Abstract
G quadruplex structures have been predicted by bioinformatics to form in the 5′- and 3′-untranslated regions (UTRs) of several thousand mature mRNAs and are believed to play a role in translation regulation. Elucidation of these roles has primarily been focused on the 3′-UTR, with limited focus on characterizing the G quadruplex structures and functions in the 5′-UTR. Investigation of the affinity and specificity of RNA binding proteins for 5′-UTR G quadruplexes and the resulting regulatory effects have also been limited. Among the mRNAs predicted to form a G quadruplex structure within the 5′-UTR is the survival motor neuron domain containing 1 (SMNDC1) mRNA, encoding a protein that is critical to the spliceosome. Additionally, this mRNA has been identified as a potential target of the fragile X mental retardation protein (FMRP), whose loss of expression leads to fragile X syndrome. FMRP is an RNA binding protein involved in translation regulation that has been shown to bind mRNA targets that form G quadruplex structures. In this study we have used biophysical methods to investigate G quadruplex formation in the 5′-UTR of SMNDC1 mRNA and analyzed its interactions with FMRP. Our results show that SMNDC1 mRNA 5′-UTR forms an intramolecular, parallel G quadruplex structure comprised of three G quartet planes, which is bound specifically by FMRP both in vitro and in mouse brain lysates. These findings suggest a model by which FMRP might regulate the translation of a subset of its mRNA targets by recognizing the G quadruplex structure present in their 5′-UTR, and affecting their accessibility by the protein synthesis machinery.
TOC image
SMNDC1 mRNA adopts a 5′-UTR G quadruplex structure recognized specifically by FMRP, potentially affecting spliceosome assembly in FXS.
INTRODUCTION
A variety of protein activity regulation strategies occur post-transcriptionally, mainly at the level of mRNA translation. The 5′- and 3′- untranslated regions (5′- and 3′-UTRs) of mRNA templates contain cis-acting structural moieties and this regulation may involve both sequence and secondary structure(s) of these non-protein coding regions.1, 2 Among these structures involved in translation regulation is the mRNA G quadruplex.1–14
Biochemical and biophysical studies have revealed that certain guanine-rich mRNA sequences fold into the G quadruplex structure.15–17 Four guanine residues form a planar G quartet conformation that is stabilized by Hoogsteen base pairing, with sets of G quartets stacking upon each other and stabilized by central K+ ions, to form the G quadruplex structure.16, 17 In addition to 5′-UTR activities such as translation regulation, the G quadruplex structure has been implicated in other location-based processes, such as regulation of alternative splicing when in the coding region as well as translation regulation via the microRNA pathway and/or increased polyadenylation efficiency when in the 3′UTR.1, 9, 18–26
It is known that the genetic information governing translation is primarily localized in the 5′- and 3′-UTRs of mRNA.2 Based on the G3+N1–7G3+N1–7G3+N1–7G3+ RNA sequence (where N is any nucleotide), bioinformatics analyses have predicted that 2334 and 3530 G quadruplexes occur in the 5′- and 3′-UTRs, respectively, of human mRNA transcripts from known protein-coding genes.1 Despite their potentially major roles in mRNA translation regulation, only a limited number of these predicted G quadruplexes have been structurally and thermodynamically characterized.4–6, 8, 9, 19, 24, 27–30 The prevalence of G quadruplexes in the 5′-UTR suggests a role in the regulation of translation considering the advantageous control of the process at or near its point of initiation.1 Additionally, some 5′-UTR G quadruplex structures have been shown to inhibit translation while others promote translation, suggesting a complex form of regulation that requires further investigation to elucidate.1, 4, 6–8, 30 Moreover, these mechanisms likely have different layers of complexity, as the regulatory function of the G quadruplex structure in translation could also be mediated by its interactions with specific RNA binding proteins.
Among such potential regulatory proteins is the fragile X mental retardation protein (FMRP), an RNA binding protein implicated in translation regulation and specific binding to G quadruplex-forming RNAs.27–41 Loss of FMRP expression typically leads to a constitutive increase in the synthesis of certain dendritic proteins and results in the most prevalent inherited intellectual disability in humans: fragile X syndrome (FXS).30, 36, 42 FMRP is primarily expressed in neurons and testes, and contains an arginine-glycine-glycine box (RGG box) domain, two K-homology domains, a nuclear localization signal (NLS), and a nuclear export signal (NES). Additionally, FMRP uses its NLS and NES to shuttle between the nucleus and cytoplasm, acting to transport mRNAs throughout the cell.43–45 The role of FMRP in translation regulation, and other processes, remains to be fully elucidated. FMRP normally represses the translation of target mRNAs, with relief of inhibition triggered by synaptic input to enable local protein synthesis.46, 47 However, FMRP has also been demonstrated to promote the translation of human achaete-scute homologue-1 (hASH1) mRNA via binding its 5′-UTR G quadruplex, supporting the idea that G quadruplex structures in mRNA regulation have multiple layers of complexity involving RNA binding proteins.48 In general, the RGG box domain is characterized as having two or three RGG repeats, with five to nine residues separating the repeats, and has been identified in 44 human proteins.49, 50 FMRP has been shown to use its RGG box domain to bind mRNA targets that form G quadruplex structures (Table 1).19, 24, 27–29, 33, 34, 51, 52 However, with the exception of the 5′-UTRs from microtubule associated protein 1B (MAP1B) mRNA,27, 40 the protein phosphatase 2A catalytic subunit mRNA,39 and the hASH1 mRNA 5′-UTR,48 characterization of the FMRP RGG box binding G quadruplex structures has focused on coding region and 3′-UTR locations.19, 24, 28, 29, 34
Table 1.
