Abstract
Protein ubiquitylation is an important post-translational modification, regulating aspects of virtually every biochemical pathway in eukaryotic cells. Hundreds of enzymes participate in the conjugation and deconjugation of ubiquitin, as well as the recognition, signaling functions, and degradation of ubiquitylated proteins. Regulation of ubiquitylation is most commonly at the level of recognition of substrates by E3 ubiquitin ligases. Characterization of the network of E3–substrate relationships is a major goal and challenge in the field, as this expected to yield fundamental biological insights and opportunities for drug development. There has been remarkable success in identifying substrates for some E3 ligases, in many instances using the standard protein–protein interaction techniques (e.g., two-hybrid screens and co-immunoprecipitations paired with mass spectrometry). However, some E3s have remained refractory to characterization, while others have simply not yet been studied due to the sheer number and diversity of E3s. This review will discuss the range of tools and techniques that can be used for substrate profiling of E3 ligases.
Keywords: E3 ubiquitin ligases, RING E3s, HECT E3s, RBR E3s, Cullin-Ring ligases
Introduction
Ubiquitylation has many essential functions in cell cycle control, DNA damage responses, protein trafficking, protein turnover, the disposal of misfolded and damaged proteins, and many other biochemical pathways. The strict control of the substrate selection process is, therefore, imperative. A hierarchal system of enzymes, culminating in the recognition of substrates by E3 ligases, coordinates the conjugation of ubiquitin to substrates. The first step toward conjugation is ATP-dependent activation of ubiquitin by an E1 enzyme. The E1 first catalyzes the formation of a ubiquitin–adenylate intermediate, and then, the active-site cysteine of the E1 forms a thioester with the C-terminus of ubiquitin, with the release of AMP. Ubiquitin is then transferred from the E1 enzyme to the active-site cysteine of an E2 enzyme in a transthiolation reaction. E3 ligases bind ubiquitin-charged E2s and substrates and promote the transfer of ubiquitin from the E2 to a substrate lysine residue or a lysine of a previously conjugated ubiquitin molecule, forming an isopeptide bond linkage. In unusual cases, ubiquitin can be linked to the free N-terminal amino group of a substrate or of ubiquitin, or even to serine or threonine side chains (forming ester bonds). In humans, there are two E1 enzymes and 32 E2s that are known to facilitate ubiquitin conjugation [1], while Saccharomyces cerevisiae has a single E1 and 11 E2s [2]. There are over 600 E3s encoded in the human genome and at least 51 in S. cerevisiae [2], and these enzymes collectively coordinate the ubiquitylation of thousands of substrates. There are several types of E3s, varying substantially in their mode of ubiquitin transfer to substrates, as described in the following paragraphs. E3s generally target more than one substrate and multiple E3s may target the same substrate.
There are two main mechanistic classes of E3s, RING (Really Interesting New Gene) and HECT (Homologous to the E6-AP Carboxyl Terminus) E3s, as well as a third class, RBR (RING-between-RING) E3s, which have characteristics of both RING and HECT E3s. The vast majority of E3s are RING E3s, with over 600 annotated in humans [3], while there are only 28 human HECT E3s [4] and 14 RBR E3s [5]. RING E3s range from 13 kDa to over 500 kDa and adopt a common structure through coordination of two zinc ions with eight conserved Cys/His residues. The RING domain recruits the ubiquitin–E2 conjugate and a substrate recognition domain binds the substrate, allowing the transfer of ubiquitin to the substrate from the E2. U-box domain E3s (e.g., CHIP) are E3s that form a RING-like structure without the coordination of zinc ions [6]. RING E3s may function as a singular entity (e.g., MDM2) or as part of a modular complex (Fig. 1).
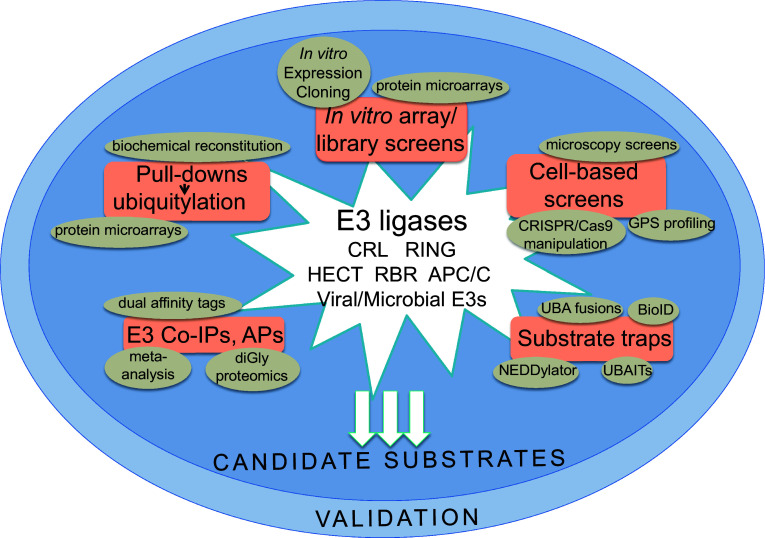
Fig. 1.
Methods to identify substrates of E3 ligase. Classes of E3s are indicated in the center, and techniques and approaches for the identification of substrates are categorized as follows (orange boxes): pull-downs coupled with in vitro ubiquitylation reactions; cell-based screens; co-immunoprecipitations (Co-IPs) or affinity purification (AP) of the E3; and substrate traps. Examples of techniques covered under these subcategories are circled
The largest family of RING E3s, the Cullin-RING ubiquitin Ligases (CRLs), exist as multi-subunit complexes. One of six cullin proteins (CUL1, CUL2, CUL3, CUL4A/CUL4B, CUL5, and CUL7) forms the scaffold for over 200 CRLs. SCF (SKP1-CUL1-F-box protein) and ECS (Elongin C–CUL2/5–SOCS box) complexes are the best-characterized CRLs [7]. For SCF ligases, the cullin, CUL1 (activated by NEDD8) binds the RING protein (RBX1 or RBX2), the RING protein recruits the E2, and the substrate receptor proteins (F-box proteins) recruit the substrate. SKP1 serves as an adaptor that connects the cullin to the F-box protein. An ECS ubiquitin ligase consists of a cullin (CUL2/5), a RING protein (RBX1 or RBX2), and a substrate receptor (e.g., VHL or SOCS box proteins), and adaptor proteins (e.g., Elongin B and Elongin C) that bridge the substrate receptor to the cullin protein [8]. The wide array of substrate receptors, which includes 69 different interchangeable F-box proteins, account for the extensive substrate profiles of CRL E3s. Subdivided by their protein interaction domains, there are three families of F-box proteins, FBXW, FBXL, and FBXO. FBXL proteins contain leucine-rich repeats (LRRs); FBXW proteins have WD repeats; and FBXO proteins may or may not contain an additional motif [9]. Another type of CRL, the Anaphase Promoting Complex/Cyclosome (APC/C), is structurally very distinct from other CRLs and consists of 14 subunits, including multiple substrate adaptors. APC/C plays a critical role in mitosis, triggering the transition from metaphase to anaphase [10].
