Significance
High-risk neuroblastoma remains a therapeutic challenge, and adjuvant retinoic acid (RA) treatment shows poor efficacy. We demonstrate that combined treatment with 5-Aza-deoxycytidine (AZA) and RA impedes neuroblastoma growth and induces a transcriptional response characterized by high levels of the HIF2α transcription factor. This approach targets high-risk neuroblastoma that responds poorly to RA. In addition, genome-wide analysis of treated tumors and patient data links HIF2α to tumor suppression, which is supported by a HIF2α-specific small molecule inhibitor-mediated block of the tumor response to AZA+RA treatment.
Keywords: neuroblastoma, differentiation, retinoic acid, 5-Aza-dC, HIF2a
Abstract
Neuroblastoma is a pediatric cancer characterized by variable outcomes ranging from spontaneous regression to life-threatening progression. High-risk neuroblastoma patients receive myeloablative chemotherapy with hematopoietic stem-cell transplant followed by adjuvant retinoid differentiation treatment. However, the overall survival remains low; hence, there is an urgent need for alternative therapeutic approaches. One feature of high-risk neuroblastoma is the high level of DNA methylation of putative tumor suppressors. Combining the reversibility of DNA methylation with the differentiation-promoting activity of retinoic acid (RA) could provide an alternative strategy to treat high-risk neuroblastoma. Here we show that treatment with the DNA-demethylating drug 5-Aza-deoxycytidine (AZA) restores high-risk neuroblastoma sensitivity to RA. Combined systemic distribution of AZA and RA impedes tumor growth and prolongs survival. Genome-wide analysis of treated tumors reveals that this combined treatment rapidly induces a HIF2α-associated hypoxia-like transcriptional response followed by an increase in neuronal gene expression and a decrease in cell-cycle gene expression. A small-molecule inhibitor of HIF2α activity diminishes the tumor response to AZA+RA treatment, indicating that the increase in HIF2α levels is a key component in tumor response to AZA+RA. The link between increased HIF2α levels and inhibited tumor growth is reflected in large neuroblastoma patient datasets. Therein, high levels of HIF2α, but not HIF1α, significantly correlate with expression of neuronal differentiation genes and better prognosis but negatively correlate with key features of high-risk tumors, such as MYCN amplification. Thus, contrary to previous studies, our findings indicate an unanticipated tumor-suppressive role for HIF2α in neuroblastoma.
Neuroblastoma (NB) arises from precursor cells of the sympathetic nervous system and is the most frequent extracranial solid childhood cancer (1). It exhibits a high degree of clinical heterogeneity ranging from spontaneous regression to swift progression accompanied by a fatal outcome despite intense treatment intervention (1). High-risk tumors are characterized by MYCN amplification (MYCNamp) and chromosomal aberrations, such as hemizygous loss of the distal part of chromosome 1p (1p36) (1). Several studies have also reported that high levels of the hypoxia-induced protein HIF2α is a hallmark of aggressive NB (2–5), implying that it acts as an oncogene in NB. An additional feature of high-risk NB is high levels of DNA methylation of promoters of putative tumor suppressors (6). Although several studies have shown that this CpG island (CGI) methylator phenotype (CIMP) can serve as a predictive factor for poor prognosis, little is known if and how it actually contributes to the biology of NB (7–9). Retinoids are strong drivers of neuronal differentiation and are used as adjuvant therapy for high-risk NB patients, after myeloablative chemotherapy with hematopoietic stem-cell transplant (10). However, the efficacy is limited, and the outcome for these patients often remains fatal (10); thus, the development of alternative approaches is of vital importance. The poor response of high-risk NB to retinoic acid (RA) treatment (10) together with a highly methylated genome provide a conceptual basis for a combined therapy, wherein DNA demethylation is combined with the differentiation-promoting effects of RA. In this study, we show that treatment with the demethylating agent 5-Aza-deoxycytidine (AZA) can restore RA responsiveness in high-risk NB. Thus, growth of xenografted NB tumors, which fail to respond to RA treatment, can be thwarted with a combined systemic delivery of two FDA-approved drugs (AZA+RA) (10, 11). We show that the AZA+RA-dependent inhibition of tumor growth rapidly induces a HIF2α-associated hypoxia-like response in the treated xenografts. Upon simultaneous delivery of the HIF2α-specific small-molecule inhibitor PT2385 (12), the tumor response to AZA+RA treatment is diminished. This is paralleled by our genome-wide analysis of large NB patient datasets, which imply a hitherto unanticipated role for HIF2α as a tumor suppressor in NB. In addition, by combining genome-wide methylome and transcriptome analysis, we show that even though AZA treatment efficiently leads to global demethylation, it is a poor activator of gene expression.
Results
AZA Restores Sensitivity to RA Treatment in High-Risk 1p36− NB Cells.
Several NB cell lines respond to RA treatment with the acquisition of neuronal features such as neurite extension, expression of neuronal markers, and cessation of proliferation (1). We treated 1p36+ SK-N-SH, SH-SY5Y, and SK-N-BE (1) NB cell lines for 8 d with 10 μM RA, resulting in an increase of expression of the neuronal marker TUJ1 in combination with a pronounced neuronal morphology in all cell lines after RA treatment (Fig. 1 A–B′′). Cell counting over 10 d of RA treatment showed a marked loss of cellular growth (Fig. S1 A–C). This was underscored by cell-cycle analysis combining propidium iodide (PI) with BrdU staining, which showed that there was a significant decrease of cells in the S phase upon RA treatment (Fig. 1 C–C′′). To investigate the responsiveness to RA delivery in vivo, we transplanted SK-N-SH cells to the flanks of immunodeficient nude mice and treated a cohort with RA, leading to decreased tumor growth (Fig. 1D). In contrast to this response, RA treatment of 1p36− NB cell lines without (SK-N-AS) or with (LAN-1 and CHP-212) MYCN amplification did not efficiently promote the acquisition of neuronal features (Fig. 1 E–F′′).
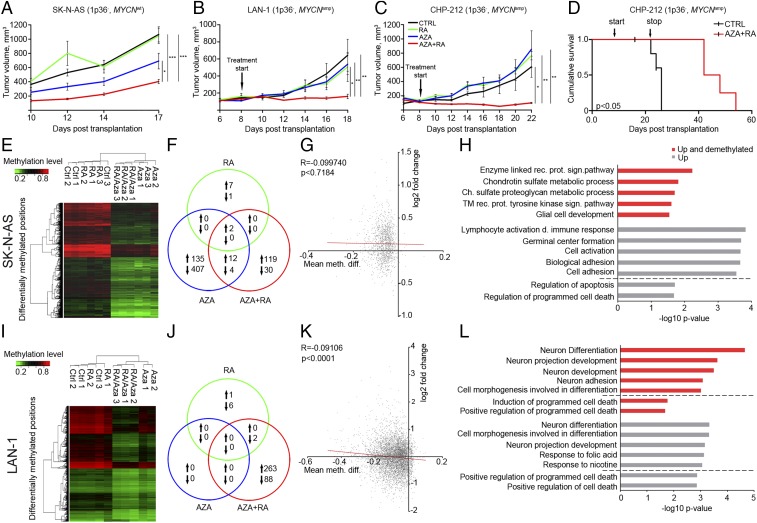
Fig. 1.
Pretreatment with AZA restores response to RA treatment in 1p36− NB cells. (A–C) Treatment of SK-N-BE (1) (A–C), SK-N-SH (A′–C′), and SH-SY5Y (A′′–C′′) NB cells for 8 d with 10 μM RA promoted acquisition of neuronal morphology with increased expression of TUJ1 (A and B) and a reduction of cells in the S phase (C). (D) In mice xenografted with SK-N-SH cells, daily treatment with RA perturbed tumor growth. (E–I′′) 10 μM RA or 2 μM AZA treatment alone in the 1p36− NB cells SK-N-AS (E–G), LAN-1 (E′–G′), and CHP-212 (E′′–G′′) did not promote neuronal features (E–G′′), however pretreatment with AZA for 4 d followed by combined AZA+RA exposure for 8 d promoted neuronal features and inhibited cellular growth (H–H′′). Both RA and AZA treatment reduced the number of cells in the S phase but not as efficiently as the combined treatment (I–I′′). Cell-cycle data are represented as means ± SEM; P value was calculated with unpaired Student’s t test in C–C′′ and one-way ANOVA with a Bonferroni posttest in I–I′′. Tumor growth in D is represented as mean tumor volume ± SEM; **P < 0.01; two-way ANOVA (n = 6–7 per group).
Fig. S1.
(A–C) Cell counts of 1p36+ NB cell lines SK-N-BE (1) (A), SK-N-SH (B), and SH-SY5Y (C) treated for 10 d. (D–F) Cell counts of 1p36− NB cell lines SK-N-AS (D), LAN-1 (E), and CHP-212 (F) treated for 10 d. (G–G′′) LAN-1 cells treated with vehicle (CTRL) for 12 d (G), 12 d + 8 d without treatment (G′), and 12 d + 18 d without treatment (G′′). (H–H′′) LAN-1 cells treated with RA for 12 d (H), 12 d RA + 8 d without treatment (H′), and 12 d RA + 18 d without treatment (H′′). (I–I′′) LAN-1 cells treated with AZA for 12 d (I), 12 d AZA + 8 d without treatment (I′), and 12 d AZA + 18 d without treatment (I′′). (J–J′′) LAN-1 cells treated with AZ+RA for 12 d (J), 12 d AZA+RA + 8 d without treatment (J′), and 12 d AZA+RA + 18 d without treatment (J′′). Black line, CTRL; blue line, RA; green line, AZA; red line, AZA+RA. *P < 0.05, ***P < 0.001; two-way ANOVA with a Bonferroni posttest.
The impaired response to RA stimulation in the 1p36− NB cells, combined with a limited clinical efficacy of RA treatment (10) and the CIMP signature of high-risk NB, prompted us to test whether demethylation before RA treatment could augment the RA responsiveness of these NB cells. We treated the 1p36− CHP-212, LAN-1, and SK-N-AS NB cells with the demethylating drug AZA for 4 d before addition of RA. Notably, NB cells treated with a combination of AZA+RA acquired neuronal morphology, up-regulated expression of the TUJ1, and exhibited decreased cell growth (Fig. 1 H–H′′). Cell-cycle analysis showed that cells undergoing combined AZA+RA treatment exhibited a substantial decline of cells in the S phase and an increase of G1 and G2/M phase cells (Fig. 1 I–I′′). Single treatment with RA or AZA alone also caused a reduction in cells in the S phase and cell number (Fig. 1 I–I′′ and Fig. S1 D–F), albeit to a lesser extent. Taken together, in vitro pretreatment of these 1p36− NB cell lines with AZA sensitizes them for RA treatment, efficiently limiting proliferation while promoting features of neuronal differentiation. To understand whether this response to AZA+RA treatment was reversible or permanent, we terminated AZA+RA treatment in LAN-1 cells at D10 and allowed them to grow in media without AZA and RA. Eighteen days after ending the treatment, the AZA+RA-treated cells still retained a pronounced neuronal morphology and showed no signs proliferation (Fig. S1 G–J′′).
In Vivo Treatment of RA-Resistant 1p36− NB with AZA+RA Impedes Tumor Growth and Prolongs Survival While Inducing Expression of Neuronal Differentiation and Apoptosis Genes.
