Abstract
Increased abundance of the prostate-specific membrane antigen (PSMA) on prostate epithelium is a hallmark of advanced metastatic prostate cancer (PCa) and correlates negatively with prognosis. However, direct evidence that PSMA functionally contributes to PCa progression remains elusive. We generated mice bearing PSMA-positive or PSMA-negative PCa by crossing PSMA-deficient mice with transgenic PCa (TRAMP) models, enabling direct assessment of PCa incidence and progression in the presence or absence of PSMA. Compared with PSMA-positive tumors, PSMA-negative tumors were smaller, lower-grade, and more apoptotic with fewer blood vessels, consistent with the recognized proangiogenic function of PSMA. Relative to PSMA-positive tumors, tumors lacking PSMA had less than half the abundance of type 1 insulin-like growth factor receptor (IGF-1R), less activity in the survival pathway mediated by PI3K-AKT signaling, and more activity in the proliferative pathway mediated by MAPK-ERK1/2 signaling. Biochemically, PSMA interacted with the scaffolding protein RACK1, disrupting signaling between the β1 integrin and IGF-1R complex to the MAPK pathway, enabling activation of the AKT pathway instead. Manipulation of PSMA abundance in PCa cell lines recapitulated this signaling pathway switch. Analysis of published databases indicated that IGF-1R abundance, cell proliferation, and expression of transcripts for antiapoptotic markers positively correlated with PSMA abundance in patients, suggesting that this switch may be relevant to human PCa. Our findings suggest that increase in PSMA in prostate tumors contributes to progression by altering normal signal transduction pathways to drive PCa progression and that enhanced signaling through the IGF-1R/β1 integrin axis may occur in other tumors.
INTRODUCTION
Prostate cancer (PCa) is the most commonly diagnosed cancer in men in the United States and the second leading cause of cancer deaths in American men over 50 [after lung cancer (1)]. Although the precise cause of PCa remains unknown, clinical and experimental observations suggest that hormonal, genetic, and environmental factors may each play a role. It is well established that androgens play a crucial role in PCa development. In most of the early cases, PCa can be effectively treated by either surgery or targeted pharmacological manipulation of the androgen receptor (AR) and its target genes (2). Unfortunately, once metastatic disease develops, this therapy is no longer effective, in part related to the loss or mutation of AR in late-stage PCa, suggesting an activation or shift to alternate prosurvival cancer signaling pathways (3). Although the relationship between these signaling pathways most likely varies within different tumors, elucidation of the global molecular mechanisms responsible for this prosurvival switch is necessary to improve treatment strategies for patients with advanced-stage PCa.
One common mechanism promoting the malignant cell behavior involves changes in the activation status of receptor tyrosine kinases (RTKs). The type 1 insulin-like growth factor receptor (IGF-1R) has often been implicated in cancer progression (4, 5), and increased abundance of IGF-1R have been observed in most primary and metastatic prostate tumors (6). Evidence suggests that increased and sustained IGF-1R signaling underlies persistent PCa survival and growth as tumors progress to androgen independence (7). However, results from recent clinical trials targeting the IGF-1R pathway have been disappointing (8), suggesting that a more complex mechanism promotes malignant cell behavior. Alternatively, cross-talk between critical signal transduction pathways and aberrant activation of distinct pathways have both been implicated in cancer promotion (9), typified by the cross-talk between the IGF-1R and the β integrin signaling pathways, which has been implicated in breast cancer (10–21) and PCa (22–26), neuroblastoma (27, 28), and multiple myeloma (29), among others (30). Identification of the shared and unique participants of these pathways and their contribution to disease progression are necessary to improve therapeutic strategies for cancers of numerous etiologies.
PSMA is a 750–amino acid type II transmembrane peptidase enzyme that is encoded by the folate hydrolase 1 (FOLH1) gene. Although PSMA is also known as glutamate carboxypeptidase II, N-acetyl-l-aspartyl-l-glutamate peptidase I, and N-acetylaspartylglutamate peptidase, those studying PCa or general oncology commonly use the term PSMA, which will be used here. It has been shown that PSMA is present in low amounts on prostate epithelial cells and is progressively up-regulated during disease progression in prostate tumors, in which it correlates negatively with prognosis (31–33) and consequently may be a promising tool for the diagnosis, detection, localization, and treatment of PCa. Currently, PSMA is used as an immunoscintigraphic target in the clinic to direct therapy to androgen-independent prostate tumors. RNA aptamers selectively targeting PSMA enzymatic activity have also been successful in slowing primary tumor growth in murine models (34, 35). Although we have previously shown that endothelial-expressed PSMA regulates angiogenesis (36, 37) and retinal neovascularization (38) primarily via β1 integrin–mediated cell adhesion, an important functional role for PSMA in PCa has not been demonstrated. To directly investigate the role of PSMA in PCa progression, we crossed FOLH1 global knockout mice (hereafter called PSMA knockout) (39) with the well-characterized transgenic adenocarcinoma of the mouse prostate (TRAMP) murine model (40, 41) to study PCa tumor progression in the absence of PSMA expression.
Here, we report that expression of PSMA in prostatic epithelial cells directly underlies prostate tumor progression in vivo. We found that tumors in wild-type animals were larger and of higher grade with a higher microvessel density as compared to tumors in the PSMA knockout animals, which is consistent with our previous results implicating PSMA as an angiogenic regulator (36, 37, 42). In addition, PSMA-positive tumor cells were viable at greater distances from the vasculature than their PSMA knockout counterparts, suggesting that cell-intrinsic survival components also contribute to tumor growth. Accordingly, wild-type tumors expressed relatively greater amounts of IGF-1R and exhibited greater activation of the phosphatidylinositol 3-kinase (PI3K)–AKT pathway, whereas tumors lacking PSMA not only had decreased IGF-1R expression but also had diverted signaling downstream of PI3K-AKT to the mitogen-activated protein kinase (MAPK)–extracellular signal–regulated kinases 1 and 2 (ERK1/2) pathway, consistent with a PSMA-dependent signaling switch. Moreover, manipulation of PSMA expression in mouse TRAMP-C1 cell lines and human PCa cell lines recapitulated this change in signaling. Analysis of publically available gene expression data sets from PCa samples confirmed that high PSMA expression was predictive of a high Gleason score. In addition, patient samples with high PSMA expression and high Gleason scores displayed a prosurvival gene expression signature with increased expression of the antiapoptotic marker survivin and IGF-1R, consistent with a role for PSMA in the regulation of signal transduction in human PCa disease as well. Therefore, in addition to its role as a PCa marker and target, our results indicate that increasing amounts of PSMA in prostate tumor epithelium serve to drive prosurvival mechanisms and thus identify it as a functional regulator of prostate tumor progression. These findings also suggest that PSMA-positive tumors may be more sensitive to PI3K pathway inhibitors and less sensitive to MAPK pathway inhibitors.
RESULTS
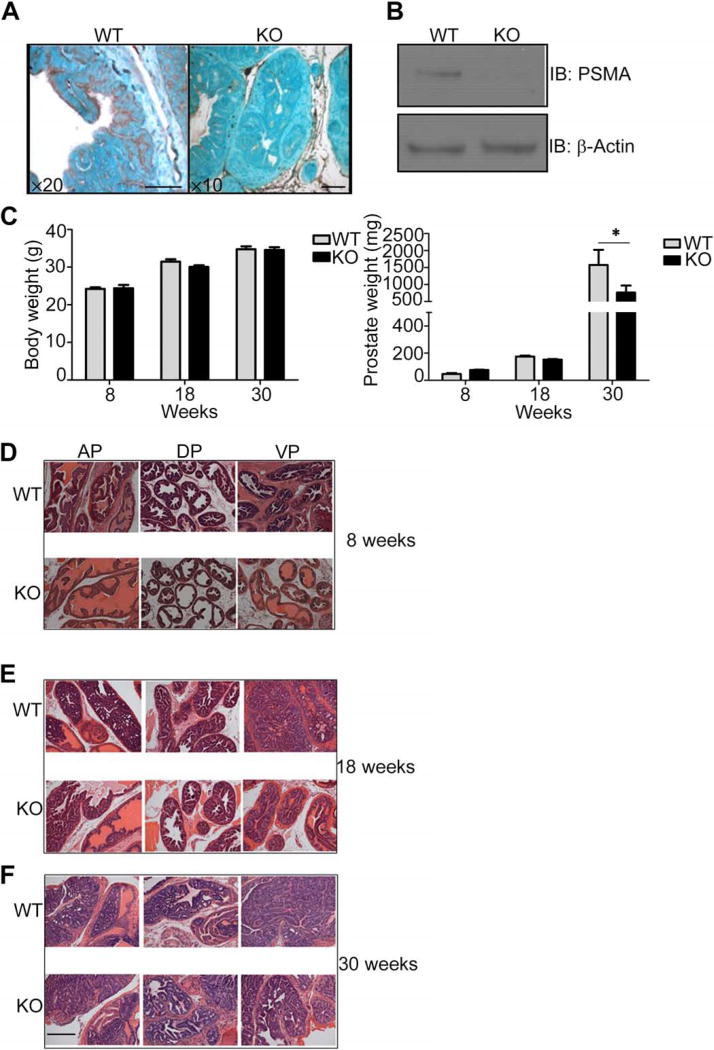
Previous studies of PSMA in PCa progression have primarily involved manipulation of PSMA expression in established tumor cell lines. To genetically address the contribution of PSMA to PCa development and progression, we crossed PSMA knockout mice with TRAMP transgenic mice. The TRAMP murine model of PCa has been extensively characterized and closely mimics PCa progression seen in patients from onset of hyperplasia to adenoma and, eventually, to adenocarcinoma (40, 41). TRAMP mice uniformly and spontaneously develop autochthonous prostate tumors after the onset of puberty consequent to the expression of the SV40 T antigen from the rat probasin promoter (41, 43). Disease onset occurs by 8 weeks of age, adenoma occurs by 18 weeks, and most tumors are poorly differentiated and highly invasive by 30 weeks (40, 41). Considered within the range of their validity, TRAMP mouse models have been predictive of clinical outcome. To directly assess the contribution of PSMA to PCa initiation and progression, we initially determined that PSMA was expressed in tumors from the TRAMP model. F2 progeny of PSMA knockout or wild-type mice bred to TRAMP transgenic animals produced tumors that were either positive or negative for PSMA expression (Fig. 1A). Western blot analysis of tumor lysates derived from PSMA knockout animals verified the complete loss of PSMA protein expression (Fig. 1B). Analysis of body and total prostate weights of wild-type and PSMA knockout animals at the established time points of 8, 18, and 30 weeks of age showed that although overall body weight at 18 weeks of age was significantly different between genotypes, this discrepancy was not seen at 8, 30, or >33 weeks of age (Fig. 1C). Conversely, the total prostate weights of wild-type mice were substantially higher than those of the PSMA knockout mice older than 30 weeks.
Fig. 1. Characterization of tumors from WT and PSMA KO TRAMP mice.
(A) Immunohistological characterization of PSMA distribution (brown strain) in prostate tumors from 18-week-old PSMA wild-type (WT) or knockout (KO) TRAMP mice. Tissues were counterstained with methyl green. Magnification, ×20 (WT) and ×10 (KO); scale bars, 100 µm. (B) Western blot (IB) analysis of PSMA abundance (monoclonal antibody 3E2) in whole prostate tumor lysates from 18-week-old PSMA WT or KO TRAMP mice. (C) Total body and prostate weights in WT and PSMA KO TRAMP mice at 8,18, and 30 weeks of age. Data are means ± SE from 10 WT and 10 KO mice per group. *P < 0.05, paired Student’s t test. (D to F) Representative images of H&E-stained sections of anterior prostate (AP), ventral prostate (VP), and dorsal prostate (DP) lobes from PSMA WT and PSMA KO mice at 8 (D), 18 (E), and 30 (F) weeks of age. n = 30 for each experimental group. Magnification, ×25; scale bar, 100 µm.
Considering the differences between mouse and human prostate tissue and the differences in rates at which lesions can progress in each lobe of the prostate (44), we assessed the dorsal, ventral, and anterior lobes separately at each time point using a double-blind, numerical scoring system based on histologically distinguishable patterns of growth and disease (45). Briefly, on the basis of a histological grading scheme for TRAMP mice developed by Suttie et al. (45), hematoxylin and eosin (H&E)–stained tissues were graded as hyperplasia (grades 1 to 3), adenoma (grades 4 to 5), or adenocarcinoma (grade 6). Within each grade, a distribution was assigned as focal, multifocal, or diffuse (Table 1).
