Abstract
Albinism, a phenomenon characterized by pigmentation deficiency on the ocular side of Japanese flounder (Paralichthys olivaceus), has caused significant damage. Limited mRNA and microRNA (miRNA) information is available on fish pigmentation deficiency. In this study, a high-throughput sequencing strategy was employed to identify the mRNA and miRNAs involved in P. olivaceus albinism. Based on P. olivaceus genome, RNA-seq identified 21,787 know genes and 711 new genes by transcripts assembly. Of those, 235 genes exhibited significantly different expression pattern (fold change ≥2 or ≤0.5 and q-value≤0.05), including 194 down-regulated genes and 41 up-regulated genes in albino versus normally pigmented individuals. These genes were enriched to 81 GO terms and 9 KEGG pathways (p≤0.05). Among those, the pigmentation related pathways-Melanogenesis and tyrosine metabolism were contained. High-throughput miRNA sequencing identified a total of 475 miRNAs, including 64 novel miRNAs. Furthermore, 33 differentially expressed miRNAs containing 13 up-regulated and 20 down-regulated miRNAs were identified in albino versus normally pigmented individuals (fold change ≥1.5 or ≤0.67 and p≤0.05). The next target prediction discovered a variety of putative target genes, of which, 134 genes including Tyrosinase (TYR), Tyrosinase-related protein 1 (TYRP1), Microphthalmia-associated transcription factor (MITF) were overlapped with differentially expressed genes derived from RNA-seq. These target genes were significantly enriched to 254 GO terms and 103 KEGG pathways (p<0.001). Of those, tyrosine metabolism, lysosomes, phototransduction pathways, etc., attracted considerable attention due to their involvement in regulating skin pigmentation. Expression patterns of differentially expressed mRNA and miRNAs were validated in 10 mRNA and 10 miRNAs by qRT-PCR. With high-throughput mRNA and miRNA sequencing and analysis, a series of interested mRNA and miRNAs involved in fish pigmentation are identified. And the miRNA-mRNA regulatory network also provides a solid starting point for further elucidation of fish pigmentation deficiency.
Introduction
Albinism, characterized by pigmentation deficiency in the skin, hair and eyes of mammals, has been extensively studied since the early 19th century [1, 2]. Visible pigmentation in mammals results from the biogenesis, transport and transfer of melanin, a type of pigment granule synthesized in the melanocyte [3]. During this process, more than 171 genes have been identified with a list at http://www.espcr.org/micemut, and their mutations have been reported to cause three main types of disorders: hyperpigmentation, hypopigmentation (albinism), or mixed hyper- /hypopigmentation [4–6]. There have been five enzymes identified as being required for human melanin pigmentation, namely tyrosinase (TYR), tyrosinase-related protein-1 (TYRP1), P (OCA2), solute carrier family 45 member 2 (SLC45A2), and G-protein coupled receptor 143 (GPR143), and defects in any of these enzymes could result in albinism [7]. Besides these important enzymes, three melanocyte-specific transcription factors also play central roles in the development, migration and differentiation of melanocytes: microphthalmia-associated transcription factor (MITF), paired box gene 3 (PAX3) and SRY-box containing gene 10 (SOX10) [7–10].
MicroRNAs (miRNAs), a type of non-coding RNA with a length between 22–24 nt, could act as post-transcriptional negative regulators of mRNA stability or translation [11, 12]. Since their initial discovery in the 1990s, miRNAs have been proven to extensively regulate cell differentiation, development and diseases [13–15]. In mammals, over 60% of mRNAs are thought to be regulated by miRNAs [16].
Since the first evidence of miRNA involvement in melanocyte biology was verified in 2010 [17], an increasing number of miRNAs have been found to participate in melanocyte development and diseases of mammals. For example, miR-140, miR-143, and miR-145 play important roles in targeting Dicer, a miRNA processing protein, during the development of the neural crest, from which melanocytes are derived [18, 19]. Additionally, miRNAs including miR-125b, miR-25, miR-137, miR-192, and miR-194 have been shown to regulate melanogenesis by reducing the expression level of MITF [20–23]. Recent studies have also revealed more than 30 miRNAs involved in the development and progression of melanoma, a type of skin cancer that originates in melanocytes [24–27].
Six types of chromatophores (melanophores, xanthophores, iridophores, leucophores, erythrophores and cyanophores) were discovered in fishes [28], suggesting that fish may possess more complicated pigmentation regulatory mechanisms than mammals. Previous studies of zebrafish (Danio rerio), medaka (Oryzias latipes), Mexican tetra (Astyanax mexicanus), and common carp (Cyprinus carpio) have identified that more than 100 genes participate in pigmentation [29–31], while only one miRNA has been confirmed to regulate pigmentation [32].
To date, limited studies are available reviewing pigmentation deficiencies of benthic marine fish such as Japanese flounder (P. olivaceus), a flatfish living in the coastal areas of China, Korea and Japan. Since the 1980s, an increasing frequency of albinism characterized by pigment deficiency on the ocular side of P. olivaceus has significantly reduced the commercial value and seriously hindered sustainable aquaculture development [33]. Although numerous reports have revealed the participation of miRNAs in the immune defences [34], metamorphosis [35–37], gonad development [38] and muscle development [39] of P. olivaceus, it remains unclear whether miRNAs play a role in pigmentation and albinism of P. olivaceus.
In present study, a high-throughput sequencing strategy was employed to screen differentially expressed mRNA and miRNAs between albino and normally pigmented P. olivaceus. Furthermore, the potential target genes of differentially expressed miRNAs were predicted by in silico analysis. The enrichment analysis of the GO term and KEGG pathway for these predicted genes and differentially expressed genes were subsequently conducted in order to gain insight into the function of mRNAs and miRNAs in P. olivaceus pigmentation deficiency.
Materials and methods
Ethical statement
The collection and handling of the animals used in this study was approved by the Animal Care and Use Committee at the Chinese Academy of Fishery Sciences, and all the experimental procedures were performed in accordance with the guidelines for the Care and Use of Laboratory Animals at the Chinese Academy of Fishery Sciences.
Sample collection and RNA isolation
At the age of 5 months, P. olivaceus individuals from a full-sib family were obtained from Haiyang Yellow Sea Aquatic Product Co., Ltd. The fish were kept in seawater maintained at 20°C for one week. Skin tissue samples with an area of 2×2 cm2 from the ocular side of three normal and three albino individuals were collected and denoted as PO_con1, PO_con2, PO_con3, PO_alb1, PO_alb2, and PO_alb3, respectively. The samples were stored in liquid nitrogen for subsequent miRNA analysis.
The total RNA from each sample was extracted using Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. Total RNA integrity was assessed using the RNA Nano 600 Assay Kit of the Bioanalyzer 2100 system (Agilent, CA, USA) for quality control.
RNA-seq sequencing, gene annotation and novel gene identification
A total amount of 3 μg RNA (RIN>7.0) per sample was used for cDNA paired-end libraries using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations. In order to select cDNA fragments of 150-200bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). 3 μl USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. After cluster generation on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina), the libraries were sequenced on an Illumina Hiseq2000 platform by Novogene Bioinformatics Technology Co. Ltd (Beijing, China) and 100 bp paired-end reads were generated.
The clean reads were obtained by removing reads containing adapter, reads containing Poly-N and low quality reads from raw reads. At the same time, Q20, Q30 and GC content of the clean reads were calculated.
The index of Japanese flounder genome was built using Bowtie v2.0.6 and clean reads were aligned to Japanese flounder genome using TopHat v2.0.9 for gene mapping and annotation.
The Cufflinks v2.1.1 Reference Annotation Based Transcript (RABT) assembly method was used to construct and identify both known and novel transcripts from TopHat alignment results.
Differentially expressed genes identification and GO/KEGG enrichment analysis
The reads numbers mapped to each gene were counted using HTSeq v0.6.1. Then, RPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene [40].
Differential expression analysis of samples was performed using the DESeq R package (1.10.1). The resulting p-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted p-value <0.05 found by DESeq were assigned as differentially expressed genes involved in P. olivaceus pigmentation.
Subsequently, based on the pigmentation genes list at http://www.espcr.org/micemut, previous publications [4, 7] and present study, the overlapped information were analysed and listed in a table.
Gene Ontology functional and KEGG pathway enrichment analysis of GeneCodis [41] was used to decipher the major biological processes and pathways involved by the differentially expressed genes. And GO terms and KEGG pathways with corrected p-value less than 0.05 were considered significantly enriched.
Small RNA library construction, sequencing, and miRNA prediction
Approximately 1 μg RNA of an RNA integrity number (RIN) >7.0 from each sample was used for the subsequent cDNA library construction following the protocol of TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA). Single-end sequencing (36 bp) was then performed on an Illumina HiSeq 2500 at the LC-BIO (Hangzhou, China) according to the vendor’s recommended protocol.
Initially, the raw reads were subjected to the Illumina pipeline filter (Solexa 0.3), and the datasets were further processed with an in-house program, ACGT101-miR (LC Sciences, Houston, Texas, USA), to remove adapter dimers, low complexity, common RNA families (rRNA, tRNA, snRNA, snoRNA) and repeats. Subsequently, the remaining clean reads were used to search against miRBase 21.0 (http://www.mirbase.org/search.shtml) for known miRNA prediction. During the alignment, length variation at both the 5’ and 3’ ends and one mismatch inside of the sequence were allowed. For the miRNAs identified in this study, the conservation analysis was conducted through comparisons with 40 species listed in S1 Table.
The unmapped sequences were further searched against P. olivaceus genomes [42] using RNAfold software (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) for novel miRNA prediction. The criteria for secondary structure prediction were similar to a previous study [34].
