Overexpression of OsHAD1 enhances P accumulation in rice by activation of Pi starvation responses such as ATP depletion, APase secretion, organic acid production, and induction of Pi transporters.
Abstract
Phosphorus (P) deficiency limits plant growth and yield. Since plants can absorb only the inorganic form of P (Pi), a large portion of soil P (organic and inorganic P complexes) remains unused. Here, we identified and characterized a PHR2-regulated, novel low-Pi-responsive haloacid dehalogenase (HAD)-like hydrolase, OsHAD1. While OsHAD1 is a functional HAD protein having both acid phosphatase and phytase activities, it showed little homology with other known low-Pi-responsive HAD superfamily members. Recombinant OsHAD1 is active at acidic pH and dephosphorylates a broad range of organic and inorganic P-containing substrates, including phosphorylated serine and sodium phytate. Exogenous application of recombinant OsHAD1 protein in growth medium supplemented with phytate led to marked increases in growth and total P content of Pi-deficient wild-type rice (Oryza sativa) seedlings. Furthermore, overexpression of OsHAD1 in rice resulted in enhanced phosphatase activity, biomass, and total and soluble P contents in Pi-deficient transgenic seedlings treated with phytate as a restricted Pi source. Gene expression and metabolite profiling revealed enhanced Pi starvation responses, such as up-regulation of multiple genes involved in Pi uptake and solubilization, accumulation of organic acids, enhanced secretory phosphatase activity, and depletion of ATP in overexpression lines as compared with the wild type. To elucidate the underlying regulatory mechanisms of OsHAD1, we performed in vitro pull-down assays, which revealed the association of OsHAD1 with protein kinases such as OsNDPKs. We conclude that, besides dephosphorylation of cellular organic P, OsHAD1 in coordination with kinases may regulate the phosphorylation status of downstream targets to accomplish Pi homeostasis under limited Pi supply.
Rice (Oryza sativa) is one of the most consumed cereals in the world. Therefore, to sustain a growing population, especially in rice-consuming areas, it is necessary to increase crop yield in coming years (Papademetriou, 2000; Tilman et al., 2011). Phosphorus (P) is an essential macronutrient for plant growth and development. It is an integral part of nucleic acids, phospholipid membranes, and many metabolic pathways. Essentially, most of the signal transduction pathways are regulated by phosphorylation and dephosphorylation events. Widespread soil inorganic phosphate (Pi) deficiency is known to limit rice yield, particularly in upland acidic soils (Kirk et al., 1998). Since most of the modern nutrient-exhaustive high-yielding rice varieties are low Pi sensitive (Wissuwa and Ae, 2001; Mehra et al., 2015, 2016), it is imperative to develop Pi-efficient high-yielding rice varieties to accomplish high rice production and reduce extensive application of phosphate fertilizers. Notably, unrestrained use of Pi fertilizers has serious ecological impacts, such as heavy metal contamination of soils (Chien et al., 2011) and eutrophication of water bodies through soil erosion or surface runoff (Ha and Tran, 2014). Additionally, fast-depleting Pi fertilizer sources (high-quality rock phosphate) are finite and nonrenewable in nature (Cordell and Neset, 2014). Therefore, improvement of the Pi use efficiency of rice could be an efficient and sustainable means for crop production.
Roots absorb P in the form of Pi from soil solution. Unfortunately, ∼80% of the applied Pi fertilizers are rapidly fixed in the soil in organic/inorganic insoluble complexes due to microbial assimilation and adsorption. Therefore, up to 65% of the total soil P exists in the organic form (Harrison, 1987), which needs mineralization before root uptake (Dobermann et al., 1998). To release Pi from these bound sources, plants produce several intracellular and extracellular acid phosphatases (APases), secrete organic acids into the rhizosphere, undergo symbiotic associations with mycorrhizal fungi, and develop specialized proteoid roots (Raghothama, 1999). Among these strategies, the production of extracellular and intracellular APases is important, as these enzymes can hydrolyze a broad range of organic P sources to release Pi under low-Pi conditions. Intracellular APases maintain internal Pi homeostasis by releasing Pi from organic P compounds in the cytoplasm and vacuole, whereas extracellular APases hydrolyze organic P sources present in the extracellular matrix (Tran et al., 2010a). Various strategies have been employed to enhance Pi availability from organic resources by engineering phytases and APases from microbial or plant origin (Richardson et al., 2001; Zimmermann et al., 2003; Lung et al., 2005; George et al., 2007; Liu et al., 2012; Ma et al., 2012; Singh and Satyanarayana, 2012; Mehra et al., 2017). While a growing body of literature emphasizes the roles of extracellular phytases and APases in P solubilization and acquisition, the potential of intracellular phytases/APases in low-Pi tolerance has remained unrecognized.
APases such as haloacid dehalogenase (HAD) superfamily members are regulated by Pi deficiency (Baldwin et al., 2001; Hur et al., 2007; May et al., 2011). The HAD superfamily of enzymes is composed of several ATPases, epoxide hydrolases, dehalogenases, phosphonatases, phosphomutases, phosphoserine, and other phosphatases (Burroughs et al., 2006; Kuznetsova et al., 2006). LePS2 was the first low-Pi-inducible HAD superfamily gene characterized in tomato (Solanum lycopersicum; Baldwin et al., 2001, 2008). The protein phosphatase activity of overexpressed LePS2 led to increased APase activity, anthocyanin accumulation, and delayed flowering in tomato (Baldwin et al., 2008). Similarly, another HAD gene, PvHAD1, showed low-Pi-specific induction and encodes a functional Ser/Thr phosphatase (Liu et al., 2010). In Arabidopsis (Arabidopsis thaliana), two low-Pi-responsive HADs, AtPPsPase1 and AtPECP1, were reported to encode functional pyrophosphatase and phosphoethanolamine phosphatase, respectively (May et al., 2011, 2012). Later, PvPS2:1, a plasma membrane-localized homolog of LePS2, was reported to increase Pi acquisition and root growth in Arabidopsis overexpressing transgenics (Liang et al., 2012). These studies suggest the important functions of HAD superfamily members in regulating Pi homeostasis. In rice, only one HAD, OsACP1 (also a homolog of LePS2), was shown to be up-regulated under Pi deficiency (Hur et al., 2007). However, the physiological roles of any other HAD are largely unknown in rice. Therefore, in this study, we identified and characterized a novel low-Pi-inducible HAD gene, OsHAD1, in rice. Biochemical assays revealed that OsHAD1 is a functional APase with broad substrate specificities and possesses substantial protein phosphatase and phytase activities. Furthermore, overexpression of OsHAD1 led to increased APase/phytase activity, P accumulation, and improved growth of rice under restricted Pi supply. Here, to our knowledge for the first time, we provide a detailed investigation on a HAD superfamily member in rice and its efficacy in improving Pi homeostasis.
RESULTS
OsHAD1 Is Induced under Pi Deficiency
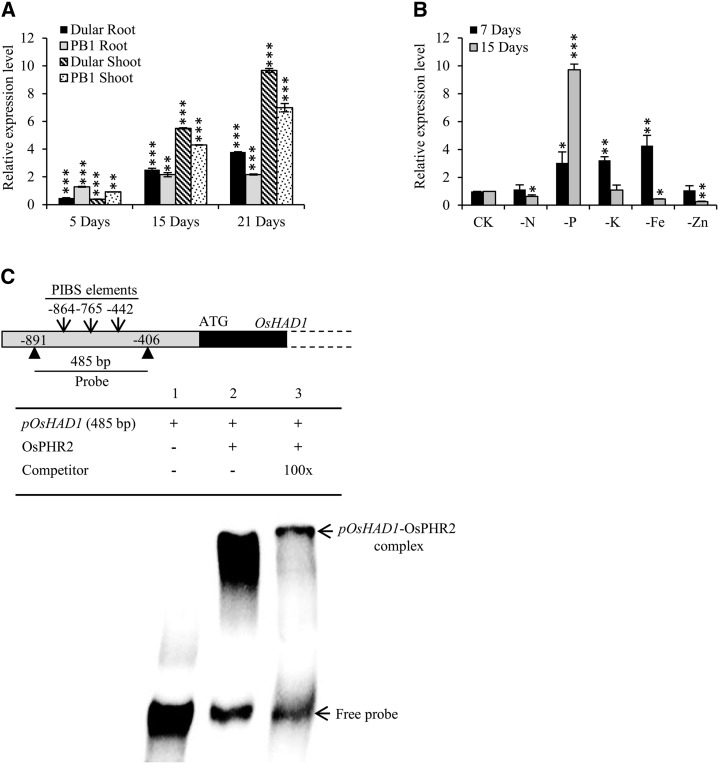
We earlier identified several low-Pi-responsive genes using a comparative transcriptomic approach in low-Pi-tolerant and -sensitive rice genotypes (Mehra et al., 2016). Among these, a HAD (Os03g61829), designated here as OsHAD1, was found to be differentially induced in root and shoot under low Pi. OsHAD1 was highly up-regulated in the low-Pi-tolerant genotype Dular as compared with PB1, a low-Pi-sensitive modern high-yielding genotype. To further assess the low Pi responsiveness of OsHAD1, gene expression was analyzed after 5, 15, and 21 d of Pi deficiency. We found increased transcript levels of OsHAD1 with increasing duration of low-Pi stress, preferentially in shoot (Fig. 1A). Notably, up-regulation of OsHAD1 was higher in Dular as compared with PB1, except at 5 d. Furthermore, OsHAD1 showed significant up-regulation in P, K, and Fe deficiencies at 7 d of stress treatment. However, after prolonged stress exposure for 15 d, OsHAD1 was specifically up-regulated only in low Pi (Fig. 1B). Additionally, expression analysis of OsHAD1 at different developmental stages and tissues showed that it is expressed in almost all organs to varying levels (Supplemental Fig. S1). OsHAD1 expression was higher in shoot and floral organs. These results revealed that OsHAD1 is low-Pi responsive, with preferential expression in shoot and floral organs.
Figure 1.
Transcriptional regulation of OsHAD1. A, Relative expression of OsHAD1 in Dular and PB1 roots and shoot under Pi-deficient (1 µm) conditions with respect to corresponding Pi-sufficient (320 µm) conditions at 5, 15, and 21 d. B, Relative expression of OsHAD1 under N-, P-, K-, Fe-, and Zn-deficient conditions in roots after 7 and 15 d of the respective deficiency treatments. Relative expression levels under deficient conditions were calculated with respect to expression levels under the sufficient nutrient supply condition (CK). Expression profiling was carried out with qRT-PCR. Significant differences between deficient versus sufficient treatments were evaluated by Student’s t test. Asterisks indicate P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***). C, Electrophoretic mobility shift assay showing physical binding of OsPHR2 with the OsHAD1 promoter. A 485-bp radiolabeled promoter of OsHAD1 (pOsHAD1) containing three P1BS cis-elements at −864, −765, and −442 bp was used as a probe. Lane 1, Radiolabeled free promoter probe of OsHAD1 (12.74 ng); lane 2, [α-32P]CTP-labeled probe of OsHAD1 (12.74 ng) plus OsPHR2 protein (1.5 µg); lane 3, OsPHR2 protein (1.5 µg) plus labeled promoter probe of OsHAD1 (12.74 ng) and a 100-fold excess of unlabeled OsHAD1 probe as a competitive inhibitor.
