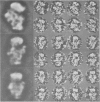
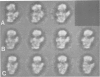
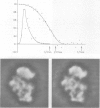
Abstract
Large sets of electron microscopic images of the 30S ribosomal subunits of Bacillus stearothermophilus (914 molecules) and Escherichia coli (422 molecules) were analysed with image processing techniques. Using computer alignment and a new multivariate statistical classification scheme, three predominant views of the subunit were found for both species. These views, which together account for approximately 90% of the population of images, were determined to a reproducible resolution of up to 1.7 nm, thus elucidating many new structural details. The angular spread of the molecular orientations around the three main stable positions is remarkably small (less than 8 degrees). Some of the current models for the small ribosomal subunit are incompatible with our new results.
Full text
PDF

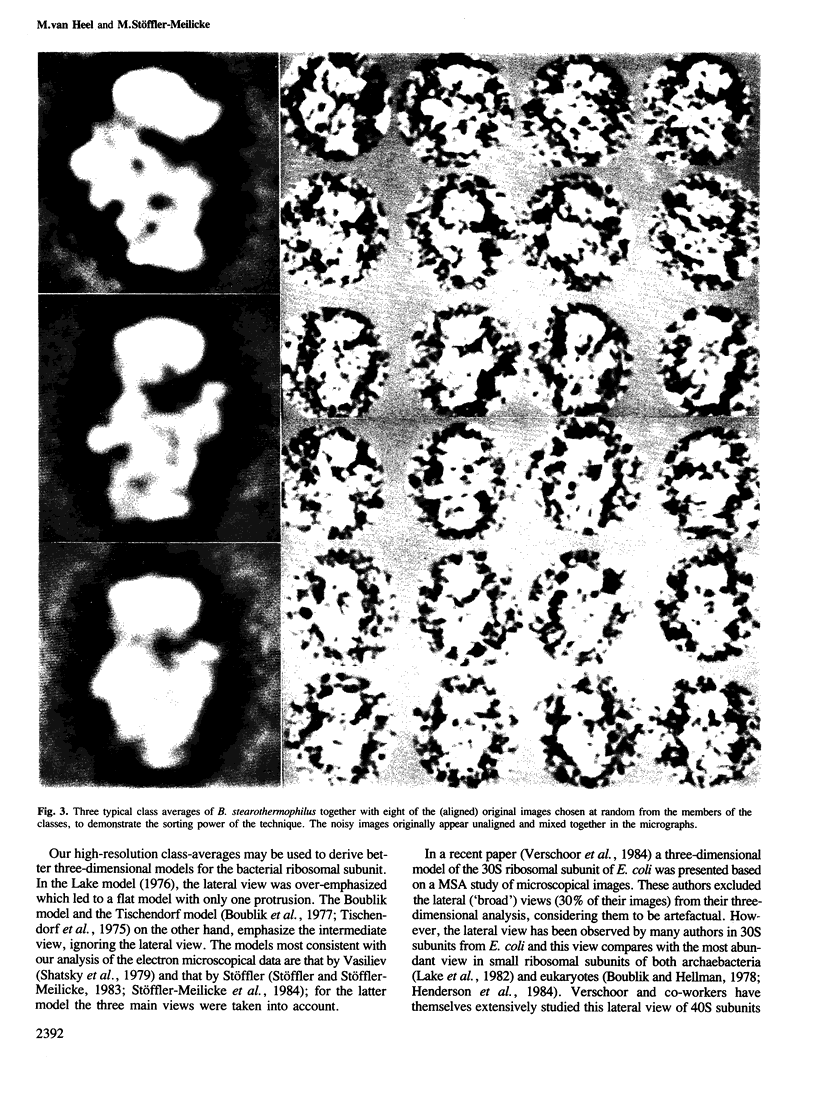
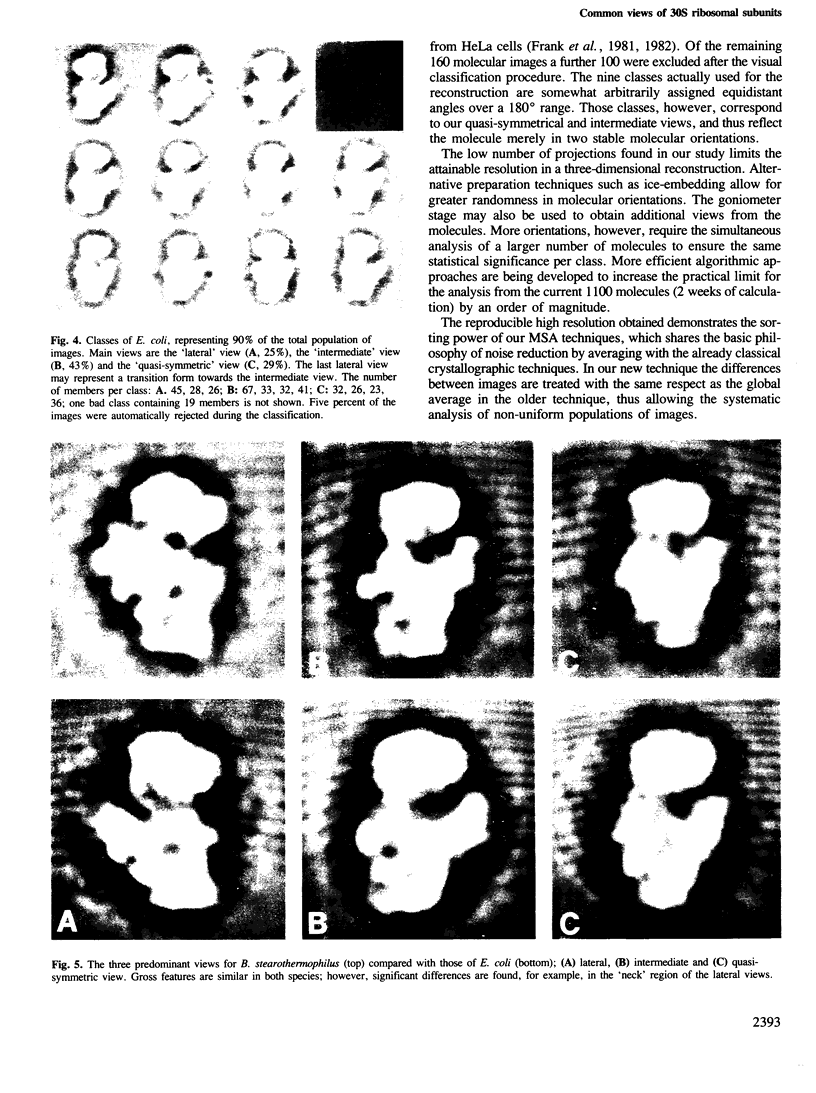
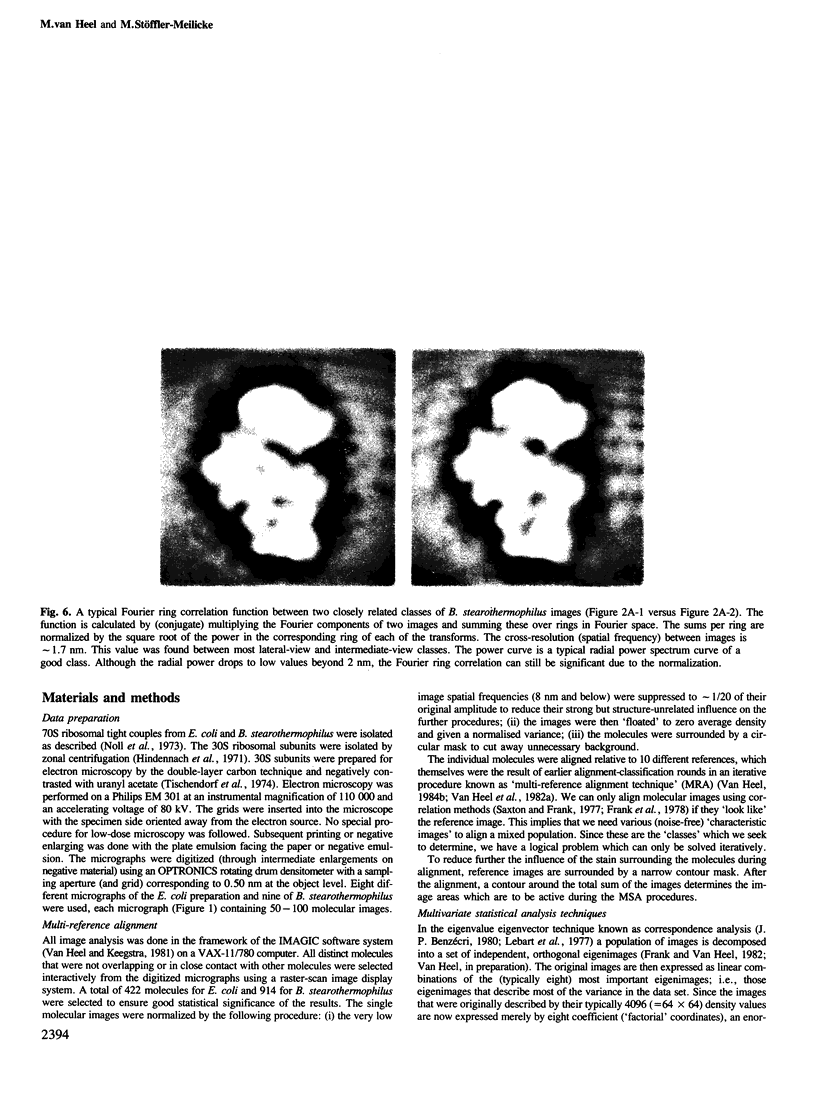

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arad T., Leonard K., Wittmann H. G., Yonath A. Two-dimensional crystalline sheets of Bacillus stearothermophilus 50S ribosomal particles. EMBO J. 1984 Jan;3(1):127–131. doi: 10.1002/j.1460-2075.1984.tb01772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E., Guinto E., Kuhl W., Matsumoto F. Existence of only a single functional pool of adenosine triphosphate in human erythrocytes. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2825–2828. doi: 10.1073/pnas.75.6.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. W., Leonard K., Lake J. A. Ribosomal crystalline arrays of large subunits from Escherichia coli. Science. 1982 May 28;216(4549):999–1001. doi: 10.1126/science.7043735. [DOI] [PubMed] [Google Scholar]
