover, high temperature worsened and low temperature ameliorated pex1-2 PTS2 processing (Fig. 6C) and GFP-PTS1 import (Fig. 6D). These temperature-dependent defects in peroxisome physiology and import were accompanied by reduced and elevated PEX1 and PEX6 protein levels at high and low temperatures, respectively (Fig. 6C), suggesting that pex1-2 instability leads to PEX6 degradation and peroxisomal defects.
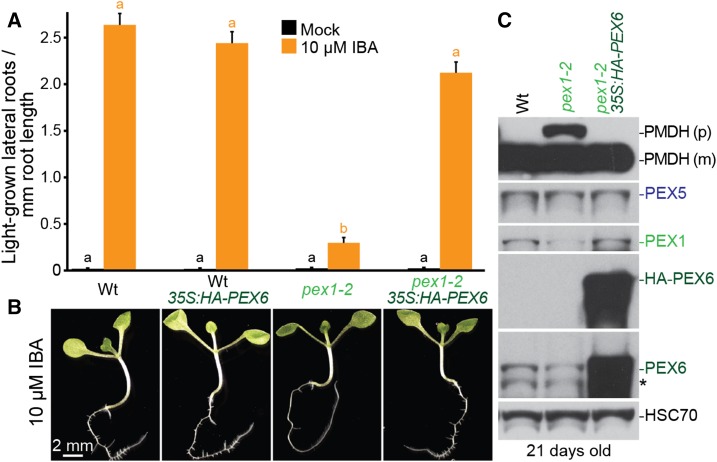
PEX6 Overexpression Rescues pex1-2 Defects
Our observation that PEX6 levels were reduced in pex1-2 but not in PEX1/pex1-3 (Figs. 5, B and C, and 6C) suggested that PEX6 stability decreases when PEX1 levels decline. To assess whether decreased PEX6 levels contributed to pex1-2 defects, we expressed HA-PEX6 from the 35S promoter in pex1-2. We found that HA-PEX6 expression restored pex1-2 IBA sensitivity in lateral root promotion (Fig. 7, A and B) and PTS2 processing (Fig. 7C). Moreover, HA-PEX6 expression increased pex1-2 protein levels to resemble wild-type PEX1 levels (Fig. 7C). These results suggest that increased PEX6 levels counteract the pex1-2 instability caused by the pex1-2 missense mutation and that reduced PEX6 and/or PEX1 levels contribute to the defects observed in the pex1-2 mutant.
Figure 7.
HA-PEX6 expression rescues IBA resistance in lateral root production, PTS2-processing defects, and low PEX1 levels in pex1-2. A, Seedlings were grown in the absence of hormone for 4 d and then transferred to either medium without hormone or supplemented with IBA for an additional 4 d. The numbers of lateral roots and root lengths were measured, and the ratio is shown. Error bars show se (n = 20). Significant differences (one-way ANOVA, P < 0.001) for each growth condition are marked with different letters above the bars. Data are representative of three replicates. B, Micrographs of representative 8-d-old IBA-treated seedlings from A. Bar = 2 mm. C, Twenty-one-day-old plants were prepared for immunoblot analysis and serially probed with antibodies recognizing the indicated proteins. PMDH is translated as a precursor (p) with a PTS2 that is cleaved inside the peroxisome to yield mature (m) protein. HSC70 is a loading control. The asterisk indicates a cross-reacting band. Wt, Wild type.
Because HA-PEX6 expression rescued pex1-2 defects, we attempted the reciprocal experiment to test whether PEX1 overexpression could rescue pex6-1. We crossed plants carrying PEX1-overexpressing constructs to pex6-1. However, we were unable to isolate homozygous pex6-1 carrying 35S:PEX1 or 35S:HA-PEX1, suggesting that PEX1 overexpression did not rescue pex6-1 defects.
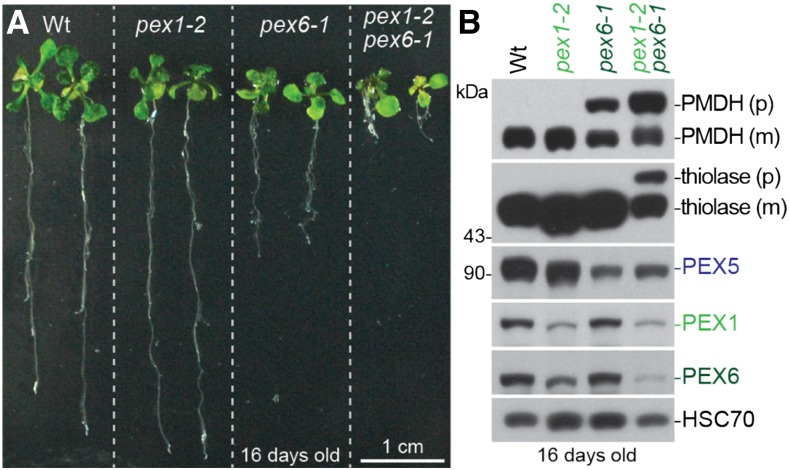
pex1-2 and pex6-1 Defects Are Synergistic
To further explore the genetic interactions between PEX1 and PEX6, we constructed a pex1-2 pex6-1 double mutant. pex1-2 pex6-1 seedlings were smaller than either single mutant (Fig. 8A) and displayed worsened PTS2-processing defects (Fig. 8B). The pex1-2 pex6-1 double mutants died as seedlings without leaving progeny, indicating that the peroxisomal defects of pex1-2 and pex6-1 were synergistic.
Figure 8.
pex1-2 exacerbates pex6-1 defects in growth and PTS2 processing. A, Photographs of 16-d-old seedlings grown on agar-based medium. Bar = 1 cm. B, Sixteen-day-old seedlings were prepared for immunoblot analysis and serially probed with antibodies recognizing the indicated proteins. PMDH and thiolase are translated as precursors (p); the PTS2 region is cleaved inside the peroxisome to yield mature (m) proteins. HSC70 is a loading control, and the positions of molecular mass markers are indicated on the left. Wt, Wild type.
We also attempted to isolate pex1-3 pex6-1 double mutants. Among progeny from a PEX1/pex1-3 PEX6/pex6-1 heterozygote, we found one PEX1/pex1-3 pex6-1/pex6-1 individual and one pex1-3/pex1-3 pex6-1/pex6-1 individual that were both nongerminated seeds. However, we were unable to isolate PEX1/pex1-3 pex6-1/pex6-1 or pex1-3/pex1-3 pex6-1/pex6-1 seedlings. This lethality suggests that peroxisomal defects of pex1-3 and pex6-1 also were synergistic.