HECT E3s are structurally and mechanistically distinct from RING E3s in that they directly catalyze substrate ubiquitylation. This involves a second transthiolation reaction, with ubiquitin being transferred from the E2 to the catalytic cysteine of the HECT domain, before being transferred to substrate lysine residues. HECT E3s are relatively large monomeric E3 proteins (>90 kDa, up to ~500 kDa), with the HECT domain always located at the C-terminus. Outside of the HECT domain, these E3s are structurally diverse and the lack of easily identifiable substrate recognition domains for many HECT E3s (e.g., E6AP, HECTD1, and HECTD2) has hindered progress in pairing substrates to enzymes. In some cases, there are obvious substrate recognition domains, such as the WW domains in the Rsp5/NEDD4 subfamily of HECT E3s, and these are in fact the most thoroughly characterized HECT E3s [11]. It should also be noted that while some HECT E3s have clearly defined motifs or domains (e.g., RCC1 repeats in the HERC family of HECT E3s), in the majority of cases, these have not been demonstrated to be direct substrate interaction motifs, even though they may be required for the function of the E3 [12].
RBR E3s contain a RING1 domain that, like a conventional RING E3, binds ubiquitin-charged E2s; however, they also contain an active-site cysteine (within the RING2 domain) that, like an HECT E3, forms a ubiquitin–E3 thioester intermediate [5]. There are 14 human RBR E3s [5] and two in S. cerevisiae [1]. PARKIN, associated with the neurological condition Parkinson’s disease, is one of the best-studied RBR E3s [13]. Viruses and bacteria have exploited the ubiquitin system in many ways, including the expression of viral and microbial ubiquitin ligases [14–16]. Some of these fall into recognizable classes of E3s (e.g., RING E3), and others are completely novel (e.g., IpaH family of Shigella ligases).
E3 substrates can be modified by a single ubiquitin at one or more sites (monoubiquitylated or multi-monoubiquitylated, respectively). More typically, a target is conjugated with chains of ubiquitin molecules at one or more sites (polyubiquitylation). Since polyubiquitin chain formation can occur through any one of the seven lysines on ubiquitin or the terminal amino group, polyubiquitin chains of different structural topology can be generated, which has important consequences for the target protein. For example, targets covalently conjugated with K48-linked chains are typically recognized by the proteasome and degraded [17]. Linear ubiquitin chains play an important role in innate and adaptive immune signaling [18], while those with K63-linked chains are involved in many different intracellular signaling events, including the internalization of plasma membrane proteins. Specific functions of most other chain types are unclear; however, this is a very active area of research in the field and is reviewed elsewhere [19]. Although the molecular determinates for chain-type formation are not well understood, for RING E3s, it is generally the E2 that determines chain type, while determinants of chain-type specificity of HECT E3-catalyzed reactions are a function of the HECT domain itself [20]. Most E3s result in the same type of ubiquitylation for all of their substrates. For example, substrates of RING-like E3 Rad5 typically have K63-linked chains, while substrates of SCF ligases often display K48-linked chains. There are exceptions to this, generally as a result of an E3 interacting with multiple E2s that catalyze different types of chain linkages [21–25]. Chain-type information can prove very useful for E3 substrate identification. For example, it may be possible to enrich for substrates by adding proteasome inhibitor to cells if K48 chains are typically generated by the E3. Alternatively, chain-specific antibodies or affinity matrices can be used to enrich for substrates with specific polyubiquitin linkages.
Substrate abundance and turnover rates vary substantially, which can make co-immunoprecipitation (Co-IP) strategies for identifying relevant E3s particularly challenging. This has fueled the use of proteasome inhibitors in conjunction with Co-IPs and the development of specialized tools, such as TUBES (Tandem Ubiquitin-Binding Entities) [26], for the enrichment of ubiquitylated substrates. TUBES consist of artificial arrays of Ubiquitin-Binding-Associated (UBA) domains, which have a high avidity and affinity for specific polyubiquitin chain types. Such enrichment reagents were originally used to define the ubiquitinome; however, they can also be useful in a combinatorial approach for defining the substrates of a specific E3.
The class of E3, whether it is a monomeric or multimeric enzyme, and knowledge of previously characterized interacting proteins, functions, and intracellular localization are important considerations when choosing a suitable approach for pairing substrates to the E3 of interest. To date, protein–protein interaction techniques such as yeast two-hybrid and standard Co-IPs coupled with mass spectrometry have successfully identified many E3 substrates [27–30]. It is unlikely that any one technique can capture the full range of substrates for a particular E3 ligase, given a likely wide range of enzyme–substrate affinities and the potential complexity of regulatory mechanisms that govern enzyme–substrate interactions.
Other post-translational modifications can also influence ubiquitylation events. Neddylation of cullins activates the CRLs [31]. Phosphorylation can regulate ubiquitylation enzymes to influence E3–substrate interactions [32]. For example, phosphorylation can inhibit the E3 from interacting with its cognate E2 [33–35], which can profoundly affect the dynamics and specificity of the E3. The PINK1-induced phosphorylation of both ubiquitin and PARKIN is required for the complete activation of PARKIN [36, 37]. In addition, phosphorylation activates the HECT E3 ITCH [38]. Phosphorylation also directly influences the ubiquitylation of substrates; phosphorylation of C-Jun promotes its ubiquitylation [39], while phosphorylation results in the recognition of IκBα by the SCFβ-TrCP E3 ligase and ultimately its degradation [40]. Proline hydroxylation of HIF1α, dependent on molecular oxygen concentration, stimulates the recognition of this substrate by the VHL E3 [41]. Considering this interplay of post-translational modifications, it is to be expected that some E3–substrate binding events might only be captured under certain experimental conditions. The following sections will focus on the techniques and resources for the identification of substrates of E3 ligases.
Pull-downs coupled with ubiquitylation reactions
Non-specifically bound proteins are the bane of co-immunoprecipitations and GST fusion protein pull-downs. In the case of E3s, substrates and non-specifically associated proteins can be potentially distinguished by essentially performing a ubiquitylation reaction following a pull-down; the assumption is that true substrates will be ubiquitylated, whereas non-specifically bound proteins will not be ubiquitylated. While this is not always a safe assumption, this approach has proven useful in some cases. For example, bacterially expressed glutathione Sepharose-bound GST-Rsp5 (a yeast HECT E3) was mixed with 35S-methionine-labeled yeast extracts and after pull-down and washing of the matrix, purified ubiquitin, E1, and E2 enzymes, and ATP were added directly to the beads [42]. It was predicted that Rsp5 substrates would be ubiquitylated by wild-type Rsp5, but not when pulled-down with the active-site cysteine-to-alanine mutant Rsp5 (C–A). SDS-PAGE and autoradiography were performed to identify bands that bound to both the GST-Rsp5 and the C–A mutant, but were only ubiquitylated by WT GST-Rsp5. After identification of candidate substrate bands, the pull-down was repeated on a large scale (with unlabeled cell extract), and bands were excised from the gel and identified by protein sequencing. This approach identified Rpb1, the large subunit of RNA polymerase II, as an Rsp5 substrate [43, 44].
A similar approach was employed by Pagano and colleagues to identify targets of the human F-box protein SCFβ-TrCP2 [45]. HA-βTrCP2 was expressed in human 293T cells, and following an anti-HA IP, all of the components necessary for a typical in vitro ubiquitylation reaction, including FLAG-ubiquitin, were mixed with the resin. An anti-FLAG IP was then performed to isolate ubiquitylated proteins, followed by LC–MS/MS. Claspin was identified as a new substrate by this approach. The specificity of the interaction between Claspin and SCFβ-TrCP2 was bolstered by the inability of 19 other F-box proteins to Co-IP with Claspin.