Table 1. Grade and distribution of prostate lesions.
Histopathological categorization of H&E-stained prostatic lobes from WT and PSMA KO tumors. Number of lesions in each lobe are assigned by grade [hyperplasia (grades 2 to 3), adenoma (grades 4 to 5), or adenocarcinoma (grade 6)] and distribution (focal, multifocal, or diffuse).
| Treatment group and lobe |
Grade 2
|
Grade 3
|
Grade 4
|
Grade 5
|
Grade 6
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Focal | Multifocal | Diffuse | Focal | Multifocal | Diffuse | Focal | Multifocal | Diffuse | Focal | Multifocal | Diffuse | Focal | Multifocal | Diffuse | |
| Anterior lobe | |||||||||||||||
| Week 8 | |||||||||||||||
| WT (n = 26) | 3 | 8 | 3 | 7 | 5 | ||||||||||
|
| |||||||||||||||
| KO (n = 19) | 4 | 7 | 3 | 2 | 2 | ||||||||||
|
| |||||||||||||||
| Week 18 | |||||||||||||||
| WT (n = 27) | 3 | 2 | 8 | 5 | 5 | 1 | 3 | ||||||||
|
| |||||||||||||||
| KO (n = 23) | 7 | 8 | 4 | 4 | |||||||||||
|
| |||||||||||||||
| Week 30 | |||||||||||||||
| WT (n = 16) | 4 | 3 | 5 | 2 | 2 | ||||||||||
|
| |||||||||||||||
| KO (n = 16) | 1 | 2 | 4 | 4 | 2 | 2 | |||||||||
|
| |||||||||||||||
| Dorsal lobe | |||||||||||||||
| Week 8 | |||||||||||||||
| WT (n = 27) | 7 | 12 | 5 | 2 | 1 | ||||||||||
|
| |||||||||||||||
| KO (n = 21) | 3 | 5 | 3 | 7 | 2 | 1 | |||||||||
|
| |||||||||||||||
| Week 18 | |||||||||||||||
| WT (n = 27) | 1 | 13 | 9 | 2 | 22 | ||||||||||
|
| |||||||||||||||
| KO (n = 23) | 1 | 8 | 6 | 5 | 3 | ||||||||||
|
| |||||||||||||||
| Week 30 | |||||||||||||||
| WT (n = 16) | 4 | 3 | 5 | 2 | |||||||||||
|
| |||||||||||||||
| KO (n = 17) | 2 | 4 | 2 | 1 | 2 | 4 | |||||||||
|
| |||||||||||||||
| Ventral lobe | |||||||||||||||
| Week 8 | |||||||||||||||
| WT (n = 29) | 2 | 3 | 2 | 6 | 13 | 1 | 2 | ||||||||
|
| |||||||||||||||
| KO (n = 21) | 3 | 5 | 3 | 7 | 2 | 1 | |||||||||
|
| |||||||||||||||
| Week 18 | |||||||||||||||
| WT (n = 27) | 1 | 3 | 7 | 8 | 8 | ||||||||||
|
| |||||||||||||||
| KO (n = 23) | 3 | 14 | 4 | 1 | 1 | ||||||||||
|
| |||||||||||||||
| Week 30 | |||||||||||||||
| WT (n = 18) | 2 | 3 | 4 | 2 | 1 | 4 | 1 | 1 | |||||||
|
| |||||||||||||||
| KO (n = 19) | 1 | 6 | 3 | 1 | 2 | 3 | 1 | 2 | |||||||
At 8 weeks of age (the initial time point), in the anterior lobe, the minimum lesion grade was the same for both the wild-type and PSMA knockout tumors (grade 2, focal), and the most severe grade was higher in the wild-type mouse tumor (grade 3, diffuse). In the dorsal lobe, the highest grade observed (grade 4, diffuse) was once again in the wild-type tumor, and the lowest grade (grade 2, multifocal) was observed in the PSMA knockout tumor. Finally, in the ventral lobe, the highest grade was seen in the wild-type tumor (grade 4, multifocal), and the lowest grade observed was identical for both experimental conditions (Fig. 1D).
At 18 weeks of age (the intermediate time point), in the anterior lobe, although the minimum lesion grade was the same for the wild type and PSMA knockout (grade 3, focal), the most severe grade was significantly higher in the wild-type mouse tumor (grade 5, multifocal) than in the PSMA knockout (grade 4, focal). In the dorsal lobe, the minimum lesion grade was slightly higher in the wild type (grade 3, diffuse) than in the PSMA knockout (grade 3, multifocal). However, the maximum grade was significantly higher in the wild type (grade 5, multifocal) than in the PSMA knockout (grade 4 diffuse). Finally, in the ventral lobe, the maximum and minimum grades were identical for both conditions; however, the distribution of the grades was significantly different with most wild-type tumors having a higher grade (Fig. 1E).
At 30 weeks of age (the final time point), in the anterior lobe, the minimum grade of the wild type (grade 4, focal) remained slightly higher than that of the PSMA knockout (grade 3, diffuse). However, the highest grade was identical for both conditions (grade 6, diffuse). In both the dorsal lobe and anterior lobe, the maximum and minimum grades were identical (Fig. 1F).
To resolve the difference in grade and distribution between the wild type and PSMA knockout at the 8, 18, and 30 weeks of age, the data were combined and represented by a “distribution-adjusted lesion grade,” which was calculated by assigning a number to each successive rank (for example, 0 = normal; 1 = grade 1, focal; 2 = grade 2, multifocal; … and so on, whereby 18 = grade 6, diffuse) (Table 2). Proliferative lesions were present in all mice by 8 weeks; however, the PSMA knockout clearly demonstrated less severe pathology at 8 and 18 weeks. There was no statistical difference in tumor score between wild-type and PSMA knockout tumors by 30 weeks, likely indicating that the tumors had progressed to a point beyond the ability to distinguish any differences. Therefore, we chose to focus our subsequent experiments on whole prostate tissues at the time point of 18 weeks of age.
Table 2. Prostate mean distribution–adjusted lesion grades.
The mean distribution–adjusted lesion grades for the WT and PSMA KO groups from Table 1 are summarized here. Data are means ± SE.
| Lobe | Age in weeks | WT | KO |
|---|---|---|---|
| Anterior | 8 | 6.8 ± 0.20 | 5.7 ± 0.16 |
| 18 | 10.4 ± 0.21 | 8.1 ± 0.17 | |
| 30 | 12.2 ± 0.26 | 13.3 ± 0.04* | |
| Dorsal | 8 | 8.5 ± 0.16 | 7.3 ± 0.03* |
| 18 | 11.7 ± 0.07 | 10.1 ± 0.05* | |
| 30 | 13.3 ± 0.36 | 13.6 ± 0.21 | |
| Ventral | 8 | 7.3 ± 0.15 | 6.4 ± 0.07 |
| 18 | 10.7 ± 0.10 | 9.1 ± 0.03* | |
| 30 | 12.3 ± 0.12 | 13.4 ± 0.15 |
P < 0.05.
To quantitatively confirm our observation that the lack of PSMA delayed tumor development, we measured expression of established markers of cancer progression. Survivin is a member of the inhibitor of apoptosis family that is highly abundant in most solid tumors where it supports tumor cell survival but is absent in normal, nonmalignant cells (46). Relevant to our study, increased survivin abundance has been associated with resistance to antiandrogen therapy in advanced PCa (46). Survivin prevents the proteolytic cleavage of procaspase-3 to its activated form to inhibit apoptosis, and thus, a reduction in full-length caspase-3 levels is an independent marker of tumor progression (46). Assessment of the 18-week-old tumor lysates showed significantly increased abundance of survivin (Fig. 2A) and concomitantly decreased cleaved caspase-3 (Fig. 2B) in the wild-type prostate tumors, consistent with increased disease progression and supporting our observation that PSMA promotes a more aggressive PCa phenotype.
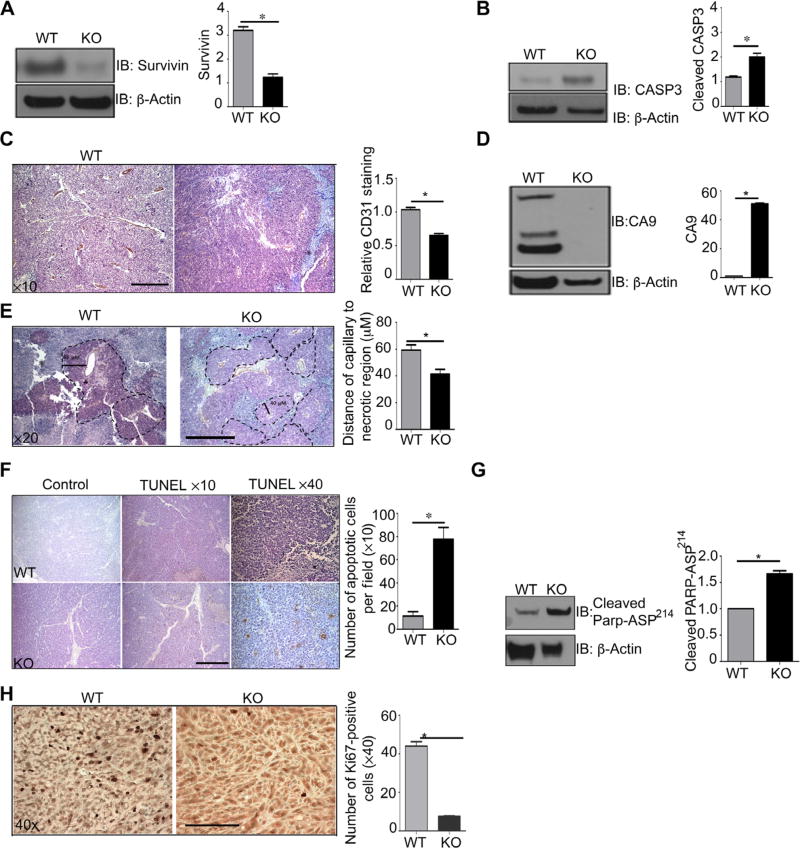
Fig. 2. PSMA WT prostate tumors display a more vascularized, hypoxia-tolerant, antiapoptotic phenotype.
(A and B) Western blotting for survivin and caspase-3 (CASP3) in whole prostate tumor lysates from 18-week-old PSMA WT and KO TRAMP mice. (C) Immunohistochemical analysis of tumor vasculature by CD31 staining, quantified for CD31+ pixel area sum from five nonoverlapping fields per sample using Image-Pro Plus software (hematoxylin counterstain). Scale bar, 50 µm. (D) Western blot analysis for CA9, a marker of hypoxia, in whole prostate tumor lysate from 18-week-old PSMA WT and KO mouse tumors. (E) Thickness of viable layer of tumor cells from CD31-stained capillaries. PSMA WT (65 µM) and KO (40 µM). Tissue was counterstained with hematoxylin, and images are shown at ×20 magnification. Scale bar, 100 µm. (F) Representative TUNEL stain to detect apoptotic cells (brown) in PSMA WT and KO tumor sections from 18-week-old mice. Sections were counterstained with hematoxylin. Analysis is representative of the number of apoptotic cells per field. Images are shown at ×10 and ×40 magnifications. Scale bar, 50 µm. (G) Western blot anaylis of cleaved PARP Asp214, a marker of apoptosis, in PSMA WT tumors compared to PSMA KO tumors. (H) Immunohistochemical analysis of cell proliferation (Ki67 staining) in PSMA WT and PSMA KO tumor sections. Scale bar, 100 µm. All Western blots were normalized to β-actin and are presented as fold change relative to WT. Data are means ± SE from n = 4 animals for each experimental group, with at least three experimental replicates. *P < 0.05, paired Student’s t test.