Differential analysis of miRNAs
To analyse the expression patterns of miRNAs among the control and albino groups, the frequency of miRNA counts was normalized with a modified global normalization method. Based on the normalization counts, the fold change was calculated as PO_alb versus PO_con, and the expression difference was measured with Student’s t test. The miRNAs with fold change ≥1.5 or fold change ≤0.67 and p ≤0.05 were considered significant. The pheatmap program of R software was utilized to display the differentially expressed miRNAs.
Prediction and analysis of the target genes of miRNAs
The putative target genes were predicted utilizing two computational target prediction algorithms: TargetScan 6.2 (http://www.targetscan.org) and miRanda (http://www.microrna.org). The parameters of TargetScan and miRanda were set as score>50 and free energy<-10 kcal/mol, respectively. The overlap genes between the two algorithms were regarded as the target genes, whose enrichment analysis was conducted using GO terms (http://www.geneontology.org) and KEGG pathways (http://www.genome.jp/kegg). Based on these results, the miRNA-GO-network and miRNA-KEGG-network were generated. Cytoscape was used for displaying a visualized interaction between miRNAs and mRNAs.
Real-time quantification of mRNA and miRNA
In order to validate the confidence of high-throughput transcriptome sequencing, 10 differentially expressed genes are selected and analyzed by quantitative real-time PCR assay. P. olivaceus β-actin gene was used as the internal reference. Primers were listed in S2 Table. The total RNAs for high-throughput transcriptome sequencing were reverse transcripted into cDNA by PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan). Then qRT-PCR were conducted using SYBR Premix Ex Taq (Takara, Japan) in 20 μl reaction solution containing 10 μl SYBR Premix Ex Taq (2X), 0.4 μl forward primer and 0.4 μl reverse primer, 0.4 μl ROX reference dye, 1μl cDNA and 8.2 μl ddH2O. The PCR amplification procedure was carried out at 95°C for 10 s; 40 cycles of 95°C for 5 s and 60°C for 34 s; followed by disassociation curve analysis in a ABI 7500 fast real-time PCR system (Applied Biosystems, USA). The amplification reaction without the template was used as a no template control. All reactions were performed in triplicate. The relative gene expression was calculated using the 2-ΔΔCt method [43]. Statistical comparison of the levels detected at different time points is carried out by SPSS, and p<0.05 were considered as significant.
To validate the differential expression results from miRNA sequencing, 10 miRNAs were randomly selected for qRT-PCR. Firstly, the total small RNAs from the normal and albino pigmented skins were extracted using a miRcute miRNA Isolation Kit (TianGen, Beijing, China). The poly(A) tail addition and reverse transcription for 2 μg RNA were performed using the miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China). The qRT-PCR was then performed by the miRcute miRNA qPCR Detection Kit (SYBR) (Tiangen, Beijing, China). Next, PCRs consisting of 20 μl PCR reaction solution containing 1 μl cDNA, 10 μl 2×miRcute miRNA premix, 1.6 μl 50×ROX Reference Dye, 0.4 μl forward primer, 0.4 μl reverse primer, and 6.6 μl ddH2O were carried out on an ABI Prism 7500 Fast Sequence Detection System (ABI, Carlsbad, CA, USA) at 94°C for 2 min, followed by 40 cycles of 94°C for 20 s and 60°C for 34 s. Forward primers were designed based on mature miRNA sequences (S3 Table), and the reverse primer was provided by the miRcute miRNA qPCR Detection Kit (SYBR) (Tiangen, Beijing, China). The 5s rRNA of P. olivaceus was used as the internal control for the normalization of expression levels. The results were analysed using the 2-ΔΔCt method, with the threshold cycle (Ct) determined by the default threshold settings. The experiment was conducted independently three times.
Results
RNA-seq sequencing and analysis
In order to better understand P. olivaceus albinism mechanism, we conducted a comparative transcriptomic analysis among three normal and albino pigmented individuals. A total of six cDNA libraries, named as PO_con1, PO_con2, PO_con3, PO_alb1, PO_alb2 and PO_alb3, were constructed and sequenced. A mean of 31,655,702 clean reads with Q20 higher than 94.17% were obtained from each library (S4 Table, S1 Fig). Further genome mapping analysis revealed that over 79.13% of reads were mapped with P. olivaceus genome. Subsequently, a total of 21787 know genes and 711 new genes were identified by further transcripts assembly based on P. olivaceus genome.
Differentially expressed genes identification and validation
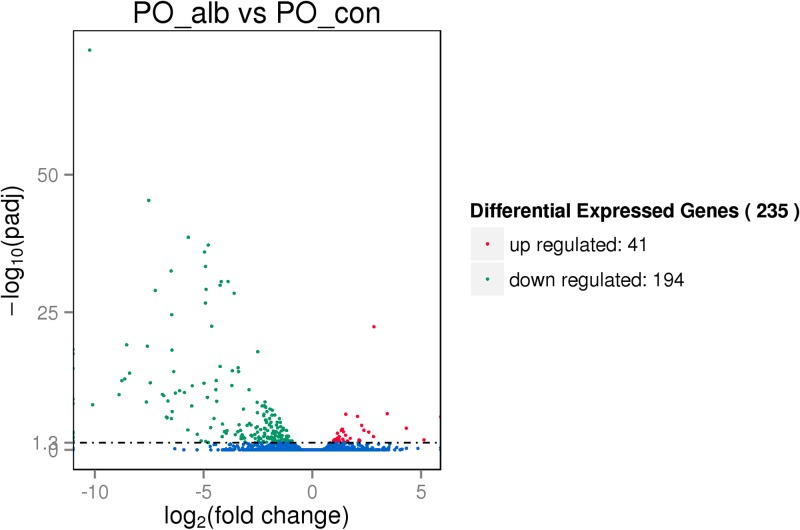
Based on the expression level of each gene in samples, the correlation coefficient was calculated to evaluate the repeatability. The result showed that the pearson correlation coefficient between three replicas was 95.0~97.5% (S2 Fig). The subsequent differential expression showed that 235 genes exhibited significantly different (Fig 1, S5 Table) (fold change≥ 2 or ≤ 0.5 and q-value ≤ 0.05), including 194 down-regulated genes and 41 up-regulated genes in PO_alb vs PO_con.
Fig 1. The volcanoplot of differentially expressed genes in RNA-seq.
The differentially expressed genes including 41 up-regulated genes and 194 down-regulated genes in PO_alb vs PO_con were illustrated in a volcanoplot (fold change≥2 and q-value≤0.05).
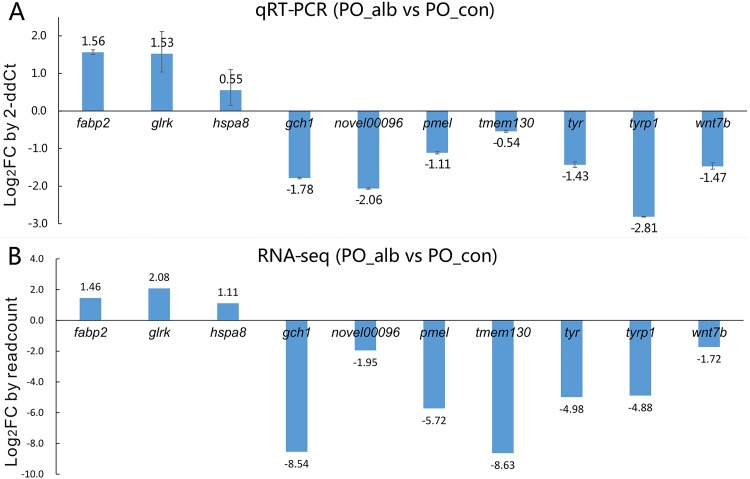
Subsequently, 10 genes, including up-regulated genes fatty acid-binding protein, intestinal (fabp2), probable glutamate receptor (glrk), heat shock cognate 71 kDa protein (hspa8), and down-regulated genes GTP cyclohydrolase 1 (gch1), novel00096, melanocyte protein (pmel), transmembrane protein 130 (tmem130), tyr, tyrp1, wnt7b were selected for qRT-PCR validation experiment. The relative expression levels of these genes were similar with those of high-throughput transcriptome results (Fig 2).
Fig 2. The expression levels of ten genes in qRT-PCR and RNA-seq.
The expression fold change (FC) 10 P. olivaceus genes in PO_alb versus PO_con detected by qRT-PCR and RNA-seq were calculated by 2-ddCt and readcount, respectively. And these genes’ log2FC values of qRT-PCR and RNA-seq are shown in A and B, respectively. In qRT-PCR analysis, β-actin expression levels were used for the internal control, and values are indicated as means ± standard error (SE) derived from triplicate experiments.
The overlapped pigmentation related genes between previous publications and present study
In order to further confirm the fidelity of the differentially expressed genes presented in this study, we compared the overlapped pigmentation related genes between previous publications and present study. And the comparison revealed that 30 genes including Endothelin B receptor (EDNRB), GCH1, Mast/stem cell growth factor receptor (KIT), melanocortin 1 receptor (MC1R), MC5R, MITF, OCA2, PAX7, SLC45A2, SOX10, TYR, TYRP1, WNT7B were overlapped, which all showed down-regulated expression pattern in PO_alb vs PO_con (Table 1).