To investigate how systemic delivery of RA and AZA alone or in combination affects 1p36− NB tumor growth in vivo we, transplanted MYCN nonamplified (MYCNWT) SK-N-AS cells to immunodeficient mice, which then received daily i.p. injections of vehicle (CTRL), RA only (RA), AZA only (AZA), or AZA combined with RA (AZA+RA). Tumor growth in the RA group exhibited no significant difference compared with the CTRL group (Fig. 2A). The AZA-treated tumors appeared smaller, albeit the difference was not statistically significant (Fig. 2A). The most profound reduction in tumor growth was evident in the AZA+RA group, with an average tumor volume four times smaller than the CTRL group (Fig. 2A). To understand whether AZA+RA treatment could show efficacy in already formed tumors derived from NB cells with molecular characteristics distinct from SK-N-AS cells, we transplanted MYCN-amplified (MYCNamp) LAN-1 NB cells and allowed tumors to grow for 8 d, reaching a size of ∼100 mm3 before initiating treatment. With this approach, the only responsive group was the one receiving combined AZA+RA treatment (Fig. 2B). The LAN-1 cells are mutant for P53, and to test if AZA+RA treatment could affect tumor growth in 1p36−, MYCNamp NB cells that express wild-type P53, we repeated the xenograft experiments with CHP-212 NB cells. CHP-212 xenografts treated with combined AZA+RA exhibited a significant inhibition of tumor growth, whereas xenografts treated with RA or AZA alone did not differ significantly from the control group (Fig. 2C). Thus, in three xenograft experiments, with cells derived from different 1p36− NB tumors that do not respond well to RA treatment and harbor distinct combinations of genetic lesions, the AZA+RA treatment significantly impeded tumor growth. To investigate if there was any posttreatment survival benefit, we terminated treatment of the CHP212 xenografted mice at D22. Even after termination of AZA+RA treatment, there was a significant increase in survival, with an almost doubling in survival time compared with the CTRL group (Fig. 2D).
Fig. 2.
In vivo treatment of xenografted RA-resistant 1p36− tumors with AZA+RA impedes tumor growth and prolongs survival. (A) In mice xenografted with SK-N-AS cells, only daily AZA+RA injections starting at D1 significantly inhibited tumor growth (n = 6–7 per group). (B and C) In mice xenografted with MYCN-amplified LAN-1 (B) and CHP-212 (C) cells wherein treatment started at D8, only AZA+RA delivery significantly impeded tumor growth (n = 4–5 per group). (D) Significant prolonged survival (tumor volume < 1,000 mm3) of CHP-212 xenografted mice treated with AZA+RA was evident after termination of treatment at D22 (n = 5 per group). (E) Unsupervised clustering of differentially methylated positions 2 kb upstream or downstream of TSS from retrieved SK-N-AS tumors. (F) Venn diagram depicting coregulated transcripts between the different treatment groups compared with the CTRL group in SK-N-AS tumors. (G) Graph showing how promoter methylation changes correlates with gene expression in AZA+RA- vs. CTRL-treated SK-N-AS tumors. Plotted on the x axis is the log-twofold change in gene expression; plotted on the y axis is the mean difference in means across all sites (mean meth. diff.) in a given promoter region. Only promoters with a mean meth. diff. adjusted P value of <0.25 are plotted. (H) GO analysis of the transcripts both demethylated and up (red) and up only (gray) of SK-N-AS tumors. The diagram represents the five most significant GO categories and GO categories pertaining to cell death (below dashed line). (I) Unsupervised clustering of differentially methylated positions 2 kb upstream or downstream of TSS from retrieved LAN-1 tumors. (J) Venn diagram depicting coregulated transcripts between the different treatment groups compared with the CTRL group in LAN-1 tumors. (K) Graph showing how promoter methylation changes correlate with gene expression in AZA+RA- vs. CTRL-treated LAN-1 tumors. Plotted on the x axis is the log-twofold change in gene expression; plotted on the y axis is the mean difference in means across all sites (mean meth. diff.) in a given promoter region. Only promoters with a mean meth. diff. adjusted P value of <0.05 are plotted. (L) GO analysis of the transcripts both demethylated and up (red) and up only (gray). The diagram represents the five most significant GO categories and GO categories pertaining to cell death (below dashed line). In tumor growth curves (A–C), Kaplan–Meier curve (D), and Venn diagrams (G and K), the different treatment protocols are depicted as follows: black lines, CTRL; green lines, RA; blue lines, AZA; red lines, AZA+RA. Tumor growth is represented as mean tumor volume ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; two-way ANOVA with Bonferroni’s posttest for multiple comparisons. Survival P value calculated with Log-rank test (Mantel Cox); R, Spearman coefficient.
At the end point (EP) of the treatment period, SK-N-AS and LAN-1 tumors were retrieved for genome-wide analysis of DNA methylation with an Infinium Human Methylation 450 BeadChip array (450K array) following bisulfate conversion. Unsupervised clustering of differentially methylated positions within 2 kb of transcriptional start sites (TSSs) showed a clear grouping according to treatment in both SK-N-AS and LAN-1 xenografts (Fig. 2 E and I). RNA-sequencing (RNA-seq) analysis showed that a substantial number of genes were differentially regulated in the SK-N-AS tumors treated with AZA or AZA+RA. In the LAN-1 tumors, this pertains only to the AZA+RA-treated group, reflecting the lack of effect of AZA alone in the LAN-1 tumors (Fig. 2 F and J).
Even though both SK-N-AS– and LAN-1–derived tumors exhibited significant demethylation upon AZA+RA treatment, the correlation with increased gene expression of transcripts with demethylated promoters was weak (LAN-1) or nonexistent (SK-N-AS) (Fig. 2 G and K). Gene ontology (GO) analysis showed that in tumors derived from MYCNamp NB cells (CHP-212 and LAN-1), the main transcriptional response to combined treatment involved GO categories related to neuronal differentiation (Fig. 2L and Fig. S2A). In tumors derived from SK-N-AS cells lacking MYCN amplification, the transcriptional response was related to immune response, cell adhesion, and cell death (Fig. 2H).
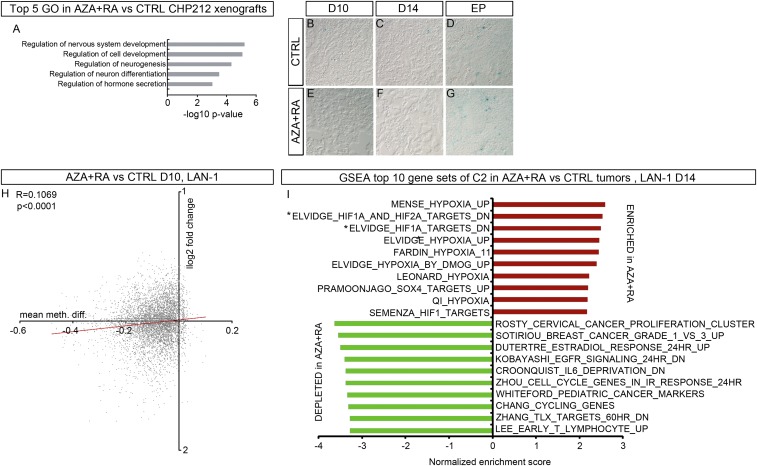
Fig. S2.
(A) The top five GO categories derived from DEG up in AZA+RA vs. CTRL in CHP-212 xenografts. (B–G) Representative β-gal staining in CTRL- (B–D) or AZA+RA-treated (E–G) tumors at the indicated time points. (H) Graph showing how promoter methylation changes correlate with gene expression in AZA+RA- vs. CTRL-treated LAN-1 tumors at D10. Plotted on the x axis is the log-twofold change in gene expression; plotted on the y axis is the mean difference in means across all sites (mean meth. diff.) in a given promoter. Only promoters with a mean meth. diff. adjusted P value of <0.05 are plotted. (I) The top 10 GSEA C2 categories enriched for (in red) and depleted (in green) in AZA+RA-treated LAN-1 tumors at D14; all categories have an FDR of <0.00001. The two gene sets marked by * are defined as genes down-regulated in MCF7 cells (breast cancer) after knockdown of both HIF1A and HIF2A and genes down-regulated in MCF7 cells (breast cancer) after knockdown of HIF1A. R and two-tailed P value in H calculated by Pearson coefficient analysis, two-tailed Student’s t test.
The Early Cellular Response to AZA+RA Treatment Is Characterized by Apoptosis and Cell-Cycle Exit.
To understand the cellular and transcriptional effects of AZA+RA treatment as the treatment actually starts to have an effect (i.e., when the tumor growth curves diverge), we again transplanted LAN-1 cells and initiated treatment with daily injections of AZA+RA or vehicle (CTRL) on day 8 (D8). One cohort of tumors was harvested after 2 d (D10) and one cohort 6 d (D14) after initiation of treatment (D8). Already at D10, there was a decrease of mitotic cells, which also was evident in D14 in AZA+RA-treated tumors (Fig. 3 A and B). In addition, an increase in apoptosis was detected at D14 (Fig. 3 C and D). We stained for β-gal+–senescent cells, but the amount of β-gal+ cells in both CTRL- and AZA+RA-treated tumors were so low that senescence was ruled out as the cause of reduced tumor size in the AZA+RA-treated tumors (Fig. S2 B–G).
Fig. 3.
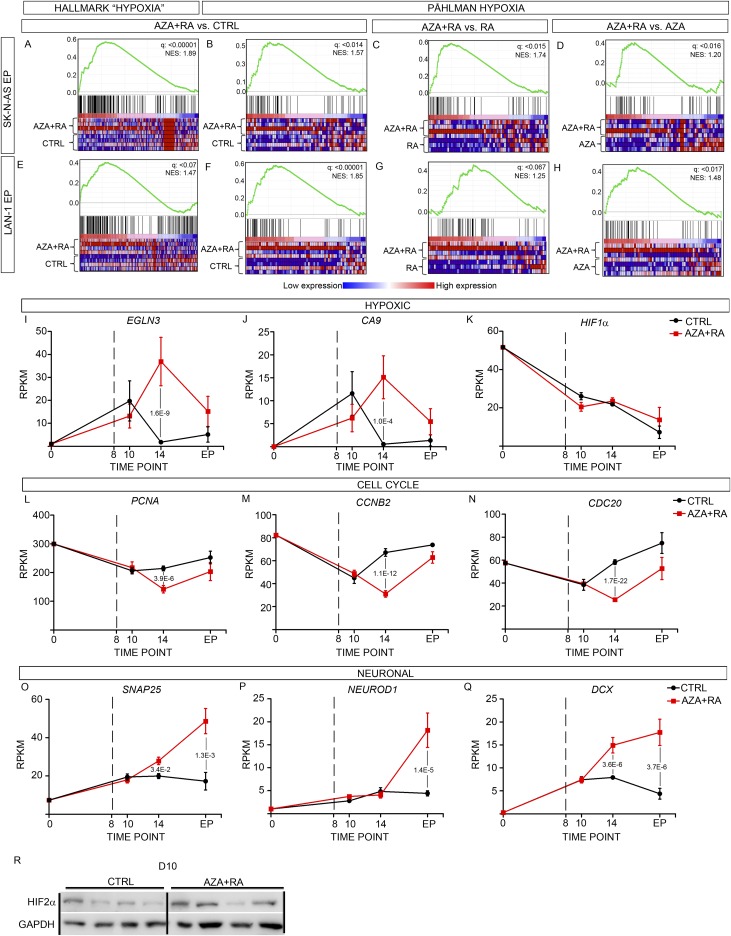
In vivo treatment of LAN-1 xenografts with AZA+RA induces a robust hypoxic-like and a global reduction in transcripts promoting proliferation. (A and B) Percentage of mitotic cells (PH3+) at D10 (A) and D14 (B) in the CTRL- and AZA+RA-treated LAN-1 tumors. (C and D) Number of activated Caspase 3+ D10 (C) and D14 (D) in the CTRL- and AZA+RA-treated LAN-1 tumors. (E) Numbers of significantly differentially expressed genes (DEGs) 2 d (D10) and 6 d (D14) after treatment initiation show a delayed response with more than 3,000 DEGs at D14 compared with 102 DEGs at D10. The number of DEGs at the EP of the experiment described in Fig. 2 is shown as a comparison. Proportions of DEGs also demethylated are highlighted in red. (F) Graph showing how promoter methylation changes correlate with gene expression in AZA+RA- vs. CTRL-treated LAN-1 tumors at D14. Plotted on the x axis is the log-twofold change in gene expression; plotted on the y axis is the mean difference in means across all sites (mean meth. diff.) in a given promoter. Only promoters with a mean meth. diff. adjusted P value of <0.05 are plotted. (G) GSEA of D14 transcriptome with the Hallmarks collection of curated gene sets showing the enriched, “UP” (red) and depleted, “DOWN” (green) gene sets in AZA+RA-treated tumors. Only gene sets with a FDR q < 0.01 are shown. (H) Log-twofold changes for transcripts belonging to the top enriched Hypoxia and depleted “E2F_TARGETS” in the AZA+RA-treated tumors compared with CTRL tumors. (I and J) GSEA of the Fardin_Hypoxia_11 (I) and Påhlman Hypoxia” (J) NB-derived gene sets. (K and L) GSEA of the “Manalo_Hypoxia_Down” (K) and “Whiteford_Pediatric_Cancer_Markers” (L) gene sets. ES, enrichment score; NES, normalized enrichment score. Data in A–D are represented as mean ± SD; *P < 0.05, **P < 0.01; unpaired Student’s t test. In F, R is the Spearman coefficient. Data in H are represented as log-twofold AZA+RA vs. CTRL; **adj. P < 0.01, ***adj. P < 0.001 as calculated by Deseq2.