We have previously shown that PSMA regulates endothelial activation in angiogenesis (36) and in retinal neovascularization (38). To assess the effects of PSMA on the formation of tumor vasculature, we quantified endothelial-specific CD31 abundance by immunohistochemistry in the 18-week-old tumors and saw a significant decrease in CD31 staining in prostate tumors from PSMA knockout mice in agreement with our previous studies (Fig. 2C). However, a closer examination revealed marked structural and morphological differences in vessel structure between genotypes. Vessels in wild-type tumors were irregularly branched, tortuous, random, and dilated, a phenotype consistent with tumor vessel angiogenesis. By contrast, vessels in tumors lacking PSMA appeared more regular and organized, a phenotype referred to as normalized, suggesting that PSMA also contributes to the dysregulated vessel growth characteristic of tumor angiogenesis. Tumor vessel “normalization” results in better perfusion, reduced hypoxia, and presumably increased tumor growth (47), which is in contrast to our findings. To functionally confirm that vasculature lacking PSMA is more normalized, we assessed relative hypoxia levels by measuring abundance of the enzyme carbonic anhydrase IX (CA9), which is strongly induced in response to hypoxia, particularly in hypoxic solid tumors that are refractory to conventional therapies (48). Consistent with our observation of more normalized blood vessels, lysates from PSMA knockout tumors exhibited significantly lower amounts of CA9 (Fig. 2D), indicating that these tumors are indeed less hypoxic than their wild-type counterparts. Further histologic examination indicated that both wild-type and PSMA knockout tumors contained defined areas of viable cells immediately adjacent to capillaries surrounded by a perimeter of necrotic cells (granular area). However, the viable cell area (measured as the distance from the capillary to the necrotic region) was more than 30% greater in the wild-type versus PSMA knockout tumors (Fig. 2E). Thus, wild-type cells apparently are able to remain viable at oxygen concentrations that are toxic for normal cells and can survive at greater distances from the vasculature than cells lacking PSMA despite increased tissue hypoxia. TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) staining indicated that cell death by apoptosis was markedly less in the wild-type compared to the PSMA knockout tumors (Fig. 2F), which is supported by an increased amount of cleaved poly(adenosine 5′-diphosphate-ribose) polymerase (PARP) Asp241, a marker of apoptosis (Fig. 2G) (9). Consequently, immunohistochemical analysis to assess cell proliferation exhibited decreased abundance of the cell proliferation marker Ki67 in PSMA knockout tumors (Fig. 2H). Together, these observations indicate that although vessels are more normalized and tumor tissues are less hypoxic in the absence of PSMA, PSMA expression in tumor cells appears to confer an intrinsic survival advantage that is more beneficial to tumor growth than a favorable vessel phenotype.
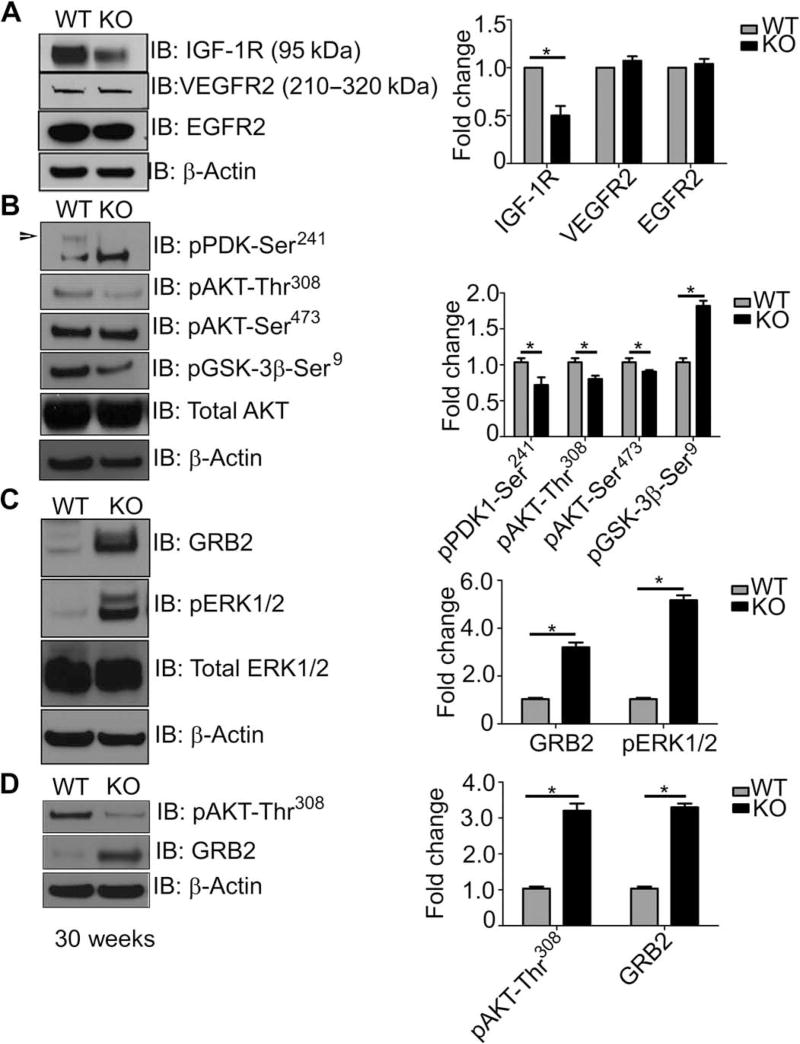
Tumor cells often escape apoptosis by modifying the expression or activation status of RTKs to induce hyperactivation of prosurvival signal transduction pathways and preserve cell integrity (49). To determine whether the presence of PSMA in advanced PCa may induce similar prosurvival mechanisms, we assayed the activation status of a panel of RTKs that are commonly altered in solid tumors (Fig. 3A) (50). Whereas protein abundance of the RTKs VEGFR2 (vascular endothelial growth factor receptor 2) and EGFR2 (epidermal growth factor receptor 2) was unchanged in both the wild-type and PSMA knockout PCa tumors, wild-type PCa tumors displayed higher amounts of total IGF-1R, consistent with our prosurvival phenotype. In addition, elements of the PI3K-AKT prosurvival pathway PDK1-Ser241, and AKT-Thr308 were stimulated and amounts of GSK-3β-Ser309 were decreased (Fig. 3B). Surprisingly, signal transduction in the PSMA knockout prostate tumors was completely reversed, where the MAPK-ERK1/2 pathway is clearly enhanced, as illustrated by increased growth factor receptor–bound protein 2 (GRB2) adaptor protein levels and ERK phosphorylation in PSMA knockout tumors compared to wild type (Fig. 3C).
Fig. 3. PSMA within the prostate tumor epithelium shifts cell signaling from an “active GRB2-ERK1/2 pathway–inactive PI3K-AKT pathway” state to an “active PI3K-AKT pathway–inactive GRB2-ERK1/2 pathway” state.
(A to C) Western blotting of whole prostate tumor lysates from 18-week-old PSMA WT or KO TRAMP mice for various RTKs (A), AKT pathway markers (B), and MAPK pathway markers (C). (D) Blotting for the indicated markers in tumors from 30-week-old PSMA WT and KO mice. Blots are representative of three experiments from n = 3 WT and 3 KO mice, normalized to β-actin and presented as fold change relative to WT. *P < 0.05, paired Student’s t test.
As a relatively slow-growing tumor, primary PCa can be present for years before detection, and latency to metastasis can also be quite protracted. However, a percentage of patients present with a more rapidly growing, aggressive form of PCa that has a markedly worse diagnosis. We observed that tumor progression and growth are delayed in PSMA knockout animals, although tumors from wild-type and PSMA knockout animals are histologically indistinguishable at the extended 30-week time point, suggesting either that the pathway switch we see at 18 weeks of age is eventually reversed, that a small percentage of cells do not switch and eventually outgrow the slower growing population, or that tumors with low levels of PSMA may correlate with the slower, less aggressive tumor type. To determine the signaling status of PSMA knockout tumors at 30 weeks of age, we assessed activation levels of the AKT versus ERK signaling pathways. Western blot analysis determined that AKT phosphorylation Thr308 is maintained in the long term in PSMA wild-type tumors but not in PSMA knockout (Fig. 3D), consistent with a permanent pathway switch in the absence of PSMA and perhaps indicating that tumors presenting with low PSMA abundance may be a marker of the slower, less aggressive form of PCa.
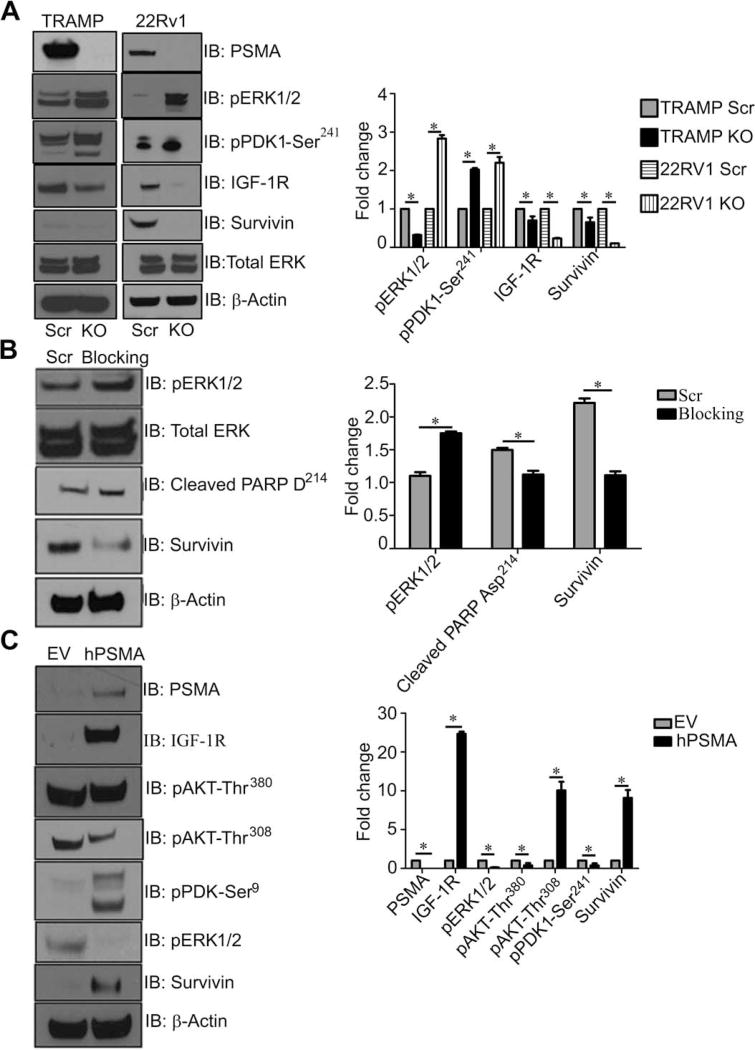
To confirm that this pathway switch is strictly dependent on PSMA, we functionally manipulated PSMA by CRISPR (clustered regularly interspaced short palindromic repeats) deletion in both the human 22Rv1 and murine TRAMP-C1 cell lines (Fig. 4A and figs. S1 and S2) or small interfering RNA (siRNA) knockdown or by disrupting intracellular interactions with a PSMA-specific N-terminal blocking peptide in TRAMP-C1 cells (fig. S3) (36), which consistently resulted in effects on signaling via the AKT pathway (Fig. 4, A and B). Overexpression of human PSMA in the PSMA knockout human PC-3 PCa cell line altered signaling pathways similar to that we observed in wild-type tumors, demonstrating that these mechanisms are operative in human PCa (Fig. 4C). Together, our data suggest that in both the in vivo murine TRAMP model and the in vitro tumor cell lines, PSMA directly contributes to tumor progression by affecting IGF-1R signal transduction to result in a signaling pathway switch that promotes tumor survival, growth, and progression.
Fig. 4. Manipulation of PSMA in both mouse TRAMP-C1 and human 22RV1 and PC-3 PCa cell lines mimic pathway switch.
(A) CRISPR knockdown of PSMA in both the mouse TRAMP-C1 cell line (TRAMP-PSMAKO) and the human 22Rv1 cell line (22RV1-PSMAKO). Western blot analysis of both CRISPR cell lines to examine changes in PDK-Ser241, IGF-1R, survivin, and pERK1/2 compared to controls (Scr). (B) TRAMP-C1 cells transiently transfected with peptides blocking the PSMA NH2-terminal cytoplasmic tail. Western blot at 24 hours for pERK1/2, cleaved PARP Asp214, and survivin at 24 hours compared to the control (Scr). (C) Western blot analysis of PC-3 cells expressing human PSMA (hPSMA) and empty vector (EV) control cell lysates to investigate changes in the previously identified signaling pathways (PSMA, IGF-1R, AKT-Ser380, AKT-Thr308, PDK1-Ser241, pERK1/2, survivin, and β-actin). All data are representative images from the mean ± SE of n = 3 for each experimental condition and three experimental replicates normalized to β-actin and presented as fold change, where WT is equal to 1. *P < 0.05, paired Student’s t test.