Blocking Autophagy Partially Rescues PEX1/pex1-3 Defects
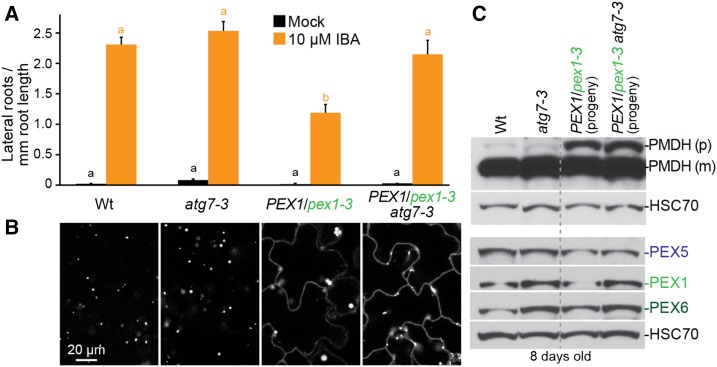
The enlarged GFP-PTS1 puncta observed in PEX1/pex1-3 (Figs. 1B, 3A, and 6D) were evocative of the enlarged peroxisome-targeted GFP puncta observed when the peroxisomal protease LON2 is dysfunctional (Farmer et al., 2013). Because preventing autophagy can restore peroxisomal functions to lon2 mutants, we combined the PEX1/pex1-3 mutation with atg7-3, a null mutation in an essential autophagy component (Doelling et al., 2002). We found that preventing autophagy rescued PEX1/pex1-3 IBA responsiveness (Fig. 9A) and GFP-PTS1 puncta size (Fig. 9B). Interestingly, these phenotypic ameliorations were not accompanied by notable improvements in PTS1 import (Fig. 9B) or PTS2 processing (Fig. 9C). These results indicate that intact autophagy machinery is required to develop enlarged GFP-PTS1 puncta in PEX1/pex1-3 but that the defects in matrix protein import in this mutant do not result from excessive autophagy of peroxisomes.
Figure 9.
Blocking autophagy partially rescues PEX1/pex1-3. A, Seedlings were grown in the absence of hormone for 4 d and then transferred to either medium without hormone or supplemented with IBA for an additional 4 d. The numbers of lateral roots and root lengths were measured, and the ratio is shown. Error bars show se (n ≥ 12). Significant differences (one-way ANOVA, P < 0.001) for each growth condition are marked with different letters above the bars. B, Confocal micrographs of cotyledon epidermal cells of 8-d-old seedlings expressing peroxisome-targeted fluorescence (GFP-PTS1). Data are representative of three replicates. Bar = 20 µm. C, Eight-day-old seedlings were prepared for immunoblot analysis and serially probed with antibodies recognizing the indicated proteins. Progeny of PEX1/pex1-3 or PEX1/pex1-3 atg7-3 individuals were pooled for immunoblotting. PMDH is translated as a precursor (p) with a PTS2 that is cleaved inside the peroxisome to yield mature (m) protein. HSC70 is a loading control. Wt, Wild type.
DISCUSSION
PEX1 Is Essential for Embryogenesis and Contributes to Peroxisome Function in Plants
PEX1 was among the first peroxins identified in yeast and mammals. However, nearly two decades after the first reports of Arabidopsis peroxin mutants (Hayashi et al., 1998; Zolman et al., 2000), viable mutants have been reported for all single-copy peroxin genes except for PEX1 (for review, see Bartel et al., 2014). This deficit is perhaps because null mutations in peroxins can be lethal in plants. In fact, PEX14 is the only Arabidopsis peroxin for which reported null alleles are viable (Monroe-Augustus et al., 2011). In contrast, lethality results from eliminating PEX26, the peroxisomal tether of the PEX1-PEX6 heterohexamer (Goto et al., 2011; Li et al., 2014), PEX13, which aids in docking of cargo-loaded PEX5-PEX7 to the peroxisome (Boisson-Dernier et al., 2008; Woodward et al., 2014), or any of the three peroxins implicated in PEX5 ubiquitination: PEX2 (Hu et al., 2002), PEX10 (Schumann et al., 2003; Sparkes et al., 2003), and PEX12 (Fan et al., 2005). In agreement with previous reports (http://www.seedgenes.org/; Muralla et al., 2011), we found that PEX1 also is essential in Arabidopsis: a homozygous pex1-3 missense mutation conferred lethality (Fig. 2, B–D) that was rescued by PEX1 overexpression (Fig. 4, F and G).
The pex1-2 and pex1-3 mutations both altered conserved residues in D2, the second AAA domain (Fig. 1D). An Ile-989-to-Thr mutation in the human PEX1 residue that is analogous to the Leu-993 residue mutated in pex1-2 (Fig. 1D) results in peroxisome biogenesis disorders when compounded with other PEX1 mutations (Maxwell et al., 2005; Smith et al., 2016). PEX1 D2 mutations can confer general peroxisomal defects (Birschmann et al., 2005) and decrease ATPase activity (Gardner et al., 2015) and PEX5 export in yeast (Platta et al., 2005), reduce PEX1 binding to PEX6 in yeast (Birschmann et al., 2005) and humans (Tamura et al., 2006), and impair PEX26 disassociation from PEX14 in humans (Tamura et al., 2014). The peroxisomal defects found in our two Arabidopsis PEX1 D2 mutants indicate that this domain also is important for peroxisome function in plants.
In contrast to the marked growth defects of a PEX1 RNAi line (Nito et al., 2007) and pex6-1 seedlings (Zolman and Bartel, 2004), pex1-2 and PEX1/pex1-3 seedlings grew similarly to the wild type even on medium lacking Suc (Fig. 2, E and F), indicating that sufficient PEX1 function remains in these mutants to metabolize enough fatty acids to achieve robust seedling growth. pex1-2 defects became more apparent with age: young pex1-2 seedlings displayed slight or no PTS2-processing defects (Figs. 3B, 5B, and 8B), whereas older pex1-2 plants displayed clear PTS2-processing defects (Figs. 3B, 4D, 5C, and 7C). This age dependence might explain the pex1-2 Suc independence and variability in GFP-PTS1 import that we observed in pex1-2 seedlings (Fig. 3A), as some seedlings might develop defects more quickly than others.
