Abstract
Background
Childhood asthma is likely the result of gene-by-environment (G×E) interactions. Dust mite is a known risk factor for asthma morbidity. Yet, there have been no genome-wide G×E studies of dust mite allergen on asthma-related phenotypes.
Objective
To identify genetic variants whose effects on lung function in children with asthma are modified by level of dust mite allergen exposure.
Methods
A genome-wide interaction analysis of dust mite allergen level and lung function was performed in a cohort of Puerto Rican children with asthma (PRGOAL). Replication was attempted in two independent cohorts, the Childhood Asthma Management Program (CAMP) and the Genetics of Asthma in Costa Rica Study.
Results
SNP rs117902240 showed a significant interaction with dust mite allergen level on FEV1 in PRGOAL (interaction P=3.1×10−8), and replicated in the same direction in CAMP White children and CAMP Hispanic children (combined interaction P=0.0065 for replication cohorts and 7.4×10−9 for all cohorts). Rs117902240 was positively associated with FEV1 in children exposed to low dust mite allergen levels, but negatively associated with FEV1 in children exposed to high levels. This SNP is on chromosome 8q24, adjacent to a binding site for CEBPβ, a transcription factor that forms part of the IL-17 signaling pathway. None of the SNPs identified for FEV1/FVC replicated in the independent cohorts.
Conclusions
Dust mite allergen exposure modifies the estimated effect of rs117902240 on FEV1 in children with asthma. Analysis of existing data suggests this SNP may have transcription factor regulatory functions.
Keywords: Childhood asthma, lung function, dust mite allergen, genome-wide interaction study, gene-by-environment interaction, CEBPβ
Graphical abstract

INTRODUCTION
Asthma, the most common non-communicable chronic illness among children1, has an estimated heritability of up to 50–90%2,3. Although dozens of genes that confer susceptibility to asthma or asthma morbidity have been recently identified4, single nucleotide polymorphisms (SNPs) in such genes account for as little as 10–50% of the disease’s heritability. One potential reason for this “missing heritability” is gene-by-environment (G×E) interactions, as certain genetic variants may confer risk only in the presence of certain environmental exposures.
One of the best characterized environmental risk factors for asthma morbidity is exposure to indoor allergens. In particular, dust mite allergen is known to increase the risk of asthma exacerbations, and to increase airway responsiveness while reducing lung function5–7. The cysteine proteases in dust mites’ fecal particles are potent inducers of allergic sensitization8, and exposure to house dust mite can also trigger a direct, non-allergic, inflammatory reaction9. Both interventions to reduce dust mite exposure10,11 and trials of immune therapy to induce dust mite tolerance12–14 have been shown to improve asthma symptoms or reduce morbidity from asthma.
Few studies have looked at the interaction between genetic variants and allergens on asthma. Gene-by-allergen interactions have been reported for dust mite and several candidate genes for asthma, including IL1015, TGFB116, or the purinergic receptor P2RY1217. More recently, a study using genome-wide expression in vitro responses to dust mite allergen reported that IL9 may interact with environmental dust mite allergen and increase severe asthma exacerbations18. In the present study, we performed a genome-wide study interaction study (GWIS) of house dust mite allergen (Dermatophagoides pteronyssinus) level on lung function in Puerto Rican children with asthma, with replication studies in two independent cohorts of children with asthma.
METHODS
Study population
As part of a case-control study of childhood asthma in Puerto Ricans (PRGOAL), we recruited 618 children with asthma (defined as physician-diagnosed asthma and at least one episode of wheeze in the prior year) living in Hartford (CT) and San Juan (PR). The details of subject recruitment have been published previously19–21.
In brief, from September 2003 to July 2008, flyers were distributed to all parents of children in grades K-8 in 15 public elementary/middle schools in Hartford, CT, that enroll a high proportion (42% to 94%) of Puerto Rican children. Of 640 children whose parents were interested in the study, 585 (91.4%) were eligible after screening; parents of 449 (76.7%) of these 585 children (including 267 children with asthma and 182 control subjects) agreed to participate. There were no significant differences in age, gender, or area of residence between eligible children who did (n = 449) and did not (n = 136) participate. From March 2009 to June 2010, children were recruited from households in metropolitan San Juan, PR, selected using a multistage probability sample design. Primary sampling units (PSUs) were randomly selected neighborhood clusters based on the 2000 U.S. census, and secondary sampling units were randomly selected households within each PSU. A household was eligible if ≥1 resident was a child 6–14 years old. In households with more than one eligible child, one child was selected for screening; in households with multiple eligible children, one child was randomly selected using Kish tables. Of 6,401 contacted households, 1,111 had ≥1 child within the study age range who met other inclusion criteria. In order to reach our target sample size (≥700 children), we attempted to enroll a random sample (n = 783) of these 1,111 children. Parents of 105 (13.4%) of these households refused to participate or could not be reached. There were no significant differences in age, gender, or area of residence between eligible children who did (n = 678, including 351 children with asthma and 327 control subjects) and did not (n = 105) participate. At both study sites, participants had to have four Puerto Rican grandparents and had to have lived in the same household for ≥1 year. Of the 618 children with asthma at both study sites, 520 (287 from San Juan and 233 from Hartford) had blood samples and sufficient DNA for genome-wide genotyping. Of these 520 children, 440 (84.6%, 249 from San Juan and 191 from Hartford) also had data on indoor dust mite allergen level and lung function, and were thus included in the current analysis.
Ethics statement
Written consent was obtained from parents of participating children, from whom written assent was also obtained. The study was approved by the Institutional Review Boards (IRBs) of Connecticut Children’s Medical Center (Hartford, CT [Protocol #135503]), the University of Puerto Rico (San Juan, PR [Protocol #0160507]), Brigham and Women’s Hospital (Boston, MA [Protocol #2007P-001174), and the University of Pittsburgh (Pittsburgh, PA [Protocol #PRO10030498]).