Primary sequence of the FMRP RGG box domain peptide that targets mRNA G quadruplex structures, with the characteristic RGG repeats underlined. The four asymmetrically dimethylated arginines are the only arginines located within and between the RGG repeats.
| RRGDGRRRGGGGRGQGGRGRGGGFKGNDDHSR |
We are expanding these studies to analyze neuronal mRNAs that have the potential to form G quadruplexes in their 5′-UTR and be regulated by FMRP. Among these is the survival motor neuron domain containing 1 mRNA (SMNDC1, also known as survival of motor neuron-related splicing factor 30, SPF30), which has been predicted by bioinformatics to form a G quadruplex structure located at position 163 in its 333 nucleotide (nt) 5′-UTR (Figure 1A, B).1, 53 Furthermore, from the 12 predicted mature SMNDC1 mRNA transcripts, four contain this predicted G quadruplex forming sequence at relatively similar locations near the beginning of the 5′-UTR.54 It has been proposed that these locations near the 5′ end of the mature transcript 5′UTR suggest a role in translation initiation.1 Despite evidence that the 5′-UTR G quadruplex inhibits translation via reporter gene assays, the translation initiation activity of the structure in the 5′-UTR is more consistent with the FXS phenotype since the absence of FMRP typically results in overexpression of target proteins.
Figure 1.
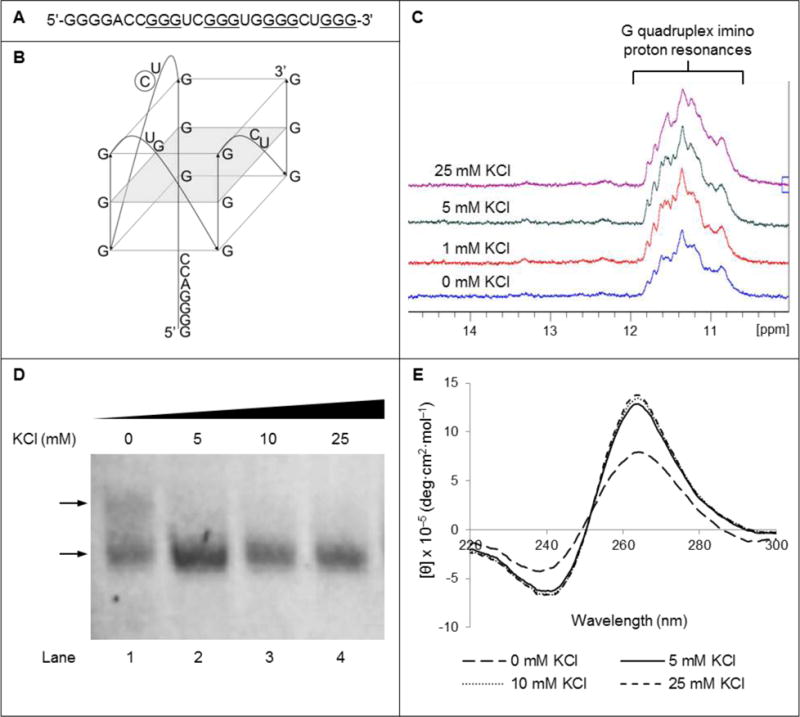
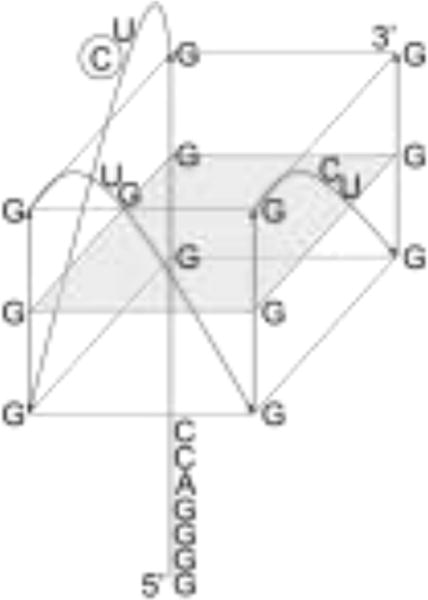
(A) Sequence of the 25 nt segment from the SMNDC1 mRNA 5′-UTR predicted to form the G quadruplex structure. Based on QGRS mapper analysis,53 the guanines predicted to be involved in the most stable G quadruplex structure are underlined. (B) Proposed parallel, intramolecular G quadruplex structure formed by the 25 nt fragment from the 5′ UTR of SMNDC1 mRNA. The intramolecular and parallel nature of the given structure was based on our experimental evidence while the guanines incorporated in our model are based on the single highest G score of 42 from QGRS mapper analysis,53 thus predicted to be the most stable G quadruplex. Seven additional G quadruplex conformers were predicted with G scores ranging from 39 to 41, indicating similar stability. Broad NMR resonances at low KCl concentration may indicate a sample population containing multiple conformers, some of which are predicted to incorporate the 5′ G tract while others exclude it. Upon titration of K+, increased resonance sharpness in the NMR spectra would indicate stabilization of a single conformation, which we believe would be the G quadruplex shown in our model, which excludes the 5′ G tract and incorporates the 3′ G tract. The naturally occurring cytidine at position 12 substituted by the PC fluorescent analog is circled. (C) Imino proton resonance region of the 1D 1H NMR spectra of 230 μM SMNDC1 mRNA in 10 mM cacodylic acid, pH 6.5, 40°C, in the presence of increasing KCl concentrations. (D) EMSA of 10 μM SMNDC1 mRNA using 20% native PAGE in the presence of varying KCl concentrations from 0 mM (lane 1) to 25 mM (lane 4). Next to lane 1, the lower arrow indicates the dominant conformation while the upper arrow indicates the minor conformation. Gel was visualized via UV shadowing at 254 nm. (E) CD spectra of 10 μM SMNDC1 RNA in 10 mM cacodylic acid, pH 6.5, at 25°C, in the presence of increasing KCl concentrations.
SMNDC1 mRNA has been shown to be constitutively transcribed in skeletal muscle, as well as in fetal and adult brain and spinal cord.55 Furthermore, SMNDC1 mRNA has been identified as a potential FMRP target via PAR-CLIP analysis.56 The corresponding SMNDC1 protein has been implicated in pre-mRNA splicing and is critical for spliceosome assembly in the nucleus.55, 57–60 A typical outcome of FXS is that proteins are overexpressed if their corresponding mRNAs are a target of FMRP. Depletion of FMRP associated with FXS could affect the levels of SMNDC1 protein, thus also affect splicing, via dis-regulation of SMNDC1 mRNA translation. SMNDC1 overexpression has also been implicated in apoptosis, thus if SMNDC1 mRNA is a target of FMRP then this outcome could supplement the current knowledge of FXS etiology.55 SMNDC1 is a partial paralog of the survival motor neuron protein (SMN), with both proteins having seemingly similar functions.55, 57 Low levels of SMN caused by deletions or mutations in the smn1 gene leads to the spinal muscular atrophy neuromuscular disease.61, 62 Being overshadowed by SMN in spinal muscular atrophy, the biological function of SMNDC1 and any effects from its synthesis dis-regulation have not been fully elucidated.