PEX1 Limits the Autophagy of Peroxisomes
Autophagy is a bulk degradation pathway to recycle cell constituents (Li and Vierstra, 2012). Peroxisome-specific autophagy (pexophagy) was discovered recently in plants (for review, see Young and Bartel, 2016); autophagy-defective seedlings have increased numbers of peroxisomes (Kim et al., 2013; Shibata et al., 2013). We isolated PEX1/pex1-3 from a microscopy-based screen for mutants with altered GFP-PTS1 patterning that were visible under relatively low magnification (Rinaldi et al., 2016). Although this screen yielded mutations in 15 genes that conferred aberrantly large or clustered peroxisomes (Rinaldi et al., 2016), PEX1/pex1-3 was the only peroxin mutant recovered. Because PTS1 import defects are common to numerous pex mutants (for review, see Bartel et al., 2014) that were not recovered in this screen, we presumably isolated PEX1/pex1-3 because of its enlarged GFP-PTS1 puncta (Figs. 1B and 3A), a phenotype that is not generally reported for pex mutants. The enlarged peroxisomes in PEX1/pex1-3 are reminiscent of pxn peroxisomes (Mano et al., 2011), which are defective in a peroxisomal transporter (Agrimi et al., 2012; Bernhardt et al., 2012; van Roermund et al., 2016), and lon2 peroxisomes (Farmer et al., 2013; Goto-Yamada et al., 2014), which are defective in a peroxisomal protease needed for sustained peroxisomal function (Lingard and Bartel, 2009). Indeed, four pxn and three lon2 loss-of-function alleles were recovered from the same GFP-PTS1-based screen (Rinaldi et al., 2016) from which we recovered PEX1/pex1-3. The large GFP-PTS1 puncta in pxn mutants are not reduced when autophagy is prevented (Rinaldi et al., 2016), whereas preventing autophagy in lon2 eliminates enlarged puncta (Farmer et al., 2013; Goto-Yamada et al., 2014), indicating that enlarged puncta can be pexophagy intermediates. As in lon2, we found that preventing autophagy in PEX1/pex1-3 eliminated the enlarged puncta (Fig. 9B), suggesting that these PEX1/pex1-3 structures were autophagy intermediates. Preventing autophagy in lon2 restores not only peroxisome size and IBA responsiveness but also matrix protein import and PTS2 processing (Farmer et al., 2013; Goto-Yamada et al., 2014), indicating that lon2 defects are largely due to the heightened pexophagy of otherwise functioning peroxisomes. As in lon2, PEX1/pex1-3 IBA responsiveness was restored when autophagy was prevented (Fig. 9A). Unlike in lon2, however, atg7-3 did not restore matrix protein import or PTS2 processing in PEX1/pex1-3 seedlings (Fig. 9, B and C). These differences suggest that, although PEX1 dysfunction triggers the pexophagy of peroxisomes that are still capable of IBA-to-indole-3-acetic acid conversion, preventing pexophagy is not the sole peroxisomal function of PEX1.
As in Arabidopsis PEX1/pex1-3, pexophagy is induced in yeast pex1 and pex6 mutants (Nuttall et al., 2014), perhaps because the ubiquitinated substrates that are not efficiently retrotranslocated from the peroxisomal membrane attract the autophagy machinery. Similarly, RNAi-mediated depletion of PEX1 or PEX26 in mammalian cells leads to the accumulation of ubiquitinated PEX5 and elevated pexophagy, and autophagy inhibitors improve peroxisome functioning in cells carrying the pex1-G843D missense allele (Law et al., 2017). Thus, preventing pexophagy appears to be a conserved function of the peroxisomal AAA ATPase complex.
PEX1 and PEX6 Are Interdependent
The pex1-3 mutation changes a conserved Gly residue in the PEX1 D2 intersubunit signaling region to a Glu residue (Fig. 1D). In related ATPases, this region interacts with the neighboring subunit near the ATP-binding site (Augustin et al., 2009). Mutation of the analogous residue (Gly-610) in the mammalian AAA homohexamer p97 to Ala, Val, or Cys reduces p97 ATPase activity by more than 50% (Huang et al., 2012). The similarity between PEX1 and p97 (Fig. 1D) suggests that the pex1-3 substitution may impair the ability of PEX1 to appropriately influence the ATPase activity of the neighboring PEX6 subunit.
The pex1-3 mutation conferred lethality when homozygous (Fig. 2, B–D) and semidominant peroxisome-related defects when heterozygous (Figs. 2, E–G, and 3, A and B). Some human PEX1 mutations also confer mild symptoms even when heterozygous (Majewski et al., 2011). Yeast PEX1 and PEX6 form a heterohexamer with three subunits of each protein (Saffian et al., 2012; Blok et al., 2015; Ciniawsky et al., 2015; Gardner et al., 2015), providing a rationale for the semidominance of a pex1 mutation. In a PEX1/pex1-3 heterozygote, most (seven of eight) PEX1-PEX6 heterohexamers would be expected to have at least one pex1-3 unit that might decrease the activity or stability of the complex, a phenomenon known as subunit poisoning.
Interestingly, PEX1/pex1-3 defects became less apparent with age. Eight-day-old PEX1/pex1-3 seedlings were partially IBA resistant in lateral root promotion (Figs. 2G and 4, B and C), had a slight PTS2-processing defect (Figs. 3B, 4E, and 5B), and displayed enlarged GFP-PTS1 puncta and cytosolic GFP-PTS1 mislocalization (Figs. 1B and 3A). In contrast, PEX1/pex1-3 displayed normal adult morphology (Figs. 2A and 4G) and fully processed PTS2 proteins in older seedlings (Fig. 5C). Perhaps pex1-3 is degraded over time, allowing hexamers consisting of wild-type PEX1 and PEX6 to accumulate. Although PEX1 overexpression prevented the lethality of homozygous pex1-3 mutants (Fig. 4, F and G), peroxisomal defects persisted in pex1-3 35S:PEX1 plants (Fig. 4, B, C, and E), presumably because pex1-3 imparts defects even in the presence of wild-type PEX1.
In spite of a clear reduction in matrix protein import in pex1 mutants (Fig. 3A), increasing PEX5 levels did not ameliorate pex1 mutant defects, suggesting that these defects are not caused solely by reduced delivery of PEX5 cargo to the peroxisome. The exacerbation of PTS2-processing defects upon PEX5 overexpression in pex1-2 (Fig. 5C) also is observed in pex4-1 (Kao and Bartel, 2015), suggesting that PEX5 that is not recycled or degraded lingers in the peroxisomal membrane to the detriment of peroxisome function.
Although PEX1 and PEX6 are related proteins (Fig. 1D) that cooperate to retrotranslocate PEX5 from the membrane (Platta et al., 2005), they also have distinct functions. For example, PEX6, but not PEX1, binds PEX26 to tether the PEX1-PEX6 complex to the peroxisome (Goto et al., 2011). We found that PEX1, but not PEX6, overexpression in wild-type plants conferred growth defects (Fig. 4, G and H), even though PEX1 accumulated at lower levels relative to endogenous PEX1 (Fig. 4, D and E) than PEX6 (Fig. 7C). These observations suggest that PEX1 levels cannot exceed a particular level or ratio relative to PEX6. Human PEX1 can form homotrimers in the cytosol (Tamura et al., 2006) that bind PEX5 (Tamura et al., 2014), suggesting that excessive PEX1 may sequester PEX5 in the cytosol and prevent cargo delivery, leading to the observed defects. Additionally, because PEX1 was expressed from the constitutive 35S promoter, defects could be due to ectopic expression in tissues that usually do not have high PEX1 levels.