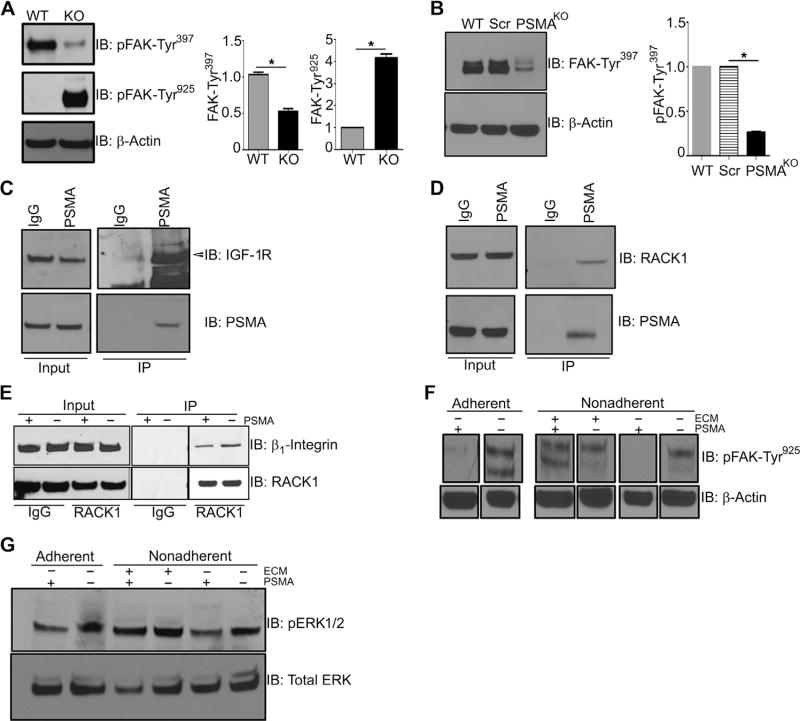
Aberrant or disrupted interactions between components of signaling pathways are frequently responsible for alterations in downstream signaling pathways during tumorigenesis (51). One such example is cooperation between IGF-1R and β1 integrin that regulates cancer cell growth, survival, and invasion (52). We have previously shown that PSMA regulates angiogenesis by modulating β1 integrin-dependent focal adhesion kinase (FAK)–Tyr925 phosphorylation and signal transduction in endothelial cells (36, 37), and here, we demonstrate that PSMA affects IGF-1R signaling pathways in PCa cell lines (Fig. 5), suggesting that PSMA may affect IGF-1R/β1 integrin/FAK cross-talk. To further investigate PSMA in this capacity, we examined the aspects of the complex containing scaffolding protein RACK1 (receptor for activated C kinases) that facilitates cooperation between β1 integrin and RTKs (18, 19, 21, 53). Stable IGF-1R/RACK1/β1 integrin complex formation leads to activation of the ERK pathway, and disruption of this complex stimulates the AKT survival pathway (53–56), analogous to the PSMA-dependent pathway switch. Initial evaluation of potential PSMA effects on FAK status in PCa cells showed that whereas FAK is markedly phosphorylated on FAK-Tyr397 in the wild-type 22Rv1 cell lines and tumors, elimination of PSMA expression in 22Rv1-Crispr-PSMAknockout cells or PSMA knockout tumors clearly switches FAK tyrosine phosphorylation to FAK-Tyr925 (Fig. 5, A and B). This result confirms a role for PSMA in integrin activation in PCa cells and is consistent with observations of FAK-Tyr397 hyperphosphorylation in cells where the IGF-1R/RACK1/β1 integrin complex is disrupted by IGF-1R mutation (21). Furthermore, immunoprecipitation of PSMA followed by Western blot analysis demonstrated that PSMA physically associates with RACK1 and IGF-1R in prostate cell lysates (Fig. 5, C and D) and that the association between RACK1 and β1 integrin is increased when PSMA is deleted (Fig. 5E). In further support of this notion, ligation of integrins by the addition of fibronectin, which is reported to enhance complex formation and FAK-Tyr925 activation in breast cancer cells (18), leads to increased FAK-Tyr925 and ERK activation in control 22Rv1 PCa cells expressing PSMA (Fig. 5, F and G). Together, these results support a model where the progressive up-regulation of PSMA expression in PCa tumors serves to alter the IGF-1R/RACK1/β1 integrin complex, leading to alterations in FAK phosphorylation, AKT activation, and enhanced PCa progression (Fig. 6). The dependence of TRAMP tumors on the AKT pathway is supported by studies in which systemic treatment of TRAMP transgenic mice with AKT inhibitors invariably inhibits tumor growth and progression (57–61).
Fig. 5. PSMA expression markedly alters FAK phosphorylation, and PSMA is in a complex with RACK1 and IGF-1R.
(A) Western analysis for FAK-Tyr397 and FAK-Tyr925 in tumor lysates from 18-week-old WT and KO mice. (B) Western blot analysis for FAK-Tyr397 in parental WT, control scramble 22Rv1 cells (Scr), and 22Rv1-PSMAKO cells. (C) Immunoprecipitation (IP) of PSMA in WT 22Rv1 cells using PSMA monoclonal antibody or rabbit immunoglobulin G (IgG) control and Western blot for IGF-1R (arrow indicates band). Input refers to unbound fraction. (D) Immunoprecipitation of PSMA in WT 22Rv1 cells using PSMA rabbit monoclonal antibody or rabbit IgG control and Western blot for RACK1. (E) Immunoprecipitation of RACK1 in both 22Rv1-PSMAScr and 22Rv-PSMAKO cells and Western blot analysis for β1 integrin. (F and G) Both 22Rv1-PSMAScr and 22Rv-PSMAKO cells were put either not in suspension (adherent) or in suspension (nonadherent) for 2 hours and assayed by Western blot for the direct activation of FAK-Tyr925 (F) and ERK (G) by addition of extracellular matrix (ECM) for 30 min. All data are representative images from the means ± SE of n = 3 for each experimental conditions and three experimental replicates normalized to β-actin and presented as fold change, where WT is equal to 1. *P < 0.05, paired Student’s t test.
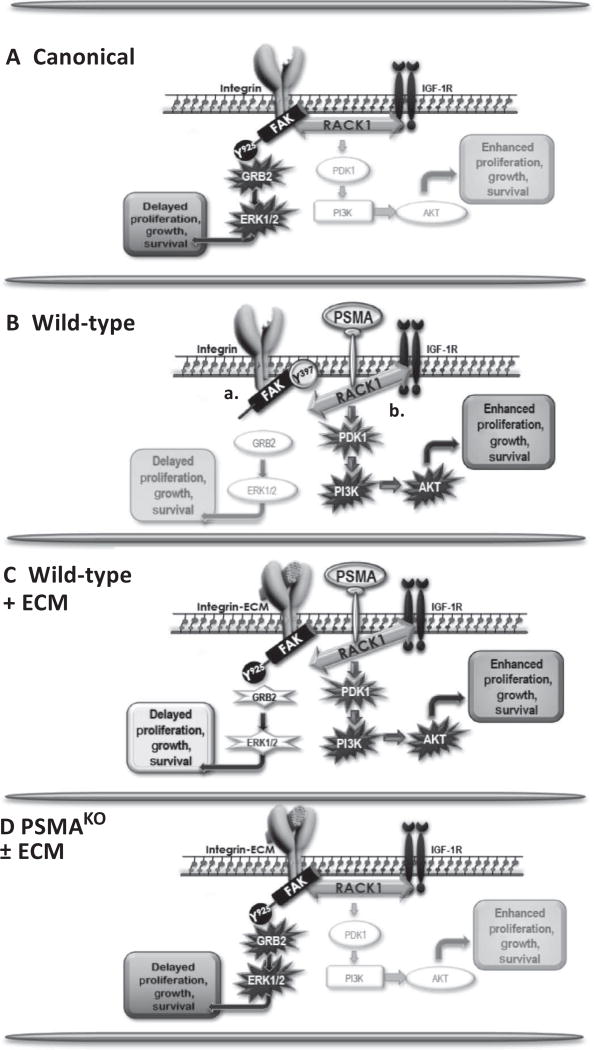
Fig. 6. Schematic of PSMA regulation of PCa signaling.
(A) In the canonical pathway, a stable scaffolding complex containing β1 integrin, RACK1, and IGF-1R activates the FAK-Tyr925/GRB2/ERK pathway, leading to tumor cell proliferation, growth, and migration. (B) In WT cells, PSMA expression disrupts two pathways, both of which contribute to activation of the more aggressive, protumor PI3K-AKT pathway. PSMA physically associates with IGF-1R and RACK1, consistent with PSMA disrupting the scaffolding complex, which leads to FAK-Tyr397 hyperphosphorylation (“a”), and PSMA expression reverses the phosphorylation of FAK-Tyr925, which is required for FAK/GRB2 interactions, thus inactivating ERK signal transduction (“b”). (C) Bypassing PSMA interference by directly activating FAK-Tyr925 with extracellular matrix (ECM) rescues activation of FAK-Tyr925 and the GRB2/ERK signaling pathway. (D) The absence of PSMA allows stable IGF-1R/RACK1/β1 integrin complex formation and activation of the FAK-GRB2-ERK canonical pathway, producing less aggressive tumors.
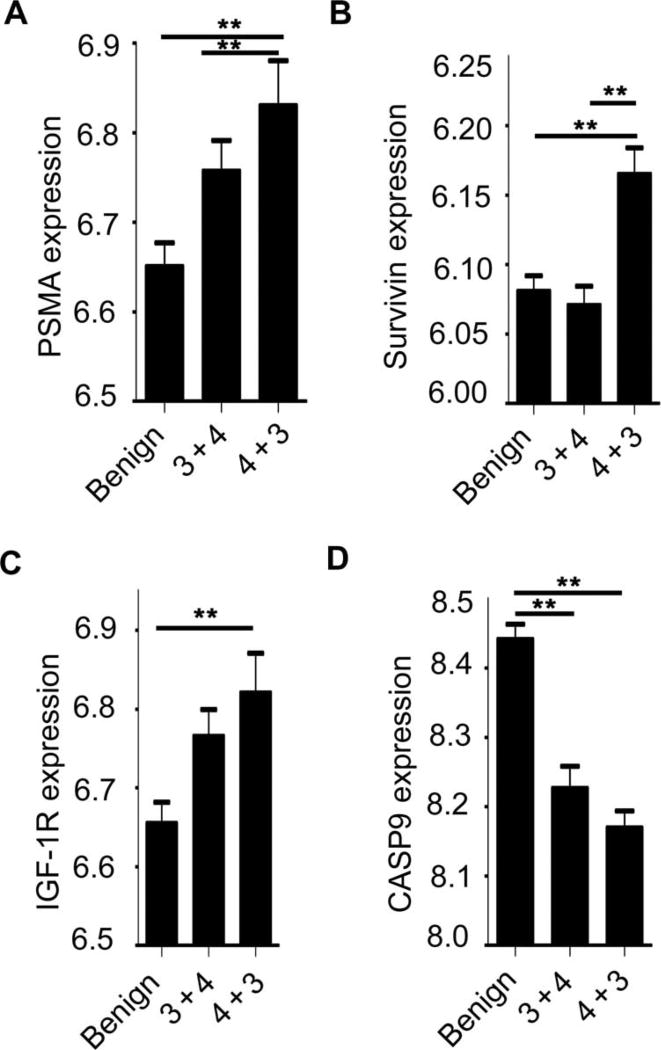
To further investigate the potential link between PSMA expression levels and tumor progression in human PCa, we examined publically available PCa gene expression data sets [National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo/) and SEEK (http://seek.princeton.edu)] to determine (i) whether PSMA abundance is predictive of increased Gleason score and (ii) whether samples with high PSMA and Gleason score display a more prosurvival gene expression phenotype, as we observed in mouse tumors (Fig. 7). A transcriptional analysis of 59 PCa and 39 matched benign tissue samples in the NCBI GEO data set GSE32571 compared tumor samples with a higher Gleason score (4 + 3 and higher) against those with a lower score (3 + 4 and below). Further analysis using GEO2R (data file S1) showed that high PSMA abundance was significantly associated with a higher Gleason score when compared to benign tissue (Fig. 7, A and B). Furthermore, changes in IGF-1R, survivin, and caspase-9, which is involved in the activation of caspases responsible for the initiation of apoptosis, support our murine data (Fig. 7, C and D). Together, these observations support the translational relevance of our findings regarding the role of PSMA in PCa progression and highlight that the identification of novel oncogenic and druggable pathways in patient subgroups with poor prognosis may promote the development of tumor-specific, targeted therapeutic approaches.