OsPHR2 Regulates the Expression of OsHAD1
The PHR1 transcription factor is a central regulator of low-Pi-responsive genes in various plants. OsPHR2 is a functional ortholog of AtPHR1 (Zhou et al., 2008). We found three P1BS (PHR2-binding site) elements at positions −864, −765, and −442 in a 2-kb promoter region of OsHAD1 (Fig. 1C). Our electrophoretic mobility shift assay showed that OsPHR2 binds physically to the OsHAD1 promoter (Fig. 1C). Furthermore, this interaction was reduced significantly when a 100× excess of unlabeled OsHAD1 promoter probe was used as a competitive inhibitor. All this confirmed that OsHAD1 is a component of the rice low-Pi-responsive machinery regulated by OsPHR2.
OsHAD1 Is a Novel Functional Acid Phosphatase
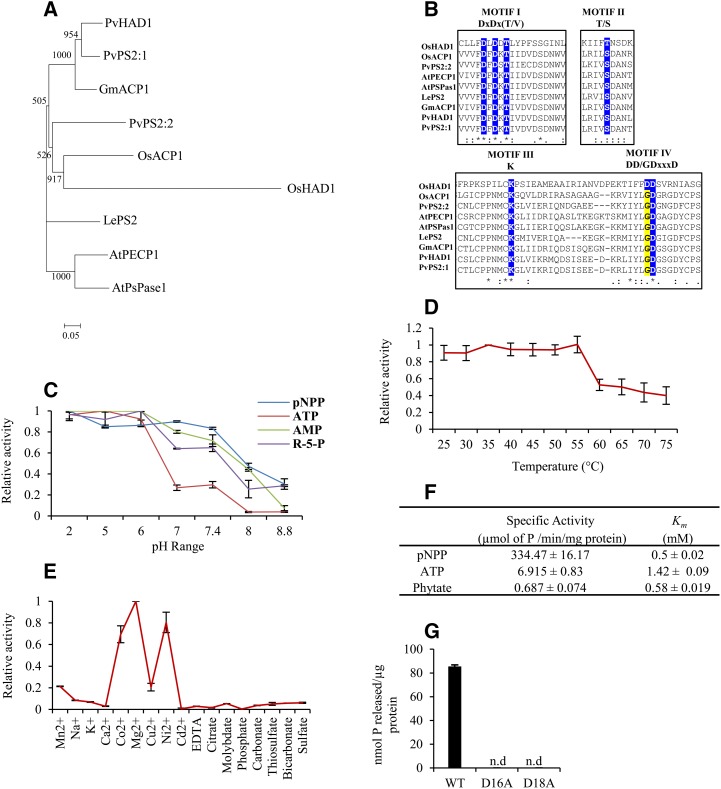
Protein sequence alignment revealed the divergence of OsHAD1 from other known plant HAD domain-containing proteins (Fig. 2A). However, as expected for the HAD family, homology was restricted to strictly conserved residues only (Fig. 2B). Furthermore, OsHAD1 contains four conserved motifs (DxDxT/V, S/T, K, and DD) in which the first motif (DxDxT/V) is essential for APase activity (Fig. 2B). It is reported that the first Asp residue of the first conserved motif (D1LD2DT) acts as a nucleophile and that the second Asp residue acts as acid or base, which protonates the leaving groups in most of the phosphatases from the HAD superfamily (Burroughs et al., 2006). Thus, this motif can play an important role in releasing Pi from various P-containing compounds.
Figure 2.
Phylogenetic relationships and biochemical properties of OsHAD1. A, Phylogenetic relationships of OsHAD1 with known HAD proteins from rice (OsACP1), Arabidopsis (AtPsPase1 and AtPECP1), tomato (LePS2), soybean (Glycine max; GmACP1), and common bean (Phaseolus vulgaris; PvHAD1, PvPS2:1, and PvPS2:2), shown by a neighbor-joining phylogenetic tree. The scale bar represents the rate of amino acid substitutions. Bootstrap values are mentioned at the branches. B, Alignment of the amino acid sequence of OsHAD1 with protein sequences of LePS2, OsACP1, PvPS2:1, PvPS2:2, AtPECP1, AtPsPase1, GmACP1, and PvHAD1. Conserved HAD motifs (I–IV) are highlighted. C, Effects of different pH values on the activity of OsHAD1 using 10 mm pNPP, ATP, AMP, and Rib-5-P (R-5-P) as substrates in sodium acetate buffer (pH range 2–6) and Tris-Cl buffer (pH range 7–8.8). D, Relative activity of OsHAD1 at different temperatures. E, Effects of different activators and inhibitors on the activity of OsHAD1. F, Specific activity and Km values of OsHAD1 with pNPP, ATP, and sodium phytate as substrates. G, Phosphatase activity of wild-type (unmutated; WT) and mutated (D16A and D18A) OsHAD1 recombinant proteins. n.d. indicates no activity detection. Relative activity was calculated with respect to the highest activity, which was taken as 1. Error bars in all graphs indicate sd from three independent measurements.
To test whether OsHAD1 encodes an active protein, we purified recombinant OsHAD1 protein with a 6xHis tag from Escherichia coli (Supplemental Fig. S2). Purified recombinant OsHAD1 showed phosphatase activity with pNPP as a substrate. We next used various other P-containing substrates to elucidate the substrate specificity of OsHAD1 (Table I). This assay showed that free nucleotides like ATP, ADP, and AMP are the preferred substrates of OsHAD1. However, OsHAD1 also exhibited moderate phosphatase activity with sodium phytate, Glc-6-P, Fru-6-P, Rib-5-P, and other P-containing compounds (Table I). Maximum phosphatase activity was obtained with ATP (40.2%) as compared with pNPP (100%). Notably, OsHAD1 also was able to hydrolyze phosphorylated Ser and, to some extent, phosphorylated Tyr. These results confirmed the protein phosphatase activity of OsHAD1.
Table I. Relative activity of recombinant OsHAD1 with different organic and inorganic P compounds.
Relative percentage activity was calculated with respect to the activity with pNPP as substrate.
| Substrate | Percentage Activity |
|---|---|
| pNPP | 100.00 |
| ATP | 40.20 |
| ADP | 17.28 |
| AMP | 25.21 |
| Inorganic pyrophosphate | 2.69 |
| Phytic acid | 1.70 |
| O-Phosphoserine | 2.29 |
| O-Phosphothreonine | 0.96 |
| Glc-6-P | 6.23 |
| Fru-6-P | 4.14 |
| Man-6-P | 4.73 |
| Rib-5-P | 10.48 |
| dRib-5-P | 6.26 |
| α-d-Man-1-P | 3.68 |
A pH kinetics study revealed that OsHAD1 is an APase, as it could hydrolyze substrates (pNPP, ATP, AMP, and Rib-5-P) at very low pH 2 to 5 (Fig. 2C). OsHAD1 was found to be moderately thermostable (25°C–55°C), with maximum phosphatase activity at 37°C (Fig. 2D). Furthermore, different metal ions could enhance the OsHAD1 activity (Mg2+ > Ni2+ > Co2+ > Mn2+); Mg2+ turned out to be the preferential cofactor. High Pi concentrations, Cd2+, citrate, EDTA, and Ca2+, seemed to be inhibitors of OsHAD1 phosphatase activity (Fig. 2E). Furthermore, OsHAD1 showed specific activity: 334.47, 6.95, and 0.687 μmol P released min−1 mg−1 protein with pNPP, ATP, and sodium phytate, respectively. Lastly, OsHAD1 showed higher affinity (Km = 0.5 and 0.58 mm) with pNPP and sodium phytate and lower affinity with ATP (Km = 1.42 mm; Fig. 2F).
Site-Directed Mutagenesis Revealed That Asp Residues Are Critical for OsHAD1 Activity
To confirm the role of motif 1, which is critical for phosphatase activity, we independently modified two key Asp residues, D16 and D18, to Ala from motif I (D16LD18DT) of OsHAD1 (Fig. 2B). Notably, these mutations (D16A and D18A) led to the absolute loss of phosphatase activity in OsHAD1 (Fig. 2G). These results confirmed the importance of Asp residues for the catalytic activity of OsHAD1.
Exogenous Application of OsHAD1 Enhances the Growth of Pi-Deficient Plants on Phytate-Supplemented Medium
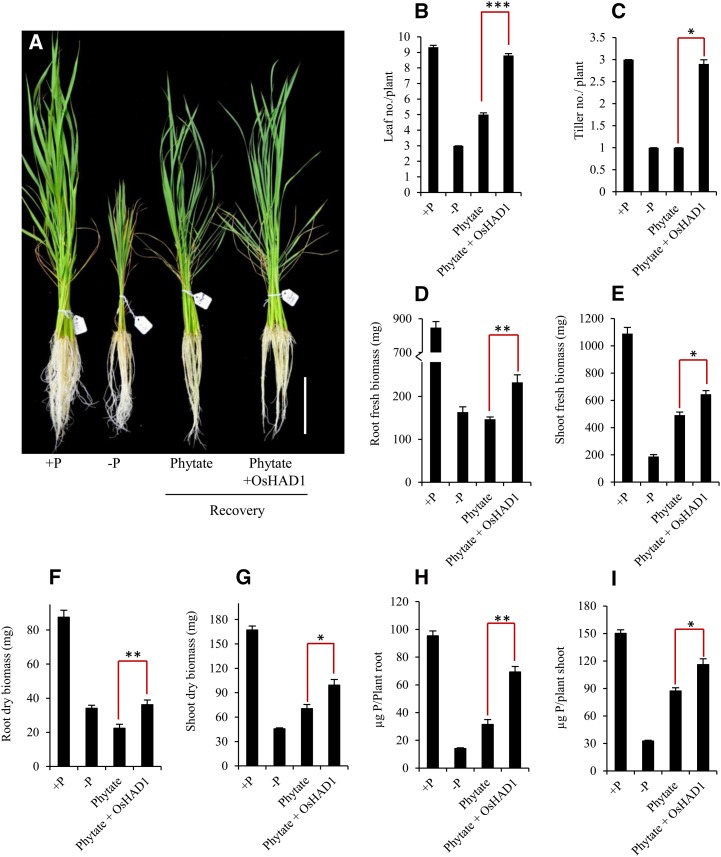
To measure the impact of phytase activity of OsHAD1, 15-d-old Pi-starved seedlings were supplemented with 100 µm sodium phytate with or without 125 ng mL−1 recombinant OsHAD1 in Yoshida medium for 7 d. The overall growth of phytate + OsHAD1-supplemented plants was superior to that of plants supplemented with phytate only (Fig. 3A). Fresh root and shoot biomass of phytate + OsHAD1-supplied plants was increased up to 58.8% and 31.2%, respectively, as compared with phytate-supplemented plants (Fig. 3, D and E). Increased shoot biomass was in good concordance with the increased number of tillers and leaf numbers of phytate + OsHAD1-supplemented plants (Fig. 3, B and C). Similar trends also were observed in dry biomass gain (Fig. 3, F and G). To understand the reason for the enhanced growth after exogenous application of OsHAD1, we quantified the total P content of root and shoot separately. Phytate + OsHAD1-supplied plants showed significantly higher P content (120% in roots and 33% in shoots) as compared with phytate-supplemented plants (Fig. 3, H and I). Moreover, exogenous application of a mutagenized protein of OsHAD1 (D16A) in phytate-supplemented medium resulted in no significant growth advantage to wild-type seedlings as compared with Pi-deficient seedlings treated with phytate alone (Supplemental Fig. S3). Overall, these results showed the efficacy of OsHAD1 in the solubilization of phytate and, thus, improved plant growth.
Figure 3.