Screening protein arrays or protein expression libraries by in vitro ubiquitylation
Rotin and colleagues performed large scale in vitro ubiquitylation assay on protein microarrays to identify new substrates of Rsp5 [46]. The protein microarrays were loaded with 4,000 GST and HIS-tagged S. cerevisiae proteins and incubated with fluorescently-labeled ubiquitin along with E1, E2, Rsp5, and ATP. Ubiquitylation was allowed to proceed for 3 h and was then detected with a fluorescent laser scanner. Candidates with the strongest signal relative to the positive control were deemed high-confidence hits. This approach confirmed many previously identified substrates of Rsp5 and led to the identification of several new substrates, including Sna4 and Rcr1. Simply incubating the microarray slides with fluorescently-labeled Rsp5 protein also identified substrate proteins and interacting proteins, however, this may not be a generally useful approach for other E3s as WW domains interact directly with small linear epitopes of substrates (“PY” motifs). Both data sets—direct Rsp5 interactors and proteins ubiquitylated by Rsp5—were enriched for known Rsp5-interacting proteins as well as proteins with PY motifs [11]. In a further variation of the approach, an in vitro assay for Rsp5 was developed using a bank of GST-tagged yeast proteins that were deemed potential substrates, because they contained PY motifs (consensus sequence being PPxY) [47]. Fifty-eight PY-motif-containing yeast proteins were selected for screening, and a larger set of randomly chosen proteins were analyzed. To screen for potential Rsp5 substrates, an in vitro ubiquitylation assay was performed using a GST-tagged set of candidate proteins and biotin-labeled ubiquitin. A mix of streptavidin and GST beads specifically isolated ubiquitylated substrates, which were detected by AlphaScreen™ Technology luminescence detection. The fluorescent detection of ubiquitylation increased the sensitivity of detection and offered a reduced workload and processing time compared to the standard western blot detection. Nine positive hits were identified and most of these had PY motifs. Such an approach is obviously useful when substrates are predicted to share consensus E3 recognition domains, but is not useful when dealing with an E3 that does not contain previously characterized protein–protein interaction domains.
The principles described previously have also been applied to find substrates of the APC/C in an Extract-based Functional Assay (EFA) [48]. Here, a human protein microarray (Human ProtoArray microarrays; Invitrogen) of 8000 proteins was incubated with synchronized HeLa cell extracts. Since APC/C is activated at cell cycle transitions, extracts from two cycle stages were compared for ubiquitylation using an anti-ubiquitin antibody and fluorescent detection. As a preliminary experiment, extracts were mixed with the 35S methionine-labeled Securin, a known APC/C target, to verify the activity of APC/C in the extracts before proceeding with the screen. 132 candidates were identified, and at the time, there were 16 known substrates for APC/C, 11 of which were among the positive hits. Many of the putative substrates were associated with mitosis (based on GO terms) and were chosen for validation. Substrates were validated by incubating 35S-labeled proteins with extracts containing active or inactive APC/C. Five of the six mitosis-associated candidates were shown to be substrates of APC/C. The protein microarray used in this study consisted of N-terminally GST-tagged proteins; however, the nature of the tag and its location (N- or C-terminal) may be an important consideration in applying this to other E3s.
In 1997, Kirchner and colleagues developed the in vitro expression cloning (IVEC) technique [49] as a way of identifying the key players in the process of mitosis and, thereafter, the targets of the APC/C [50]. In this approach, pools of cDNAs (50–100 cDNAs per pool) were in vitro transcribed and translated in the presence of 35S-labeled methionine. The pools of translated proteins were then either left untreated or a ubiquitylation reaction was initiated with the E3 and supporting E1 and E2 enzymes, ATP, and ubiquitin. The translation products were compared, with and without ubiquitylation components, to determine if any of the translated proteins from a given pool of cDNAs were substrates of the E3 (based on a molecular weight shift or degradation). If there was a positive hit with a pool, the pool was sorted and reanalyzed to identify the single cDNA that encodes the putative substrate. IVEC led to the discovery of Geminin as an APC/C substrate [51], which plays a critical role in cell cycle regulation. IVEC has been successfully used to identify other APC/C substrates as well, including Securin, Xkid, Tome-1, Sororin, and Mcph1-B [52–56], and has also been successfully applied for identifying substrates of kinases [49, 57], caspases [58], and p53 interacting proteins [59]. IVEC is likely to work well for E3–substrate interactions, where smaller well-defined epitopes of substrates are recognized by the E3 (e.g., PY motifs recognized by NEDD4 family E3s or D-box motifs by the APC/C). However, IVEC has its limitations, since many of the translation products may come from incomplete cDNAs and translated proteins may, therefore, not be folded correctly and not be recognized by the E3. Another limitation is that it may not be possible to reconstitute regulated ubiquitylation (e.g., in response to substrate phosphorylation) in the in vitro translation system.
Cell-based genome-wide screens
The advantages of cell-based screening approaches include the fact that (1) the results may have a greater chance of being biologically relevant and (2) E3s that are difficult to express in an active form in vitro (e.g., multi-subunit E3s) may be more amenable to study in a cell-based system.
SCF ligases influence a wide range of cellular processes, mediated by the diverse array of F-box proteins. Toczyski and colleagues used a large-scale microscopy screening method in S. cerevisiae to identify direct targets of Grr1, an F-box protein required for glucose sensing in yeast [60]. To develop the technique, a grr1 deletion strain was constructed, in which grr1 was replaced with Red Fluorescent Protein (RFP). The grr1 (RFP-positive) deletion strain was crossed with a GFP-Open Reading Frame (ORF) fusion library of 4000 yeast strains and mixed haploid populations of Δgrr1 (RFP-positive) and GRR1 (RFP-negative) cells expressing an individual GFP-tagged protein were examined by immunofluorescence microscopy. An increase in median fluorescence intensity of the GFP signal in the Δgrr1 cells relative to the GRR1 cells (as distinguished by the RFP signal) was indicative of that GFP fusion protein being a target of the Grr1-associated ligase. This large-scale screen was conducted in a 96-well plate format and over 100 putative targets were identified. After secondary screening, 24 of these proved to be indirect targets and were affected by grr1 deletion at the mRNA level, while seven were stabilized by grr1 deletion in a cycloheximide chase assay, confirming them to be direct ubiquitylation targets of Grr1. One of the main benefits of this technique is the precise comparative analysis of the wild type (WT) and mutant strains.