Lung function
Spirometry was performed in participating children using an EasyOne spirometer (NDD Medical Technologies, Andover, MA). Subjects had to be free of respiratory infections for four or more weeks before testing, and they were also instructed (when possible) to avoid use of inhaled short-and long-acting bronchodilators for ≥4 and ≥12 hours before testing, respectively. Expiratory maneuvers were judged acceptable if they met or exceeded American Thoracic Society criteria for children22. The best FEV1 and FVC from each test were selected for data analysis.
Dust mite allergen measurement
Dust samples were obtained from three areas in the home: the one in which the child slept (usually his/her bedroom), the living room/television room, and the kitchen. The dust was sifted through a 50μ-mesh metal sieve, and the fine dust was reweighed, extracted, and aliquoted for analysis of dust mite allergen (D. pteronyssinus [Der p 1], hereinafter referred to as “dust mite”) using monoclonal antibody Multiplex assays that used the same reagents as the established ELISA23,24. Dust mite allergen level was categorized as <2 μg/g vs. ≥2 μg/g, because of known effects on allergic sensitization, distribution of allergen levels across the study cohorts (see Table 1), and ease of exposition6.
Table 1.
Participant characteristics
| PRGOAL | CAMP Whites | CAMP Hispanics | GACRS | |
|---|---|---|---|---|
| N | 440 | 497 | 56 | 549 |
| Age (years) | 9.9 (2.6) | 8.9 (2.1) | 9.7 (2.1) | 9.0 (1.8) |
| Male sex | 246 (55.9%) | 297 (59.8%) | 39 (69.6%) | 321 (58.5%) |
| Race/ethnicity | Puerto Rican | Caucasian | Hispanic | Costa Rican |
| FEV1 (L) | 1.87 (0.65) | 1.67 (0.47) | 1.85 (0.50) | 1.73 (0.49) |
| FEV1/FVC (%) | 81.5 (8.7) | 79.4 (8.2) | 79.3 (8.5) | 82.4 (7.3) |
| Der p 1 ≥ 2μg/g | 213 (48.4%) | 108 (21.7%) | 16 (28.6%) | 504 (91.8%) |
Numbers are means (SD) for continuous variables, or N (%) for binary variables. Final numbers included in the analysis may have varied slightly due to different missing rates for each SNP. Dust mite allergen dichotomized using the median level for our cohort (<2 vs ≥2 micrograms of Der p 1 per gram of dust).
Genotyping
Details of genotyping in PRGOAL have been previously described21,25. In brief, genotyping was performed using the Illumina HumanOmni2.5 BeadChip platform (Illumina, Inc., San Diego, CA), which includes ~2.5 million SNPs. Subjects with a call rate <95% were removed from the analysis. SNPs were removed if they were not in Hardy–Weinberg equilibrium (P<10−6) in control subjects, had minor allele frequency lower than 1%, or had a failure rate greater than 5%. After all subject and marker quality control steps were completed, we had ~1.83 million SNPs available for data analysis. In addition, SNPs not directly genotyped in the top region(s) of interest were imputed using all populations from the 1,000 Genomes Project (phase I v3)26 as reference for phasing and imputation with SHAPE-IT27 and IMPUTE228, respectively.
Replication populations
We attempted replication in two cohorts of children with asthma: the Childhood Asthma Management Program (CAMP) and the Genetics of Asthma in Costa Rica Study (GACRS). CAMP was a clinical trial of asthma therapy in 1,041 children. Details on study population, subject recruitment, and study procedures have been previously published29,30. In this analysis, we included 497 non-Hispanic white children and 56 Hispanic children who had genome-wide genotypic data, Der p 1 level in house dust, and lung function. Genotyping in CAMP was performed using the Illumina HumanHap550v3 BeadChip, and imputation was performed based on the 1,000 Human Genomes Project26 as previously described31. The GACRS was a cross-sectional study of 616 children with asthma and their parents. Details on study population, subject recruitment, and study procedures have been previously published32. In this analysis, we included 549 children who had genome-wide genotypic data, Der p 1 level in house dust, and lung function. Genotyping in the GACRS was performed using the Illumina BeadStation 500G platform, and imputation was performed based on the 1,000 Human Genomes Project26, using the same methodology as in CAMP. In both CAMP and the GACRS, Der p 1 level in house dust was measured using the same approach as in the discovery cohort (PRGOAL, see above).
Statistical analysis
Our main outcomes were FEV1 and FEV1/FVC. FEV1 was analyzed as absolute value in liters (adjusted for sex, age, and height); we decided not to utilize percent-predicted values because there are no validated predictive equations for Puerto Ricans or Costa Ricans. For CAMP, we used pre-randomization lung function in order to avoid any potential treatment arm effects. Dust mite allergen level was dichotomized using the median level for our cohort (<2 vs ≥2 micrograms of Der p 1 per gram [μg/g] of dust), a cutoff that has been previously associated with allergic sensitization to dust mite6. All multivariate analyses were conducted under the assumption of an additive genetic model, and included the main effect of the SNP, the main effect of dust mite allergen level, an interaction term (genotype*allergen), age, gender, study site (two sites for PRGOAL and eight sites for CAMP), and cohort-specific principal components to account for potential population stratification (eight for PRGOAL, six for CAMP, four for the GACRS); models for FEV1 also included height and height squared.
Genome-wide statistical significance was set at P <5×10−8. The top five SNPs from the PRGOAL analysis for FEV1 and FEV1/FVC were carried forward for replication in CAMP and the GACRS. For each cohort, replication was defined as a one-tailed P-value <0.05, in the same direction of interaction as in the discovery cohort. Fisher combined p-values (sum of logs method) were obtained for all cohorts and for the replication cohorts only. All genome-wide interaction analyses for dust mite allergen were performed using PLINK v1.0733. All other analyses were performed in R (version 3.1.3), using R-Studio (version 0.98.1103).