Effects of exogenously supplied OsHAD1 with phytate on the growth of Pi-starved wild-type rice plants. A, Improvement in growth of 15-d-old Pi-deficient wild-type rice seedlings after 7 d of recovery with phytate (100 µm) or phytate + OsHAD1 (125 ng of OsHAD1 per 1 mL of Yoshida medium supplemented with 100 µm phytate). Each stack consists of five seedlings. Bar = 10 cm. B to I, Leaf number (B), tiller number (C), fresh root biomass (mg plant−1; D), fresh shoot biomass (mg plant−1; E), root dry biomass (F), shoot dry biomass (G), and total P content (µg plant shoot−1; H) in dried shoot as well as total P content (µg plant root−1; I) in dried roots of wild-type rice seedlings grown under Pi sufficient (+P), Pi deficient (−P), phytate recovered (Phytate), and phytate plus OsHAD1 (Phytate+OsHAD1) recovered conditions. –P and +P plants were provided 1 µm Pi (NaH2PO4) and 320 µm Pi, respectively, for 22 d in Yoshida medium. Error bars indicate se (n = 20). Student’s t test was used to calculate significance. Asterisks indicate P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***).
Overexpression of OsHAD1 Enhances Pi Accumulation in Transgenic Rice
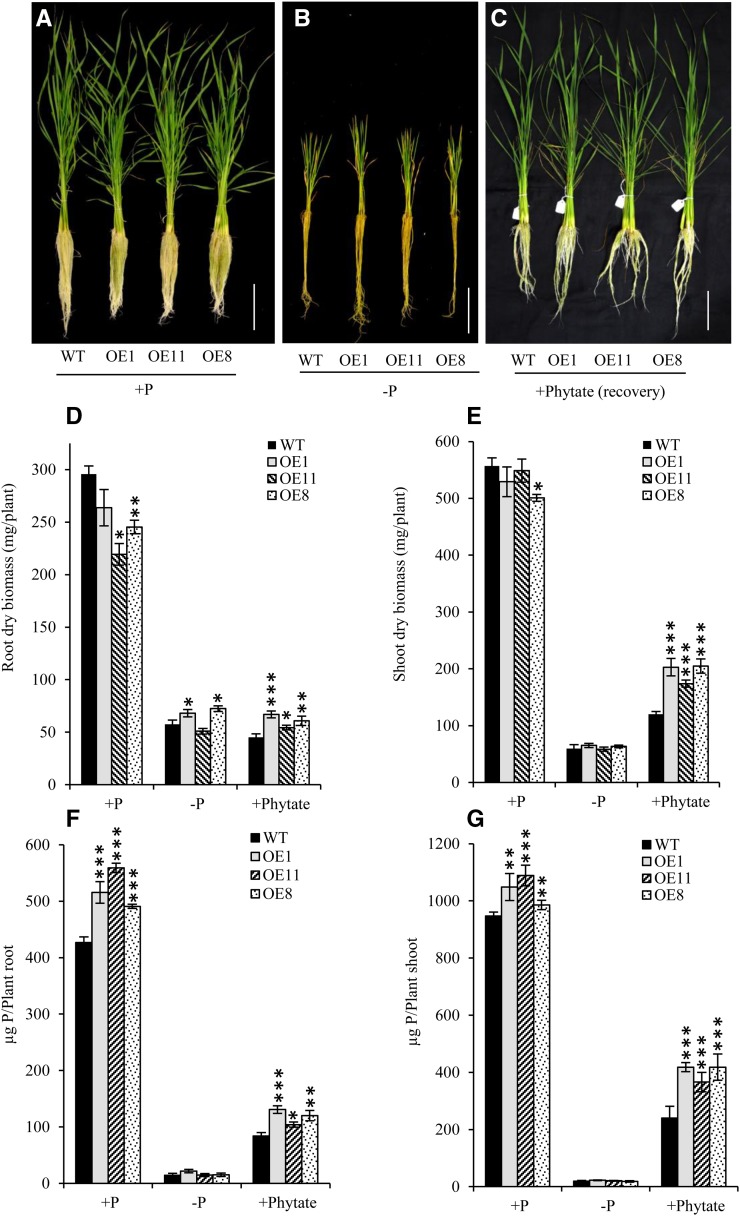
Our results so far revealed OsHAD1 as a low-Pi-responsive APase with broad substrate specificities. Therefore, to study the effect of its phosphatase activity on low-Pi tolerance, we cloned OsHAD1 under the control of a constitutive promoter (ZmUbi1) and raised overexpression lines of OsHAD1 in rice. Five homozygous lines were used for the initial screening, and the three highest overexpressing (OE) lines were selected for further study. Transgenics were confirmed for gene integration and overexpression using PCR, qRT-PCR, western blotting, and GUS histochemical assay (Supplemental Fig. S4). Immunoblotting of total plant protein with anti-OsHAD1 antibody confirmed the overexpression of OsHAD1 in all three OE lines; however, no band was detected in wild-type seedlings, indicating very low expression of OsHAD1 under the native condition (Supplemental Fig. S4B). To test low-Pi tolerance of OE lines, we grew OE lines along with the wild type under Pi-deficient (−P; 1 µm NaH2PO4) and Pi-sufficient (+P; 320 µm NaH2PO4) conditions for 30 d (Fig. 4, A and B). Additionally, we treated 15-d-old Pi-deficient wild-type and transgenic lines with 100 µm phytate (+Phytate) as a restricted P source for the next 15 d (Fig. 4C). Interestingly, dry root and shoot biomass of OsHAD1 overexpressed lines increased significantly (48.5% and 68.8%, respectively) as compared with the wild type under phytate treatment (Fig. 4, D and E). Similarly, the root biomass of OE1 and OE8 was significantly higher (18.5% and 25.9%, respectively) than in the wild type under Pi deficiency (Fig. 4D). Furthermore, total P content was significantly higher in OE lines under Pi-sufficient and phytate recovery conditions in root and shoot (Fig. 4, F and G). However, under Pi deficiency, none of the lines showed significant change in P accumulation, as a very low amount of Pi was supplied. Intriguingly, transgenics grown under Pi-sufficient conditions showed some growth retardation, which could be due to high P accumulation.
Figure 4.
Effects of OsHAD1 overexpression on biomass and total P content in hydroponics. A to C, Plant phenotypes are shown for 30-d-old hydroponically raised wild-type (WT) and OsHAD1 overexpressed lines (OE1, OE11, and OE8) grown under 320 µm Pi (+P) for 30 d (A), 1 µm Pi (−P) for 30 d (B), or 1 µm Pi (−P) for 15 d followed by recovery with 100 µm phytate (+Phytate) for another 15 d (C). Each stack consists of three seedlings. Bars = 10 cm. D to G, Dry root biomass (D), dry shoot biomass (E), total P content per plant root (F), and total P content per plant shoot (G) of wild-type and OsHAD1 overexpression lines under control conditions (+P; 30 d), Pi deficiency (−P; 30 d), and phytate recovery (15 d of −P treatment followed by recovery with 100 µm phytate for another 15 d). Error bars in all graphs indicate se (n = 10). Significant differences in overexpression lines with respect to the wild type were determined by Student’s t test. Asterisks indicate P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***).
We further extended our analysis to wild-type and OsHAD1 transgenic plants raised in a soil system using PVC pipes filled with sand, soilrite, and vermiculite (Supplemental Fig. S5). Atomic absorption spectroscopy measurements showed that a formulated mixture had a very low amount of P (0.1 µm). These pipes were supplied with Pi-sufficient, Pi-deficient, and phytate-containing growth media (see “Materials and Methods”). Notably, phytate-supplemented OE lines had significantly higher shoot biomass (15%–40%) as compared with the wild type (Supplemental Fig. S6B). Furthermore, Pi-deficient plants also showed 38% more shoot biomass as compared with controls (Supplemental Fig. S6B). Total P in all the OE lines was significantly higher in both root and shoot than in the wild type under +P and phytate treatments (Supplemental Fig. S6, C and D). Moreover, a significant increase in total P also was found in shoot of OsHAD1 OE lines under the Pi-deficient condition (Supplemental Fig. S6D). These results confirmed that overexpression of OsHAD1 leads to increased P accumulation in plants under both hydroponics and soil conditions.
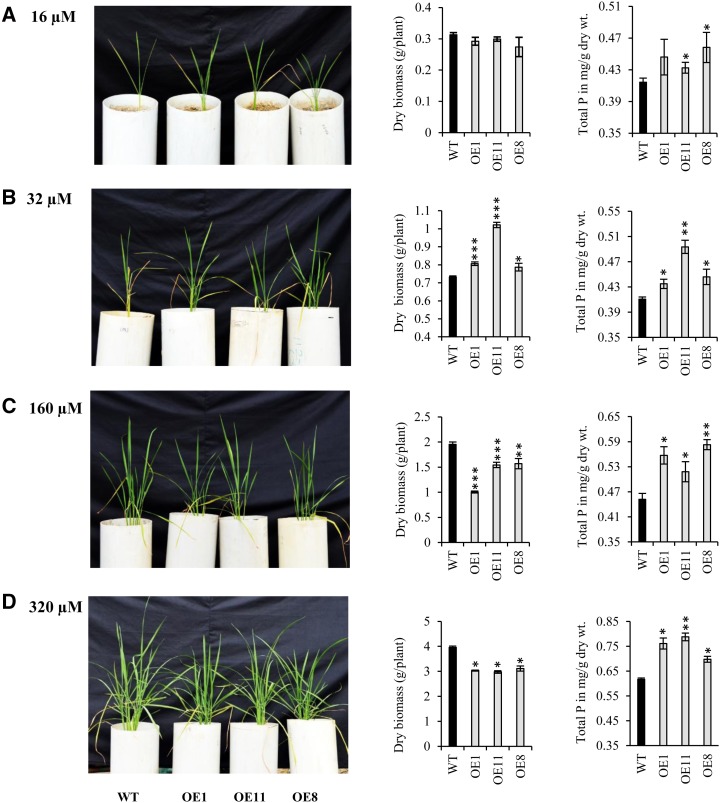
Similar to hydroponics, growth retardation also was seen in overexpression lines in the soil system as compared with wild-type plants under high-Pi conditions. To investigate this further, we performed a dose-dependent experiment consisting of different Pi concentrations (16, 32, 160, and 320 µm) for 50 d in soil. Although the biomass of OsHAD1 OE lines was significantly higher at 32 µm Pi as compared with the wild type (Fig. 5B), the same was significantly decreased at higher dosages of Pi (160 and 320 µm) than in the wild type (Fig. 5, C and D). Consistent with previous observations, total P content of OsHAD1 OE lines was significantly higher at all concentrations of external Pi as compared with wild-type plants (Fig. 5). To this end, we conclude that OsHAD1 overexpression led to increased P accumulation in rice and that transgenics perform better in low-Pi conditions.
Figure 5.
Growth performance and P accumulation in wild-type (WT) and OsHAD1 overexpressed lines under soil conditions at different Pi concentrations. Plant phenotype, dry biomass, and total P content are shown at 16 µm (A), 32 µm (B), 160 µm (C), and 320 µm (D) Pi. The experiment was set up in three biological replicates. Student’s t test was used to evaluate significant differences in overexpression lines with respect to the wild type. Asterisks indicate P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***).
OsHAD1 Is a Cytosolic and Membrane-Localized Protein
It is possible that OsHAD1 is a secretory APase and hydrolyzes external Pi compounds in the rhizosphere of transgenics. Therefore, we studied its subcellular localization using an OsHAD1:YFP fusion protein to determine its secretory or nonsecretory nature. OsHAD1:YFP was found to be localized in cytosol and either cell wall or plasma membrane in onion (Allium cepa) and Nicotiana benthamiana epidermal cells (Supplemental Figs. S7A and S8A). Interestingly, plasmolysis of OsHAD1-overexpressing cells confirmed that OsHAD1 is a cytosolic protein and, therefore, an intracellular or nonsecretory APase (Supplemental Figs. S7B and S8B).