Transcriptional Changes Induced by AZA+RA at Early Time Points Correlate Poorly with Promoter Demethylation.
RNA was extracted from tumors at D10 and D14, and libraries were generated for RNA-sequencing analysis. When incorporating the expression changes from EP tumors, it was clear that AZA+RA treatment had the most pronounced effects on gene expression at D14 (Fig. 3E). Interestingly, as with the EP tumors (Fig. 2K), no clear correlation between demethylation and increased expression was evident at D10 or D14 (Fig. 3 E and F and Fig. S2H). The previously reported correlation of hypermethylated CGIs and high-risk NB prompted us to scrutinize if there was a correlation between demethylation of CIMP factor promoters and increased gene expression upon AZA+RA treatment at any of the time points (Fig. S3). Even though in the LAN-1 tumors there was a significant reduction in methylation levels of 31 genes out of 72 genes previously described as CIMP factors (6–9, 13), only 3 of those 31 genes (NAV2, NCAM2, and PRPH) exhibited a significant, but low, increase in expression at any time point (Fig. S3). Three other CIMP factors (SPRED3, ZSCAN30, and FER1L4) showed increased expression levels without any decrease in methylation (Fig. S3). A similar lack of clear correlation between demethylation and expression of CIMP factors was evident in the SK-N-AS tumors (Fig. S3).
Fig. S3.
Table showing DEG and methylation changes in AZA+RA-treated LAN-1 tumors at D10, D14, and EP and in SK-N-AS tumors at EP of previously described CIMP factors. Red color denotes increased gene expression, and green color denotes decreased gene expression. NC, no change.
The Principal Transcriptional Response to AZA+RA Is Induction of Hypoxia-Associated Transcripts and a Reduction in Transcripts Promoting Proliferation.
Because only 2 transcripts (HBB and HBE1) out of the 102 significantly altered genes (Fig. 3E) were altered more than twofold at D10, we turned our attention to the effects of AZA+RA treatment at D14. Gene set enrichment analysis (GSEA) of the D14 tumor transcriptome using the “Hallmark” collection of gene sets from Broad Institute (14, 15) showed a strong negative correlation, in the AZA+RA-treated tumors, with terms associated with proliferation, MYC targets, DNA repair, and oxidative phosphorylation (Fig. 3 G and H). In addition, there was an unanticipated and highly significant enrichment of the “HYPOXIA” gene set in the AZA+RA-treated tumors (Fig. 3 G and H). To acquire a more detailed view of the transcriptional response, we performed GSEA on the C2-curated 3,738 gene sets from which several of the founder sets of Hallmark are derived. In the AZA+RA-treated group, 9 out of the top 10 gene sets enriched for were directly related to hypoxia, all with reported FDR q values of 0 (Fig. S2I). This included the NB-specific “FARDIN_HYPOXIA” (Fig. 3I) (16). To validate the NB-specific hypoxic response, we created a gene set, “PÅHLMAN_HYPOXIA,” from a study on hypoxia in NB wherein a group of hypoxia-induced genes was defined (17). This gene set was also significantly enriched in the AZA+RA-treated tumors (Fig. 3J). Transcripts that were depleted in the AZA+RA-treated tumors were enriched for several cancer signatures as well as gene sets associated with proliferation and for genes that are down-regulated upon hypoxia (Fig. 3 K and L and Fig. S2I). To understand whether this hypoxic expression profile also was present in AZA+RA-treated SK-N-AS–derived tumors and if it was maintained in the AZA+RA-treated LAN-1–derived xenografts at the EP, we performed GSEA using the SK-N-AS and LAN-1 EP xenograft transcriptomes. In SK-N-AS xenografts, the Hallmark term HYPOXIA was enriched for in AZA+RA vs. CTRL (Fig. S4A). We also used the NB-specific gene set PÅHLMAN_HYPOXIA and compared the SK-N-AS tumors treated with AZA+RA with CTRL-, RA-, or AZA-treated tumors; this showed enrichment in the AZA+RA group compared with all other treatment groups (Fig. S4 B–D). Similar analysis showed that the hypoxic response in LAN-1–derived tumors was still present in the AZA+RA-treated group at EP, albeit slightly diminished (Fig. S4 E–H). In CHP-212 tumors where treatment was terminated at D22 and the tumors were retrieved when they had reached 1000 mm2, there was no enrichment for Hallmark HYPOXIA in any of the treatment groups.
Fig. S4.
(A–D) Enrichment of HALLMARK HYPOXIA in SK-N-AS tumors treated with AZA+RA vs. CTRL (A), enrichment of PÅHLMAN_HYPOXIA in AZA+RA vs. CTRL (B), AZA+RA vs. RA (C), and AZA+RA vs. AZA (D). (E–H) Enrichment of HALLMARK HYPOXIA in LAN-1 tumors treated with AZA+RA vs. CTRL (E), enrichment of PÅHLMAN_HYPOXIA in AZA+RA vs. CTRL (F), AZA+RA vs. RA (G), and AZA+RA vs. AZA (H). (I–K) Time curves show how expression levels (RPKMs) of hypoxic-responsive genes EGLN3 (I), CA9 (J), and HIF1α (K) vary from normoxic in vitro conditions (0) to different time points (D8, D10, and EP) in the CTRL (black line) and AZA+RA (red line) tumors. (L–N) Time curves show how expression levels (RPKMs) of cell cycle-associated genes PCNA (L), CCNB2 (M), and CDC20 (N) vary from normoxic in vitro conditions (0) to different time points (D8, D10, and EP) in the CTRL (black line) and AZA+RA (red line) tumors. (O–Q) Time curves show how expression levels (RPKMs) of neuronal genes SNAP25 (P), NEUROD1 (Q), and DCX (R) vary from normoxic in vitro conditions (0) to different time points (D8, D10, and EP) in the CTRL (black line) and AZA+RA (red line) tumors. Dashed lines at D8 denotes initiation of treatment in I–Q. Significant P values are indicated in I–Q. (R) Western blot showing HIF2α protein level in CTRL- and AZA+RA-treated tumors at D10; GAPDH is shown as the loading control.
The Induction of Hypoxic Genes Is Linked to EPAS1 but Not to HIF1α.
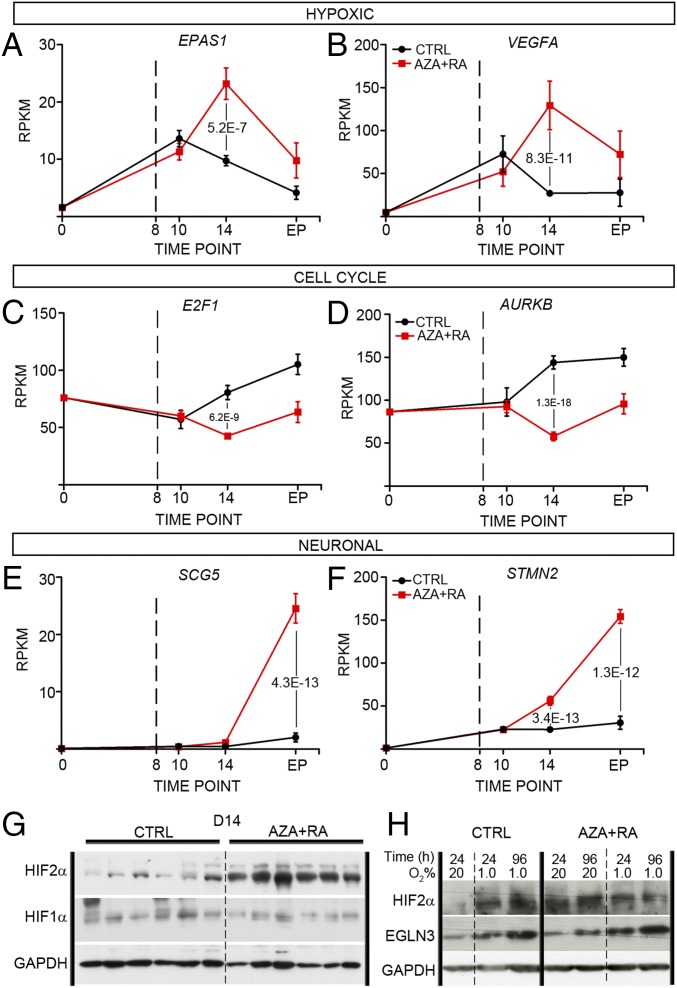
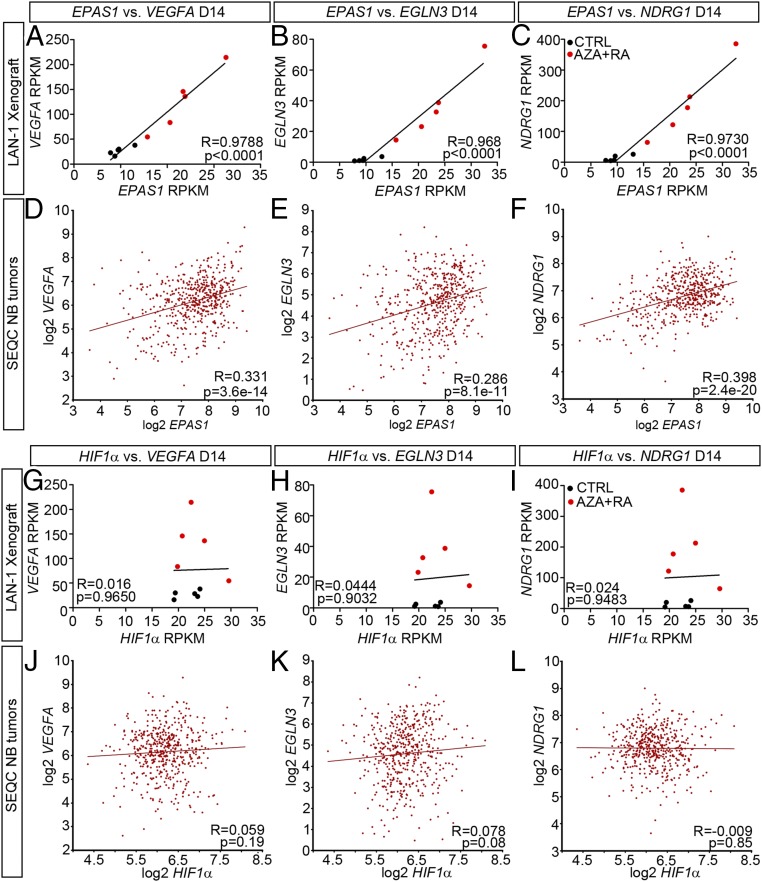
To acquire an overview of transcriptional changes associated with LAN-1 tumor growth, we plotted expression levels of key hypoxic factors and genes associated with cell cycle or neuronal maturation over time (Fig. 4 A–F and Fig. S4 I–Q). This showed that compared with untreated cells grown in vitro under normoxia (D0), there was a clear increase in expression of hypoxic factors in both CTRL- and AZA+RA-treated xenografts at D10 (Fig. 4 A and B and Fig. S4 I and J). In the CTRL xenografts, this response was blunted at D14, whereas in the AZA+RA-treated xenografts, there instead was a distinct increase (Fig. 4 A and B and Fig. S4 I and J). When plotting levels of transcripts involved in cell-cycle progression, there was a clear divergence at D14, as AZA+RA-treated tumors expressed significantly lower levels of such factors (Fig. 4 C and D and Fig. S4 L–N). In the AZA+RA-treated tumors, transcripts associated with neuronal differentiation exhibited a slight increase already at D14, and at the EP, this effect was substantially amplified (Fig. 4 E and F and Fig. S4 O–Q), mirroring the strong enrichment of neuronal-associated GO terms in the EP tumors (Fig. 2L). The AZA+RA-induced hypoxic factors included the EPAS1 transcript coding for the hypoxic response factor HIF2α but not the HIF1α transcript (Fig. 4A and Fig. S4K). Because HIF2α protein levels have been shown to be largely dependent on the balance between degradation through VHL/EGLN-dependent proteosomal degradation and hypoxia-induced stabilization (18), we also performed Western blots of D10 and D14 tumors probing for HIF2α. Slightly increased HIF2α protein levels were evident already at D10 (Fig. S4R), when no significant difference in EPAS1 RNA levels