Table 1. The overlapped pigmentation related genes between previous study and present study.
| Index | Gene_ID | Gene symbol, description and chromosome location | PO_con (readcount) | PO_alb (readcount) | log2 fold change (PO_alb vs PO_con) | padj |
|---|---|---|---|---|---|---|
| 1 | GS_012501 | BMP1, Bone morphogenetic protein 1, chro13 | 16.47 | 1.06 | -3.96 | 0.000954 |
| 2 | GS_012647 | EDNRB, Endothelin B receptor, chro11 | 418.19 | 166.60 | -1.33 | 0.000218 |
| 3 | GS_018359 | EDNRB, Endothelin B receptor, chro12 | 561.53 | 263.69 | -1.09 | 0.003513 |
| 4 | GS_005903 | GCH1, GTP cyclohydrolase 1, chro24 | 284.36 | 0.00 | Inf | 1.65E-15 |
| 5 | GS_008748 | GCH1, GTP cyclohydrolase 1, chro4 | 803.68 | 2.16 | -8.54 | 8.45E-20 |
| 6 | GS_005902 | GCH1, GTP cyclohydrolase 1, chro24 | 1056.71 | 3.11 | -8.41 | 1.18E-14 |
| 7 | GS_013228 | GCH1, GTP cyclohydrolase 1, chro3 | 187.56 | 1.07 | -7.45 | 6.61E-13 |
| 8 | GS_009046 | KIT, Mast/stem cell growth factor receptor, chro5 | 76.74 | 17.25 | -2.15 | 3.79E-06 |
| 9 | GS_018838 | K1C13, Keratin, type I cytoskeletal 13, chro10 | 117.72 | 30.17 | -1.96 | 2.05E-06 |
| 10 | GS_014679 | K1C13, Keratin, type I cytoskeletal 13, chro5 | 29391.00 | 10648.78 | -1.46 | 0.018738 |
| 11 | GS_012545 | K1C18, Keratin, type I cytoskeletal 18, chro13 | 144.15 | 5.82 | -4.63 | 3.57E-23 |
| 12 | GS_007378 | MC1R, Melanocortin 1 receptor, chro7 | 27.32 | 3.99 | -2.78 | 0.003166 |
| 13 | GS_011397 | MC5R, Melanocortin 5 receptor, chro6 | 27.61 | 0.00 | Inf | 6.14E-10 |
| 14 | GS_018288 | MCHR2, Melanin-concentrating hormone receptor 2, chro12 | 23.23 | 2.35 | -3.30 | 0.000297 |
| 15 | GS_002043 | MITF, Microphthalmia-associated transcription factor, chro1 | 33.44 | 0.33 | -6.68 | 1.54E-06 |
| 16 | GS_000869 | MAR1, Melanoma antigen recognized by T-cells 1, chro13 | 277.19 | 14.69 | -4.24 | 1.26E-30 |
| 17 | GS_004597 | MREG, Melanoregulin, chro10 | 154.35 | 33.53 | -2.20 | 1.01E-08 |
| 18 | GS_001946 | MYO5A, Myosin-Va, chro7 | 296.08 | 151.63 | -0.97 | 0.047734 |
| 19 | GS_017193 | OCA2, P protein, chro24 | 21.26 | 1.40 | -3.92 | 0.000309 |
| 20 | GS_003876 | PAX7, Paired box protein Pax-7, chro17 | 88.93 | 40.55 | -1.13 | 0.041166 |
| 21 | GS_003590 | PMEL, Melanocyte protein, chro1 | 210.22 | 3.98 | -5.72 | 0.000246 |
| 22 | GS_012916 | SLC45A2, Membrane-associated transporter protein, chro13 | 35.13 | 5.84 | -2.59 | 0.000102 |
| 23 | GS_012661 | SOX10, Transcription factor SOX-10, chro10 | 270.45 | 81.67 | -1.73 | 4.55E-07 |
| 24 | GS_014478 | TBX19, T-box transcription factor 19, chro14 | 12.77 | 0.33 | -5.29 | 0.001515 |
| 25 | GS_020582 | TRPM1, Transient receptor potential cation channel subfamily M member 1, chro19 | 222.04 | 57.56 | -1.95 | 3.80E-08 |
| 26 | GS_020975 | TYR, Tyrosinase, chro21 | 50.90 | 1.61 | -4.98 | 7.81E-13 |
| 27 | GS_009038 | TYRP1, Tyrosinase-related protein 1, chro5 | 289.28 | 9.33 | -4.95 | 1.16E-36 |
| 28 | GS_012029 | TYRP1, Tyrosinase-related protein 1, chro9 | 292.57 | 9.91 | -4.88 | 6.72E-30 |
| 29 | GS_021046 | WNT7B, Protein Wnt-7b, chro16 | 40.65 | 12.32 | -1.72 | 0.02056 |
| 30 | GS_014490 | XDH, Xanthine dehydrogenase/oxidase, chro14 | 91.58 | 7.17 | -3.68 | 4.31E-15 |
The GO term and KEGG pathway enrichment analysis of RNA-seq
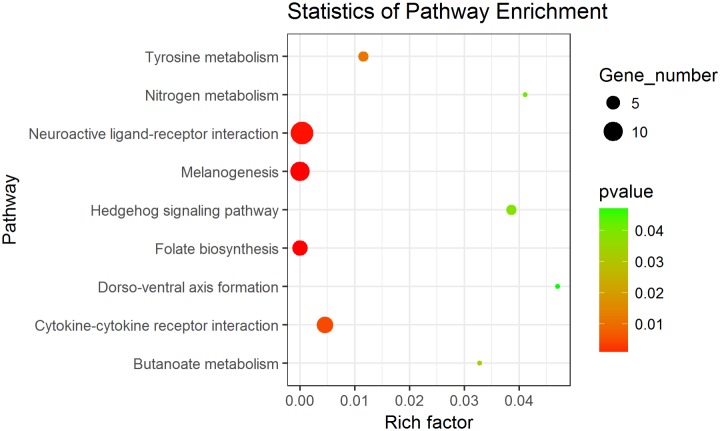
In order to further understand the function of these differentially expressed genes, GO term and KEGG pathway were subsequently carried out. GO term enrichment analysis revealed that 81 GO terms with p≤0.05 were enriched, including extracellular region, transmembrane signalling receptor activity, G-protein coupled receptor signalling pathway, signal transducer activity, molecular transducer activity, cell surface receptor signalling pathway, G-protein coupled receptor activity, heme oxygenase (decyclizing) activity, heme oxidation, receptor activity and so on (S6 Table). While, no GO terms with q≤0.05 were enriched. The KEGG pathway enrichment analysis showed that 9 pathways with p≤0.05 were significantly enriched, including Folate biosynthesis, Melanogenesis, Neuroactive ligand-receptor interaction, Tyrosine metabolism, and so on (Fig 3, S7 Table). Of those, the top three pathways exhibited a more significant difference with q≤0.05.
Fig 3. KEGG pathways enrichment in RNA-seq (p<0.05).
Gene number: number of target genes in each pathway. Rich factor: the ratio of the number of target genes divided by the number of all the genes in each pathway.
MicroRNA library construction and identification
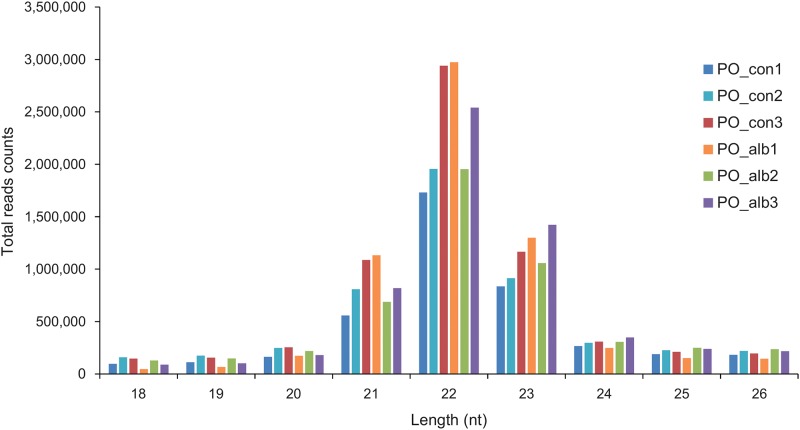
To identify miRNAs involved in the albinism of P. olivaceus, six small RNA libraries, PO_con1, PO_con2, PO_con3, PO_alb1, PO_alb2 and PO_alb3, were constructed and sequenced using Illumina HiSeq 2500. A total of 9,960,534, 10,957,008, 11,060,368, 10,559,741, 9,702,427, and 10,974,117 raw reads were acquired, respectively. The quality scores across all bases before and after trimming showed high accuracy (>99.9%) in all six libraries (S3 Fig). These reads were first adjusted to remove the sequencing artefacts: reads without a 3’ adaptor, reads that were <18 nt and >26 nt, and junk reads (≥2N, ≥7A, ≥8C, ≥6G, ≥7T, ≥10Dimer, ≥6Trimer, or ≥5Tetramer). Furthermore, these sequences were mapped to the RFam database (http://rfam.janelia.org), P. olivaceus mRNA database [42], and Repbase (http://www.girinst.org/repbase) for filtering the non-coding RNA families except miRNA, mRNA, and repetitive sequences. As a result of data treatment, 4,134,252 (41.51%), 5,004,942 (45.68%), 6,467,034 (58.47%), 6,238,636 (59.08%), 4,989,758 (51.43%), and 5,955,979 (54.27%) clean reads were obtained from the total reads of PO_con1, PO_con2, PO_con3, PO_alb1, PO_alb2 and PO_alb3, respectively (S8 Table). The length analysis indicated that 84–89% of clean read sequences were distributed between 20 and 24 nt (Fig 4), in accordance with the typical characteristics of Dicer processing products [44].
Fig 4. Length distribution of small RNAs.
The clean reads of small RNAs were distributed from 18 nt to 26 nt in six libraries including PO_con1, PO_con2, PO_con3, PO_alb1, PO_alb2, and PO_alb3. The reads of 20–24 nt accounted for 84–93% of small RNAs.
The biological replicate quality check revealed that one albino sample (PO_alb1) exhibited a poor correlation with the other two samples, with R = 0.648–0.668 (S4 Fig). Hence, PO_alb1 and PO_con1 were excluded from subsequent miRNA identification and expression pattern analysis.