OsHAD1 Overexpressed Lines Showed Increased APase Activity and Soluble P Content
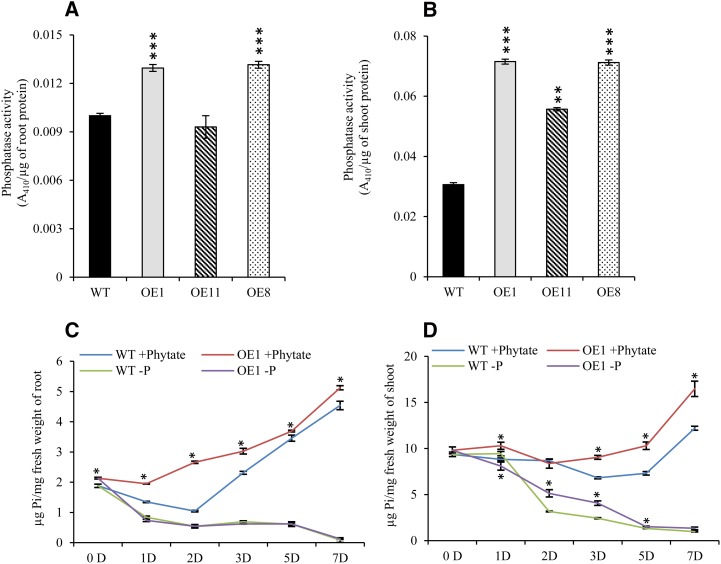
Since OsHAD1 is an intracellular APase, we tested its activity in 30-d-old hydroponically raised OE lines. Enhanced intracellular APase activity would increase Pi levels in a cell system. We found significantly increased phosphatase activity in roots and shoots of OE lines as compared with the wild type (Fig. 6, A and B). Notably, OE lines also showed significantly higher phytase activity under phytate-supplied conditions (Supplemental Fig. S9).
Figure 6.
Increased phosphatase activity and soluble P content in OsHAD1 overexpression lines. A and B, Total acid phosphatase activity in root (A) and shoot (B) of 30-d-old hydroponically raised wild-type (WT) and OsHAD1 overexpression (OE1, OE11, and OE8) lines raised under +Phytate (100 µm) treatment (15-d-old Pi-deficient plants recovered with 100 µm phytate for another 15 d). Phosphatase activity was quantified with 5 µg of root and 10 µg of shoot protein, respectively, using 10 mm pNPP substrate at 37°C. C and D, Root (C) and shoot (D) soluble P content of the wild type and OE1 at different time points under Pi deficiency and phytate supply. For soluble P estimation, 15-d-old wild-type and OsHAD1 OE1 plants raised hydroponically under +P conditions were placed in Pi-deficient or phytate-supplied (100 µm) medium. Student’s t test was used to evaluate significant differences in overexpression lines with respect to the wild type. Asterisks indicate P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***).
To further validate the consequence of elevated intracellular APase activity (Pi solubilization from cellular organic P compounds) in OE lines, Pi content was measured temporally in +P seedlings transferred to −P or phytate-supplemented medium. Transgenics showed higher soluble Pi content than the wild type under both conditions (Fig. 6, C and D). This indicates that enhanced cellular Pi levels in OsHAD1 OE lines could be due to increased intracellular APase activity.
Increased Accumulation of P in OsHAD1 OE Lines Is Mediated by an Enhanced Phosphate Starvation Response
Increased Expression of Pi Transporters and Secretory APases in OE Lines
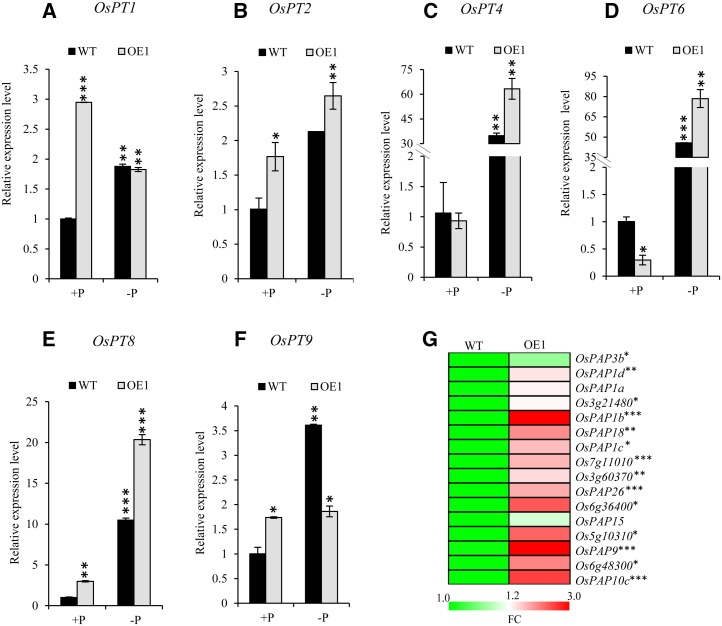
Next, we studied the expression patterns of phosphate starvation response (PSR) Pi transporters (OsPT1, OsPT2, OsPT4, OsPT6, OsPT8, and OsPT9) to determine the reason for the greater P accumulation in transgenics. Notably, OsPT8, OsPT2, OsPT9, and OsPT1 were significantly up-regulated in roots of OsHAD1 plants under the +P condition as compared with the wild type. Their expression was further induced under −P with higher magnitude in OE1 (Fig. 7). Interestingly, OsPT4 and OsPT6 were highly up-regulated in overexpressed lines under −P as compared with the wild type (Fig. 7, C and D). We further determined the expression of several predicted secretory phytases and low-P-responsive purple acid phosphatases (PAPs) in 30-d-old hydroponically raised wild-type and OE lines. Interestingly, most of the phosphatases were consistently up-regulated in OE lines as compared with the wild type (Fig. 7G). In concordance with this, overexpression of OsHAD1 led to increased secretory APase activity as compared with the wild type (Supplemental Fig. S10). Therefore, the increased growth and P accumulation in transgenics could be additive effects of enhanced hydrolysis of cellular Pi, activity of Pi transporters, and secretory phosphatases/phytases.
Figure 7.
Expression profiling of Pi transporters in wild-type (WT) and OsHAD1 transgenic plants. A to F, Relative expression levels are shown for OsPT1 (A), OsPT2 (B), OsPT4 (C), OsPT6 (D), OsPT8 (E), and OsPT9 (F) in roots of 30-d-old wild-type and OsHAD1 overexpression (OE1) plants raised under +P (320 µm) and −P (1 µm) conditions. qRT-PCR was used to determine gene expression, and relative expression level was calculated with respect to the expression level in roots of wild-type +P plants. Each bar represents an average of three replicates with sd. G, Heat map representing relative expression levels (fold change [FC]) of predicted secretory phosphatases/phytases in roots of 30-d-old OsHAD1 overexpression lines (OE1 and OE11) with respect to the wild type, raised under +P (320 µm). The heat map was generated with MEV 4.6.0. software. Significant changes were determined by Student’s t test. Asterisks indicate P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***).
OsHAD1 OE Leads to Increased Organic Acids and Decreased ATP Accumulation
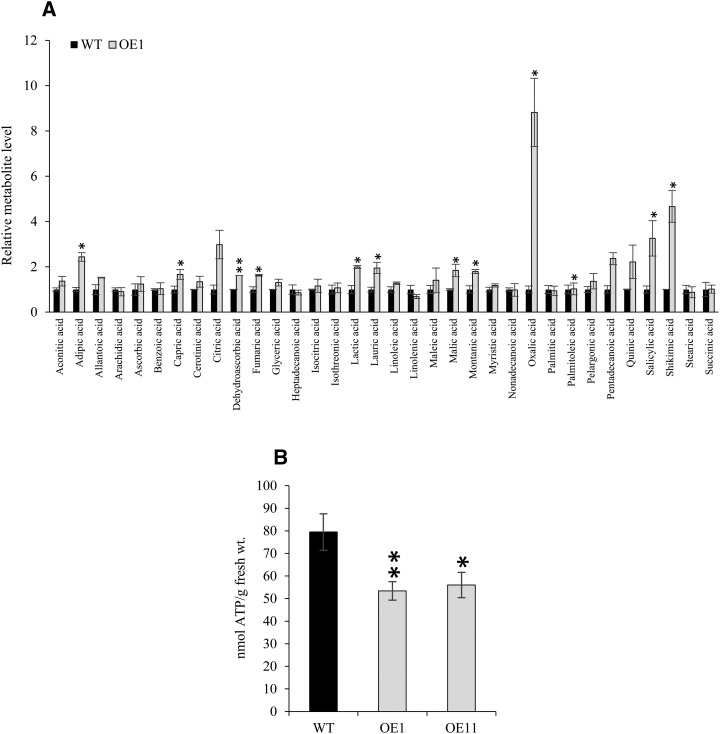
To further understand the increased PSR in OsHAD1 OE plants at the metabolome level, we performed metabolite profiling of the wild type and OE1. A total of 319 metabolites were detected through gas chromatography-time of flight-mass spectrometry (GC-TOF-MS), of which 90 identified compounds were categorized further into different classes, such as P-containing metabolites, sugars, acids, amino acids, and others (Supplemental Table S1). Overexpression of OsHAD1 significantly increased levels of several amino acids and organic acids. Interestingly, most of organic acids, such as oxalic acid, citric acid, malic acid, and succinic acid, were increased significantly in OE1 as compared with the wild type (Fig. 8A; Supplemental Table S1). Elevated levels of these organic acids can help to catalyze faster release of soluble Pi from fixed P sources in the rhizosphere. Furthermore, two of the phosphorylated metabolites, glycerol-α-phosphate and Glc-6-P, were increased significantly in OE1 as compared with the wild type. On the other hand, another P compound, inositol-4-monophosphate, was reduced significantly in OE1 as compared with the wild type (Supplemental Table S1). However, ATP, the most preferred organic substrate of OsHAD1, could not be quantitated in GC-TOF-MS analysis; therefore, we quantitated ATP levels in wild-type and OE lines colorimetrically. Notably, OsHAD1 OE lines (OE1 and OE11) showed striking depletion of 41% to 48% in ATP levels as compared with the wild type (Fig. 8B). This observation indicates that the increased hydrolysis of intracellular ATP could add to the enhanced P pool in OE lines and subsequent phenotypic alterations. To this end, we conclude that OsHAD1 activates PSR on overexpression in transgenics, which leads to the observed phenotypes.
Figure 8.
Organic acid accumulation and ATP depletion in OsHAD1 overexpression lines. A, Relative metabolite levels (ratio of peak heights) in OsHAD1 OE lines with respect to the wild type (WT). Error bars indicate se from three biological replicates. B, ATP content in shoots of the wild type and OsHAD1 overexpression lines (OE1 and OE11). Analyses were carried out in 30-d-old hydroponically raised plants grown under +P conditions (320 µm NaH2PO4). Error bars indicate se from eight replicates. Significant differences in overexpression lines with respect to the wild type were determined by Student’s t test. Asterisks indicate P ≤ 0.05 (*) and P ≤ 0.01 (**).