Fig. 7. PSMA levels in human prostate tumors are significantly associated with an increase in Gleason score.
(A) GEO data set GSE32571 analyzed 59 PCa and 39 matched benign tissue samples for transcriptional differences between tumor samples with higher Gleason score (4 + 3 and higher) against samples of lower Gleason score (3 + 4 and lower). High PSMA gene expression is concurrent with high Gleason score in human tumor samples. (B and C) Abundances of survivin and IGF-1R are high in more aggressive tumors. (D) Abundance of caspase-9 (CASP9) decreases with tumor aggressiveness. Subgroup labels are along the bottom of the chart. Benign: n = 39; “3 + 4”: tumors with a low Gleason score, n = 32; “4 + 3”: tumors with a high Gleason score, n = 27. Groups that reach **P < 0.005 are indicated on the graph. Gene expression on the y axis is presented as the relative expression of the gene of interest compared to all the other genes in the array. A distribution analysis of all selected samples determined that all selected samples were suitable for comparison. Data analysis was completed using GEO2R, for which the R script is provided in data file S1.
DISCUSSION
The striking up-regulation of PSMA expression during PCa progression and its correlation with patient outcome have been recognized for many years (62). Numerous immunohistochemical investigations have demonstrated extensive PSMA expression in prostate adenocarcinomas and metastases (63–65), but its functional contribution to any aspect of PCa has remained enigmatic. Studies manipulating PSMA concentrations in PCa cell lines have produced contradictory results; one determined that the enzymatic activity of PSMA inhibited PCa cell invasion (66), whereas others found that PSMA expression increased PCa cell invasion in vitro (67). Neither study addressed the underlying mechanism. An interesting in vivo model that used transplanted prostate tissue from PSMA-overexpressing transgenic mice found that PSMA expression correlated with the development of small atypical glands representative of adenocarcinoma, but these PIN-like lesions never progressed to cancer and, again, molecular mechanism was not investigated (68). Here, we directly address whether the increased abundance of PSMA in advanced prostate tumors is indicative of a functional role for this cell surface peptidase in PCa progression. Using the well-characterized TRAMP model of PCa (69) and global PSMA knockout mice (39), we demonstrate that PSMA promotes dysregulation of IGF-1R signal transduction, resulting in a shift in downstream signal transduction pathways that promote PCa tumor progression in vivo.
The IGF-1R has emerged as a molecule with important roles in cancer biology (7) and is overexpressed in a high percentage of primary and metastatic tumors, including PCa (4, 5, 7, 70). Most of the studies on IGF-1R signaling have focused on its role in the differential activation of two important intracellular signaling pathways: the MAPK-ERK pathway, which regulates cell proliferation, differentiation, and tissue homeostasis, or the PI3K-AKT cascade, which promotes cell survival, metabolism, antiapoptosis, and differentiation (6, 71). It is known that ligand binding to IGF-1R triggers the phosphorylation of multiple tyrosine sites within the IGF-1R activation loop, creating a docking site for the signaling molecules IRS-1 and Shc. These interactions subsequently activate the PI3K-AKT and MAPK-ERK cascades through IGF-1R recruitment of either the P85 subunit of PI3K or the Ras-activating Grb2/ SOS complex (72).
We see a high abundance of IGF-1R in wild-type tumors, which is markedly reduced upon loss of PSMA. A retrospective analysis of next-generation sequencing data from about 900 breast cancer samples showed that a large majority of tumors contained genetic alterations that resulted in either the activation of PI3K pathway or the repression of the MAPK pathway, but rarely contained both (73). A third group of tumors harbored amplifications or mutations resulting in increased expression of genes encoding the receptor-type tyrosine kinases IGF-1R, EGFR, or ERB-B2, which were accompanied by PI3K activation and MAPK repression, whereas the genes encoding components of the PI3K and MAPK pathways were intact. The authors interpreted this mutually exclusive, “mirror” pattern of genetic alterations to illustrate the intimate interregulation of the RTK, PI3K, and MAPK pathways and hypothesized that the primary role of RTK up-regulation in tumors may be to regulate these kinases, thereby affecting the tumor cell balance among proliferation, self-renewal, and differentiation (73). Our study show a very similar signaling signature in PCa with increased IGF-1R abundance, activation of the PI3K, and down-regulation of MAPK pathways in the presence of PSMA. However, in our model, rather than IGF-1R up-regulation by mutation or amplification, we demonstrate that IGF-1R abundance is PSMA-dependent and is potentially responsible for PI3K activation, thus controlling primary tumor advancement. Amounts of IGF-1R–mediated signaling do not change significantly during primary tumor progression in the TRAMP mice on the wild-type (PSMA+) background (74), and treatment of wild-type TRAMP mice with various AKT inhibitors reliably reduces tumor size and progression (57–61), further implicating PSMA as a driver of IGF-1R/AKT signaling and prostate tumor progression. Therefore, although the IGF-1R–dependent pathway switch we observe in advancing tumors is not unprecedented, its connection with this highly verified marker of PCa progression provides novel mechanisms, whereby PSMA expression promotes a more aggressive, apoptosis-resistant prostate tumor phenotype.
During cancer development, frequent cross-talk between receptor-driven signal transduction pathways often occurs and results in alterations in downstream signal transduction pathways (51). We have previously shown that PSMA regulates angiogenesis by modulating integrin signal transduction and β1 integrin–dependent FAK-Tyr925 phosphorylation in endothelial cells (36, 37, 42) and show here that its expression alters FAK phosphorylation and IGF-1R signaling pathways in PCa cells (Fig. 6). A large body of evidence has suggested that cooperation between β1 integrin and RTKs regulates cancer cell growth, survival, and invasion in many tumor types (10–30, 52) and has been shown to involve the multifunctional scaffolding protein RACK1 (18–21). RACK1 physically interacts with multiple partners, thereby influencing their function by modifying stability, activity, or interactions with other proteins [reviewed in (75)]. Pertinent to this study, in breast and prostate tumor cells, RACK1 has been shown to promote migration and cell survival by scaffolding IGF-1R and β1 integrin to integrate signaling via recruitment of signaling proteins such as SHC and SHP2 (18–20, 26), whereas interruption of the IGF-1R/RACK1 interaction leads to activation of AKT (21). In prostate tumor cells, we find that RACK1 and IGF-1R associate with PSMA and that deletion of PSMA in PCa cell lines enhances the β1 integrin/RACK1 interaction, suggesting that PSMA may regulate integrin/IGF-1R cross-talk via its interaction with RACK1. Because binding of some proteins to RACK1 is competitive, our data are consistent with a model where the PSMA/RACK1 interaction disrupts or alters the IGF-1R/RACK1/β1 integrin complex, leading to aberrant cross-talk between β1 integrin and IGF-1R that favors PI3K-AKT activation in our PSMA wild-type tumor cells. Finally, although PSMA is expressed primarily in prostate and neuroendocrine cancers, the IGF-1R/β1 integrin axis has been implicated in many tumor types, and RACK1 has been demonstrated to interact with numerous, disparate proteins from cytoplasmic (PP2A, p85 PI3K, SHP-2, SRC, smoothened, Stat3, and TROP-2) to various cell surface proteins (PSMA, IGF-1R, and IR) (18–21, 23, 26, 53, 76). Thus, our findings regarding the regulation of signal transduction by interaction of PSMA with RACK1/IGF-1R will likely extend to other tumor types and cell surface molecules.
Historically, in oncology, PSMA has been studied as a reliable clinical biomarker for the diagnosis, detection, localization, and management of PCa. A number of PSMA-targeted small and low–molecular weight inhibitors and antibodies have proven valuable in providing high image quality in diagnostic imaging and delivering high local doses of radiopharmaceuticals [reviewed in (77–79)]. Alternatively, targeting the PSMA-dependent mechanisms driving PCa tumor progression may provide a novel paradigm for the development of therapeutic strategies for treatment by targeting both progression and vascularization because we have shown that inhibition of PSMA with the inhibitor 2-PMPA impairs integrin activation, endothelial cell adhesion, and angiogenesis (36, 37, 42). Ligand engagement of PSMA induces its internalization, and therefore, it also presents an effective delivery system for therapeutic molecules (80). Recent preclinical characterization of PSMA-targeted RNA aptamers that bind to and block the enzymatic activity of PSMA has been described that specifically target therapies to primary tumors and disseminated PCa cell lines in vivo (35, 81, 82). Finally, TRAMP mice lacking PSMA develop more slowly growing tumors but eventually proceed to high-grade pathologies, likely because of their dependence on the weaker but still oncogenic Ras/MAPK pathway. Together with these observations, our study suggests the potential for design of combination therapies that exploit the PCa-dependent increase in PSMA expression, the proven ability of PSMA-targeting agents to deliver payloads, and the PSMA-dependent pathway switch by agents designed to deliver Ras/MAPK/ERK inhibitors or siRNA directly to PSMA-expressing tumor cells, thereby inducing the switch to the less aggressive pathway while silencing the tumors’ escape mechanism. Such targeted, biology-based therapy has the potential to provide novel, less-toxic, and effective therapies that could have a broad positive impact on outcomes in PCa and in other tumors that rely on the IGF-1R/integrin axis for their oncogenicity.
MATERIALS AND METHODS
Mice
All procedures used in handling mice were approved by the Animal Care Committee of UCONN Health in compliance with Animal Welfare Assurance. TRAMP mice on a C57BL6 background have been described (83) and were obtained from The Jackson Laboratory. C57BL6 PSMA knockout mice have been described (84) and were a donation from W. Heston at the Cleveland Clinic. Sibling female TRAMP mice were mated to wild-type and PSMA knockout sibling males. The resultant female TRAMP-hemizygote PSMA heterozygote females were bred with PSMA knockout males to yield TRAMP-hemizygote PSMA knockout females, which were then bred to produce TRAMP-hemizygote PSMA knockout males for the study. In addition, mice from the original cohort were bred with PSMA wild-type males to obtain TRAMP-hemizygote PSMA wild-type males. Overall, male mice from the same cohort of animals (same original founders) hemizygous for the TRAMP transgene, either PSMA wild type or PSMA knockout (20 mice per genotype per time point; a total of 120 mice), were be bred and held until they reach the desired time points (8, 18, and 30 weeks). Mice were provided food and water ad libitum. Littermates were compared in all experiments.
Genotyping
Mice were genotyped for both PSMA and TRAMP by isolating DNA from tail biopsies [50 mM tris (pH 8.8), 1 mM EDTA (pH 8.0), 0.5% Tween 20, and 3 µg of proteinase K; 50°C overnight and then 100°C for 10 min]. Genotype for PSMA and the TRAMP transgene has been described previously (83, 84).
Necropsy
Animals were euthanized by CO2 asphyxiation, in accordance with the guidelines given by the Animal Care Committee of UCONN Health, followed by cervical dislocation to ensure death. Prostate was removed as part of the whole male urogenital system and then dissected away from nonprostatic tissue.
Cell culture
TRAMP-C1, 22RV1, and PC-3 cells were obtained from the American Type Culture Collection (ATCC). Unless otherwise noted, TRAMP-C1 were cultured in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal bovine serum (FBS), 5% Nu-Serum IV, 1% penicillin-streptomycin (pen-strep), bovine insulin (0.005 mg/ml), and 10 nM de-hydroisoandrosterone. 22RV1 were cultured in RPMI supplemented with 10% FBS, and 1% pen-strep. PC-3 cells were cultured in F-12K supplemented with 10% FBS and 1% pen-strep. All cells were maintained at 37°C, 5% CO2.
Apoptosis evaluation by TUNEL assay
For the evaluation of apoptosis, deparaffinized and rehydrated 6-µm-thick sections of whole prostate tumor sections from PSMA wild-type and PSMA knockout mice at 18 weeks of age were incubated with proteinase K (20 µg/µl) in 10 mM tris-HCl (pH 7.4) at 37°C for 20 min, and apoptotic cells were detected by the TUNEL method by using the ApopTag Peroxidase In Situ Apoptosis Detection system (EMD Millipore) according to the manufacturer’s protocol. Negative controls were prepared by incubating slides without terminal deoxynucleotidyl transferase. Mouse small intestine was used as a positive control. To visualize the immunoreaction products, sections were incubated with a mixture of DAB (3,3′-diaminobenzidine) and H2O2. Slides were examined and TUNEL-positive cells were counted with hematoxylin. Images were acquired using a Zeiss LSM 510 META based on an Axiovert 200 microscope and processed using Zeiss AxioVision software.
