Abstract
Chromatin is the complex of eukaryotic DNA and proteins required for the efficient compaction of the nearly two-meter long human genome into a roughly ten-micron diameter cell nucleus. The fundamental repeating unit of chromatin is the nucleosome: 147bp of DNA wrapped about an octamer of histone proteins. Nucleosomes are stable enough to organize the genome yet must be dynamically displaced and reassembled to allow access to the underlying DNA for transcription, replication, and DNA damage repair. Histone chaperones are a non-catalytic group of proteins that are central to the processes of nucleosome assembly and disassembly, and thus the fluidity of the ever-changing chromatin landscape. Histone chaperones are responsible for binding the highly basic histone proteins, shielding them from non-specific interactions, facilitating their deposition onto DNA, and aiding in their eviction from DNA. Though most histone chaperones perform these common functions, recent structural studies of many different histone chaperones reveal that there are few commonalities in their folds. Importantly, sequence-based predictions show that histone chaperones are highly enriched in intrinsically disordered regions (IDRs) and acidic stretches. In this review, we focus on the molecular mechanisms underpinning histone binding, selectivity, and regulation of these highly dynamic protein regions. We highlight new evidence suggesting that IDRs are often critical for histone chaperone function and play key roles in chromatin assembly and disassembly pathways.
Graphical abstract
INTRODUCTION
The nucleosome is the fundamental repeating unit of chromatin: the physiological form of the eukaryotic genome. Early studies of chromatin revealed this repeating unit of histones and DNA [1,2], and higher resolution structural studies that followed yielded exquisite insights into the arrangement and interactions of nucleosomal components [3,4]. Nucleosomes do not form spontaneously, therefore the construction and destruction of a single nucleosome requires the coordinated actions of many proteins within the cell [5–11]. Central to this process are both ATP-dependent chromatin remodeling complexes and ATP-independent histone chaperones (reviewed in [12,13]). ATP-dependent chromatin remodelers can be divided into four families: the SWI/SNF, the NuRD/Mi-2/CHD, the ISWI, and the INO80/SWR1 family (reviewed in [14]). Despite differences in their mechanisms and subunit compositions, all chromatin remodeling complexes contain a structurally similar catalytic ATPase core. This catalytic core is necessary to convert the chemical energy of ATP into rotational movement leading to nucleosome sliding, eviction or exchange.
The large family of histone chaperones are key players in chromatin assembly and remodeling processes, despite them not consuming ATP [9,15–21]. Histone chaperones are defined as a group of proteins that bind to histones and stimulate their transfer onto DNA or to other proteins [5,22,23]. Histone chaperones perform many functions in the cell, including: binding to histones [5], shielding charge and preventing aggregation [24], using their nuclear localization signals (NLSs) along with histone NLSs to aid in cytoplasmic-nuclear shuttling [25,26], maintaining a soluble pool of histones [27], regulating their deposition onto DNA [28], and aiding in histone sliding, nucleosome destabilization and histone eviction [29,30] (Figure 1). As histone chaperones are non-catalytic, they rely solely on multiple energetic minima to guide histone through the deposition pathways and onto DNA [24]. Surprisingly, histones chaperones have evolved a multitude of highly divergent molecular structures to perform relatively similar tasks [31–34].
Figure 1. Roles of Histone Chaperones in the Cell.
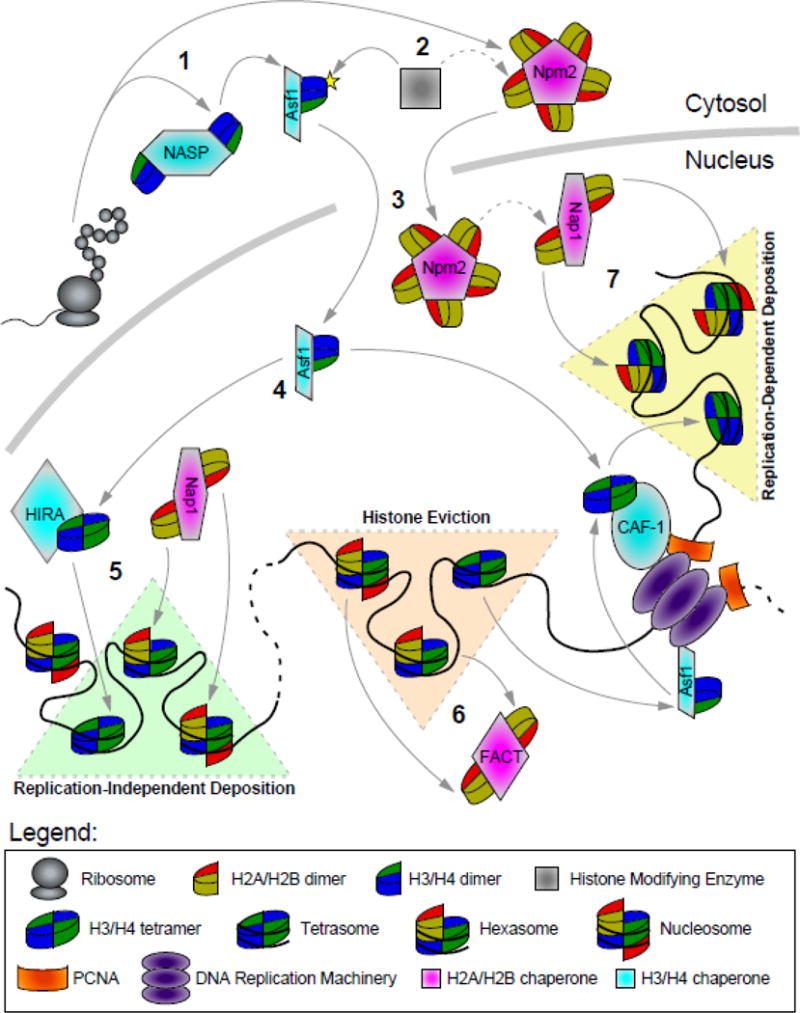
1. Heat-shock proteins (HSPs) along with histone chaperones, such as NASP, Npm2, and Asf1 bind to the newly translated histones and shield them from non-specific interactions in the cytosol.
2. Histone chaperones, such as Asf1 and RbAp46, provide a scaffold for pre-deposition histone modifications, such as HAT1-mediated acetylation of H4 tails. These modifications are crucial for nuclear import and transfer of H3/H4 to other chaperones. Less is known about the function of pre-deposition H2A/H2B modifications (dashed arrow).
3. Histone chaperones, such as Npm2, NASP, Nap1, and Asf1 use their nuclear localization signals (NLS) to interact with importins to aid in shuttling histones between the cytosol and nucleus. Histones also interact with import factors via NLS’s on their disordered tails. Chaperones also store and provide a soluble pool of histones.
4. Histone chaperones transfer histones to other chaperones. Asf1 transfers H3.1/H4 dimers to CAF-1 and H3.3/H4 dimers to HIRA, DAXX, or DEK for deposition. CAF-1 tetramerizes H3.1/H4 prior to deposition, whereas HIRA, DAXX and DEK may bind and deposit both the dimeric and tetrameric forms of H3.3/H4 depending on the chaperone bound and chromatin context (only the tetramer deposition model is shown for simplicity). Less is known about transfer among H2A/H2B chaperones (dashed arrow).
5. Replication-independent histone deposition of H3.3/H4 by HIRA, DAXX and DEK, and H2A/H2B by Nap1 and FACT. Histone variants can be incorporated by other variant-specific histone chaperones, such as YL1, HJURP and APLF.
6. Chaperones, along with ATP-dependent chromatin remodelers, mediate histone eviction prior to DNA replication. H2A/H2B is displaced by FACT and Nap1. H3/H4 is displaced by Asf1 and recycled by CAF-1. Large-scale histone eviction occurs during DNA replication (shown), transcription, and DNA repair. The MCM2 subunit of the replisome aids in Asf1-mediated chaperoning at the replication fork, and CAF1 also interacts with the sliding clamp PCNA. Histone variants can be displaced by variant-specific chaperones, such as ANP32E and ATP-dependent chromatin remodelers such as SWR1.
7. Replication-dependent histone deposition of H3.1/H4 by CAF1 and H2A/H2B by Nap1.
Histones are extremely basic proteins, with predicted isoelectric points (pI) of human histones ranging from 10.3–11.4. Histones are also very small, contain a high degree of hydrophobic amino acids, and are prone to interact non-specifically and aggregate with many other macromolecules within the cell. One of the most important responsibilities of a histone chaperone is to partially neutralize the charge of histone proteins to prevent non-functional and detrimental interactions. To do this, histone chaperones are thought to rely on their acidic nature to bind histones and neutralize charge [5]. Interestingly, acidic amino acids are not evenly distributed throughout most histone chaperones, but instead tend to cluster into “acidic stretches” in their primary sequences. The importance of acidic stretches appears to vary between different histone chaperones, with some chaperones requiring acidic stretches to function (such as Npm) [35,36], and other chaperones in which the deletion of acidic stretches does not have an effect on in vitro histone deposition activity (such as yeast Nap1) [37], although the in vivo necessity is still unclear. It has long been assumed these regions in histone chaperones only serve to increase affinity via non-specific electrostatic interactions with histones. In this “electrostatic steering” model, disordered acidic stretches of a chaperone would interact with the basic histone proteins non-specifically to bring them into proximity of the folded core of the chaperone, which would then make more specific contacts such as hydrogen bonds and van der Waals interactions (concept reviewed in [38]). Consistent with this hypothesis, deletion of acidic stretches in many chaperones lead to an overall decrease in affinity for histone proteins [36,39–41]. However, more recently many acidic stretches have also been shown to make very specific contacts with histones and, in some cases, may actually act as drivers of chaperone specificity [42,43].
Intrinsically disordered regions (IDRs) are also highly prevalent on histone chaperones. Intrinsically disordered proteins (IDPs) and IDRs are defined as extremely flexible and dynamic proteins, or regions of proteins, resulting from a lack of stable secondary structure (reviewed in [44,45]). IDRs do not fold spontaneously into a single stable structure, but rather sample an ensemble of different conformations. Many disordered proteins and proteins with disordered regions fall into the “dark proteome”: the regions of proteins whose conformation remains unknown [46]. IDRs are generally very highly charged and contain few hydrophobic amino acids [47,48], therefore these regions can be accurately predicted directly from the primary sequence of a protein [49]. Proteome wide surveys show that predicted IDRs are highly enriched on many nuclear and chromatin binding proteins [50,51], suggesting that these highly dynamic regions may play key roles in histone binding and chromatin organization. Furthermore, the enrichment of IDRs in nuclear and chromatin proteins, and their lack of enrichment in mitochondrial and more ancient classes of proteins, suggests that functional disorder is largely a eukaryotic innovation.
IDRs in proteins confer many advantages compared to well-ordered proteins, particularly in protein:protein interactions. Many IDRs are able to extend far from the protein core and have large capture radii to interact with potential ligands [52]. These regions also have intrinsic kinetic advantages compared to ordered proteins whose association rates are often diffusion limited. For example, highly charged IDRs can make many transient contacts with potential ligands to form an encounter complex with fast on and off rates, and can often fold only upon binding to their specific ligand. This “fly casting” mechanism facilitates fast association rates and highly specific binding between IDRs and their protein targets, such as the interaction between the intrinsically disordered p53 transactivation domain (TAD) and CREB [53,54]. Additionally, by adopting different secondary structures upon binding different ligands, one IDR can facilitate interactions with multiple structurally different ligands (reviewed in [55]). This unique property of IDRs allows for a greater number of potential interactions compared to what a well-ordered protein can achieve and is essential for highly complex protein interaction networks. IDRs are also enriched in known protein hubs, consistent with their importance in multivalent protein interactions [56]. Finally, many IDRs can remain disordered even when bound to a ligand. These “fuzzy complexes” are often formed using multivalent, transient interactions between protein and ligand and are of great importance in many processes such as FGNUP-mediated nuclear import [57,58] and Npm1-mediated local phase separation during nucleolus formation [59,60].
Despite the high prevalence of acidic stretches and IDRs in histone chaperones, their roles in histone binding, deposition, and regulation of chaperone function have remained elusive. Due to their dynamic nature, IDRs are notoriously difficult to study at the molecular level. Despite these difficulties, in this review we highlight recently published structural and functional analyses of acidic stretches and IDRs in histone chaperones showing that these regions are not only important for increasing affinity, but play a multitude of specific and diverse regulatory roles. This review is not an exhaustive list detailing all known chaperone:histone interactions and complete deposition pathways, which have been recently surveyed elsewhere (reviewed in [61–63]). Rather, the studies highlighted in this review are prime examples from the recent literature that shed light on the functions and regulation of highly dynamic regions in human histone chaperones and chromatin-binding proteins, including how these regions may influence chromatin structure, gene expression and epigenetic inheritance.
Conservation of IDRs and Acidic Stretches in Histone Chaperones
Due to their structural plasticity and ability to bind specifically to multiple ligands, IDRs are thought to be critical for establishing and fine-tuning highly complex, multivalent protein interaction and complex signaling networks [56,64]. Consistent with this hypothesis, previous comparative proteome-wide disorder predictions have shown that IDRs are more prevalent among higher eukaryotes compared to both bacteria and archaea [65]. In addition, IDRs are predicted to be unequally distributed throughout the human proteome [50]. Classification of predicted disordered regions by gene ontology and subcellular localization show that IDRs are, not surprisingly, enriched in processes specific to eukaryotic cellular function including nuclear and chromatin binding proteins, as well as those involved in nucleosome assembly [50,51]. On the other hand, proteins involved in processes that function in all three kingdoms of life, such as metabolic enzymes, are predicted to be highly ordered. These predictions have led to the hypothesis of an active molecular evolution of protein disorder (reviewed in [66]).
Using the Regression-based Accurate Prediction of Protein Intrinsic Disorder content (RAPID) [51], we compared the disorder content of 28 human histone chaperones against annotated human chromosomal, nuclear, cytoplasmic, mitochondrial, and transmembrane proteins by one-way ANOVA followed by multiple comparisons. We find that, as a group, histone chaperones have a high content of intrinsic disorder (mean of 38%) and are significantly more disordered than nuclear (27%), cytoplasmic (23%), mitochondrial (16%), and transmembrane (11%) proteins, but are not significantly different compared to chromosomal phosphoproteins (35%), chromosomal proteins (33%), and nuclear phosphoproteins (29%) (Figure 2). We next used DISOPRED3 software to predict the by-residue disorder content of these 28 human histone chaperones [49,67]. We find that IDRs greater than 25 amino acids are present in 26 out of 28 human histone chaperones, with the WD-repeat proteins RbAp48 and RbAp46 being the two exceptions (Figure 3). However, both of these proteins are known to be constituents of chaperone complexes and have not been shown to transfer histones when isolated. Together, these results indicate that histone chaperones as a group are highly enriched in predicted disordered regions, and that these regions likely play important roles in their function.
Figure 2. Histone Chaperones are Predicted to be Highly Disordered.
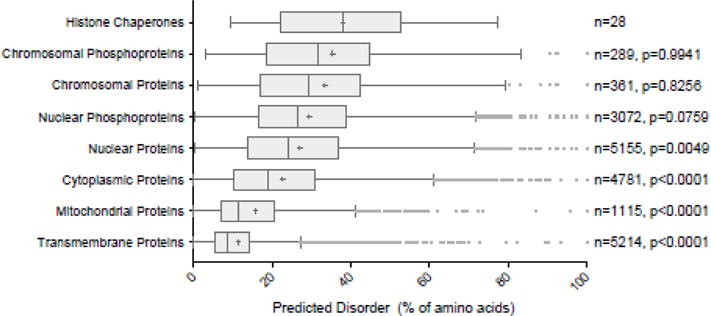
A. Box-and-whiskers plot of RAPID disorder content predictions from 28 human histone chaperones (top) compared to annotated chromosomal phosphoproteins, chromosomal proteins, nuclear phosphoproteins, nuclear proteins, cytoplasmic proteins, mitochondrial proteins, and transmembrane proteins. Mean of each group is shown by +. Number of proteins in each group is shown by n value. p values shown from one-way ANOVA and multiple comparisons of each group compared to histone chaperones. Note: many histone chaperones are also included in the groups of nuclear proteins, nuclear phosphoproteins, chromosomal proteins, and chromosomal phosphoproteins.