could be detected. This implies that protein stabilization of HIF2α precedes the main transcriptional response to AZA+RA treatment, while coinciding with a reduction in mitotic cells but preceding increased apoptosis and reduced tumor volume (Fig. 3 A–D). At D14, HIF2α protein levels were substantially higher in AZA+RA-treated tumors, whereas HIF1α protein levels were unaltered compared with tumors in the CTRL group (Fig. 4G). To understand whether the induction of HIF2α was dependent on decreased oxygen levels, we cultivated LAN-1 cells under normoxia (20% O2) and hypoxia (1% O2) either with untreated CTRL or with AZA+RA for 24 h and 96 h and then performed Western blot analysis, probing for HIF2α. In contrast to the CTRL group, the AZA+RA group exhibited high levels of HIF2α protein under normoxia (Fig. 4H). Similar results were obtained for the HIF2α target EGLN3 (Fig. 4H). This suggests that the HIF2α increase in the treated cells is not just a consequence of lowered oxygen levels but a response to combined AZA+RA treatment. This is further supported by the increase in expression of EPAS1 and a large cohort of hypoxia-induced genes, despite these tumors being smaller than those of the CTRL group. The strong hypoxic component and its link to EPAS1 expression in the transcriptional response at D14 is further underscored by analysis of D14 tumors, which shows a near to perfect correlation between EPAS1 expression and downstream hypoxic factors (Fig. 5 A–C). Importantly, this association is present also in a patient dataset (SEQC) containing 498 sequenced NB tumors (Fig. 5 D–F) (19, 20), whereas HIF1α shows no correlation with these factors, neither in the treated LAN-1 tumors nor in the SEQC dataset (Fig. 5 G–L).
Fig. 4.
AZA+RA treatment induces expression of the response associated with high EPAS1/HIF2α levels. (A and B) Time curves show how expression levels (RPKMs) of key hypoxic responsive genes EPAS1 (A) and VEGFA (B) vary from untreated normoxic in vitro conditions (0) to different time points (D8, D10, and EP) in the CTRL (black line) and AZA+RA (red line) tumors. Dashed lines at D8 denotes initiation of treatment. (C and D) Time curves show how expression levels (RPKMs) of cell cycle-associated genes E2F1 (C) and AURKB (D) vary from normoxic in vitro conditions (0) to different time points (D8, D10, and EP) in the CTRL (black line) and AZA+RA (red line) tumors. Dashed lines at D8 denotes initiation of treatment. (E and F) Time curves show how expression levels (RPKMs) of neuronal transcripts SCG5 (E) and STMN2 (F) vary from untreated normoxic in vitro conditions (0) to different time points (D8, D10, and EP) in the CTRL (black line) and AZA+RA (red line) tumors. Dashed lines at D8 denotes initiation of treatment. (G) Western blot analysis of AZA+RA and CTRL tumors at D14 with HIF2α- and HIF1α-specific antibodies; GAPDH is shown as a loading control. (H) Western blot analysis of HIF2α and EGLN3 in LAN-1 cells cultivated in 20% or 1.0% oxygen untreated (CTRL) or treated with AZA+RA. GAPDH is shown as a loading control. Adjusted P values in A–F as calculated by Deseq2 (Benjamini–Hochberg adjusted P value); error bars in A–F denote SEM.
Fig. 5.
EPAS1 but not HIF1α expression levels correlate with expression of key hypoxia-inducible genes in LAN-1 tumors and in patient-derived NB. (A–C) In LAN-1 xenografts at D14, EPAS expression correlates with expression of VEGFA (A), EGLN3 (B), and NDRG1 (C). (D–F) In NB patient-derived tumors, EPAS expression correlates with expression of VEGFA (D), EGLN3 (E), and NDRG1 (F). (G–I) In LAN-1 xenografts at D14, there is no correlation between HIF1α expression and expression of VEGFA (G), EGLN3 (H), and NDRG1 (I). (J–L) In NB patient-derived tumors, there is no correlation between HIF1α expression and expression of VEGFA (J), EGLN3 (K), and NDRG1 (L).
Inspection of the EPAS1 promoter region revealed that there was a significant reduction of methylated groups. The magnitude of the reduction was, however, minute (∼7%) and thus unlikely to be responsible for the increased expression levels. The strong correlation between 1p36 loss of heterozygosity and worse prognosis (1) implies that 1p36 harbors one or several tumor suppressors (21). We therefore investigated whether genes located to the smallest region of deletion at 1p36.3, as defined by Brodeur and colleagues (21), exhibited increased demethylation followed by increased expression upon AZA+RA treatment. Of the 27 genes located within this region, spanning roughly 4 mb, only one, TNFRSF9, exhibited both significant promoter demethylation (combined adjusted FDR P value < 0.05) and an increase in gene expression (adjusted P value < 0.05). The increase, only evident at D14, as measured in reads per kilobase per million mapped reads (RPKM) was, however, modest, from an average of 1.13 in CTRL- to 2.01 in AZA+RA-treated tumors.
The HIF2α-Specific Small-Molecule Inhibitor PT2385 Diminishes the Effects of AZA+RA Treatment.
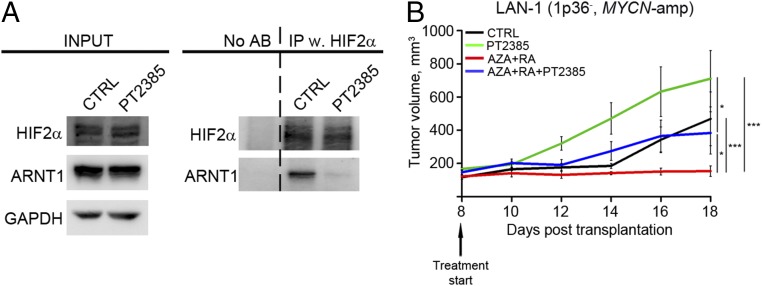
To understand whether the increase in HIF2α levels upon AZA+RA treatment is of importance for the stunted tumor growth, we performed a loss-of-function experiment using a recently developed HIF2α inhibitor, PT2385 (12). PT2385 inhibits the dimerization of HIF2α with ARNT1 and thus prevents activation of HIF2α target genes (12). To validate that PT2385 retained this activity in LAN-1 cells, cells were subjected to 24 h of hypoxia and then treated for 1 h with 10 μM of PT2385. Disruption of the HIF2α/ARNT1 interaction was assayed through immunoprecipitation with HIF2α followed by immunoblotting with an ARNT1-specific antibody (Fig. 6A). Cells exposed to PT2385 exhibited a pronounced decrease in HIF2α/ARNT1 binding (Fig. 6A) in a similar fashion as previously reported for clear cell renal cell carcinoma (ccRCC) cells (12). To test if inhibition of HIF2α activity would impede the in vivo tumor response to AZA+RA, we injected mice with LAN-1 cells, and as in the previously described experiment (Fig. 2B), daily treatment was initiated at D8. The four groups of treatment consisted of DMSO (CTRL), 30 mg/kg HIF2α inhibitor (PT2385), AZA+RA, and AZA+RA together with the inhibitor (AZA+RA+PT2385). Addition of PT2385 diminished the growth inhibitory effect of AZA+RA treatment (Fig. 6B). Thus, LAN-1 cells lost their AZA+RA responsiveness upon block of HIF2α function, indicating that the effects of AZA+RA on tumor growth is at least partly dependent on the increase in HIF2α levels. Notably, the PT2385 group exhibited rapid initial growth and the tumors were significantly (P < 0.01) larger than the CTRL at D14 (Fig. 6B). Even though size difference remained at the last day of measurement (D18), the effect was no longer significant (Fig. 6B).
Fig. 6.
The HIF2α-specific small-molecule inhibitor PT2385 diminishes the effects of AZA+RA treatment. (A) Immunoprecipitation of HIF2α with lysates from LAN-1 cells grown at hypoxia (1% O2) with or without PT2385 treatment, followed by Western blotting with HIF2α- and ARNT1-specific antibodies, shows a reduction of HIF2α and ARNT1 interaction upon treatment with PT2385. (B) In mice xenografted with MYCN-amplified LAN-1 cells, treatment with PT2385 diminishes the response to AZA+RA treatment (n = 6 per group). Tumor growth is represented as mean tumor volume ± SEM; *P < 0.05, ***P < 0.001; two-way ANOVA with Bonferroni’s posttest for multiple comparisons.
High Levels of EPAS1/HIF2α, but Not HIF1α, Are Strongly Linked with Favorable Prognosis of NB Patients and Negatively Correlated with Hallmarks of High-Risk NB.