In this study, we have shown by biophysical and biochemical methods that a guanine-rich 25 nt stretch within the SMNDC1 mRNA 5′-UTR folds into a stable G quadruplex structure which is recognized with high affinity and specificity by the full-length recombinant FMRP ISO1 as well as by endogenous FMRP in mouse brain lysates. Altogether, the results of this study show for the first time that FMRP interacts with the SMNDC1 mRNA 5′-UTR, via recognition of a G quadruplex structure by its RGG box domain. Our findings expand the short list of 5′-UTR G quadruplexes whose binding by the RGG box domains have been characterized. While it is postulated that FMRP interacts with the microRNA (miRNA) pathway to regulate the translation of specific mRNAs via direct interactions with 3′-UTR sequences, the mechanism(s) by which it regulates translation via interactions within the mRNA’s 5′-UTR remains largely unknown and we show that binding can occur without mediation. It is expected that the results of this study will spur further research into elucidating such mechanism(s).
MATERIALS AND METHODS
RNA oligonucleotides
The synthetic DNA template, based on the GenBank BC039110.11 SMNDC1 mRNA sequence (Figure 1A), and primer were purchased from TriLink Biotechnologies, Inc. The mutant DNA template was also purchased from TriLink Biotechnologies, Inc, corresponding to the RNA in which the G quadruplex structure was abolished (Supplemental Figure 1A). Synthetic templates were used to in vitro transcribe RNA oligonucleotides via T7 RNA polymerase produced in-house.63 The RNA oligonucleotides were purified by 20% 8 M urea denaturing 19:1 PAGE, electrophoretic elution of the excised target band, followed by extensive dialysis in 10 mM cacodylic acid, pH 6.5.
The fluorescent labeled SMNDC1_12PC RNA 25mer was designed with the fluorescent cytosine analog, pyrrolo C (PC), in place of the cytidine at position 12 (Figure 1B) and chemically synthesized by GE Healthcare. The biotinylated SMNDC1 RNA was synthesized by Dharmacon, Inc.
Annealed RNA samples were incubated at 95°C for 5 min, followed by slow cooling to room temperature for 15 min, in 10 mM cacodylic acid, pH 6.5, and in the presence or absence KCl concentrations.
Peptide synthesis
The FMRP RGG box peptide (Table 1) and the hepatitis C virus (HCV) core peptide (combined sequences of amino acids 2–23 and 38–74)24, 64 were chemically synthesized and purified by the Peptide Synthesis Unit at the University of Pittsburgh, Center for Biotechnology and Bioengineering.
1H NMR spectroscopy
The 1D 1H NMR spectra were acquired at 40°C using a 500 MHz Bruker AVANCE spectrometer to analyze a 230 μM SMNDC1 RNA sample annealed in a 90% H2O/10% D2O ratio in 10 mM cacodylic acid, pH 6.5 solution. G quadruplex formation was observed by titrating KCl from a 2 M stock, equilibrating for 20 min, and monitoring the imino proton resonance region (10–12 ppm). Water suppression was accomplished via the Watergate pulse sequence.65
Circular Dichroism (CD) spectroscopy
The CD spectra were collected using a Jasco J-810 spectropolarimeter at 25°C, with scans from 180 nm to 400 nm, using a 200 μL quartz cuvette (Starna Cells) with a 1 mm path-length. Each set of conditions was scanned three times with a 1 sec response time and a 2 nm bandwidth. All spectra were corrected for solvent contributions and dilutions.
For the K+-dependent G quadruplex formation analysis, a 10 μM SMNDC1 RNA sample was annealed in the absence of KCl, and KCl was titrated from a 2 M stock solution and allowed to equilibrate for 10 min at 25°C.
UV spectroscopy thermal denaturation
The UV spectroscopy thermal denaturation curves were obtained using a Varian Cary 3E spectrophotometer equipped with a Peltier temperature control cell. Experiments were performed in a 200 μL quartz cuvette (Starna Cells) with a 10 mm path length, up to a final volume of 200 μL, and changes in absorbance at 295 nm were monitored. Sample and reference cells were covered with 200 μL mineral oil to prevent evaporation. The RNA samples were heated during analysis from 25°C to 95°C at a rate of 0.2°C/min, recording absorbance every 1°C, monitoring the absorbance changes at 295 nm, wavelength sensitive to G quadruplex dissociation.66
To determine if an intramolecular or intermolecular G quadruplex structure is formed the melting temperature (Tm) was determined at RNA concentrations ranging from 3 μM to 25 μM, in the presence of 5 mM KCl. The Tm is indicated by the midpoint of the hypochromic transition during the thermal denaturation (Figure 2A), and the Tm for an intermolecular species containing n number of strands is dependent on the total RNA concentration (cT), the gas constant (R), and the van’t Hoff thermodynamic parameters (ΔH°vH and ΔS°vH) (Equation 1).67
Figure 2.
