Abstract
Epigenetics is now emerging as a key regulation in response to various stresses. We herein identified the Drosophila histone methyltransferase G9a (dG9a) as a key factor to acquire tolerance to starvation stress. The depletion of dG9a led to high sensitivity to starvation stress in adult flies, while its overexpression induced starvation stress resistance. The catalytic domain of dG9a was not required for starvation stress resistance. dG9a plays no apparent role in tolerance to other stresses including heat and oxidative stresses. Metabolomic approaches were applied to investigate global changes in the metabolome due to the loss of dG9a during starvation stress. The results obtained indicated that dG9a plays an important role in maintaining energy reservoirs including amino acid, trehalose, glycogen, and triacylglycerol levels during starvation. Further investigations on the underlying mechanisms showed that the depletion of dG9a repressed starvation-induced autophagy by controlling the expression level of Atg8a, a critical gene for the progression of autophagy, in a different manner to that in cancer cells. These results indicate a positive role for dG9a in starvation-induced autophagy.
Introduction
Appropriate responses to stressful conditions are crucial for the adaptation of an organism to environmental challenges. Epigenetic regulation in response to these conditions has been attracting increasing attention due to its rapid and long-lasting effects on gene expression in response to environmental changes without alterations in DNA sequences1. In a wide variety of organisms, epigenetic regulation has been found to mediate long-term effects on gene expression or the transgenerational inheritance of phenotypic changes caused by various stresses2. In terms of starvation stress, prenatal starvation was initially reported to alter DNA methylation marks on the imprinted gene IGF2 and this change persisted throughout the human life-span3. Other studies also suggested that Sirtuin 1 (Sirt1), a histone deacetylase, plays a role in responses to starvation stress in Drosophila 4. However, except for Sirt1, the epigenetic factors that are critical in responses to starvation stress have not yet been identified. The histone methyltransferase G9a was recently reported to mediate autophagy by regulating the expression of autophagy-related genes including LC3B, WIPI, and DOR in starved human pancreatic cancer cells5. Since autophagy is an intracellular degradation system that responds to starvation, the signaling pathways mediating starvation-induced autophagy have been elucidated in detail6. However, gene regulation of the components in these signaling pathways has not been fully examined in vivo.
G9a was initially reported in 2001 as a mammalian histone lysine methyltransferase accounting for methylation at lysine 9 (K9) of histone H3 (H3K9) in vitro 7. In the last decade, the epigenetic regulation of gene expression via post-translational modifications by G9a in diverse biological processes has been a rapidly growing research subject. G9a may specifically associate with euchromatin and catalyze the mono-, di-, and trimethylation of H3K9, which is involved in transcriptional gene silencing8–11. This function of G9a was found to be important for early embryogenesis in mice12–15, the propagation of imprints16, and control of DNA methylation17, 18. Moreover, G9a is functional in the survival of mammalian cells under hypoxic stress19. These findings demonstrate the importance of G9a during development as well as in stress tolerance in mammals. However, the embryonic lethal phenotype has made it difficult to study the function of mammalian G9a at later developmental stages. Thus, the role of G9a on longevity under stress conditions in living organisms currently remains unknown.
In Drosophila melanogaster, G9a (dG9a) has been shown to play an important role in neural development as well as behavioral processes such as learning and memory20. A previous study using a dG9a null mutant strain (dG9a RG5) demonstrated that dG9a was not essential for Drosophila viability or fertility21; however, embryogenesis was delayed22. It is important to note that while there are various environmental stresses in the wild, Drosophila is always maintained under optimal conditions in the laboratory. Therefore, dG9a may play a critical role in environmental stress tolerance. On the other hand, a relationship has been proposed between gene regulation, nutrition, and metabolism because many enzymes involved in epigenetic gene regulation require co-substrates generated by cellular metabolism23, 24. Similarly, environmentally-induced epigenetic responses may induce changes in metabolism in an organism in order to support adaptation or stress tolerance25, 26. Hence, metabolomics may be used as a fundamental method to assess changes in the metabolic pathways of an organism and provide an insight into the dynamics of cellular functions that contribute to the survival of an organism in nature27.
In the present study, we exposed flies lacking dG9a to various stress conditions. We found that dG9a-depleted flies were specifically sensitive to starvation, but not heat or oxidative stress. In order to explain these results, the global metabolic profiling of fasted wild-type and dG9a-depleted flies was performed to elucidate the metabolic changes that occur when dG9a is removed.
The main changes in cellular metabolites accounting for energy generation under starvation stress were observed in dG9a-depleted flies. Further investigations showed that the loss of dG9a repressed starvation-induced autophagy by controlling the expression level of Atg8a in a methyltransferase-independent manner. This regulation by dG9a appears to be opposite to that reported previously in mammalian pancreatic cancer cells in which human G9a negatively regulated autophagic cell death5.
Materials and Methods
Fly stocks
All fly stocks were reared at 25 °C on standard food (0.7% agar, 10% glucose, 4% dry yeast, 5% cornmeal, 3% rice bran). Canton S was used as the wild-type. dG9a RG5 and dG9a del34 flies were kindly provided by Dr. P. Spierer and Dr. C. Seum. dG9a RG5 flies were backcrossed 10 times with Canton S to adjust the genetic background to Canton S. A mutant allele was recovered with the adjacent w 1118 marker. We confirmed that thew 1118 marker did not reduce fly viability, life span, or survival (data not shown). The UAS-FLAG-IR dG9a (strain #79) fly stock was produced previously28. The FB-GAL4 (y w; GAL4 fat) strain was kindly provided by Ronald P. Kühnlein29. We generated a fly strain carrying GAL80 ts and FB-GAL4 (y w; P(GAL4)fat; GAL80 ts.αTub84B) by crossing. The Atg8a d4 strain is kindly provided by Gabor Juhasz30. The Atg8a d4 mutant lacks the first 25 codons of Atg8a30. All other stocks used in this study were obtained from the Drosophila Genetic Resource Center in Kyoto and Bloomington Drosophila Stock Center.
Starvation assay
In the starvation assay, 3–5-day-old adult flies were placed into vials including a piece of paper soaked in 1.0 mL PBS and the number of living flies was monitored until all had died. All assays were performed under non-crowded conditions (<20 flies per vial) and the results from several independent assays were combined. The median survival time was calculated and graphs were generated with the Kaplan-Meier method by GraphPad Prism 6 software. Significance was calculated with Log-rank tests using GraphPad Prism 6 software. In rescue assays with glucose, glucose was dissolved in PBS in vials.
In the starvation assay utilizing temperature-sensitive Gal80, flies were grown at 18 °C on normal food as described previously31. Three-five-day-old adult male flies were placed into vials including a piece of paper soaked in 1.0 mL PBS in a temperature controlled incubator at 29 °C and the number of living flies was monitored until all had died.
Stress assays
In the oxidative stress assay, newly eclosed adult flies were transferred on instant food (Carolina) with 10 mM paraquat (Nacalai Tesque). Every 3 days, they were transferred to new vials containing fresh instant food and the number of living flies was monitored until all had died. In the heat stress assay, 3–5-day-old adult flies in vials containing fresh instant food were placed into a temperature controlled incubator at 36 °C and the number of living flies was monitored until all had died. All assays were performed under non-crowded conditions (20 flies per vial).
GC-MS and LC-MS analyses
Samples were collected after 0, 12, 24, and 36 h of fasting. Samples were washed, freeze-dried, and kept at −80 °C until analyzed. A 5-mg dry weight of each sample was homogenized using a ball mill (5 min, 20 Hz). There were 4–5 biological replicates for each condition. One milliliter of extraction solvent (2.5 methanol: 1 ultra-pure water: 1 chloroform) was used for each sample to extract a wide range of low molecular hydrophilic metabolites. After centrifugation (16,000 x g for 3 min), 900 µL supernatant was transferred to a 1.5-mL micro tube and mixed with 400 µL ultra-pure water (Wako). Centrifugation was performed again to separate the polar and non-polar phases. The polar phase was transferred into two fresh 1.5-mL Eppendorf tubes (400 µL/tube) for GC-MS and LC-MS analyses.