Identification of conserved miRNAs
The clean reads from PO_con2, PO_con3, PO_alb2, and PO_alb3 were aligned with miRbase 21.0 and the P. olivaceus genome & EST, and 411 conserved miRNAs with normalized reads ≥10 were identified in P. olivaceus. According to the mapping information, four groups were clustered: gp1a, gp1b, gp2, and gp3. Group 1, gp1a, included 31 unique miRNAs mapping to P. olivaceus miRNA/pre-miRNAs in miRbase, and the pre-miRNAs further mapped to the genome & EST (S9 Table). Group 2, gp1b, included 240 unique miRNAs mapping to selected species’ (except for P. olivaceus) miRNAs/pre-miRNAs in miRbase, and the pre-miRNAs further mapped to the genome & EST (S10 Table). Group 3, gp2, included 75 unique miRNAs mapping to selected species’ (except for P. olivaceus) miRNAs/pre-miRNAs in miRbase, while only miRNA (not pre-miRNA) was mapped to the genome (S11 Table). Group 4, gp3, included 65 unique miRNAs mapping to selected species’ (except for P. olivaceus) miRNAs/pre-miRNAs in miRbase, but neither pre-miRNA nor miRNA was mapped to the genome (S12 Table).
The conservation of the identified miRNAs with other species
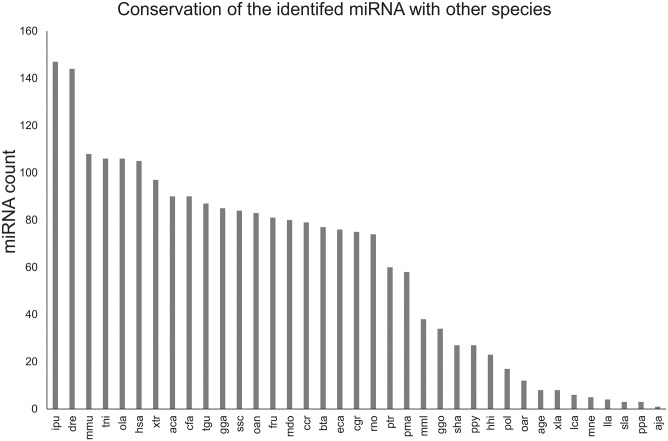
The frequency with which 411 conserved miRNAs occurred in other species was calculated, and the result (Fig 5) showed that the top five species were channel catfish (Ictalurus punctatus), zebrafish (Danio rerio), house mouse (Mus musculus), Tetraodon pufferfish (Tetraodon nigroviridis), and Japanese killifish (Oryzias latipes), with 147, 144, 108, 106 and 106 conserved miRNAs, respectively.
Fig 5. Conservation profile of the identified miRNA.
The frequency of identified miRNAs is calculated by mapping with 37 species (horizontal axis) in miRbase, and the miRNA counts are shown on the vertical axis.
Identification of putative novel miRNAs
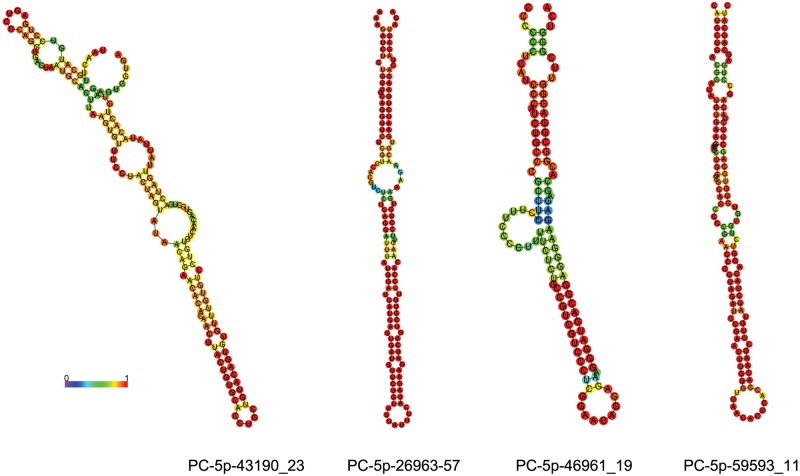
Following the identification of conserved miRNAs, the unannotated sequences were matched to P. olivaceus genomes [42] and analysed using RNAfold software. Consequently, 64 putative novel miRNAs were identified (S13 Table), and the hairpin structures formed by the precursor sequences of four novel miRNAs are illustrated in Fig 6.
Fig 6. The illustration of four novel miRNAs in P. olivaceus.
The secondary structures formed by four novel miRNA precursors are illustrated by RNAfold. The colour bar represents base-pair probabilities from 0 (blue) to 1 (red).
miRNA differential expression profiles
By combining the conserved and novel miRNAs, a total of 475 miRNAs were identified. The subsequent differential expression revealed 33 significantly differentially expressed miRNAs in albino versus normally pigmented P. olivaceus (fold change ≥1.5 or ≤0.67 and p≤0.05), including 13 up-regulated miRNAs and 20 down-regulated miRNAs (Table 2).
Table 2. Differential expression of miRNAs in albino and normal Japanese flounder (p<0.05).
| Index | miRNA name | miRNA sequence | up/down (PO_alb vs PO_con) | fold change (PO_alb vs PO_con) | pvalue (t_test) | PO_con (mean) | PO_alb (mean) |
|---|---|---|---|---|---|---|---|
| 1 | dre-miR-25-3p | CATTGCACTTGTCTCGGTCTGA | up | 1.70 | 4.22E-02 | 10,546 | 17,961 |
| 2 | hsa-miR-205-5p_R+2 | TCCTTCATTCCACCGGAGTCTGTT | up | 1.79 | 3.23E-02 | 527 | 942 |
| 3 | hsa-miR-27a-3p_R+1 | TTCACAGTGGCTAAGTTCCGCG | up | 2.55 | 3.75E-03 | 11 | 28 |
| 4 | mmu-miR-143-5p_R+2 | GGTGCAGTGCTGCATCTCTGGTC | up | 1.93 | 4.19E-02 | 78 | 151 |
| 5 | oan-miR-139-3p_R-1_1ss8AC | TGGAGACCCAGCTCTGTTGGA | up | 2.42 | 4.09E-02 | 20 | 48 |
| 6 | ola-miR-30c_1ss21AG | TGTAAACATCCTACACTCTCGGC | up | 1.61 | 3.33E-02 | 1,262 | 2,031 |
| 7 | ola-miR-99_R-1 | CAAGCTCGCCTCTGTGGGTCT | up | 2.75 | 3.08E-02 | 283 | 778 |
| 8 | PC-5p-46961_19 | CCCCTTTTCTCTGCCGTCGTCCC | up | 1.98 | 4.45E-02 | 10 | 20 |
| 9 | PC-5p-59593_11 | GGACCCGTAGATCCGAACTTGT | up | 2.60 | 1.66E-02 | 7 | 19 |
| 10 | pol-miR-199a-5p_R+1 | CCCAGTGTTCAGACTACCTGTTC | up | 2.71 | 2.91E-02 | 129,672 | 351,541 |
| 11 | tni-miR-205 | TCCTTCATTCCACCGGAGTCTG | up | 1.82 | 4.64E-02 | 60,815 | 110,934 |
| 12 | xtr-miR-222_R-1 | AGCTACATCTGGCTACTGGGTCT | up | 1.94 | 3.53E-02 | 3,066 | 5,950 |
| 13 | xtr-miR-92a_R+4 | TATTGCACTTGTCCCGGCCTGTTTT | up | 2.41 | 2.84E-02 | 370 | 893 |
| 14 | dre-miR-18b-5p_1ss11TC | TAAGGTGCATCTAGTGCAGATA | down | 0.39 | 6.23E-03 | 127 | 50 |
| 15 | dre-miR-202-5p_R-1 | TTCCTATGCATATACCTCTTT | down | 0.07 | 1.37E-02 | 13 | 1 |
| 16 | dre-miR-203b-3p_L-1R+2_1ss11CT | TGAAATGTTTAGGACCACTTGAT | down | 0.67 | 3.87E-02 | 90 | 60 |
| 17 | dre-miR-204-5p_L+1 | TTTCCCTTTGTCATCCTATGCCT | down | 0.20 | 3.73E-02 | 358 | 70 |
| 18 | dre-miR-204-5p_R+2 | TTCCCTTTGTCATCCTATGCCTGT | down | 0.30 | 8.67E-03 | 61 | 18 |
| 19 | dre-miR-20a-3p_2ss7GA11GA | ACTGCAATGTAAGCACTTGAAG | down | 0.54 | 3.79E-02 | 88 | 47 |
| 20 | dre-miR-26a-5p_R-1_1ss21CT | TTCAAGTAATCCAGGATAGGT | down | 0.48 | 3.89E-02 | 632 | 302 |