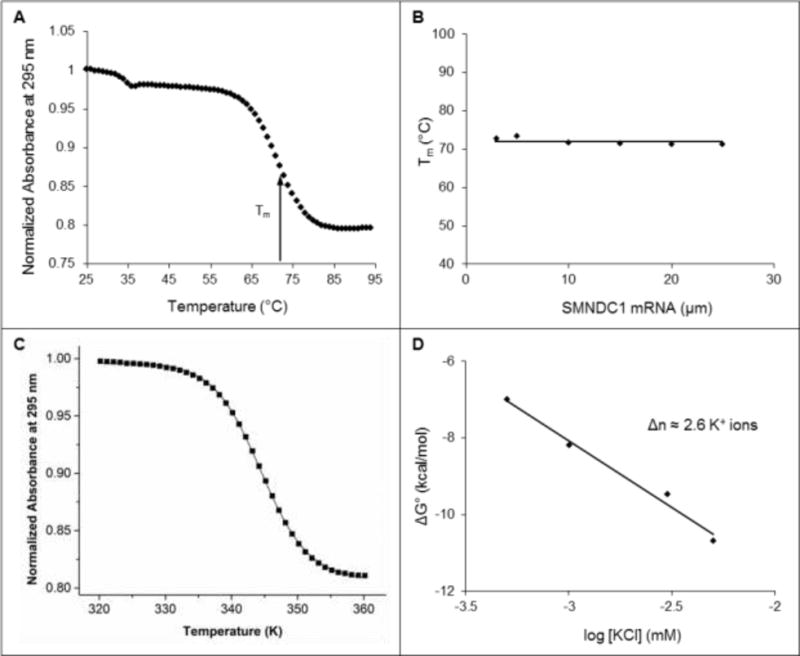
(A) UV spectroscopy thermal denaturation profile of 10 μM SMNDC1 mRNA in 10 mM cacodylic acid, pH 6.5, 5 mM KCl. The melting temperature (Tm) of the main hypochromic transition is indicated by the arrow. (B) Melting temperature Tm as a function of the SMNDC1 RNA concentration, in 10 mM cacodylic acid, pH 6.5, 5 mM KCl. (C) Fit of the main hypochromic transition of 10 μM SMNDC1 mRNA in 10 mM cacodylic acid, pH 6.5, 5 mM KCl to Equation 3 (Materials and Methods) that assumes a two-state model. (D) Free energy of folding for the SMNDC1 G quadruplex as a function of KCl concentration.
| (1) |
However, the Tm of an intramolecular G quadruplex for which n = 1 is independent of cT, thus, equation 1 reduces to Equation 2:
| (2) |
The hypochromic transition of the UV thermal spectrum was fit to determine the thermodynamic parameters of folding, assuming an independent two-state model, where AU and AF represent the absorbance of the unfolded and folded G quadruplex, respectively, and R is the universal gas constant (Figure 2C) (Equation 3).
| (3) |
To determine the number of coordinating K+ ion equivalents, the Tm of the G quadruplex was determined at a fixed RNA concentration (10 μM) and at K+ concentrations ranging from 0.5 to 25 mM, titrated from a 2 M KCl stock. Assuming a simple model for the folded-to-unfolded G quadruplex structure during which Δn K+ ion equivalents are released, Δn can be determined from the negative slope of the plot of ΔG° as a function of the logarithm of K+ ion concentration (Figure 2D) (Equation 4).19, 27
| (4) |
Native gel electrophoresis analysis
The RNA samples were resolved using 20% non-denaturing 19:1 PAGE in 0.5X Tris/borate/EDTA buffer, run at 4°C and 75 V, in a final volume of 15 μL. RNA samples were at a 10 μM concentration, annealed in KCl concentrations ranging from 0 mM to 25 mM. The electromobility shift assay (EMSA) with the FMRP RGG box peptide used 10 μM RNA annealed in 5 mM KCl. The RNA was incubated with FMRP RGG box peptide at increasing ratios and allowed to equilibrate for 20 min at 25°C. The free RNA and RNA:peptide complexes were visualized by UV-shadowing at 254 nm using an AlphaImager HP (AlphaInnotech, Inc.).
Fluorescence spectroscopy
Full-length FMRP ISO1 was expressed and purified using previously described methods.35 The fluorescence spectroscopy experiments using previously described methods19, 24, 27–29, 34, 35, 51 were performed on a J.Y. Horiba Fluoromax-3 equipped fitted with a 150 W ozone-free xenon arclamp and with a variable temperature control sample chamber. Experiments were performed in 150 μL sample final volume in a quartz cuvette (Starna Cells) with a 3 mm path-length. The excitation wavelength was set at 350 nm and the emission spectra were recorded from 400 nm to 530 nm at 25°C, with a bandpass of 5 mm for both the excitation and emission monochromators. FMRP ISO1 or FMRP RGG box peptide was titrated at certain increments into 150 nM SMNDC1_12PC RNA annealed in the presence of 150 mM KCl, incubated for 10 min at 25°C, and then monitored for the steady-state fluorescence emission of the PC reporter. 750 nM of a peptide derived from the HCV core protein or bovine serum albumin (BSA) was added to the RNA sample prior to titrating RGG box peptide or FMRP ISO1, respectively, to prevent non-specific binding. The complex dissociation constant (Kd) was determined by fitting the resultant binding curve of normalized SMNDC1_12PC fluorescence emission intensity at 445 nm as a function of protein concentration using Origin 8.0 software, where IB and IF are the steady-state fluorescence intensities of the bound and free SMNDC1_12PC RNA respectively, [RNA]t is the total concentration of SMNDC1_12PC, [P]t is the total concentration of either RGG box or FMRP ISO1 (Figures 3B, C) (Equation 5).
Figure 3.
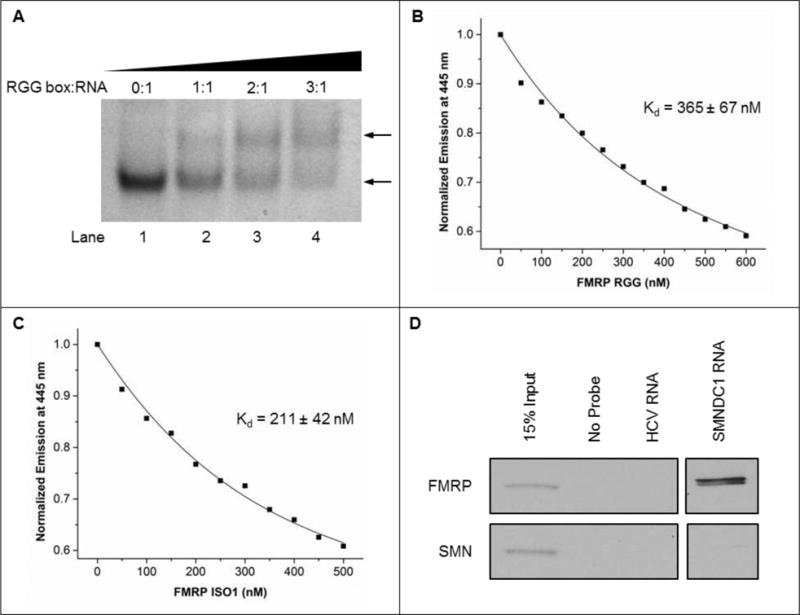
(A) EMSA using 20% native PAGE of 10 μM SMNDC1 mRNA in the presence of 5 mM KCl, with ratios of FMRP RGG box peptide:SMNDC1 mRNA from 0:1 (lane 1) to 3:1 (lane 4). Gels were visualized by UV shadowing at 254 nm. The upper arrow shows the emerging band representing the RGG box:RNA complex, the lower arrow indicates the free RNA band. Representative fluorescence spectroscopy fitted curves of (B) FMRP RGG box peptide and (C) full-length FMRP ISO1 individually titrated into 150 nM SMNDC1_12PC RNA in the presence of 150 mM KCl, 10 mM cacodylic acid, pH 6.5, at 25°C with 5:1 BSA protein to screen non-specific interactions. (D) Binding of 5′ biotinylated SMNDC1 mRNA by FMRP after incubation with E17 mouse brain lysates. RNA probes were precipitated and potential co-purified FMRP and SMN proteins were assessed by immunoblot. Precipitated SMNDC1 mRNA was simultaneously exposed to immunoblot by FMRP and SMN with all other samples. However, the gel contained mRNA targets unrelated to this study, thus the SMNDC1 samples are shown in a separate window.