In the GC-MS analysis, samples were spun using centrifugal concentration (VCe36S, Taitec Co., Tokyo, Japan) for 2 h in order to remove the solvent and then freeze-dried for 16 h. Samples underwent derivatization by methoxyamine hydrochloride (Sigma Aldrich, St. Louis, MO, USA) in pyridine (50 µL, 10 mg/mL) at 30 °C for 90 min, and then by 50 µL of N-methyl- trimethylsilyltrifluoroacetamide (MSTFA) (GL Sciences, Tokyo, Japan) at 37 °C for 30 min. The GC-MS analysis was performed using the same conditions described in our previous study on Drosophila embryos32.
In the LC-MS analysis, samples were concentrated into 50 µL by centrifugal concentration and analyzed within 24 h of extraction. The analysis was performed using the Shimadzu Nexera UHPLC system coupled with LCMS 8030 Plus (Shimadzu, Kyoto, Japan) using the conditions described in our previous study33.
Data analysis for metabolic profiling
A principle component analysis (PCA), Partial Least Square projection to the latent structure discriminant analysis (PLS-DA), and orthogonal projections to latent structures discriminant analysis (OPLS-DA) were performed by utilizing SIMCA-P+ version 13 (Umetrics, Umea, Sweden). Unit variance was used as the scaling method and no transformation was performed. The good quality of the OPLS-DA model was designated by goodness-of-fit (R 2) and goodness-of-prediction (Q 2) values greater than 0.9.
The PLS-DA model was considered to be valid after permutations when the R 2 Y-intercepts and Q 2-intercepts did not exceed 0.3–0.4 and 0.05, respectively34.
The OPLS-DA model was then validated using CV-ANOVA (Analysis of variance testing of cross-validated residuals) and two groups of samples were significantly different when p(CV-ANOVA) < 0.05. Important metabolites from this model were defined based on the combination of VIP (Variable Importance in the Projection) score and p(corr) (loading scaled as a correlation coefficient between the model and original data). The criteria for the cut-off point were VIP > 1.0 and |p(corr)| > 0.535.
An independent two-way ANOVA test was used to assess whether the candidates obtained from OPLS-DA were significantly different. The two-way ANOVA test was performed using the “Time series Analysis” function of Metabolyst 3.036, a web-based metabolomic data processing tool (available for free at http://www.metaboanalyst.ca).
A Hierarchical Cluster Analysis (HCA) was conducted using Multiexperiment View Version 4.937 (Dana-Farber Cancer Institute, Boston, MA, USA, available for free at the website http://www.tm4.org/mev.html). In this study, similarities between two objects were obtained without losing generality based on the Euclidean distance.
Measurement of glycogen and triacylglycerol (TAG)
Glycogen and TAG levels were measured as described previously38–41. In the glycogen assay, 5 flies were homogenized in 200 μL of a solution containing 2% sodium sulfate and 800 μL of a chloroform/methanol (1:1) mixture was added to the homogenate. After centrifugation at 6,000 x g for 3 min, the pellet was dissolved in 1 mL of anthrone reagent (1.4 g of anthrone in 1 L of 72% H2SO4). The solution was heated at 100 °C for 17 min and analyzed in a spectrometer at 625 nm. In the TAG assay, 10 adult flies were homogenized in 200 μL of a solution containing 0.1% Tween 20 and the homogenate was heated at 70 °C for 5 min. After centrifugation at 15,000 x g for 5 min, 200 μl of TAG reagent (Sigma) was added to 50 μL of the supernatant and incubated at 37 °C for 30 min. The incubated solution was analyzed in a spectrophotometer at 540 nm. The remaining homogenate was used for protein quantification by the Bradford assay with bovine serum albumin as a standard. The amount of TAG was normalized to the protein amount.
Immunostaining analysis
Three to five adult flies were dissected in PBS and fixed for 20 min in 4% paraformaldehyde at 25 °C. After permeabilization with 0.3% Triton X-100 in PBS, samples were incubated with primary antibodies specific for Atg8a (1:200, Novus Biologicals), dG9a (1:500, produced previously40), or H3K9me2 (1/400, previously produced and used for Drosophila cells22, 42) at 4 °C for 16 h. After washing with 0.3% Triton X-100 in PBS, samples were incubated with the secondary antibody, Alexa Fluor 594 anti-mouse IgG (1:400, Molecular Probes) at 25 °C for 2 h. Samples were mounted with VECTASHIELD mounting media with DAPI (Vector Laboratories, Inc.). Intensity was only measured from nuclei in interphase and with a z-axis clearly present in their centers. These nuclei were selected with DAPI signals. We measured the intensities of signals from 10 nuclei derived from at least 3 adult flies. Preparations were examined using confocal microscopy (Olympus FV10i). The images obtained were analyzed with MetaMorph (Molecular Devices).
Flip-out clonal analysis
We generated mosaic gene expression in larval fat body with flip-out clone generation system43. hsFLP; Act5C > DC2 > GAL4, UAS-GFP flies were crossed with Atg8a RNAi strain Atg8a HMS01328. 0–36 h embryos were collected and heat-shocked for 30 min at 37 °C at 36 h after egg laying. Third instar larva were put into a dish containing a piece of paper and 1 mL PBS for 6 h and immunostained as described above after dissection.
Western immunoblot analysis
Fifteen adult flies were homogenized in 100 μL of 2x SDS sample buffer (2% SDS, 10% glycerol, 0.002% BPB, and 0.063 M Tris-HCl), heated at 95 °C for 2 min, and centrifuged at 15,000 x g at 4 °C for 10 min. Five microliters of supernatants was fractionated by 15% SDS-PAGE and transferred to PVDF membranes (Biorad). The membranes were blocked with 0.3% dry milk in Tris-buffered saline containing Tween 20 (TBS-T) (137 mM NaCl, 2.7 mM KCl, 25 mM Tris-HCl, and 0.1% Tween 20, pH 7.6) and incubated with an anti-Atg8a antibody (1:1000, Cell Signaling) or anti-α-tubulin antibody (1:10000, DSHB) at 4 °C for 16 h. Membranes were washed with TBS-T and incubated with HRP-conjugated anti-rabbit or mouse IgGs (Thermo Scientific) for 60 min. After washing membranes with TBS-T, visualization was performed with ECL-select (GE Healthcare). Signal intensity measurements were performed with a CS analyzer (ATTO).
Reverse transcription-quantitative PCR (RT-qPCR)
RNAs were isolated using Trizol® reagent (Invitrogen) from adult whole bodies. cDNA was synthesized using the PrimeScript RT reagent kit (TaKaRa) according to the manufacturer’s instructions. qPCR was performed with SYBR® Premix Ex TaqTM II (TaKaRa) using CFX96 touchTM (Biorad), and data were analyzed with the ΔΔCt method. β-tubulin was used as an internal control. The primer sequences of the genes examined are listed below.
dG9a forward 5′-TCAGATGGCCTATCTCCTTC-3′ (used in Fig. S1B)
dG9a reverse 5′-CAGTCCGCAGTTCATAATCC-3′ (used in Fig. S1B)
dG9a forward 5′-ACCGATGACAGCTACTACTTTG-3′ (used in Fig. 2F)
Figure 2.