RESULTS
Table 1 summarizes the baseline characteristics of all cohorts. PRGOAL and CAMP White participants were slightly older than those in CAMP Hispanics and the GACRS. There was a higher proportion of boys in CAMP Hispanics (~69%) than in the other study groups (~56–60%). All children had similar lung function measures after adjusting for sex and age. Compared to PRGOAL, CAMP had a lower proportion of households with dust mite allergen level >2 μg/g, whereas the GACRS had a higher proportion of households with dust mite allergen >2 μg/g than the other cohorts.
FEV1
Interaction P-values for the GWIS of dust mite allergen in PRGOAL are shown in the Manhattan plot in Figure 1. The top five SNPs for FEV1 are shown in Table 2. SNP rs117902240, located on chromosome 8q24.13, had a significant interaction with dust mite allergen level (P for interaction= 3.1×10−8), while the P-values for the other four SNPs ranged from 1.3×10−7 to 5.4×10−7. Minor allele frequencies for the top five SNPs ranged from 1.3% to 9%.
Figure 1. Manhattan plot of GWIS interaction p-values in PRGOAL for FEV1.

Plot depicts the –log10(interaction P-value) for FEV1 on the y-axis vs SNPs in each chromosome on the x-axis. Red line: genome-wide significance level (P<5×10−8). Blue line: genome-wide suggestive level (P<1×10−7).
Table 2.
GWIS results for dust mite allergen level and FEV1 in PRGOAL
| SNP | CHR | BP | A1 | A2 | BETA | P-VAL | MAF | n(A1) |
|---|---|---|---|---|---|---|---|---|
| rs117902240 | 8 | 122930641 | C | A | −1.156 | 3.114E-08 | 0.015 | 4/7 |
| rs1404568 | 3 | 143210189 | G | A | 0.391 | 1.294E-07 | 0.09 | 34/29 |
| rs72716237 | 9 | 26458550 | A | G | 0.935 | 4.228E-07 | 0.013 | 8/5 |
| rs7541272 | 1 | 108211005 | G | A | 0.476 | 4.447E-07 | 0.06 | 29/22 |
| rs35312258 | 3 | 143193111 | G | A | 0.371 | 5.358E-07 | 0.09 | 33/31 |
Models included the main effects of genotype (additive) and dust mite allergen level (dichotomized <2 vs ≥2 μg/g) and the interaction term (genotype*dust mite allergen level), and were additionally adjusted for age, sex, study site (San Juan, PR, or Hartford, CT), height, and height2, and principal components. SNP=single nucleotide polymorphism; CHR=chromosome; BP=base position; A1=minor allele; A2=major allele; BETA=beta coefficient for interaction term; P-VAL=interaction p-value; MAF=minor allele frequency; n(A1)=number of subjects with at least one copy of the minor allele and low/high dust mite exposure.
Table 3 shows the results of the replication efforts for FEV1. Of the five SNPs carried forward for replication, SNP rs117902240 was associated with FEV1 among non-Hispanic whites (P value=0.05) and Hispanics (P=0.01) in CAMP, but not in the GACRS. The combined P-value for all three replication cohorts (in CAMP and the GACRS) was 0.0048, and the combined P-value for interaction with dust mite in all four cohorts was 9.75×10−9. Given differences in sample sizes and effect estimates among cohorts, we also combined P-values using a weighted approach: when weighted by the square root of the sample sizes, the combined P-value was 1.1×10−4; when weighted by the effect sizes, the combined P-value was 3.0×10−11. Of the remaining four SNPs, three were associated with FEV1 in the GACRS (rs1404568 and rs35312258 in chr.3, and rs72716237 in chr.9), but the direction of the estimated interactions with dust mite allergen was opposite that found in PRGOAL.
Table 3.
GWIS replication results for FEV1 in CAMP and the GACRS
| CAMP Whites | CAMP Hispanics | GACRS | COMBINED P-VAL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | A1 | BETA | P-VAL | MAF | n | A1 | BETA | P-VAL | MAF | n | A1 | BETA | P-VAL | MAF | n | All cohorts | Replication only |
| rs117902240 | C | −0.43 | 0.051 | 0.01 | 8/1 | C | −0.61 | 0.011 | 0.03 | 2/1 | C | 0.26 | 0.15 | 0.01 | 2/5 | 7.4E-09 | 0.0065 |
| rs 1404568 | G | −0.0078 | 0.45 | 0.11 | 76/23 | G | 0.048 | 0.32 | 0.17 | 13/3 | G | −0.12 | 0.048 | 0.18 | 20/167 | 1.6E-06 | 0.13 |
| rs72716237 | A | 0.067 | 0.33 | 0.02 | 22/3 | A | NA | NA | NA | NA | A | −0.38 | 0.046 | 0.01 | 2/14 | 1.3E-06 | 0.079 |
| rs7541272 | G | −0.105 | 0.25 | 0.02 | 16/3 | G | NA | NA | NA | NA | G | 0.24 | 0.21 | 0.02 | 1/20 | 4.1E-06 | 0.21 |
| rs35312258 | G | 0.031 | 0.32 | 0.09 | 63/19 | G | 0.048 | 0.33 | 0.17 | 13/3 | G | −0.12 | 0.046 | 0.17 | 19/159 | 3.8E-06 | 0.098 |
Models included the main effects of genotype (additive) and dust mite allergen level (dichotomized <2 vs ≥2 μg/g) and the interaction term (genotype*dust mite allergen level), and were additionally adjusted for age, sex, study site, height, height2, and principal components. SNP=single nucleotide polymorphism; A1=minor allele; BETA=beta coefficient for interaction term; P-VAL=interaction p-value; MAF=minor allele frequency; n=number of subjects with at least one copy of the minor allele and low/high dust mite exposure; NA=not available (genotyped or imputed).