Antibodies
Antibody against PSMA for immunohistochemistry was purchased from Zymed (517–3294). PSMA monoclonal antibody 3E2 was a kind gift from Memorial Sloan Kettering. Rabbit monoclonal PSMA (D718E; cat. no. 12815) and RACK1 (D59D5; cat. no. 5432) were purchased from Cell Signaling Technologies (CST). Antibody against CD31/PECAM-1 was purchased from Santa Cruz Biotechnology (SCB; sc-1506). Antibody against Ki67 was purchased from Abcam (15580). Antibody against GRB2 was purchased from SCB (sc-503). The following antibodies were purchased from CST: IGF-1R (cat. no. 9750), β1 intergrin (cat. no. 34971), survivin (cat. no. 71G4B7), caspase-3 (cat. no. 9665), CA9 (cat. no. 5648), PDK1-Ser241 (cat. no. 3438), AKT-Thr308 (cat. no. 13038), AKT-Ser473 (cat. no. 4060), GSK-3β-Ser9 (cat. no. 5558), total AKT (cat. no. 4691), pERK1/2 (cat. no. 197G2), total ERK (cat. no. 9102), and β-actin (cat. no. 3700).
Western blot analysis
Mouse prostate tumor tissues were removed, weighed, and then washed three times in cold phosphate-buffered saline (PBS). A portion of the tumor was the incubated in 1× RBC lysis buffer for 10 min (Thermo Fisher Scientific), and the remaining portion of the tumor was saved for RNA extraction and/or paraffin embedding. Samples were then centrifuged at 1000 rpm for 2 min, and the supernatant was removed. The tumor tissue was then homogenized with a PowerGen 125 tissue homogenizer (Thermo Fisher Scientific) in lysis buffer containing 50 mM tris (pH 7.4), 150 mM NaCl, 1% NP-40, plus the tyrosine phosphatase inhibitor Na3VO4 (1 mM), and the protease inhibitors phenylmethyl-sulfonyl fluoride (PMSF) (1 mM), pepstatin (1 mM), and aprotinin (1.5 mg/ml). After incubation at 4°C for 20 min, nuclear and cellular debris were removed by microcentrifugation at 14,000 rpm for 10 min at 4°C. Total protein concentrations were determined by the BCA Assay kit (Thermo Fisher Scientific), and 30 to 50 µg of each sample was electrophoresed on a 4 to 20% tris-Hepes SDS gradient gel (Pierce) and then electroblotted onto Protran nitrocellulose membranes (Thermo Fisher Scientific) using a semidry transfer apparatus. Membranes were blocked with tris-buffered saline, 5% bovine serum albumin (BSA) for 30 min, washed, and incubated with indicated primary and secondary antibodies for 12 hours and 30 min, respectively. The membranes were washed extensively with tris-buffered saline and 0.1% Tween 20 after secondary antibody incubation and detected using the ECL Western blotting kit (Thermo Fisher Scientific) according to the manufacturer’s suggested protocol.
Immunohistochemistry
Mouse prostate tissues were fixed overnight in 4% paraformaldehyde at 4°C and were paraffin-processed. Slides containing 6-µm-thick sections were deparaffinized and rehydrated. Antigen retrieval was conducted using 10 mM sodium citrate (pH 6.0) in a pressure cooker. Endogenous peroxidase activity was quenched by incubating slides for 15 min in 0.3% H2O2. Slides were blocked in 1% BSA for 30 min at room temperature in a humidified chamber and then incubated with the specified primary antibody overnight in a humidified chamber. Slides were washed in PBS. Biotinylated secondary antibody (Vector Laboratories) in 1% BSA was applied, and slides were incubated for 1 hour at room temperature in a humidified chamber. Slides were washed again, and Vectastain Elite ABC kit (Vector Laboratories) was applied for 30 min. Slides were developed with NovaRED (Sigma-Aldrich) and counterstained with hematoxylin (Vector Laboratories) and then dehydrated and mounted under Cytoseal 60. Images were acquired using a Zeiss LSM 510 META based on an Axiovert 200 microscope and processed using the Zeiss AxioVision software.
PSMA knockdown in TRAMP-C1 by siRNA
siRNA constructs were designed from the mouse Folh1 GenBank sequence BC119604.1 using the Oligoengine 2.0 software and cloned into the mammalian expression vector pSUPER.retro.puro at Bg lII/ Hind III (OligoEngine) according to the manufacturer’s directions. To generate the retrovirus, plasmid DNA (10 µg) was transfected into the Phoenix Amphotropic retrovirus packaging cell line (ATCC) by lipid transfection. Medium containing virus was collected 48 hours after transfection. TRAMP-C1 cells were infected and selected in puromycin (5 µg/ml; Sigma-Aldrich). Reduction of PSMA transcript level was evaluated by reverse transcription polymerase chain reaction (PCR) and real-time PCR followed by Western blot evaluation of PSMA protein expression.
PSMA siRNA primers were as follows: 581, 5′-GATCCCCGAA-GATCAGTTGTTCTGGGTTCAAGAGACCCAGAACAACT-GATCTTCTTTTTA-3′ (forward) and 5′-AGCTTAAAAAG-AAGATCAGTTGTTCTGGGTCTCTTGAACCCAGAACAACT-GATCTTCGGG-3′ (reverse); 472, 5′-GATCCCCATGTAGTGCCAC-CATACAGTTCAAGAGACTGTATGGTGGCACTACATTTTTTA-3′ (forward) and 5′-AGCTTAAAAAATGTAGTGCCACCATA-CAGTCTCTTGAACTGTATGGTGGCACTACATGGG-3′ (reverse); 348, 5′-GATCCCCGTCTTGCTGTCCTATCCAATTCAAGAGATTG-GATAGGACAGCAAGACTTTTTA-3′ (forward) and 5′-AGCT-TAAAAAGTCTTGCTGTCCTATCCAATCTCTTGAATTGGATAG-GACAGCAAGACGGG-3′ (reverse).
CRISPR knockout of PSMA in PCa cell lines TRAMP-C1 and 22RV1
PSMA CRISPR/Cas9 constructs were created in both TRAMP-C1 and 22RV1 cell lines using the lentiCRISPRv2 plasmid #52961 (Addgene) (85,86). To guard against genome-wide off-target effects and still maintain high target specificity, 20–base pair (bp) guide RNA (gRNA) was designed through the CRISPR Design (http://crispr.mit.edu) using the target sequence from either the mouse Folh1 GenBank sequence BC119604.1 or the human FOLH1 GenBank sequence NG_029170.1 as input; scramble gRNA was also designed as a control: gRNA for PSMA 22RV1, 5′-TCACGAAACCGACTCGGCTG-3′; gRNA scramble 22RV1, 5′-CAGTCGGGCGTCATCATGAT-3′; gRNA for PSMA TRAMP, 5′-GCAGGACAGAGACTCCGCGG-3′; and gRNA scramble TRAMP, 5′-CAGTCGGGCGTCATCATGAT-3′. In the genome, gRNA sequences were flanked on the 3′ end by a 3-bp NGG protospacer adjacent motif sequence. The gRNAs were phosphorylated, annealed, and cloned into a dephosphorylated lentiCRISPRv2 at BSMB1 according to the manufacturer’s directions (Addgene). To generate the lentivirus, the plasmid lentiCRISPRv2 containing either the human or the mouse PSMA or scramble gRNA was cotransfected (Lipofectamine 2000, Thermo Fisher Scientific) into HEK293(F)T cells along with the packaging plasmids pMD2.G (#12259, Addgene) and psPAX2 (#12260, Addgene). The culture medium containing virus was collected 48 h after transfection. 22RV1 and TRAMP-C1 cells were infected and selected in puromycin (2 or 5 µg/ml, respectively) (Sigma-Aldrich). Validation of genetic modification in individual TRAMP-CRISPR-PSMAknockout and 22RV1-CRISPR-PSMAknockout clones was assessed by PCR amplification of the targeted loci and subsequent sequencing. Chromatographs were compared to wild-type cells using TIDE (Tracking of Indels by Decomposition; https://tide-calculator.nki.nl/) analysis to identify the mutations in the two alleles (figs. S1 and S2) (87).
TaqMan (real-time quantitative reverse transcription PCR) analysis
Total RNA from cultured cells was isolated using RNeasy (Qiagen). RNA was reverse-transcribed by standard methods using reverse transcriptase (Invitrogen). For TaqMan real-time PCR, PSMA and 18S primers, TaqMan Gene Expression Assay probe, and primer sets were purchased from Applied Biosystems. An Applied Biosystems Prism 7500 Fast Real-time PCR system (Applied Biosystems) was used with the default thermal cycling program (95°C for 20 s, followed by 40 cycles at 95°C for 3 s and 60°C for 30 s). Reactions were performed in triplicate and normalized to the level of 18S RNA transcript.
Primers
PSMA primers were as follows: forward IntA, 5′-ATTCAATCCTGCT-CAGACCC-3′; forward Neo-S, 5′-AGCAGGCATGCTGGGGATGC-3′; reverse S49, 5′-GTAGAAGAGGAACTGCTGAGGA-3′. TRAMP primers were as follows: forward TRAMP SV5, 5′-CAGAGCAGA-ATTGTGGAGTGG-3′; reverse TRAMP SV1, 5′-GGACAAACCA-CAACTAGAATGCAGTG-3′.
PSMA blocking peptide
A peptide containing the C-terminal domain of PSMA was synthesized by Invitrogen. A C-terminal free acid and an N-terminal free amine were added to the completed peptides to aid in transfection. Mouse PSMA, MWNALQDRDSAEVLGHRQR; scramble, ESAMTWVRLRNP-TADRLAH. For peptide transfections, 1 mg of the PSMA and scrambled peptides were transfected into 2×105 to 4×105 TRAMP C-1 cells using BioPORTER (Qiagen) protein delivery system according to the manufacturer’s recommendations. Cotransfection of a positive control fluorescein isothiocyanate (FITC)–containing protein ensured appropriate transfection efficiency (fig. S4). Cells were incubated for 24 hours, washed in PBS, and lysed in 1% NP-40, 150 mM NaCl, 50 mM tris-HCl (pH 7.4) buffer plus the tyrosine phosphatase inhibitor Na3VO4 (1 mM), and the protease inhibitors PMSF (1 mM), pepstatin (1 mM), and aprotinin (1.5 mg/ml). After incubation at 4°C for 20 min, nuclear and cellular debris were removed by microcentrifugation at 14,000 rpm for 10 min at 4°C.
Stimulation of adherent and nonadherent cells
For analysis of signaling responses affected by the ligation of integrins in 22RV1-CRISPR-PSMAknockout adherent cells, the cells and scramble control were washed with PBS and starved from serum for 4 hours before being stimulated with culture medium containing 10% FBS. For analysis of signaling responses affected by the ligation of integrins in nonadherent cells, confluent 22RV1-CRISPR-PSMAknockout cells and scramble control were detached with trypsin and then washed with PBS. Cells were resuspended in serum-free medium containing 5% ECM Gel (Sigma-Aldrich) and maintained in suspension for 4 hours before stimulation with culture medium containing 10% FBS.
Preparation of cellular protein extracts and immunoprecipitation
To study the interaction of PSMA, RACK-1, IGFR-1B, and β1 integrin, cellular protein extracts from either 22RV1-CRISPR-PSMAknockout, scramble, or wild-type cells were prepared by washing cells with PBS and lysed in buffer consisting of tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40 plus the tyrosine phosphatase inhibitor Na3VO4 (1 mM), and the protease inhibitors PMSF (1 mM), pepstatin (1 mM), and aprotinin (1.5 mg/ml). After incubation at 4°C for 20 min, nuclear and cellular debris were removed by microcentrifugation at 14,000 rpm for 10 min at 4°C. Protein (700 µg) was immunoprecipitated with either 1 µg of RACK1 (D59D5) or PSMA 3E2 antibodies and immunoglobulin G antibodies, with 5 µg of goat anti-rabbit immunoglobulin G Fab fragment as a control (Cell Signaling Technologies), and attached to protein G Dynabeads (Invitrogen) at 4°C with either of the prepared beads overnight. Beads were washed while attached to a DynaMag and eluted according to the manufacturer’s directions (Invitrogen).
Statistical analysis
All experiments in this study were repeated for a minimum of three independent experiments. Differences between means were analyzed using either the two-tailed Student’s t test or analysis of variance (ANOVA), where appropriate, and significance was set at a P < 0.05.