Figure 3. Many Histone Chaperones Contain Long Intrinsically Disordered Regions (IDRs) and Acidic Stretches.
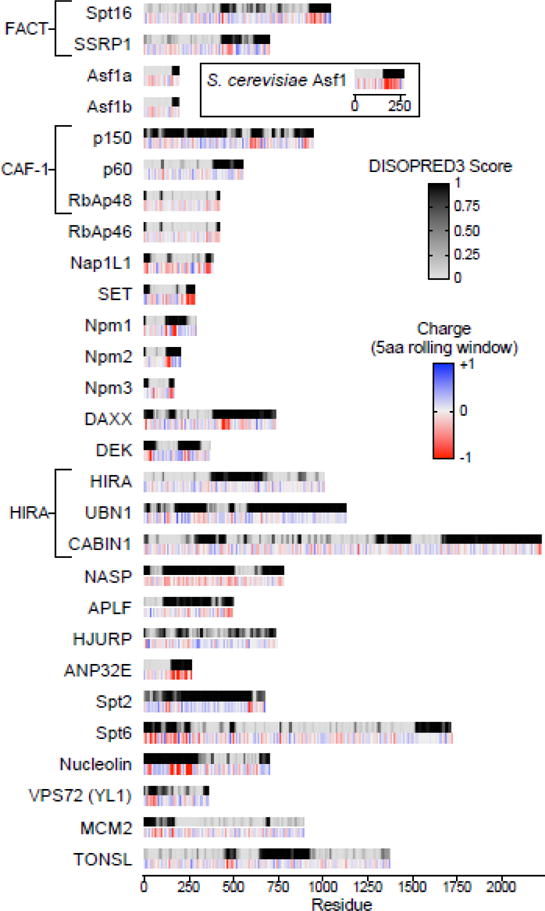
A. Top: By-residue DISOPRED3 disorder predictions for 28 human histone chaperones. Primary sequences colored light gray to black from 0 to 1 by DISOPRED3 score. 0=predicted to be highly ordered, 1=predicted to be highly disordered. Disordered regions >25aa are found in 26/28 human histone chaperones listed. Bottom: EMBOSS-calculated charge of the primary sequences of human histone chaperones, colored red to blue from −1 to +1. Charge calculated using a 5-amino acid sliding window. Acidic stretches are found in 23/28 human histone chaperones and cluster in predicted disordered regions. Insert: DISOPRED3 and EMBOSS charge analysis of yeast Asf1 showing that the yeast Asf1 has a longer and more acidic disordered C-terminus compared to human Asf1a and Asf1b.
These regions of histone chaperones, like most IDRs, are enriched in polar and charged amino acids with very few hydrophobic amino acids. We note that regions of different charge tend to cluster into acidic and basic stretches. We define an acidic stretch as a region in the primary sequence of a protein at least 8 amino acids in length and composed of at least 75% glutamate or aspartate residues, though stretches of more than 15 tandem acidic residues exist in many histone chaperones. Based on this definition, acidic stretches, predicted to be important mediators of histone interactions, are found in 23 out of the 28 human histone chaperones listed (Table 1) and are consistently found in regions that are predicted to be disordered (Figure 3). Despite little conservation in the primary sequences, there has likely been selective pressure to maintain these highly charged IDRs in histone chaperones. Additionally, many of the five chaperones that don’t have acidic stretches in their primary sequence do have acidic regions on their folded structures, further indicating acidic regions as key mediators in histone interactions [34,68–70]. Below, we present recent evidence for roles of IDRs and acidic stretches, in relationship to previously characterized ordered domains, within individual chaperone families.
Table 1.
Intrinsic Disorder and Acidic Stretches in Human Histone Chaperones
| Histone Chaperone | Uniprot Entry | Apparent Histone Specificity | % Disordered (RAPID prediction) | % Disordered (DISOPRED3 prediction) | Theoretical Isoelectric Point (pI) | Acidic Stretch? | Key References |
|---|---|---|---|---|---|---|---|
| FACT Spt16 | Q9Y5B9 | H2A/H2B | 22 | 34 | 5.50 | Yes | [71,73,83] |
| FACT SSRP1 | Q08945 | H3/H4 | 37 | 38 | 6.44 | Yes | [71,73,79] |
| Asf1a | Q9Y294 | H3/H4 | 14 | 22 | 4.29 | No | [32,132,146] |
| Asf1b | Q9NVP2 | H3/H4 | 9 | 21 | 4.46 | No | [32,132,145,146] |
| CAF-1 p150 | Q13111 | H3.1/H4 | 45 | 72 | 5.69 | Yes | [135,138,216] |
| CAF-1 p60 | Q13112 | H3.1/H4 | 24 | 34 | 7.18 | No | [134,135] |
| CAF-1 RbAp48 | Q09028 | H3.1/H4 | 16 | 13 | 4.74 | No | [34,157–159,162] |
| RbAp46 | Q16576 | H3.1/H4 | 12 | 13 | 4.89 | No | [17,34,159] |
| Nap1L1 | P55209 | H2A/H2B | 39 | 30 | 4.36 | Yes | [85,88,89,93] |
| SET | Q01105 | H3/H4 and H2A/H2B | 60 | 35 | 4.22 | Yes | [86,217,218] |
| Npm1 | P06748 | H3/H4 and H2A/H2B | 53 | 48 | 4.64 | Yes | [40,108,110,111] |
| Npm2 | Q86SE8 | H2A/H2B | 68 | 49 | 4.97 | Yes | [5,31,35,36,95] |
| Npm3 | O75607 | H3/H4 and H2A/H2B | 30 | 36 | 4.55 | Yes | [113,114] |
| DAXX | Q9UER7 | H3.3/H4 | 41 | 63 | 4.79 | Yes | [41,176,177] |
| DEK | P35659 | H3.3/H4 | 77 | 55 | 8.69 | Yes | [178,219] |
| HIRA | P54198 | H3.3/H4 | 16 | 36 | 8.40 | Yes | [23,69,207] |
| UBN1 | Q9NPG3 | H3.3/H4 | 50 | 76 | 9.37 | Yes | [168,172,173] |
| CABIN1 | Q9Y6J0 | H3.3/H4 | 22 | 49 | 5.70 | Yes | [169,220] |
| NASP | P49321 | H3/H4 | 49 | 75 | 4.26 | Yes | [27,221,222] |
| APLF | Q8IW19 | Macro-H2A/H2B | 49 | 69 | 4.98 | Yes | [127–130] |
| HJURP | Q8NCD3 | CENP-A/H4 | 29 | 47 | 9.40 | Yes | [33,192,196] |
| ANP32E | Q9BTT0 | H2A.Z/H2B | 56 | 44 | 3.76 | Yes | [30,121] |
| Spt2 | Q68D10 | H3/H4 | 67 | 83 | 9.79 | Yes | [163,166,167] |
| Spt6 | Q7KZ85 | H3/H4 | 27 | 31 | 4.81 | Yes | [165,223] |
| Nucleolin | P19338 | H2A/H2B | 62 | 58 | 4.60 | Yes | [224,225] |
| VPS72 (YL1) | Q15906 | H2A.Z/H2B | 51 | 46 | 6.09 | Yes | [122–124] |
| MCM2 | P49736 | H3/H4 | 16 | 43 | 5.34 | Yes | [137,148,149] |
| TONSL | Q96HA7 | H3.1/H4 | 23 | 32 | 5.99 | Yes | [226,227] |
Examples of IDRs and Acidic Stretch Function in H2A/H2B Chaperones
FACT
Facilitates Chromatin Transcription (FACT) is a heterodimeric H2A/H2B specific chaperone composed of Spt16 and SSRP1 (known as Pob3 in yeast) subunits [71,72]. FACT was originally identified as a factor that enhances RNA Pol II transcription through chromatin templates [73]. Subsequent analyses demonstrated that FACT is a histone chaperone responsible for disassembly and reassembly of nucleosomes during transcription and DNA replication [74,75]. More recent single molecule FRET studies show that FACT binds to mono-nucleosomes and can induce partial uncoiling of nucleosomal DNA from histones, consistent with a role for FACT in activating transcription elongation through chromatinized DNA templates [76]. Structural studies on the FACT complex show that it is composed of four distinct domains: Spt16 N-terminal domain (Figure 4A), the dimerization domains of Spt16 and SSRP1, the middle domains of Spt16 and SSRP1 (Figures 4B and 4C, respectively), and the long C-terminal domains (CTD) of Spt16 and SSRP1, which are predicted to be disordered and have acidic stretches (Figure 3) [77–79]. Structural work has shown that the middle domain of Spt16 is a major site of H2A/H2B interaction [80], whereas SSRP1 is thought to interact mainly with H3/H4 and DNA to aid in H2A/H2B displacement from intact nucleosomes [74,79]. Comprehensive quantitative histone binding studies using Spt16, SSRP1, and the FACT heterodimer show that FACT binds specifically to histones H2A/H2B mainly through its Spt16 subunit. This interaction is promoted by the presence of histone tails, and deletion of the acidic CTD of the Spt16 subunit decreases histone affinity by ~6 fold though loss of the CTD did not completely abolish binding [81]. Recently, it was also shown that FACT can bind to, and is critical for the deposition of, the H2A/H2B-like CENP-T/-W complex at centromeres [82]. The disordered CTD of the Spt16 subunit is also necessary for this interaction, possibly indicating that there may be alternative ligands that can be recognized by acidic regions of many histone chaperones. This “ligand switching” mechanism may provide additional levels of cellular regulation by acidic stretches of histone chaperones.
Figure 4. Diversity in the Structures of Various Histone Chaperones.
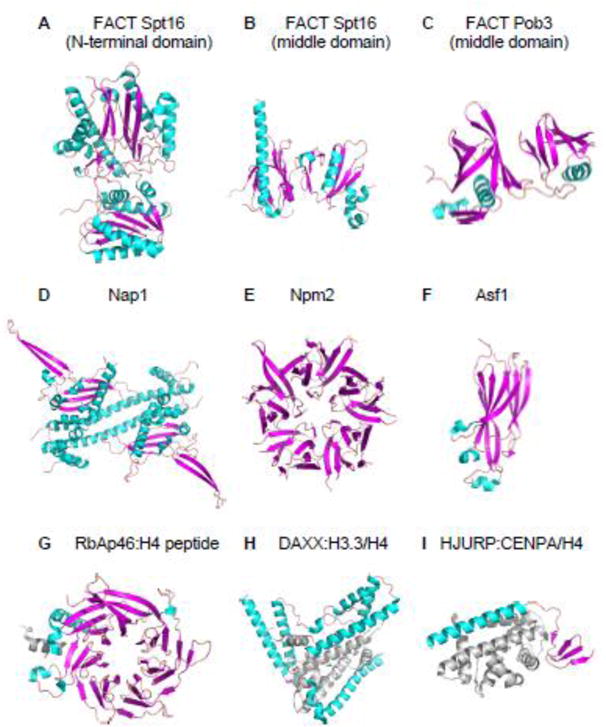
A. Crystal structure of the N-terminal domain (residues 1–451) of yeast Spt16 subunit of FACT (PDB: 3BIT). Residues 452–1035 are not included. Helices, sheets and loops of histone chaperones are colored cyan, magenta, and light pink, respectively, throughout this figure.
B. Crystal structure of the middle domain (residues 653–944) of yeast Spt16 subunit of FACT (PDB: 4KHO). Residues 1–652 and 945–1035 are not included.
C. Crystal structure of the middle domain (residues 237–474) of yeast Pob3 (homologue of human SSRP1) subunit of FACT (PDB: 2GCL). Residues 1–236 and 474–552 are not included.
D. Crystal structure of the core domain (residues 74–365) of yeast Nap1 dimer (PDB: 2Z2R). Residues 1–73 and 366–417 are not included.
E. Crystal structure of the core domain (residues 16–118) of Xenopus laevis Nucleoplasmin pentamer (PDB: 1K5J). Residues 1–15 and 119–195 are not included.
F. NMR structure of the core domain (residues 1–156) of human Asf1a (PDB: 1TEY). Residues 157–204 are not included.
G. Crystal structure of human RbAp46 (residues 7–410) in complex with H4 peptide (residues 28–42) (PDB: 3CFV). Residues 1–6 and 411–425 of RbAp46 are not included. H4 peptide colored gray.
H. Crystal structure of the core domain (residues 181–387) of human DAXX in complex with a H3.3/H4 dimer (PDB: 4HGA). Residues 1–180 and 388–740 of DAXX are not included. H3.3/H4 dimer colored gray.
I. Crystal structure of human HJURP (residues 14–74) in complex with a CENP-A/H4 dimer (PDB: 3R45). Residues 1–13 and 75–748 of HJURP are not included. CENP-A/H4 dimer colored grey.
A recent study used scanning alanine mutations in yeast Spt16 and Pob3 (homologue of vertebrate SSRP1) to identify regions of the disordered, acidic CTDs that are important for cell proliferation and histone binding [83]. Interestingly, the authors identify not only acidic residues, but also conserved aromatic residues adjacent to acidic stretches as key mediators of histone interactions. A crystal structure determination of the major Spt16 peptide (966EEEVSEY972) bound to H2A/H2B shows a strikingly conserved mode of interaction; with Tyr972 anchored into a hydrophobic groove of the H2A/H2B dimer created by H2B residues Tyr45 and Met62. In addition, Glu968 of Spt16 makes electrostatic and hydrogen bond contacts capping the basic dipole created by the N-terminus of α2 of H2B (Figure 5A). This “anchoring and capping” mechanism appears to be extremely well conserved among at least H2A/H2B interacting proteins, as the Spt16 peptide overlays nearly perfectly with previously solved crystal structures of peptides from the H2A.Z/H2B chaperone ANP32E (Figure 5B) and the chromatin remodeler SWR1 (Figure 5C). Importantly, all of these peptides bind sites that are at the histone:DNA interface in the nucleosome structure, suggesting that these acidic peptides directly compete with DNA for binding histones. Furthermore, the authors identify a region in the CTD of Pob3 critical for binding H2A/H2B that has an extremely similar aromatic-adjacent-to-acidic composition (507SEVEDF512), and that mutation of many of these residues in Spt16 and Pob3 showed moderate Spt null phenotypes in vivo. Taken together with structural studies of other chaperone:histone complexes, these results strongly suggest a conserved mode of interaction. Furthermore, the anchoring of single aromatic residues within the chaperone may provide an efficient means of achieving histone specificity, otherwise not achievable by low-complexity acidic stretches. The structures presented in Figure 5 represent small regions of histone chaperone IDRs that become ordered upon binding histones. There may also exist histone chaperone IDRs that remain disordered in the functional complex, but this has yet to be described in detail.
Figure 5. Recent Structural Insights into the Roles of Disordered Regions in Binding Histones.