Using imputed SNP data based on the 1,000 Human Genomes Project, we performed fine mapping of the region surrounding the top SNP (rs117902240): several SNPs showed R2 ~0.4 with this SNP; the top interaction P value for these imputed SNPs was 2.6×10−6 (rs186935901 and rs10108856), with others ranging from 1.3×10−4 to 9.5×10−4 (Figure 2). Figure S1 (online supplementary data) shows the LD plots for the other SNPs: seven SNPs showed R2 ~0.75 with rs72716237. SNPs rs13078996 and rs1404568 were in high LD with rs35312258; their interaction P values were 6.01×10−6 and 1.29×10−7, respectively.
Figure 2. Regional association plot for SNP rs117902240.
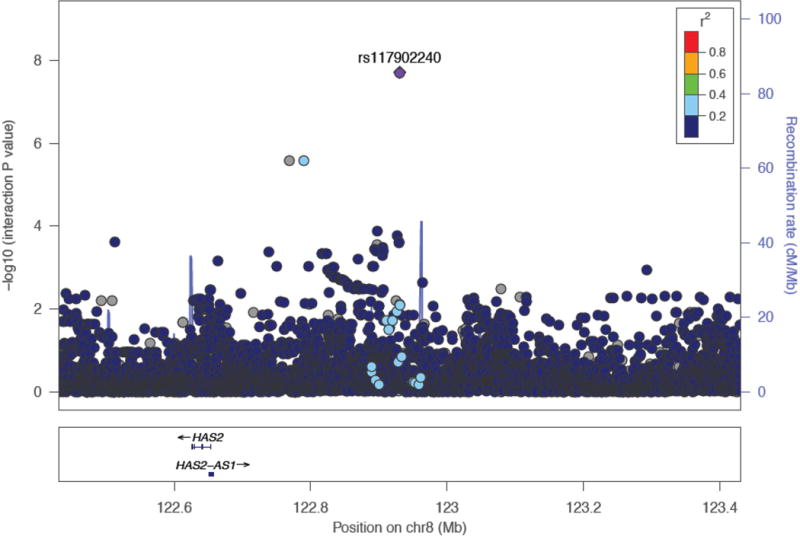
Regional association plot generated using the LocusZoom tool from the Abecasis lab at the University of Michigan (http://locuszoom.sph.umich.edu/locuszoom/)52 with data imputed from the 1,000 Genomes populations (phase I v3).
We then repeated the analysis for rs117902240 in PRGOAL, before and after stratification by dust mite allergen level. When evaluated in PRGOAL as a whole, rs117902240 was not significantly associated with FEV1 (β=0.07 [SD=0.10], p-value=0.48). However, after stratifying by dust mite allergen level, SNP rs117902240 was positively associated with FEV1 (β=+0.84 [0.15], p=1.2×10−7) in children exposed to low dust mite allergen levels, but negatively associated with FEV1 in children exposed to high dust mite allergen levels (β=−0.41 [0.13], p=0.003). Figure 3 shows the boxplot for FEV1, stratified by dust mite level and rs117902240 genotype. Table S1 (online supplementary data) shows further details for the stratified analysis of rs117902240 and the other top SNPs.
Figure 3. FEV1 stratified by rs117902240 genotype and dust mite allergen level.
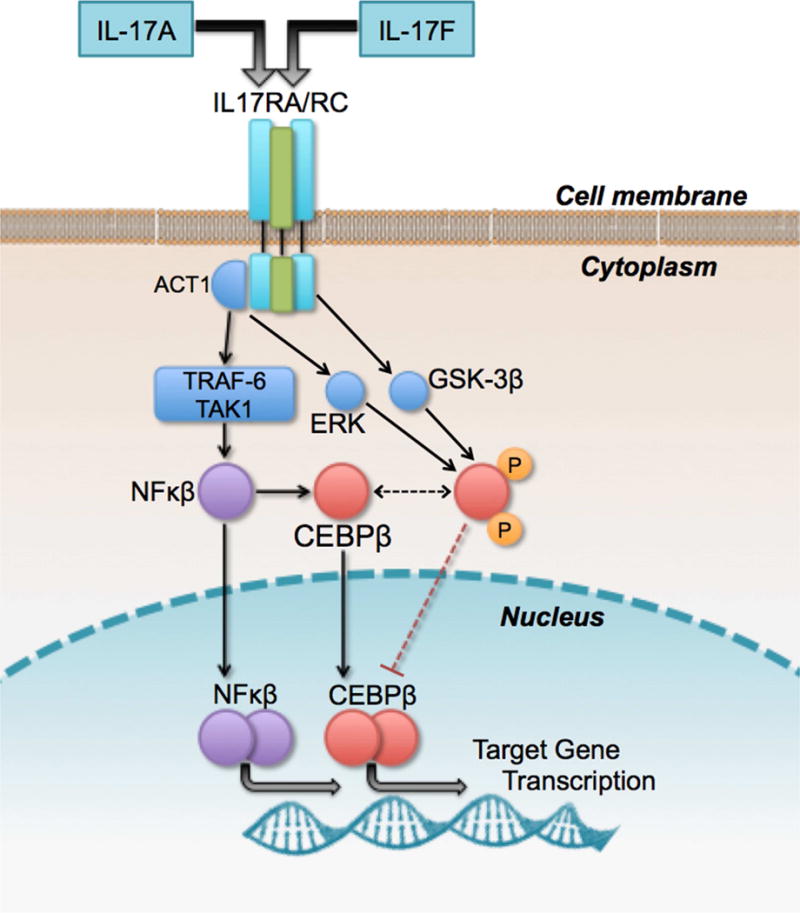
FEV1 in PRGOAL stratified by dust mite allergen level (low in blue vs. high in red) and rs117902240 genotype (AA=no minor alleles vs. AC=one minor allele; there were no subjects homozygous for the minor allele [CC]). Model adjusted for age, sex, and height.