- Frank J., Goldfarb W., Eisenberg D., Baker T. S. Reconstruction of glutamine synthetase using computer averaging. Ultramicroscopy. 1978;3(3):283–290. doi: 10.1016/s0304-3991(78)80038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Verschoor A., Boublik M. Computer averaging of electron micrographs of 40S ribosomal subunits. Science. 1981 Dec 18;214(4527):1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- Frank J., Verschoor A., Boublik M. Multivariate statistical analysis of ribosome electron micrographs. L and R lateral views of the 40 S subunit from HeLa cells. J Mol Biol. 1982 Oct 15;161(1):107–133. doi: 10.1016/0022-2836(82)90281-9. [DOI] [PubMed] [Google Scholar]
- Frank J., van Heel M. Correspondence analysis of aligned images of biological particles. J Mol Biol. 1982 Oct 15;161(1):134–137. doi: 10.1016/0022-2836(82)90282-0. [DOI] [PubMed] [Google Scholar]
- Henderson E., Oakes M., Clark M. W., Lake J. A., Matheson A. T., Zillig W. A new ribosome structure. Science. 1984 Aug 3;225(4661):510–512. doi: 10.1126/science.6429855. [DOI] [PubMed] [Google Scholar]
- Higo K., Held W., Kahan L., Nomura M. Functional correspondence between 30S ribosomal proteins of Escherichia coli and Bacillus stearothermophilus. Proc Natl Acad Sci U S A. 1973 Mar;70(3):944–948. doi: 10.1073/pnas.70.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindennach I., Stöffler G., Wittmann H. G. Ribosomal proteins. Isolation of the proteins from 30S ribosomal subunits of Escherichia coli. Eur J Biochem. 1971 Nov 11;23(1):7–11. doi: 10.1111/j.1432-1033.1971.tb01584.x. [DOI] [PubMed] [Google Scholar]