| (5) |
The experiment was performed in triplicate, fitted individually, and the reported error is the standard uncertainty of the data from the best-fit theoretical curves since they are larger than the standard uncertainty of the measurements.
SMNDC1 RNA-based affinity pull-down assay
The biotinylated SMNDC1 RNA probe was denatured at 95°C for 5 min and cooled at room temperature for 15 minutes. The negative control biotinylated HCV RNA probe24, 68, 69 (5′-GCC AGC CCC CUG AUG GGG GCG ACA CUC CAC CAU GAA UCA CUC CCC UG-3′) was denatured at 95°C for 3 min and cooled quickly in a dry ice-ethanol bath. 5 μM of probe was incubated with E17 mouse brain lysate for 20 min at room temperature and NeutrAdivin agarose (Thermo Scientific, Inc.) pre-blocked with BSA was used to precipitate the probes. After extensive washing, proteins were detected by immunoblot against FMRP (1:1250, Sigma) and negative control SMN (1:500, BD Transduction Laboratories & San Jose). All experiments were performed in compliance with relevant laws and institutional guidelines. Emory University IACUC approved the experiments.
RESULTS AND DISCUSSION
Structural and thermodynamic characterization of the SMNDC1 mRNA G quadruplex
The G quadruplex predicting software QGRS Mapper53 predicted the formation of a G quadruplex structure within the 5′-UTR of the SMNDC1 mRNA (BC039110.1)1 (Figure 1A). To experimentally confirm the existence of this G quadruplex structure, we have produced a 25 nt fragment predicted to fold into the G quadruplex structure (Figure 1B) by in vitro transcription from a synthetic DNA template.
We first used one-dimensional (1D) 1H NMR spectroscopy to analyze G quadruplex structure formation within SMNDC1 mRNA in the presence of increasing KCl concentrations, monitoring the imino proton resonance region. Even in the absence of KCl, imino proton resonances were present in the 10–12 ppm region, which correspond to guanine imino protons involved in G quartet formation through Hoogsteen base pairs, indicating that the 25 nt SMNDC1 mRNA fragment forms a G quadruplex structure (Figure 1C). While K+ ions are required for DNA G quadruplex formation, RNA sequences are able to fold into G quadruplexes even in their absence.70 A few broad resonances are present in the 12–14.5 ppm region, where imino protons involved in Watson-Crick base pairs resonate, indicating the formation of a minor alternate structure.
The increase in resonance intensities in the signature 10–12 ppm range while increasing K+ concentrations indicates G quadruplex structure stabilization by K+ ions, while the broadness of these resonances suggests the existence of alternative structure conformers. To further explore this possibility, we have employed native gel electrophoresis using a constant RNA concentration while increasing KCl concentrations from 0 mM to 25 mM (Figure 1D). Two RNA bands are present in the absence of KCl (Figure 1D, lane 1), corresponding to at least two distinct conformations. At KCl concentrations of 5 mM and greater, one conformation becomes predominant, evident by the increase in the intensity of the lower band with the concomitant decrease of the intensity of the upper band (Figure 1D, lanes 2–4). These findings indicate that though multiple conformations are possible and two are demonstrated to exist, in the presence of K+ ions, the 25 nt SMNDC1 mRNA fragment adopts only one predominant structure.
Next, we employed CD spectroscopy to analyze the fold of the SMNDC1 mRNA G quadruplex.16 Positive and negative bands were observed at 265 nm and 240 nm, respectively, even in the absence of K+ ions, yielding the characteristic CD spectrum of a parallel G quadruplex structure (Figure 1E).16 This parallel strand orientation is also consistent with previous reports for various intramolecular RNA G quadruplexes.4–6, 9, 19, 20, 24, 27–29 The band intensities increased in the presence of 5 mM KCl, supporting the conclusion that this G quadruplex is stabilized by K+ and consistent with the 1H NMR spectroscopy and native gel electrophoresis results.
UV spectroscopy thermal denaturation analysis was performed to determine the overall stability as well as the intramolecular or intermolecular composition of the SMNDC1 mRNA G quadruplex structure, by monitoring the change in absorption at 295 nm66 while increasing the temperature in the range 25°C to 95°C. A major hypochromic transition is present, with a Tm of ~71°C, which we attribute to the unfolding of the SMNDC1 mRNA predominant G quadruplex structure (Figure 2A). Additionally, a second minor transition with a Tm of ~34°C is present, likely corresponding to the unfolding of the minor alternate structure formed by SMNDC1 mRNA. To determine if the major G quadruplex structure formed by SMNDC1 mRNA is intramolecular or intermolecular we have measured the Tm values as a function of the RNA concentration (Materials and Methods, Equations 1–2). Tm is independent of the RNA concentration for the major hypochromic transition (Figure 2B), indicating that the dominant SMNDC1 mRNA G quadruplex structure is intramolecular (Materials and Methods, Equation 2).67 Conversely, the melting temperature of the minor hypochromic transition increases with the RNA concentration (data not shown), indicative of an intermolecular species (Materials and Methods, Equation 1), which we therefore attribute to quadruplex stacking.