Identification of OsHAD1-Interacting Proteins
To further understand the molecular regulation by OsHAD1, we performed a pull-down assay to find its interacting partners using OsHAD1 OE plants. This analysis led to the identification of seven novel putative interactive partners (Table II). One nucleoside diphosphate kinase, OsNDPK3, showed good protein coverage (12%) and high MASCOT scores (131) in liquid chromatography-mass spectrometry analysis. This kinase is known to transfer a Pi group from its His residue to phosphorylate S/T/Y residues in various nucleoside diphosphates. A yeast two-hybrid assay also confirmed the strong interaction of OsHAD1 with OsNDPK3 (Supplemental Fig. S11). Moreover, OsNDPK3 (also found to be low-Pi inducible) was significantly up-regulated in OsHAD1 OE lines as compared with the wild type (Supplemental Fig. S12). This kinase also was found to coexpress with OsHAD1 under cytokinin treatment (Supplemental Fig. S13). We also performed interaction studies of OsHAD1 with other rice NDPKs. Interestingly, OsHAD1 also showed strong interaction with OsNDPK1 (Supplemental Fig. S11). The pull-down assay additionally identified one receptor-like kinase (Os10g04730) as an interactive partner of OsHAD1. Notably, it has been shown that the first Asp residue of the DFxDxT/V motif undergoes transient phosphorylation (Baldwin et al., 2001). Thus, these kinases may invariably regulate the OsHAD1 activity by a phosphorylation event. Interestingly, the NetPhos 2.0 prediction server revealed that OsHAD1 possesses 13 Ser, four Tyr, and three Thr residues as phosphorylation sites. Of these, Ser-117, Ser-160, Ser-167, Ser-182, Ser-189, Ser-213, and Tyr-10 are highly predicted (score > 0.93) potential phosphorylation sites. OsHAD1 in close association with these kinases also may regulate the phosphorylation status of downstream targets to activate PSR.
Table II. List of putative interacting partners of OsHAD1 identified by liquid chromatography-tandem mass spectrometry analysis combined with electrospray ionization.
| Locus Identifier | Score | Putative Function | Nominal Mass | Calculated pI | Protein Coverage |
|---|---|---|---|---|---|
| Os05g51700 | 131 | Nucleoside diphosphate kinase | 26,124 | 8.88 | 12 |
| Os08g34280 | 41 | Cinnamoyl-CoA reductase | 47,493 | 6.34 | 20 |
| Os02g06770 | 41 | Expressed protein | 10,571 | 9.34 | 10 |
| Os10g04730 | 35 | TKL_IRAK_DUF26-la.6 | 47,348 | 5.38 | 3 |
| Os02g57140 | 35 | Expressed protein | 104,855 | 5.57 | 0 |
| Os03g04990 | 29 | Expressed protein | 99,020 | 5.76 | 1 |
| Os02g10794 | 19 | Expressed protein | 207,557 | 5.32 | 0 |
DISCUSSION
Given the indispensable role of P in plant growth and the limited availability of phosphate rocks, enhancing P acquisition and utilization in crops is the key target for sustainable agriculture. Rice is one of most important cereal crops for the developing world. At the same time, rice cultivation requires the highest consumption of Pi fertilizers in countries like India (FAI, 2008; http://www.faidelhi.org/). Therefore, improving the low-P tolerance of rice is a major concern in rice-growing developing nations. Notably, a larger proportion of P exists in the form of organic compounds within cells and the rhizosphere. However, little effort has been devoted to enhancing Pi utilization by releasing bound P from these compounds in rice. Low-Pi-inducible phosphatases play important roles in releasing Pi from bound organic compounds, both intracellularly and extracellularly. The plant HAD superfamily comprises a large number of phosphatases. However, very few low-Pi-inducible phosphatases from the HAD superfamily have been identified and characterized in crop plants (Baldwin et al., 2008; Liang et al., 2012; Zhang et al., 2014). Here, we show that a low-Pi-inducible haloacid dehalogenase, OsHAD1 encodes a functional acid phosphatase and plays a key role in Pi homeostasis in rice.
This study revealed that OsHAD1 is a novel low-Pi-responsive APase in rice. Notably, the protein sequence of OsHAD1 does not show significant homology with any other known HAD superfamily phosphatase in plants (Supplemental Fig. S14). However, a multiple sequence alignment showed that OsHAD1 possesses the highly conserved motif I, which is essential for the phosphatase activity of LePS2, OsACP1, PvPS2:2, and GmACP1 phosphatases of the HAD superfamily (Hur et al., 2007; Liang et al., 2012; Zhang et al., 2014). Furthermore, only the four nearest homologs of OsHAD1 in rice shared conserved motifs with OsHAD1 (Supplemental Fig. S15); two of these also showed transcript accumulation under low Pi, although lower than OsHAD1 (Supplemental Fig. S16). This shows that OsHAD1 is unrelated to previously characterized low-Pi-responsive HAD superfamily members. This may be due to the fact that there exists a great degree of sequence divergence among HAD superfamily members (Kuznetsova et al., 2006). This sequence divergence also has been linked to the substrate diversity of HAD proteins (Caparrós-Martín et al., 2013). Although reported members of the HAD superfamily were induced by low Pi, any evidence of their transcriptional regulation was missing. Notably, the expression of many PSR genes is mediated by a MYB transcription factor, OsPHR2, which is a functional ortholog of AtPHR1 in rice (Zhou et al., 2008; Wu et al., 2013; Guo et al., 2015).The physical interaction of OsPHR2 with an OsHAD1 probe in a gel-shift assay proved that OsHAD1 acts under OsPHR2-mediated transcriptional regulation.
OsHAD1 possesses both APase and phytase activities and shows a broad substrate specificity. It is important to mention here that most of the PAPs and APases show wide substrate specificity (Kuang et al., 2009; Liang et al., 2010; Tran et al., 2010b; Wang et al., 2011; Cabello-Díaz et al., 2012). It is likely that broad substrate specificity would help to release Pi from various intracellular or extracellular P sources to alleviate Pi starvation. However, to date, no known HAD in rice has been characterized to have broad substrate specificity. Moreover, biochemical properties of OsHAD1, such as high activity in acidic pH and high thermostability, further make it a suitable candidate for improving the low-P tolerance of rice and other crops that are cultivated under higher temperature and acidic ecosystems. Since OsHAD1 is an intracellular and membrane-bound protein, OsHAD1 also can be overexpressed under a potential signal peptide and targeted in an extracellular milieu for the solubilization of bound P, including phytate. Contradictory to most of the highly low-Pi-responsive PAPs, in silico analysis (using SignalP 4.1, SMART, and PrediSi) of known low-P-responsive HAD superfamily members (such as LePS2, OsACP1, PvPS2:1, PvPS2:2, GmACP1, PvHAD1, AtPECP1, and AtPSPase1) revealed the lack of a potential signal peptide. Additionally, PvPS2:1, PvPS2:2, and LePS2 have been shown to be intracellular protein phosphatases. This indicates that low-Pi-responsive HAD superfamily proteins are destined to play some unknown cellular signaling roles under Pi deficiency.
OsHAD1 OE lines showed higher P accumulation than the wild type under +P, −P, and restricted P supply (+Phytate) in hydroponics, soil conditions, as well as in aseptic agar (Figs. 4–6; Supplemental Figs. S6 and S17). This indicates high Pi acquisition in OE lines irrespective of the degree/type of P supply. However, under high-Pi conditions, OE lines showed retarded growth as compared with the wild type, which could be due to enhanced P accumulation. Previous studies also have reported growth retardation due to high Pi accumulation in rice. For instance, it is known that OsPHR2 overexpression resulted in excessive P accumulation in rice shoots, leading to retarded growth under sufficient Pi (Zhou et al., 2008). Higher Pi accumulation in OsPHR2 OE lines was attributed to increased expression of several PSR genes, such as OsPTs (Zhou et al., 2008). Likewise, overexpression/suppression of many PSR genes, like OsMyb2P-1, OsSPX1, OsmiR399, and ltn1, showed overaccumulation of P and retarded growth under sufficient Pi (Wang et al., 2009; Hu et al., 2011; Dai et al., 2012). Overexpression of OsHAD1 activated the phosphate starvation response in rice seedlings. Elevated expression of secretory phytases/phosphatases and PSR phosphate transporters in OsHAD1 OE lines further strengthens the signaling role for OsHAD1 in Pi homeostasis. Several studies have shown that increased expression of PSR transporters and phosphatases/phytases is involved directly in the higher P accumulation (Zhou et al., 2008; Tran et al., 2010a; Kong et al., 2014). Furthermore, overexpression of OsPT8 (also up-regulated in OsHAD1 OE1) showed excessive P accumulation under high-P conditions (Jia et al., 2011). OsPT8 is membrane localized, and OsHAD1 also showed signal in membrane. Coexpression and qRT-PCR analysis of OsHAD1 showed that both OsHAD1 and OsPT8 are down-regulated in shoot when treated with cytokinin (Supplemental Fig. S13), a known suppressor of PSR genes under low-P conditions (Martín et al., 2000; Hou et al., 2005; Lai et al., 2007).
Notably, like overexpression of intracellular HAD, PvPS2:1 also was found to up-regulate the transcripts of a PSR Pi transporter, leading to enhanced P content and superior P efficiency in Arabidopsis OE lines as compared with the wild type (Liang et al., 2012). Apart from these findings, well-known low-Pi responses such as increased secretory APase activity, accumulation of organic acids (Pant et al., 2015), and decreased levels of ATP (Mikulska et al., 1998) in OE lines indicate that OsHAD1 is involved in regulating PSR. Reduction in ATP content also could be caused by increased phosphatase activity of OsHAD1, as ATP was found to be the most preferred substrate of OsHAD1. Increased ATP hydrolysis by ATP can add to the cellular Pi pool and enhance internal Pi utilization efficiency.
Reversible phosphorylation, mediated by PSR phosphatases and kinases, is one of the prominent posttranslational modifications that regulate Pi homeostasis (Lan et al., 2013; Li et al., 2014). Regulation of the phosphorylation status of proteins can affect their activity, stability, structure, localization, and interactions (Lan et al., 2013). Some of the proteins, such as AtPHT1;1 and Pho4, are reported to be regulated by reversible phosphorylation under Pi deficiency in Arabidopsis and yeast, respectively (Springer et al., 2003; Bayle et al., 2011). Several kinases and phosphatases are differentially regulated by Pi deficiency (Wu et al., 2003; Fragoso et al., 2009; Gamuyao et al., 2012; Lan et al., 2012). Some of these also are suggested to play pivotal roles in maintaining Pi homeostasis through modifying the phosphorylation status of downstream targets. For instance, overexpression of a protein kinase-encoding PSR gene, OsPSTOL1, was hypothesized to regulate the phosphorylation of unknown targets, leading to the induction of numerous genes involved in root proliferation and, thus, Pi uptake (Gamuyao et al., 2012). Unlike protein kinases, protein phosphatases form a small group of proteins but can associate themselves with a huge number of target proteins and regulate their functions (Wang et al., 2007; Plaxton and Shane, 2015). Overexpression of a Ser/Thr phosphatase-encoding HAD gene, LePS2, resulted in delayed plant development and increased APase activity and anthocyanin content in transgenic tomato (Baldwin et al., 2008). The altered phosphorylation of unknown targets by LePS2 was proposed to initiate an unknown signaling cascade that led to distinct changes in plant morphology. OsHAD1 showed protein phosphatase activity, cytoplasmic location, and regulation by OsPHR2; therefore, it might play key signaling roles leading to the enhanced PSR observed in our study.