In an exploratory analysis, we assessed the biological plausibility of SNP rs117902240 as an allelic variant interacting with dust mite allergen. Using data from ENCODE34 and is-rSNP35, a software designed to predict whether a SNP has a regulatory effect on transcription factor binding sites in silico, we found that SNP rs117902240 is <1 kb away from a transcription factor binding island identified in ENCODE using ChIP-seq. Transcription factor CEBPβ (CCAAT/Enhancer Binding Protein Beta) binds in this region in human lung fibroblasts (ENCODE accession ENCSR000EFM). CEBPβ forms part of the signaling pathway of interleukin (IL)-17, IL-6, nuclear factor Kappa-B (NF-κβ), and interferon (IFN-)γ (see Figure 4)36,37. Details for the other top SNPs may be found in the online supplement.
Figure 4. CEBPβ in the IL-17 signaling pathway.
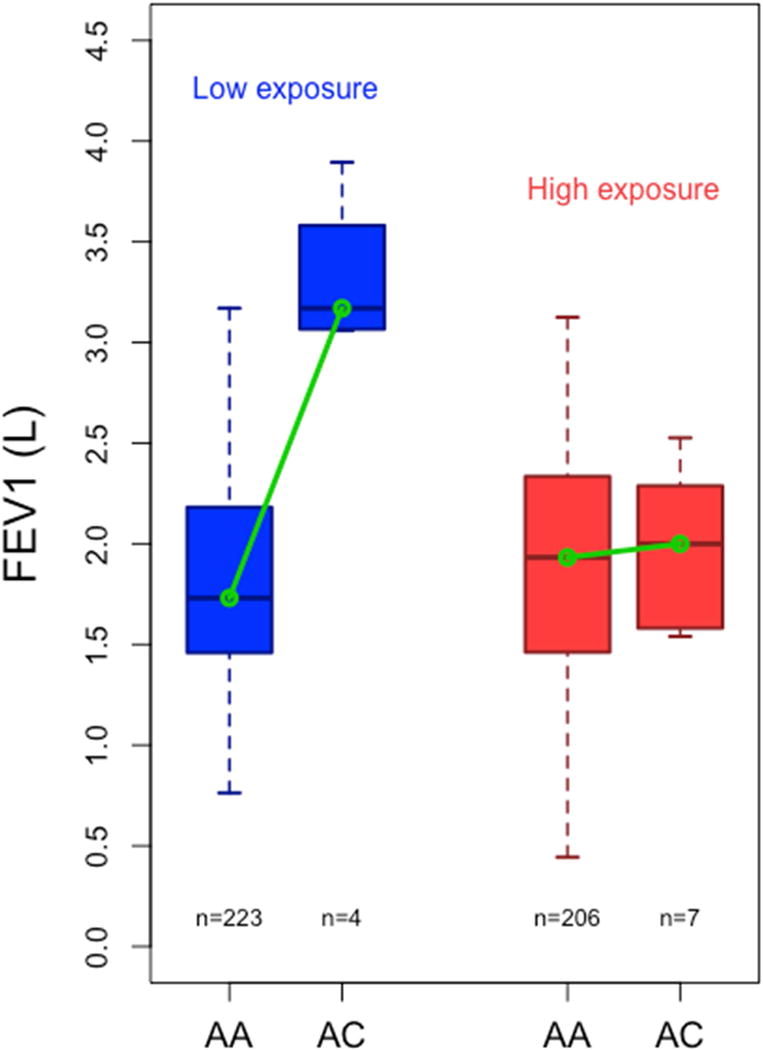
IL-17 pathway (simplified). CEBPβ = CCAAT Enhancer Binding Protein Beta. CEBPβ is one of the mediators of IL-17A and IL-17F signaling pathways, as well as NF-κβ signaling36,37. CEBPβ is also involved in IL-4, IL-6, and other signaling pathways (not shown).
Finally, we explored SNPs within previously reported genes (IL-9, IL-10, TGF-β1, P2RY2) for nominal association (one-tailed P-value<0.05) with FEV1. Several SNPs within those genes showed nominal associations, either for the main effect of genotype or for the genotype*dust mite interaction (see Table S2 in the online supplementary data).
FEV1/FVC
The top 5 SNPs for FEV1/FVC are shown in Table S3 (online supplementary data) and the replication results are shown in Table S4. One SNP (rs115997623) was associated in the GACRS and one (rs78930606) in CAMP Whites, but both in opposite directions as in PRGOAL. None of the top SNPs for FEV1/FVC had significant combined P-values for the replication cohorts.
DISCUSSION
In this study, we report that dust mite allergen level significantly modifies the estimated effect of one SNP (rs117902240) on FEV1 in children with asthma. To our knowledge, this is the first study to examine a gene-by-allergen interaction on an asthma-related phenotype (in this case, lung function) using a genome-wide approach.
Dust mite exposure and sensitization have been widely reported as risk factors for asthma symptoms and exacerbations in affected individuals5,6,38,39. Previous G×E studies of candidate genes have reported that dust mite exposure modifies the effects of IL-9 and IL-10 polymorphisms on asthma exacerbations15,18, TGF-β1 variants on asthma severity16, or purinergic receptor gene P2RY12 haplotypes on lung function17. Our results extend current knowledge on the topic by identifying novel polymorphisms using a genome-wide approach.