Missense mutations can lead to degradation of the mutant protein. Certain human pex1 missense mutants can be ameliorated by molecular chaperones that restore PEX1 levels (Zhang et al., 2010) or by lower growth temperatures that decrease degradation (Imamura et al., 1998a, 1998b; Zhang et al., 2010) and restore PEX1 levels (Tamura et al., 2001; Maxwell et al., 2002). Similarly, Arabidopsis PEX1 levels were reduced in pex1-2 (Figs. 4D, 5, B and C, 6C, 7C, and 8B). Lowering the growth temperature ameliorated this reduction, and increasing temperature exacerbated this reduction (Fig. 6C), suggesting that the Leu-993-to-Phe mutation destabilizes the pex1-2 protein. Interestingly, PEX6 levels also were reduced in pex1-2 (Figs. 3B, 4D, 5, B and D, 6C, 7C, and 8B), especially at elevated growth temperature (Fig. 6C), suggesting that PEX1 promotes PEX6 stability. Moreover, PEX6 overexpression restored pex1-2 accumulation (Fig. 7C), suggesting that increasing pex1-2-PEX6 complex formation stabilized the mutant pex1-2 protein. IBA responsiveness and PTS2 processing in pex1-2 were worsened by growth at high temperature (Fig. 6, A and B) and restored by overexpressing PEX6 (Fig. 7), indicating that the Leu-993-to-Phe mutation does not prevent PEX1 function if sufficient pex1-2 protein is present. Similarly, PEX6 overexpression rescues a pex1 missense mutation but not a pex1 null mutation in humans (Geisbrecht et al., 1998), and PEX6 overexpression rescues a temperature-sensitive P. pastoris pex1 mutation (Faber et al., 1998).
Despite the proximity of the pex1-2 and pex1-3 mutations (Fig. 1D), the distinct mutant phenotypes indicate different effects on the encoded proteins. pex1-2 decreased PEX1 levels, and the various pex1-2 defects all appear to stem from reduced PEX1 (and PEX6) levels (Figs. 6 and 7). In contrast, PEX1 and PEX6 protein levels resembled wild-type levels in PEX1/pex1-3, suggesting that pex1-3 might be stable. Indeed, the semidominance of pex1-3 implies that the pex1-3 protein is sufficiently stable to incorporate into and poison the PEX1-PEX6 complex, at least in young seedlings.
Unlike the rescue seen when overexpressing PEX6 in pex1-2 (Fig. 7), PEX1 overexpression fails to rescue pex6-1, as we failed to isolate pex6-1 plants overexpressing PEX1. This result contrasts with findings in human cells, where PEX1 overexpression can rescue a pex6 missense mutation (Geisbrecht et al., 1998). However, these findings are probably allele specific; PEX6 levels are not low in pex6-1 (Figs. 3B, 5B, and 6C; Ratzel et al., 2011), suggesting that pex6-1 is stable and, thus, not aided by additional PEX1. The detrimental effects of PEX1 overexpression observed in the wild type may exacerbate pex6-1 defects, preventing our isolation of pex6-1 plants overexpressing PEX1.
Our data are consistent with the model of a PEX1-PEX6 heterohexamer that sustains peroxisomal function by retrotranslocating PEX5 for recycling and preventing precocious pexophagy, suggesting that PEX1 and PEX6 functions are conserved in Arabidopsis. The unique pex1 alleles described here represent valuable tools with which to further elucidate both evolutionarily conserved and plant-specific PEX1 functions in a multicellular organism.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) wild type and mutants were in the Columbia-0 (Col-0) background. pex1-2 was isolated in a screen for IBA resistance in dark-grown hypocotyl elongation (Strader et al., 2011) from the progeny of seeds mutagenized by soaking for 16 h in 0.05% (v/v) methyl methanesulfonate. Whole-genome sequencing of three pooled backcrossed lines displaying IBA resistance and PTS2-processing defects revealed pex1-2 (Supplemental Fig. S1A; Supplemental Data Set S1). A PEX1/pex1-3 heterozygote was isolated from the progeny of seeds mutagenized by soaking in 0.24% (v/v) ethyl methanesulfonate for 16 h in a microscopy-based screen for seedlings displaying cotyledon GFP-PTS1 mislocalization (Rinaldi et al., 2016). Recombination mapping using individuals selected for IBA resistance in hypocotyl elongation that had progeny with GFP-PTS1 mislocalization, 2,4-DB resistance, and PTS2-processing defects suggested a causal heterozygous mutation at the top of chromosome 5 (Fig. 1C). We sequenced genomic DNA of two backcrossed lines displaying segregating cytosolic GFP-PTS1 mislocalization. Among mutations common to both backcrossed lines, we found a heterozygous pex1-3 mutation (Supplemental Fig. S1B; Supplemental Data Set S1). pex1-2 and PEX1/pex1-3 were backcrossed at least once prior to phenotypic analysis.
To visualize peroxisomes, we used a line expressing GFP-PTS1 driven by the constitutive cauliflower mosaic virus 35S promoter (35S:GFP-PTS1). This line and 35S:PEX5, pex6-1, pex6-1 35S:GFP-PTS1, pex6-1 35S:PEX5 (Zolman and Bartel, 2004), and atg7-3 GFP-PTS1 (Lai et al., 2011; Farmer et al., 2013) were described previously. We crossed 35S:GFP-PTS1 to pex1-2 and isolated a homozygous line of pex1-2 35S:GFP-PTS1 by following the construct in the progeny using fluorescence and PCR-based genotyping (Table I). We crossed atg7-3 GFP-PTS1 to PEX1/pex1-3 to obtain PEX1/pex1-3 atg7-3/atg7-3 GFP-PTS1.
Table I. PCR-based markers used to genotype identified mutants, reference lines, and transgenes.
| Mutant or Transgene | Primer Name (5′ to 3′ Sequence) | Enzyme | Amplicon Size |
|
|---|---|---|---|---|
| Wild Type | Mutant | |||
| bp | ||||
| pex1-2 | PEX1-RsaI-F (ACTCAATTAACAGTAGACCAGATGTA)a | RsaI | 151, 27 | 178 |
| PEX1-RsaI-R (TGAGGACAAAAATGATTAGCAGAA) | ||||
| pex1-3 | R405-p1-3 (AGCTCCATGCATACTCTTTTTC) | BccI | 183, 157 | 340 |
| R405-p1-4 (TATTATTAACTATCAACTCAG) | ||||
| pex6-1 | F10O3-7 (CAGACTTTACTGGCAAAAGCTGTGGCG) | XhoI | 270, 115 | 385 |
| F10O3-T (GCTTGCACCTATAATAAACAGATCCTGGG) | ||||
| 35S:PEX1 and 35S:HA-PEX1 | PEX1-F1 (CATTCGTTGCTTCTGTCCA) | – | 206 | 206, 107 |
| PEX1-E2 (GCCCCACGTTCCGAGGTAG) | ||||
| 35S:PEX5 | PEX5-38 (TGAAGACCAACAGATAAGG) | – | 264 | 264, 168 |
| PEX5-39 (CCCATTGGAGGCATAGG) | ||||
| 35S:HA-PEX6 | PEX6-3F1-BstNI (AACAGACCTGACTTGATTGAT) | – | 252 | 252, 176 |
| PEX6-3R1 (GAGGGACACTTCTTTGCTAC) | ||||
The underlined base indicates a difference from the original sequence to insert an RsaI site in the wild-type amplicon.