Supplementary Material
Fig. S1. CRISPR knockout of PSMA in 22RV1 cells.
Fig. S2. CRISPR knockout of PSMA in TRAMP-C1 cells.
Fig. S3. PSMA knockdown increases the phosphorylation of ERK1/2 and decreases that of PDK in TRAMP-C1 cells.
Fig. S4. Cotransfection of TRAMP-C1 cells with FITC and PSMA blocking peptide.
Data file S1. R script document for analysis of GEO data set GSE32571.
Acknowledgments
We thank the UCONN Health Histology Core for the processing and sectioning of samples as well as for the use of their microscopes, and the UCONN Health Human Genome Editing Core for CRISPR construction and sequencing.
Funding: This work was supported by NIH NCI Mentored Research Scientist Development Award to Promote Diversity (K01CA188412) to L.A.C. and the Prostate Cancer Foundation and the Department of Defense Prostate Cancer Program (PC073976) to L.H.S. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions: L.A.C. and L.H.S. designed the project; L.A.C., K.D., M.M.R., and C.L.G. performed experiments; C.S. was responsible for the CRISPR construction and sequencing; F.A.F provided clinical and urological expertise; and L.A.C. and L.H.S. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, Lin C, Leach C, Cannady RS, Cho H, Scoppa S, Hachey M, Kirch R, Jemal A, Ward E. Cancer treatment and survivorship statistics, 2012. CA Cancer J. Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr. Opin. Pharmacol. 2008;8:440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roudier MP, True LD, Higano CS, Vesselle H, Ellis W, Lange P, Vessella RL. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum. Pathol. 2003;34:646–653. doi: 10.1016/s0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 4.Jones HE, Goddard L, Gee JMW, Hiscox S, Rubini M, Barrow D, Knowlden JM, Williams S, Wakeling AE, Nicholson RI. Insulin-like growth factor-I receptor signalling and acquired resistance to gefitinib (ZD1839; Iressa) in human breast and prostate cancer cells. Endocr. Relat. Cancer. 2004;11:793–814. doi: 10.1677/erc.1.00799. [DOI] [PubMed] [Google Scholar]
- 5.Warshamana-Greene GS, Litz J, Buchdunger E, García-Echeverría C, Hofmann F, Krystal GW. The insulin-like growth factor-I receptor kinase inhibitor, NVP-ADW742, sensitizes small cell lung cancer cell lines to the effects of chemotherapy. Clin. Cancer Res. 2005;11:1563–1571. doi: 10.1158/1078-0432.CCR-04-1544. [DOI] [PubMed] [Google Scholar]
- 6.Hellawell GO, Turner GDH, Davies DR, Poulsom R, Brewster SF, Macaulay VM. Expression of the type 1 insulin-like growth factor receptor is up-regulated in primary prostate cancer and commonly persists in metastatic disease. Cancer Res. 2002;62:2942–2950. [PubMed] [Google Scholar]
- 7.Krueckl SL, Sikes RA, Edlund NM, Bell RH, Hurtado-Coll A, Fazli L, Gleave ME, Cox ME. Increased insulin-like growth factor I receptor expression and signaling are components of androgen-independent progression in a lineage-derived prostate cancer progression model. Cancer Res. 2004;64:8620–8629. doi: 10.1158/0008-5472.CAN-04-2446. [DOI] [PubMed] [Google Scholar]
- 8.Chen HX, Sharon E. IGF-1R as an anti-cancer target—Trials and tribulations. Chin. J. Cancer. 2013;32:242–252. doi: 10.5732/cjc.012.10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu S-W, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loughran G, Healy NC, Kiely PA, Huigsloot M, Kedersha NL, O’Connor R. Mystique is a new insulin-like growth factor-I-regulated PDZ-LIM domain protein that promotes cell attachment and migration and suppresses Anchorage-independent growth. Mol. Biol. Cell. 2005;16:1811–1822. doi: 10.1091/mbc.E04-12-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deevi RK, Cox OT, O’Connor R. Essential function for PDLIM2 in cell polarization in three-dimensional cultures by feedback regulation of the β1-integrin-RhoA signaling axis. Neoplasia. 2014;16:422–431. doi: 10.1016/j.neo.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sureshbabu A, Okajima H, Yamanaka D, Tonner E, Shastri S, Maycock J, Szymanowska M, Shand J, Takahashi S-I, Beattie J, Allan G, Flint D. IGFBP5 induces cell adhesion, increases cell survival and inhibits cell migration in MCF-7 human breast cancer cells. J. Cell Sci. 2012;125:1693–1705. doi: 10.1242/jcs.092882. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Kamaraju S, Hakuno F, Kabuta T, Takahashi S-I, Sachdev D, Yee D. Motility response to insulin-like growth factor-I (IGF-I) in MCF-7 cells is associated with IRS-2 activation and integrin expression. Breast Cancer Res. Treat. 2004;83:161–170. doi: 10.1023/b:brea.0000010709.31256.c6. [DOI] [PubMed] [Google Scholar]
- 14.Huang C, Park CC, Hilsenbeck SG, Ward R, Rimawi MF, Wang Y-c, Shou J, Bissell MJ, Osborne CK, Schiff R. β1 integrin mediates an alternative survival pathway in breast cancer cells resistant to lapatinib. Breast Cancer Res. 2011;13:R84. doi: 10.1186/bcr2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park CC, Zhang HJ, Yao ES, Park CJ, Bissell MJ. β1 integrin inhibition dramatically enhances radiotherapy efficacy in human breast cancer xenografts. Cancer Res. 2008;68:4398–4405. doi: 10.1158/0008-5472.CAN-07-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin KJ, Patrick DR, Bissell MJ, Fournier MV. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLOS ONE. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park CC, Zhang H, Pallavicini M, Gray JW, Baehner F, Park CJ, Bissell MJ. β1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Res. 2006;66:1526–1535. doi: 10.1158/0008-5472.CAN-05-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiely PA, O’Gorman D, Luong K, Ron D, O’Connor R. Insulin-like growth factor I controls a mutually exclusive association of RACK1 with protein phosphatase 2A and β1 integrin to promote cell migration. Mol. Cell. Biol. 2006;26:4041–4051. doi: 10.1128/MCB.01868-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermanto U, Zong CS, Li W, Wang L-H. RACK1, an insulin-like growth factor I (IGF-I) receptor-interacting protein, modulates IGF-I-dependent integrin signaling and promotes cell spreading and contact with extracellular matrix. Mol. Cell. Biol. 2002;22:2345–2365. doi: 10.1128/MCB.22.7.2345-2365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch L, Vodyanik PI, Boettiger D, Guvakova MA. Insulin-like growth factor I controls adhesion strength mediated by α5β1 integrins in motile carcinoma cells. Mol. Biol. Cell. 2005;16:51–63. doi: 10.1091/mbc.E04-05-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiely PA, Leahy M, O’Gorman D, O’Connor R. RACK1-mediated integration of adhesion and insulin-like growth factor I (IGF-I) signaling and cell migration are defective in cells expressing an IGF-I receptor mutated at tyrosines 1250 and 1251. J. Biol. Chem. 2005;280:7624–7633. doi: 10.1074/jbc.M412889200. [DOI] [PubMed] [Google Scholar]
- 22.Goel HL, Sayeed A, Breen M, Zarif MJ, Garlick DS, Leav I, Davis RJ, FitzGerald TJ, Morrione A, Hsieh C-C, Liu Q, Dicker AP, Altieri DC, Languino LR. β1 integrins mediate resistance to ionizing radiation in vivo by inhibiting c-Jun amino terminal kinase 1. J. Cell. Physiol. 2013;228:1601–1609. doi: 10.1002/jcp.24323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trerotola M, Li J, Alberti S, Languino LR. Trop-2 inhibits prostate cancer cell adhesion to fibronectin through the β1 integrin-RACK1 axis. J. Cell. Physiol. 2012;227:3670–3677. doi: 10.1002/jcp.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayeed A, Alam N, Trerotola M, Languino LR. Insulin-like growth factor 1 stimulation of androgen receptor activity requires β1A integrins. J. Cell. Physiol. 2012;227:751. doi: 10.1002/jcp.22784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goel HL, Underwood JM, Nickerson JA, Hsieh C-C, Languino LR. β1 Integrins mediate cell proliferation in three-dimensional cultures by regulating expression of the sonic hedgehog effector protein, GLI1. J. Cell. Physiol. 2010;224:210–217. doi: 10.1002/jcp.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goel HL, Breen M, Zhang J, Das I, Aznavoorian-Cheshire S, Greenberg NM, Elgavish A, Languino LR. β1A integrin expression is required for type 1 insulin-like growth factor receptor mitogenic and transforming activities and localization to focal contacts. Cancer Res. 2005;65:6692–6700. doi: 10.1158/0008-5472.CAN-04-4315. [DOI] [PubMed] [Google Scholar]
- 27.Meyer A, van Golen CM, Kim B, van Golen KL, Feldman EL. Integrin expression regulates neuroblastoma attachment and migration. Neoplasia. 2004;6:332–342. doi: 10.1593/neo.03445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Golen CM, Soules ME, Grauman AR, Feldman EL. N-Myc overexpression leads to decreased β1 integrin expression and increased apoptosis in human neuroblastoma cells. Oncogene. 2003;22:2664–2673. doi: 10.1038/sj.onc.1206362. [DOI] [PubMed] [Google Scholar]
- 29.Tai Y-T, Podar K, Catley L, Tseng Y-H, Akiyama M, Shringarpure R, Burger R, Hideshima T, Chauhan D, Mitsiades N, Richardson P, Munshi NC, Kahn CR, Mitsiades C, Anderson KC. Insulin-like growth factor-1 induces adhesion and migration in human multiple myeloma cells via activation of β1-integrin and phosphatidylinositol 3′-kinase/AKT signaling. Cancer Res. 2003;63:5850–5858. [PubMed] [Google Scholar]
- 30.Edderkaoui M, Hong P, Lee JK, Pandol SJ, Gukovskaya AS. Insulin-like growth factor-I receptor mediates the prosurvival effect of fibronectin. J. Biol. Chem. 2007;282:26646–26655. doi: 10.1074/jbc.M702836200. [DOI] [PubMed] [Google Scholar]
- 31.Heston WDW. Characterization and glutamyl preferring carboxypeptidase function of prostate specific membrane antigen: A novel folate hydrolase. Urology. 1997;49:104–112. doi: 10.1016/s0090-4295(97)00177-5. [DOI] [PubMed] [Google Scholar]
- 32.Israeli RS, Powell CT, Fair WR, Heston WDW. Molecular cloning of a complementary DNA encoding a prostate-specific membrane antigen. Cancer Res. 1993;53:227–230. [PubMed] [Google Scholar]
- 33.Pinto JT, Suffoletto BP, Berzin TM, Qiao CH, Lin S, Tong WP, May F, Mukherjee B, Heston WD. Prostate-specific membrane antigen: A novel folate hydrolase in human prostatic carcinoma cells. Clin. Cancer Res. 1996;2:1445–1451. [PubMed] [Google Scholar]
- 34.Eder M, Eisenhut M, Babich J, Haberkorn U. PSMA as a target for radiolabeled small molecules. Eur. J. Nucl. Med. Mol. Imaging. 2013;40:819–823. doi: 10.1007/s00259-013-2374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dassie JP, Hernandez LI, Thomas GS, Long ME, Rockey WM, Howell CA, Chen Y, Hernandez FJ, Liu XY, Wilson ME, Allen L-A, Vaena DA, Meyerholz DK, Giangrande PH. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol. Ther. 2014;22:1910–1922. doi: 10.1038/mt.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conway RE, Petrovic N, Li Z, Heston W, Wu D, Shapiro LH. Prostate-specific membrane antigen regulates angiogenesis by modulating integrin signal transduction. Mol. Cell. Biol. 2006;26:5310–5324. doi: 10.1128/MCB.00084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conway RE, Joiner K, Patterson A, Bourgeois D, Rampp R, Hannah BC, McReynolds S, Elder JM, Gilfilen H, Shapiro LH. Prostate specific membrane antigen produces pro-angiogenic laminin peptides downstream of matrix metalloprotease-2. Angiogenesis. 2013;16:847–860. doi: 10.1007/s10456-013-9360-y. [DOI] [PubMed] [Google Scholar]