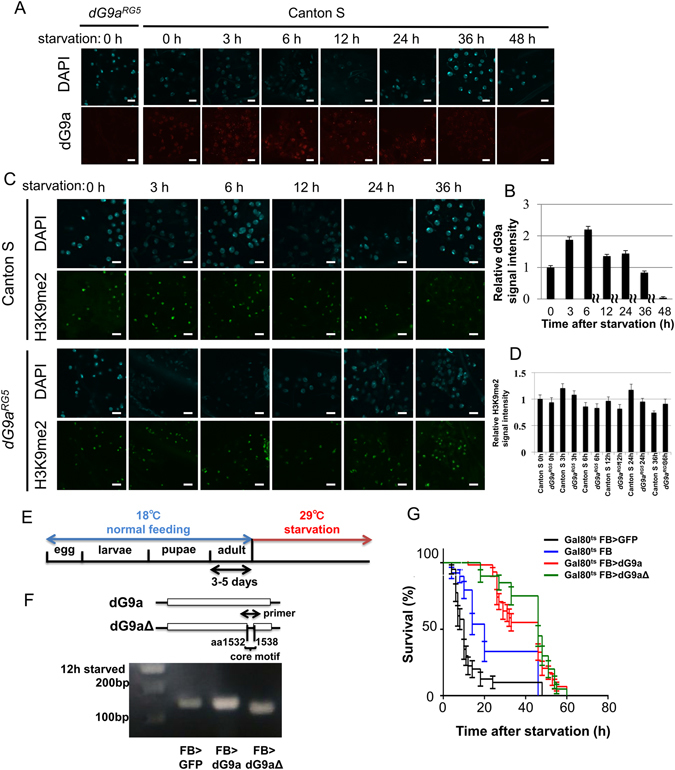
dG9a function in the fat body is critical for adult fly viability under starvation stress. (A) Transient induction of dG9a in the adult fat body under starvation conditions. Fat bodies of the wild-type and dG9aRG5 mutant were immunostained with an anti-dG9a antibody (red) and stained with DAPI (cyan). The dG9aRG5 mutant was used as a negative control. (B) Quantification of dG9a signals in nuclei of the wild-type fat body. Values were adjusted by subtracting background fluorescence n = 10. (C) Quantification of H3K9me2 signals in nuclei of the wild-type and dG9aRG5 mutant fat bodies. Values were adjusted by subtracting background fluorescence (n = 10). (D) H3K9me2 signal in the adult fat body under starvation conditions. Fat bodies of the wild-type and dG9aRG5 mutant were immunostained with an anti-H3K9me2 antibody (green) and stained with DAPI (cyan). (E) The experimental design of dG9a overexpression under starvation conditions. (F) Confirmation of dG9a and dG9aΔ1532–1538 overexpression by semi-quantitative RT-PCR after 12 h of starvation. The primer included the core motif of the SET domain of dG9a. dG9a was more strongly expressed in FB > dG9a (FB-GAL4/GAL80ts.αTub84B; UAS-dG9a/+) than in FB > GFP (a control: FB-GAL4/GAL80ts.αTub84B; UAS-GFP/+). The amplicon size is 142 bp in FB > dG9a and FB > GFP. The amplicon size of FB > dG9aΔ(FB-GAL4/GAL80ts.αTub84B; UAS-dG9aΔ1532–1538/+) is 21 bp shorter because of the lack of the core motif of the SET domain. The full-length gel image is shown in Fig. S6A. (G) The results of a viability assay under starvation conditions using males of the control (GAL80ts FB > GFP: FB-GAL4/+; GAL80ts.αTub84B/UAS-GFP) (n = 40 from 2 independent experiments), another control (GAL80ts FB: FB-GAL4/+; GAL80ts.αTub84B/+) (n = 39 from 2 independent experiments), a dG9a overexpression line (GAL80ts FB > dG9a: FB-GAL4/+; GAL80ts.αTub84B/UAS-dG9a) (n = 58 from 3 independent experiments), and dG9aΔ1532–1538 overexpression line (GAL80ts FB > dG9aΔ: FB-GAL4/+; GAL80ts.αTub84B/UAS-dG9aΔ1532–1538) (n = 40 from 2 independent experiments). Significant differences were observed between GAL80ts FB > GFP and GAL80ts FB > dG9a (P < 0.0001) and between GAL80ts FB > GFP and GAL80ts FB > dG9aΔ (P < 0.0001). Significant differences were also noted between GAL80ts FB and GAL80ts FB > dG9a (P < 0.0001) and between GAL80ts FB and GAL80ts FB > dG9aΔ (P < 0.0001). No significant differences were found between GAL80ts FB > dG9a and GAL80ts FB > dG9aΔ (P = 0.23). (B,C,G) Error bars represent SE.
dG9a reverse 5′-TGTAGTCCTGATGCTCGTAGA-3′ (used in Fig. 2F)
β-tubulin forward 5′-ATACGGTGACCTGAACCATC-3′
β-tubulin reverse 5′-TACTCGGACACCAGATCG-3′
CG6262 forward 5′-GGATCGGGTACACAGCTATTC-3′
CG6262 reverse 5′-CGGGAACGAGAAGGTGAAA-3′
G6P forward 5′-CTGGGAAGTTACCTGGGAATTAG-3′
G6P reverse 5′-AAACAGCTGAGACCGCATAG-3′
HEXA forward 5′-GGATCGGGTACACAGCTATTC-3′
HEXA reverse 5′-CGGGAACGAGAAGGTGAAA-3′
Pgi forward 5′-TCGAGAAGAATGCTCCTGTTATC-3′
Pgi reverse 5′-GCAAGTACTGATCGTAGGGAAG-3′
fbp forward 5′-GCCGGAGAAGGGAAAGATATAC-3′
fbp reverse 5′-TCTTGGCCGCAATGTAGTT-3′
Gapdh1 forward 5′-ATGTCTCCGTTGTGGATCTTAC-3′
Gapdh1 reverse 5′-CCTCGACCTTAGCCTTGATTT-3′
Pgk forward 5′-GCTGAACAAGGAGCTGAAGTA-3′
Pgk reverse 5′-TCTCAATCAGCTGGATCTTGTC-3′
Atg8a forward 5′-TACCAGGAACATCACGAGGA-3′
Atg8a reverse 5′-CGACCGGAGCAAAGTTAGTTA-3′
Results
dG9aRG5 null mutant flies are sensitive to starvation
Previous studies using the dG9a null mutant strain (dG9a RG5) showed that dG9a was not essential for Drosophila viability or fertility21; however, dG9a has been shown to regulate the expression of a wide variety of genes in vivo. In order to investigate epigenetic regulation by dG9a in response to various environmental changes, the adult wild-type (Canton S) and dG9a RG5 null mutant strain were examined under several challenging environmental conditions including heat exposure, oxidative stress, and starvation. We found that dG9a RG5 mutant flies were exclusively sensitive to fasting conditions, but not heat or oxidative stress (Fig. 1A–D). The median survival times of dG9a RG5 mutant males (48 h) and females (78 h) were 47% and 32% shorter, respectively, than those of wild-type males (90 h) and females (114 h) under starvation stress (Fig. 1A,B). However, the viability of the larval dG9a RG5 mutant under starvation stress was not significantly less than that of the wild-type (Fig. S1A). Under heat shock or oxidative stress conditions, the viability of the dG9a RG5 mutant was not significantly different from that of the wild-type (Fig. 1C,D). These results indicate that dG9a plays an important role in the survival of adult flies during starvation stress conditions.
Figure 1.
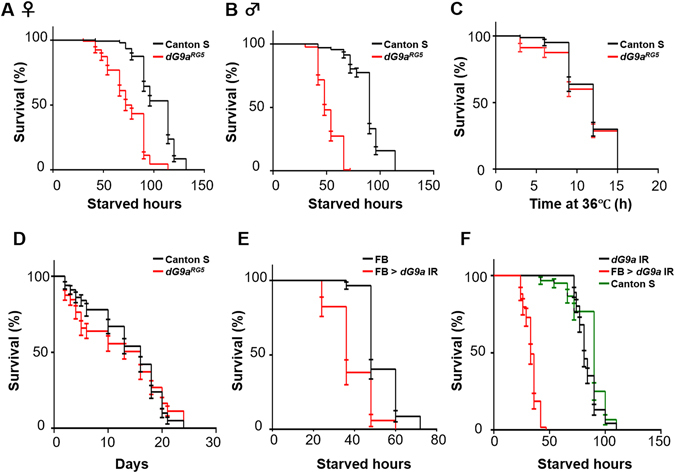
dG9a is critical for survival under starvation stress. (A) The results of a viability assay under starvation conditions using females of the wild-type (Canton S) (n = 153 from 8 independent experiments) and dG9a null mutant (dG9aRG5) (n = 135 from 7 independent experiments) P < 0.0001 (B). The results of a viability assay under starvation conditions using males of the wild-type (n = 131 from 7 independent experiments) and dG9a null mutant (dG9aRG5) (n = 138 from 7 independent experiments) P < 0.0001. (C) The results of a viability assay under heat stress using males of the wild-type (n = 80 from 4 independent experiments) and dG9a null mutant (dG9aRG5) (n = 80 from 4 independent experiments) P = 0.41. (D) The results of a viability assay under oxidative stress using males of the wild-type (n = 100 from 5 independent experiments) and dG9a null mutant (dG9aRG5) (n = 97 from 5 independent experiments) P = 0.80. (E) The results of a viability assay under starvation conditions using males of the FB (+; FB-GAL4; +) (n = 57 from 3 independent experiments) and FB > dG9a IR (w; UAS-dG9a IR/FB-GAL4; +) (n = 34 from 2 independent experiments) P < 0.0001. (F) The results of a viability assay under starvation conditions using males of the FB > dG9a IR (w; UAS-dG9a IR/FB-GAL4; +) (n = 59 from 3 independent experiments), males of the dG9a IR (w; UAS-dG9a IR/+; +) (n = 92 from 5 independent experiments) and males of Canton S (n = 60 from 3 independent experiments). dG9a IR and Canton S: P > 0.05. (A–F) Error bars represent standard errors (SE).