| 21 | dre-miR-9-3p_R+1 | TAAAGCTAGATAACCGAAAGTA | down | 0.38 | 3.35E-02 | 10 | 4 |
| 22 | hhi-miR-26_R+1 | TTCAAGTAATCCAGGATAGGCT | down | 0.55 | 2.92E-02 | 31,645 | 17,508 |
| 23 | oan-miR-16b-5p_1ss22GA | TAGCAGCACGTAAATATTGGTA | down | 0.38 | 3.00E-02 | 39 | 15 |
| 24 | ola-miR-106a_R+2 | TAAAGTGCTTACAGTGCAGGTAG | down | 0.43 | 2.68E-02 | 1,524 | 649 |
| 25 | ola-miR-135b_R+3 | TATGGCTTTTTATTCCTACGTG | down | 0.21 | 2.39E-02 | 55 | 11 |
| 26 | ola-miR-16 | TAGCAGCACGTAAATATTGGC | down | 0.47 | 6.27E-03 | 6,999 | 3,285 |
| 27 | ola-miR-205_L-1R+1_1ss20TA | GATTTCAGTGGTGTGAAGAGTA | down | 0.57 | 4.18E-02 | 257 | 146 |
| 28 | PC-5p-26963_57 | TATGGATCATGAGTGATGTTCT | down | 0.45 | 1.15E-02 | 24 | 11 |
| 29 | PC-5p-43190_23 | TAACAGAACACACAACTTACTG | down | 0.60 | 3.46E-02 | 15 | 9 |
| 30 | pma-miR-1c-3p_1ss2GT | TTGAATGTAAAGAAGTATGTAC | down | 0.43 | 3.62E-02 | 74 | 32 |
| 31 | tni-miR-135b_R+1 | TATGGCTTTTTATTCCTATCTGA | down | 0.19 | 3.14E-02 | 35 | 7 |
| 32 | tni-miR-15a_R-1 | TAGCAGCACGGAATGGTTTGT | down | 0.46 | 4.88E-02 | 9,843 | 4,536 |
| 33 | tni-miR-16_R-1 | TAGCAGCACGTAAATATTGGA | down | 0.43 | 3.59E-02 | 11,582 | 4,964 |
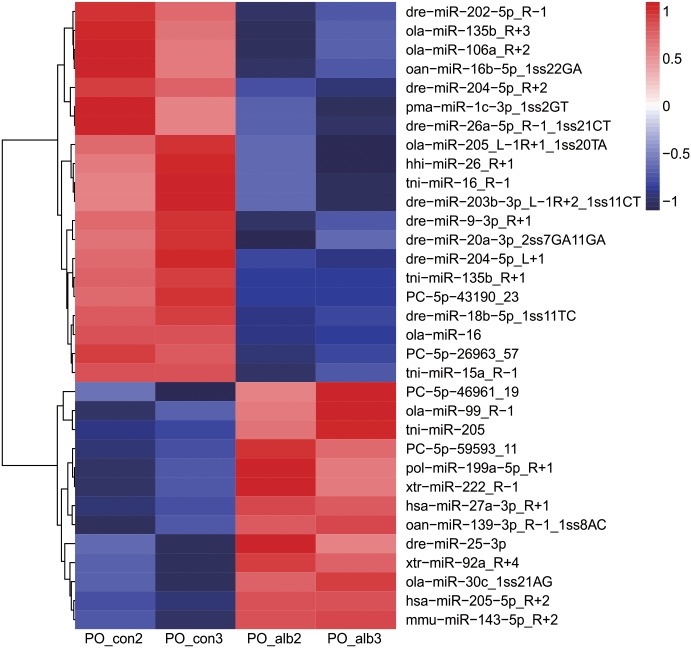
A heat map based on the miRNA differential expression pattern (Fig 7) showed two clusters of miRNAs, 13 up-regulated miRNAs and 20 down-regulated miRNAs. Further clustering revealed that two sub-clusters were identified within the down-regulated miRNAs: one contained 7 miRNAs, and the other contained 13 miRNAs (Fig 7). Additionally, the up-regulated miRNAs also contained two sub-clusters, one consisting of 3 miRNAs and the other consisting of 10 miRNAs (Fig 7).
Fig 7. Clustering of expression patterns of 33 differentially expressed miRNAs.
The expression patterns of 33 differentially expressed miRNAs (p<0.05) in the PO_con2, PO_con3, PO_alb2, and PO_alb3 libraries are displayed in a pheatmap. Each column represents one library, and the colour bar indicates relative expression level from high (red) to low (green).
Real-time quantitative RT-PCR
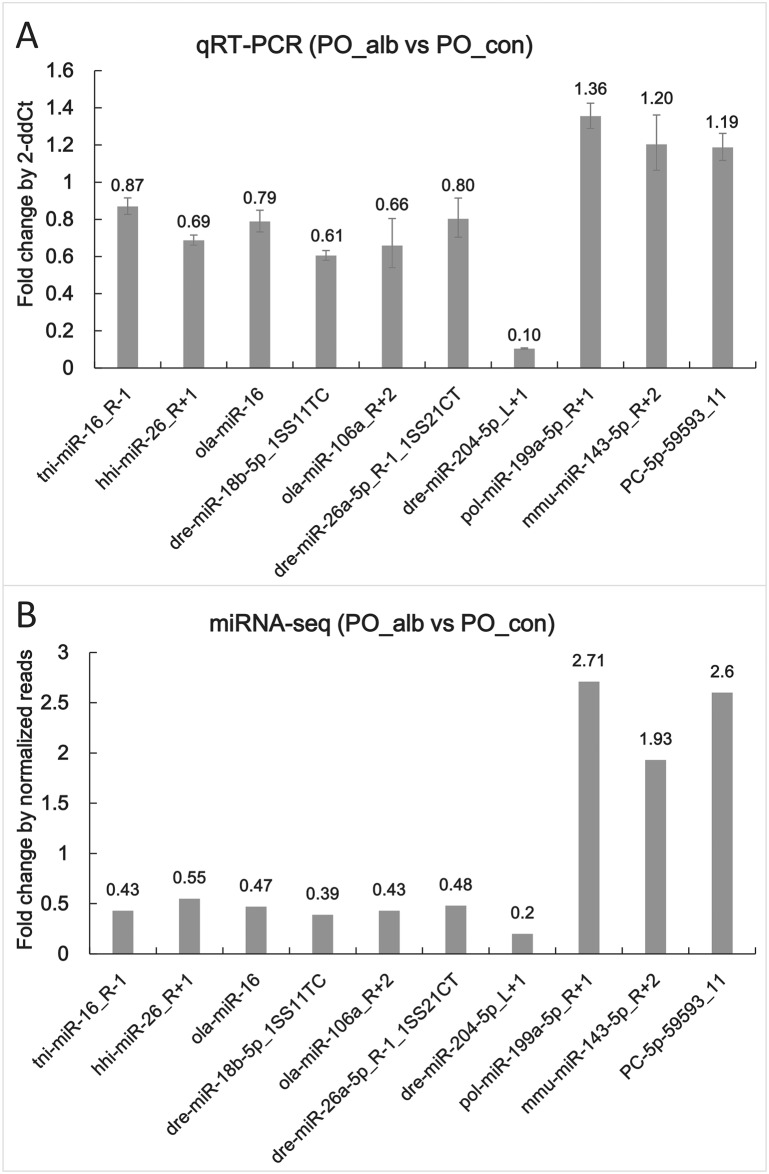
To validate the expression patterns of the miRNAs, qRT-PCR was utilized to detect the expression of 10 randomly selected miRNAs in which both up-regulated and down-regulated miRNAs were included. The three up-regulated miRNAs were pol-miR-199a-5p_R+1, mmu-miR-143-5p_R+2 and PC-5p-59593_11. The seven down-regulated miRNAs were tni-miR-16_R-1, ola-miR-26_R+1, ola-miR-16, dre-miR-18b-5p_1ss11TC, ola-miR-106a_R+2, dre-miR-26a-5p_R-1_1ss21CT and dre-miR-204-5p_L+1. These results verified the similar expression pattern trends of all 10 miRNAs between qRT-PCR and miR-seq (Fig 8), although there were slight differences in fold-change.
Fig 8. The expression levels of 10 randomly selected miRNAs in qRT-PCR and miRNA-seq.
The expression fold changes of 10 P. olivaceus miRNAs in PO_alb versus PO_con detected by poly (A)-tailed qRT-PCR and high-throughput sequencing are shown in A and B, respectively. In qRT-PCR analysis, 5s rRNA expression levels were used for the internal control, and values are indicated as means ± standard error (SE) derived from triplicate experiments.
Target prediction for significantly differentially expressed miRNAs and function analysis
To provide clues for the possible roles of 33 differentially expressed miRNAs, target prediction analysis was conducted by TargetScan 6.2 and miRanda algorithms. These algorithms identified 14655 putative target genes, including genes that participate in the trafficking of melanosomes such as melanophilin (MELPH) and melanotransferrin (MFI2); genes coding various proteins involved in melanin synthesis such as TYR, TYRP1, TYRP2, MC1R, and tyrosine 3-monooxygenase (TH); and melanocyte-specific transcription factors such as MITF, transcription factor 7-like 2 (TCF7L2), and paired box protein Pax-3-B (PAX3B) [7].
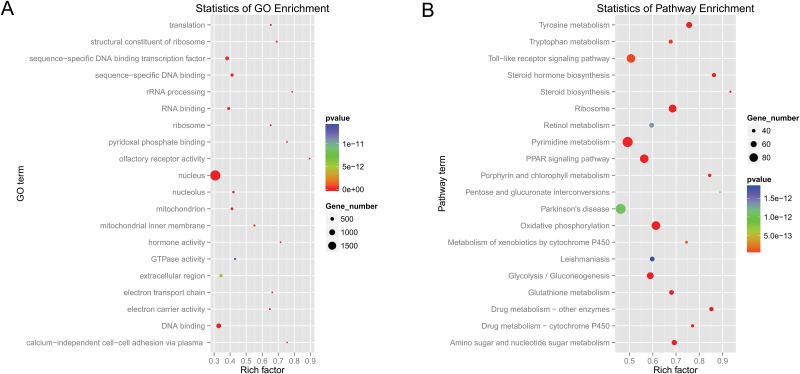
To further interpret the possible physiological processes and pathways regulated by these identified miRNAs, their putative target genes were subjected to GO term and KEGG pathway analysis. GO term analysis indicated that a total of 254 out of the 7263 clustered GO terms were significantly enriched (p<0.001) (S14 Table). The top 20 enriched terms were mainly involved in biological processes (translation, rRNA processing, electron transport chain, calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules), cellular components (mitochondrion, mitochondrial inner membrane, ribosome, nucleus, nucleus, nucleolus, extracellular complex) and molecular functions (sequence-specific DNA binding, sequence-specific DNA binding transcription factor activity, structural constituent of ribosome, electron carrier activity, olfactory receptor activity, hormone activity, RNA binding, pyridoxal phosphate binding, DNA binding, GTPase activity) (Fig 9A).