To generate overexpressing lines, a PEX1 cDNA in pCR8/GW/TOPO (Goto et al., 2011) and a PEX6 cDNA in pENTR223 (stock G21748) from the Arabidopsis Biological Resource Center at Ohio State University were cloned into pEarlyGate destination vectors pEG100 and pEG201 (Earley et al., 2006) from the Arabidopsis Biological Resource Center using Clonase (Invitrogen). The resultant 35S:PEX1, 35S:HA-PEX1, and 35S:HA-PEX6 plasmids were used to transform Agrobacterium tumefaciens GV3101 (pMP90) (Koncz and Schell, 1986) by electroporation. Transformed A. tumefaciens strains were used to transform pex1-2 plants using the floral dip method (Clough and Bent, 1998) to obtain pex1-2 35S:PEX1, pex1-2 35S:HA-PEX1, and pex1-2 35S:HA-PEX6. We crossed pex1-2 35S:HA-PEX1 and pex1-2 35S:PEX1 to the wild type and obtained wild-type 35S:HA-PEX1 and wild-type 35S:PEX1, respectively. We also crossed pex1-2 35S:PEX1 to PEX1/pex1-3 and obtained pex1-3 35S:PEX1 and crossed wild-type 35S:PEX5 to pex1-2 and to PEX1/pex1-3 to obtain pex1-2 35S:PEX5 and PEX1/pex1-3 35S:PEX5, respectively. Homozygous lines were selected from the progeny of crosses using glufosinate ammonium (Basta) resistance and PCR-based genotyping (Table I).
Growth Conditions and Phenotypic Assays
Seeds were surface sterilized and stratified at 4°C for 1 d before sowing on plates containing plant nutrient medium (Haughn and Somerville, 1986) solidified with 0.6% (w/v) agar and supplemented with 0.5% (w/v) Suc. Plates were sealed with gas-permeable tape and incubated at 22°C under continuous white fluorescent light unless noted otherwise. Light was filtered through yellow long-pass filters when treating with hormones to slow indolic compound breakdown (Stasinopoulos and Hangarter, 1990). The same volume of ethanol was added to control medium when using ethanol-dissolved hormones (IBA, 2,4-DB, and 1-naphthaleneacetic acid). Hypocotyl lengths were measured on seedlings grown on plates that were incubated under light for 1 d and then wrapped in aluminum foil and incubated for 4 d. Lateral roots that protruded from the epidermis were counted on seedlings that were grown on medium without hormone for 4 d and then transferred to medium with or without hormone and grown for an additional 4 d. For assays that measured single individuals, progeny of PEX1/pex1-3 heterozygous plants were measured and then individually genotyped to identify heterozygotes. Plants were transferred to soil (Sun Gro Metro-Mix 366) after 14 to 16 d on plates and grown at 22°C under continuous white fluorescent light.
DNA Analysis
PCR was performed on DNA that was prepared as described (Celenza et al., 1995). For recombination mapping of pex1-3, progeny from an outcross to the Arabidopsis Landsberg erecta (Ler) accession were genotyped using PCR-based markers that exploit polymorphisms between Col-0 and Ler DNA (Table II).
Table II. PCR-based markers used for recombination mapping of pex1-3.
| Marker | Nearest Gene | Primer Name (5′ to 3′ Sequence) | Amplicon Size |
|
|---|---|---|---|---|
| Col-0 | Ler | |||
| bp | ||||
| CIW18 | At5g05170 | CIW18-1 (AACACAACATGGTTTCAGT) | 135 | 129 |
| CIW18-2 (GCCGTTTGTCTCTTCAC) | ||||
| F7K24 | At5g19270 | F7K24-1 (TGCAAAATTCTAGCTATCG) | 125 | 141 |
| F7K24-2 (ACTTTTGTATGGCTAATGAG) | ||||
| MWD9 | At5g22420 | MWD9-1 (TAGGGTCGTGGTTGGTTG) | 133 | 118 |
| MWD9-2 (CTGGCCTCTCTATCTGATAC) | ||||
| GA3 | At5g25900 | GA3-1 (CGGAGGCTATCATGTCCCTGC) | 156 | 144 |
| GA3-2 (CCCAACGCTTCTTATCCATGTTGC) | ||||
Genomic DNA for whole-genome sequencing was prepared as described previously (Thole et al., 2014) and sent to the Genome Technology Access Center at Washington University in St. Louis for sequencing with Illumina HiSeq 2000 sequencers. The Arabidopsis Col-0 genome from TAIR (build 10) was used with Novoalign (Novocraft; http://novocraft.com) to align the sequences, and SAMtools (Li et al., 2009) and snpEFF (Cingolani et al., 2012) were used to identify mutations. Mutations that were found previously in our laboratory Col-0 line (Farmer et al., 2013) were disregarded. Sequencing coverage for pex1-2 was 10× or greater for 71% of the genome and 5× or greater for 85% of the genome; coverage for pex1-3 was 10× or greater for 86% to 88% of the genome and 5× or greater for 99% of the genome.
Microscopy
Seedpods, seeds, and seedlings were imaged with a Leica ZM10 F dissecting microscope equipped with a Leica DFC295 camera.
To visualize GFP-PTS1 localization, cotyledons of light-grown seedlings were mounted in water and imaged with a Carl Zeiss LSM 710 laser scanning confocal microscope using a 63× oil-immersion objective, a Meta detector, and the ZEN 2010 version 6.0.0.485 software. A 488-nm argon laser was used for excitation, and GFP fluorescence emission was collected between 493 and 526 nm through a 24-µm pinhole, corresponding to a 1-µm optical slice. Each image is an average of four exposures. Fluorescence was imaged at two different planes in epidermal cells: midway through the cell (midcell), where cytosolic fluorescence outlines the cell, and immediately below the plasma membrane (subcortical), where cytosolic fluorescence is distributed across the plane of acquisition.