The fat body functions as a major store in response to nutrition demand in insects44. In order to clarify whether the function of dG9a in the fat body is critical for fly viability under starvation stress, we performed a viability assay utilizing fat body-specific dG9a knockdown flies generated with the GAL4-UAS targeted expression system45. The efficient knockdown of dG9a was confirmed by RT-qPCR (Fig. S1B). We also confirmed that the fat body-specific driver (FB-GAL4) properly induced the expression of the target protein in the adult fat body by immunostaining with the anti-GFP antibody utilizing the strain carrying the FB-GAL4 and UAS-GFP constructs (Fig. S1C). The median survival time of fat body-specific dG9a knockdown flies (FB > dG9a IR in Fig. 1E, median survival time: 36 h) was 25% shorter than that of flies that have only FB-GAL4 driver (FB in Fig. 1E, median survival time: 48 h). The median survival time of FB-GAL4/+ flies (48 h) is much shorter than that of Canton S (90 h). The generation of GAL4 protein induces rapid energy consumption, which may result in the decrease of starvation stress tolerance in FB-GAL4/+ flies. Furthermore, FB-GAL4 transgene is inserted in the genomic region of Pyruvate carboxylase (PCB) gene. The insertion may result in the decrease of starvation stress tolerance in FB-GAL4/+ flies, since PCB is required for gluconeogenesis and lipogenesis. We also performed starvation assay utilizing the fat body-specific dG9a knockdown flies (FB > dG9a IR in Fig. 1F), flies that have only UAS-dG9a IR transgene (dG9a IR in Fig. 1F) and wild type flies. The median survival time of dG9a-IR flies was not significantly different from that of Canton S (Fig. 1F), therefore we concluded that the mutations of dG9a-IR flies do not affect the survival time. These results indicate that the function of dG9a in the fat body is critical for fly viability under starvation stress. These results also confirmed that the decrease observed in the viability of the dG9a RG5 mutant under starvation stress, shown in Fig. 1A,B, was not due to a possible background mutation in the dG9a RG5 mutant. We further confirmed that the trans-heterozygous combination of dG9a RG5 and dG9a del34 alleles (dG9a RG5/dG9a del34) shows severe reduction of starvation tolerance compared to wild type under fasting conditions (Fig. S1D). The data support the conclusion that the reduction of starvation tolerance of dG9a RG5 is not due to the background mutations of dG9a RG5 flies. However, the rescue of the dG9a RG5 mutant by a genomic DNA fragment containing the whole dG9a gene may be necessary to further confirm this conclusion.
In order to reveal the localization of dG9a in the fat bodies of starved adult flies, we immunostained fat bodies with the anti-dG9a antibody (Fig. 2A). dG9a signals were detected in nuclei and signal intensity increased up to 6 h after starvation (Fig. 2B). These results indicate that dG9a localizes in the nuclei of fat body cells and may play a role in gene expression under starvation stress. However, the overall intensity of H3K9me2 in the nuclei of fat body cells under starvation was not significantly affected by the loss of dG9a (Fig. 2C,D). These results suggest that dG9a regulates gene expression in fat body nuclei in a H3K9 methyltransferase activity-independent manner. However, we cannot exclude the possibility that the absence of a significant difference in H3K9me2 levels was due to the insufficient detection thresholds of immunostaining analyses. More sensitive analyses, such as a chromatin immunoprecipitation sequencing analysis, will be required in order to investigate this possibility.
In an attempt to clarify whether the overexpression of dG9a extends survival times under starved conditions, we performed a viability assay utilizing a temporal control system of dG9a overexpression by temperature-sensitive GAL8031. dG9a or GFP, as a control, was only overexpressed after the initiation of starvation (Fig. 2E). In order to examine whether the methyltransferase activity of dG9a is critical, we also overexpressed dG9aΔ1532–1538 lacking the catalytic core motif of the SET domain, which is necessary for the histone methyltransferase activity of dG9a28. The overexpression of dG9a and dG9a Δ1532–1538 was confirmed by semi-quantitative RT-PCR (Fig. 2F). The overexpression of dG9a or dG9aΔ1532–1538 extended survival times under starvation stress over those of the control strains (GAL80ts FB > GFP and GAL80ts FB strains in Fig. 2G) (P < 0.001), and no significant difference was observed in the viabilities of the dG9a and dG9a Δ1532–1538 overexpression lines (P = 0.23) (Fig. 2G). The shorter survival times of FB and FB > GFP strains (Fig. 2G) than that of the wild-type Canton S (Fig. 1) may have been due to the higher temperature used for the GAL80/GAL4 system since the temperature directly affects the central metabolism of Drosophila 46. Another possibility could be the production of additional proteins such as GAL80, GAL4, and GFP. We also performed same starvation assays utilizing flies that carry only UAS construct. No significant difference in survival time was observed among these flies (Fig. S1E), therefore we concluded that the mutations of these flies did not affect the survival time. In any event, these results suggest that the catalytic activity of dG9a is not required for the acquisition of starvation stress resistance by dG9a.
Differences between wild-type and dG9aRG5 mutant flies under starvation could be explained by the composition of the metabolome
In order to obtain a general perspective on changes caused by the loss of dG9a, we performed metabolic profiling of fasted wild-type and dG9a RG5 mutant by employing a set of targeted and non-targeted analytical methods using IP-LC-MS/MS and GC-Q/MS, respectively. Since male flies of the dG9a RG5 mutant were more sensitive to starvation stress (Fig. 1A,B), only unmated male flies were used and samples were collected after 0, 12, 24, and 36 h of fasting. With our platforms, 83 low-molecular-weight hydrophilic metabolites belonging to central metabolic pathways were detected (Fig. 2A).
HCA was constructed to provide a global view of the metabolic state of fasted control and dG9a null mutant flies (Fig. 3A). Due to the horizontal axis of the HCA results, it was clear that the two strains shared similar metabolic profiles before starvation (at 0 h); therefore, they were grouped into the same grey cluster. However, during starvation, the expression patterns of metabolites varied between the two strains, resulting in the separation of Canton S and dG9a RG5 into two different clusters, blue and orange, respectively. A similar discrimination was again observed in the supervised discriminant analysis PLS-DA (Fig. 3B). In the PLS-DA model with the eight different groups (wild-type and dG9a RG5 mutant at four different time points), we found that samples were mainly discriminated into two groups based on genotypes. Nevertheless, within each group, samples at 0 h, representing samples not exposed to starvation, were completely separated from those after 12, 24 and 36 h of fasting. PLS-DA model was considered valid after permutation since the R 2 Y-intercepts and Q 2-interceps did not exceed 0.3–0.4 and 0.05, respectively (Fig. 3C).
Figure 3.