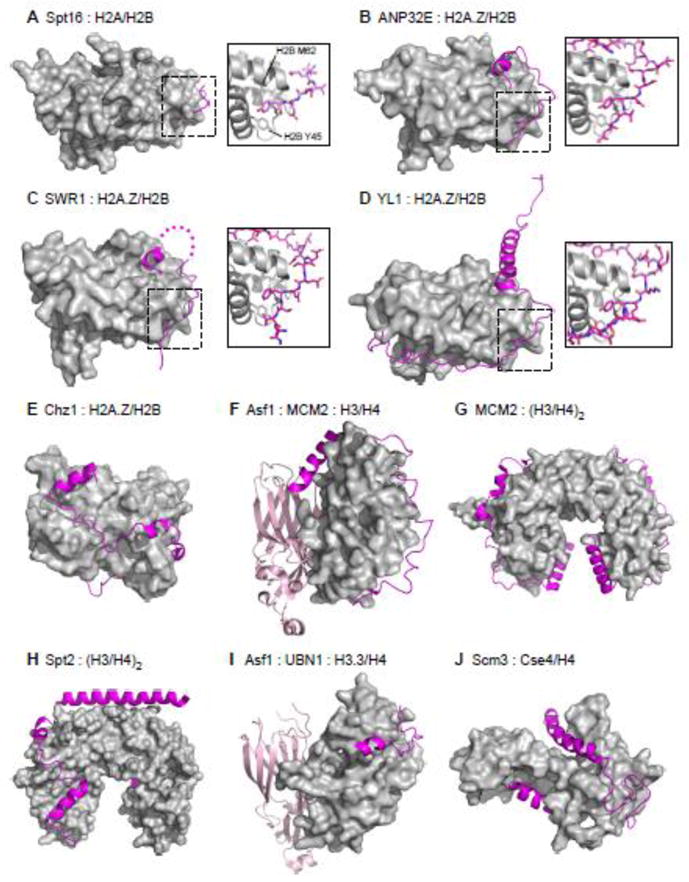
A. Crystal structure of a peptide derived from the acidic disordered C-terminus of yeast Spt16 (residues 967–972) bound to a H2A/H2B dimer (PDB: 4WNN). Chaperone peptide represented as a magenta ribbon, histones represented as grey surface. Boxed: Details of the interaction, with Spt16 peptide represented as magenta sticks and histones shown as gray ribbon with H2B residues Tyr45 and Met62 shown as sticks. Spt16 anchors a conserved tyrosine into the hydrophobic pocket created by H2B Tyr45 and Met62, and uses neighboring acidic residues to cap helix 2 of H2B.
B. Crystal structure of a peptide derived from the acidic disordered C-terminus of human ANP32E (residues 215–237) bound to a H2A.Z/H2B dimer (PDB: 4CAY). Same coloring and representation used as in A. ANP32E also shows conserved “anchoring and capping” binding mechanism to the same site on the H2A.Z/H2B dimer. Additional contacts are made between H2A.Z specific residues and ANP32E, and are necessary for chaperone specificity.
C. Crystal structure of a peptide derived for the Z-domain of yeast SWR1 chromatin remodeler (residues 598–628) bound to a H2A.Z/H2B dimer (PDB: 4M6B). Same coloring and representation used as in A. SWR1 also shows conserved “anchoring and capping” binding mechanism to the same site on the H2A.Z/H2B dimer.
D. Crystal structure of a peptide derived from the acidic disordered N-terminus of Drosophila YL1 histone chaperone component of the SCRAP/SWR1 chromatin remodeling complex (residues 4–71) bound to a H2A.Z/H2B dimer (PDB: 5CHL). Same coloring and representation used as in A. YL1 also shows conserved “anchoring and capping” binding mechanism to the same site on the H2A.Z/H2B dimer.
E. NMR structure of yeast Chz1 histone chaperone core (residues 1–62) bound to a H2A.Z/H2B dimer (PDB: 2JSS). Same coloring and representation used as in A. Chz1 binds to H2A.Z/H2B via a long irregular extended conformation capped by short N and C-terminal helices. No clear anchoring a capping motif is apparent in this structure.
F. Crystal structure of the quaternary complex of human histone chaperone Asf1a core (residues 2–154) and replicative helicase MCM2 (residues 69–121) bound to a H3/H4 dimer (PDB: 5C3I). Same coloring and representation used as in A, but with Asf1 core represented as a light pink ribbon. MCM2 adopts an extended conformation capped by a C-terminal helix. Tyr90 of MCM2 is anchored into a hydrophobic pocket created by H3 helices α1 and α2, with adjacent acidic residues making electrostatic contacts with histone surface residues similar to conserved “anchoring and capping” binding motifs.
G. Crystal structure of human replicative helicase subunit MCM2 (residues 69–121) bound to a H3/H4 tetramer (PDB: 5BNV). Same coloring and representation used as in A. MCM2 adopts a very similar conformation as in F.
H. Crystal structure of the human histone chaperone Spt2 (residues 606–675) bound to a H3/H4 tetramer (PDB: 5BS7). Same coloring and representation used as in A. Spt2 binds to the H3/H4 tetramer in an extended conformation capped by helices, most notably a long acidic C-terminal helix which bridges the dimer-dimer interface in the H3/H4 tetramer. No clear anchoring a capping motif is apparent in this complex structure.
I. Crystal structure of the quaternary complex of yeast histone chaperone Asf1 core (residues 5–154) and a peptide from UBN1 (residues 122–142) bound to a H3.3/H4 dimer (PDB: 4ZBJ). Same coloring and representation used as in A, but with Asf1 core represented as a light pink ribbon. The UBN1 peptide adopts an extended conformation capped by a small N-terminal helix. Tyr132 and Phe138 of UBN1 are anchored into hydrophobic grooves of the H3.3/H4 dimer, and Asp133, Asp136, and Asp140 form electrostatic contacts similar to other anchoring and capping motifs.
J. NMR structure of the Cse4-binding domain of yeast Scm3 (residues 90–169) bound to a Cse4/H4 dimer. Same coloring and representation used as in A. Scm3 binds to Cse4/H4 in an extended conformation with N and C-terminal helices. The N-terminal helix of Scm3 contains residues Trp107, Ile110, Ile111, Tyr114, and I117 that make hydrophobic contacts with many Cse4-specific residues, and many acidic residues on the middle loop and C-terminus likely make electrostatic contacts.
Nap1 and Nap1L1
Nucleosome Assembly Protein 1 (Nap1) is one of the major H2A/H2B and linker histone chaperones first identified in yeast [29,84]. The crystal structure of the central ordered region of yeast Nap1 (residues 70–370) was solved over a decade ago [85] (Figure 4D). This structure shows that Nap1 exists as a stable dimer held together by an unusual non-coiled coil motif, similar to the later solved structures of the histone chaperones SET and yeast Vps75 [86,87]. Absent from the structure of Nap1 are the N-terminal 70 and C-terminal 47 amino acids, both of which are predicted to be disordered. Deletions of both the N and C-terminal disordered regions independently and in combination suggest that both regions contribute to binding H2A/H2B and likely act synergistically [24]. The C-terminal region, in particular, is highly acidic and has thus been named the C-terminal acidic domain (CTAD). Though truncations of the CTAD of yeast Nap1 did not completely abolish its histone binding or deposition activity in vitro [37], the CTAD is necessary for chaperone dependent dissociation of H2A/H2B from nucleosomes and nucleosome sliding [29], indicating that this region may be important in competing for histone:DNA interactions. Consistent with this hypothesis, mechanistic studies of yeast Nap1 indicate that it thermodynamically favors proper nucleosome formation by disfavoring non-nucleosomal histone:DNA interactions and that in vivo deletion of Nap1 caused increased H2A/H2B occupancy on DNA and altered gene expression [24,88]. This mechanism of disfavoring non-nucleosomal histone:DNA interactions may be a general feature of non-catalytic chaperones. While an atomic-resolution structure of the Nap1:H2A/H2B complex has not yet been solved, hydrogen-deuterium exchange mass spectrometry (HDX-MS) and low resolution crystallography studies indicate direct interactions between H2A/H2B and acidic residues on both the central region of Nap1 and CTAD, though these studies conflict on the oligomerization state of the complex [89,90].
Compared to yeast Nap1, the human homologue Nap1-like 1 (Nap1L1) contains a very similar domain layout, with a highly conserved and ordered central region between two disordered regions (Figure 3). Nap1L1 has been demonstrated to be a histone chaperone for both H2A/H2B and linker histone H1 [91,92]. Recent binding studies comparing human Nap1L1 with yeast Nap1 indicate that deletion of the CTAD has a greater negative effect on the human protein than on the yeast homologue [93]. The same study showed that two distinct regions in the Nap1L1 CTAD can each independently bind to an H2A/H2B dimer, and that these regions are both composed of acidic residues adjacent to a single tyrosine (349DDDDDYDE356 and 372ENDPDYDP379), similar to anchoring and capping motifs in peptides from Spt16, ANP32E and SWR1 [30,83,94], again suggesting a conserved mechanism of histone recognition.
Npm Family
Nucleoplasmin (Npm2) is the archetypal molecular chaperone and the first protein to be called a chaperone [5,95]. Npm2 is the major H2A/H2B storage chaperone present in vertebrate oocytes and early embryos, but is not found in any other somatic cells [96]. Xenopus laevis Npm2 contains a highly stable core domain (residues 15–118), whose structure has been solved and assembles into a homopentamer composed of antiparallel beta-sheets (Figure 4E) [31], extremely similar to the structure of the human protein [97]. This homopentameric core domain fold is also found in a variety of other chromatin-related proteins in Drosophila, such as FKBP39 and many others with unknown function [98]. In Npm2, the core domain is flanked by the short N-terminal tail (residues 1–14) and longer C-terminal tail domain (residues 119–195), both of which are predicted to be disordered (Figure 3). The C-terminal tail contains multiple acidic and basic stretches, including the longest acidic stretch in X. laevis Npm2 (called “A2” and comprised of 18 glutamates and aspartate residues in a 25 amino acid stretch). Negative stain electron microscopy studies have suggested the importance of the Npm2 C-terminal tail domain in histone interactions [42,99]. Quantitative binding studies using C-terminal truncations of Npm2 have shown that though it is not required for binding, the C-terminal tail domain dramatically enhances affinity of Npm2 for H2A/H2B and H3/H4 [36,100]. Interestingly, truncation mutants that retain A2, but lose the remainder of the tail domain seem to have increased affinity toward histones and more robust deposition in in vitro chromatin assembly assays, suggesting that the extreme C-terminal region negatively regulates the function of Npm2 [35,101]. The molecular mechanisms underlying this regulatory function of the disordered tail domain of Npm2 remain unknown.
Npm1 (nucleophosmin/numatrin/B23/NO38) is an Npm-family member found in somatic cells and is the most commonly mutated gene in normal karyotype acute myeloid leukemia (AML) [102]. Npm1 has a similar domain layout as Npm2, including an N-terminal core pentamerization domain and a disordered C-terminal tail domain with acidic stretches (Figure 3) [103]. Npm1 also contains an extreme C-terminal 3-helix bundle with a nucleolar localization signal (NoLS), which is not found in other Npm family members (reviewed in [104]). AML-associated mutations in this 3-helix bundle lead to unfolding and loss of nucleolar localization of Npm1 [105,106]. The disordered C-terminus of Npm1 has also been implicated in organizing subcompartments of the nucleolus through phase-separation [59,60]. Npm1 was originally described as a histone chaperone, but has a multitude of other functions in the cell including nucleic acid binding, ribosome biogenesis, transcriptional regulation, and regulation of the ARF/p53 pathway [104,107–109]. Pull down assays show that Npm1 interacts with both H3/H4 and H2A/H2B, and C-terminal truncations show that the tail is necessary for binding H2A/H2B, linker histone H1, and histone deposition, whereas the core domain is sufficient for H3/H4 binding [40,110]. A separate study showed that C-terminal truncation mutants can act in a dominant negative fashion to inhibit rRNA transcription and cell proliferation [111]. Npm1 was also shown to regulate its RNA binding activity through intra and intermolecular interactions in the tail domain, implicating this disordered region as a site of regulation potentially similar to Npm2 [112].
The related Npm3 protein has a conserved core domain, but a shorter C-terminal disordered tail domain than either Npm1 or Npm2, and contains a single acidic stretch (Figure 3). Few studies have probed the function of the somatic Npm3 protein, but it binds to both H2A/H2B and H3/H4 with a modest preference for H3/H4 in vitro, and its domain arrangement suggests a similar model of histone binding to the disordered acidic stretch [113]. Interestingly, Npm3 was also shown to form hetero-oligomers with Npm1, which can function to suppress the RNA binding activity of Npm1 [114]. Further studies into the roles of Npm3 are necessary to delineate the function of this protein and its hetero-oligomers with family members Npm1 and Npm2.
Examples of IDRs and Acidic stretch Function in H2A Variant Chaperones
Histone chaperones are generally specific for either H2A/H2B dimers, H3/H4 dimers or H3/H4 tetramers, however histone variants pose a unique challenge. Histone variants serve highly specific functions on chromatin, but often only differ from their canonical counterparts by a few amino acids. Variant-specific chaperones recognize, ensure accurate deposition and aid in the removal of these non-canonical histone variants. Below are examples of how disordered regions and acidic stretches of histone chaperones can aid in specifically recognizing, depositing, or displacing these variants.
H2A.Z Chaperones
H2A.Z is a variant of canonical H2A that is incorporated into nucleosomes at promoters of actively transcribing genes. H2A.Z has been shown to alter nucleosome stability and is thought to facilitate more open chromatin forms [115,116]. H2A.Z incorporation and DNA methylation are found in mutually exclusive areas of the genome of Arabidopsis and are antagonistic to one another, indicating a complex mechanism for maintenance of gene expression by histone variant incorporation [117]. Though similar to canonical H2A (64% identity), the cell specifically recognizes and incorporates H2A.Z in the proper genomic location, ensuring accurate and stable gene expression.
The chaperone Chz1 was identified in yeast as the major H2A.Z/H2B specific chaperone [118]. The core domain of Chz1 has a bipolar charge distribution, with acidic residues near the N-terminus and basic residues near the C-terminus, and is predicted to be almost completely disordered. The NMR structure of the Chz1:H2A.Z/H2B complex shows a largely extended conformation for Chz1 draped over the H2A.Z/H2B dimer and capped by 2 short helices (Figure 5E), similar to the structure of the MCM2:(H3/H4)2 complex (Figure 5G) [119]. This structural study and follow up biophysical studies indicate that Chz1 relies predominantly on its bimodal charge distribution to make electrostatic contacts with both basic and acidic surfaces of the H2A.Z/H2B dimer for its high affinity [43,120]. Some of these surface residues are only present in the H2A.Z/H2B dimer, but not in canonical H2A, and are likely important for the specificity of the chaperone.
A metazoan homologue for Chz1 has not been identified, however it was recently demonstrated that the protein ANP32E is a histone chaperone that can specifically remove H2A.Z/H2B dimers from chromatin [30,121]. In these studies, the authors independently solve the crystal structure of a short acidic peptide derived from ANP32E bound to H2A.Z/H2B (Figure 5B). This complex is strikingly similar to the Spt16 peptide bound to H2A/H2B dimers [83], with a single tyrosine anchor and multiple acidic residues capping the H2A.Z/H2B dimer, along with additional non-polar contributions from Leu218 and Met222 of ANP32E. Compared to the Chz1:H2A.Z/H2B complex (Figure 5E), the ANP32E peptide binds to a different site on the H2A.Z/H2B dimer, specifically recognizing H2A.Z/H2B through a single glycine residue (Gly92) in the C-terminal helix of H2A.Z. Binding of ANP32E also leads to a conformational extension of the C-terminal helix of H2A.Z, promoting further contacts between the histone variant and chaperone. Comparison of this structure with the structure of a nucleosome containing H2A.Z/H2B indicates that this chaperone-induced helix extension likely hinders nucleosomal interactions between the C-terminus of H2A.Z and the C-terminus of H4, consistent with ANP32E assisting in the removal of H2A.Z/H2B dimers from nucleosomes [115]. Interestingly, a peptide derived from the catalytic subunit of the chromatin remodeler SWR1 was also crystallized in complex with a H2A.Z/H2B dimer, where it adopts an almost identical conformation and binding site as ANP32E (Figure 5C) [94]. This study suggests that certain ATP-dependent chromatin remodelers interact with histones in a similar fashion to chaperones.