Using data from ENCODE34 and is-rSNP35, we found that rs117902240 is closely adjacent to a transcription factor island that binds CEBPβ, which forms part of the IL-17 signaling pathway (Figure 4). IL-17A or IL-17F can stimulate the NF-κβ pathway, which in turn acts with CEBPβ to activate target gene transcription in the nucleus37. IL-17 has been amply implicated in the pathogenesis of asthma40–42, and studies in dust-mite sensitized subjects have shown that specific immunotherapy can lead to reductions in IL-17 levels that correlate with symptom reduction43. However, CEBPβ also participates in several other pathways, including IL-4, IL-6, TNFα and IFN-γ signaling pathways, and it has been implicated in airway epithelium differentiation during lung development44. Thus, functional studies will be needed to determine the precise role of CEBPβ in the interaction between dust mite and genotype on lung function in asthma.
Children with more severe or poorly controlled asthma have worse lung function45. Moreover, poor lung function in children with asthma predicts exacerbations over subsequent follow-up46, and is associated with worse outcomes in adulthood47. Several studies (including GWAS and candidate-gene studies) have been performed to identify genetic variants associated with lung function in subjects with asthma48–50, regardless of allergen exposure. Of note, however, the top SNP reported here would have been missed in a “regular” GWAS, but had significant (and opposite) effects when stratified by level of dust mite allergen exposure. This has potential implications, both in terms of improving our understanding of the underlying mechanisms by which dust mite impacts lung function, but also in terms of developing new approaches to identify susceptibility variants for asthma-related phenotypes.
Our study has several strengths, including the overall sample size of ~1,550 children with asthma and data on dust mite allergen and lung function, and the biological plausibility of the top SNP based on ENCODE data. It also has several limitations: some of the SNPs examined had low minor allele frequencies and thus we had small sample size within some of the genotype*allergen level strata, which may have led to low power to detect modest interactions. Post hoc analysis based on results from SNP rs117902240 shows we had sufficient statistical power (see Table S5 in the online supplementary data) and the SNP*dust mite exposure level interaction replicated in the same direction in three out of four populations; nonetheless, results based on small strata must be interpreted with caution and should be further validated in future, prospective studies. In addition, we used statistical methods to adjust for population stratification but these may not be sufficient and some residual stratification may remain unaccounted for. Moreover, the choice of method for combining P-values had an important impact on the results; different approaches yielded combined P-values ranging from 1.1×10−4 (weighted by sample size), to 7.4×10−9 (Fisher’s unweighted method), to 3.0×10−11 (weighted by effect size). Finally, the timing and duration of dust mite exposure may be important factors in the pathogenesis of asthma, in addition to the level of the allergen51.
In summary, we show that dust mite allergen exposure modifies the estimated effect of a SNP on a key measure of lung function (FEV1) in children with asthma. Analysis of existing data suggests possible transcription factor regulatory functions for this SNP; future studies should focus on the functional validation of these pathways.
Supplementary Material
Capsule Summary.
Dust mite allergen exposure modifies the effect of certain genetic polymorphisms on lung function in children with asthma.
Clinical Implications.
The effect of certain genetic polymorphisms on lung function in children with asthma varies depending on their dust mite allergen exposure level. Future studies should assess whether personalized interventions for these children have a greater impact on lung function.
Acknowledgments
Sources of support: Dr. Forno’s contribution was supported by grant HL125666 from the U.S. National Institutes of Health (NIH) and by a grant from Children’s Hospital of UPMC. Dr. Celedón’s contribution was supported by grants HL079966 and HL117191 from the U.S. NIH, and by an endowment from the Heinz Foundation.
Abbreviations
- CEBPβ
CCAAT/Enhancer binding protein beta
- ChIP-seq
Chromatin immunoprecipitation and DNA sequencing
- DNA
Deoxyribonucleic acid
- ERK3
Extracellular signal-regulated kinase 3
- FEV1
Forced expiratory volume in the first second
- FVC
Forced vital capacity
- GSK3β
Glycogen synthase kinase 3 beta
- GWAS
Genome-wide association study
- GWIS
Genome-wide interaction study
- G×E
Gene-by-environment interaction
- IL
Interleukin
- IRB
Institutional review board
- P2RY12
Purinergic receptor P2Y, G-protein coupled, 12
- PSU
Primary sampling unit
- SNP
Single nucleotide polymorphism
- TAK1
Transforming growth factor beta 1-activated kinase 1
- TGFβ1
Transforming growth factor beta 1
- TRAF6
Tumor necrosis factor receptor-associated factor 6
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Asthma. WHO. 2014 Available from http://www.who.int/mediacentre/factsheets/fs307/en/
- 2.Palmer LJ, Burton PR, James AL, Musk AW, Cookson WO. Familial aggregation and heritability of asthma-associated quantitative traits in a population-based sample of nuclear families. Eur J Hum Genet. 2000;8:853–60. doi: 10.1038/sj.ejhg.5200551. [DOI] [PubMed] [Google Scholar]