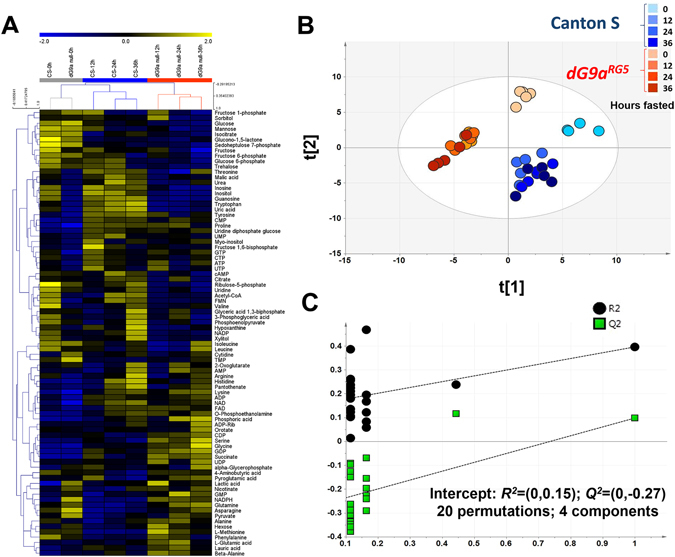
Fasted dG9aRG5 mutant flies show a distinct metabolic profile. (A) HCA data showing changes in the cellular metabolites of wild-type and dG9aRG5 mutant flies under starvation; the color scale is plotted on the top of the figure. Before starvation, flies with both genotypes shared similar profiles and were grouped into one cluster (grey). During starvation, flies with each genotype showed different profiles and were discriminated hierarchically into two clusters, wild-type (blue) and dG9aRG5 mutant (red). A similar discrimination based on genotypes was observed on the score plot of the supervised analysis PLS-DA (B). In each genotype, samples collected before fasting were clearly separated from samples collected during fasting. (C) Statistical validation by the permutation test with 20 permutations of PLS-DA (R2Y-intercepts = (0,0.15); Q2-interceps = (0, −0.27)).
Collectively, these results indicated that prior to starvation, both strains shared similar metabolic profiles. However, the loss of dG9a caused the altered cellular metabolic profile in Drosophila during starvation.
The loss of dG9a causes major changes in amino acid metabolism
In an attempt to identify the metabolites affected by the loss of function of dG9a, OPLS-DA was generated to maximize differences between wild-type and dG9a RG5 mutant flies. This model was constructed with one predictive and two orthogonal components, which showed a clear separation between the groups along the predictive components (Fig. 4A). In this model, important metabolites related to the function of dG9a under fasting conditions were proposed (Fig. 4B; Table S1), and were found to be related to amino acid metabolism.
Figure 4.
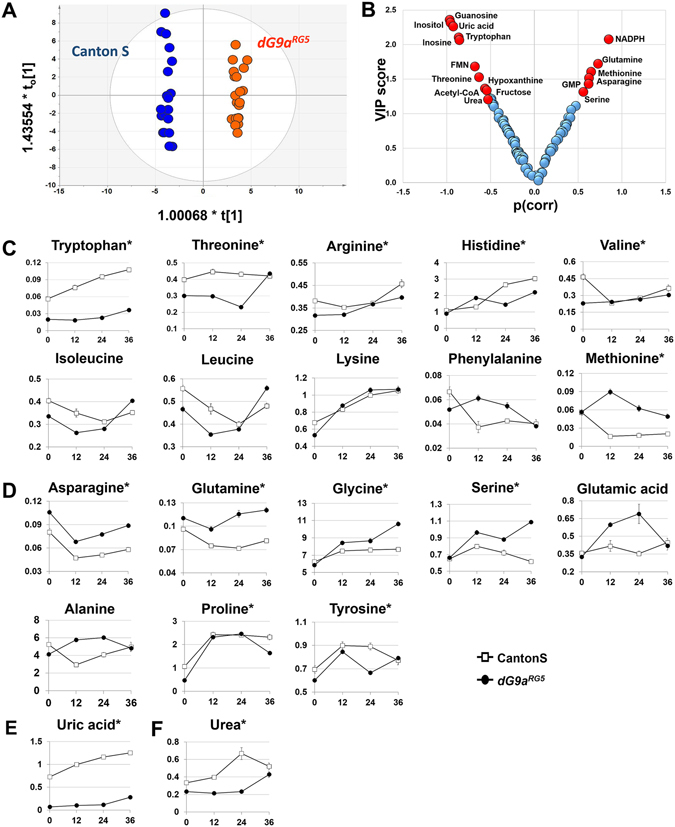
Amino acid metabolism. A general view of the metabolites detected belonging to amino acids including: (A) The OPLS-DA score plot showed a clear separation between two genotypes with p(CV-ANOVA) = 1.99925E-23. (B) A plot formed by VIP scores and p(corr) revealed using OPLS-DA. The criteria VIP > 1.0 and |p(corr)|>0.5 were used to select potential metabolites (red) showing marked changes due to the loss of dG9a. (C) Essential amino acids (D) Non-essential amino acids (E) Uric acid (F) Urea. 4–5 replications for each time point. *The metabolites were significantly different in “genotype” between the wild-type and dG9aRG5 mutant in a two-way ANOVA with P < 0.05.
Similar to mammals, 20 amino acids found in the proteins of Drosophila may also be classified into essential and non-essential amino acids47. We herein demonstrated that the levels of many essential amino acids in dG9a-depleted flies were lower than in those in the wild-type during 36 h of fasting (Fig. 4C). Significant differences were observed in the expression levels of tryptophan, threonine, arginine, histidine, and valine between the fasted wild-type and dG9a RG5 mutant with the p-value for the genotype being less than 0.05 in the two-way ANOVA. Since there was no nutrient supplement during fasting, the only source of free essential amino acids was from protein degradation. The levels of non-essential amino acids do not reflect the actual rate of proteolysis because they may be synthesized or derived from essential amino acids. Moreover, the expression levels of uric acid and urea, the waste products of amino acid and nitrogen metabolism, increased overtime and were significantly higher in the wild-type than in the dG9a RG5 mutant (Fig. 4E,F). Thus, these results suggested that in contrast to the wild-type strain, dG9a-depleted flies were unable to maintain an appropriate protein degradation rate, which is an essential process for recycling amino acids for translation or energy generation under fasting. On the other hand, asparagine, glutamine, and methionine levels were significantly higher in dG9a-depleted flies (Fig. 4C,D). These amino acids were reported to be inhibitors of the autophagic pathway in studies using isolated rat hepatocytes48. A high level of L-glutamine was previously reported to be important for the activation of the target of rapamycin (TOR) pathway, which suppresses autophagy49.
Collectively, these results suggest that the loss of dG9a affects the ability to recycle amino acids via protein degradation in starvation-induced autophagic pathways, which may be the reason why the viability of the mutants was lower than that of the wild-type strain under starvation stress.
dG9a plays an important role in maintaining energy reservoirs during starvation
The responses of cells to nutrient deprivation via autophagy involve a non-selective process of bulk cytoplasmic degradation50, 51. Hence, if our hypothesis is correct, these changes will also appear in metabolites related to lipid and carbohydrate sources. In Drosophila, TAG and glycogen are the main primary forms of energy sources stored in the fat body, analogous to the mammalian liver and white adipose tissues. In response to the energy needs of the body, trehalose will be synthesized in the fat body, excreted into the hemolymph, and utilized to produce glucose52. As predicted, when we measured TAG and glycogen levels, we found that they were lower in dG9a-depleted than in wild-type flies after 12 h of fasting (Fig. 5A,B). Metabolic profiling results revealed that trehalose levels were also significantly lower in the dG9a RG5 mutant with a p-value = 0.01 for the genotype in the two-way ANOVA throughout starvation stress (0–36 h) (Fig. 5C). If these variations were caused by the mechanisms used to consume the reservoirs, the level of ATP may differ between the two strains. However, no significant difference was detected in the levels of glucose, the initial material for glycolysis, the major energy currency molecule of cells produced from glycolysis and the TCA cycle (Fig. 5D). When the levels of metabolites together with the mRNA levels of enzymes related to glycolysis were measured, no significant difference was observed between wild-type and dG9a-depleted flies (Fig. S3). Collectively, these results suggested that the loss of dG9a did not affect the ability to use resources to generate energy via glycolysis, whereas it may have affected the capability to conserve energy sources, which quickly drains the body’s energy and leads to weak tolerance under starvation stress.