Another H2A.Z/H2B chaperone, YL1, was recently identified in complex with H2A.Z/H2B dimers in the soluble portion of cell extracts [122]. While ANP32E and SWR1 appear specifically aid in the removal of H2A.Z from chromatin, YL1 acts as a depositor of this variant histone. Two independently solved crystal structures of YL1 (human and Drosophila proteins) bound to H2A.Z/H2B (Figure 5D) share many similarities to the ANP32E:H2A.Z/H2B complex structure (Figure 5B), including the same anchoring and capping mechanism, as well as a similar extension of the C-terminal helix of H2A.Z [123,124]. YL1 also has some unique features not seen in the ANP32E:H2A.Z/H2B structure, such as more extensive hydrophobic interaction with the C-terminal helix, particularly Tyr30 of YL1 that is positioned at the interface of the extended C-terminal helix and interacts directly with the H2A.Z specific residue Asp97. YL1 continues to extend around the H2A.Z/H2B dimer, following the path of nucleosomal DNA and ending with its C-terminal region making contacts with the α1 helix of H2A.Z. The positioning of this C-terminal region is similar to that of the N-terminal region of Chz1 (Figure 5E), suggesting that YL1 may use a combination of the binding mechanisms observed for both ANP32E and Chz1. Together, these four structures of H2A.Z/H2B specific chaperones (Chz1, ANP32E, SWR1, and YL1) give key insights into commonalities and differences in molecular mechanisms used by chaperones to specifically recognize, remove and deposit this variant histone.
APLF
MacroH2A is a histone variant of H2A that has a C-terminal macro domain linked to a H2A homology domain and is generally found in transcriptionally repressed regions of the genome, including throughout the inactive X chromosome [125,126]. Aprataxin-PNK-like Factor (APLF) is an acidic nuclear protein that is recruited to sites of DNA damage via interactions with PARP-1 catalyzed poly-ADP ribose chains [127–129]. APLF participates in chaperoning histones at DNA damage sites [130]. In the same study the authors use a variety of truncations to identify the C-terminal acidic domain, which shares homology to the Nap1L1 CTAD, as a major interaction site for both H2A/H2B and H3/H4. Interestingly, the C-terminal acidic domain is predicted to be disordered and has acidic and aromatic residues (Figure 3). In particular, its 480DEDSDWE486 region is reminiscent of anchoring and capping motifs present in Spt16, ANP32E and SWR1 peptide structures [30,83,94]. The authors of this study further show that APLF deficient cells are unable to recruit macroH2A to sites of DNA damage, suggesting APLF is the first identified chaperone specific to macroH2A. Pull-down experiments using truncations of the C-terminal acidic domain lose the ability to bind all histones, including macroH2A. The molecular mechanisms underlying the specificity of APLF toward macroH2A remain largely unknown.
Examples of IDRs and Acidic Stretch Function in H3/H4 Chaperones
Asf1 and MCM2
Antisilencing Factor 1 (Asf1) is a somatic H3/H4 chaperone that was originally identified in yeast as a protein which, upon overexpression, could de-repress silent loci [131]. Asf1 is responsible for binding the dimeric form of H3/H4 [132], aiding in histone acetylation prior to nuclear import [133], and coordinating with either CAF-1 for replication-dependent deposition of H3.1/H4, or HIRA, DAXX or DEK for replication-independent deposition of H3.3/H4 [134–137]. CAF-1 likely tetramerizes H3.1/H4 prior to deposition, whereas it is less clear if H3.3/H4 are deposited as dimers or tetramers [23,41,135,138]. In mammals, Asf1 is present in cells in two distinct isoforms, Asf1a (also known as CIA) and Asf1b. Structurally, human Asf1a and Asf1b are composed of an N-terminal core of antiparallel beta-sheets (residues 1–156) (Figure 4F) and a C-terminal disordered domain (residues 157–204). Multiple NMR and x-ray crystallography studies of Asf-1 N-terminal core bound to H3/H4 or the H3 C-terminal helix yield insights into how Asf-1 blocks the tetramerization of two H3/H4 dimers by using both hydrophobic and acidic residues to compete for the dimer:dimer interaction surface [32,132,139–141]. Though human Asf1a and Asf1b lack distinct acidic stretches, the C-terminal disordered domains are slightly acidic (Figure 3), whereas the yeast Asf1 C-terminus is both longer and contains multiple acidic stretches (Figure 3 insert). Genetic disruption of the acidic C-terminus in yeast shows phenotypes consistent with loss of H3/H4 binding [142]. Binding studies using serial truncations of the yeast Asf1 C- terminal domain showed a ~200-fold reduction in affinity upon loss of the entire C-terminal domain, but no effect upon truncation of the C-terminal 69 amino acids [39], indicating a major site of H3/H4 interaction within residues 155–210. Further cross-linking experiments in the same study implicate a direct interaction between Glu210 and H3/H4. In contrast, introduction of a truncated Asf1 lacking the C-terminal acidic stretch into a S. pombe Asf1 null strain was sufficient to rescue the lethality phenotype, suggesting that this region may be dispensable in vivo for S. pombe yeast [143].
The two human isoforms of Asf1 share 71% identity. Interestingly, the core of Asf1a and Asf1b share a high degree of homology (84% identity), whereas their C-terminal tails are far more divergent (28% identity). Despite not having an acidic stretch, human Asf1a has histone chaperone activity in vitro [144]. It was also recently shown that Asf1b, but not Asf1a, is necessary for cellular proliferation, and that Asf1b levels correlate with breast cancer disease outcomes [145]. Further evolutionary analyses showed that Asf1a and Asf1b arose from a duplication in the ancestor of jawed vertebrates, and that the C-terminus of both Asf1a and Asf1b have undergone positive selection consistent with distinct functional roles for the C-termini in the cell [146]. More complete experiments are necessary to understand the differences in the histone chaperone functions of these two isoforms at the molecular level.
Another question that remains is why the human Asf1 C-terminal domains have lost so much of their acidic character compared to yeast Asf1 (Figure 3 insert). One hypothesis is that another disordered acidic protein in human cells can compensate for this lack of acidic stretches to aid in Asf1-mediated chaperoning of H3/H4. In support of this hypothesis, previous functional analyses show that Asf-1 cooperates with MCM2-7 subunits of the human DNA helicase to bind histones at the replication fork, and this mechanism can regulate replication fork progression by histone supply and demand in human cells [137]. In addition, the direct interaction between human MCM2 and histone H3 has been known for 20 years [147]. More recent NMR and crystallography studies have yielded molecular insights into the mechanism of cooperation between the MCM2 subunit of human DNA helicase and Asf1 in chaperoning H3/H4 at the replication fork [148,149]. The heterotetrameric (Asf1 Core, MCM2, and H3/H4 dimer) crystal structures reveal cooperativity between the ordered region of Asf1 and the highly extended and acidic MCM2 in binding opposite faces of the H3/H4 dimer (Figure 5F). MCM2 makes extensive contacts with H3/H4 using mainly electrostatic interactions and with a single tyrosine (Tyr90) anchored into a hydrophobic pocket created by H3 helices α1 and α2, reminiscent of anchoring and capping motifs in other H2A/H2B chaperones. These studies also support a model in which parental H3/H4 tetramers are dissociated into dimers and chaperoned separately onto the DNA of daughter strands, suggesting that histone chaperones may play important roles in the processes of epigenetic inheritance. However, MCM2 can also bind the tetrameric form of H3/H4 (Figure 5G) [148], and other studies suggest that H3.1/H4 tetramers are largely not split into dimers during replication-dependent deposition whereas H3.3/H4 tetramers are split during both replication-independent and replication-dependent deposition [150,151]. Together these studies suggest that H3/H4 splitting decisions during replication may be dependent on functionally distinct chromatin regions. A better understanding of the network of chaperone:histone interactions during DNA replication is necessary to give mechanistic insight into these conflicting observations.
CAF-1
Chromatin Assembly Factor 1 (CAF-1) is the major H3.1/H4 chaperone that acts synergistically with Asf1 to deposit H3.1/H4 during DNA replication [135,152,153]. CAF-1 is a heterotrimeric chaperone composed of p150, p60 and RbAp48 (also known as p48) subunits, also known as Cac1, Cac2, and Cac3 in yeast, respectively. Early studies suggested that human CAF-1 binds to and deposits H3.1/H4 dimers in vivo [23], however more recent evidence suggests that yeast CAF-1 forms tetramers of two H3/H4 dimers and deposits the tetrameric form onto DNA [154,155], though the mechanisms of tetramerization in the human protein remains an active area of research. The Cac1/p150 subunit is largely disordered and contains a large acidic stretch (the glutamate/aspartate or ED domain) in the middle of the protein sequence (Figure 3). Pull-down studies using GST-tagged individual subunits show that Cac1 is the only subunit able to pull-down endogenous H3/H4 from HeLa cells [156]. Recent quantitative binding studies using individual subunits show that Cac1 has the highest affinity for H3/H4, though this affinity was an order of magnitude weaker than the trimeric CAF-1 complex indicating that other subunits likely aid in coordinating H3/H4 [138]. In the same study the authors show that Cac1 alone is sufficient to promote H3/H4 tetramerization and assemble tetrasomes on DNA, and by using truncation mutants they go on to show that the C-terminus, including the structured domain and disordered acidic stretch, is sufficient to induce H3/H4 tetramerization. Though a complete structure of the CAF-1:H3/H4 complex remains elusive, this study utilizes a hybrid of x-ray crystallography, hydrogen-deuterium exchange mass spectrometry (H/DX-MS) and chemical cross linking to provide novel information about the architecture of the complex.
Though Cac1/p150 is the largest subunit and independently binds histones and forms tetrasomes, the two smaller subunits (p60 and RbAp48) also aid in histone chaperone function of CAF-1. RbAp48 is the smallest subunit of the CAF-1 heterotrimer and is a highly ordered WD-repeat protein that forms a seven bladed β-propeller structure. RbAp48 is not an obligate subunit of CAF-1, as it can be found in complex with a multitude of other proteins [157,158]. The structure of RbAp46, a homologue of RbAp48 with 90% sequence identity, was solved along with a peptide derived from residues 25–41 of H4 (Figure 4G) [34]. This very ordered complex structure shows α1 of H4 inserted into a pocket on the side of the donut-shaped β-propeller forming extensive contacts with mainly hydrophobic and acidic residues from helices at the N and C-terminal regions and an acidic loop in RbAp46. Importantly, these key interacting residues in RbAp46 are 100% conserved in the CAF-1 subunit RbAp48, and it was previously shown that both RbAp46 and RbAp48 bind to the first helix of H4, making it very likely that the interaction observed in the RbAp46:H4 crystal structure is conserved in RbAp48 [159]. Additionally, the Drosophila protein Nurf55/p55 is nearly identical to mammalian RbAp46/48 and functions as a non-catalytic member of multiple chromatin remodeling complexes, including Polycomb Repressive Complex 2 (PRC2), to increase affinity toward histones [160]. Nurf55 was also crystallized with the first helix of H4 where it forms very similar contacts compared to RbAp46 [70,161]. In the same studies, the authors demonstrate that an acidic surface of Nurf55 can also interact specifically with the N-terminal tail of H3, and that the strength of this interaction is modulated by lysine 4 trimethylation (H3K4me3), demonstrating that these WD repeat proteins can bind multiple targets to increase affinity and specificity of chaperone and chromatin modifying enzyme complexes. These studies and others show that RbAp48 mainly binds the dimeric form of H3/H4 and that there is a conformational change in the H3/H4 dimer upon binding RbAp48 which inhibits tetramerization, suggesting a mechanism for exchange between Asf-1 and CAF-1 during H3/H4 deposition [162]. Taken together with studies of subunit p150, these studies indicate that there is likely cooperativity between ordered and disordered regions of the CAF-1 heterotrimer to bind to, tetramerize, and deposit H3/H4.
Spt Family of Transcriptional Regulators
Much of the research that identified new histone chaperones was originally done in yeast. Classic examples are experiments using Ty transposons to disrupt gene expression followed by forward genetic screens for secondary mutations that rescue gene expression. These suppressor of Ty (Spt) mutants are often involved in eukaryotic transcription processes, such as Spt15 (now known as TATA-binding protein), Spt3, 7, and 8 (members of the SAGA chromatin modifying complex), and Spt11 and 12 (H2A and H2B, respectively). Other Spt genes, such as Spt16 of FACT, Spt2, and Spt6 have been identified as histone chaperones in yeast [71,163–165].
More recent pull-down studies using H3/H4 conjugated beads and HeLa cell extracts identified a novel protein as a possible histone chaperone in metazoan cells [166]. Though this protein as a whole is basic (pI=9.79), pull-down studies using truncation mutants implicate the acidic C-terminus in interactions with histones H3/H4. This C-terminal region also shares homology to known yeast histone chaperone Spt2, and since no other known motifs were identified it was named human Spt2 (hSpt2). This study showed that hSpt2 can also assembly nucleosomes in vitro and is critical for RNA PolI-mediated transcription of rRNA, suggesting it functions as a histone chaperone in the nucleolus. The C-terminal Spt2 homology domain of hSpt2 was later crystallized bound to a H3/H4 tetramer (Figure 5H) [167]. The structure shows hSpt2 C-terminal domain bound around the periphery of a H3/H4 tetramer, mimicking the trajectory of nucleosomal DNA, and making contacts using both acidic and hydrophobic amino acids. The structure of Spt2 is mainly composed of short helices and loops, similar to structures of MCM2 and Chz1 bound to histones. In addition, hSpt2 also has a long C-terminal helix that makes key contacts with the four-helix bundle formed by the H3-H3 interface in the tetramer, consistent with this chaperone specifically recognizing H3/H4 tetramers.
Examples of IDRs and Acidic Stretch Function in H3 Variant Chaperones
H3.3 Chaperones
H3.3 is a H3 histone variant that is 96% identical to canonical H3.1. H3.3 incorporation into nucleosomes occurs largely in a replication-independent fashion and at gene promoters is generally associated with transcriptional activation, however it is also specifically deposited in heterochromatic regions. It is therefore the responsibility of a network of histone chaperones specific to this histone variant to incorporate it into these distinct regions of chromatin to ensure proper and stable gene expression.
HIRA is a highly conserved histone chaperone that was originally identified in yeast and was the first chaperone demonstrated to be specific to H3.3/H4 [23]. In humans, the HIRA protein was shown to be a member of a functional complex, along with proteins Ubinuclein-1 (UBN1) and Calcineurin Binding Protein 1 (CABIN1) [168,169]. The human HIRA complex (HIRA/UBN1/CABIN1) is known to cooperate with both Asf1a and RNA PolII, as well as a variety of transcription factors and chromatin remodelers to mediate replication-independent deposition of H3.3/H4 mainly into gene bodies, intergenic and regulatory regions of actively transcribing genes [69,170,171]. Though the HIRA protein is necessary for the function of the HIRA complex, UBN1 and CABIN both play critical roles in histone recognition and deposition by the HIRA complex. UBN1, in particular, was demonstrated to be necessary for the stability and function of the HIRA complex [172]. More recent x-ray crystallography of a small peptide of UBN1 (residues 122–142) bound to the Asf1:H3.3/H4 complex showed that UBN1 makes contacts with a number H3.3 specific residues, especially Gly90 (Figure 5I), and shares some similarities with the previously solved DAXX:H3.3/H4 structure (Figure 4H) [173,174]. Mutation and quantitative binding analyses in the same study showed that UBN1 is the major determinant of H3.3 specificity of the HIRA complex, and that mutation of Gly90 to the H3.1 residue Met90 caused loss of specificity. Though this region of UBN1 is not predicted to be fully disordered (Figure 3), it contains aromatic residues Tyr132 and Phe138 which are inserted into a hydrophobic groove in the H3.3/H4 dimer and polar residues Asp133, Asp136, Asp140, and Asn141 that make key contacts to the basic histone surface, similar to anchoring and capping motifs observed in many H2A/H2B chaperones [30,83,94,123].