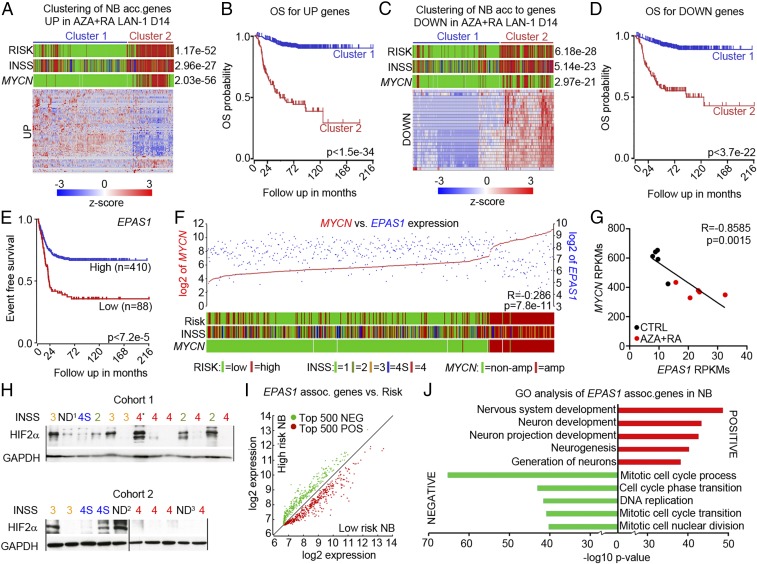
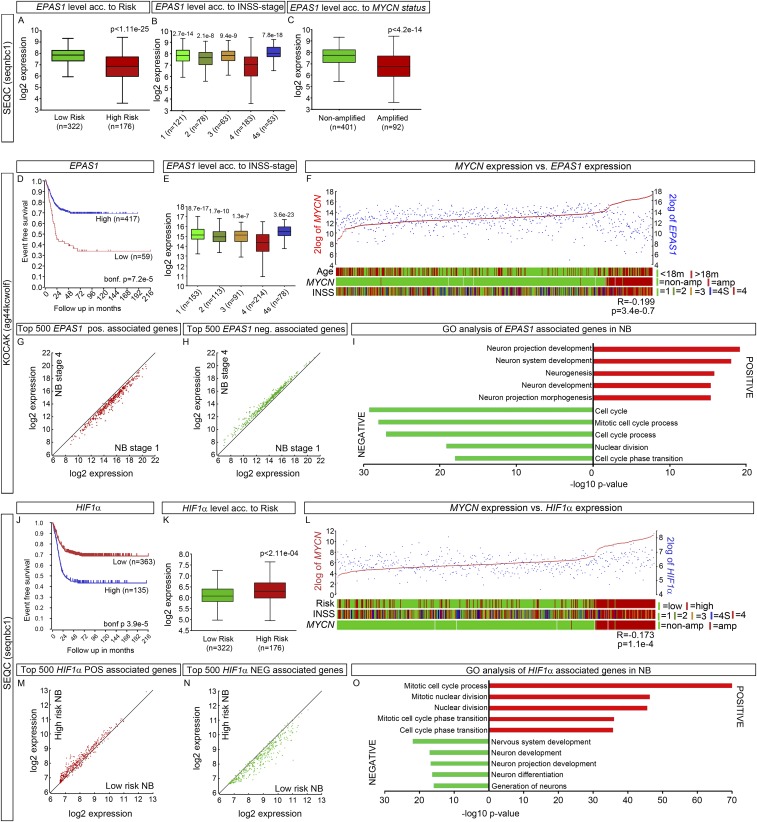
To investigate how genes regulated by AZA+RA treatment at D14 correlate with clinical parameters, we performed k-means clustering on the SEQC 498 patient dataset (19, 20). Genes significantly (adjusted P < 0.05) increased (UP) or decreased (DOWN) with twofold difference upon AZA+RA treatment at D14 were used to group the NB dataset. The k-means clustering revealed two distinct clusters of tumors both in the UP and DOWN analysis (Fig. 7 A and C). In cluster 1, a majority of genes from the UP group were expressed at high levels, whereas in cluster 2 these genes were expressed at lower levels (Fig. 7A). The k-means clustering with DOWN genes showed the opposite pattern, with genes in cluster 1 lowly expressed, whereas cluster 2 expressed these genes at high levels (Fig. 7C). Cluster 1 clearly delineated a cohort of NB patients with better overall survival both for UP and DOWN genes (Fig. 7 B and D). Because several studies have described HIF2α as a tumor-promoting factor associated with increased NB tumor grade and worse patient outcome (3–5, 17), we were intrigued by how AZA+RA-induced inhibition of tumor growth was associated with both a robust increase in EPAS1 levels and a downstream hypoxia-like response. However, the main body of literature describing high levels of HIF2α as a hallmark of high-risk NB depends on HIF2α immunostaining of sections obtained from cohorts of NB patients (4, 17) or microarrays from a limited number of patients (3, 16, 22). We analyzed the 498 patient NB SEQC dataset (20) and an additional 649 patient NB set examined by microarray (KOCAK) (23), both of which showed that high levels of EPAS1 were significantly associated with higher event free survival (Fig. 7E and Fig. S5D), overall survival, low risk, and lower tumor grade (Fig. S5 A, B, and E). We also plotted expression levels of the strongest predictor of high-risk NB, the oncogene MYCN vs. EPAS1. This showed a clear negative correlation (Fig. 7F and Fig. S5 C and F) that also was evident in the LAN-1 AZA+RA-treated tumors at D14 (Fig. 7G). To investigate whether HIF2α protein levels were associated with tumor grade, we performed Western blot analysis in two cohorts of NB patient samples. In the grade 4 tumors, only one out of nine was clearly positive for HIF2α, whereas in the lower grade 1–3 and 4 tumors, 8 out of 11 tumors showed HIF2α immunoreactivity (Fig. 7H). Three tumors that were not determined (ND) according to International Neuroblastoma Staging System (INSS) stage criteria were also included in the analysis. Among these, two tumors expressed HIF2α. One (ND1) lacked MYCNamp, and there was no evidence of disease after more than 60 mo. The other (ND2) was determined as 1p36+ and as a low-risk NB according to CGH classification (Fig. 7H). In addition, the grade 4 tumor that was positive for HIF2α (4*) also had high levels of the neuronal marker Tyrosine Hydroxylase (TH) (24).
Fig. 7.
High levels of EPAS1/HIF2α are associated with favorable NB prognosis. (A) The k-means analysis showed that transcripts increased 2× (UP) in AZA+RA-treated LAN-1 tumors at D14 could separate the patient tumor dataset according to Risk, INSS, and MYCN status. (B) Kaplan–Meier analysis showed that the UP genes from A predict increased overall survival. (C) The k-means analysis showed that transcripts decreased 2× (DOWN) in AZA+RA-treated LAN-1 tumors at D14 could separate the patient tumor dataset according to Risk, INSS, and MYCN status. (D) Kaplan–Meier analysis showed that the DOWN genes from C predict decreased overall survival. (E) Kaplan–Meier analysis of 498 NB tumor samples showed longer event free survival for patients with high EPAS1 levels. (F) EPAS1 expression levels (blue) plotted against MYCN expression levels (red). The lower bars show Risk, INSS, and MYCN status. Color legend for RISK, INSS, and MYCN status is shown in A, C, and F. (G) Correlation of EPAS1 expression levels with MYCN expression in the D14 LAN-14 tumors. (H) Western blot showing HIF2α protein levels in NB of different grades; GAPDH is shown for protein normalization. The ND1 tumor lacks INSS staging but is MYCNWT, and the patient had no evidence of disease after more than 60 months. ND2 denominates a tumor without INSS staging but is determined as a 1p36+ localized (L1) tumor lacking MYCNamp and as not being a high-risk tumor according to CGH classification. ND3 denominates a tumor lacking clinical information. Tumor 4* was classified as grade 4 but expresses high levels of TH. (I) Analysis of the top 500 genes positively (POS, red) correlated or negatively (NEG, green) regulated with the EPAS1 transcript showed that all of the POS genes were enriched for in the low-risk NB and all of the NEG genes were enriched for in the high-risk NB. (J) Enrichment of neuronal-associated GO terms for the genes showing a positive correlation with EPAS1 expression and enrichment of cell cycle-associated GO terms for the genes showing a negative correlation with EPAS1 expression. ND, not determined.
Fig. S5.
(A) In the 498 SEQC NB patient cohort, high EPAS1 levels were associated with low-risk tumors. (B) High EPAS1 levels were associated with low-grade tumors, whereas grade 4 tumors exhibit low-expression EPAS1 levels. (C) Low EPAS1 expression was evident in MYCNamp tumors. (D) Kaplan–Meier analysis of the KOCAK NB dataset containing 649 tumor samples showed longer event free survival for patients with high EPAS1 levels. (E) In the KOCAK NB dataset, high EPAS1 levels were associated with low-grade tumors, whereas grade 4 tumors exhibit low expression EPAS1 levels. (F) In the KOCAK NB dataset, EPAS1 expression levels (blue) were plotted against MYCN expression levels (red). The lower bars show Risk, INSS, and MYCN status. (G and H) Analysis in the KOCAK dataset of the top 500 genes positively (POS, red) correlated or negatively (NEG, green) correlated with the EPAS1 transcript showed that POS genes were enriched in the grade 1 NB (G) and NEG genes were enriched for in grade 4 NB (H). (I) In the KOCAK dataset, enrichment of neuronal-associated GO terms for the genes showed positive correlation with EPAS1 expression, and enrichment of cell cycle-associated GO terms for the genes showed negative correlation with EPAS1 expression. (J) Kaplan–Meier analysis of the 498 NB SEQC tumor samples dataset showed shorter event free survival for patients with high levels of HIF1α. (K) In the same patient cohort, high HIF1α levels were associated with low-risk tumors. (L) HIF1α expression levels (blue) plotted against MYCN expression levels (red). The lower bars show Risk, INSS, and MYCN status. (M and N) Analysis of the top 500 genes positively (POS, red) correlated or negatively (NEG, green) correlated with the HIF1α transcript showed that POS genes were enriched for in the high-risk NB (M) and NEG genes were enriched for in the low-risk NB (N). (O) Enrichment of cell cycle-associated GO terms for the genes showing positive correlation with HIF1α expression and enrichment of neuronal-associated GO terms for the genes showing a negative correlation with HIF1α expression.
We furthermore correlated the genes positively (POS) or negatively (NEG) associated with EPAS1 expression in the 498 SEQC NB patient dataset and then plotted the expression levels of the 500 most significantly POS- or NEG-associated genes in high- vs. low-risk tumors. This revealed that all 500 POS genes were enriched for in low-risk NB, whereas all of the 500 NEG genes were enriched for in high-risk tumors (Fig. 7I). Analysis of the 649 NB KOCAK dataset revealed a similar pattern (Fig. S5 G and H). GO analysis of the POS vs. NEG EPAS1-correlated genes exposed that in the POS group there are several categories involving neuronal differentiation but not any cell cycle category, whereas the opposite is true for the NEG group (Fig. 7J and Fig. S5I). In contrast, high HIF1α levels correlated with worse prognosis, increased risk, high MYCN levels, and an opposite pattern of POS and NEG genes (Fig. S5 J–N). In addition, GO analysis of the HIF1α-associated genes showed a strong correlation with cell-cycle genes in the POS group and neuronal differentiation in the NEG group (Fig. S5O)—that is, the opposite of EPAS1-associated genes (Fig. 7J and Fig. S5I).
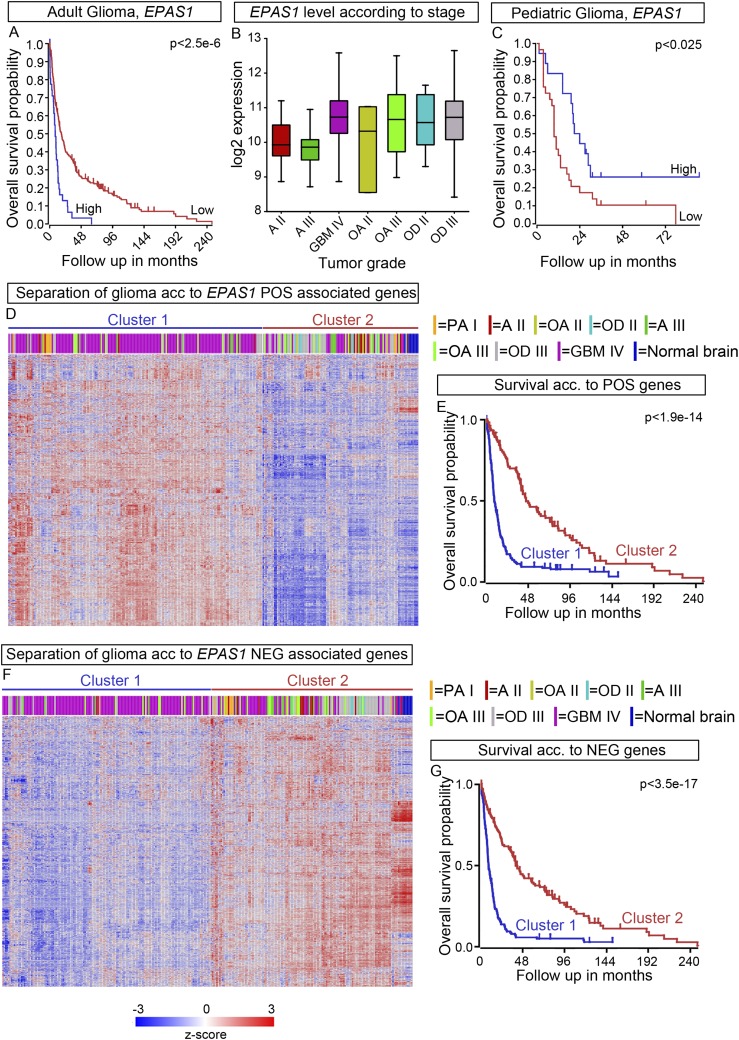
High Levels of EPAS1 Expression Correlate with Worse Outcomes in Adult but Not in Pediatric Glioma.