Figure 5.
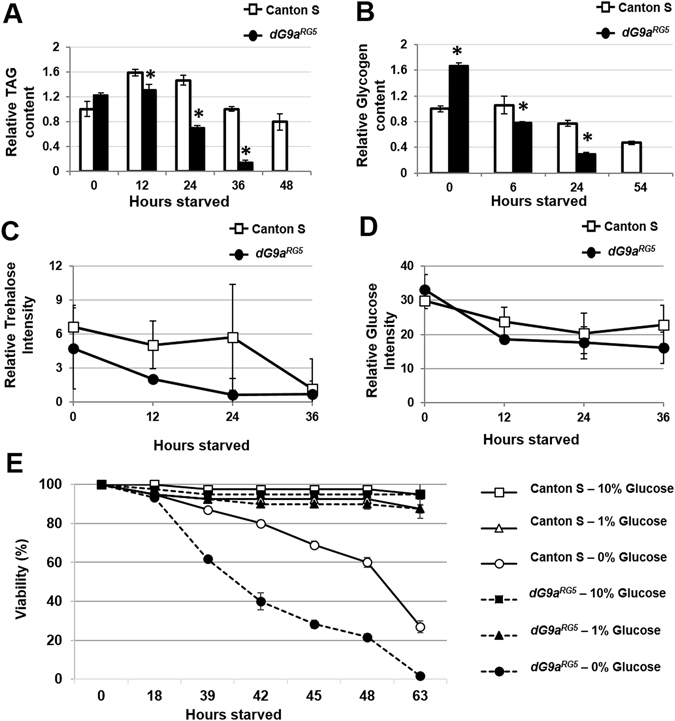
The loss of dG9a affects energy homeostasis in Drosophila during fasting. (A) Relative levels of TAG in the wild-type and dG9aRG5 mutant under starvation stress n = 3. *P < 0.05. (B) Relative levels of glycogen in the wild-type and dG9aRG5 mutant under starvation stress n = 5. *P < 0.05. The levels of trehalose (C) and glucose (D) from the GC-MS analysis. Only the level of trehalose showed a significantly different “genotype” between the wild-type and dG9aRG5 mutant in a two-way ANOVA with P = 9.17E-03. (E) The results of a viability assay with glucose supply (n = 40). When dG9aRG5 mutant flies were fed 1% and 10% glucose in PBS, they recovered their sensitivity and had similar viability to the wild-type.
We then investigated whether glucose supplements have the ability to rescue the viability of dG9a-depleted flies. After hatching, adult flies of the wild-type and dG9a RG5 mutant were maintained on standard food for 48 h and then shifted to either PBS or 1% and 10% glucose in PBS. As we predicted, the addition of glucose completely rescued the sensitivity of dG9a-depleted flies to starvation stress; they showed similar viability to the wild-type (Fig. 5E). These results confirmed that dG9a plays an important role in maintaining not only amino acid catabolism, but also overall energy reservoirs, which may be used to generate glucose for the body during starvation.
dG9a controls starvation-induced autophagy by regulating the expression of Atg8a
Previous studies utilizing a human pancreatic cancer cell line showed that G9a regulates the expression of some autophagy genes in vitro 5. Autophagy occurs in the fat body in Drosophila under starvation stress53. Moreover, our metabolomic results indicated that the loss of dG9a affected the ability to recycle amino acids via protein degradation in starvation-induced autophagic pathways. Therefore, we focused on autophagy activity in dG9a RG5 mutant under starvation stress. We immunostained fat bodies with anti-Atg8a IgG, a widely used marker for detecting autophagy activity. The inactive form of Atg8a (Atg8a-I) may be processed into its active form (Atg8a-II), which accumulates to form a dotted signal at the phagophore, an initial membrane for autophagosome membrane formation, the assembly site54. The antibody used in this study was more sensitive to Atg8a-II55. In the wild-type, the number of dotted signals of Atg8a increased after 6, 12, and 24 h of fasting (Fig. 6Aa–d). These dotted signals decreased in the fat bodies of fat body-specific Atg8a knockdown flies, confirming the specificity of anti-Atg8a IgG (Fig. 6Ad, e).
Figure 6.
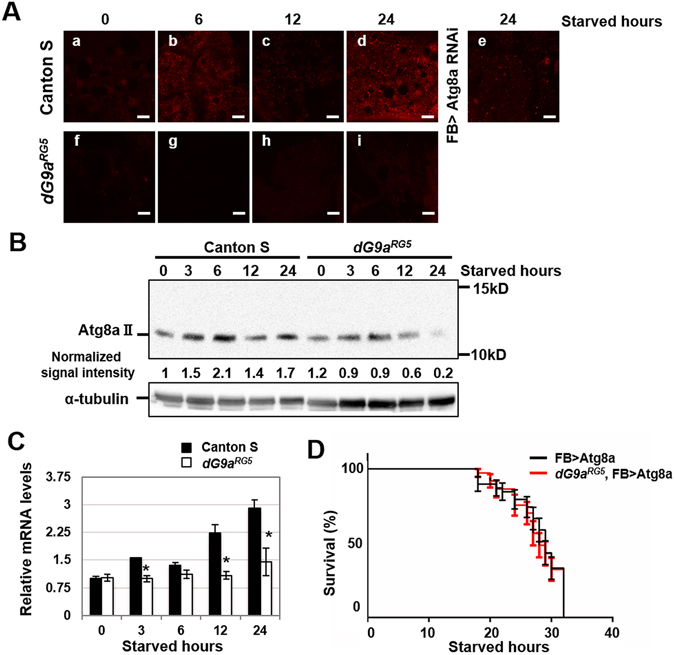
dG9a is responsible for the induction of autophagy under starvation stress. (A) Immunostaining of wild-type and dG9aRG5 mutant fat bodies under starvation conditions with an anti-Atg8a antibody. Strains: (a–d) Canton S (e) w; FB-GAL4/+; Atg8aHMS01328/+ (f–i) dG9aRG5. Starved hours: (a,f) 0 h (b,g) 6 h (c,h) 12 h (d,i) 24 h. Scale bars, 10 μm. (B) A western blot analysis of extracts from the starved wild-type and dG9aRG5 mutant. Blots were probed with anti-Atg8a and anti-α-tubulin antibodies. Signal intensity normalized with that of α-tubulin is shown. The full-length image of the blot is shown in Fig. S6B. (C) Quantification of mRNA levels by a RT-qPCR analysis of Atg8a in the starved wild-type and dG9aRG5 mutant. Results were normalized to α-tubulin and displayed as relative values to that of the 0-h starved wild-type n = 3. *P < 0.05. (D) The results of a viability assay under starvation conditions using the males of dG9aRG5, FB > Atg8a (dG9aRG5; FB-GAL4/Atg8aScer/UAS.P/T.T:Avic/GFP-EGFP,T:Disc/RFP-mCherry; +) (n = 39 from 2 independent experiments) and FB > Atg8a (+; FB-GAL4/Atg8aScer/UAS.P/T.T:Avic/GFP-EGFP,T:Disc/RFP-mCherry; +) (n = 38 from 2 independent experiments) strains P = 0.79. (C,D) Error bars represent SE.
We confirmed the specificity of the anti-Atg8a antibody by generating somatic clones marked by GFP that express Atg8a dsRNA in larval fat body. The number of dot signals was clearly decreased in the RNAi clone area (Fig. S4A), which support the Atg8a staining specificity. In the dG9a RG5 mutant, the number of dotted signals of Atg8a was less than that in the wild-type under starvation conditions (Fig. 6Aa–d,f–i). A Western blot analysis is widely used to evaluate autophagy activity. Atg8a-II(12 kDa) may be easily distinguished from Atg8a-I (15 kDa) by SDS-PAGE54. The western blot analysis further confirmed that the protein level of Atg8a-II was lower in the dG9a RG5 mutant than in the wild-type (Fig. 6B). The mRNA level of Atg8a was significantly lower after 3, 12, and 24 h of fasting in the dG9a RG5 mutant than in the wild-type (Fig. 6C). The Atg8a-II band was not detected in the Atg8a mutant (Atg8a d4) (Fig. S4B), which validate the antibody specificity in the Western blot analysis.