Immunoblotting
Frozen plant tissue was ground in 2 volumes of 2× NuPAGE sample buffer (Invitrogen) and centrifuged at 16,100g for 5 min. DTT was added to the supernatants to a final concentration of 50 mm, and samples were incubated at 100°C for 5 min. A total of 15 µL of each sample was loaded on NuPAGE or BOLT 10% Bis-Tris gels (Invitrogen) alongside molecular mass markers (sc-2035; Santa Cruz Biotechnology) and prestained protein markers (P7708; New England Biolabs) and subjected to electrophoresis using 1× MOPS running buffer (50 mm MOPS, 50 mm Tris base, 0.1% SDS, and 1 mm EDTA). Gels were transferred to Hybond nitrocellulose membranes (Amersham Pharmacia Biotech) using NuPAGE transfer buffer (Invitrogen). Membranes were blocked in 8% nonfat dry milk in 20 mm Tris, pH 7.5, 150 mm NaCl, and 0.1% Tween 20 for 1 h at 4°C with rocking. After blocking, membranes were incubated overnight at 4°C with primary antibodies in blocking solution with 0.1% sodium azide at the indicated dilutions: rabbit antibodies against PMDH2 (1:1,500; Pracharoenwattana et al., 2007), the PED1 isoform of thiolase (1:10,000; Lingard et al., 2009), PEX1 (1:200; raised to the first 400 amino acids of Arabidopsis PEX1 and affinity purified by Proteintech Group), PEX5 (1:100; Zolman and Bartel, 2004), and PEX6 (1:1,000; Ratzel et al., 2011); mouse antibodies against HSC70 (1:50,000; Stressgen SPA-817) and GFP (1:100; sc-9996; Santa Cruz Biotechnology); or a rat antibody against HA (1:100; Roche 3F10). Membranes were washed with 20 mm Tris, pH 7.5, 150 mm NaCl, and 0.1% Tween 20 and incubated for 4 h at 4°C with secondary horseradish peroxidase-linked goat antibodies against rabbit, mouse, or rat (1:5,000; sc-2030, sc-2031, and sc-2032, respectively; Santa Cruz Biotechnology) diluted in blocking solution. Membranes were washed and incubated with WesternBright ECL (Advansta) to visualize horseradish peroxidase activity by exposing membranes to autoradiography film. Films were imaged using a scanner. Various primary antibodies were used sequentially to probe membranes without stripping.
Fractionation
Cell fractionation was performed as described previously (Ratzel et al., 2011). Approximately 500 mg of tissue was chopped with scissors in 1 mL of ice-cold fractionation buffer (150 mm Tris, pH 7.6, 100 mm Suc, 10 mm KCl, 1 mm EDTA, 1 mm DTT, 1 mm N-ethylmaleimide, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail [Sigma P9599]), processed with a Dounce homogenizer (20 strokes), and filtered through Miracloth (Millipore). Samples were centrifuged for 10 min at 4,300g, and one-third of the supernatant (around 150 µL) was used as the homogenate fraction (H). The rest of the volume (around 300 µL) was centrifuged for 20 min at 13,400g, and most of the new supernatant (around 240 µL) was collected as the supernatant fraction (S). The pellet was resuspended in twice the homogenate volume (around 300 µL) of fractionation buffer, centrifuged for 20 min at 13,400g, and most of the new supernatant (around 240 µL) was collected as the wash fraction (W). The remaining supernatant was discarded, and the pellet was resuspended in twice the homogenate volume (around 300 µL) of fractionation buffer to be used as the pellet fraction (P). Volumes were adjusted according to the volume obtained in the first centrifugation to maintain a ratio of H:S:P:W of 5:8:10:8. Each fraction was mixed with one-third volume of NuPAGE 4× loading buffer (Invitrogen), and 40 µL was used for immunoblotting as described above.
Statistical Analysis
When more than two samples were compared, the SPSS Statistics software (version 22.0.0.1) was used to analyze differences among samples in a particular treatment group using one-way ANOVA followed by Duncan’s test. Mean values that were significantly different from each other (P < 0.001) are marked with different letters above the bars.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Arabidopsis PEX1 (At5g08470), Arabidopsis PEX6 (At1g03000), human PEX1 (NP_000457.1), human PEX6 (NP_000278.3), and mouse p97 (NP_033529.3).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Mutations identified through whole-genome sequencing of pex1 mutant DNA.
Supplemental Figure S2. PEX5 is distributed like the wild type between cytosolic and organellar fractions in pex1 mutants.
Supplemental Data Set S1. Mutations identified through whole-genome sequencing.
Acknowledgments
We thank Steven Smith (University of Western Australia) for the PMDH2 antibody, Shino Goto (National Institute for Basic Biology, Japan) for the PEX1 cDNA, Lucia Strader (Washington University) for originating the mutant screens, Andrew Woodward (University of Mary Hardin-Baylor) for assistance in optimizing methyl methanesulfonate for use as an Arabidopsis mutagen, as well as Yun-Ting Kao, Roxanna Llinas, Zachary Wright, and Pierce Young for critical comments on the article.
Glossary
- IBA
indole-butyric acid
- 2,4-DB
2,4-dichlorophenoxybutyric acid
- RNAi
RNA interference
- Col-0
Columbia-0
- SRH
second region of homology
Footnotes
This work was supported by the National Institutes of Health (NIH; R01GM079177) and the Robert A. Welch Foundation (C-1309). K.L.G. was supported by a National Science Foundation Graduate Research Fellowship (DGE-0940902) and by the National Science Foundation IRISE training program at Rice University (EHR-0966303). J.P. and M.J.V. were partially supported by a Howard Hughes Medical Institute Professors Grant (52005717 to B.B.). Confocal microscopy was performed on equipment obtained through a Shared Instrumentation Grant from the National Institutes of Health (S10RR026399-01). Whole-genome sequencing at the Genome Technology Access Center at Washington University in St. Louis was supported by the NIH National Center for Research Resources (UL1 RR024992).
Articles can be viewed without a subscription.