HIF2α has been ascribed a role as a tumor-promoting factor in several other types of tumors (18, 25), including glioma (26). We performed a comparative analysis of EPAS1 in a dataset (FRENCH) derived from 248 adult glioma patients (27). In contrast to the NB datasets, in the glioma dataset, high EPAS1 levels correlated with lower overall survival and higher tumor grade (Fig. S6 A and B). In addition, k-means clustering showed that genes whose expression was positively correlated with EPAS1 expression delineate higher tumor grade and worse prognosis, whereas negatively correlated genes define lower tumor grade and better prognosis (Fig. S6 D–G). Interestingly, in pediatric glioma (PAUGH dataset) (28), high EPAS1 levels correlate with better prognosis in a similar fashion as in the NB datasets (Fig. S6C), suggesting that HIF2α might act as a tumor suppressor in pediatric nervous system tumors, potentially due to a capacity to promote neuronal differentiation.
Fig. S6.
(A) Kaplan–Meier analysis of 284 adult glioma tumor samples (FRENCH) showed shorter overall survival for patients with high levels of EPAS1. (B) In glioma EPAS1 expression is significantly higher in grade IV (GBM IV) tumors than in astrocytoma grade II (AII) and grade III (AIII). (C) Kaplan–Meier analysis of 53 pediatric glioma tumor samples (PAUGH) showed longer overall survival for patients with high levels of EPAS1. (D) The k-means analysis of genes positively (POS) correlated with EPAS1 in adult glioma. (E) Kaplan–Meier analysis showed that the POS genes from D predict decreased overall survival. (F) The k-means analysis of genes negatively (NEG) correlated with EPAS1 in adult glioma. (G) Kaplan–Meier analysis showed that the NEG genes from F predict decreased overall survival.
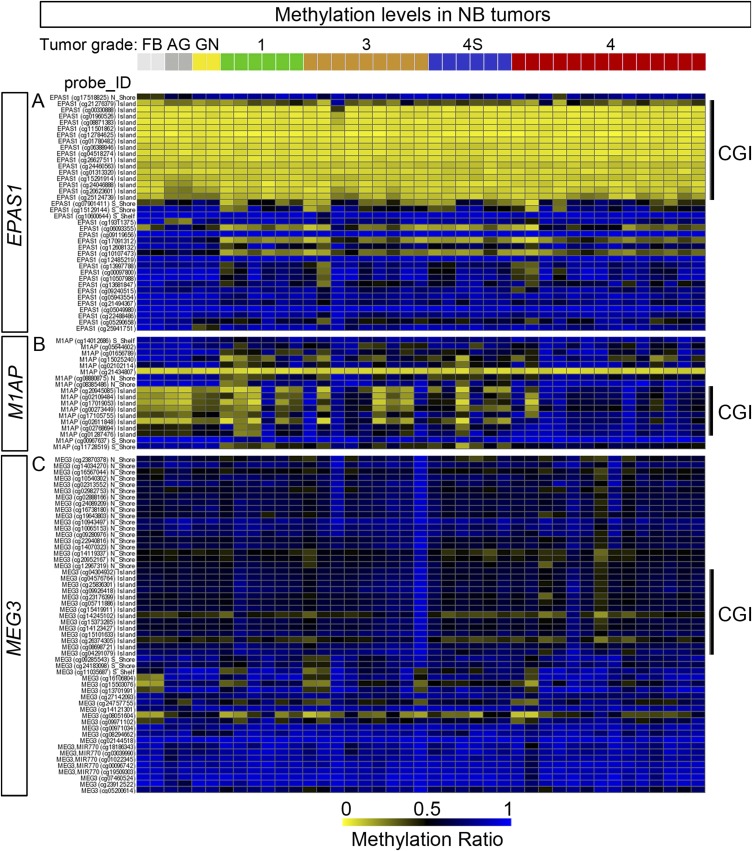
Methylation Levels of CGIs in the EPAS1 Promotor Are Low in NB Tumors and Do Not Vary with Tumor Grade.
Considering the effects of AZA+RA treatment on EPAS1 expression in the NB cells/xenografts and the decreased expression levels in high-stage NB, we also wished to investigate whether CGI methylation levels of the EPAS1 locus differ between tumor stages in primary NB. Thus, we investigated a dataset (LAVARINO) from 42 NB patients analyzed by 450K array technology (8). In these tumors, levels of CGI methylation of the EPAS1 locus are generally low compared with imprinted genes, such as MEG3, or to genes selectively displaying high CGI methylation in high-grade tumors (e.g., M1A) (Fig. S7 A–C). Furthermore, they do not differ between tumor stages (Fig. S7 A–C), implying that the difference in expression levels between stages (Fig. 6 F and H and Fig. S5 B and E) does not depend on alterations in CGI methylation.
Fig. S7.
(A–C) 450K Illumina methylation array probes associated with three different genes: EPAS1 (A), M1AP displaying high levels of CGI methylation in high-grade NB tumors (B), and the imprinted gene MEG3 exhibiting overall high CGI methylation (C) derived from the LAVARINO dataset consisting of 41 NB tumors, ganglioneuroma (GN), adrenal gland (AG), and fetal brain (FB). The probes defined as CpG islands are marked as CGI.
Discussion
Our study shows that combined systemic treatment with the FDA-approved drugs RA and AZA induces high levels of EPAS1 expression and a cohort of hypoxia-associated genes while significantly inhibiting NB growth. The weak correlation between AZA-induced promoter demethylation and increased gene expression implies that the mechanisms through which AZA potentiates RA are not exclusively dependent on its demethylation activity. Previous reports imply that off-target effects of AZA include the accumulation of reactive oxygen species (ROS) (29) and DNA damage (30). However, in the AZA+RA-treated tumors, there was rather a reduction of genes governing the response to oxidative phosphorylation and DNA repair in the AZA+RA-treated tumors (Fig. 3G), lessening the probability that these processes are responsible for treatment response. Unlike the majority of NB cells, both LAN-1 and SK-N-AS cells have been shown to be mutated for TP53 (31, 32). Despite almost undetectable P53 protein levels in LAN-1 cells and a crippled P21 response in SK-N-AS (31, 32), both cell lines exhibit increased apoptosis and stunted growth upon AZA+RA treatment. This implies that an intact P53 pathway is not critical to respond to AZA+RA treatment. Thus, combined AZA+RA treatment shows efficacy in NB cells with different combinations of severe genetic aberrations associated with high-risk NB: 1p36−, mutated P53, and normal MYCN status (SK-N-AS); 1p36−, P53 null mutation, and amplified MYCN (LAN-1); and 1p36−, intact P53, and amplified MYCN (CHP-212).
AZA is already in clinical use for treatment of myelodysplasive syndrome (11) and in several clinical trials for different types of tumors (33). In a phase I case study, a patient who suffered from relapsed stage 4 NB received AZA treatment combined with dendritic cell vaccine, which resulted in complete remission (34). In addition, clinical studies combining AZA with RA for treating Acute Myeloid Leukemia are underway (35, 36). However, despite the well-known demethylating effects of AZA and the clinical efficacy, no single definitive mechanism through which AZA confers its antitumorigenic effects has been proven. Nevertheless, our study shows that growth of high-risk NB can be efficiently inhibited in a preclinical in vivo tumor model, by combining AZA with RA.
The transcriptional response to AZA+RA treatment combined with how PT2835-mediated inhibition of HIF2α activity abrogates the effects of AZA+RA treatment on tumor growth question whether EPAS1/HIF2α can be characterized as an NB oncogene. Our analysis of large NB tumor datasets further undermines this hypothesis. Instead, our analysis reveals that increased EPAS1/HIF2α levels are linked to genes associated with neuronal differentiation and to NB tumors of lower grade with favorable outcomes. Previous studies on the association of EPAS1/HIF2α with high-risk NB and worse patient outcome have relied on smaller collections of sequenced/microarrayed tumors or on immunohistochemical staining of tumor sections (3–5, 16, 17, 22). The two publically available patient datasets with 498 and 649 cases (20, 23) that we used provide a foundation for a more robust statistical analysis of several clinical parameters (e.g., MYCN expression, tumor grade, high- vs. low-risk tumor, and overall survival). This revealed a strong association between high EPAS1 expression levels and low-risk tumors with increased patient survival. The different roles of HIF1α and HIF2α suggested by our tumor analysis are supported in a study by Simon and colleagues (37), wherein they show that high levels of HIF1α are preferentially present in MYCNamp tumors, whereas HIF2α levels usually are high in non–MYCN-amplified tumors (37). Western blot analysis of actual protein levels in 21 patient-derived NB tumors supports the NB expression dataset analysis with decreased protein levels in grade 4 tumors (Fig. 7H). Our analysis is in line with studies of other tumor types wherein HIF2α but not HIF1α is associated with decreased tumor growth and better patient outcome (24, 38–42). This further underscores how tumor context determines if HIF2α acts as an oncogene (e.g., ccRCC and adult glioma) (12, 26, 43, 44) or can actually have tumor-suppressive properties (e.g., soft tissue sarcomas, hepatocellular carcinoma, and colon cancer) (24, 38–42). Furthermore, in nonsmall lung cancer, HIF2α has been described both as an oncogene and a tumor suppressor (45, 46). The xenograft model used for our study is a straightforward way to test simple parameters such as tumor growth and response to systemic drug delivery. However, other preclinical models are more suitable to investigate how AZA+RA treatment could affect additional key properties of tumor biology, such as metastasis, angiogenesis, and tumor microenvironment. Orthotopic xenografts wherein human NB cells are transplanted beneath the renal capsule (47) and transgenic mouse models of NB (48, 49) would provide more appropriate experimental settings for future investigation of such parameters.
The biology of NB is intertwined with several aspects of normal sympathoadrenal development, such as migration, cell death, and differentiation (50, 51), and indeed, there is a strong association between neuronal differentiation genes and high EPAS1 levels in NB tumors (Fig. 7J). Moreover, the contrast between adult and pediatric glioma with regards to the association between high EPAS1 levels and overall survival (Fig. S6 A and C) implies that in pediatric tumors the role of HIF2α is different from its role in adult tumors. Interestingly, studies show that during development HIF2α is necessary for differentiation of neural crest-derived catecholamine-producing cells in mice (52) and for neural differentiation in the vertebrate CNS (53). During development, the proto-oncogene MYCN regulates expansion and ventral migration of neural crest cells, but levels decline as cells undergo sympathoadrenal differentiation (54). Furthermore, when overexpressed in sympathoadrenal precursors, MYCN impinges on differentiation and promotes the formation of NB (49, 55). Notably, in the AZA+RA-treated tumors, there is a pronounced decline in MYCN expression at D14 (Fig. 7G) preceding the up-regulation of markers of differentiated sympathoadrenal neurons (Fig. 4 E and F and Fig. S4 O–Q), further underscoring that combined treatment and EPAS1 up-regulation are linked to key events promoting differentiation.