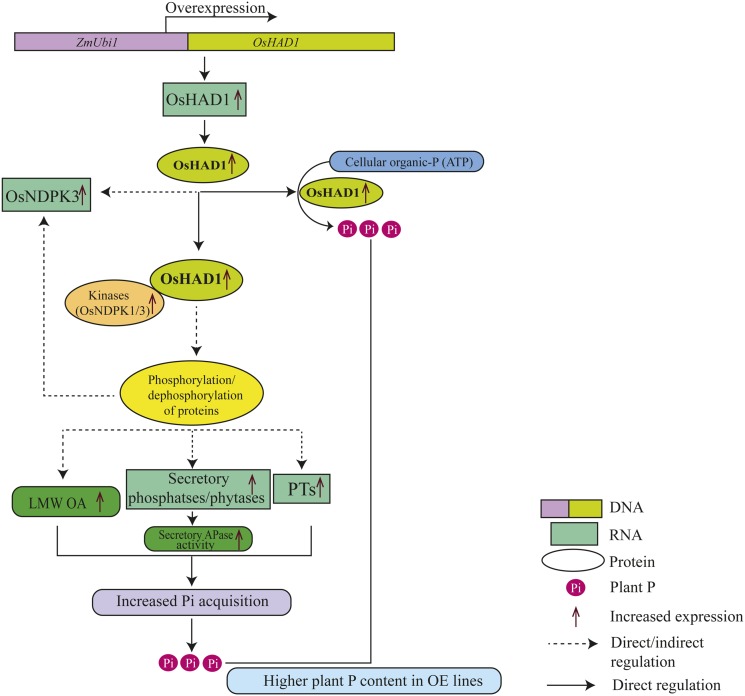
Overexpression of OsHAD1 in rice also might have influenced the phosphorylation status of several target proteins, leading to the induction of Pi transporters, phytases, protein phosphatases, and genes involved in organic acid production. Interestingly, phosphatases of the HAD superfamily have been reported to share similar active site geometries, folds, and reaction chemistry to the response regulator signaling protein of a two-component signaling system (His kinase phosphatase) in bacteria (Ridder and Dijkstra, 1999; Immormino et al., 2015). Moreover, OsHAD1 showed an association with kinases. Of these, NDPKs play important roles in several signal transduction pathways and play important roles in various abiotic and biotic stresses (for review, see Dorion and Rivoal, 2015). Thus, OsHAD1 in coordination with protein kinases may regulate the phosphorylation status of downstream targets to regulate Pi homeostasis in plants (Fig. 9). The effect of this signaling is apparent in the form of the induction of several genes and alterations in metabolite profile that ultimately led to increased plant P content (Fig. 9).
Figure 9.
Model for enhanced P accumulation in OsHAD1 overexpression lines. Constitutive expression of OsHAD1 under the control of the maize (Zea mays) ubiquitin promoter (ZmUbi1) in rice leads to increased P content by direct or indirect routes. Increased expression of OsHAD1 (protein phosphatase) along with interacting kinases affects the phosphorylation status of unknown downstream targets, which increases the expression of PSR Pi transporters (PTs), phosphatases/phytases, and the accumulation of low-molecular-weight organic acids (LMW OA). Overexpression of OsHAD1 also leads to increased expression of its interacting kinase, OsNDPK3. Increased phosphatase activity of OsHAD1 remobilizes P from cellular ATP. These events ultimately lead to increased Pi acquisition in OsHAD1 overexpression lines.
Taken together, our work provides a novel resource to improve the Pi utilization ability of rice. Since only ∼20% of applied P is assimilated by crops in a year and the rest becomes fixed in soil, sustainable agriculture using crops with improved Pi use efficiency would be helpful in reducing the use of Pi fertilizers and their escape in the environment.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of the rice (Oryza sativa) genotypes Dular and PB1 were surface sterilized and germinated as described earlier (Mehra et al., 2016). Uniformly germinated seeds were transferred to Pi-sufficient (320 µm) and Pi-deficient (1 µm) Yoshida media as described (Mehra et al., 2015). Seedlings were raised in growth chambers at 30°C (16 h)/28°C (8 h), ∼70% relative humidity, and 280 to 300 µmol photons m−2 s−1. For OsHAD1 expression analysis, root and shoot from +P and –P seedlings were profiled after 5, 15, and 21 d of treatment. For expression analysis under different nutrient deficiency conditions (−N, −P, −K, −Fe, and −Zn), seedlings were raised hydroponically as described (Singh et al., 2015). Root tissues were collected after 7 and 15 d of the respective deficiency treatments.
To analyze the effect of cytokinin on gene expression, 15-d-old hydroponically raised seedlings were given cytokinin (2 µm BAP) treatment for variable times (0.5, 1, and 2 h). Seedlings were harvested and frozen in liquid nitrogen after the respective time points.
For the analysis of OsHAD1 OE lines in hydroponics, the wild type and three independent T3 homozygous transgenic lines were grown hydroponically under +P and –P for 15 d as explained above. Thereafter, half of the –P seedlings were supplied with 100 µm phytate as a restricted P source for an additional 15 d. Thirty-day-old wild-type and transgenic plants were harvested for downstream phenotypic, biochemical, and molecular analyses.
For soil-based experiments, seedlings were raised in hydroponics under +P and –P for 20 d. Afterward, seedlings were transferred to one-end-closed PVC pipes (42 cm × 9.5 cm) filled with washed sand:soilrite:vermiculite (2:1:1). One seedling per pipe was maintained. Eight replicates of both the wild type and three overexpression lines were arranged randomly in 4 × 8 arrays. The whole experiment was performed with three such arrays. The first array was planted with seedlings raised in +P medium, while the other two arrays were planted with –P seedlings. All three arrays were fed with 1 L of the respective +P and –P Yoshida solutions per day per pipe. After 5 d, one array of −P-raised seedlings was recovered with 100 µm phytate-supplemented Yoshida medium for the next 25 d. The other two arrays were continually supplied with their respective +P and –P Yoshida solutions for 25 d. Thus, 50-d-old plants from all three arrays were harvested for phenotypic analysis and total P content. This experiment was carried out in a greenhouse maintained at 30°C (16 h)/28°C (8 h) and ∼70% relative humidity.
For P content analysis under the aseptic condition, germinated seeds were placed in one-half-strength Murashige and Skoog medium containing 100 µm phytate for 7 d. For soluble Pi content analysis, 15-d-old hydroponically raised +P plants of the wild type and OE1 were subjected to Pi-deficient medium and 100 µm phytate-supplemented medium separately. Root and shoot tissues were frozen at different time points (0, 1, 2, 3, 5, and 7 d) of Pi deficiency and phytate treatment, respectively.
RNA extraction and qRT-PCR were performed as described (Mehra and Giri, 2016) using gene-specific primers (Supplemental Table S2). Total and soluble P estimation was performed as described (Mehra et al., 2016, 2017). The statistical significance of relative expression levels (fold changes) was evaluated using Student’s t test.
Gel Retardation Assay
The coding region of OsPHR2 was amplified using gene-specific forward and reverse primers (Supplemental Table S2) carrying NdeI and BamHI restriction sites, respectively, and cloned in pET28a expression vector (Novagen). 6xHis-tagged OsPHR2 was purified using Ni2+ affinity chromatography as described (Mehra and Giri, 2016). The 485-bp promoter region of OsHAD1 was amplified using gene-specific primers (Supplemental Table S2) and used as a probe. The promoter probe was labeled with [α-32P]CTP using the Megaprime DNA labeling system kit (Amersham Biosciences) according to the manufacturer’s protocol. Three different reactions were set: the first reaction contained 12.74 ng of [α-32P]CTP-labeled OsHAD1 promoter probe only; the second reaction contained 1.5 µg of OsPHR2 protein with 12.74 ng of labeled probe; and the third reaction contained 1.5 µg of OsPHR2 protein, 12.74 ng of labeled probe, with a 100× concentration of unlabeled probe (competitor). All three reactions were incubated with 1 μg of poly(dI-dC), 30 mm KCl, 15 mm HEPES (pH 8), 0.02 mm DTT, 1 mm MgCl2, 0.2 mm EDTA, and 0.6% glycerol at 37°C for 30 min. Thereafter, all reactions were electrophoresed on a 4% polyacrylamide gel for 8 h at 100 V. The gel was exposed to a phosphor screen overnight. The image was captured with the Typhoon 9210 phosphor imager (GE Healthcare).
Expression, Purification, and APase Assays of Recombinant OsHAD1 (Wild Type and Mutated)
The coding region of OsHAD1 was amplified with gene-specific terminal primers (OsHAD1_pET28a F and OsHAD1_pET28a R; Supplemental Table S2). For site-directed mutagenesis, the D16A and D18A mutant versions of OsHAD1 were generated by an overlap extension PCR method as described (Ho et al., 1989). Mutated PCR amplicons were generated using terminal primers (OsHAD1_pET28a) and overlapping primers (OsHAD1_SDM_overlap) with desired mutations (Supplemental Table S2). Wild-type and mutagenized OsHAD1 coding sequences were cloned into BamHI and EcoRI sites of the 6xHis expression vector pET28a. Recombinant wild-type/mutagenized OsHAD1 protein was induced in Escherichia coli strain BL21(DE3)pLysS with 100 mm isopropylthio-β-galactoside and purified by Ni2+ affinity chromatography (Mehra and Giri, 2016). Five micrograms of purified D16A, D18A, and wild-type OsHAD1 proteins was used for the APase assay in a 100-µL reaction containing 10 mm pNPP as substrate, 5 mm MgCl2, and 50 mm sodium acetate (pH 6). Reactions were incubated at 37°C for 30 min. The amount of Pi released was quantified by adding 100 µL of ammonium molybdate reagent (Kitson and Mellon, 1944). Released Pi was estimated by measuring A410 using POLARstar Omega (BMG Labtech) in at least three replicates.
Biochemical Characterization of OsHAD1
pH and Temperature Optima of OsHAD1
The pH optimum of OsHAD1 was determined at various pH levels (2, 5, 6, 7, 7.4, 8, and 8.8) in a reaction containing 10 mm substrate (pNPP, ATP, AMP, and Rib-5-P), 5 mm MgCl2, and 10 µg of recombinant OsHAD1 protein at 37°C for 30 min. A 50 mm concentration of sodium acetate and Tris-Cl buffer were used to set the pH range from pH 2 to 6 and pH 7 to 8.8, respectively. To determine the optimum temperature of OsHAD1, activity assays of OsHAD1 were performed for 30 min at different temperatures (25°C–75°C) in a reaction containing 10 μg of purified OsHAD1 protein, 5 mm MgCl2, 50 mm sodium acetate buffer (pH 6), and 10 mm pNPP. Released Pi was measured by the yellow vanadomolybdate method.
Substrate Specificity and OsHAD1 Reaction Kinetics
To determine the substrate specificity of OsHAD1, various P-containing compounds (pNPP, ATP, ADP, AMP, inorganic pyrophosphate, sodium phytate, O-phosphoserine, O-phosphothreonine, Glc-6-P, Fru-6-P, Man-6-P, Rib-5-P, dRib-5-P, and α-d-Man-1-P) were used. Activity assays were performed with 10 mm of different substrates as described above. For the kinetics study, activity assays were carried out with ATP and pNPP as substrates over a wide range of concentrations (0.1, 0.15, 0.2, 0.25, 0.5, 1, 1.25, 2, 5, 10, 15, 20, 25, 50, 75, and 100 mm). Specific activity and Km were calculated from a Lineweaver-Burk plot. All reactions were performed in triplicate.