References
- Agrimi G, Russo A, Pierri CL, Palmieri F (2012) The peroxisomal NAD+ carrier of Arabidopsis thaliana transports coenzyme A and its derivatives. J Bioenerg Biomembr 44: 333–340 [DOI] [PubMed] [Google Scholar]
- Augustin S, Gerdes F, Lee S, Tsai FT, Langer T, Tatsuta T (2009) An intersubunit signaling network coordinates ATP hydrolysis by m-AAA proteases. Mol Cell 35: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Burkhart SE, Fleming WA (2014) Protein transport in and out of plant peroxisomes. In Brocard C, Hartig A, eds, Molecular Machines Involved in Peroxisome Biogenesis and Maintenance. Springer, Vienna, pp 325–345 [Google Scholar]
- Bernhardt K, Wilkinson S, Weber AP, Linka N (2012) A peroxisomal carrier delivers NAD+ and contributes to optimal fatty acid degradation during storage oil mobilization. Plant J 69: 1–13 [DOI] [PubMed] [Google Scholar]
- Birschmann I, Rosenkranz K, Erdmann R, Kunau WH (2005) Structural and functional analysis of the interaction of the AAA-peroxins Pex1p and Pex6p. FEBS J 272: 47–58 [DOI] [PubMed] [Google Scholar]
- Birschmann I, Stroobants AK, van den Berg M, Schäfer A, Rosenkranz K, Kunau WH, Tabak HF (2003) Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6p to peroxisomal membranes. Mol Biol Cell 14: 2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok NB, Tan D, Wang RY, Penczek PA, Baker D, DiMaio F, Rapoport TA, Walz T (2015) Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc Natl Acad Sci USA 112: E4017–E4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Frietsch S, Kim TH, Dizon MB, Schroeder JI (2008) The peroxin loss-of-function mutation abstinence by mutual consent disrupts male-female gametophyte recognition. Curr Biol 18: 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman N, Dodt G, Gould SJ, Valle D (1998) An isoform of pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum Mol Genet 7: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Burkhart SE, Kao YT, Bartel B (2014) Peroxisomal ubiquitin-protein ligases peroxin2 and peroxin10 have distinct but synergistic roles in matrix protein import and peroxin5 retrotranslocation in Arabidopsis. Plant Physiol 166: 1329–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart SE, Lingard MJ, Bartel B (2013) Genetic dissection of peroxisome-associated matrix protein degradation in Arabidopsis thaliana. Genetics 193: 125–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL Jr, Grisafi PL, Fink GR (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Charlton WL, Johnson B, Graham IA, Baker A (2005) Non-coordinate expression of peroxisome biogenesis, beta-oxidation and glyoxylate cycle genes in mature Arabidopsis plants. Plant Cell Rep 23: 647–653 [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciniawsky S, Grimm I, Saffian D, Girzalsky W, Erdmann R, Wendler P (2015) Molecular snapshots of the Pex1/6 AAA+ complex in action. Nat Commun 6: 7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collins CS, Kalish JE, Morrell JC, McCaffery JM, Gould SJ (2000) The peroxisome biogenesis factors pex4p, pex22p, pex1p, and pex6p act in the terminal steps of peroxisomal matrix protein import. Mol Cell Biol 20: 7516–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammai V, Subramani S (2001) The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105: 187–196 [DOI] [PubMed] [Google Scholar]
- Dodt G, Gould SJ (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol 135: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Erdmann R, Veenhuis M, Mertens D, Kunau WH (1989) Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 86: 5419–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Heyman JA, Subramani S (1998) Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol Cell Biol 18: 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Quan S, Orth T, Awai C, Chory J, Hu J (2005) The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol 139: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE, Bartel B (2013) Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25: 4085–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Chowdhury S, Lander GC, Martin A (2015) The Pex1/Pex6 complex is a heterohexameric AAA+ motor with alternating and highly coordinated subunits. J Mol Biol 427: 1375–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht BV, Collins CS, Reuber BE, Gould SJ (1998) Disruption of a PEX1-PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc Natl Acad Sci USA 95: 8630–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Mano S, Nakamori C, Nishimura M (2011) Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell 23: 1573–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto-Yamada S, Mano S, Nakamori C, Kondo M, Yamawaki R, Kato A, Nishimura M (2014) Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol 55: 482–496 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S (1989) A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol 108: 1657–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm I, Saffian D, Platta HW, Erdmann R (2012) The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae. Biochim Biophys Acta 1823: 150–158 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Hayashi M, Nito K, Toriyama-Kato K, Kondo M, Yamaya T, Nishimura M (2000) AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J 19: 5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M (2005) Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J Biol Chem 280: 14829–14835 [DOI] [PubMed] [Google Scholar]
- Helm M, Lück C, Prestele J, Hierl G, Huesgen PF, Fröhlich T, Arnold GJ, Adamska I, Görg A, Lottspeich F, et al. (2007) Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA 104: 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405–409 [DOI] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Li G, Lennarz WJ (2012) Dynamic flexibility of the ATPase p97 is important for its interprotomer motion transmission. Proc Natl Acad Sci USA 109: 9792–9797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura A, Tamura S, Shimozawa N, Suzuki Y, Zhang Z, Tsukamoto T, Orii T, Kondo N, Osumi T, Fujiki Y (1998a) Temperature-sensitive mutation in PEX1 moderates the phenotypes of peroxisome deficiency disorders. Hum Mol Genet 7: 2089–2094 [DOI] [PubMed] [Google Scholar]
- Imamura A, Tsukamoto T, Shimozawa N, Suzuki Y, Zhang Z, Imanaka T, Fujiki Y, Orii T, Kondo N, Osumi T (1998b) Temperature-sensitive phenotypes of peroxisome-assembly processes represent the milder forms of human peroxisome-biogenesis disorders. Am J Hum Genet 62: 1539–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YT, Bartel B (2015) Elevated growth temperature decreases levels of the PEX5 peroxisome-targeting signal receptor and ameliorates defects of Arabidopsis mutants with an impaired PEX4 ubiquitin-conjugating enzyme. BMC Plant Biol 15: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YT, Fleming WA, Ventura MJ, Bartel B (2016) Genetic interactions between PEROXIN12 and other peroxisome-associated ubiquitination components. Plant Physiol 172: 1643–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karata K, Inagawa T, Wilkinson AJ, Tatsuta T, Ogura T (1999) Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases: site-directed mutagenesis of the ATP-dependent protease FtsH. J Biol Chem 274: 26225–26232 [DOI] [PubMed] [Google Scholar]
- Khan BR, Zolman BK (2010) pex5 mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol 154: 1602–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee H, Lee HN, Kim SH, Shin KD, Chung T (2013) Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kragt A, Voorn-Brouwer T, van den Berg M, Distel B (2005) The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J Biol Chem 280: 7867–7874 [DOI] [PubMed] [Google Scholar]
- Lai Z, Wang F, Zheng Z, Fan B, Chen Z (2011) A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J 66: 953–968 [DOI] [PubMed] [Google Scholar]
- Law KB, Bronte-Tinkew D, Di Pietro E, Snowden A, Jones RO, Moser A, Brumell JH, Braverman N, Kim PK (2017) The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy 13: 868–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XR, Li HJ, Yuan L, Liu M, Shi DQ, Liu J, Yang WC (2014) Arabidopsis DAYU/ABERRANT PEROXISOME MORPHOLOGY9 is a key regulator of peroxisome biogenesis and plays critical roles during pollen maturation and germination in planta. Plant Cell 26: 619–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Bartel B (2009) Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol 151: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Monroe-Augustus M, Bartel B (2009) Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA 106: 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A (2000) Stress induces peroxisome biogenesis genes. EMBO J 19: 6770–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Wang Z, Lopez I, Al Humaid S, Ren H, Racine J, Bazinet A, Mitchel G, Braverman N, Koenekoop RK (2011) A new ocular phenotype associated with an unexpected but known systemic disorder and mutation: novel use of genomic diagnostics and exome sequencing. J Med Genet 48: 593–596 [DOI] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Fukao Y, Araki M, Matsuda A, Kondo M, Nishimura M (2011) A defect of peroxisomal membrane protein 38 causes enlargement of peroxisomes. Plant Cell Physiol 52: 2157–2172 [DOI] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Nito K, Kondo M, Nishimura M (2006) The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J 47: 604–618 [DOI] [PubMed] [Google Scholar]