- Kiselev N. A., Stel'mashchuk VYa, Orlova E. V., Platzer M., Noll F., Bielka H. On the fine structure of rat liver ribosome small subunits. Mol Biol Rep. 1982 Nov 30;8(4):185–189. doi: 10.1007/BF00776578. [DOI] [PubMed] [Google Scholar]
- Lake J. A., Henderson E., Clark M. W., Matheson A. T. Mapping evolution with ribosome structure: intralineage constancy and interlineage variation. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5948–5952. doi: 10.1073/pnas.79.19.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. Ribosome structure determined by electron microscopy of Escherichia coli small subunits, large subunits and monomeric ribosomes. J Mol Biol. 1976 Jul 25;105(1):131–139. doi: 10.1016/0022-2836(76)90200-x. [DOI] [PubMed] [Google Scholar]
- Noll M., Hapke B., Schreier M. H., Noll H. Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J Mol Biol. 1973 Apr 5;75(2):281–294. doi: 10.1016/0022-2836(73)90021-1. [DOI] [PubMed] [Google Scholar]
- Saxton W. O., Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc. 1982 Aug;127(Pt 2):127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Saxton W. O., Frank J. Motif detection in quantum noise-limited electron micrographs by cross-correlation. Ultramicroscopy. 1977 Apr;2(2-3):219–227. doi: 10.1016/s0304-3991(76)91385-1. [DOI] [PubMed] [Google Scholar]