HIRA is largely responsible for depositing H3.3/H4 into actively transcribing regions of the genome, however another chaperone pathway must exist to mediate deposition into telomeres and pericentric heterochromatin. To fulfill this role, the death-associated protein 6 (DAXX) cooperates with the alpha-thalassemia X-linked mental retardation (ATRX) chromatin remodeler in chaperoning H3.3/H4 independent of HIRA at telomeres and pericentric DNA repeats [175,176]. DAXX is composed of a core domain (residues 183–417) that is predicted to be ordered and sufficient to interact with H3.3/H4 [176], a partially ordered N-terminal domain (residues 1–182), and a long disordered C-terminal domain (residues 418–740) that contains a large acidic stretch (residues 433–488) (Figure 3). Pull-down experiments using DAXX truncations show that the central domain of DAXX that includes the acidic stretch (residues 302–495) interacts with H3.3/H4 more strongly than the N-terminal region (residues 1–302) [41], suggesting that the acidic stretch is an important mediator of H3.3/H4 interactions. The structure of the DAXX core domain in complex with a H3.3/H4 dimer shows that the core of the chaperone is highly helical and envelopes the histone dimer covering ~40% of the histone surface area (Figure 4H) [174]. This structure and mutational analysis show that DAXX also specifically recognizes Gly90 of H3.3, similar to the later solved Asf1:UBN1:H3.3/H4 structure. Though this structure is highly ordered, more recent HDX-MS analysis of DAXX alone and in complex with H3.3/H4 indicate that both DAXX and H3.3/H4 likely co-fold upon binding, and that though the core domain of DAXX is predicted to be ordered, it is likely highly dynamic in solution [177]. The exact contribution from the disordered, acidic C-terminal domain in binding to and specifically recognizing H3.3/H4 remains unclear.
Genetic screens in Drosophila identified the transcriptional coactivator DEK as a histone chaperone specific for H3.3/H4 upon phosphorylation by CK2 [178]. Staining of polytene chromosomes showed that DEK is associated with more open chromatin, suggesting that DEK deposits histones in transcriptionally active regions of the genome, in contrast to DAXX. Further studies show that H3.3/H4 dimers are normally recruited to PML nuclear bodies by Asf1 and DAXX [179], where they are stored by PML and DEK proteins prior to deposition, and that loss of DEK causes a global redistribution of H3.3 from telomeres to chromosome arms [180]. These studies indicate that there are multiple independent pathways to mediate H3.3/H4 deposition in functionally distinct chromatin regions, H3.3/H4 can be stored in PML body “triage” centers, and supply of this histone variant can be mediated by “gatekeeper” chaperones such as DEK. This may be a general phenomenon of histone chaperones, as many other histone variants, such as macroH2A, are found in functionally distinct domains of chromatin [181].
CENP-A Chaperones
CENP-A is a more divergent H3 variant histone (roughly 50% identity with canonical H3 in humans) that is incorporated specifically at the centromere [182]. Studies in S. cerevisiae have shown that the centromere is dictated by a ~125bp sequence of DNA that is necessary for the incorporation of Cse4 (the yeast homologue of CENP-A) into nucleosomes [183,184]. Cse4, in turn, is necessary for the recruitment of key complexes that make up the kinetochore to bridge mitotic spindles to centromeric chromatin during mitosis [185]. In metazoans, this process is much more complex, with much larger centromeric regions (0.3–5Mbp DNA) composed of highly variable repeated arrays of a 171bp sequence known as α satellite DNA, which is interspersed with both H3 and CENP-A containing nucleosomes [186]. Further biophysical analyses showed that CENP-A/H4 tetramers are more compact than canonical H3/H4 tetramers, and impart significant rigidity when incorporated into nucleosomes at the centromere, possibly important for maintaining centromeric identity in metazoans [187].
How CENP-A is specifically targeted to and deposited in these highly variable regions remains an area of active investigation. Analyses in yeast showed that the acidic domain of protein Scm3 is necessary to recruit Cse4 to centromeres and assemble centromere-specific nucleosomes [188,189]. NMR structural analysis of the histone-binding domain of Scm3 shows that this region is intrinsically disordered in solution and, by making a single chain Cse4-Scm3-H4 fusion protein, the structure was solved (Figure 5J) [190]. The structure shows that Scm3 wraps the Cse4/H4 dimer via a long irregular structure capped by a long N-terminal and shorter C-terminal helices, similar to the structure of Chz1 (Figure 5E). The chaperone packs it’s longer N-terminal helix against α2 of Cse4 and makes largely hydrophobic contacts to multiple Cse4-specific residues, though there are many acidic residues present in the loop and C-terminus of the Scm3. The structure also shows that Scm3 binding induces a major conformational change in the Cse4/H4 dimer that blocks a DNA binding site of Cse4/H4.
Bioinformatic and functional analyses showed that the protein Holliday Junction Recognition Protein (HJURP) is the human homologue of yeast Scm3 and is also necessary for assembly of CENP-A containing nucleosomes at the centromere [191–193]. Previous structural and mutational analyses showed that loop 1 and α2 of the histone fold region form a CENP-A targeting domain (CATD) and are necessary to direct CENP-A to centromeres, suggesting that this region is specifically recognized by the chaperone HJURP [194,195]. Truncation analysis showed that the N-terminal 80 amino acids of HJURP are necessary and sufficient for preferential recognition of CENP-A/H4 [196]. The crystal structure of the N-terminal region of HJURP bound to a CENP-A/H4 dimer was solved and shows that HJURP forms a long N-terminal helix, followed by a 15 amino acid loop and a three-stranded antiparallel β-sheet (Figure 4I) [33]. The amphipathic N-terminal helix packs against α2 of CENP-A in an antiparallel arrangement, in a similar position to the N-terminal helix of yeast Scm3 (Figure 5J), making contacts with many acidic residues on the CENP-A surface. The loop and 3-stranded β-sheet of HJURP make largely hydrophobic and van der Waals contacts with both CENP-A and H4, which help to prevent tetramerization of two CENP-A/H4 dimers. In addition, the authors find a number of contacts outside of the previously described CATD of CENP-A, particularly Ser68, that are important for the specificity of HJURP. It’s also worth noting that, in contrast to many other chaperone structures, this region of HJURP is very basic (predicted pI=10.5) and makes contacts with many acidic residues of the histone variant. This may represent a unique mechanism of this variant chaperones function, though many of these acidic residues are found in both CENP-A and canonical H3.1.
Finally, more recent functional analyses showed another protein, acidic nucleoplasmic DNA-binding protein 1 (And-1), interacts with both CENP-A and HJURP and is necessary for assembly of CENP-A containing nucleosomes at the centromere in mammalian cells [197]. This protein is highly acidic and contains WD40 and HMG domains similar to those found in other histone chaperones RbAp46/48 and the SSRP1 subunit of FACT, suggesting that it may function as a novel CENP-A variant chaperone. Further analysis of these CENP-A chaperone pathways may help delineate new mechanisms of centromere establishment and maintenance.
Post-Translational Modifications and the Functional Regulation of Histone Chaperones
Structural states of IDRs/IDPs are easily perturbed due to their extreme structural plasticity and shallow energy barriers between multiple different states [198,199]. These unique features allow IDRs/IDPs to act as responsive sensors to even subtle changes in environment or to post-translational modifications (PTMs). One of the most common, and the most thoroughly studied, protein PTMs is phosphorylation. Charge-shifting addition of a phosphate group is an exquisite mechanism for controlling the function of IDRs on many proteins. Many histone chaperones are also regulated by cellular kinases by acting as substrates for phosphorylation on their IDRs. We and others previously showed that Xenopus Npm2 is heavily phosphorylated on its disordered N and C-terminal tails during development, and that this modification increases acidic stretch solvent accessibility leading to increased histone affinity and sequestration [35,200–202]. The family member Npm1 is also heavily modified by phosphorylation and acetylation, which alter its histone chaperone activity, RNA binding activity, oligomerization state, and subcellular localization [40,203–206]. Additionally, the H3.3/H4 chaperone HIRA was shown to be dynamically phosphorylated by Akt1 on its disordered region during muscle cell development [207]. This phosphorylation leads to a suppression of H3.3 incorporation into chromatin, and disappears in response to differentiation cues, leading to increased H3.3 deposition and activation of muscle genes. Phosphorylation may also act to up regulate the activity of a histone chaperone. This is the case for DEK, another H3.3/H4 chaperone, whose chaperone activity is dependent on CK2 phosphorylation on multiple sites along its predicted disordered regions [178,208]. Altering the stability of the protein is another mechanism by which phosphorylation can control histone chaperone function. This was demonstrated for the H3/H4 chaperone Asf1 in a study that showed that Tousled-like kinases (TLKs) phosphorylate its disordered C-terminus leading to increased stability of the protein and decreased proteosomal degradation [209]. Together, these studies demonstrate mechanistically distinct ways by which histone chaperones can be regulated via phosphorylation.
Another lesser-known PTM commonly found on histone chaperones is glutamylation. Glutamylation is the isopeptide addition of one or more glutamates onto a backbone glutamate of a protein. This unique PTM was originally discovered on the disordered acidic tails of α and β tubulin, and is catalyzed by the tubulin tyrosine ligase-like (TTLL) family of enzymes [210,211]. Glutamylation on tubulin tails is known to regulate the binding of microtubule associating proteins (MAPs), including Tau [212]. More recently, it was shown that this modification is not unique to tubulin, but that many acidic nuclear proteins are subject to post-translational glutamylation [213]. In this study, the authors identify many known histone chaperones as substrates for TTLL4 and TTLL5-mediated glutamylation, including Nap1&2, Npm1, and ANP32E. Our mass spectrometry of Xenopus oocyte and egg Npm2 also identified multiple glutamylation sites on its largest acidic stretch in the intrinsically disordered C-terminal tail domain [35].
Though the precise function of histone chaperone glutamylation remains unknown, a recent study shows that glutamylation of the CTAD of Xenopus Nap1 increases maternal linker histone deposition and alters H1 dynamics on chromatin [214]. It is likely that histone chaperone glutamylation alters histone affinity, specificity and/or deposition rates, as this unique PTM adds both additional negative charge and bulk onto acidic stretches of multiple histone chaperones. It is also possible that glutamylation of non-histone chaperones could provide additional negative charge needed to activate their histone chaperone function. This would be similar to the chromatin modifying protein PARP-1, which activates its own histone chaperone function via auto poly-ADP ribosylation (PARylation) [215]. Given the abundance and importance of acidic stretches in histone chaperones, further studies of this unique acidic modification are needed to delineate its function. Other modifications such as lysine methylation, lysine acetylation, arginine methylation, and ubiquitination also likely regulate histone chaperone function.
Common Themes, Open Questions and Perspectives
The recent studies highlighted in this review have begun to uncover diverse roles for IDRs and acidic stretches in histone chaperones and have illuminated some common themes. These include:
The cooperation between ordered and disordered regions of histone chaperones in binding to and chaperoning histones.
Acidic stretches in histone chaperones shielding histone charge and often hijacking nucleosomal histone:DNA interaction sites.
The use of aromatic residues adjacent to acidic stretches in a conserved “anchoring and capping” histone binding mechanism.
The use of IDRs as optimal sites for post-translational modification and chaperone regulation.
Despite IDRs and acidic stretches being common to most histone chaperones, many questions remain unanswered. The exact molecular mechanisms underlying histone transfer from chaperone to DNA in vivo remain largely unexplored. Additionally, histone handoff pathways between many chaperones remain unclear or completely undefined, particularly in the case of H2A/H2B chaperones. What roles do chaperones play in the process of epigenetic inheritance of histone PTMs and variants? How do modifications to IDRs and acidic stretches of chaperones affect their activity and global chromatin organization?
Though high-resolution studies of IDRs are inherently difficult to due to their dynamic nature, x-ray crystallography of small peptides derived from these regions bound to histones have provided exquisite insights into mechanisms of histone recognition. In addition, NMR is poised to provide information on even transient structural states of histone chaperone IDRs at the atomic level in solution, as well as probe the potential for these regions to adopt different structures upon binding different histone ligands, though this mode of regulation has currently not been shown for a histone chaperone. Hydrogen-deuterium exchange mass spectrometry (H/DX-MS) and chemical cross-linking have also emerged as powerful solution techniques to gain valuable insights into the architecture and dynamics of chaperone:histone complexes. Cryo-EM is another emergent technology poised to yield high-resolution insights into how chaperones cooperate with larger complexes, such as chromatin remodeling complexes or the DNA replication machinery. Quantitative histone binding studies using truncation mutants have, and will continue to, provide new insights into the functional roles of these regions. The emergent CRISPR/Cas9 genetic engineering technology will provide an efficient means to create targeted knockouts, truncations, and even point mutations in living cells, which will deepen our understanding of the roles of histone chaperones, their IDRs and acidic stretches in vivo. In-depth genetic analyses of these highly divergent regions of histone chaperones may also help to provide further insights into their roles and illuminate their evolutionary history. Finally, further investigations into the functions of PTMs that occur on histone chaperones will undoubtedly deepen our understanding of the many diverse mechanisms of regulation. By applying combinations of these biochemical, biophysical, structural, and genetic techniques, the field is more prepared than ever to delineate the functional roles of these dynamic regions in histone chaperones, and to gain a more complete understanding of the pathways governing histone deposition and chromatin maintenance.
HIGHLIGHTS.
Intrinsically disordered regions (IDRs) and acidic stretches are common features among the structurally diverse histone chaperone family.
These regions are often directly involved in histone binding and regulation of chaperone function.
Recent studies have begun to shed light on both common and unique mechanisms of IDRs and acidic stretches in mediating chaperone:histone interactions.