After determining that the main G quadruplex structure formed by SMNDC1 is intramolecular, we fit the hypochromic transition corresponding to its unfolding using an independent two-state model to determine the thermodynamic parameters of G quadruplex structure formation (Materials and Methods, Equation 3) (Figure 2C). The structure formation was enthalpically-driven as demonstrated by the enthalpy value of −64.0 ± 0.1 kcal/mol and free energy value of −8.6 ± 0.1 kcal/mol for 10 μM RNA in 5 mM KCl. Considering that the enthalpy of formation of a single G quartet plane in an intramolecular G quadruplex ranges from −18 kcal/mol to −25 kcal/mol under similar conditions,53, 67 these findings demonstrate that the determined enthalpy of folding is consistent with the predicted three G quartet planes in the SMNDC1 mRNA G quadruplex (Figure 1B).
To obtain additional information about the predominant SMNDC1 mRNA G quadruplex, UV spectroscopy thermal denaturation experiments were performed at a fixed RNA concentration and variable KCl concentration to determine the number of coordinating K+ ion equivalents coordinating the structure.19, 27 Assuming a simple model for the folded-to-unfolded G quadruplex structure during which Δn K+ ion equivalents are released, Δn can be determined from the negative slope of the plot of ΔG° as a function of the logarithm of K+ ion concentration (Materials and Methods, Equation 4) (Figure 2D).19, 27 These analyses reveal that the SMNDC1 G quadruplex coordinates ~2.6 K+ ion equivalents, consistent with the proposed three-plane structure.
In summary, by using various biophysical methods, we characterized the structural and thermodynamic properties of the 25 nt fragment from the SMNDC1 mRNA 5′-UTR. Our results demonstrate that this mRNA fragment adopts a dominant G quadruplex structure that is intramolecular, has a parallel strand orientation, comprised of three G quartet planes, coordinates ~2.6 K+ ions, and has high stability at the physiologic temperature (Figure 1B).
FMRP interactions with the SMNDC1 mRNA G quadruplex
FMRP is an RNA binding protein primarily found in neurons, where SMDNC1 expression has been demonstrated.55 Though the regulatory mechanism is not well understood, FMRP is proposed to use its NLS and NES to bind specific neuronal mRNA targets in the nucleus and shuttle them throughout the cell, and regulate translation in response to synaptic input.33, 36, 71 The activity of FMRP is critical for humans since its absence leads to FXS, the most common inheritable intellectual disability.36, 42 The RGG box domain within FMRP has been shown to bind G quadruplex forming mRNAs with high affinity and specificity, including those within the 5′-UTR of other neuronal targets and, like SMNDC1 mRNA, those devoid of a stem structure.19, 24, 27–29, 33–35, 37, 51 The G quadruplex itself has also been shown to regulate mRNA translation1–8, 14, 30 and it has been proposed that FMRP binding the G quadruplex structure enables an additional level of translation regulation.27, 32, 41, 72–74
Since SMNDC1 mRNA has been identified as a potential FMRP target via PAR-CLIP analysis56 we investigated if the 5′-UTR SMNDC1 mRNA G quadruplex is bound by FMRP. We used a 32mer peptide consisting of the FMRP RGG box domain (Table 1) that has been previously shown to bind G quadruplex RNA targets with comparable affinity and specificity to the FMRP isoforms whose binding have been similarly characterized.19, 24, 27–29, 34, 51 Native EMSA PAGE performed at constant RNA and KCl concentrations, with 0:1, 1:1, 2:1, and 3:1 FMRP RGG box peptide:SMNDC1 mRNA ratios, demonstrated the formation of a higher molecular weight complex via the emergence of an upper band in the presence of the FMRP RGG box peptide, with the concomitant decrease in intensity of the lower band that corresponds to the free RNA (Figure 3A, compare lanes 1 vs. 2–4). To demonstrate that the G quadruplex structure formed in the SMNDC1 mRNA segment is specifically targeted by FMRP we synthesized a mutant sequence having strategic G-to-C substitutions designed to abolish the G quadruplex structure (Supplemental Figure 1A). 1D 1H NMR spectroscopy experiments were performed to probe the structure of the mutant construct. Previously observed resonances in the signature 10–12 ppm G quadruplex imino proton region were replaced by resonances in the 12–14 ppm region corresponding to Watson-Crick base pairs, thus demonstrating that the G quadruplex formed in the SMNDC1 wild-type sequence is abolished in the mutant construct and that a hairpin structure is dominant in the mutant construct even at increasing KCl concentrations (Supplemental Figure 1B). Native EMSA PAGE was then performed using the mutant RNA construct, and as FMRP RGG peptide concentration was increased a single band corresponding to the free RNA remained (Supplemental Figure 1C), demonstrating that elimination of the G quadruplex structure in the mutant construct ablates binding. Altogether, these findings further support the conclusion that the RGG box domain of FMRP specifically binds to the G quadruplex structure within the 5′-UTR of SMNDC1 mRNA.