- Shatsky I. N., Mochalova L. V., Kojouharova M. S., Bogdanov A. A., Vasiliev V. D. Localization of the 3' end of Escherichia coli 16 S RNA by electron microscopy of antibody-labelled subunits. J Mol Biol. 1979 Oct 9;133(4):501–515. doi: 10.1016/0022-2836(79)90404-2. [DOI] [PubMed] [Google Scholar]
- Tischendorf G. W., Zeichhardt H., Stöffler G. Architecture of the Escherichia coli ribosome as determined by immune electron microscopy. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4820–4824. doi: 10.1073/pnas.72.12.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischendorf G. W., Zeichhardt H., Stöffler G. Location of proteins S5, S13 and S14 on the surface of the 3oS ribosomal subunit from Escherichia coli as determined by immune electron microscopy. Mol Gen Genet. 1974;134(3):209–223. doi: 10.1007/BF00267716. [DOI] [PubMed] [Google Scholar]
- Unwin P. N., Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol. 1975 May 25;94(3):425–440. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]
- Verschoor A., Frank J., Radermacher M., Wagenknecht T., Boublik M. Three-dimensional reconstruction of the 30 S ribosomal subunit from randomly oriented particles. J Mol Biol. 1984 Sep 25;178(3):677–698. doi: 10.1016/0022-2836(84)90245-6. [DOI] [PubMed] [Google Scholar]
- van Heel M., Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6(2):187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- van Heel M. Multivariate statistical classification of noisy images (randomly oriented biological macromolecules). Ultramicroscopy. 1984;13(1-2):165–183. doi: 10.1016/0304-3991(84)90066-4. [DOI] [PubMed] [Google Scholar]