Acknowledgments
We are grateful to Maxim Maron, Benjamin Lorton, and Michael Brenowitz for careful reading of this manuscript and for providing helpful comments and edits. We also acknowledge all the contributions to the field that could not be included in this review. Funding for this work was from The American Cancer Society – Robbie Sue Mudd Kidney Cancer Research Scholar Grant (124891-RSG-13-396-01-DMC) and NIH R01GM108646 (all to D.S.), as well as NIH training grants T32GM007491 and F31GM116536 (both to C.W).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg RD, Thomas JO. Chromatin Structure: Oligomers of the Histones. Science (80-) 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 2.Thomas JO, Kornberg RD. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci USA. 1975;72:2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 Å Resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. doi: http://dx.doi.org/10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 5.Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- 6.Laskey RA, Earnshaw WC. Nucleosome assembly. Nature. 1980;286:763–767. doi: 10.1038/286763a0. http://dx.doi.org/10.1038/286763a0. [DOI] [PubMed] [Google Scholar]
- 7.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC452741/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/MCB.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome Assembly by a Complex of CAF-1 and Acetylated Histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. doi: http://dx.doi.org/10.1016/S0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-Containing and ATP-Utilizing Chromatin Assembly and Remodeling Factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. doi: http://dx.doi.org/10.1016/S0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 11.Böhm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Tóth K, Luger K, Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyler JK. Chromatin assembly. Cooperation between histone chaperones and ATP-dependent nucleosome remodeling machines. Eur J Biochem. 2002;269:2268–2274. doi: 10.1046/j.1432-1033.2002.02890.x. [DOI] [PubMed] [Google Scholar]
- 13.Li B, Carey M, Workman JL. The Role of Chromatin during Transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. doi: http://dx.doi.org/10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Clapier CR, Cairns BR. The Biology of Chromatin Remodeling Complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 15.Fotedar R, Roberts JM. Multistep pathway for replication-dependent nucleosome assembly. Proc Natl Acad Sci U S A. 1989;86:6459–63. doi: 10.1073/pnas.86.17.6459. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=297863&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haigney A, Ricketts MD, Marmorstein R. Dissecting the Molecular Roles of Histone Chaperones in Histone Acetylation by Type B Histone Acetyltransferases (HAT-B) J Biol Chem. 2015;290:30648–30657. doi: 10.1074/jbc.M115.688523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knapp AR, Wang H, Parthun MR. The yeast histone chaperone hif1p functions with RNA in nucleosome assembly. PLoS One. 2014;9:e100299. doi: 10.1371/journal.pone.0100299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adkins MW, Tyler JK. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J Biol Chem. 2004;279:52069–74. doi: 10.1074/jbc.M406113200. [DOI] [PubMed] [Google Scholar]
- 20.Zabaronick SR, Tyler JK. The Histone Chaperone Anti-Silencing Function 1 Is a Global Regulator of Transcription Independent of Passage through S Phase. Mol Cell Biol. 2005;25:652–660. doi: 10.1128/MCB.25.2.652-660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shintomi K, Takahashi TS, Hirano T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat Cell Biol. 2015;17:1014–1023. doi: 10.1038/ncb3187. http://dx.doi.org/10.1038/ncb3187. [DOI] [PubMed] [Google Scholar]
- 22.Kleinschmidt Ja, Fortkamp E, Krohne G, Zentgraf H, Franke WW. Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J Biol Chem. 1985;260:1166–76. http://www.ncbi.nlm.nih.gov/pubmed/2981836. [PubMed] [Google Scholar]
- 23.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 Complexes Mediate Nucleosome Assembly Pathways Dependent or Independent of DNA Synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. doi: http://dx.doi.org/10.1016/S0092-8674(03)01064-X. [DOI] [PubMed] [Google Scholar]
- 24.Andrews AJ, Downing G, Park Y, Luger K. A Thermodynamic Model for Nap1-Histone Interactions. J Biol Chem. 2008;283:32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falces J, Arregi I, Konarev PV, Urbaneja MA, Svergun DI, Taneva SG, Banuelos S. Recognition of Nucleoplasmin by its Nuclear Transport Receptor Importin α/β: Insights Into a Complete Import Complex. Biochemistry. 2010;49:9756–69. doi: 10.1021/bi101179g. [DOI] [PubMed] [Google Scholar]
- 26.Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF. Nuclear Import of Histone H2a and H2b Is Mediated by a Network of Karyopherins. J Cell Biol. 2001;153:251–262. doi: 10.1083/jcb.153.2.251. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2169462/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook AJL, Gurard-Levin ZA, Vassias I, Almouzni G. A Specific Function for the Histone Chaperone NASP to Fine-Tune a Reservoir of Soluble H3-H4 in the Histone Supply Chain. Mol Cell. 2011;44:918–927. doi: 10.1016/j.molcel.2011.11.021. doi: http://dx.doi.org/10.1016/j.molcel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 28.Jackson V, Chalkley R. A reevaluation of new histone deposition on replicating chromatin. J Biol Chem. 1981;256:5095–5103. [PubMed] [Google Scholar]
- 29.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome Assembly Protein 1 Exchanges Histone H2A-H2B Dimers and Assists Nucleosome Sliding. J Biol Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 30.Obri A, Ouararhni K, Papin C, Diebold ML, Padmanabhan K, Marek M, Stoll I, Roy L, Reilly PT, Mak TW, Dimitrov S, Romier C, Hamiche A. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature. 2014;505:648–653. doi: 10.1038/nature12922. http://dx.doi.org/10.1038/nature12922. [DOI] [PubMed] [Google Scholar]
- 31.Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol Cell. 2001;8:841–53. doi: 10.1016/s1097-2765(01)00354-9. http://www.ncbi.nlm.nih.gov/pubmed/11684019. [DOI] [PubMed] [Google Scholar]
- 32.Mousson F, Lautrette A, Thuret JY, Agez M, Courbeyrette R, Amigues B, Becker E, Neumann JM, Guerois R, Mann C, Ochsenbein F. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc Natl Acad Sci U S A. 2005;102:5975–5980. doi: 10.1073/pnas.0500149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, Li G, Xu RM. Structure of a CENP-A–histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46. Struct England1993) 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onikubo T, Nicklay JJ, Xing L, Warren C, Anson B, Wang W, Burgos ES, Ruff SE, Shabanowitz J, Cheng RH, Hunt DF, Shechter D. Developmentally Regulated Post-translational Modification of Nucleoplasmin Controls Histone Sequestration and Deposition. CellReports. 2015;10:1–14. doi: 10.1016/j.celrep.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taneva SG, Bañuelos S, Falces J, Arregi I, Muga A, Konarev PV, Svergun DI, Velázquez-Campoy A, Urbaneja Ma. A mechanism for histone chaperoning activity of nucleoplasmin: thermodynamic and structural models. J Mol Biol. 2009;393:448–63. doi: 10.1016/j.jmb.2009.08.005. http://www.ncbi.nlm.nih.gov/pubmed/19683001 (accessed March 19, 2014) [DOI] [PubMed] [Google Scholar]
- 37.Mcbryant SJ, Park Y, Abernathy SM, Laybourn PJ, Nyborg JK, Luger K. Preferential Binding of the Histone (H3-H4)2 Tetramer by NAP1 Is Mediated by the Amino-terminal Histone Tails. J Biol Chem. 2003;278:44574–44583. doi: 10.1074/jbc.M305636200. [DOI] [PubMed] [Google Scholar]
- 38.Moreira IS, Fernandes PA, Ramos MJ. Hot spots—A review of the protein–protein interface determinant amino-acid residues. Proteins Struct Funct Bioinforma. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 39.Dennehey BK, Noone S, Liu WH, Smith L, Churchill MEA, Tyler JK. The C Terminus of the Histone Chaperone Asf1 Cross-Links to Histone H3 in Yeast and Promotes Interaction with Histones H3 and H4. Mol Cell Biol. 2013;33:605–621. doi: 10.1128/MCB.01053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swaminathan V, Kishore AH, Febitha KK, Kundu TK. Human Histone Chaperone Nucleophosmin Enhances Acetylation-Dependent Chromatin Transcription. Mol Cell Biol. 2005;25:7534–7545. doi: 10.1128/MCB.25.17.7534-7545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos I, Fernández-Rivero N, Arranz R, Aloria K, Finn R, Arizmendi JM, Ausió J, Valpuesta JM, Muga A, Prado A. The intrinsically disordered distal face of nucleoplasmin recognizes distinct oligomerization states of histones. Nucleic Acids Res. 2014;42:1311–25. doi: 10.1093/nar/gkt899. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3902905&tool=pmcentrez&rendertype=abstract (accessed March 26, 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu X, Wang Y, Gan L, Bai Y, Han W, Wang E, Wang J. Importance of electrostatic interactions in the association of intrinsically disordered histone chaperone Chz1 and histone H2A.Z-H2B. PLoS Comput Biol. 2012;8:e1002608. doi: 10.1371/journal.pcbi.1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Oldfield CJ, Dunker AK. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu Rev Biochem. 2014;83:553–84. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 46.Perdigão N, Heinrich J, Stolte C, Sabir KS, Buckley MJ, Tabor B, Signal B, Gloss BS, Hammang CJ, Rost B, Schafferhans A, O’Donoghue SI. Unexpected features of the dark proteome. Proc Natl Acad Sci. 2015;112:15898–15903. doi: 10.1073/pnas.1508380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct Funct Bioinforma. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::AID-PROT130>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 48.Müller-Späth S, Soranno A, Hirschfeld V, Hofmann H, Rüegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. http://dx.doi.org/10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 50.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 51.Yan J, Mizianty MJ, Filipow PL, Uversky VN, Kurgan L. RAPID: Fast and accurate sequence-based prediction of intrinsic disorder content on proteomic scale. Biochim Biophys Acta - Proteins Proteomics. 2013;1834:1671–1680. doi: 10.1016/j.bbapap.2013.05.022. doi: http://dx.doi.org/10.1016/j.bbapap.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–5. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 54.Huang Y, Liu Z. Kinetic Advantage of Intrinsically Disordered Proteins in Coupled Folding– Binding Process: A Critical Assessment of the “Fly-Casting” Mechanism. J Mol Biol. 2009;393:1143–1159. doi: 10.1016/j.jmb.2009.09.010. doi: http://dx.doi.org/10.1016/j.jmb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 55.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. doi: http://dx.doi.org/10.1016/S0959-440X(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 56.Patil A, Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 57.Milles S, Mercadante D, Aramburu IV, Jensen MR, Banterle N, Koehler C, Tyagi S, Clarke J, Shammas SL, Blackledge M, Gräter F, Lemke EA. Plasticity of an Ultrafast Interaction between Nucleoporins and Nuclear Transport Receptors. Cell. 2015;163:734–745. doi: 10.1016/j.cell.2015.09.047. doi: http://dx.doi.org/10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raveh B, Karp JM, Sparks S, Dutta K, Rout MP, Sali A, Cowburn D. Slide-and-exchange mechanism for rapid and selective transport through the nuclear pore complex. Proc Natl Acad Sci U S A. 2016;113:E2489–E2497. doi: 10.1073/pnas.1522663113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. doi: http://dx.doi.org/10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife. 2016;5:e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammond CM, Stromme CB, Huang H, Patel DJ, Groth A. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol advance online publication. 2017 doi: 10.1038/nrm.2016.159. http://dx.doi.org/10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed]
- 62.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Das C, Tyler JK, Churchill ME. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci. 2010;35 doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim PM, Sboner A, Xia Y, Gerstein M. The role of disorder in interaction networks: a structural analysis. Mol Syst Biol. 2008;4 doi: 10.1038/msb.2008.16. http://msb.embopress.org/content/4/1/179.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lobanov MY, Galzitskaya OV. How Common Is Disorder? Occurrence of Disordered Residues in Four Domains of Life. Int J Mol Sci. 2015;16:19490–19507. doi: 10.3390/ijms160819490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.James LC, Tawfik DS. Conformational diversity and protein evolution – a 60-year-old hypothesis revisited. Trends Biochem Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. doi: http://dx.doi.org/10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 67.Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinforma. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daganzo SM, Erzberger JP, Lam WM, Skordalakes E, Zhang R, Franco AA, Brill SJ, Adams PD, Berger JM, Kaufman PD. Structure and Function of the Conserved Core of Histone Deposition Protein Asf1. Curr Biol. 2003;13:2148–2158. doi: 10.1016/j.cub.2003.11.027. doi: https://doi.org/10.1016/j.cub.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 69.Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, Canutescu A, Dunbrack R, Adams PD, Marmorstein R. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol. 2006;13:921–929. doi: 10.1038/nsmb1147. http://dx.doi.org/10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. http://dx.doi.org/10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 72.Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ. Spt16–Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001;20:3506–3517. doi: 10.1093/emboj/20.13.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orphanides G, Leroy G, Chang C, Luse DS, Reinberg D. FACT, a Factor that Facilitates Transcript Elongation through Nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 74.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT Facilitates Nucleosome Alteration. Science (80-) 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 75.Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A, Tada S, Nakayama T, Murofushi H, Okumura K, Takeda S, Horikoshi M, Seki M, Enomoto T. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem. 2011;286:30504–12. doi: 10.1074/jbc.M111.264721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valieva ME, Armeev GA, Kudryashova KS, Gerasimova NS, Shaytan AK, Kulaeva OI, McCullough LL, Formosa T, Georgiev PG, Kirpichnikov MP, Studitsky VM, Feofanov AV. Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat Struct Mol Biol. 2016;23:1111–1116. doi: 10.1038/nsmb.3321. http://dx.doi.org/10.1038/nsmb.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kemble DJ, Whitby FG, Robinson H, McCullough LL, Formosa T, Hill CP. Structure of the Spt16 Middle Domain Reveals Functional Features of the Histone Chaperone FACT. J Biol Chem. 2013;288:10188–10194. doi: 10.1074/jbc.C113.451369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marcianò G, Huang DT. Structure of the human histone chaperone FACT Spt16 N-terminal domain. Acta Crystallogr Sect F, Struct Biol Commun. 2016;72:121–128. doi: 10.1107/S2053230X15024565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang W, Zeng F, Liu Y, Shao C, Li S, Lv H, Shi Y, Niu L, Teng M, Li X. Crystal Structure of Human SSRP1 Middle Domain Reveals a Role in DNA Binding. Sci Rep. 2015;5:18688. doi: 10.1038/srep18688. http://dx.doi.org/10.1038/srep18688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hondele M, Stuwe T, Hassler M, Halbach F, Bowman A, Zhang ET, Nijmeijer B, Kotthoff C, Rybin V, Amlacher S, Hurt E, Ladurner AG. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature. 2013;499:111–114. doi: 10.1038/nature12242. http://dx.doi.org/10.1038/nature12242. [DOI] [PubMed] [Google Scholar]
- 81.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286:41883–92. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prendergast L, Müller S, Liu Y, Huang H, Dingli F, Loew D, Vassias I, Patel DJ, Sullivan KF, Almouzni G. The CENP-T/-W complex is a binding partner of the histone chaperone FACT. Genes Dev. 2016;30:1313–26. doi: 10.1101/gad.275073.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kemble DJ, McCullough LL, Whitby FG, Formosa T, Hill CP. FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs. Mol Cell. 2015;60:294–306. doi: 10.1016/j.molcel.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kepert JF, Mazurkiewicz J, Heuvelman GL, Toth KF, Rippe K. Nap1 modulates binding of linker histone H1 to chromatin and induces an extended chromatin fiber conformation. J Biol Chem. 2005;280 doi: 10.1074/jbc.M507322200. [DOI] [PubMed] [Google Scholar]