Next, we employed steady-state fluorescence spectroscopy to obtain quantitative information regarding the interactions of the FMRP RGG box peptide and of the full-length FMRP ISO1 protein with the 5′-UTR SMNDC1 mRNA G quadruplex. We designed SMNDC1_12PC RNA, which was formed by the labeling of the 25 nt G quadruplex forming SMNDC1 mRNA fragment with the fluorescent cytosine analog PC in place of the cytidine at position 12 (Figure 1B, circled C). The fluorescence emission of PC is sensitive to changes in its local microenvironment, including those induced by protein binding.75 Binding curves were obtained by titrating increasing concentrations of either the RGG box peptide or FMRP ISO1 into a fixed RNA concentration. To ensure that the observed PC quenching is not due to non-specific protein aggregation, similar control experiments were performed by titrating BSA and, as expected, the steady state fluorescence of the PC reporter did not change (Supplemental Figure 1D). All experiments were performed in triplicate and the Kd for either the RGG peptide or FMRP ISO1 were determined by fitting the binding curves with Equation 5 (Materials and Methods) (Figures 3B, C). Though the full length FMRP ISO1 binds with greater affinity (Kd = 211 ± 42 nM) than the RGG box peptide (Kd = 365 ± 67 nM), both values are consistent with other nM affinities demonstrated by either the FMRP RGG box peptide or various FMRP isoforms binding to other G quadruplex forming RNAs.19, 24, 27–29, 34, 35, 51 Additionally, though the affinities between FMRP ISO1 and the RGG box peptide are statistically different, they correspond to comparable free energies of binding of −9.1 ± 0.2 kcal/mol and −8.8 ± 0.2 kcal/mol respectively. Furthermore, these free energy values are consistent with the recently characterized affinity of FMRP for two G quadruplex structures from the 3′-UTR of Shank1 as well as other mRNAs.19, 24, 34, 35
Lastly, using a biotin-RNA pull-down assay, we demonstrated binding of endogenous brain FMRP to the SMNDC1 mRNA G quadruplex.24 Biotin-labeled SMNDC1 RNA probes were incubated with E17 mouse brain lysates followed by precipitation via NeutrAdivin agarose. Subsequent Western blot analysis demonstrated that the SMNDC1 RNA probe is bound by endogenous FMRP but not the negative control, SMN (Figure 3D, Supplemental Figure 2), thus indicating SMNDC1 mRNA is specific for FMRP and devoid of non-specific interactions with other proteins.
G quadruplex structures have been identified in the 5′-UTR of a number of genes1 and, for the few that have been characterized, their importance as translation regulators has been demonstrated, either inhibiting or promoting translation.10–13 Our results expand upon this small list and show for the first time that SMNDC1 mRNA forms a G quadruplex structure in its 5′-UTR, thus suggesting a possible translation regulation function for this structure. FMRP is proposed to be a translation repressor of specific mRNA targets at distal sites and, until responding to synaptic input, local protein translation will be triggered. The exact mechanisms by which FMRP exerts its translation regulator function are not known. However, it has been shown that, for mRNA targets that are bound by FMRP in their 3′-UTR, this might be mediated by interactions with the miRNA pathway.9, 76, 77 Though it has been shown that translation increased in the absence of FMRP for two mRNAs that each have at least one potential G quadruplex structure in their 5′-UTR,39, 40 only one study analyzed the FMRP RGG box directly binding the G quadruplex structure.27 Our findings that FMRP directly interacts with a G quadruplex in the SMNDC1 5′-UTR expands this short list. In addition, FMRP, having demonstrated neuronal expression, could play an indirect role in splicing by modulating the levels of the SMNDC1 protein, a critical component of the spliceosome. The SMNDC1 protein has been found in Cajal bodies, implicated in the splicing of mRNA in cells with elevated metabolic activity such as neurons and proliferating cells, and overexpression of SMNDC1 has been associated with apoptosis.55, 59, 78
CONCLUSION
Overall, our results provide further evidence that G quadruplex structures with consistent structural and thermodynamic properties exist in neuronal mRNAs, which was implied by other indirect methods.79 We show for the first time that a G quadruplex structure forms in the 5′-UTR of SMNDC1 mRNA and that this structure is sufficient for recognition by FMRP. This study provides the foundation of potential relationships between levels of proteins, translation activity, and splicing activity that are mediated by G quadruplex mRNAs, FMRP and its RGG box domain, and SMNDC1. These findings merit further investigation of these relationships, further support the role of FMRP in FXS, and suggest that the molecular mechanism of impairment involves the loss of regulation for neuronal mRNA localization and/or translation.
Supplementary Material
Acknowledgments
This work was supported by the President’s Mentorship Fund and College Research Council grants of the University of Pittsburgh at Johnstown to T.L.E., NIH grants 9R15HD078017–03A1 to M.R.M and 1R21NS089080 to G.J.B, and A.M.H., C.N.L., and T.L.E. were supported by the NSF REU and MRI grants CHE-1263279 and CHE-1126465.
Footnotes
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
References
- 1.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. Nucleic acids research. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- 3.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. Nature chemical biology. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shahid R, Bugaut A, Balasubramanian S. Biochemistry. 2010;49:8300–8306. doi: 10.1021/bi100957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez D, Guedin A, Mergny JL, Salles B, Riou JF, Teulade-Fichou MP, Calsou P. Nucleic acids research. 2010;38:7187–7198. doi: 10.1093/nar/gkq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halder K, Wieland M, Hartig JS. Nucleic acids research. 2009;37:6811–6817. doi: 10.1093/nar/gkp696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumari S, Bugaut A, Balasubramanian S. Biochemistry. 2008;47:12664–12669. doi: 10.1021/bi8010797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefanovic S, Bassell GJ, Mihailescu MR. RNA. 2015;21:48–60. doi: 10.1261/rna.046722.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agarwala P, Pandey S, Maiti S. Biochimica et biophysica acta. 2014;1840:3503–3510. doi: 10.1016/j.bbagen.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Agarwala P, Pandey S, Mapa K, Maiti S. Biochemistry. 2013;52:1528–1538. doi: 10.1021/bi301365g. [DOI] [PubMed] [Google Scholar]
- 12.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. J Am Chem Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 13.Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats AC, Vagner S. The Journal of biological chemistry. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wieland M, Hartig JS. Chem Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Neidle S, Balasubramanian S. 2006 [Google Scholar]