Effects of Different Ions and Inhibitors on the Phosphatase Activity of OsHAD1
The phosphatase activity of OsHAD1 was analyzed with 10 mm chloride salts of different cations (Mn2+, Na+, K+, Ca2+, Co2+, Mg2+, Cu2+, and Ni2+) and 10 mm sodium salts of different anions (EDTA, citrate, molybdate, phosphate, thiosulfate, bicarbonate, and sulfate). Reactions were performed in three replicates with 10 mm pNPP as described above.
Exogenous Application of OsHAD1 to Wild-Type Seedlings
Rice seedlings were raised hydroponically in five separate containers in three biological replicates. One container was supplied with 320 µm NaH2PO4 (+P) while four others were supplied with 1 µm NaH2PO4 (−P) for 15 d. Each container contained 20 seedlings. After 15 d, Pi-starved seedlings in three of the containers were recovered with 100 µm phytate (Sigma; P8810) as the P source. Out of these three containers, two were supplemented with 125 ng mL−1 purified recombinant OsHAD1 wild-type or mutated protein (D16A). Seedlings were recovered for another 7 d. The rest of the seedlings in +P and –P medium-containing containers continued to be raised in their respective nutrient media for another 7 d. After this, the seedlings were harvested for phenotypic and total P content analyses.
Vector Construction and Development of Transgenic Lines
Full-length cDNA (AK240756) of OsHAD1 was amplified using gene-specific primers (Supplemental Table S2) and cloned under the control of the ZmUbi1 promoter in a Gateway-compatible overexpression vector, pANIC6B (Mann et al., 2012). The overexpression construct was transformed into Agrobacterium tumefaciens strain EHA105 and introduced into rice calli by the A. tumefaciens-mediated transformation method as described (Toki et al., 2006). Regenerated plants were selected on hygromycin (50 µg mL−1) and screened by PCR with hptII gene-specific primers and GUS histochemical staining.
Phosphatase and Phytase Activity Assay in Wild-Type and Transgenic Plants
Total protein was extracted from 250 mg of frozen roots and shoots of 30-d-old hydroponically raised (+Phytate) wild-type and OsHAD1 transgenic plants in chilled extraction buffer (100 mm potassium acetate, 20 mm CaCl2, 2 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, and 20% glycerol, pH 6) as described (Mehra et al., 2017). For total phosphatase activity, 20 μg of extracted protein was incubated with 10 mm pNPP, 5 mm MgCl2, and 50 mm sodium acetate buffer (pH 6) in a total volume of 100 µL. Reactions were performed at 37°C for 15 min. Released Pi was measured by the yellow vanadomolybdate method (Kitson and Mellon, 1944). For phytase activity, reactions were performed with 40 μg of total protein and 10 mm sodium phytate as substrate.
For the estimation of secretory APase activity, wild-type and OE1 seedlings were raised hydroponically under +P and –P conditions for 12 d. After this, 10 plants each of the wild type or OE1 were kept in 100 mL of +P or –P medium in a glass tube. After 3 d, liquid medium was collected from each tube and filtered. The secretory APase activity assay was performed as described previously (Mehra et al., 2017). A total of 800 µL of filtered medium was incubated with 5 mm pNPP at 37°C for 1 h. Reactions were stopped by 100 µL of 0.4 m NaOH. APase activity was quantitated spectrophotometrically at 410 nm. The whole experiment was performed in three replicates.
Subcellular Localization of the OsHAD1:YFP Fusion Protein
The coding sequence of OsHAD1 was fused in frame with the coding sequence of YFP by cloning in pSITE-3CA using the Gateway cloning system (Invitrogen). Particle bombardment assays were performed on onion (Allium cepa) epidermal cells as described earlier (Mehra et al., 2017). For transient expression in Nicotiana benthamiana, the OsHAD1 construct in pSITE-3CA was transformed into A. tumefaciens strain GV3101. Agroinfiltration of N. benthamiana was carried out according to Walter et al. (2004). For plasmolysis, epidermal cells were exposed to 5% NaCl solution for a few seconds. Images were analyzed using the AOBS TCS-SP2 (Leica) confocal microscope.
Metabolite Profiling and ATP Estimation
For metabolite profiling and ATP estimation, 30-d-old +P-grown wild-type and OsHAD1 OE lines were used. For metabolite extraction, 1 mg of lyophilized leaves was ground in 1 mL of prechilled solvent (methanol:chloroform:water, 5:2:2, v/v/v) for 30 s at 1,500 rpm using a Retsch ball mill. Samples were vortexed for 6 min at 4°C followed by centrifugation at 14,000 rcf for 2 min. Clear supernatant was collected and dried in a Labconco Centrivap cold trap concentrator. Extracts were derivatized and prepared for GC-TOF-MS using the ALEX-CIS GC-TOF-MS apparatus as described (Fiehn et al., 2008). The experiment was performed in three replicates.
ATP estimation was performed according to Zeng et al. (2013). Briefly, 100 mg of leaf samples of 30-d-old +P-grown wild-type and OsHAD1 OE1 plants was crushed in liquid nitrogen. Ground samples were mixed immediately with 400 µL of 0.0005% (v/v) perchloric acid and boiled for 10 min in a water bath. The whole mixture was centrifuged at 13,000 rpm for 15 min at 4°C, and a clear supernatant was collected for ATP estimation using an ATP colorimetric assay kit (Sigma; MAK190). Quantitation was done from eight independent replicates.
Immunoblotting of OsHAD1
A total of 40 μg of protein extract was electrophoresed on 12% SDS-PAGE. Subsequently, proteins were transferred to a polyvinylidene difluoride membrane (Millipore) in a western-blot tank according to the manufacturer’s protocol. The membrane was blocked with 5% (w/v) skim milk in PBST buffer overnight. Immunodetection of OsHAD1 was carried out by incubating with primary antibody (anti-OsHAD1 raised in rabbit) in 5% skim milk for 2 h followed by three gentle washings with PBST buffer for 5, 10, and 5 min. After washing, the membrane was incubated with secondary antibody diluted in 5% skim milk for 2 h. Following two washings, each for 10 min, OsHAD1 protein was detected by incubating the membrane in BCIP-NBT (Sigma; B1911) substrate for 2 min.
Pull-Down and Yeast Two-Hybrid Assays
The coding region of OsHAD1 was fused with GST tag by cloning in pGEX4T-1 expression vector (Amersham Biosciences). Recombinant OsHAD1 and empty pGEX4T-1 vector were independently transformed and induced in E. coli strain BL21(DE3)pLysS as described above. Recombinant OsHAD1 was immobilized on GST beads (Sigma; G4510) according to the manufacturer’s protocol. Interactome analysis through GST pull down was performed as described (Datta et al., 2015). Briefly, total protein from 30-d-old +P-grown OsHAD1 OE1 was extracted using extraction buffer (1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA [pH 8], in 1× phosphate-buffered saline, pH 7.4). Total plant protein was incubated with OsHAD1-GST-bound agarose beads at 4°C overnight. Furthermore, the beads were washed with 1× phosphate-buffered saline three times for 10 min with gravity flow. Proteins bound with GST beads were eluted with 20 mm glutathione at pH 8. Eluted proteins were electrophoresed on 12% SDS-PAGE. Separated protein bands were identified with liquid chromatography-mass spectrometry combined with electrospray ionization as described earlier (Jaiswal et al., 2013).
Yeast two-hybrid assays were performed using the Matchmaker Gold Yeast two-hybrid system (Clontech) according to the manufacturer’s protocol. The coding sequence of OsHAD1 was cloned into the bait vector pGBKT7 (BD). The coding sequences of OsNDPK1, OsNDPK2, and OsNDPK3 were cloned into the prey vector pGADT7 (AD). AD and BD clones were cotransformed in yeast strain Y2H Gold (Clontech) and selected on minimal medium, His, Leu, and Trp, and adenine, His, Leu, and Trp.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression profiling of OsHAD1 in different tissues and developmental stages.
Supplemental Figure S2. Induction and purification of OsHAD1 recombinant protein.
Supplemental Figure S3. Asp residues of motif I are essential for the activity of OsHAD1.
Supplemental Figure S4. Confirmation of OsHAD1 overexpression lines for transgenic nature.
Supplemental Figure S5. Phenotype of OsHAD1 overexpressed lines grown in soil under +P, −P, and phytate recovery conditions.
Supplemental Figure S6. Effects of OsHAD1 overexpression on biomass and total P content of plants grown in soil.
Supplemental Figure S7. Transient expression of YFP-OsHAD1 in onion epidermal cells.
Supplemental Figure S8. Transient expression of YFP-OsHAD1 in N. benthamiana.
Supplemental Figure S9. Phytase activity of the wild type and OsHAD1 overexpression lines raised under phytate recovery.
Supplemental Figure S10. Overexpression of OsHAD1 increased secretory APase activity in rice.
Supplemental Figure S11. Yeast two-hybrid assay of OsHAD1 with OsNDPK1/2/3.
Supplemental Figure S12. Expression profile of OsNDPK3.
Supplemental Figure S13. Coexpression of OsHAD1 with OsPT8 and OsNDPK3 under cytokinin treatment.
Supplemental Figure S14. Sequence alignment of OsHAD1 with known low-P-responsive HAD superfamily members.
Supplemental Figure S15. Amino acid sequence alignment of OsHAD1 with its nearest rice homologs.
Supplemental Figure S16. Expression profile of OsHAD1 and its nearest homologs in rice under the P-deficient condition in Dular and PB1 roots and shoots.
Supplemental Figure S17. P profile and dry biomass of the wild type and OsHAD1 overexpression lines grown under low-P or phytate supply.
Supplemental Table S1. List of metabolites identified in OsHAD1 OE lines and the wild type using ALEX-CIS GC-TOF-MS.
Supplemental Table S2. List of primers used in this study.
Acknowledgments
We acknowledge the phytotron facility at the National Institute of Plant Genome Research. B.K.P. and J.B. acknowledge research fellowships from DBT. P.M and L.V. acknowledge research fellowships from CSIR.
Glossary
- Pi
inorganic phosphate
- PSR
phosphate starvation response
Footnotes
This work was supported by DBT (grant no. BT/PR3299/AGR/2/813/2011), Government of India, and by NIPGR core grants.