- 85.Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muto S, Senda M, Akai Y, Sato L, Suzuki T, Nagai R. Relationship between the structure of SET/TAF-I$β$/INHAT and its histone chaperone activity. Proc Natl Acad Sci USA. 2007;104 doi: 10.1073/pnas.0603762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hammond CM, Sundaramoorthy R, Larance M, Lamond A, Stevens MA, El-Mkami H, Norman DG, Owen-Hughes T. The histone chaperone Vps75 forms multiple oligomeric assemblies capable of mediating exchange between histone H3–H4 tetramers and Asf1–H3–H4 complexes. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone NAP1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37 doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.D’Arcy S, Martin K, Panchenko T, Chen X, Bergeron S, Stargell L, Black B, Luger K. Chaperone Nap1 Shields Histone Surfaces Used in a Nucleosome and Can Put H2A-H2B in an Unconventional Tetrameric Form. Mol Cell. 2013;51:662–677. doi: 10.1016/j.molcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aguilar-Gurrieri C, Larabi A, Vinayachandran V, Patel NA, Yen K, Reja R, Ebong IO, Schoehn G, Robinson CV, Pugh BF, Panne D. Structural evidence for Nap1-dependent H2A– H2B deposition and nucleosome assembly. EMBO J. 2016;35:1465–1482. doi: 10.15252/embj.201694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okuwaki M, Kato K, Nagata K. Functional characterization of human nucleosome assembly protein 1-like proteins as histone chaperones. Genes Cells. 2010;15 doi: 10.1111/j.1365-2443.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Q, Giebler HA, Isaacson MK, Nyborg JK. Eviction of linker histone H1 by NAP-family histone chaperones enhances activated transcription. Epigenetics {&} Chromatin. 2015;8:30. doi: 10.1186/s13072-015-0022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohtomo H, Akashi S, Moriwaki Y, Okuwaki M, Osakabe A, Nagata K, Kurumizaka H, Nishimura Y. C-terminal acidic domain of histone chaperone human NAP1 is an efficient binding assistant for histone H2A-H2B, but not H3-H4. Genes to Cells. 2016 doi: 10.1111/gtc.12339. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 94.Hong J, Feng H, Wang F, Ranjan A, Chen J, Jiang J, Ghirlando R, Xiao TS, Wu C, Bai Y. The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer. Mol Cell. 2014;53:498–505. doi: 10.1016/j.molcel.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Earnshaw WC, Honda BM, Laskey RA, Thomas J. Assembly of Nucleosomes: The reaction involving X. laevis Nucleoplasmin. Cell. 1980;21:373–383. doi: 10.1016/0092-8674(80)90474-2. [DOI] [PubMed] [Google Scholar]
- 96.Wedlich D, Dreyer C. The distribution of nucleoplasmin in early development and organogenesis of Xenopus laevis. Cell Tissue Res. 1988;254:295–300. doi: 10.1007/BF00225802. [DOI] [PubMed] [Google Scholar]
- 97.Platonova O, Akey IV, Head JF, Akey CW. Crystal Structure and Function of Human Nucleoplasmin (Npm2): A Histone Chaperone in Oocytes and Embryos. Biochemistry. 2011;50:8078–8089. doi: 10.1021/bi2006652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Edlich-Muth C, Artero JB, Callow P, Przewloka MR, Watson AA, Zhang W, Glover DM, Debski J, Dadlez M, Round AR, Forsyth VT, Laue ED. The Pentameric Nucleoplasmin Fold Is Present in Drosophila FKBP39 and a Large Number of Chromatin-Related Proteins. J Mol Biol. 2015;427:1949–1963. doi: 10.1016/j.jmb.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ramos I, Martín-Benito J, Finn R, Bretaña L, Aloria K, Arizmendi JM, Ausió J, Muga A, Valpuesta JM, Prado A. Nucleoplasmin binds histone H2A-H2B dimers through its distal face. J Biol Chem. 2010;285:33771–8. doi: 10.1074/jbc.M110.150664. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2962476&tool=pmcentrez&rendertype=abstract (accessed March 19, 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fernández-rivero N, Franco A, Velázquez-Campoy A, Alonso E, Muga A, Prado A. A Quantitative Characterization of Nucleoplasmin/Histone Complexes Reveals Chaperone Versatility. Sci Rep. 2016;6 doi: 10.1038/srep32114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hierro A, Arizmendi JM, Banuelos S, Prado A, Muga A. Electrostatic Interactions at the C-Terminal Domain of Nucleoplasmin Modulate Its Chromatin Decondensation Activity. Biochemistry. 2002;41:6408–6413. doi: 10.1021/bi020002r. [DOI] [PubMed] [Google Scholar]
- 102.Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, Ehninger G. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011 LP-4020. doi: 10.1182/blood-2005-08-3167. http://www.bloodjournal.org/content/107/10/4011.abstract. [DOI] [PubMed] [Google Scholar]
- 103.Namboodiri VMH, Akey IV, Schmidt-Zachmann MS, Head JF, Akey CW. The Structure and Function of Xenopus NO38-Core, a Histone Chaperone in the Nucleolus. Structure. 2004;12:2149–2160. doi: 10.1016/j.str.2004.09.017. doi: http://dx.doi.org/10.1016/j.str.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 104.Frehlick LJ, Eirín-López JM, Ausió J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. BioEssays. 2007;29:49–59. doi: 10.1002/bies.20512. http://www.ncbi.nlm.nih.gov/pubmed/17187372 (accessed March 19, 2014) [DOI] [PubMed] [Google Scholar]
- 105.Grummitt CG, Townsley FM, Johnson CM, Warren AJ, Bycroft M. Structural Consequences of Nucleophosmin Mutations in Acute Myeloid Leukemia. J Biol Chem. 2008;283:23326–23332. doi: 10.1074/jbc.M801706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chiarella S, Cola A De, Scaglione GL, Carletti E, Graziano V, Barcaroli D, Lo Sterzo C, Di Matteo A, Di Ilio C, Falini B, Arcovito A, Laurenzi V De, Federici L. Nucleophosmin mutations alter its nucleolar localization by impairing G-quadruplex binding at ribosomal DNA. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lindstrom MS. NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochem Res Int. 2011;2011:1–16. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu Y, Maggi LB, Brady SN, Apicelli AJ, Dai MS, Lu H, Weber JD. Nucleophosmin Is Essential for Ribosomal Protein L5 Nuclear Export. Mol Cell Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bertwistle D, Sugimoto M, Sherr CJ. Physical and Functional Interactions of the Arf Tumor Suppressor Protein with Nucleophosmin/B23. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gadad SS, Senapati P, Syed SH, Rajan RE, Shandilya J, Swaminathan V, Chatterjee S, Colombo E, Dimitrov S, Pelicci PG, Ranga U, Kundu TK. The Multifunctional Protein Nucleophosmin (NPM1) Is a Human Linker Histone H1 Chaperone. Biochemistry. 2011;50:2780–2789. doi: 10.1021/bi101835j. [DOI] [PubMed] [Google Scholar]
- 111.Murano K, Okuwaki M, Hisaoka M, Nagata K. Transcription Regulation of the rRNA Gene by a Multifunctional Nucleolar Protein. B23/Nucleophosmin, through Its Histone Chaperone Activity. Mol Cell Biol. 2008;28:3114–3126. doi: 10.1128/MCB.02078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hisaoka M, Nagata K, Okuwaki M. Intrinsically disordered regions of nucleophosmin/B23 regulate its RNA binding activity through their inter- and intra-molecular association. Nucleic Acids Res. 2014;42:1180–95. doi: 10.1093/nar/gkt897. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3902904&tool=pmcentrez&rendertype=abstract (accessed March 17, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gadad SS, Shandilya J, Kishore AH, Kundu TK, NPM3, a Member of the Nucleophosmin/Nucleoplasmin Family Enhances Activator-Dependent Transcription. Biochemistry. 2010;49:1355–1357. doi: 10.1021/bi9021632. [DOI] [PubMed] [Google Scholar]
- 114.Okuwaki M, Sumi A, Hisaoka M, Saotome-Nakamura A, Akashi S, Nishimura Y, Nagata K. Function of homo- and hetero-oligomers of human nucleoplasmin/nucleophosmin family proteins NPM1, NPM2 and NPM3 during sperm chromatin remodeling. Nucleic Acids Res. 2012;40:4861–4878. doi: 10.1093/nar/gks162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Mol Biol. 2000;7:1121–1124. doi: 10.1038/81971. http://dx.doi.org/10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 116.Placek BJ, Harrison LN, Villers BM, Gloss LM. The H2A. Z/H2B dimer is unstable compared to the dimer containing the major H2A isoform. 2005:514–522. doi: 10.1110/ps.041026405.proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. http://dx.doi.org/10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, Wu C. Chz1, a Nuclear Chaperone for Histone H2AZ. Mol Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 119.Zhou Z, Feng H, Hansen DF, Kato H, Luk E, Freedberg DI, Kay LE, Wu C, Bai Y. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat Struct Mol Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. http://dx.doi.org/10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hansen DF, Zhou Z, Feng H, Jenkins LMM, Bai Y, Kay LE. Binding kinetics of histone chaperone Chz1 and variant histone H2A.Z-H2B by relaxation dispersion NMR spectroscopy. J Mol Biol. 2010;387:1–9. doi: 10.1016/j.jmb.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mao Z, Pan L, Wang W, Sun J, Shan S, Dong Q, Liang X, Dai L, Ding X, Chen S, Zhang Z, Zhu B, Zhou Z. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 2014;24:389–399. doi: 10.1038/cr.2014.30. http://dx.doi.org/10.1038/cr.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. The Mammalian YL1 Protein Is a Shared Subunit of the TRRAP/TIP60 Histone Acetyltransferase and SRCAP Complexes. J Biol Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 123.Latrick CM, Marek M, Ouararhni K, Papin C, Stoll I, Ignatyeva M, Obri A, Ennifar E, Dimitrov S, Romier C, Hamiche A. Molecular basis and specificity of H2A.Z-H2B recognition and deposition by the histone chaperone YL1. Nat Struct Mol Biol. 2016;23:309–316. doi: 10.1038/nsmb.3189. http://dx.doi.org/10.1038/nsmb.3189. [DOI] [PubMed] [Google Scholar]
- 124.Liang X, Shan S, Pan L, Zhao J, Ranjan A, Wang F, Zhang Z, Huang Y, Feng H, Wei D, Huang L, Liu X, Zhong Q, Lou J, Li G, Wu C, Zhou Z. Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1. Nat Struct Mol Biol. 2016;23:317–323. doi: 10.1038/nsmb.3190. http://dx.doi.org/10.1038/nsmb.3190. [DOI] [PubMed] [Google Scholar]
- 125.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. http://dx.doi.org/10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 126.Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S. Mechanism of Polymerase II Transcription Repression by the Histone Variant macroH2A. Mol Cell Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) Is a Novel Human Protein Involved in the Cellular Response to Chromosomal DNA Strand Breaks. Mol Cell Biol. 2007;27:3793–3803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW. APLF (C2orf13) Is a Novel Component of Poly(ADP-Ribose) Signaling in Mammalian Cells. Mol Cell Biol. 2008;28:4620–4628. doi: 10.1128/MCB.02243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rulten SL, Fisher AEO, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF Function Together to Accelerate Nonhomologous End-Joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. doi: http://dx.doi.org/10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 130.Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, Owen-Hughes T, Ahel I. DNA repair factor APLF is a histone chaperone. Mol Cell. 2011;41:46–55. doi: 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Le S, Davis C, Konopka JB, Sternglanz R. Two New S-Phase-Specific Genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 132.Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. http://dx.doi.org/10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 133.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of Lysine 56 of Histone H3 Catalyzed by RTT109 and Regulated by ASF1 Is Required for Replisome Integrity. J Biol Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 134.Ray-Gallet D, Quivy J-P, Silljé HWW, Nigg EA, Almouzni G. The histone chaperone Asf1 is dispensable for direct de novo histone deposition in Xenopus egg extracts. Chromosoma. 2007;116:487–496. doi: 10.1007/s00412-007-0112-x. [DOI] [PubMed] [Google Scholar]
- 135.Liu WH, Roemer SC, Port AM, Churchill MEA. CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 2012;40:11229–11239. doi: 10.1093/nar/gks906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schulz LL, Tyler JK. The histone chaperone ASF1 localizes to active DNA replication forks to mediate efficient DNA replication. FASEB J. 2006 doi: 10.1096/fj.05-5020fje. [DOI] [PubMed] [Google Scholar]
- 137.Groth A, Corpet A, Cook AJL, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of Replication Fork Progression Through Histone Supply and Demand. Science (80-) 2007;318:1928 LP-1931. doi: 10.1126/science.1148992. http://science.sciencemag.org/content/318/5858/1928.abstract. [DOI] [PubMed] [Google Scholar]
- 138.Liu WH, Roemer SC, Zhou Y, Shen ZJ, Dennehey BK, Balsbaugh JL, Liddle JC, Nemkov T, Ahn NG, Hansen KC, Tyler JK, Churchill MEA. The Cac1 subunit of histone chaperone CAF-1 organizes CAF-1-H3/H4 architecture and tetramerizes histones. Elife. 2016;5:e18023. doi: 10.7554/eLife.18023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Agez M, Chen J, Guerois R, Van Heijenoort C, Thuret J, Mann C, Ochsenbein F. Structure of the Histone Chaperone Asf1 Bound to the Histone H3 C-Terminal Helix and Functional Insights. Structure. 2007;15:191–199. doi: 10.1016/j.str.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 140.English CM, Adkins MW, Carson JJ, Churchill MEA, Tyler JK. Structural Basis for the Histone Chaperone Activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. doi: https://doi.org/10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Antczak AJ, Tsubota T, Kaufman PD, Berger JM. Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct Biol. 2006;6:26. doi: 10.1186/1472-6807-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tamburini BA, Carson JJ, Linger JG, Tyler JK. Dominant Mutants of the Saccharomyces cerevisiae ASF1 Histone Chaperone Bypass the Need for CAF-1 in Transcriptional Silencing by Altering Histone and Sir Protein Recruitment. Genetics. 2006;173:599–610. doi: 10.1534/genetics.105.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Umehara T, Chimura T, Ichikawa N, Horikoshi M. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes to Cells. 2002;7:59–73. doi: 10.1046/j.1356-9597.2001.00493.x. [DOI] [PubMed] [Google Scholar]
- 144.Munakata T, Adachi N, Yokoyama N, Kuzuhara T, Horikoshi M. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes to Cells. 2000;5:221–233. doi: 10.1046/j.1365-2443.2000.00319.x. [DOI] [PubMed] [Google Scholar]
- 145.Corpet A, De Koning L, Toedling J, Savignoni A, Berger F, Lemaître C, O’Sullivan RJ, Karlseder J, Barillot E, Asselain B, Sastre-Garau X, Almouzni G. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011;30:480–493. doi: 10.1038/emboj.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Abascal F, Corpet A, Gurard-Levin ZA, Juan D, Ochsenbein F, Rico D, Valencia A, Almouzni G. Subfunctionalization via Adaptive Evolution Influenced by Genomic Context: The Case of Histone Chaperones ASF1a and ASF1b. Mol Biol Evol. 2013;30:1853–1866. doi: 10.1093/molbev/mst086. [DOI] [PubMed] [Google Scholar]
- 147.Ishimi Y, Ichinose S, Omori A, Sato K, Kimura H. Binding of Human Minichromosome Maintenance Proteins with Histone H3. J Biol Chem. 1996;271:24115–24122. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- 148.Richet N, Liu D, Legrand P, Velours C, Corpet A, Gaubert A, Bakail M, Moal-raisin G, Guerois R, Almouzni G, Compper C, Besle A, Curie M. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. 2015;43:1905–1917. doi: 10.1093/nar/gkv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang H, Wang M, Yang N, Xu RM. Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2. Protein Cell. 2015;6:693–697. doi: 10.1007/s13238-015-0190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Annunziato AT. Split Decision: What Happens to Nucleosomes during DNA Replication? J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 151.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of Histone H3-H4 Tetramers During DNA Replication–Dependent Chromatin Assembly. Science (80-) 2010;328:94 LP-98. doi: 10.1126/science.1178994. http://science.sciencemag.org/content/328/5974/94.abstract. [DOI] [PubMed] [Google Scholar]