Global protein stability (GPS) profiling was developed as a means of tracking the abundance of 8000 human proteins in response to environmental cues or cellular perturbations [61]. Dual fluorescent tags, DsRed and GFP, were transcribed from a single mRNA separated by an internal ribosome binding site (IRES), driven by a constitutive promoter which was integrated into the genome by retroviral transduction. GFP was fused to the ORF of interest and the IRES allowed DsRed to serve as a control for protein expression, since alterations to protein stability due to interactions with an E3 ligase should only affect the levels of GFP. In addition, this ensured that transcriptional regulation does not affect protein levels, since both tags are transcribed from the same mRNA. Using GPS profiling and the human ORFeome library, the relative abundance of close to 8000 proteins was assessed at the single cell level. The total pool of cells, representing all 8000 ORFs, was subdivided into 7 populations using Fluorescence-activated cell sorting (FACS), according to the GFP/DsRed ratio, which indicated the relative stability of each protein. The DNA representing the ORFs from each subpopulation, as well as the total population, was then quantified by microarray hybridization. The relative enrichment of ORFs was assessed and, correspondingly, the levels of each protein in the library. To test the capabilities of GPS profiling, the proteasome inhibitor MG132 was added to cells. The effect was profound, with >80% of proteins showing increased stability. The subsequent validation of all 85 randomly chosen samples demonstrates the robustness of GPS analysis and the potential for this technique in detecting subtle cellular perturbations in response to expression or depletion of a particular E3 in the cell.
Elledge and colleagues used GPS for the identification of substrates for SCF ligases [62]. SCF ligases were inhibited by lentiviral delivery of dominant negative CUL1, the core component of SCF E3 ligases. Putative targets were those with increased ratio of GFP/DsRed when CUL1 was inhibited. The scope and sensitivity of GPS profiling are exceptional: 73% of SCF substrates were identified, and in addition, 31 of the 66 newly identified substrates were independently validated. Concerns for interference from N-terminal tags were addressed by testing ORF-HA fusions for 19 candidates; there was 89% correlation with the GFP-ORF results.
The technique was further improved upon with a more efficient lentivirus and an updated human ORFeome collection [63]. The updated ORFeome collection covered more ORFs and almost 13,000 genes. The capacity of the GPS technique was once again interrogated, this time by manipulating SCF ligases with the addition of MLN4924. MLN4924 is an NEDD8-activating enzyme inhibitor and inhibits the activity of all CRLs by preventing the activation of CUL1 and would be expected to increase the abundance of substrates. Indeed, half of the 190 known cullin-interacting proteins were identified in this screen. A quantitative proteomics approach was developed and used in parallel to the GPS profiling [63]. Quantitative ubiquitylation interrogation (QUAINT) analysis is essentially a variant of quantitative mass spectrometry. Ubiquitylated proteins were specifically co-immunoprecipitated using a diGly (diGlycine) antibody (discussed further in the following). Peptides that were enriched in response to MLN4924 treatment were identified using a SILAC (Stable Isotope Labeling with Amino acids in Cell culture)-based mass spectrometry approach. GPS and QUAINT appear to be good complementary techniques, since both identified known substrates that would have been missed with a single approach and putative substrates present in both data sets can be pursued with higher confidence.
Co-IP and affinity purification of E3s to identify substrates
Affinity purification coupled with mass spectrometry (AP/MS) is a widely used global approach for characterizing protein–protein interactions, in general. By expressing an affinity-tagged version of an E3 ligase in cells, substrates will often co-immunoprecipitate and can be identified by mass spectrometry. In the design of individual experiments, the size of the tag and its positioning at the N- or C-terminus of the protein may be important considerations, especially if functional domains are close to either end of the protein. Just as the design of AP experiments is essential to the generation of interpretable data, the ability to decipher genuine interactors from false positives is crucial to the success of AP/MS. Computational tools that can help here include HGSCore, CompPASS, and SAINT [64–66]. CompPASS, for example, assigns a score to bait-prey pairs across multiple experiments by integrating the abundance, uniqueness, and reproducibility of an interacting protein. In this manner, high-confidence candidate interacting proteins (HCIPs) are sorted from the common technical artifacts and false positives [67].
Dual affinity tags such as His6-FLAG allow for a two-step purification which can reduce background binding, and moreover, an His6 tag can tolerate denaturing conditions. Compared to His6-FLAG, an His6-biotin affinity purification allows for stringent conditions throughout the purification process due to the high affinity of biotin and streptavidin used for the IP [68]. Tagwerker et al. used formaldehyde as a crosslinking reagent and His6-biotin-tagged Skp1 (adaptor component of SCF ligases) to identify substrates. In preparing the samples for MS, an on-bead tryptic digestion was used to circumvent the difficulty in removing the bound proteins from the beads.
Global approaches to map the ubiquitinome have been a major goal for the field and using epitope-tagged ubiquitin, close to 1100 conjugates had been identified [69]. In 2011, Gygi, Harper, and colleagues used a new approach called diGly proteomics to identify ~5000 ubiquitylated proteins in human cells [65]. In the process of preparing protein samples for diGly mass spectrometry analysis, the sample is trypsinized, which leaves a diglycine (diGly) remnant on the side chain of ubiquitylated lysine residues. DiGly remnant antibodies were developed [65, 70] that can be used to immunoprecipitate these peptides, which are then identified by mass spectrometry.
The capacity of diGly proteomics for detecting subtle changes in ubiquitylation was tested in the initial study by Gygi and coworkers, by treating cells with proteasome inhibitor and/or MLN4924, compared to untreated cells. In the process, known and novel CRL substrates were identified and the specific detection of the ubiquitin-modified version of H2B validated this candidate as a CRL substrate. DiGly proteomics has since been used for the discovery of many substrates for E3s [71–74]. Using siRNAs to perturb E3 ligase activity and diGly proteomics, new substrates for HRD1 [72] and HUWE1 [74] have been identified and the PARKIN interaction network was significantly expanded using an SILAC and diGly proteomics approach [71]. A new approach developed by the Gygi lab combines isobaric labeling of peptides and diGly proteomics [75]. By interrogating the PARKIN-PINK1 pathway, the authors successfully demonstrated the capacity of this technique for increased sensitivity and reproducibility in identifying candidate substrates for E3 ligases.
E6AP/UBE3A, an HECT E3, was one of the first E3 ligases to be characterized. E6AP cooperates with the HPV E6 protein in the degradation of p53 to promote HPV-associated cervical cancer [76, 77]. Mutations in the E6AP/UBE3A gene or alterations in expression are also the cause of Angelman syndrome, a severe neurologic disease [78]. Despite its clinical importance, few natural (E6-independent) substrates have been identified. A comprehensive approach to identify new substrates of E6AP compared the interactomes of the multiple isoforms of E6AP, both wild-type and a dominant negative (DN) mutant [79]. Based on previous work, it was hypothesized that a DN mutant may lead to accumulation of E6AP substrates in the cell [80]. Tagged proteins were inducibly expressed in Flp-In T-REx 293 cells and affinity-purified. Mass spectrometry data were processed through CompPASS to identify high-confidence candidate interactors. To assist the filtering process, NEDD4, another HECT E3, was also included in the experiment as a negative control. Despite no significant differences between isoforms of E6AP or between WT and the DN mutant, several interactors were identified. As confirmation, MAPK6, HIF1AN, and NEURL4 pulled down when E6AP was affinity purified from cells. Moreover, these proteins were more readily detected in the DN mutant; however, the physiological relevance and function of these interacting proteins remain unclear.