HIF2α has previously been characterized as an NB tumor-promoting factor (3–5, 16), a view that both our experimental data and our analysis of large cohorts of NB patient material call into question. The small-molecule HIF2α inhibitor PT2385 is already in clinical trials for ccRCC, and inhibition of HIF2α has been suggested as a tractable strategy to treat NB (56). However, a deeper understanding of the role played by HIF2α in NB biology is clearly warranted, before HIF2α inhibitors are administered to children with NB. Nonetheless, our study shows that high-risk NB that responds poorly to RA can be efficiently treated by systemic delivery of AZA combined with RA. This could potentially be used as a novel strategy to treat high-risk NB patients who fail to respond well to RA treatment after myeloablative chemotherapy with hematopoietic stem-cell transplant.
Materials and Methods
Ethical Considerations.
All animal experiments were performed according to Swedish guidelines and regulations; the ethical permits N110/13 (with amendment 2823–2017) and N575/11 were granted by “Stockholms Norra djurförsöksetiska nämnd, Sweden.” NB primary tumors came from the Swedish NB Registry, and ethical permission (DN03-736) was granted by Karolinska Institutets Forskningsetikommitté Nord. Informed consent from families of the subjects was obtained for samples.
Mouse Xenografts.
The 2 × 106 NB cells in 200 μL PBS:matrigel (BD, 354248) (1:1) were injected s.c. into the right flank of adult female Crl:Nu (Ncr)-Foxn1 nu (nude mice). Treatments started 1 d or 8 d after injection. The control group was injected i.p. with 50 µL DMSO per mouse per day. RA treatment of 10 mg⋅kg⋅d and Aza treatment of 0.1 mg⋅kg⋅d were diluted to the right concentrations in DMSO, aliquated, and stored at –80 °C until used; no rethawing/freezing occurred. Xenografted mice treated with PT2385 received daily i.p. injections at a concentration of 30 mg/kg starting 8 d after transplantation. Mice were treated with PT2385 alone or in combination with RA (10 mg/kg) and Aza (0.1 mg/kg), and tumors were measured every 2 d. Tumors were measured externally using a digital caliper every other day; the greatest longitudinal diameter (length) and the greatest transverse diameter (width) were determined, and tumor volume was calculated using the modified ellipsoidal formula [(length × width2)/2]. Overall health was monitored during treatment, and weight was measured weekly. Mice were injected i.p. with 50 mg/kg BrdU (Sigma, B5002) 2 h before euthanasia. Upon allocation of mice to the different treatment groups, sibling groups were broken up and mixed to avoid batch effects. Additional methods are described in SI Materials and Methods.
SI Materials and Methods
Cell Culture.
NB cell lines SK-N-SH, SHSY5Y, SK-N-BE (1), SK-N-AS, LAN1, and CHP212 were maintained at 37 °C in a saturating humidity atmosphere containing 95% air and 5% CO2. Cells were routinely grown in T75 flasks in cell culture medium containing RPMI medium 1640 (Gibco, 31870) supplemented with 2 mM l-glutamine (Gibco, 25030), penicillin (100 units per mL), streptomycin (100 mg/mL) (Gibco, 15140), and 10% FBS (HyClone, SV30180). Cells were obtained from ATCC (SK-N-AS and SH-SY5Y) and from colleagues [LAN-1, SK-N-BE (1), CHP-212, and SK-N-SH].
Treatment with RA and AZA.
Cultured cells were treated with 10 µM AZA (Sigma, A3656) and/or 2 µM RA (Sigma, R2625). 1p36+ cell lines were cultured in medium supplemented with RA for 8 d and then harvested for analysis or fixed for staining and compared with untreated cells. The 1p36– cell lines were divided into four treatment groups: untreated, RA, AZA, or AZA/RA. The first three groups were treated for 12 d with each respective treatment. The combined treatment group (AZA/RA) was treated for 4 d with only AZA and afterward for 8 d with both AZA and RA. On day 12, cells were harvested for analysis or fixed for staining. Medium was changed every other day.
Quantification of Cell Number.
To analyze the cell number, cultured cells were counted every second day using Countess automated cell counter (Invitrogen, C10281) 1:1 medium:trypan blue stain 0.4% (T10282) in a cell-counting chamber slide (T10283).
Cell-Cycle Analysis Using BrdU and PI in FACS.
Cells were treated with the respective treatment and incubated for 1 h with 30 mM BrdU (Sigma, B5002) before fixing cells with 70% EtOH at 4 °C. After washing with PBS, cells were incubated with 2N HCl/Triton-X 0.5% for 30 min and then neutralized with 0.1 M Na2B4O7. Incubation was overnight with PBS/0.5% Tween/1% BSA, anti-BrdU–FITC (BD, 556028), and RNase before analysis by FACS (BD, FACSCanto II) and PI (Sigma, P4864) against BrdU–FITC. Compensation was performed and all samples were made in triplicates, and three time points were analyzed: 4, 8, and 12 d of treatment.
Western Blot Analysis.
Cultured cells or xenografted tumors were lysed using EBC buffer (50 mM Tris, pH 8.0, 120 mM NaCl, 0.5% Nonidet P-40) supplemented with a protease inhibitor mixture (Roche, 04693116001). The lysates were rotated in a cold room for 10 min and cleared by centrifugation at 14,000 × g for 10 min at 4 °C. Protein concentration was determined by a Bradford Assay (Bio Rad, 500–0006). Loading buffer was added to the lysates, and an equal amount of protein was loaded on a gradient 12–8% SDS/PAGE gel together with a protein ladder (Pageruler, ThermoScientific, 26616). Subsequently, proteins were transferred to a nitrocellulose blotting membrane (Amersham 0.45 µm, 10600016) and blocked in 5% milk in PBS/0.1% Tween (Sigma, P1379) followed by primary antibody incubation overnight in a cold room. Anti-mouse or anti-rabbit horseradish peroxidase-coupled secondary antibody (Vector Laboratories, WB-1000 WB-2000) was added in 1:5,000 dilution and incubated for 1 h. ECL immobilon western (Millipore, WBKLS0500) was used as the chemical substrate.
Immunoprecipitation.
Lan1 cells were incubated in hypoxia chamber with 1% O2 and 5% CO2 for 24 h before treatment with 10 μM PT2385 for 1 h. Cells were lysed in lysis buffer containing 10 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 0.5% Nonidet P-40, and protease mixture inhibitor (Roche). We used 5 mg total protein lysate to immunoprecipitate with HIF2 alpha antibody (Abcam, ab199) using the standard protocol according to the manufacturer’s instructions (Dynabeads Protein G from ThermoFisher Scientific). Bound proteins were detected with immunoblotting using the following antibodies: HIF2 alpha (Novusbio, NB100-122, 1:500), ARNT1 (Santa Cruz, sc-55526, 1:100), and GAPDH (Abcam, ab22555, 1:1,000).
Immunostainings.
Tumors were fixed for 24 h in 4% PFA and placed in 30% sucrose, after which they were frozen and cryostat-sectioned at 12 µM. Cultured cells were washed with PBS and fixed with 4% PFA at room temperature for 20 min. Tumor sections or culture cells were after fixation blocked (PBS containing 5% BSA, 0.2% Tween, 0.02% NaAz) for 1 h at room temperature and incubated with primary antibody overnight in a cold room, washed with PBS/Tween, and incubated with secondary antibody for 1 h at room temperature, and the coverslips were mounted and analyzed using confocal microscopy.
Antibodies Used for Western Blot and Immunostaining.
The following antibodies were used: ARNT1 (Santa Cruz, Sc55526, 1:100), TUJ1 (Covance, MMS-435P, 1:1,000), MAP2 (Millipore, AB5622, 1:500), NeuN (Millipore, MAB377, 1:1,000), NCAM (Millipore, MAB5324, 1:500), Ki67 (Abcam, ab16667, 1:500), GAPDH (Abcam, ab22555, 1:2,000), Tubulin (Sigma, T9026, 1:2,000), HIF-2 alpha/EPAS1 (Novusbio, NB100-131, 1:1,000), HIF2α (Novusbio, NB100-22, 1:500), human nuclei (Millipore, MAB1281, 1:250), PH3 (1:500), aPHD/EGLN3 (1:20), Cleaved caspase-3 (Cell signaling, Asp175, 1:500), and DAPI (Roche, 10236276001, 1:1,000).
Mitotic Cell Counting.
Tumor sections were stained as described above with an antibody specific for PH3 and with DAPI to visualize nuclei. Images was taken by confocal microscopy (5×; zoom 0.5×). For quantification, the images were coded and PH3+ nuclei were manually counted by a researcher who had not participated in staining and image acquisition. The number of nuclei in each section was quantified using “CellProfiler” software. The identity of each analyzed section was revealed after all quantifications were complete.
Annexin Analysis.
Cells were analyzed by FACS (BD, FACSCanto II) for apoptosis using FITC Annexin V apoptosis detection kit I (BD Pharmingen, 556547) according to the manufacturer’s protocol. Compensation was performed, and all samples were made in triplicates. Two time points were analyzed: 24 and 96 h of treatment.
Detection of Senescence.
Cells were stained for beta-gal activity to detect senescence (Abcam, Senescence Detection Kit, ab65351) according to the manufacturer’s protocol, and the result was analyzed by light microscopy.
Hypoxia Experiment.
For the hypoxia experiment, the NB cells were cultured in the hypoxia chamber INVIVO2300 at 37 °C in a saturating humidity atmosphere containing 1% O2 and 5% CO2. Cells were placed in a hypoxia chamber 24 h before the start of treatment and stayed there during the duration of the treatment and were lysed.
Analysis of Primary Tumors.
The tumors were categorized according to the INRG criteria (57) and INSS (1). Analysis of these tumors has been described previously (58).
High-Throughput Sequencing.
Total RNA was extracted from cultured cells or from xenografted tumors (at D10 and D14, five tumors from each treatment group, and at EP, four tumors for each treatment group) using the five prime-perfect RNA tissue kit or culture cell kit (2900317, 2900319), Illumina TruSeq RNA Library Preparation Kit v2.
Methylation Array Analysis.
DNA methylation profile of tumor samples was assessed on the Infinium HumanMethylation450 BeadChip (Illumina, WG-314-1003) from Illumina. Methylation data were analyzed by the R package “RnBeads” (59). Normalization of the DNA methylation data was performed using “BMIQ” (60).
RNA-Seq Analysis.
For analysis of LAN-1 and CHP-212 xenografts, Raw 50-bp single end reads were aligned to the human genome (hg19 assembly) using STAR v2.3.0 (61) with default settings. Gene expression was calculated using rpkmforgenes.py (62) with default settings. For analysis of SK-N-AS xenografts, raw reads from RNA-seq data were mapped to the human genome assembly 19 using “Subread” (63). Mapped reads were summarized using “featureCounts” function in “Subread.” Differential gene expression analysis was performed by the R package “DESeq2” (64).
Analysis of Patient Datasets in the R2 Online Database Tool.
Analysis was performed in the online R2 platform (19) on the datasets referred to in the text as SEQC (20), KOCAK (23), LAVARINO (8), FRENCH (27), and PAUGH (28).
GO Analysis.
GO in Fig. 2 H and L and Fig. S2A was performed in DAVID (65). GO analysis in Fig. 6H and Fig. S5 I and O was performed in the R2 platform (19).
Statistical Analysis.