- 152.Mello JA, Silljé HHW, Roche DMJ, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF‐1 interact and synergize in a repair‐coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329 LP-334. doi: 10.1093/embo-reports/kvf068. http://embor.embopress.org/content/3/4/329.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shibahara K, Stillman B. Replication-Dependent Marking of DNA by PCNA Facilitates CAF-1-Coupled Inheritance of Chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. doi: http://doi.org/10.1016/S0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 154.Mattiroli F, Gu Y, Yadav T, Balsbaugh JL, Harris MR, Findlay ES, Liu Y, Radebaugh CA, Stargell LA, Ahn NG, Whitehouse I, Luger K. DNA-mediated association of two histone-bound complexes of yeast Chromatin Assembly Factor-1 (CAF-1) drives tetrasome assembly in the wake of DNA replication. Elife. 2017;6:e22799. doi: 10.7554/eLife.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sauer PV, Timm J, Liu D, Sitbon D, Boeri-Erba E, Velours C, Mücke N, Langowski J, Ochsenbein F, Almouzni G, Panne D. Insights into the molecular architecture and histone H3-H4 deposition mechanism of yeast Chromatin assembly factor 1. Elife. 2017;6:e23474. doi: 10.7554/eLife.23474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of Histone H3 Lysine 56 Regulates Replication-Coupled Nucleosome Assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. doi: http://dx.doi.org/10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. RbAp48 Belongs to the Histone Deacetylase Complex That Associates with the Retinoblastoma Protein. J Biol Chem. 2000;275:9797–9804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]
- 158.Zhang Q, Vo N, Goodman RH. Histone Binding Protein RbAp48 Interacts with a Complex of CREB Binding Protein and Phosphorylated CREB. Mol Cell Biol. 2000;20:4970–4978. doi: 10.1128/MCB.20.14.4970-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. doi: http://dx.doi.org/10.1016/S0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 160.Martínez-Balbás MA, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci U S A. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC18150/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nowak AJ, Alfieri C, Stirnimann CU, Rybin V, Baudin F, Ly-Hartig N, Lindner D, Müller CW. Chromatin-modifying Complex Component Nurf55/p55 Associates with Histones H3 and H4 and Polycomb Repressive Complex 2 Subunit Su(z)12 through Partially Overlapping Binding Sites. J Biol Chem. 2011;286:23388–23396. doi: 10.1074/jbc.M110.207407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang W, Tyl M, Ward R, Sobott F, Maman J, Murthy AS, Watson AA, Fedorov O, Bowman A, Owen-Hughes T, Mkami H El, Murzina NV, Norman DG, Laue ED. Structural plasticity of histones H3–H4 facilitates their allosteric exchange between RbAp48 and ASF1. Nat Struct Mol Biol. 2013;20:29–35. doi: 10.1038/nsmb.2446. http://dx.doi.org/10.1038/nsmb.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Roeder GS, Beard C, Smith M, Keranen S. Isolation and characterization of the SPT2 gene, a negative regulator of Ty-controlled yeast gene expression. Mol Cell Biol. 1985;5:1543–1553. doi: 10.1128/MCB.5.7.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nourani A, Robert F, Winston F. Evidence that Spt2/Sin1, an HMG-Like Factor. Plays Roles in Transcription Elongation, Chromatin Structure, and Genome Stability in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:1496–1509. doi: 10.1128/MCB.26.4.1496-1509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bortvin A, Winston F. Evidence That Spt6p Controls Chromatin Structure by a Direct Interaction with Histones. Science (80-) 1996;272:1473 LP-1476. doi: 10.1126/science.272.5267.1473. http://science.sciencemag.org/content/272/5267/1473.abstract. [DOI] [PubMed] [Google Scholar]
- 166.Osakabe A, Tachiwana H, Takaku M, Hori T, Obuse C, Kimura H, Fukagawa T, Kurumizaka H. Vertebrate Spt2 is a novel nucleolar histone chaperone that assists in ribosomal DNA transcription. J Cell Sci. 2013;126:1323 LP-1332. doi: 10.1242/jcs.112623. http://jcs.biologists.org/content/126/6/1323.abstract. [DOI] [PubMed] [Google Scholar]
- 167.Chen S, Rufiange A, Huang H, Rajashankar KR, Nourani A, Patel DJ. Structure–function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2. Genes Dev. 2015;29:1326–1340. doi: 10.1101/gad.261115.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Banumathy G, Somaiah N, Zhang R, Tang Y, Hoffmann J, Andrake M, Ceulemans H, Schultz D, Marmorstein R, Adams PD. Human UBN1 Is an Ortholog of Yeast Hpc2p and Has an Essential Role in the HIRA/ASF1a Chromatin-Remodeling Pathway in Senescent Cells. Mol Cell Biol. 2009;29:758–770. doi: 10.1128/MCB.01047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rai TS, Puri A, McBryan T, Hoffman J, Tang Y, Pchelintsev NA, van Tuyn J, Marmorstein R, Schultz DC, Adams PD. Human CABIN1 Is a Functional Member of the Human HIRA/UBN1/ASF1a Histone H3.3 Chaperone Complex. Mol Cell Biol. 2011;31:4107–4118. doi: 10.1128/MCB.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LET, Almouzni G. Dynamics of Histone H3 Deposition In Vivo Reveal a Nucleosome Gap-Filling Mechanism for H3.3 to Maintain Chromatin Integrity. Mol Cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. doi: http://dx.doi.org/10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 171.Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G, Adams PD. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 2013;3:1012–1019. doi: 10.1016/j.celrep.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tang Y, Puri A, Ricketts MD, Rai TS, Hoffmann J, Hoi E, Adams PD, Schultz DC, Marmorstein R. Identification of an Ubinuclein 1 Region Required for Stability and Function of the Human HIRA/UBN1/CABIN1/ASF1a Histone H3.3 Chaperone Complex. Biochemistry. 2012;51:2366–2377. doi: 10.1021/bi300050b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Daniel Ricketts M, Frederick B, Hoff H, Tang Y, Schultz DC, Rai T Singh, Grazia Vizioli M, Adams PD, Marmorstein R. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat Commun. 2015;6:7711. doi: 10.1038/ncomms8711. http://dx.doi.org/10.1038/ncomms8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Elsässer SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. DAXX envelops a histone H3.3–H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Denizio JE, Elsässer SJ, Black BE. DAXX co-folds with H3.3/H4 using high local stability conferred by the H3.3 variant recognition residues. Nucleic Acids Res. 2014:1–14. doi: 10.1093/nar/gku090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, Ueda T, Umetsu D, Ito T, Takeyama K, Kato S. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Delbarre E, Ivanauskiene K, Küntziger T, Collas P. DAXX-dependent supply of soluble (H3.3–H4) dimers to PML bodies pending deposition into chromatin. Genome Res. 2013;23:440–451. doi: 10.1101/gr.142703.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ivanauskiene K, Delbarre E, McGhie JD, Küntziger T, Wong LH, Collas P. The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res. 2014;24:1584–1594. doi: 10.1101/gr.173831.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gamble MJ, Kraus WL. Multiple facets of the unique histone variant macroH2A: From genomics to cell biology. Cell Cycle. 2010;9:2568–2574. doi: 10.4161/cc.9.13.12144. [DOI] [PubMed] [Google Scholar]
- 182.Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, Poirier GG, Earnshaw WC. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. doi: http://dx.doi.org/10.1016/S0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 183.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. http://genesdev.cshlp.org/content/9/5/573.abstract. [DOI] [PubMed] [Google Scholar]
- 184.Krassovsky K, Henikoff JG, Henikoff S. Tripartite organization of centromeric chromatin in budding yeast. Proc Natl Acad Sci. 2012;109:243–248. doi: 10.1073/pnas.1118898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Blower MD, Sullivan BA, Karpen GH. Conserved Organization of Centromeric Chromatin in Flies and Humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. doi: https://doi.org/10.1016/S1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. http://dx.doi.org/10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 188.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and Histones CenH3-H4 Assemble the Core of Centromere-Specific Nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. doi: https://doi.org/10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 189.Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 Is Essential to Recruit the Histone H3 Variant Cse4 to Centromeres and to Maintain a Functional Kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. doi: https://doi.org/10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 190.Zhou Z, Feng H, Zhou B-R, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, Bai Y. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. http://dx.doi.org/10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common Ancestry of the CENP-A Chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP Is a Cell-Cycle-Dependent Maintenance and Deposition Factor of CENP-A at Centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. doi: http://dx.doi.org/10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 194.Black BE, Jansen LET, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere Identity Maintained by Nucleosomes Assembled with Histone H3 Containing the CENP-A Targeting Domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. doi: https://doi.org/10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 195.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, Kimura H, Kurumizaka H. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. http://dx.doi.org/10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 196.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jaramillo-Lambert A, Hao J, Xiao H, Li Y, Han Z, Zhu W. Acidic Nucleoplasmic DNA-binding Protein (And-1) Controls Chromosome Congression by Regulating the Assembly of Centromere Protein A (CENP-A) at Centromeres. J Biol Chem. 2013;288:1480–1488. doi: 10.1074/jbc.M112.429266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Higo J, Nishimura Y, Nakamura H. A Free-Energy Landscape for Coupled Folding and Binding of an Intrinsically Disordered Protein in Explicit Solvent from Detailed All-Atom Computations. J Am Chem Soc. 2011;133:10448–10458. doi: 10.1021/ja110338e. [DOI] [PubMed] [Google Scholar]
- 199.Burger MV, Gurry T, Stultz MC. Intrinsically Disordered Proteins: Where Computation Meets Experiment. Polym. 2014;6 doi: 10.3390/polym6102684. [DOI] [Google Scholar]
- 200.Cambridge HC, Cotten M, Scaly L, Chalkley R. Massive Phosphorylation Distinguishes Xenopus Zaevis Nucleoplasmin Isolated from Oocytes or Unfertilized Eggst. 1986:5063–5069. doi: 10.1021/bi00366a014. [DOI] [PubMed] [Google Scholar]
- 201.Vancurova I, Paine TM, Lou W, Paine PL. Nucleoplasmin associates with and is phosphorylated by casein kinase II. 1995;787:779–787. doi: 10.1242/jcs.108.2.779. [DOI] [PubMed] [Google Scholar]
- 202.Bañuelos S, Hierro A, Arizmendi JM, Montoya G, Prado A, Muga A. Activation Mechanism of the Nuclear Chaperone Nucleoplasmin: Role of the Core Domain. J Mol Biol. 2003;334:585–593. doi: 10.1016/j.jmb.2003.09.067. doi: http://dx.doi.org/10.1016/j.jmb.2003.09.067. [DOI] [PubMed] [Google Scholar]
- 203.Chou YH, Yung BYM. Cell Cycle Phase-Dependent Changes of Localization and Oligomerization States of Nucleophosmin / B23. Biochem Biophys Res Commun. 1995;217:313–325. doi: 10.1006/bbrc.1995.2779. doi: http://dx.doi.org/10.1006/bbrc.1995.2779. [DOI] [PubMed] [Google Scholar]
- 204.Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. Specific Phosphorylation of Nucleophosmin on Thr199 by Cyclin- dependent Kinase 2-Cyclin E and Its Role in Centrosome Duplication. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- 205.Okuwaki M, Tsujimoto M, Nagata K. The RNA Binding Activity of a Ribosome Biogenesis Factor. Nucleophosmin/B23, Is Modulated by Phosphorylation with a Cell Cycle-dependent Kinase and by Association with Its Subtype. Mol Biol Cell. 2002;13:2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mitrea DM, Grace CR, Buljan M, Yun M-K, Pytel NJ, Satumba J, Nourse A, Park C-G, Madan Babu M, White SW, Kriwacki RW. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc Natl Acad Sci. 2014;111:4466–4471. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yang JH, Song TY, Jo C, Park J, Lee HY, Song I, Hong S, Jung KY, Kim J, Han JW, Youn HD, Cho EJ. Differential regulation of the histone chaperone HIRA during muscle cell differentiation by a phosphorylation switch. Exp Mol Med. 2016;48:e252. doi: 10.1038/emm.2016.68. http://dx.doi.org/10.1038/emm.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. Phosphorylation by Protein Kinase CK2 Changes the DNA Binding Properties of the Human Chromatin Protein DEK. Mol Cell Biol. 2004;24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pilyugin M, Demmers J, Verrijzer CP, Karch F, Moshkin YM. Phosphorylation-Mediated Control of Histone Chaperone ASF1 Levels by Tousled-Like Kinases. PLoS One. 2009;4:e8328. doi: 10.1371/journal.pone.0008328. http://dx.doi.org/10.1371%252Fjournal.pone.0008328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Eddé B, Rossier J, Le Caer JP, Desbruyères E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–5. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 211.Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub J-M, Temurak N, Dijk J van, Boucher D, van Dorsselaer A, Suryavanshi S, Gaertig J, Eddé B. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–62. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 212.Bonnet C, Boucher D, Lazereg S, Pedrotti B, Islam K, Denoulet P, Larcher JC. Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B, and MAP2 by tubulin polyglutamylation. J Biol Chem. 2001;276:12839–48. doi: 10.1074/jbc.M011380200. [DOI] [PubMed] [Google Scholar]
- 213.van Dijk J, Miro J, Strub JM, Lacroix B, van Dorsselaer A, Edde B, Janke C. Polyglutamylation is a post-translational modification with a broad range of substrates. J Biol Chem. 2008;283:3915–22. doi: 10.1074/jbc.M705813200. [DOI] [PubMed] [Google Scholar]
- 214.Miller KE, Heald R. Glutamylation of Nap1 modulates histone H1 dynamics and chromosome condensation in Xenopus. J Cell Biol. 2015;209:211–220. doi: 10.1083/jcb.201412097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Muthurajan UM, Hepler MRD, Hieb AR, Clark NJ, Kramer M, Yao T, Luger K. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc Natl Acad Sci U S A. 2014;111:12752–12757. doi: 10.1073/pnas.1405005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Marheineke K, Krude T. Nucleosome assembly activity and intracellular localization of human caf-1 changes during the cell division cycle. J Biol Chem. 1998;273 doi: 10.1074/jbc.273.24.15279. [DOI] [PubMed] [Google Scholar]
- 217.Adachi Y, Pavlakis GN, Copeland TD. Identification and characterization of set, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J Biol Chem. 1994;269:2258–2262. [PubMed] [Google Scholar]
- 218.Gamble MJ, Erdjument-Bromage H, Tempst P, Freedman LP, Fisher RP. The histone chaperone taf-i/set/inhat is required for transcription in vitro of chromatin templates. Mol Cell Biol. 2005;25 doi: 10.1128/MCB.25.2.797-807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 220.Adam S, Polo SE, Almouzni G. Transcription Recovery after DNA Damage Requires Chromatin Priming by the H3.3 Histone Chaperone HIRA. Cell. 2013;155:94–106. doi: 10.1016/j.cell.2013.08.029. doi: http://dx.doi.org/10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 221.Dilworth SM, Black SJ, Laskey Ra. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–18. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- 222.Wang H, Ge Z, Walsh STR, Parthun MR. The human histone chaperone sNASP interacts with linker and core histones through distinct mechanisms. Nucleic Acids Res. 2012;40:660–9. doi: 10.1093/nar/gkr781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang AH, Zare H, Mousavi K, Wang C, Moravec CE, Sirotkin HI, Ge K, Gutierrez‐Cruz G, Sartorelli V. The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. EMBO J. 2013;32:1075 LP-1086. doi: 10.1038/emboj.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongélard F, Hans F, Mietton F, Studitsky VM, Hamiche A, Dimitrov S, Bouvet P. Nucleolin is a histone chaperone with FACT‐like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669 LP-1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gaume X, Monier K, Argoul F, Mongelard F, Bouvet P. In vivo Study of the Histone Chaperone Activity of Nucleolin by FRAP. Biochem Res Int. 2011;187624:1–15. doi: 10.1155/2011/187624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Duro E, Lundin C, Ask K, Sanchez-Pulido L, MacArtney TJ, Toth R, Ponting CP, Groth A, Helleday T, Rouse J. Identification of the MMS22L-TONSL Complex that Promotes Homologous Recombination. Mol Cell. 2010;40:632–644. doi: 10.1016/j.molcel.2010.10.023. doi: http://dx.doi.org/10.1016/j.molcel.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 227.Campos EI, Smits AH, Kang YH, Landry S, Escobar TM, Nayak S, Ueberheide BM, Durocher D, Vermeulen M, Hurwitz J, Reinberg D. Analysis of the Histone H3.1 Interactome: A Suitable Chaperone for the Right Event. Mol Cell. 2015;60:697–709. doi: 10.1016/j.molcel.2015.08.005. doi: http://dx.doi.org/10.1016/j.molcel.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]