Fig 9. The top 20 enriched GO terms (A) and KEGG pathways (B).
Gene number: number of target genes in each term or pathway. Rich factor: the ratio of the number of target genes divided by the number of all the genes in each term or pathway.
Using KEGG pathway analysis, 14655 predicted genes were grouped into 256 pathways. Of these 256 pathways, 103 pathways were significantly enriched (p<0.001) (S15 Table), including tyrosine metabolism, PPAR signalling pathways, lysosomes, and phototransduction. The top 20 enriched pathway terms are shown in Fig 9B.
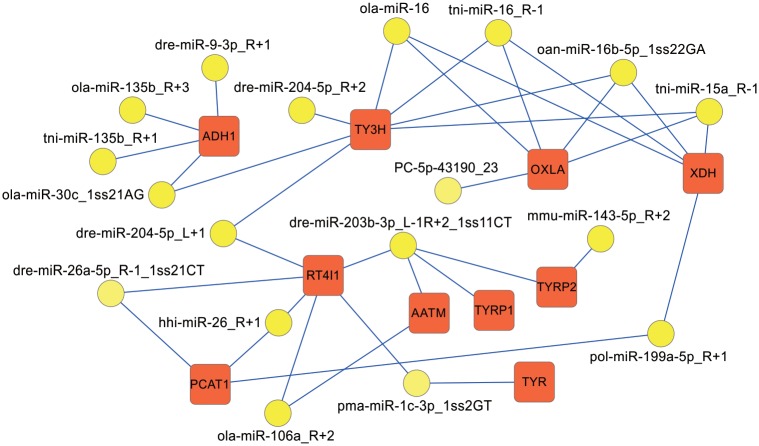
Based on these results, the relevant miRNA-GO-network and miRNA-KEGG-network were finally generated. For instance, for tyrosine metabolism, TYR was regulated by pma-miR-1c-3p-1ss2GT; TYRP1 was regulated by dre-miR-203b-3p_L-1R+2_1SS11CT; TYRP2 was regulated by dre-miR-203B-3P_L-1R+2_1SS11CT and mmu-miR-143-5p_R+2; tyrosine 3-monooxygenase (TY3H) was regulated by ola-miR-30C_1ss21AG, dre-miR-204-5p_L+1, dre-miR-204-5p_R+2, ola-miR-16, tni-miR-16_R-1, oan-miR-16b-5p_1ss22GA, tni-miR-15a_R-1; and L-amino-acid oxidase (OXLA) was targeted by ola-miR-16, tni-miR-16_R-1, oan-miR-16b-5p_1ss22GA, tni-miR-15a_R-1, and PC-5p-43190_23 (Fig 10). Tyrosine metabolism interested us due to its indispensable role in melanin biosynthesis and because its disorder has been associated with albinism in mammals [45].
Fig 10. A proposed network of putative interactions between miRNAs and mRNAs in tyrosine metabolism.
The regulation network of miRNAs and mRNAs involved in tyrosine metabolism is illustrated by Cytoscape. Yellow ellipses represent miRNAs, and red rectangles indicate their target genes associated with tyrosine metabolism. Abbreviations of the genes are detailed in the text.
The comparison of expression profiles between miRNA and the predicted miRNA targets
Based on the genes information derived from RNA-seq and miRNA targeted mRNAs in miRNA-seq, 133 predicted target genes were also found to be changed in albinism versus normal skin tissue (S16 Table). A series of pigmented related genes including MITF, MC1R, GCH1, XDH, BMP1, TRY, TYRP1, EDNRB, TBX19, and WNT7B were contained. Further, the comparison of expression profiles between miRNA and the predicted miRNA targets revealed that 206 mRNA-miRNA pairs changed in the same direction and 130 mRNA-miRNA pairs changed in the opposite direction (S17 Table).
Discussion
In this study, we have identified and investigated the expression profile of mRNAs and miRNAs from albino and normally pigmented P. olivaceus. By introducing the genome sequences of P. olivaceus as a reference, 22,498 genes containing 711 new genes and 475 miRNAs containing 64 novel miRNAs were obtained. Compared with previous fish studies, this is the largest number of miRNAs identified from a single experiment [34–38]. To date, there are 20 miRNAs known in P. olivaceus based on miRbase 21.0, derived from the miRNA libraries of the metamorphosis period [37]. The high conservation of the known 411 detected miRNAs with 37 other species’ miRNAs in miRBase 21.0 suggested the conservation of miRNA sequences in the evolution process of different species [46]. On the other hand, 316 out of 411 (76.89%) miRNAs showed differences including base deletion or addition or base substitution, which also revealed the diversity of miRNAs in different species. Additionally, the identifications of 711 novel genes and 64 novel miRNAs are important supplements to P. olivaceus genome [42] and the known pool of 1044 miRNAs of fish [47].
The expression profile of mRNA and miRNAs was further investigated to gain insight into their possible roles in P. olivaceus albinism. By comparison with previous pigmentation related genes, at least 30 genes exhibiting down-regulated expression pattern were screened in differentially expressed genes of RNA-seq. As the major enzymes participated in mammals albinism [7], TYR, TYRP1, OCA2 and SLC45A2 also exhibited down regulation in albino pigmented P. olivaceus individuals. Although no differential expression of GPR143 was observed as in mammals’ albinism [48], the identification of other G-protein coupled receptor including GPR21 and GPR61 may imply their possible involvement in fish albinism. Among the overlapped genes, more than one paralogs of EDNRB, GCH1, KRT13, and TYRP1 were observed in P. olivaceus, which is consistent with the duplication of pigmentation genes in fish [49, 50].
Although a large number of miRNAs have been identified in the melanocyte biology of mammals [51], the amount of miRNA in fish pigmentation has rarely been studied [32]. The present study discovered 33 differentially expressed miRNAs in albino versus normally pigmented P. olivaceus, which suggested that miRNAs may also be involved in flatfish pigmentation. In addition, the miRNAs identified in this study have shed light on potential regulatory mechanisms of fish pigmentation deficiency. For example, miR-25 has been identified as an important regulator of pigmentation by targeting MITF in alpaca (Lama pocos) [21]; the homologue of miR-25 (dre-miR-25-3p) was also detected in P. olivaceus, with 1.70-fold up-regulation in albino skin. Additionally, the putative target genes of dre-miR-25-3p contains pax3b, a key transcription factor involved in melanocytes [9] and MC1R, a G protein-coupled receptor involved in regulating mammalian skin and hair colour [51].
Interaction networks between miRNAs, target genes and transcription factors are critical for an appropriate balance of gene expression in the melanocytes of mammals [52]. The target gene prediction of 33 differentially expressed miRNAs in the present study revealed 14655 putative target genes. The overlapped genes list of these targeted genes and differentially expressed genes in RNA-seq, as well as the mRNA-miRNA pairs exhibited opposite expression profiles, both provided important clues for further research. One of these putative target genes, MC1R, a G protein-coupled receptor located on the plasma membrane of melanocytes, has been reported to be involved in regulating mammalian skin and hair colour by binding to adrenocorticotropic hormone (ACTH) and melanocyte-stimulating hormone (MSH) [51]. In P. olivaceus, MC1R mRNA exhibited 2.7759 log2FoldChange down-regulation in albino skin. And it was predicted to be the target gene of 10 miRNAs, five of which showed up-regulated expression pattern. As a key transcription factor in regulating melanocytes, MITF has been predicted to be an important target gene for numerous miRNAs in mammals’ pigmentation [20–23]. Our study also suggested that 3 miRNAs including one up-regulated miRNAs targeted MITF in P. olivaceus, and its mRNA correspondingly decreased by 6.6756 log2FoldChange in albino skin. Mutations in MELPH, which is known as a downstream gene regulated by MITF, result in the dilute coat colour phenotype in dogs and cats [53–55]. In this study, MELPH was identified as a potential target of 2 miRNAs. Another target gene, TY3H, is responsible for the conversion from the amino acid L-tyrosine to L-3,4-dihydroxyphenylalanine (L-DOPA), which is the substrate for melanin synthesis [56, 57]. There have been 5 miRNAs identified that regulate TY3H in P. olivaceus.
As a phenomenon of mass depigmentation on the ocular side, albinism was also observed and investigated in other flatfish, such as turbot (Scophthmlmus maximus), Southern flounder (Paralichthys lethostigma) and Summer flounder (Paralichthys dentatus). Many factors, including light and nutritional factors, have been suggested to influence the occurrence of flatfish albinism [58–60]. The feeding of diets deficient in vitamin A, docosahexaenoic acid (DHA) and phospholipids will inhibit the rhodopsin production in the retina of flatfish, which will further lead to the interruption of melanin synthesis [61]. In the present study, several phospholipid-related pathways including glycosylphosphatidylinositol and glycerophospholipid metabolism were significantly enriched, which suggested an intrinsic correlation between phospholipid metabolism and abnormal pigmentation in P. olivaceus. In invertebrates and lower vertebrates, light first activates melanopsin-expressing retinal ganglion cells, which then initiate a neuro-endocrine circuit that regulates melanin dispersion/aggregation in skin melanophores through the secretion of α-melanocyte stimulating hormone (α-MSH) [62–65]. In the present study, the phototransduction pathway was significantly enriched, including the Green-sensitive opsin-4 (OPSG4), which was targeted by 2 miRNAs: dre-miR-26a-5p_L-1_1ss21CT and hhi-miR-26_R+1. Another gene, Blue-sensitive opsin (OPSB), was targeted by 4 miRNAs: dre-miR-26a-5p_L-1_1ss21CT, hhi-miR-26_R+1, xtr-miR-222_R-1, and ola-miR-106a_R+2. These results suggest the important role of the phototransduction pathway in the pigmentation of P. olivaceus.