- 3.Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Skadhauge LR, Backer V. Increase in the heritability of asthma from 1994 to 2003 among adolescent twins. Respir Med. 2011;105:1147–52. doi: 10.1016/j.rmed.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Ortiz RA, Barnes KC. Genetics of allergic diseases. Immunol Allergy Clin North Am. 2015;35:19–44. doi: 10.1016/j.iac.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sporik R, Holgate ST, Platts-Mills T, Cogswell JJ. Exposure to house-dust mite allergen and the development of asthma in childhood. N Engl J Med. 1990;323:502–7. doi: 10.1056/NEJM199008233230802. [DOI] [PubMed] [Google Scholar]
- 6.Celedon JC, Milton DK, Ramsey CD, Litonjua AA, Ryan L, Platts-Mills TA, et al. Exposure to dust mite allergen and endotoxin in early life and asthma and atopy in childhood. J Allergy Clin Immunol. 2007;120:144–9. doi: 10.1016/j.jaci.2007.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau S, Illi S, Sommerfeld C, Niggemann B, Bergmann R, von Mutius E, et al. Early exposure to house-dust mite and cat allergens and development of childhood asthma: a cohort study. Multicentre Allergy Study Group Lancet. 2000;356:1392–7. doi: 10.1016/s0140-6736(00)02842-7. [DOI] [PubMed] [Google Scholar]
- 8.Kanchongkittiphon W, Gaffin JM, Phipatanakul W. The indoor environment and inner-city childhood asthma. Asian Pac J Allergy Immunol. 2014;32:103–10. [PMC free article] [PubMed] [Google Scholar]
- 9.De Alba J, Raemdonck K, Dekkak A, Collins M, Wong S, Nials AT, et al. House dust mite induces direct airway inflammation in vivo: implications for future disease therapy? Eur Respir J. 2010;35:1377–87. doi: 10.1183/09031936.00022908. [DOI] [PubMed] [Google Scholar]
- 10.Scott M, Roberts G, Kurukulaaratchy RJ, Matthews S, Nove A, Arshad SH. Multifaceted allergen avoidance during infancy reduces asthma during childhood with the effect persisting until age 18 years. Thorax. 2012;67:1046–51. doi: 10.1136/thoraxjnl-2012-202150. [DOI] [PubMed] [Google Scholar]
- 11.El-Ghitany EM, Abd El-Salam MM. Environmental intervention for house dust mite control in childhood bronchial asthma. Environ Health Prev Med. 2012;17:377–84. doi: 10.1007/s12199-011-0263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Yin J, Fadel R, Montagut A, de Beaumont O, Devillier P. House dust mite sublingual immunotherapy is safe and appears to be effective in moderate, persistent asthma. Allergy. 2014;69:1181–8. doi: 10.1111/all.12188. [DOI] [PubMed] [Google Scholar]
- 13.de Blay F, Kuna P, Prieto L, Ginko T, Seitzberg D, Riis B, et al. SQ HDM SLIT-tablet (ALK) in treatment of asthma–post hoc results from a randomised trial. Respir Med. 2014;108:1430–7. doi: 10.1016/j.rmed.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 14.Mosbech H, Deckelmann R, de Blay F, Pastorello EA, Trebas-Pietras E, Andres LP, et al. Standardized quality (SQ) house dust mite sublingual immunotherapy tablet (ALK) reduces inhaled corticosteroid use while maintaining asthma control: a randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol. 2014;134:568–75 e7. doi: 10.1016/j.jaci.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 15.Hunninghake GM, Soto-Quiros ME, Lasky-Su J, Avila L, Ly NP, Liang C, et al. Dust mite exposure modifies the effect of functional IL10 polymorphisms on allergy and asthma exacerbations. J Allergy Clin Immunol. 2008;122:93–8. 8 e1–5. doi: 10.1016/j.jaci.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma S, Raby BA, Hunninghake GM, Soto-Quiros M, Avila L, Murphy AJ, et al. Variants in TGFB1, dust mite exposure, and disease severity in children with asthma. American journal of respiratory and critical care medicine. 2009;179:356–62. doi: 10.1164/rccm.200808-1268OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunyavanich S, Boyce JA, Raby BA, Weiss ST. Gene-by-environment effect of house dust mite on purinergic receptor P2Y12 (P2RY12) and lung function in children with asthma. Clin Exp Allergy. 2012;42:229–37. doi: 10.1111/j.1365-2222.2011.03874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sordillo JE, Kelly R, Bunyavanich S, McGeachie M, Qiu W, Croteau-Chonka DC, et al. Genome-wide expression profiles identify potential targets for gene-environment interactions in asthma severity. J Allergy Clin Immunol. 2015;136:885–92 e2. doi: 10.1016/j.jaci.2015.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brehm JM, Ramratnam SK, Tse SM, Croteau-Chonka DC, Pino-Yanes M, Rosas-Salazar C, et al. Stress and Bronchodilator Response in Children with Asthma. Am J Respir Crit Care Med. 2015;192:47–56. doi: 10.1164/rccm.201501-0037OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs TS, Forno E, Brehm JM, Acosta-Perez E, Han YY, Blatter J, et al. Underdiagnosis of allergic rhinitis in underserved children. J Allergy Clin Immunol. 2014;134:737–9 e6. doi: 10.1016/j.jaci.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brehm JM, Acosta-Perez E, Klei L, Roeder K, Barmada MM, Boutaoui N, et al. African ancestry and lung function in Puerto Rican children. J Allergy Clin Immunol. 2012;129:1484–90 e6. doi: 10.1016/j.jaci.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, et al. ATS/ERS Task Force - Standardisation of Lung Function Testing: Standardisation of spirometry. Eur Respir J. 2005;26:319–38. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 23.Blatter J, Forno E, Brehm J, Acosta-Perez E, Alvarez M, Colon-Semidey A, et al. Fungal exposure, atopy, and asthma exacerbations in Puerto Rican children. Ann Am Thorac Soc. 2014;11:925–32. doi: 10.1513/AnnalsATS.201402-077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luczynska CM, Arruda LK, Platts-Mills TA, Miller JD, Lopez M, Chapman MD. A two-site monoclonal antibody ELISA for the quantification of the major Dermatophagoides spp. allergens, Der p I and Der f I J Immunol Methods. 1989;118:227–35. doi: 10.1016/0022-1759(89)90010-0. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Brehm JM, Lin J, Wang T, Forno E, Acosta-Perez E, et al. Expression quantitative trait loci (eQTL) mapping in Puerto Rican children. PLoS One. 2015;10:e0122464. doi: 10.1371/journal.pone.0122464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaneau O, Coulonges C, Zagury JF. Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinformatics. 2008;9:540. doi: 10.1186/1471-2105-9-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long-term effects of budesonide or nedocromil in children with asthma. The Childhood Asthma Management Program Research Group. The New England journal of medicine. 2000;343:1054–63. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 30.Forno E, Lasky-Su J, Himes B, Howrylak J, Ramsey C, Brehm J, et al. Genome-wide association study of the age of onset of childhood asthma. J Allergy Clin Immunol. 2012;130:83–90 e4. doi: 10.1016/j.jaci.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Himes BE, Jiang X, Hu R, Wu AC, Lasky-Su JA, Klanderman BJ, et al. Genome-wide association analysis in asthma subjects identifies SPATS2L as a novel bronchodilator response gene. PLoS Genet. 2012;8:e1002824. doi: 10.1371/journal.pgen.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forno E, Fuhlbrigge A, Soto-Quiros ME, Avila L, Raby BA, Brehm J, et al. Risk factors and predictive clinical scores for asthma exacerbations in childhood. Chest. 2010;138:1156–65. doi: 10.1378/chest.09-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macintyre G, Bailey J, Haviv I, Kowalczyk A. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics. 2010;26:i524–30. doi: 10.1093/bioinformatics/btq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruddy MJ, Wong GC, Liu XK, Yamamoto H, Kasayama S, Kirkwood KL, et al. Functional cooperation between interleukin-17 and tumor necrosis factor-alpha is mediated by CCAAT/enhancer-binding protein family members. J Biol Chem. 2004;279:2559–67. doi: 10.1074/jbc.M308809200. [DOI] [PubMed] [Google Scholar]
- 37.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9:556–67. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Custovic A, Taggart SC, Francis HC, Chapman MD, Woodcock A. Exposure to house dust mite allergens and the clinical activity of asthma. The Journal of allergy and clinical immunology. 1996;98:64–72. doi: 10.1016/s0091-6749(96)70227-0. [DOI] [PubMed] [Google Scholar]
- 39.Zock JP, Brunekreef B, Hazebroek-Kampschreur AA, Roosjen CW. House dust mite allergen in bedroom floor dust and respiratory health of children with asthmatic symptoms. The European respiratory journal: official journal of the European Society for Clinical Respiratory Physiology. 1994;7:1254–9. doi: 10.1183/09031936.94.07071254. [DOI] [PubMed] [Google Scholar]
- 40.Ramsey CD, Lazarus R, A CC, Jr, Weiss ST, Celedon JC. Polymorphisms in the interleukin 17F gene (IL17F) and asthma. Genes Immun. 2005;6:236–41. doi: 10.1038/sj.gene.6364170. [DOI] [PubMed] [Google Scholar]
- 41.Han YY, Forno E, Brehm J, Acosta-Perez E, Alvarez M, Colon-Semidey A, et al. Diet, Interleukin-17, and Childhood Asthma in Puerto Ricans. Ann Allergy Asthma Immunol. 2015 doi: 10.1016/j.anai.2015.07.020. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinley L, Alcorn JF, Peterson A, Dupont RB, Kapadia S, Logar A, et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol. 2008;181:4089–97. doi: 10.4049/jimmunol.181.6.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li CW, Lu HG, Chen de H, Lin ZB, Wang de Y, Li TY. In vivo and in vitro studies of Th17 response to specific immunotherapy in house dust mite-induced allergic rhinitis patients. PLoS One. 2014;9:e91950. doi: 10.1371/journal.pone.0091950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roos AB, Berg T, Barton JL, Didon L, Nord M. Airway epithelial cell differentiation during lung organogenesis requires C/EBPalpha and C/EBPbeta. Dev Dyn. 2012;241:911–23. doi: 10.1002/dvdy.23773. [DOI] [PubMed] [Google Scholar]
- 45.Rasmussen F, Taylor DR, Flannery EM, Cowan JO, Greene JM, Herbison GP, et al. Risk factors for airway remodeling in asthma manifested by a low postbronchodilator FEV1/vital capacity ratio: a longitudinal population study from childhood to adulthood. Am J Respir Crit Care Med. 2002;165:1480–8. doi: 10.1164/rccm.2108009. [DOI] [PubMed] [Google Scholar]
- 46.Kitch BT, Paltiel AD, Kuntz KM, Dockery DW, Schouten JP, Weiss ST, et al. A single measure of FEV1 is associated with risk of asthma attacks in long-term follow-up. Chest. 2004;126:1875–82. doi: 10.1378/chest.126.6.1875. [DOI] [PubMed] [Google Scholar]
- 47.Sears MR, Greene JM, Willan AR, Wiecek EM, Taylor DR, Flannery EM, et al. A longitudinal, population-based, cohort study of childhood asthma followed to adulthood. The New England journal of medicine. 2003;349:1414–22. doi: 10.1056/NEJMoa022363. [DOI] [PubMed] [Google Scholar]
- 48.Joost O, Wilk JB, Cupples LA, Harmon M, Shearman AM, Baldwin CT, et al. Genetic loci influencing lung function: a genome-wide scan in the Framingham Study. Am J Respir Crit Care Med. 2002;165:795–9. doi: 10.1164/ajrccm.165.6.2102057. [DOI] [PubMed] [Google Scholar]
- 49.Wu K, Gamazon ER, Im HK, Geeleher P, White SR, Solway J, et al. Genome-wide interrogation of longitudinal FEV1 in children with asthma. Am J Respir Crit Care Med. 2014;190:619–27. doi: 10.1164/rccm.201403-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Hawkins GA, Ampleford EJ, Moore WC, Li H, Hastie AT, et al. Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. J Allergy Clin Immunol. 2013;132:313–20 e15. doi: 10.1016/j.jaci.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simpson A, Tan VY, Winn J, Svensen M, Bishop CM, Heckerman DE, et al. Beyond atopy: multiple patterns of sensitization in relation to asthma in a birth cohort study. Am J Respir Crit Care Med. 2010;181:1200–6. doi: 10.1164/rccm.200907-1101OC. [DOI] [PubMed] [Google Scholar]
- 52.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