References
- Baldwin JC, Karthikeyan AS, Cao A, Raghothama KG (2008) Biochemical and molecular analysis of LePS2;1: a phosphate starvation induced protein phosphatase gene from tomato. Planta 228: 273–280 [DOI] [PubMed] [Google Scholar]
- Baldwin JC, Karthikeyan AS, Raghothama KG (2001) LEPS2, a phosphorus starvation-induced novel acid phosphatase from tomato. Plant Physiol 125: 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L (2011) Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L (2006) Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol 361: 1003–1034 [DOI] [PubMed] [Google Scholar]
- Cabello-Díaz JM, Quiles FA, Lambert R, Pineda M, Piedras P (2012) Identification of a novel phosphatase with high affinity for nucleotides monophosphate from common bean (Phaseolus vulgaris). Plant Physiol Biochem 53: 54–60 [DOI] [PubMed] [Google Scholar]
- Caparrós-Martín JA, McCarthy-Suárez I, Culiáñez-Macià FA (2013) HAD hydrolase function unveiled by substrate screening: enzymatic characterization of Arabidopsis thaliana subclass I phosphosugar phosphatase AtSgpp. Planta 237: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien SH, Prochnow LI, Tu S, Snyder CS (2011) Agronomic and environmental aspects of phosphate fertilizers varying in source and solubility: an update review. Nutr Cycl Agroecosyst 89: 229–255 [Google Scholar]
- Cordell D, Neset TS (2014) Phosphorus vulnerability: a qualitative framework for assessing the vulnerability of national and regional food systems to the multi-dimensional stressors of phosphorus scarcity. Glob Environ Change 24: 108–122 [Google Scholar]
- Dai X, Wang Y, Yang A, Zhang WH (2012) OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol 159: 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Kamthan A, Kamthan M (2015) A simple protocol to detect interacting proteins by GST pull down assay coupled with MALDI or LC-MS/MS analysis. Protocol Exchange doi/10.1038/protex.2015.093 [Google Scholar]
- Dobermann A, Cassman KG, Mamaril CP, Sheehy JE (1998) Management of phosphorus, potassium, and sulfur in intensive, irrigated lowland rice. Field Crops Res 56: 113–138 [Google Scholar]
- Dorion S, Rivoal J (2015) Clues to the functions of plant NDPK isoforms. Naunyn Schmiedebergs Arch Pharmacol 388: 119–132 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee DY, Lu Y, Moon S, Nikolau B (2008) Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J 53: 691–704 [DOI] [PubMed] [Google Scholar]
- Fragoso S, Espíndola L, Páez-Valencia J, Gamboa A, Camacho Y, Martínez-Barajas E, Coello P (2009) SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol 149: 1906–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539 [DOI] [PubMed] [Google Scholar]
- George TS, Simpson RJ, Gregory PJ, Richardson AE (2007) Differential interaction of Aspergillus niger and Peniophora lycii phytases with soil particles affects the hydrolysis of inositol phosphates. Soil Biol Biochem 39: 793–803 [Google Scholar]
- Guo M, Ruan W, Li C, Huang F, Zeng M, Liu Y, Yu Y, Ding X, Wu Y, Wu Z, et al. (2015) Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol 168: 1762–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Tran LS (2014) Understanding plant responses to phosphorus starvation for improvement of plant tolerance to phosphorus deficiency by biotechnological approaches. Crit Rev Biotechnol 34: 16–30 [DOI] [PubMed] [Google Scholar]
- Harrison AF. (1987) Soil Organic Phosphorus: A Review of World Literature. CAB International, Wallingford, UK [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Hou XL, Wu P, Jiao FC, Jia QJ, Chen HM, Yu J, Song XW, Yi KK (2005) Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ 28: 353–364 [Google Scholar]
- Hu B, Zhu C, Li F, Tang J, Wang Y, Lin A, Liu L, Che R, Chu C (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur YJ, Lee HG, Jeon EJ, Lee YY, Nam MH, Yi G, Eun MY, Nam J, Lee JH, Kim DH (2007) A phosphate starvation-induced acid phosphatase from Oryza sativa: phosphate regulation and transgenic expression. Biotechnol Lett 29: 829–835 [DOI] [PubMed] [Google Scholar]
- Immormino RM, Starbird CA, Silversmith RE, Bourret RB (2015) Probing mechanistic similarities between response regulator signaling proteins and haloacid dehalogenase phosphatases. Biochemistry 54: 3514–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal DK, Ray D, Choudhary MK, Subba P, Kumar A, Verma J, Kumar R, Datta A, Chakraborty S, Chakraborty N (2013) Comparative proteomics of dehydration response in the rice nucleus: new insights into the molecular basis of genotype-specific adaptation. Proteomics 13: 3478–3497 [DOI] [PubMed] [Google Scholar]
- Jia H, Ren H, Gu M, Zhao J, Sun S, Zhang X, Chen J, Wu P, Xu G (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk GJD, George T, Courtois B, Senadhira D (1998) Opportunities to improve phosphorus efficiency and soil fertility in rainfed lowland and upland rice ecosystems. Field Crops Res 56: 73–92 [Google Scholar]
- Kitson RE, Mellon MG (1944) Colorimetric determination of phosphorus as molybdivanadophosphoric acid. Ind Eng Chem Anal Ed 16: 379–383 [Google Scholar]
- Kong Y, Li X, Ma J, Li W, Yan G, Zhang C (2014) GmPAP4, a novel purple acid phosphatase gene isolated from soybean (Glycine max), enhanced extracellular phytate utilization in Arabidopsis thaliana. Plant Cell Rep 33: 655–667 [DOI] [PubMed] [Google Scholar]
- Kuang R, Chan KH, Yeung E, Lim BL (2009) Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol 151: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, et al. (2006) Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem 281: 36149–36161 [DOI] [PubMed] [Google Scholar]
- Lai F, Thacker J, Li Y, Doerner P (2007) Cell division activity determines the magnitude of phosphate starvation responses in Arabidopsis. Plant J 50: 545–556 [DOI] [PubMed] [Google Scholar]
- Lan P, Li W, Schmidt W (2012) Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol Cell Proteomics 11: 1156–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Schmidt W (2013) Genome-wide co-expression analysis predicts protein kinases as important regulators of phosphate deficiency-induced root hair remodeling in Arabidopsis. BMC Genomics 14: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Xu C, Fan W, Zhang H, Hou J, Yang A, Zhang K (2014) Phosphoproteome and proteome analyses reveal low-phosphate mediated plasticity of root developmental and metabolic regulation in maize (Zea mays L.). Plant Physiol Biochem 83: 232–242 [DOI] [PubMed] [Google Scholar]
- Liang C, Tian J, Lam HM, Lim BL, Yan X, Liao H (2010) Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol 152: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CY, Chen ZJ, Yao ZF, Tian J, Liao H (2012) Characterization of two putative protein phosphatase genes and their involvement in phosphorus efficiency in Phaseolus vulgaris. J Integr Plant Biol 54: 400–411 [DOI] [PubMed] [Google Scholar]
- Liu J, Wang X, Huang H, Wang J, Li Z, Wu L, Zhang G, Ma Z (2012) Efficiency of phosphorus utilization in phyA-expressing cotton lines. Indian J Biochem Biophys 49: 250–256 [PubMed] [Google Scholar]
- Liu JQ, Allan DL, Vance CP (2010) Systemic signaling and local sensing of phosphate in common bean: cross-talk between photosynthate and microRNA399. Mol Plant 3: 428–437 [DOI] [PubMed] [Google Scholar]
- Lung SC, Chan WL, Yip W, Wang L, Yeung EC, Lim BL (2005) Secretion of beta-propeller phytase from tobacco and Arabidopsis roots enhances phosphorus utilization. Plant Sci 169: 341–349 [Google Scholar]
- Ma XF, Tudor S, Butler T, Ge Y, Xi Y, Bouton J, Harrison M, Wang ZY (2012) Transgenic expression of phytase and acid phosphatase genes in alfalfa (Medicago sativa) leads to improved phosphate uptake in natural soils. Mol Breed 30: 377–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DG, Lafayette PR, Abercrombie LL, King ZR, Mazarei M, Halter MC, Poovaiah CR, Baxter H, Shen H, Dixon RA, et al. (2012) Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol J 10: 226–236 [DOI] [PubMed] [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24: 559–567 [DOI] [PubMed] [Google Scholar]
- May A, Berger S, Hertel T, Köck M (2011) The Arabidopsis thaliana phosphate starvation responsive gene AtPPsPase1 encodes a novel type of inorganic pyrophosphatase. Biochim Biophys Acta 1810: 178–185 [DOI] [PubMed] [Google Scholar]
- May A, Spinka M, Köck M (2012) Arabidopsis thaliana PECP1: enzymatic characterization and structural organization of the first plant phosphoethanolamine/phosphocholine phosphatase. Biochim Biophys Acta 1824: 319–325 [DOI] [PubMed] [Google Scholar]
- Mehra P, Giri J (2016) Rice and chickpea GDPDs are preferentially influenced by low phosphate and CaGDPD1 encodes an active glycerophosphodiester phosphodiesterase enzyme. Plant Cell Rep 35: 1699–1717 [DOI] [PubMed] [Google Scholar]
- Mehra P, Pandey BK, Giri J (2015) Genome-wide DNA polymorphisms in low phosphate tolerant and sensitive rice genotypes. Sci Rep 5: 13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra P, Pandey BK, Giri J (2016) Comparative morphophysiological analyses and molecular profiling reveal Pi-efficient strategies of a traditional rice genotype. Front Plant Sci 6: 1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra P, Pandey BK, Giri J (2017) Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol J 10.1111/pbi.12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulska M, Bomsel JL, Rychter A (1998) The influence of phosphate deficiency on photosynthesis, respiration and adenine nucleotide pool in bean leaves. Photosynthetica 35: 79–88 [Google Scholar]
- Pant BD, Pant P, Erban A, Huhman D, Kopka J, Scheible WR (2015) Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ 38: 172–187 [DOI] [PubMed] [Google Scholar]
- Papademetriou MK. (2000) Rice production in the Asia-Pacific region: issues and perspectives. In Papademetriou MK, Dent FJ, Herath EM, eds, Bridging the Rice Yield Gap in the Asia-Pacific Region. Food and the Agriculture Organization of the United Nations, Bangkok, Thailand, pp 4–25 [Google Scholar]
- Plaxton WC, Shane MW (2015) The role of post-translational enzyme modifications in the metabolic adaptations of phosphorus-deprived plants. Annu Plant Rev 48: 99–124 [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Richardson AE, Hadobas PA, Hayes JE (2001) Extracellular secretion of Aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant J 25: 641–649 [DOI] [PubMed] [Google Scholar]
- Ridder IS, Dijkstra BW (1999) Identification of the Mg2+-binding site in the P-type ATPase and phosphatase members of the HAD (haloacid dehalogenase) superfamily by structural similarity to the response regulator protein CheY. Biochem J 15: 223–226 [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Pandey BK, Deveshwar P, Narnoliya L, Parida SK, Giri J (2015) JAZ repressors: possible involvement in nutrients deficiency response in rice and chickpea. Front Plant Sci 6: 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Satyanarayana T (2012) Plant growth promotion by phytases and phytase-producing microbes due to amelioration in phosphorus availability. In T Satyanarayana, NB Johri, eds, Microorganisms in Sustainable Agriculture and Biotechnology. Springer, Dordrecht, The Netherlands, pp 3–15 [Google Scholar]
- Springer M, Wykoff DD, Miller N, O’Shea EK (2003) Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol 1: E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA 108: 20260–20264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC (2010a) Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci 179: 14–27 [Google Scholar]
- Tran HT, Qian W, Hurley BA, She YM, Wang D, Plaxton WC (2010b) Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ 33: 1789–1803 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H, Chevalier D, Larue C, Cho SK, Walker JC (2007) The protein phosphatases and protein kinases of Arabidopsis thaliana. The Arabidopsis Book 5: e0106, https://dx.doi.org/10.1199%2Ftab.0106 doi/10.1199/tab.0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li Z, Qian W, Guo W, Gao X, Huang L, Wang H, Zhu H, Wu JW, Wang D, et al. (2011) The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157: 1283–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P (2009) Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J Integr Plant Biol 51: 663–674 [DOI] [PubMed] [Google Scholar]
- Wissuwa M, Ae N (2001) Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breed 120: 43–48 [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Shou H, Xu G, Lian X (2013) Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr Opin Plant Biol 16: 205–212 [DOI] [PubMed] [Google Scholar]
- Zeng F, Shabala L, Zhou M, Zhang G, Shabala S (2013) Barley responses to combined waterlogging and salinity stress: separating effects of oxygen deprivation and elemental toxicity. Front Plant Sci 4: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Song H, Cheng H, Hao D, Wang H, Kan G, Jin H, Yu D (2014) The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genet 10: e1004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Zardi G, Lehmann M, Zeder C, Amrhein N, Frossard E, Bucher M (2003) Engineering the root-soil interface via targeted expression of a synthetic phytase gene in trichoblasts. Plant Biotechnol J 1: 353–360 [DOI] [PubMed] [Google Scholar]