Previous research has revealed that lysosomes share common precursors and endosomal pathways with melanosomes [66, 67], which implied that the regulation network involved in lysosomes would eventually affect melanosome biogenesis. In the present study, the observation of significant enrichment in the lysosome pathways in KEGG analysis further indicates the close relevance between lysosomes and melanosome biogenesis. The enrichment of the peroxisome proliferator-activated receptor (PPAR) signalling pathway interested us because PPARs have been shown to inhibit melanocyte growth and stimulate melanogenesis through a connection with α-MSH [67–69].
Taken together, these results provide novel insight into the albinism mechanism of P. olivaceus. The quantitative mRNA/miRNA data, network and pathway information presented here offer a solid starting point for the elucidation of detailed functions of mRNAs and miRNAs in fish albinism.
In conclusion, we constructed six cDNA libraries and six small RNA libraries of skin tissues from three albinos and three normally pigmented individuals of P. olivaceus. As a result of sequencing and data treatment, a total of 22,498 genes including 711 new genes, and a total of 475 miRNAs including 64 novel miRNAs were both identified. A total of 235 genes and 33 miRNAs exhibited differential expression patterns in albino versus normally pigmented P. olivaceus. The differentially expressed genes were significantly enriched in Folate biosynthesis, Melanogenesis, and Neuroactive ligand-receptor interaction pathways. The subsequent in silico analysis predicted 14655 putative target genes, of which a variety of genes related to melanin metabolism interested us, including MITF, TYR, MC1R, TYRP1, TYRP2, TY3H, and MELPH. The GO and KEGG analyses revealed that these target genes were significantly enriched in 254 GO terms and 103 pathways. The network mapping of albinism-related miRNAs and their target genes will serve as substantial clues for future research on the mechanism of pigmentation deficiency in P. olivaceus.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability
All RNA-seq and miRNA information are available from the NCBI database (accession numbers GSE99341,GSE99342, GSE99343).
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (NW, 31472273; SLC, 31461163005) (http://www.nsfc.gov.cn/), and the Taishan Scholar Climb Project of Shandong Province (SLC, http://www.rcsd.gov.cn/gongcheng/shengjigongcheng/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Farabee WC. Notes on negro albinism. Science. 1903; 17(419): 75 doi: 10.1126/science.17.419.75 [DOI] [PubMed] [Google Scholar]
- 2.Oetting WS, King RA. Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum Mutat. 1999; 13(2): 99–115. doi: 10.1002/(SICI)1098-1004(1999)13:2<99::AID-HUMU2>3.0.CO;2-C [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi Y, Brenner M, Hearing VJ. The regulation of skin pigmentation. J Biol Chem. 2007; 282(38): 27557–27561. doi: 10.1074/jbc.R700026200 [DOI] [PubMed] [Google Scholar]
- 4.Baxter LL, Loftus SK, Pavan WJ. Networks and pathways in pigmentation, health and disease. Wiley Interdiscip Rev Syst Biol Med. 2009; 1(3): 359–371. doi: 10.1002/wsbm.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett DC, Lamoreux ML. The color loci of mice—a genetic century. Pigment Cell Res. 2003; 16(4): 333–344. doi: 10.1034/j.1600-0749.2003.00067.x [DOI] [PubMed] [Google Scholar]
- 6.Chi A, Valencia JC, Hu ZZ, Watabe H, Yamaguchi H, Mangini NJ, et al. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. J Proteome Res. 2006; 5(11): 3135–3144. doi: 10.1021/pr060363j [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, Hearing VJ. Melanocytes and their diseases. Cold Spring Harb Perspect Med. 2014;4(5): a017046 doi: 10.1101/cshperspect.a017046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goding CR. Mitf from neural crest to melanoma signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000; 14(14): 1712–1728. [PubMed] [Google Scholar]
- 9.Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, et al. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005; 433(7028): 884–887. doi: 10.1038/nature03292 [DOI] [PubMed] [Google Scholar]
- 10.Hou L, Arnheiter H, Pavan WJ. Interspecies difference in the regulation of melanocyte development by SOX10 and MITF. Proc Natl Acad Sci U S A. 2006; 103(24): 9081–9085. doi: 10.1073/pnas.0603114103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee R C, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75(5): 843–854. doi: 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 12.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene Lin-14 by Lin-4 mediates temporal pattern-formation in C. elegans. Cell. 1993; 75(5): 855–862. [DOI] [PubMed] [Google Scholar]
- 13.Baek S, Choi H, Kim J. Ebf3-miR218 regulation is involved in the development of dopaminergic neurons. Brain Res. 2014; 1587; 23–32. doi: 10.1016/j.brainres.2014.08.059 [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435(7043): 834–838. doi: 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 15.Lin CY, Lee HC, Fu CY, Ding YY, Chen JS, Lee MH, et al. MiR-1 and miR-206 target different genes to have opposing roles during angiogenesis in zebrafish embryos. Nat Commun. 2013; 4: 2829 doi: 10.1038/ncomms3829 [DOI] [PubMed] [Google Scholar]
- 16.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010; 466(7308): 835–840. doi: 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zehir A, Hua LL, Maska EL, Morikawa Y, Cserjesi P. Dicer is required for survival of differentiating neural crest cells. Dev Biol. 2010; 340(2): 459–467. doi: 10.1016/j.ydbio.2010.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4 SOX2 and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009; 137(4): 647–658. doi: 10.1016/j.cell.2009.02.038 [DOI] [PubMed] [Google Scholar]
- 19.Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008; 40(3): 290–298. doi: 10.1038/ng.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KH, Bin BH, Kim J, Dong SE, Park PJ, Choi H, et al. Novel inhibitory function of miR-125b in melanogenesis. Pigment Cell Melanoma Res. 2014; 27(1): 140–144. doi: 10.1111/pcmr.12179 [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z, He J, Jia X, Jiang J, Bai R, Yu X, et al. MicroRNA-25 functions in regulation of pigmentation by targeting the transcription factor MITF in Alpaca Lama pacos skin melanocytes. Domest Anim Endocrinol. 2010; 38(3): 200–209. doi: 10.1016/j.domaniend.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 22.Dong C, Wang H, Xue L, Dong Y, Yang L, Fan R, et al. Coat color determination by miR-137 mediated down-regulation of microphthalmia-associated transcription factor in a mouse model. RNA. 2012; 18(9): 1679–1686. doi: 10.1261/rna.033977.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caramuta S, Egyházi S, Rodolfo M, Witten D, Hansson J, Larsson C, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010; 130(8): 2062–2070. doi: 10.1038/jid.2010.63 [DOI] [PubMed] [Google Scholar]
- 24.Nemlich Y, Greenberg E, Ortenberg R, Besser MJ, Barshack I, Jacob-Hirsch J, et al. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J Clin Invest. 2013; 123(6): 2703–2718. doi: 10.1172/JCI62980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levati L, Alvino E, Pagani E, Arcelli D, Caporaso P, Bondanza S, et al. Altered expression of selected microRNAs in melanoma antiproliferative and proapoptotic activity of miRNA-155. Int J Oncol. 2009; 35(2): 393–400. doi: 10.3892/ijo_00000352 [PubMed] [Google Scholar]
- 26.Couts KL, Anderson EM, Gross MM, Sullivan K, Ahn NG. Oncogenic B-Raf signaling in melanoma cells controls a network of microRNAs with combinatorial functions. Oncogene. 2013; 32(15): 1959–1970. doi: 10.1038/onc.2012.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009; 106(6): 1814–1819. doi: 10.1073/pnas.0808263106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujii R. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 2000; 13(5): 300–319. doi: 10.1034/j.1600-0749.2000.130502.x [DOI] [PubMed] [Google Scholar]
- 29.Kelsh RN. Zebrafish pigmentation mutations and the processes of neural crest development. Development. 1996; 123: 369–389. [DOI] [PubMed] [Google Scholar]
- 30.Fukamachi S, Asakawa S, Wakamatsu Y, Shimizu N, Mitani H, Shima A. Conserved function of medaka pink-eyed dilution in melanin synthesis and its divergent transcriptional regulation in gonads among vertebrates. Genetics. 2004; 168(3): 1519–1527. doi: 10.1534/genetics.104.030494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bilandzija H, Ma L, Parkhurst A, Jeffery WR. A potential benefit of albinism in Astyanax cavefish downregulation of the oca2 gene increases tyrosine and catecholamine levels as an alternative to melanin synthesis. PLoS One. 2013; 8(11): e80823 doi: 10.1371/journal.pone.0080823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan B, Liu B, Zhu CD, Li KL, Yue LJ, Zhao JL, et al. MicroRNA regulation of skin pigmentation in fish. J Cell Sci. 2013; 126(Pt 15): 3401–3408. doi: 10.1242/jcs.125831 [DOI] [PubMed] [Google Scholar]