- Marzioch M, Erdmann R, Veenhuis M, Kunau WH (1994) PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J 13: 4908–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N, Tamura S, Fujiki Y (2003) The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat Cell Biol 5: 454–460 [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Allen T, Solly PB, Svingen T, Paton BC, Crane DI (2002) Novel PEX1 mutations and genotype-phenotype correlations in Australasian peroxisome biogenesis disorder patients. Hum Mutat 20: 342–351 [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Leane PB, Paton BC, Crane DI (2005) Novel PEX1 coding mutations and 5′ UTR regulatory polymorphisms. Hum Mutat 26: 279. [DOI] [PubMed] [Google Scholar]
- McCollum D, Monosov E, Subramani S (1993) The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells: the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J Cell Biol 121: 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke M, Cizmowski C, Schliebs W, Krüger V, Beck S, Wagner R, Erdmann R (2010) The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol 12: 273–277 [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M, Ramón NM, Ratzel SE, Lingard MJ, Christensen SE, Murali C, Bartel B (2011) Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol Biol 77: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralla R, Lloyd J, Meinke D (2011) Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6: e28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K, Hayashi M, Nishimura M (2002) Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol 43: 355–366 [DOI] [PubMed] [Google Scholar]
- Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M (2007) Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol 48: 763–774 [DOI] [PubMed] [Google Scholar]
- Nuttall JM, Motley AM, Hettema EH (2014) Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy 10: 835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Latterich M (1998) The AAA team: related ATPases with diverse functions. Trends Cell Biol 8: 65–71 [PubMed] [Google Scholar]
- Platta HW, Brinkmeier R, Reidick C, Galiani S, Clausen MP, Eggeling C (2016) Regulation of peroxisomal matrix protein import by ubiquitination. Biochim Biophys Acta 1863: 838–849 [DOI] [PubMed] [Google Scholar]
- Platta HW, Erdmann R (2007) Peroxisomal dynamics. Trends Cell Biol 17: 474–484 [DOI] [PubMed] [Google Scholar]
- Platta HW, Grunau S, Rosenkranz K, Girzalsky W, Erdmann R (2005) Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat Cell Biol 7: 817–822 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2007) Arabidopsis peroxisomal malate dehydrogenase functions in beta-oxidation but not in the glyoxylate cycle. Plant J 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Ramón NM, Bartel B (2010) Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol Biol Cell 21: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzel SE, Lingard MJ, Woodward AW, Bartel B (2011) Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic 12: 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Bartel B (2016) Plant peroxisomes: recent discoveries in functional complexity, organelle homeostasis, and morphological dynamics. Curr Opin Plant Biol 34: 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi MA, Patel AB, Park J, Lee K, Strader LC, Bartel B (2016) The roles of beta-oxidation and cofactor homeostasis in peroxisome distribution and function in Arabidopsis thaliana. Genetics 204: 1089–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffian D, Grimm I, Girzalsky W, Erdmann R (2012) ATP-dependent assembly of the heteromeric Pex1p-Pex6p-complex of the peroxisomal matrix protein import machinery. J Struct Biol 179: 126–132 [DOI] [PubMed] [Google Scholar]
- Santos L. (2006) Molecular mechanisms of the AAA proteins in plants. In Guevara-Gonzalez I, ed, Advances in Agricultural Food Biotechnology. Trivandrum, Kerala, India, pp 1–15 [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C (2003) AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA 100: 9626–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M (2013) Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25: 4967–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Poulter JA, Levin AV, Capasso JE, Price S, Ben-Yosef T, Sharony R, Newman WG, Shore RC, Brookes SJ, et al. (2016) Spectrum of PEX1 and PEX6 variants in Heimler syndrome. Eur J Hum Genet 24: 1565–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A (2003) An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol 133: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley WA, Wilmanns M (2006) Dynamic architecture of the peroxisomal import receptor Pex5p. Biochim Biophys Acta 1763: 1592–1598 [DOI] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP (1990) Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Culler AH, Cohen JD, Bartel B (2010) Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol 153: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B (2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23: 984–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S (1991) A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J 10: 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Matsumoto N, Imamura A, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y (2001) Phenotype-genotype relationships in peroxisome biogenesis disorders of PEX1-defective complementation group 1 are defined by Pex1p-Pex6p interaction. Biochem J 357: 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Matsumoto N, Takeba R, Fujiki Y (2014) AAA peroxins and their recruiter Pex26p modulate the interactions of peroxins involved in peroxisomal protein import. J Biol Chem 289: 24336–24346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Yasutake S, Matsumoto N, Fujiki Y (2006) Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J Biol Chem 281: 27693–27704 [DOI] [PubMed] [Google Scholar]
- Thole JM, Beisner ER, Liu J, Venkova SV, Strader LC (2014) Abscisic acid regulates root elongation through the activities of auxin and ethylene in Arabidopsis thaliana. G3 (Bethesda) 4: 1259–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund CW, Schroers MG, Wiese J, Facchinelli F, Kurz S, Wilkinson S, Charton L, Wanders RJ, Waterham HR, Weber AP, et al. (2016) The peroxisomal NAD carrier from Arabidopsis imports NAD in exchange with AMP. Plant Physiol 171: 2127–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain RL, Wightman F (1954) The growth regulating activity of certain ω-substituted alkyl carboxylic acids in relation to their β-oxidation within the plant. Proc R Soc Lond B Biol Sci 142: 525–536 [DOI] [PubMed] [Google Scholar]
- Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M (2016) HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat Commun 7: 10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Ebberink MS (2012) Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim Biophys Acta 1822: 1430–1441 [DOI] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Sprenger RR, Distel B (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem 282: 22534–22543 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Fleming WA, Burkhart SE, Ratzel SE, Bjornson M, Bartel B (2014) A viable Arabidopsis pex13 missense allele confers severe peroxisomal defects and decreases PEX5 association with peroxisomes. Plant Mol Biol 86: 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
- Young PG, Bartel B (2016) Pexophagy and peroxisomal protein turnover in plants. Biochim Biophys Acta 1863: 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Chen L, Jiralerspong S, Snowden A, Steinberg S, Braverman N (2010) Recovery of PEX1-Gly843Asp peroxisome dysfunction by small-molecule compounds. Proc Natl Acad Sci USA 107: 5569–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B (2004) An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B (2005) Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]