Atg8a d4 was also used as a control for the phenotype that lack of Atg8a. As predicted, similar levels of TAG, total protein, trehalose and glucose were observed between dG9a RG5 and Atg8a d4 at 24 h after starvation (Fig. S4C–F). Of note, the total protein levels after 24 h starving of both dG9a RG5 and Atg8a d4 were not significantly different, which validated our hypothesis that the lack of dG9a affected the ability to recycle amino acids via protein degradation. These results suggested that a decrease in the expression level of Atg8a causes a shortage in Atg8a for the progression of autophagy under starvation conditions in the dG9a RG5 mutant. The specific overexpression of Atg8a in the fat body rescued the defect in viability in dG9a RG5 mutant under starvation conditions (Fig. 6D). Furthermore, the level of energy reservoirs were also recovered and similar to control strain (Fig. S5). Therefore, dG9a is responsible for the increase required in the expression of Atg8a for sufficient autophagy activity under starvation conditions.
Discussion
Previous studies revealed that G9a is important for early embryogenesis and essential for viability in mice12–15. G9a is also highly conserved among various metazoans including Drosophila, frogs (Xenopus tropicalis), fish (Danio rerio, Tetraodon nigroviridis, and Takifugu rubripes), and mammals. In Drosophila, although G9a is not essential for viability, the results of the present study suggest that G9a is conserved from the fly to mammals because of its importance in starvation stress tolerance, to which organisms are often exposed in the wild. This is also the first indication that epigenetic regulator-like G9a plays an essential role in the acquisition of starvation tolerance.
In order to clarify the underlying mechanisms by which the dG9a null mutant is more susceptible to starvation stress, “bottom up” approaches have been used. Non-targeted GC-MS-based and targeted LC-MS/MS-based metabolic profiling was performed to investigate changes in the metabolome due to the loss of dG9a. The results obtained from metabolic profiles showed that dG9a played important roles in maintaining energy homeostasis, the key factor for nutrient stress tolerance. dG9a modulated energy reservoirs including amino acid, trehalose, glycogen, and TAG levels during starvation via the autophagic process. One of the unique features of the adult dG9a RG5 mutant is its higher content of glycogen under non-starved normal conditions than that of the wild-type (Fig. 6B). A previous study reported that the deletion of G9a in mouse adipose tissues promotes adipogenesis and increases body weight56. These findings and the present results suggest that dG9a is also responsible for the suppression of adipogenesis, similar to mammalian G9a. Further analyses are needed in order to clarify this point.
The results of the present study also indicated that dG9a controlled starvation-induced autophagy by activating the expression of Atg8a; however, dG9a generally represses gene expression by dimethylating H3K9. Previous studies reported that histone and non-histone protein methylation by G9a either activated or inhibited gene expression57–62. We also found that the catalytic activity of dG9a was not required for the acquisition of starvation stress resistance by dG9a. This is consistent with the results of immunostaining showing that H3K9me2 levels in the nuclei of fat body cells under starvation were not significantly affected by the loss of dG9a. G9a has also been reported to activate gene expression as a molecular scaffold for the assembly of transcriptional co-activators, and the catalytic domain of G9a is not required for this function63. Further studies are needed in order to clarify the mechanisms by which dG9a regulates the expression of Atg8a.
Similar Atg8a mRNA levels were observed after 6 h of fasting between wild-type and dG9a RG5 mutant flies (Fig. 6C); however, Atg8a immunostaining signals was weaker in the dG9a RG5 mutant than in the wild-type (Fig. 6A). Therefore, the loss of dG9a may repress the expression of genes that control Atg8a protein stability. Further studies are needed in order to elucidate the underlying mechanisms. During the development of Drosophila, metamorphosis is also a process that flies use to tolerate starvation stress. Even though our study demonstrated that dG9a is important for starvation stress tolerance, the viability of the dG9a RG5 mutant was not significantly less than that of the wild-type during the pupal stage21, 22. Together with our results showing that the viability of the dG9a RG5 mutant at the larval stage was not affected by fasting conditions, the function of dG9a for starvation stress appears to be specific to the adult stage. Since programmed autophagy during the 3rd instar larval and pupal stages is well-known to be regulated by ecdysone through the PI3K pathway64, starvation-induced autophagy by dG9a in the adult stage may be operated by other pathways.
G9a is suggested to play a positive role in the promotion of tumorigenesis in various human cancer cells such as prostate65, leukemia60, lung66, breast67, and aggressive ovarian carcinoma68. The inhibition of G9a activity in cancer cells significantly inhibited cell proliferation by triggering cell cycle arrest, inducing apoptosis, or activating autophagic cell death5, 69–72. The novel results obtained in the present study on the role of dG9a to acquire starvation tolerance may also make it possible to explain the positive role of G9a in the promotion of tumorigenesis. Cells inside a tumor mass are exposed to starvation conditions because nutrients are not fully supplied to these cells73, 74. In order to overcome starvation stress, autophagy is induced in these cells73, 74. Therefore, G9a may play a role in the acquisition of starvation tolerance in cells in the tumor mass. In the present study, we found that the loss of dG9a led to the inactivation of starvation-induced autophagy due to a decrease in Atg8a levels. In contrast, a previous study on cancer cells showed that the loss of G9a during starvation activated the transcription of LC3B (the Atg8a ortholog in mammals) and triggered autophagy5. Taken together, these results suggest that the epigenetic gene regulation of G9a depends on cell/tissue types.
Electronic supplementary material
Acknowledgements
This study was partially supported by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) Core-to-Core Program, B. Asia-Africa Science Platforms. JSPS KAKENHI Grant Number 15J0948. We would like to thank the Vienna Drosophila RNAi Center (VDRC, Vienna, Austria), Bloomington Drosophila Stock Center (Bloomington, USA), and Kyoto Stock Center (DGRC) for fly stocks. We thank P. Spierer, C. Seum, R. P. Kühnlein and G. Juhasz for kindly gifting fly strains.
Author Contributions
P.N.T.A., K.S., R.T., H.Y., E.F., and M.Y. conceived, designed, and analyzed the experiments. P.N.T.A., K.S. and R.T. performed the experiments. P.N.T.A., K.S., and M.Y. wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Phan Nguyen Thuy An and Kouhei Shimaji contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07566-1
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eiichiro Fukusaki, Email: fukusaki@bio.eng.osaka-u.ac.jp.
Masamitsu Yamaguchi, Email: myamaguc@kit.ac.jp.