- 33.Seikai T. Reduction of occurrence frequency of albinism in juvenile flounder Paralichthys olivaceus hatchery-reared on wile zooplankton. Bull Japan Soc Sci Fish. 1985; 51(8): 1261–1267. [Google Scholar]
- 34.Zhang BC, Zhang J, Sun L. In-depth profiling and analysis of host and viral microRNAs in Japanese flounder Paralichthys olivaceus infected with megalocytivirus reveal involvement of microRNAs in host-virus interaction in teleost fish. BMC Genomics. 2014; 15: 878 doi: 10.1186/1471-2164-15-878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu Y, Shi Z, Wang G, Zhang J, Li W, Jia L. Expression of let-7 microRNAs that are involved in Japanese flounder Paralichthys olivaceus metamorphosis. Comp Biochem Physiol B Biochem Mol Biol. 2013; 165(2): 106–113. doi: 10.1016/j.cbpb.2013.03.012 [DOI] [PubMed] [Google Scholar]
- 36.Xie C, Xu S, Yang L, Ke Z, Xing J, Gai J, et al. mRNA/microRNA Profile at the Metamorphic Stage of Olive Flounder Paralichthys olivaceus. Comp Funct Genomics. 2011; 2011: 256038 doi: 10.1155/2011/256038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Y, Shi Z, Wu M, Zhang J, Jia L, Chen X. Identification and differential expression of microRNAs during metamorphosis of the Japanese flounder Paralichthys olivaceus. PLoS One. 2011; 6(7): e22957 doi: 10.1371/journal.pone.0022957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Y, Zhang L, Chen X. Differential expression analysis of Paralichthys olivaceus microRNAs in adult ovary and testis by deep sequencing. Gen Comp Endocrinol. 2014; 204: 181–184. doi: 10.1016/j.ygcen.2014.05.019 [DOI] [PubMed] [Google Scholar]
- 39.Fu YS, Shi ZY, Wang GY, Li WJ, Zhang JL, Jia L. Expression and regulation of miR-1, -133a, -206a, and MRFs by thyroid hormone during larval development in Paralichthys olivaceus. Comp Biochem Physiol B Biochem Mol Biol. 2012; 161(3): 226–232. doi: 10.1016/j.cbpb.2011.11.009 [DOI] [PubMed] [Google Scholar]
- 40.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008; 5(7):621–628. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 41.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012; 40(Web Server issue):W478–83. doi: 10.1093/nar/gks402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao C, Bao B, Xie Z, Chen X, Li B, Jia X et al. The genome and transcriptome of Japanese flounder provide insights into flatfish asymmetry. Nat Genet. 2017; 49(1):119–124. doi: 10.1038/ng.3732 [DOI] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2ΔΔCT method. Methods. 2001; 25(4): 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 44.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000; 101(1): 25–33. doi: 10.1016/S0092-8674(00)80620-0 [DOI] [PubMed] [Google Scholar]
- 45.Oetting WS, King RA. Molecular basis of albinism mutations and polymorphisms of pigmentation genes associated with albinism. Hum Mutat. 1999; 13(2): 99–115. doi: 10.1002/(SICI)1098-1004(1999)13:2<99::AID-HUMU2>3.0.CO;2-C [DOI] [PubMed] [Google Scholar]
- 46.Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009; 11(1): 50–68. doi: 10.1111/j.1525-142X.2008.00302.x [DOI] [PubMed] [Google Scholar]
- 47.Bizuayehu TT, Babiak I. MicroRNA in teleost fish. Genome Biol Evol. 2014; 6(8): 1911–1137. doi: 10.1093/gbe/evu151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sepúlveda-Vázquez HE, Villanueva-Mendoza C, Zenteno JC, Villegas-Ruiz V, Pelcastre-Luna E, García-Aguirre G. Macular optical coherence tomography findings and GPR143 mutations in patients with ocular albinism. Int Ophthalmol. 2014; 34(5):1075–1081. doi: 10.1007/s10792-014-9912-1 [DOI] [PubMed] [Google Scholar]
- 49.Braasch I, Liedtke D, Volff JN, Schartl M. Pigmentary function and evolution of tyrp1 gene duplicates in fish. Pigment Cell Melanoma Res. 2009; 22(6):839–850. doi: 10.1111/j.1755-148X.2009.00614.x [DOI] [PubMed] [Google Scholar]
- 50.Braasch I, Schartl M, Volff JN. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol Biol. 2007; 7:74 doi: 10.1186/1471-2148-7-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mione M, Bosserhoff A. MicroRNAs in melanocyte and melanoma biology. Pigment Cell Melanoma Res. 2015; 28(3): 340–354. doi: 10.1111/pcmr.12346 [DOI] [PubMed] [Google Scholar]
- 52.Abdel-Malek Z, Scott MC, Suzuki I, Tada A, Im S, Lamoreux L, et al. The melanocortin-1 receptor is a key regulator of human cutaneous pigmentation. Pigment Cell Res. 2000; 13 Suppl 8: 156–162. doi: 10.1034/j.1600-0749.13.s8.28.x [DOI] [PubMed] [Google Scholar]
- 53.Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008; 21(6):665–676. doi: 10.1111/j.1755-148X.2008.00505.x [DOI] [PubMed] [Google Scholar]
- 54.Drögemüller C, Philipp U, Haase B, Günzel-Apel AR, Leeb T. A noncoding melanophilin gene MLPH SNP at the splice donor of exon 1 represents a candidate causal mutation for coat color dilution in dogs. J Hered. 2007; 98(5): 468–473. doi: 10.1093/jhered/esm021 [DOI] [PubMed] [Google Scholar]
- 55.Ishida Y, David VA, Eizirik E, Schäffer AA, Neelam BA, Roelke ME, et al. A homozygous single-base deletion in MLPH causes the dilute coat color phenotype in the domestic cat. Genomics. 2016; 88(6): 698–705. doi: 10.1016/j.ygeno.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 56.Nagatsu T. Tyrosine hydroxylase human isoforms structure and regulation in physiology and pathology. Essays Biochem. 1995; 30: 15–35. [PubMed] [Google Scholar]
- 57.Ando H, Kondoh H, Ichihashi M, Hearing VJ. Approaches to identify inhibitors of melanin biosynthesis via the quality control of tyrosinase. J Invest Dermatol. 2007; 127(4): 751–761. doi: 10.1038/sj.jid.5700683 [DOI] [PubMed] [Google Scholar]
- 58.Estévez A, McEvoy L, Bellb J G, Sargent J R. Growth survival lipid composition and pigmentation of turbot Scophthalmus maximus larvae fed live-prey enriched in Arachidonic and Eicosapentaenoic acids. Aquaculture. 1999; 180(3–4): 321–343. doi: 10.1016/S0044-8486(99)00209-4 [Google Scholar]
- 59.Denson MR, Smith TIJ. Diet and light intensity effects on survival growth and pigmentation of southern flounder Paralichthys lethostigma. J World Aquacult Soc. 1997; 28(28): 366–373. doi: 10.1111/j.1749-7345.1997.tb00283.x [Google Scholar]
- 60.Baker EP, Alves D, Bengtson DA. Effects of rotifer and Artemia fatty-acid enrichment on survival growth and pigmentation of Summer flounder Paralichthys dentatus larvae. J World Aquacult Soc. 1998; 29(4): 494–498. doi: 10.1111/j.1749-7345.1998.tb00675.x [Google Scholar]
- 61.Kanazawa A. Nutritional mechanisms involved in the occurrence of abnormal pigmentation in hatchery-reared flatfish. J World Aquaculture Soc. 1993; 24(2): 162–166. doi: 10.1111/j.1749-7345.1993.tb00005.x [Google Scholar]
- 62.Allison WT, Hallows TE, Johnson T, Hawryshyn CW, Allen DM. Photic history modifies susceptibility to retinal damage in albino trout. Vis Neurosci. 2006; 23(1): 25–34. doi: 10.1017/S0952523806231031 [DOI] [PubMed] [Google Scholar]
- 63.Moraes MN, Ramos BC, Poletini MO, Castrucci AM. Melanopsins localization and phototransduction in Xenopus laevis melanophores. Photochem Photobiol. 2015; 91(5): 1133–1141. doi: 10.1111/php.12484 [DOI] [PubMed] [Google Scholar]
- 64.Bertolesi GE, Vazhappilly ST, Hehr CL, McFarlane S. Pharmacological induction of skin pigmentation unveils the neuroendocrine circuit regulated by light. Pigment Cell Melanoma Res. 2016; 29(2):186–198. doi: 10.1111/pcmr.12442 [DOI] [PubMed] [Google Scholar]
- 65.Bertolesi GE, Hehr CL, McFarlane S. Melanopsin photoreception in the eye regulates light-induced skin colour changes through the production of α-MSH in the pituitary gland. Pigment Cell Melanoma Res. 2015; 28(5):559–71. doi: 10.1111/pcmr.12387 [DOI] [PubMed] [Google Scholar]
- 66.Raposo G, Tenza D, Murphy DM, Berson JF, Marks MS. Distinct protein sorting and localization to premelanosomes melanosomes and lysosomes in pigmented melanocytic cells. J Cell Biol. 2001; 152(4): 809–823. doi: 10.1083/jcb.152.4.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vijayasaradhi S, Xu YQ, Bouchard B, Houghton AN. Intracellular sorting and targeting of melanosomal membrane proteins identification of signals for sorting of the human brown locus protein gp75. J Cell Biol. 1995; 130(4): 807–820. doi: 10.1083/jcb.130.4.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang HY, Chung E, Lee M, Cho Y, Kang WH. Expression and function of peroxisome proliferator-activated receptors in human melanocytes. Br J Dermatol. 2004; 150(3): 462–468. doi: 10.1111/j.1365-2133.2004.05844.x [DOI] [PubMed] [Google Scholar]
- 69.Maresca V, Flori E, Camera E, Bellei B, Aspite N, Ludovici M, et al. Linking αMSH with PPARγ in B16-F10 melanoma. Pigment Cell Melanoma Res. 2013; 26(1): 113–127. doi: 10.1111/j.1755-148X.2012.01042.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All RNA-seq and miRNA information are available from the NCBI database (accession numbers GSE99341,GSE99342, GSE99343).