For a successful AP/MS-based substrate screen, it is extremely helpful when there are known substrates for the bait protein, and defined enzyme–substrate interaction domains. The tumor suppressor and F-box protein FBW7 are known to ubiquitylate phosphorylated substrates through a conserved Cdc4 phosphodegron (CPD) motif [81–84], which is essential for the binding of FBW7 to substrates [85]. To screen for new substrates, wild-type FBW7 and a mutant that do not bind known substrates were affinity purified from human cells [86]. The MS data were filtered for proteins only present in WT and 72 proteins remained, including 5 known FBW7 targets. Analyzing the list of candidates using bioinformatics, it was noted that 26 of these proteins belonged to the same complex, which suggested that FBW7 could be pulling down indirect targets, as well as direct targets. A focus was placed on validating proteins with the highest spectral counts and determining the presence of the conserved phosphodegron (CPD) domain among candidates. This proved to be an effective approach and MED13/13L were shown to be FBW7 targets, demonstrating a role for FBW7 in broadly regulating transcription.
In 2013, Harper and colleagues applied a Parallel Adaptor Capture (PAC) technique for the cross comparison of AP/MS data from 19 FBXL proteins to systematically expand the interactome of FBXLs [87]. PAC is an approach to specifically enrich for F-box interacting proteins. Human HEK293 cells expressing HA-tagged F-box proteins were treated with either bortezomib (proteasome inhibitor) or MLN4924 (CUL1 inhibitor) and HA-tagged proteins were precipitated in parallel and subjected to mass spectrometry (PAC proteomics). High-Confidence Interacting Proteins (HCIPs) were identified using CompPASS analysis [67]. The approach proved extremely successful in identifying interactors. 80% of the candidates selected for validation by Co-IP/western blot were shown to interact with their cognate FBXL protein. In addition, almost 50% appeared to be substrates, with an increase in abundance in response to MLN4924 treatment. Interestingly, five of the candidates chosen for validation were sub-threshold HCIPs that appear to be substrates, which suggest that CompPASS filtering may have been too stringent and additional substrates could also have been missed. Similar PAC experiments were performed for 10 FBXLs in HCT116 cells (colon cancer cell line) and a significant overlap in targets identified in both cell types demonstrated the benefits of performing parallel experiments in different cell lines.
Bioinformatic analyses of E3-associated proteins can play a critical role in substrate verification. HERC2 is linked to a number of Autism-spectrum conditions and DNA damage repair pathways [88–90]. A small number of substrates have been characterized, such as NEURL4 and RNF8 [91, 92], but it has been notoriously difficult to study this very large E3 ligase and delineate its functions. Since the full-length protein is poorly expressed, Galligan et al. created six constructs, each of about 1000 bp size, collectively spanning the full-length protein [93]. They hypothesized that substrates would co-fractionate with a specific fragment by binding to its corresponding interaction domain. Using HA-GFP as a control for non-specific bead binding and filtering for peptides that were present in one sample only, 239 putative interactors were identified. With such a large list of candidates, further silico analysis was required. The Genomatrix Pathway system was applied to the data and it highlighted the abundance of proteins involved in intracellular vesicle-mediated organization and transport. Of the 40 HCIPs that were in this pathway, 27 of them were functionally linked. This approach proved successful, and of the 21 potential interactors were tested, 13 of them were shown to interact with endogenous HERC2.
Precise comparative analysis across many AP/MS experiments can eliminate background noise that is inherent to mass spectrometry data sets. A useful repository of common AP/MS contaminants has been compiled [94]. Moreover, a new interface, Spotlite provides similar scoring methods to analysis tools such as CompPASS but also integrates information on function, model organism phenotypes, and human disease relevance [95]. With an interest in substrates of the E3 ligase Keap1, Major and colleagues reanalyzed their previously published Keap1 mass spectrometry using Spotlite. A library of 44 reference data sets from previous AP experiments was comparatively analyzed to identify unique interactors. In this manner, 35 candidates were identified. As validation of their approach, eight of the candidates co-immunoprecipitated with Keap1, and one of them, MCM3 has since been confirmed as a novel substrate [96].
Tandem ubiquitin-binding entities (TUBES) are tandem UBA domain repeats consisting of UBAs from UBQLN1 and HR23A [26, 97]. TUBES have become a widely used tool in molecular biology to trap ubiquitin-bound substrates and are commercially available. New substrates for the striated muscle-specific RING E3 MuRF1 were identified with the use of TUBES, in combination with two-dimensional differential in gel electrophoresis (2D-DIGE) and matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry [98]. This technique centers on the comparative analysis of cells with differential (reduced or enhanced) E3 ligase activity. Either GFP alone or in combination with MuRF1 was overexpressed in primary cardiomyocytes and it was expected that known substrates as well as new candidates would be enriched for in response to MuRF1 overexpression. A pull-down with either TUBE or control beads was then performed. Two differentially-labeled experimental conditions were tested in a single 2D-DIGE gel. Gel 1 compared the TUBE pull-down eluate for control (GFP only) versus the experimental cells (MuRF1). Gel 2 compared the supernatants following AP with either TUBE or control beads for experimental cells. Gel 3 compared the supernatants following AP with either TUBE or control beads for control cells. Protein spots displaying greater than 1.5 fold expression changes were deemed candidates in each individual gel and the list of candidates from all three gels were aligned to determine spots that were candidates by all three comparisons. The candidates across all multiple gels were selected for further analysis and removed from the 2D gels for protein identification by MALDI-TOF mass spectrometry. In this manner, Hspd1, Tpm1, and Atp5b were identified as new substrates of MuRF1.
Trypsin Resistant (TR)-TUBES have now been created, and may be more advantageous to use for mass spectrometry-based experiments, since they prevent the TUBES and ubiquitin itself from contributing to the overall peptides detected by mass spectrometry [99]. By co-expressing the TR-TUBES along with FBXO21, an uncharacterized F-box protein, candidate substrates were identified by diGly proteomics. Only two candidates were chosen for validation; these proteins had the highest spectral counts, and they were subsequently validated as substrates. With a focus on further optimizing the TUBES-type pull-down approach, Xu and colleagues systematically tested the binding capabilities of a number of different Ubiquitin-Binding Domains (UBDs) [100]. By testing the binding capacity of five different UBAs, both alone and in combination, for ubiquitin, the authors designed Tandem hybrid UBDs (ThUBDs) that had unbiased binding to all seven types of polyubiquitin chains. Moreover, ThUBDs were more effective than naturally occurring UBDs at enriching for ubiquitylated proteins. Compared with His6-Myc-Ub in yeast, ThUBDs purified more ubiquitylated proteins. In preparing the samples for mass spectrometry, the proteins were separated by high pH reversed-phase liquid chromatography (RP-LC), instead of SDS-PAGE. This greatly improved the yield of candidates, which compared favorably with previous studies, such as diGly proteomics, in mapping the global ubiquitinome. Other fusion protein approaches include the fusion to the proteasomal subunit S5A, which was developed to trap polyubiquitylated proteins [101]. In another approach, four tandem repeats of ubiquitin-associated domain from UBQLN1 were fused to a GST tag [102]. These techniques, developed for enriching ubiquitylated proteins, may prove very useful as part of a combinatorial approach for E3–substrate pairing.