For the cell-cycle analysis in Fig. 1 C–C′′, an unpaired Student’s t test (two-tailed) was performed comparing the magnitudes of the S phases; for cell-cycle analysis in Fig. 1 J–J′′, a one-way ANOVA followed by a Bonferroni’s test for multiple comparisons comparing the magnitudes of the S phase was performed. For tumor volume comparison in Fig. 1D, two-way ANOVA was performed. For tumor volume comparison in Fig. 2 A–C, two-way ANOVA with Bonferroni’s posttest for multiple comparisons was performed. Survival P value in Fig. 2D was calculated with Log-rank test (Mantel Cox). R values in Figs. 2, 3, and 5 D–F and J–L and Fig. S1 represent Spearmann coefficients. R values in Fig. 5 A–C and G–I and 7G represent Pearson coefficients. Calculations were performed in GraphPad or Excel. Criteria for exclusion in xenograft experiments were preestablished as follows: (i) if any xenografted animal scored above the set limit for signs of reduced health and fitness according to the ethical permit, and (ii) if difference in volume of any tumor within a given group was defined as an outlier according to Grubb’s test, alpha P < 0.01. Following these criteria, a total of two animals were excluded from analysis, one in 2B (criteria 2) and one in 2C (criteria 1).
GSEA.
Analysis of the transcriptome for enriched gene groups was performed with the GSEA software (https://www.broadinstitute.org/gsea/downloads.jsp) (14, 15). To identify enriched or depleted gene signatures described in Fig. 3 G–L and Fig. S4 A–H, we used the “h.all.v5.1.symbols.gmt [Hallmarks]” gene set; for Fig. 3 J–L, we used the “c2.all.cgp.v5.1.symbols.gmt [Curated]” gene sets. To create the PÅHLMAN_HYPOXIA gene set, we used the genes listed in figure 5 from ref. 17. After trimming of duplicates and unidentified genes, 66 genes remained.
Acknowledgments
We thank Dr. J. Muhr and members of the J.H. laboratory for helpful comments on the manuscript. The J.H. laboratory is supported by StratCan, the Swedish Children Cancer Foundation, The Swedish Cancer Foundation, Knut and Alice Wallenberg Foundation, Swedish Research Council (VR), and the Ludwig Institute for Cancer Research (LICR).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE100568).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700655114/-/DCSupplemental.
References
- 1.Brodeur GM. Neuroblastoma: Biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 2.Pietras A, Johnsson AS, Påhlman S. The HIF-2α-driven pseudo-hypoxic phenotype in tumor aggressiveness, differentiation, and vascularization. Curr Top Microbiol Immunol. 2010;345:1–20. doi: 10.1007/82_2010_72. [DOI] [PubMed] [Google Scholar]
- 3.Mohlin S, Hamidian A, Påhlman S. HIF2A and IGF2 expression correlates in human neuroblastoma cells and normal immature sympathetic neuroblasts. Neoplasia. 2013;15:328–334. doi: 10.1593/neo.121706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noguera R, et al. HIF-1alpha and HIF-2alpha are differentially regulated in vivo in neuroblastoma: High HIF-1alpha correlates negatively to advanced clinical stage and tumor vascularization. Clin Cancer Res. 2009;15:7130–7136. doi: 10.1158/1078-0432.CCR-09-0223. [DOI] [PubMed] [Google Scholar]
- 5.Pietras A, et al. HIF-2alpha maintains an undifferentiated state in neural crest-like human neuroblastoma tumor-initiating cells. Proc Natl Acad Sci USA. 2009;106:16805–16810. doi: 10.1073/pnas.0904606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abe M, et al. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005;65:828–834. [PubMed] [Google Scholar]
- 7.Decock A, et al. Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers. Genome Biol. 2012;13:R95. doi: 10.1186/gb-2012-13-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gómez S, et al. DNA methylation fingerprint of neuroblastoma reveals new biological and clinical insights. Epigenomics. 2015;7:1137–1153. doi: 10.2217/epi.15.49. [DOI] [PubMed] [Google Scholar]
- 9.Kiss NB, et al. Quantitative global and gene-specific promoter methylation in relation to biological properties of neuroblastomas. BMC Med Genet. 2012;13:83. doi: 10.1186/1471-2350-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthay KK, et al. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: A children’s oncology group study. J Clin Oncol. 2009;27:1007–1013. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gore SD, Jones C, Kirkpatrick P. Decitabine. Nat Rev Drug Discov. 2006;5:891–892. doi: 10.1038/nrd2180. [DOI] [PubMed] [Google Scholar]
- 12.Wallace EM, et al. A small-molecule antagonist of HIF2α is efficacious in preclinical models of renal cell carcinoma. Cancer Res. 2016;76:5491–5500. doi: 10.1158/0008-5472.CAN-16-0473. [DOI] [PubMed] [Google Scholar]
- 13.Asada K, Abe M, Ushijima T. Clinical application of the CpG island methylator phenotype to prognostic diagnosis in neuroblastomas. J Hum Genet. 2013;58:428–433. doi: 10.1038/jhg.2013.64. [DOI] [PubMed] [Google Scholar]
- 14.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 15.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fardin P, et al. A biology-driven approach identifies the hypoxia gene signature as a predictor of the outcome of neuroblastoma patients. Mol Cancer. 2010;9:185. doi: 10.1186/1476-4598-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmquist-Mengelbier L, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 18.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koster J. 2008 R2: Genomics Analysis and Visualization Platform. Available at r2.amc.nl. Accessed January 11, 2017.
- 20.Su Z, et al. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014;15:523. doi: 10.1186/s13059-014-0523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White PS, et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- 22.Cimmino F, et al. Inhibition of hypoxia inducible factors combined with all-trans retinoic acid treatment enhances glial transdifferentiation of neuroblastoma cells. Sci Rep. 2015;5:11158. doi: 10.1038/srep11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kocak H, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4:e586. doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fell SM, et al. Neuroblast differentiation during development and in neuroblastoma requires KIF1Bb mediated transport of TRKA. Genes Dev. 31:1036–1053. doi: 10.1101/gad.297077.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakazawa MS, et al. Epigenetic re-expression of HIF-2α suppresses soft tissue sarcoma growth. Nat Commun. 2016;7:10539. doi: 10.1038/ncomms10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravendeel LA, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 28.Paugh BS, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fandy TE, et al. Decitabine induces delayed reactive oxygen species (ROS) accumulation in leukemia cells and induces the expression of ROS generating enzymes. Clin Cancer Res. 2014;20:1249–1258. doi: 10.1158/1078-0432.CCR-13-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Covey JM, D’Incalci M, Tilchen EJ, Zaharko DS, Kohn KW. Differences in DNA damage produced by incorporation of 5-aza-2′-deoxycytidine or 5,6-dihydro-5-azacytidine into DNA of mammalian cells. Cancer Res. 1986;46:5511–5517. [PubMed] [Google Scholar]
- 31.Davidoff AM, Pence JC, Shorter NA, Iglehart JD, Marks JR. Expression of p53 in human neuroblastoma- and neuroepithelioma-derived cell lines. Oncogene. 1992;7:127–133. [PubMed] [Google Scholar]
- 32.Goldschneider D, et al. Expression of C-terminal deleted p53 isoforms in neuroblastoma. Nucleic Acids Res. 2006;34:5603–5612. doi: 10.1093/nar/gkl619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Health USNIo 2016 ClinicalTrials.gov identifiers NCT01352650, NCT01882660, and NCT02664181.
- 34.Krishnadas DK, Shapiro T, Lucas K. Complete remission following decitabine/dendritic cell vaccine for relapsed neuroblastoma. Pediatrics. 2013;131:e336–e341. doi: 10.1542/peds.2012-0376. [DOI] [PubMed] [Google Scholar]
- 35.Grishina O, et al. DECIDER: Prospective randomized multicenter phase II trial of low-dose decitabine (DAC) administered alone or in combination with the histone deacetylase inhibitor valproic acid (VPA) and all-trans retinoic acid (ATRA) in patients >60 years with acute myeloid leukemia who are ineligible for induction chemotherapy. BMC Cancer. 2015;15:430. doi: 10.1186/s12885-015-1432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raffoux E, et al. Phase 2 clinical trial of 5-azacitidine, valproic acid, and all-trans retinoic acid in patients with high-risk acute myeloid leukemia or myelodysplastic syndrome. Oncotarget. 2010;1:34–42. doi: 10.18632/oncotarget.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qing G, et al. Combinatorial regulation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor HIF-1alpha. Cancer Res. 2010;70:10351–10361. doi: 10.1158/0008-5472.CAN-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun HX, et al. Hypoxia inducible factor 2 alpha inhibits hepatocellular carcinoma growth through the transcription factor dimerization partner 3/E2F transcription factor 1-dependent apoptotic pathway. Hepatology. 2013;57:1088–1097. doi: 10.1002/hep.26188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawłuszko-Wieczorek AA, Horbacka K, Krokowicz P, Misztal M, Jagodziński PP. Prognostic potential of DNA methylation and transcript levels of HIF1A and EPAS1 in colorectal cancer. Mol Cancer Res. 2014;12:1112–1127. doi: 10.1158/1541-7786.MCR-14-0054. [DOI] [PubMed] [Google Scholar]
- 40.Dai CX, et al. Hypoxia-inducible factor-1 alpha, in association with inflammation, angiogenesis and MYC, is a critical prognostic factor in patients with HCC after surgery. BMC Cancer. 2009;9:418. doi: 10.1186/1471-2407-9-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang SL, et al. The correlation of expression levels of HIF-1α and HIF-2α in hepatocellular carcinoma with capsular invasion, portal vein tumor thrombi and patients’ clinical outcome. Jpn J Clin Oncol. 2014;44:159–167. doi: 10.1093/jjco/hyt194. [DOI] [PubMed] [Google Scholar]
- 42.Imamura T, et al. HIF-1alpha and HIF-2alpha have divergent roles in colon cancer. Int J Cancer. 2009;124:763–771. doi: 10.1002/ijc.24032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho H, et al. On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature. 2016;539:107–111. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazumdar J, et al. HIF-2alpha deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci USA. 2010;107:14182–14187. doi: 10.1073/pnas.1001296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim WY, et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009;119:2160–2170. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson DM, Shohet JM, Kim ES. Preclinical models of pediatric solid tumors (neuroblastoma) and their use in drug discovery. Curr Protoc Pharmacol. 2011;Chapter 14:Unit 14.17. doi: 10.1002/0471141755.ph1417s52. [DOI] [PubMed] [Google Scholar]
- 48.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu S, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012;21:362–373. doi: 10.1016/j.ccr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang M, Stanke J, Lahti JM. The connections between neural crest development and neuroblastoma. Curr Top Dev Biol. 2011;94:77–127. doi: 10.1016/B978-0-12-380916-2.00004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheung NK, Dyer MA. Neuroblastoma: Developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian H, Hammer RE, Matsumoto AM, Russell DW, McKnight SL. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko CY, et al. Integration of CNS survival and differentiation by HIF2α. Cell Death Differ. 2011;18:1757–1770. doi: 10.1038/cdd.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmerman KA, et al. Differential expression of myc family genes during murine development. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- 55.Marshall GM, et al. The prenatal origins of cancer. Nat Rev Cancer. 2014;14:277–289. doi: 10.1038/nrc3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wigerup C, Påhlman S, Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol Ther. 2016;164:152–169. doi: 10.1016/j.pharmthera.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 57.Monclair T, et al. The International Neuroblastoma Risk Group (INRG) staging system: An INRG Task Force report. J Clin Oncol. 2009;27:298–303. doi: 10.1200/JCO.2008.16.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carén H, et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci USA. 2010;107:4323–4328. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Assenov Y, et al. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teschendorff AE, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao Y, Smyth GK, Shi W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]