References
- 1.Stankiewicz AM, Swiergiel AH, Lisowski P. Epigenetics of stress adaptations in the brain. Brain Res Bull. 2013;98:76–92. doi: 10.1016/j.brainresbull.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21:214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heijmans BT, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slade JD, Staveley BE. Extended longevity and survivorship during amino-acid starvation in a Drosophila Sir2 mutant heterozygote. Genome. 2016;59:311–318. doi: 10.1139/gen-2015-0213. [DOI] [PubMed] [Google Scholar]
- 5.Artal-Martinez de Narvajas A, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33:3983–3993. doi: 10.1128/MCB.00813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 8.Collins RE, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tachibana M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patnaik D, et al. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J Biol Chem. 2004;279:53248–53258. doi: 10.1074/jbc.M409604200. [DOI] [PubMed] [Google Scholar]
- 11.Kubicek S, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman N, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 14.Epsztejn-Litman S, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zylicz, J. J. et al. Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. Elife4, doi:10.7554/eLife.09571 (2015). [DOI] [PMC free article] [PubMed]
- 16.Xin Z, et al. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J Biol Chem. 2003;278:14996–15000. doi: 10.1074/jbc.M211753200. [DOI] [PubMed] [Google Scholar]
- 17.Ikegami K, et al. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells. 2007;12:1–11. doi: 10.1111/j.1365-2443.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 18.Estève PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009–9016. doi: 10.1158/0008-5472.CAN-06-0101. [DOI] [PubMed] [Google Scholar]
- 20.Kramer JM, et al. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol. 2011;9:e1000569. doi: 10.1371/journal.pbio.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seum C, Bontron S, Reo E, Delattre M, Spierer P. Drosophila G9a is a nonessential gene. Genetics. 2007;177:1955–1957. doi: 10.1534/genetics.107.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimaji K, et al. Genomewide identification of target genes of histone methyltransferase dG9a during Drosophila embryogenesis. Genes Cells. 2015;20:902–914. doi: 10.1111/gtc.12281. [DOI] [PubMed] [Google Scholar]
- 23.Kaelin WG, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jorge TF, et al. Mass spectrometry-based plant metabolomics: Metabolite responses to abiotic stress. Mass Spectrom Rev. 2015 doi: 10.1002/mas.21449. [DOI] [PubMed] [Google Scholar]
- 26.Alreshidi MM, et al. Metabolomic and proteomic responses of Staphylococcus aureus to prolonged cold stress. J Proteomics. 2015;121:44–55. doi: 10.1016/j.jprot.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Putri SP, et al. Current metabolomics: practical applications. J Biosci Bioeng. 2013;115:579–589. doi: 10.1016/j.jbiosc.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Kato Y, Kato M, Tachibana M, Shinkai Y, Yamaguchi M. Characterization of Drosophila G9a in vivo and identification of genetic interactants. Genes Cells. 2008;13:703–722. doi: 10.1111/j.1365-2443.2008.01199.x. [DOI] [PubMed] [Google Scholar]
- 29.Grönke S, et al. Control of Fat Storage by a Drosophila PAT Domain Protein. Curr Biol. 2003;13:603–606. doi: 10.1016/S0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- 30.Pircs K, et al. Advantages and Limitations of Different p62-Based Assays for Estimating Autophagic Activity in Drosophila. PloS One. 2012;7:e44214. doi: 10.1371/journal.pone.0044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suster ML, Seugnet L, Bate M, Sokolowski MB. Refining GAL4-driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis. 2004;39:240–245. doi: 10.1002/gene.20051. [DOI] [PubMed] [Google Scholar]
- 32.An PN, Yamaguchi M, Bamba T, Fukusaki E. Metabolome analysis of Drosophila melanogaster during embryogenesis. PLoS One. 2014;9:e99519. doi: 10.1371/journal.pone.0099519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashim Z, Mukai Y, Bamba T, Fukusaki E. Metabolic profiling of retrograde pathway transcription factors rtg1 and rtg3 knockout yeast. Metabolites. 2014;4:580–598. doi: 10.3390/metabo4030580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eriksson L, et al. Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ Health Perspect. 2003;111:1361–1375. doi: 10.1289/ehp.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheelock Å, Wheelock CE. Trials and tribulations of ‘omics data analysis: assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol Biosyst. 2013;9:2589–2596. doi: 10.1039/c3mb70194h. [DOI] [PubMed] [Google Scholar]
- 36.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Research. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howe, E. et al. In Biomedical Informatics for Cancer Research (eds Michael F. Ochs, John T. Casagrande & Ramana V. Davuluri) Ch. 15, 267–277 (Springer US, 2010).
- 38.Van Handel E. Estimation of glycogen in small amounts of tissue. Anal Biochem. 1965;11:256–265. doi: 10.1016/0003-2697(65)90013-8. [DOI] [PubMed] [Google Scholar]
- 39.Van Handel E. Rapid determination of total lipids in mosquitoes. J Am Mosq Control Assoc. 1985;1:302–304. [PubMed] [Google Scholar]
- 40.Van Handel E, Day JF. Assay of lipids, glycogen and sugars in individual mosquitoes: correlations with wing length in field-collected Aedes vexans. J Am Mosq Control Assoc. 1988;4:549–550. [PubMed] [Google Scholar]
- 41.Kaufmann C, Brown MR. Regulation of carbohydrate metabolism and flight performance by a hypertrehalosaemic hormone in the mosquito Anopheles gambiae. J Insect Physiol. 2008;54:367–377. doi: 10.1016/j.jinsphys.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 43.Sun J, Tower J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol Cell Biol. 1999;19:216–228. doi: 10.1128/MCB.19.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 46.An PNT, Yamaguchi M, Fukusaki E. Metabolic profiling of Drosophila melanogaster metamorphosis: a new insight into the central metabolic pathways. Metabolomics. 2017;13:29. doi: 10.1007/s11306-017-1167-1. [DOI] [Google Scholar]
- 47.Hinton, T., Noyes, D. T. & Ellis, J. Amino Acids and Growth Factors in a Chemi-Cally Defined Medium for Drosophila. Physiological Zoology24, 335–353, doi:10.2307/30152141 (1951).
- 48.Seglen PO, Gordon PB, Poli A. Amino acid inhibition of the autophagic/lysosomal pathway of protein degradation in isolated rat hepatocytes. Biochim Biophys Acta. 1980;630:103–118. doi: 10.1016/0304-4165(80)90141-5. [DOI] [PubMed] [Google Scholar]
- 49.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 51.Huang H, et al. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 2015;34:154–168. doi: 10.15252/embj.201489083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyes-DelaTorre, A., Riesgo-Escovar, J. R. & Peña-Rangel, M. T. Carbohydrate Metabolism in Drosophila: Reliance on the Disaccharide Trehalose. (INTECH Open Access Publisher, 2012).
- 53.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 54.Nagy P, Varga Á, Kovács AL, Takáts S, Juhász G. How and why to study autophagy in Drosophila: it’s more than just a garbage chute. Methods. 2015;75:151–161. doi: 10.1016/j.ymeth.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, et al. Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. EMBO J. 2013;32:45–59. doi: 10.1038/emboj.2012.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chin HG, et al. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res. 2007;35:7313–7323. doi: 10.1093/nar/gkm726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JS, et al. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell. 2010;39:71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JS, et al. Hypoxia-induced methylation of a pontin chromatin remodeling factor. Proc Natl Acad Sci USA. 2011;108:13510–13515. doi: 10.1073/pnas.1106106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang J, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin W, et al. Involvement of MyoD and c-myb in regulation of basal and estrogen-induced transcription activity of the BRCA1 gene. Breast Cancer Res Treat. 2011;125:699–713. doi: 10.1007/s10549-010-0876-1. [DOI] [PubMed] [Google Scholar]
- 62.Rathert P, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bittencourt D, et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci USA. 2012;109:19673–19678. doi: 10.1073/pnas.1211803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tracy K, Baehrecke EH. The role of autophagy in Drosophila metamorphosis. Curr Top Dev Biol. 2013;103:101–125. doi: 10.1016/B978-0-12-385979-2.00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kondo Y, et al. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One. 2008;3:e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen MW, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 67.Dong C, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hua KT, et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol Cancer. 2014;13:189. doi: 10.1186/1476-4598-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho HS, et al. Enhanced expression of EHMT2 is involved in the proliferation of cancer cells through negative regulation of SIAH1. Neoplasia. 2011;13:676–684. doi: 10.1593/neo.11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ren A, Qiu Y, Cui H, Fu G. Inhibition of H3K9 methyltransferase G9a induces autophagy and apoptosis in oral squamous cell carcinoma. Biochem Biophys Res Commun. 2015;459:10–17. doi: 10.1016/j.bbrc.2015.01.068. [DOI] [PubMed] [Google Scholar]
- 72.Casciello F, Windloch K, Gannon F, Lee JS. Functional Role of G9a Histone Methyltransferase in Cancer. Front Immunol. 2015;6:487. doi: 10.3389/fimmu.2015.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eng CH, Abraham RT. The autophagy conundrum in cancer: influence of tumorigenic metabolic reprogramming. Oncogene. 2011;30:4687–4696. doi: 10.1038/onc.2011.220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