Substrate traps
O’Connor et al. developed a technique to covalently capture substrates of E3 ligases. Ubiquitin-activated Interaction Traps (UBAITs) are E3 ligase fusion proteins with ubiquitin, bridged by a flexible linker region, attached to the C-terminus of the E3 [103]. The C-terminus of ubiquitin remains free for activation by the E1 enzyme, forming a thioester-linked complex, which is then transferred to an E2 enzyme, as would be expected for any N-terminally-tagged ubiquitin (although in this case, the N-terminal tag is an E3). For HECT E3s, the active-site cysteine of the E3 can attack the UBAIT ~ E2 thioester bond in an intramolecular reaction, forming a thioester-linked “lariat” protein. If a substrate then interacts with the E3, even transiently, a lysine of the substrate might react with the thioester-linked lariat ubiquitin, which will result in formation of a stable amide bond-linked UBAIT-substrate complex (i.e., E3-linker–Ub-substrate), which can be purified via an N-terminal affinity tag on the UBAIT. The approach was also predicted to be applicable to non-HECT E3s, with the only difference being that the substrate lysine would react with the UBAIT when it is in a thioester linkage to the E2; the product would again be a stable amide bond-linked complex between the UBAIT and the substrate.
To validate the approach, N-terminally tandem affinity purification (TAP)-tagged UBAITs were overexpressed in cells and affinity purified. Following SDS-PAGE, gel slices, excised from above the migration point of the UBAITs, were sent for mass spectrometry analysis. A UBAIT with the terminal glycine residues of ubiquitin mutated was used as a control for non-covalent interactors. The approach was validated in both yeast and human cells, for both HECT and RING E3s. Known substrates were trapped for the HECT E3s Rsp5 and Itch, as well as RING E3s Psh1 and RNF126. For yeast Rsp5, the UBAIT trapped 32% of known interactors, including regulators, substrates, and adaptors. The presence of known adaptors and regulators as well as substrates likely reflects the ability of the UBAIT to serve as a covalent proximity-based protein interaction trap. The high ratio of known interactors for Rsp5 as well as for Psh1 highlights the specific and selective functionality of the UBAITs. Moreover, the UBAIT was also used to identify a new target for the RING E3 RNF168, a key component of the DNA damage response pathway. The technique has applications beyond E3s, as well, and can potentially be used to profile the interactome of almost any protein of interest, with the caveat that UBAITs must have the ubiquitin moiety at the C-terminal end, which may preclude its use with certain proteins [103].
The NEDDylator is another fusion-based strategy and takes advantage of the well-defined and short list of substrates for NEDD8 [104]. NEDD8 has its own E1 and a single E2, UBC12. By fusing UBC12 to the C-terminus of an E3 and co-expressing with tagged NEDD8, targets of the E3 will be NEDDylated, rather than ubiquitylated, and these targets can then be easily isolated by affinity purification (based on the tagged NEDD8) and identified by mass spectrometry. While natural NEDD8 substrates will also be affinity purified, the natural substrates are a short list of known proteins and these can be disregarded as candidate substrates of the NEDDylator. When tested with the RING E3 XIAP (or CIAP1), different orientations (N- versus C-terminal UBC12) were comparable in vitro, but C-terminal UBC12 was used in cells, since N-terminal acetylation of UBC12 facilitated NEDDylation of certain NEDD8 substrates. A SILAC-based MS strategy identified known and unknown substrates. The data were filtered for the enrichment of peptides with the NEDDylator compared to UBC12 alone. Since NEDD8 is not degraded by the proteasome, the NEDDylator may ensure stability of the conjugates compared to overexpression strategies that employ a tagged form of ubiquitin.
As an alternative to the standard affinity purification, BioID was developed. This technique takes advantage of the biotinylation capabilities of the E. coli enzyme BirA. A mutant form of BirA promiscuously biotinylates proteins within a 10–20 nm range [105]. By stably expressing the BirA-POI (Protein of Interest) fusion protein in mammalian cells, it is expected that substrates and other binding partners can be biotinylated and subsequently identified by a streptavidin pull-down and mass spectrometry [106]. This technique was successfully applied to identify substrates of the F-box protein SCFβ-TrCP1/2 [107].
The Toczyski lab developed the Ubiquitin Ligase Substrate Trapping technique to identify substrates of SCF-F-box ligases in yeast [108]. The UBA domains of Dsk2 or Rad23 (ubiquitin receptor proteins that typically mediate the transfer of ubiquitylated proteins to the proteasome) were fused, via a 3XFLAG linker, to eight different F-box proteins. The UBA-tagged F-box substrate receptors allowed for anti-FLAG co-immunoprecipitation of the polyubiquitylated forms of the substrates with the UBA-F-box proteins. This was done in cells expressing His6-tagged ubiquitin, so that a secondary Ni-NTA purification of ubiquitylated proteins could be performed from the anti-FLAG immunoprecipitate. Artifacts of overexpression were avoided by expressing the UBA-F-box proteins from an endogenous promoter, where the bait was the only copy of the F-box protein in the cell. Both known and novel substrates of the F-box E3s were identified. An important factor for the success of this technique was the ability to compare the proteins identified with the different F-box traps, and a 250-fold enrichment was chosen as the cut-off point to filter the data for unique peptides among the different F-box traps. The Toczyski lab has expanded this technique for use in mammalian cells [109], using stable cell lines expressing doxycycline-inducible His6-tagged ubiquitin and the E3–3XFLAG–Rad23B fusion proteins.
Concluding remarks
There are clearly many options for profiling the substrate specificity of E3, and the characteristics and preexisting knowledge of any given E3 will allow some degree of prioritization of approaches (Fig. 1). Additional recent perspectives on these issues are also available [110, 111]. The development of approaches that take unique advantage of ubiquitin and Ubl biochemistry (e.g., UBAITs, Neddylators, Ubiquitin Ligase Substrate Traps) may improve the depth of coverage and help identify less abundant substrates for E3 ligases. The direct tagging of endogenous proteins for Co-IP approaches using CRISPR/Cas9 techniques may also help to eliminate artifacts of overexpression in human cell systems, and CRISPR/Cas9 approaches will also allow the investigator to infer ligase substrates by proteomic analyses in a clean deletion background. The importance of modern bioinformatics and statistical tools in analyzing output of many of the approaches described here should not be underemphasized. For example, the RBR E3 ARIH1 was recently discovered to interact with CRLs by Co-IP experiments; however, retrospective analysis of older CRL interactome studies had previously demonstrated this connection [112]. Therefore, improved bioinformatics tools and analysis of multiple data sets obtained with a variety of approaches can be useful for refining lists of candidate substrates and guiding further experimentation.