- 16.Williamson JR. Annu Rev Biophys Biomol Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 17.Williamson JR, Raghuraman MK, Cech TR. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 18.Didiot MC, Tian Z, Schaeffer C, Subramanian M, Mandel JL, Moine H. Nucleic acids research. 2008;36:4902–4912. doi: 10.1093/nar/gkn472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blice-Baum AC, Mihailescu MR. RNA. 2014;20:103–114. doi: 10.1261/rna.041442.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Nucleic acids research. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hai Y, Cao W, Liu G, Hong SP, Elela SA, Klinck R, Chu J, Xie J. Nucleic acids research. 2008;36:3320–3331. doi: 10.1093/nar/gkn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaudoin JD, Perreault JP. Nucleic acids research. 2013;41:5898–5911. doi: 10.1093/nar/gkt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Gaetano CM, Williams KR, Bassell GJ, Mihailescu MR. RNA biology. 2014;11:1364–1374. doi: 10.1080/15476286.2014.996464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin P, Alisch RS, Warren ST. Nature cell biology. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 26.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Nature neuroscience. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 27.Menon L, Mader SA, Mihailescu MR. RNA. 2008;14:1644–1655. doi: 10.1261/rna.1100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bole M, Menon L, Mihailescu MR. Molecular BioSystems. 2008;4:1212–1219. doi: 10.1039/b812537f. [DOI] [PubMed] [Google Scholar]
- 29.Menon L, Mihailescu MR. Nucleic acids research. 2007;35:5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bugaut A, Balasubramanian S. Nucleic acids research. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashley CT, Sutcliffe JS, Kunst CB, Leiner HA, Eichler EE, Nelson DL, Warren ST. Nature genetics. 1993;4:244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- 32.Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 33.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 34.Evans TL, Blice-Baum AC, Mihailescu MR. Mol Biosyst. 2012;8:642–649. doi: 10.1039/c1mb05322a. [DOI] [PubMed] [Google Scholar]
- 35.Evans TL, Mihailescu MR. Protein Expr Purif. 2010;74:242–247. doi: 10.1016/j.pep.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Donnell WT, Warren ST. Annual review of neuroscience. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- 37.Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 38.Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 39.Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel JL, Bardoni B. Hum Mol Genet. 2005;14:835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- 40.Lu R, Wang H, Liang Z, Ku L, O’Donnell WT, Li W, Warren ST, Feng Y. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westmark CJ, Malter JS. PLoS Biol. 2007;5:e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crawford DC, Acuna JM, Sherman SL. Genet Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim M, Bellini M, Ceman S. Molecular and cellular biology. 2009;29:214–228. doi: 10.1128/MCB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sittler A, Devys D, Weber C, Mandel JL. Hum Mol Genet. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 45.Tamanini F, Bontekoe C, Bakker CE, van Unen L, Anar B, Willemsen R, Yoshida M, Galjaard H, Oostra BA, Hoogeveen AT. Hum Mol Genet. 1999;8:863–869. doi: 10.1093/hmg/8.5.863. [DOI] [PubMed] [Google Scholar]
- 46.Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. J Neurosci. 2012;32:2582–2587. doi: 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fahling M, Mrowka R, Steege A, Kirschner KM, Benko E, Forstera B, Persson PB, Thiele BJ, Meier JC, Scholz H. The Journal of biological chemistry. 2009;284:4255–4266. doi: 10.1074/jbc.M807354200. [DOI] [PubMed] [Google Scholar]
- 49.Corley SM, Gready JE. Bioinformatics and Biology Insights. 2008;2:389–406. doi: 10.4137/bbi.s1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiledjian M, Dreyfuss G. The EMBO journal. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zanotti KJ, Lackey PE, Evans GL, Mihailescu MR. Biochemistry. 2006;45:8319–8330. doi: 10.1021/bi060209a. [DOI] [PubMed] [Google Scholar]
- 52.Penagarikano O, Mulle JG, Warren ST. Annual review of genomics and human genetics. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- 53.Kikin O, D’Antonio L, Bagga PS. Nucleic acids research. 2006;34:W676–682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim P, Kim N, Lee Y, Kim B, Shin Y, Lee S. Nucleic acids research. 2005;33:D75–79. doi: 10.1093/nar/gki118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talbot K, Miguel-Aliaga I, Mohaghegh P, Ponting CP, Davies KE. Hum Mol Genet. 1998;7:2149–2156. doi: 10.1093/hmg/7.13.2149. [DOI] [PubMed] [Google Scholar]
- 56.Ascano M, Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tuschl T. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meister G, Hannus S, Plottner O, Baars T, Hartmann E, Fakan S, Laggerbauer B, Fischer U. The EMBO journal. 2001;20:2304–2314. doi: 10.1093/emboj/20.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Nature genetics. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 59.Rappsilber J, Ajuh P, Lamond AI, Mann M. The Journal of biological chemistry. 2001;276:31142–31150. doi: 10.1074/jbc.M103620200. [DOI] [PubMed] [Google Scholar]
- 60.Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M. Nat Struct Mol Biol. 2011;18:1414–1420. doi: 10.1038/nsmb.2185. [DOI] [PubMed] [Google Scholar]
- 61.Fallini C, Bassell GJ, Rossoll W. Brain research. 2012;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 63.Milligan JF, Uhlenbeck OC. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 64.Ivanyi-Nagy R, Kanevsky I, Gabus C, Lavergne JP, Ficheux D, Penin F, Fosse P, Darlix JL. Nucleic acids research. 2006;34:2618–2633. doi: 10.1093/nar/gkl240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piotto M, Saudek V, Sklenar V. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 66.Mergny JL, Phan AT, Lacroix L. FEBS letters. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 67.Hardin CC, Perry AG, White K. Biopolymers. 2000;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 68.Williams KR, McAninch DS, Stefanovic S, Xing L, Allen M, Li W, Feng Y, Mihailescu MR, Bassell GJ. Molecular biology of the cell. 2016;27:518–534. doi: 10.1091/mbc.E15-07-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stefanovic S, DeMarco BA, Underwood A, Williams KR, Bassell GJ, Mihailescu MR. Mol Biosyst. 2015;11:3222–3230. doi: 10.1039/c5mb00423c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joachimi A, Benz A, Hartig JS. Bioorg Med Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 71.Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H, Bardoni B. PLoS Biol. 2009;7:e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sung YJ, Dolzhanskaya N, Nolin SL, Brown T, Currie JR, Denman RB. The Journal of biological chemistry. 2003;278:15669–15678. doi: 10.1074/jbc.M211117200. [DOI] [PubMed] [Google Scholar]
- 73.Zalfa F, Bagni C. Cell Mol Life Sci. 2005;62:251–252. doi: 10.1007/s00018-004-4521-1. [DOI] [PubMed] [Google Scholar]
- 74.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 75.Tinsley RA, Walter NG. RNA. 2006;12:522–529. doi: 10.1261/rna.2165806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Molecular cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Little JT, Jurica MS. The Journal of biological chemistry. 2008;283:8145–8152. doi: 10.1074/jbc.M707984200. [DOI] [PubMed] [Google Scholar]
- 79.Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