References
- 1.Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Nat Publ Gr. 2016;26:423–440. doi: 10.1038/cr.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae . Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Bengtson MH, Ulbrich A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 5.Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17:626–642. doi: 10.1038/nrm.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohi MD, Vander Kooi CW, Rosenberg JA, et al. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 8.Sarikas A, Hartmann T, Pan Z-Q. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 11.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 12.Dastur A, Beaudenon S, Kelley M, et al. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J Biol Chem. 2006;281:4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
- 13.Pickrell AM, Youle RJ. The roles of PINK1, Parkin, and mitochondrial fidelity in parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huibregtse J, Rohde JR, Martin G, et al. Hell’s BELs: bacterial E3 ligases that exploit the eukaryotic ubiquitin machinery. PLoS Pathog. 2014;10:e1004255. doi: 10.1371/journal.ppat.1004255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randow F, Lehner PJ. Viral avoidance and exploitation of the ubiquitin system. Nat Cell Biol. 2009;11:527–534. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]
- 16.Maculins T, Fiskin E, Bhogaraju S, Dikic I. Bacteria-host relationship: ubiquitin ligases as weapons of invasion. Cell Res. 2016;26:499–510. doi: 10.1038/cr.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu P, Duong DM, Seyfried NT, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki K, Iwai K. Roles of linear ubiquitinylation, a crucial regulator of NF-κB and cell death, in the immune system. Immunol Rev. 2015;266:175–189. doi: 10.1111/imr.12308. [DOI] [PubMed] [Google Scholar]
- 19.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 20.Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol. 2009;29:3307–3318. doi: 10.1128/MCB.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 22.Jin L, Williamson A, Banerjee S, et al. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williamson A, Wickliffe KE, Mellone BG, et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci USA. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garnett MJ, Mansfeld J, Godwin C, et al. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu T, Merbl Y, Huo Y, et al. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci USA. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hjerpe R, Hjerpe R, Aillet F, et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortier JM, Kornbluth J. NK lytic-associated molecule, involved in NK cytotoxic function, is an E3 ligase. J Immunol. 2006;176:6454–6463. doi: 10.4049/jimmunol.176.11.6454. [DOI] [PubMed] [Google Scholar]
- 28.Gray WM, Del Pozo JC, Walker L, et al. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana . Genes Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L, Shi L, Li W, et al. JFK, a Kelch domain-containing F-box protein, links the SCF complex to p53 regulation. Proc Natl Acad Sci USA. 2009;106:10195–10200. doi: 10.1073/pnas.0901864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwab M, Neutzner M, Möcker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deshaies RJ, Emberley ED, Saha A. Control of cullin-ring ubiquitin ligase activity by Nedd8. In: Groettrup M, editor. Conjugation and deconjugation of ubiquitin family modifiers. New York: Springer; 2010. pp. 41–56. [DOI] [PubMed] [Google Scholar]
- 32.Vittal V, Stewart MD, Brzovic PS, Klevit RE. Regulating the regulators: recent revelations in the control of E3 ubiquitin ligases. J Biol Chem. 2015;290:21244–21251. doi: 10.1074/jbc.R115.675165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cocklin R, Goebl M. Nutrient sensing kinases PKA and sch9 phosphorylate the catalytic domain of the ubiquitin-conjugating enzyme Cdc34. PLoS One. 2011 doi: 10.1371/journal.pone.0027099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craney A, Kelly A, Jia L, et al. Control of APC/C-dependent ubiquitin chain elongation by reversible phosphorylation. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1522423113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiao R, Weissmann F, Yamaguchi M, et al. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc Natl Acad Sci USA. 2016 doi: 10.1073/pnas.1604929113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyano F, Okatsu K, Kosako H, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 37.Kondapalli C, Kazlauskaite A, Zhang N, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallagher E, Gao M, Liu Y-C, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci USA. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu F, Choi BY, Ma WY, et al. COOH-terminal Src kinase-mediated c-Jun phosphorylation promotes c-Jun degradation and inhibits cell transformation. Cancer Res. 2006;66:5729–5736. doi: 10.1158/0008-5472.CAN-05-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strack P, Caligiuri M, Pelletier M, et al. SCF(beta-TRCP) and phosphorylation dependent ubiquitination of I kappa B alpha catalyzed by Ubc3 and Ubc4. Oncogene. 2000;19:3529–3536. doi: 10.1038/sj.onc.1203647. [DOI] [PubMed] [Google Scholar]
- 41.Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 42.Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang A, Cheang S, Espanel X, Sudol M. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:20562–20571. doi: 10.1074/jbc.M002479200. [DOI] [PubMed] [Google Scholar]
- 44.Beaudenon SL, Huacani MR, Wang G, et al. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae . Mol Cell Biol. 1999;19:6972–6979. doi: 10.1128/MCB.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peschiaroli A, Dorrello NV, Guardavaccaro D, et al. SCFTrCP-mediated degradation of claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 46.Gupta R, Kus B, Fladd C, et al. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol. 2007;3:116. doi: 10.1038/msb4100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kus B, Gajadhar A, Stanger K, et al. A high throughput screen to identify substrates for the ubiquitin ligase Rsp5. J Biol Chem. 2005;280:29470–29478. doi: 10.1074/jbc.M502197200. [DOI] [PubMed] [Google Scholar]
- 48.Merbl Y, Kirschner MW. Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc Natl Acad Sci USA. 2009;106:2543–2548. doi: 10.1073/pnas.0812892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stukenberg PT, Lustig KD, McGarry TJ, et al. Systematic identification of mitotic phosphoproteins. Curr Biol. 1997;7:338–348. doi: 10.1016/S0960-9822(06)00157-6. [DOI] [PubMed] [Google Scholar]
- 50.Ayad NG, Rankin S, Ooi D, et al. Identification of ubiquitin ligase substrates by in vitro expression cloning. Methods Enzymol. 2005;399:404–414. doi: 10.1016/S0076-6879(05)99028-9. [DOI] [PubMed] [Google Scholar]
- 51.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/S0092-8674(00)81209-X. [DOI] [PubMed] [Google Scholar]
- 52.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 53.Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/S0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- 54.Ayad NG, Rankin S, Murakami M, et al. Tome-1, a trigger of mitotic entry, is degraded during G1 via the APC. Cell. 2003;113:101–113. doi: 10.1016/S0092-8674(03)00232-0. [DOI] [PubMed] [Google Scholar]
- 55.Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase- promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18:185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 56.Hainline SG, Rickmyre JL, Neitzel LR, et al. The Drosophila MCPH1-B isoform is a substrate of the APCCdh1 E3 ubiquitin ligase complex. Biol Open. 2014;3:669–676. doi: 10.1242/bio.20148318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Armacki M, Joodi G, Nimmagadda SC, et al. A novel splice variant of calcium and integrin-binding protein 1 mediates protein kinase D2-stimulated tumour growth by regulating angiogenesis. Oncogene. 2014;33:1167–1180. doi: 10.1038/onc.2013.43. [DOI] [PubMed] [Google Scholar]
- 58.Cryns VL, Byun Y, Rana A, et al. Specific proteolysis of the kinase protein kinase C-related kinase 2 by caspase-3 during apoptosis. Identification by a novel, small pool expression cloning strategy. J Biol Chem. 1997;272:29449–29453. doi: 10.1074/jbc.272.47.29449. [DOI] [PubMed] [Google Scholar]
- 59.Lunardi A, Di Minin G, Provero P, et al. A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. Proc Natl Acad Sci USA. 2010;107:6322–6327. doi: 10.1073/pnas.1002447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benanti JA, Cheung SK, Brady MC, Toczyski DP. A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat Cell Biol. 2007;9:1184–1191. doi: 10.1038/ncb1639. [DOI] [PubMed] [Google Scholar]
- 61.Chou DM, Xu Q, Yen H-CS, et al. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 62.Yen H-CS, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 63.Emanuele MJ, Elia AEH, Xu Q, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guruharsha KG, Rual J-F, Zhai B, et al. A protein complex network of Drosophila melanogaster . Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim W, Bennett EJ, Huttlin EL, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lister AL, Pocock M, Taschuk M, Wipat A. Saint: a lightweight integration environment for model annotation. Bioinformatics. 2009;25:3026–3027. doi: 10.1093/bioinformatics/btp523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction Landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tagwerker C, Flick K, Cui M, et al. A tandem affinity tag for two-step purification under fully denaturing conditions. Mol Cell Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 69.Peng J, Schwartz D, Elias JE, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 70.Xu G, Paige J, Jaffrey S. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinty profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sarraf SA, Raman M, Guarani-Pereira V, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee KA, Hammerle LP, Andrews PS, et al. Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J Biol Chem. 2011;286:41530–41538. doi: 10.1074/jbc.M111.248856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krönke J, Udeshi ND, Narla A, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thompson JW, Nagel J, Hoving S, et al. Quantitative Lys-∊-Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 Ligase HUWE1. J Biol Chem. 2014;289:28942–28955. doi: 10.1074/jbc.M114.573352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rose CM, Isasa M, Ordureau A, et al. Highly multiplexed quantitative mass spectrometry analysis of ubiquitylomes. Cell Syst. 2016 doi: 10.1016/j.cels.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scheffner M, Werness BA, Huibregtse JM, et al. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 77.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 78.Jiang Y, Armstrong D, Albrecht U, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/S0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 79.Martinez-Noël G, Galligan JT, Sowa ME, et al. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol Cell Biol. 2012;32:3095–3106. doi: 10.1128/MCB.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Talis AL, Huibregtse JM, Howley PM. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 81.Koepp D, Schaefer L, Ye X, Keyomarsi K. Phosphorylation-dependent ubiquitination of Cyclin E by the SCF-Fbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 82.Nash P, Tang X, Orlicky S, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 83.Welcker M, Singer J, Loeb KR, et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell. 2003;12:381–392. doi: 10.1016/S1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 84.Strohmaier H, Spruck CH, Kaiser P, et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 85.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 86.Davis MA, Larimore EA, Fissel BM, et al. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with mediator. Genes Dev. 2013;27:151–156. doi: 10.1101/gad.207720.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tan MKM, Lim HJ, Bennett EJ, et al. Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol Cell. 2013;52:9–24. doi: 10.1016/j.molcel.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol. 2010;12:12–80. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- 89.Puffenberger EG, Jinks RN, Wang H, et al. A homozygous missense mutation in HERC2 associated with global developmental delay and autism spectrum disorder. Hum Mutat. 2012;33:1639–1646. doi: 10.1002/humu.22237. [DOI] [PubMed] [Google Scholar]
- 90.Harlalka GV, Baple EL, Cross H, et al. Mutation of HERC2 causes developmental delay with Angelman-like features. J Med Genet. 2013;50:65–73. doi: 10.1136/jmedgenet-2012-101367. [DOI] [PubMed] [Google Scholar]
- 91.Al-Hakim AK, Bashkurov M, Gingras AC, et al. Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture. Mol Cell Proteomics. 2012;11:M111.014233. doi: 10.1074/mcp.M111.014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Danielsen JR, Povlsen LK, Villumsen BH, et al. DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J Cell Biol. 2012;197:179–187. doi: 10.1083/jcb.201106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galligan JT, Martinez-Noël G, Arndt V, et al. Proteomic analysis and identification of cellular interactors of the giant ubiquitin ligase HERC2. J Proteome Res. 2015;14:953–966. doi: 10.1021/pr501005v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mellacheruvu D, Wright Z, Couzens AL, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goldfarb D, Hast BE, Wang W, Major MB. Spotlite: web application and augmented algorithms for predicting co-complexed proteins from affinity purification—mass spectrometry data. J Proteome Res. 2014;13:5944–5955. doi: 10.1021/pr5008416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mulvaney KM, Matson JP, Siesser PF, et al. Identification and characterization of MCM3 as a novel KEAP1 substrate. J Biol Chem. 2016 doi: 10.1074/jbc.M116.729418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aillet F, Lopitz-Otsoa F, Hjerpe R, et al. Isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. Methods Mol Biol. 2012;832:173–183. doi: 10.1007/978-1-61779-474-2_12. [DOI] [PubMed] [Google Scholar]
- 98.Rubel CE, Schisler JC, Hamlett ED, et al. Diggin’ on U(biquitin): a novel method for the identification of physiological E3 ubiquitin ligase substrates. Cell Biochem Biophys. 2013;67:127–138. doi: 10.1007/s12013-013-9624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoshida Y, Saeki Y, Murakami A, et al. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc Natl Acad Sci USA. 2015;112:4630–4635. doi: 10.1073/pnas.1422313112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao Y, Li Y, Zhang C, et al. Enhanced purification of ubiquitinated proteins by engineered tandem hybrid ubiquitin-binding domains (ThUBDs) Mol Cell Proteomics. 2016;15:1381–1396. doi: 10.1074/mcp.O115.051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Layfield R, Tooth D, Landon M, et al. Purification of poly-ubiquitinated proteins by S5a-affinity chromatography. Proteomics. 2001;1:773–777. doi: 10.1002/1615-9861(200106)1:6<773::AID-PROT773>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 102.Shi Y, Chan DW, Jung SY, et al. A data set of human endogenous protein ubiquitination sites. Mol Cell Proteomics. 2011;10:M110.002089. doi: 10.1074/mcp.M110.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O’Connor HF, Lyon N, Leung JW, et al. Ubiquitin-activated interaction traps (UBAITs) identify E3 ligase binding partners. EMBO Rep. 2015;16:1699–1712. doi: 10.15252/embr.201540620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhuang M, Guan S, Wang H, et al. Substrates of IAP ubiquitin ligases identified with a designed orthogonal E3 ligase, the NEDDylator. Mol Cell. 2013;49:273–282. doi: 10.1016/j.molcel.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roux KJ, Kim DI, Burke B. BioID: a screen for protein-protein interactions. Curr Protoc Protein Sci. 2013 doi: 10.1002/0471140864.ps1923s74. [DOI] [PubMed] [Google Scholar]
- 107.Coyaud E, Mis M, Laurent EMN, et al. BioID-based identification of Skp cullin F-box (SCF)β-TrCP1/2 E3 ligase substrates. Mol Cell Proteomics. 2015;14:1781–1795. doi: 10.1074/mcp.M114.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mark KG, Simonetta M, Maiolica A, et al. Ubiquitin ligase trapping identifies an SCFSaf1 pathway targeting unprocessed vacuolar/lysosomal proteins. Mol Cell. 2014;53:148–161. doi: 10.1016/j.molcel.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mark KG, Loveless TB, Toczyski DP. Isolation of ubiquitinated substrates by tandem affinity purification of E3 ligase-polyubiquitin-binding domain fusions (ligase traps) Nat Protoc. 2016;11:291–301. doi: 10.1038/nprot.2016.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iconomou M, Saunders DN. Systematic approaches to identify E3 ligase substrates. Biochem J. 2016;473:4083–4101. doi: 10.1042/BCJ20160719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.O’Leary CE, Lewis EL, Oliver PM. Ubiquitylation as a rheostat for TCR signaling: from targeted approaches toward global profiling. Front Immunol. 2015 doi: 10.3389/fimmu.2015.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Scott DC, Rhee DY, Duda DM, et al. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell. 2016;166(1198–1214):e24. doi: 10.1016/j.cell.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]