- 38.Grant CL, Caromile LA, Durrani K, Rahman MM, Claffey KP, Fong G-H, Shapiro LH. Prostate specific membrane antigen (PSMA) regulates angiogenesis independently of VEGF during ocular neovascularization. PLOS ONE. 2012;7:e41285. doi: 10.1371/journal.pone.0041285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bacich DJ, Ramadan E, O’Keefe DS, Bukhari N, Wegorzewska I, Ojeifo O, Olszewski R, Wrenn CC, Bzdega T, Wroblewska B, Heston WDW, Neale JH. Deletion of the glutamate carboxypeptidase II gene in mice reveals a second enzyme activity that hydrolyzes N-acetylaspartylglutamate. J. Neurochem. 2002;83:20–29. doi: 10.1046/j.1471-4159.2002.01117.x. [DOI] [PubMed] [Google Scholar]
- 40.Gingrich JR, Barrios RJ, Foster BA, Greenberg NM. Pathologic progression of autochthonous prostate cancer in the TRAMP model. Prostate Cancer Prostatic Dis. 1999;2:70–75. doi: 10.1038/sj.pcan.4500296. [DOI] [PubMed] [Google Scholar]
- 41.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, Cunha GR, Donjacour AA, Matusik RJ, Rosen JM. Prostate cancer in a transgenic mouse. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conway RE, Rojas C, Alt J, Nováková Z, Richardson SM, Rodrick TC, Fuentes JL, Richardson NH, Attalla J, Stewart S, Fahmy B, Barinka C, Ghosh M, Shapiro LH, Slusher BS. Prostate-specific membrane antigen (PSMA)-mediated laminin proteolysis generates a pro-angiogenic peptide. Angiogenesis. 2016;19:487–500. doi: 10.1007/s10456-016-9521-x. [DOI] [PubMed] [Google Scholar]
- 43.Gingrich JR, Barrios RJ, Morton RA, Boyce BF, DeMayo FJ, Finegold MJ, Angelopoulou R, Rosen JM, Greenberg NM. Metastatic prostate cancer in a transgenic mouse. Cancer Res. 1996;56:4096–4102. [PubMed] [Google Scholar]
- 44.Rojas C, Frazier ST, Flanary J, Slusher BS. Kinetics and inhibition of glutamate carboxypeptidase II using a microplate assay. Anal. Biochem. 2002;310:50–54. doi: 10.1016/s0003-2697(02)00286-5. [DOI] [PubMed] [Google Scholar]
- 45.Suttie A, Nyska A, Haseman JK, Moser GJ, Hackett TR, Goldsworthy TL. A grading scheme for the assessment of proliferative lesions of the mouse prostate in the TRAMP model. Toxicol. Pathol. 2003;31:31–38. doi: 10.1080/01926230390173842. [DOI] [PubMed] [Google Scholar]
- 46.Zhang M, Latham DE, Delaney MA, Chakravarti A. Survivin mediates resistance to antiandrogen therapy in prostate cancer. Oncogene. 2005;24:2474–2482. doi: 10.1038/sj.onc.1208490. [DOI] [PubMed] [Google Scholar]
- 47.Jain RK. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 48.Shin H-J, Rho SB, Jung DC, Han I-O, Oh E-S, Kim J-Y. Carbonic anhydrase IX (CA9) modulates tumor-associated cell migration and invasion. J. Cell Sci. 2011;124:1077–1087. doi: 10.1242/jcs.072207. [DOI] [PubMed] [Google Scholar]
- 49.Oh Y-T, Chen DWC, Dougherty GJ, McBride WH. Adenoviral interleukin-3 gene-radiation therapy for prostate cancer in mouse model. Int. J. Radiat. Oncol. Biol. Phys. 2004;59:579–583. doi: 10.1016/j.ijrobp.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 50.Takeuchi K, Ito F. Receptor tyrosine kinases and targeted cancer therapeutics. Biol. Pharm. Bull. 2011;34:1774–1780. doi: 10.1248/bpb.34.1774. [DOI] [PubMed] [Google Scholar]
- 51.Xu AM, Huang PH. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010;70:3857–3860. doi: 10.1158/0008-5472.CAN-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soung YH, Clifford JL, Chung J. Crosstalk between integrin and receptor tyrosine kinase signaling in breast carcinoma progression. BMB Rep. 2010;43:311–318. doi: 10.5483/bmbrep.2010.43.5.311. [DOI] [PubMed] [Google Scholar]
- 53.Kiely PA, Sant A, O’Connor R. RACK1 is an insulin-like growth factor 1 (IGF-1) receptor-interacting protein that can regulate IGF-1-mediated Akt activation and protection from cell death. J. Biol. Chem. 2002;277:22581–22589. doi: 10.1074/jbc.M201758200. [DOI] [PubMed] [Google Scholar]
- 54.Zhang W, Zong CS, Hermanto U, Lopez-Bergami P, Ronai Z, Wang L-H. RACK1 recruits STAT3 specifically to insulin and insulin-like growth factor 1 receptors for activation, which is important for regulating anchorage-independent growth. Mol. Cell. Biol. 2006;26:413–424. doi: 10.1128/MCB.26.2.413-424.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee HS, Millward-Sadler SJ, Wright MO, Nuki G, Al-Jamal R, Salter DM. Activation of Integrin-RACK1/PKCα signalling in human articular chondrocyte mechanotransduction. Osteoarthritis Cartilage. 2002;10:890–897. doi: 10.1053/joca.2002.0842. [DOI] [PubMed] [Google Scholar]
- 56.Liliental J, Chang DD. Rack1, a receptor for activated protein kinase C, interacts with integrin β subunit. J. Biol. Chem. 1998;273:2379–2383. doi: 10.1074/jbc.273.4.2379. [DOI] [PubMed] [Google Scholar]
- 57.Shukla S, Bhaskaran N, Babcook MA, Fu P, MacLennan GT, Gupta S. Apigenin inhibits prostate cancer progression in TRAMP mice via targeting PI3K/Akt/FoxO pathway. Carcinogenesis. 2014;35:452–460. doi: 10.1093/carcin/bgt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shukla S, Maclennan GT, Marengo SR, Resnick MI, Gupta S. Constitutive activation of PI3K-AKT and NF-κB during prostate cancer progression in autochthonous transgenic mouse model. Prostate. 2005;64:224–239. doi: 10.1002/pros.20217. [DOI] [PubMed] [Google Scholar]
- 59.Hu H, Chai Y, Wang L, Zhang J, Lee HJ, Kim S-H, Lü J. Pentagalloylglucose induces autophagy and caspase-independent programmed deaths in human PC-3 and mouse TRAMP-C2 prostate cancer cells. Mol. Cancer Ther. 2009;8:2833–2843. doi: 10.1158/1535-7163.MCT-09-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sargeant AM, Klein RD, Rengel RC, Clinton SK, Kulp SK, Kashida Y, Yamaguchi M, Wang X, Chen C-S. Chemopreventive and bioenergetic signaling effects of PDK1/Akt pathway inhibition in a transgenic mouse model of prostate cancer. Toxicol. Pathol. 2007;35:549–561. doi: 10.1080/01926230701338966. [DOI] [PubMed] [Google Scholar]
- 61.Shimizu M, Shirakami Y, Moriwaki H. Targeting receptor tyrosine kinases for chemoprevention by green tea catechin, EGCG. Int. J. Mol. Sci. 2008;9:1034–1049. doi: 10.3390/ijms9061034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wynant GE, Murphy GP, Horoszewicz JS, Neal CE, Collier BD, Mitchell E, Purnell G, Tyson I, Heal A, Abdel-Nabi H, Winzelberg G. Immunoscintigraphy of prostatic cancer: Preliminary results with 111In-labeled monoclonal antibody 7E11-C5.3 (CYT-356) Prostate. 1991;18:229–241. doi: 10.1002/pros.2990180305. [DOI] [PubMed] [Google Scholar]
- 63.Bostwick DG, Pacelli A, Blute M, Roche P, Murphy GP. Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma: A study of 184 cases. Cancer. 1998;82:2256–2261. doi: 10.1002/(sici)1097-0142(19980601)82:11<2256::aid-cncr22>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 64.Wolf P, Freudenberg N, Bühler P, Alt K, Schultze-Seemann W, Wetterauer U, Elsässer-Beile U. Three conformational antibodies specific for different PSMA epitopes are promising diagnostic and therapeutic tools for prostate cancer. Prostate. 2010;70:562–569. doi: 10.1002/pros.21090. [DOI] [PubMed] [Google Scholar]
- 65.Silver DA, Pellicer I, Fair WR, Heston WD, Cordon-Cardo C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin. Cancer Res. 1997;3:81–85. [PubMed] [Google Scholar]
- 66.Ghosh A, Wang X, Klein E, Heston WDW. Novel role of prostate-specific membrane antigen in suppressing prostate cancer invasiveness. Cancer Res. 2005;65:727–731. [PubMed] [Google Scholar]
- 67.Yao V, Bacich DJ. Prostate specific membrane antigen (PSMA) expression gives prostate cancer cells a growth advantage in a physiologically relevant folate environment in vitro. Prostate. 2006;66:867–875. doi: 10.1002/pros.20361. [DOI] [PubMed] [Google Scholar]
- 68.Yao V, Parwani A, Maier C, Heston WD, Bacich DJ. Moderate expression of prostate-specific membrane antigen, a tissue differentiation antigen and folate hydrolase, facilitates prostate carcinogenesis. Cancer Res. 2008;68:9070–9077. doi: 10.1158/0008-5472.CAN-08-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hurwitz AA, Foster BA, Allison JP, Greenberg NM, Kwon ED. The TRAMP mouse as a model for prostate cancer. Curr. Protoc. Immunol. 2001 doi: 10.1002/0471142735.im2005s45. Chapter 20, Unit 20.5. [DOI] [PubMed] [Google Scholar]
- 70.Heidegger I, Massoner P, Sampson N, Klocker H. The insulin-like growth factor (IGF) axis as an anticancer target in prostate cancer. Cancer Lett. 2015;367:113–121. doi: 10.1016/j.canlet.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 71.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat. Rev. Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 72.Belfiore A, Malaguarnera R. Insulin receptor and cancer. Endocr. Relat. Cancer. 2011;18:R125–R147. doi: 10.1530/ERC-11-0074. [DOI] [PubMed] [Google Scholar]
- 73.Guille A, Chaffanet M, Birnbaum D. Signaling pathway switch in breast cancer. Cancer Cell Int. 2013;13:66. doi: 10.1186/1475-2867-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaplan PJ, Mohan S, Cohen P, Foster BA, Greenberg NM. The insulin-like growth factor axis and prostate cancer: Lessons from the transgenic adenocarcinoma of mouse prostate (TRAMP) model. Cancer Res. 1999;59:2203–2209. [PubMed] [Google Scholar]
- 75.Li J-J, Xie D. RACK1, a versatile hub in cancer. Oncogene. 2015;34:1890–1898. doi: 10.1038/onc.2014.127. [DOI] [PubMed] [Google Scholar]
- 76.Shi S, Deng Y-Z, Zhao J-S, Ji X-D, Shi J, Feng Y-X, Li G, Li J-J, Zhu D, Koeffler HP, Zhao Y, Xie D. RACK1 promotes non-small-cell lung cancer tumorigenicity through activating sonic hedgehog signaling pathway. J. Biol. Chem. 2012;287:7845–7858. doi: 10.1074/jbc.M111.315416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haberkorn U, Eder M, Kopka K, Babich JW, Eisenhut M. New strategies in prostate cancer: Prostate-specific membrane antigen (PSMA) ligands for diagnosis and therapy. Clin. Cancer Res. 2016;22:9–15. doi: 10.1158/1078-0432.CCR-15-0820. [DOI] [PubMed] [Google Scholar]
- 78.Chatalic KLS, Heskamp S, Konijnenberg M, Molkenboer-Kuenen JDM, Franssen GM, Clahsen-van Groningen MC, Schottelius M, Wester H-J, van Weerden WM, Boerman OC, de Jong M. Towards personalized treatment of prostate cancer: PSMA I&T, a promising prostate-specific membrane antigen-targeted theranostic agent. Theranostics. 2016;6:849–861. doi: 10.7150/thno.14744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ganguly T, Dannoon S, Hopkins MR, Murphy S, Cahaya H, Blecha JE, Jivan S, Drake CR, Barinka C, Jones EF, VanBrocklin HF, Berkman CE. A high-affinity [18F]-labeled phosphoramidate peptidomimetic PSMA-targeted inhibitor for PET imaging of prostate cancer. Nucl. Med. Biol. 2015;42:780–787. doi: 10.1016/j.nucmedbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rajasekaran SA, Anilkumar G, Oshima E, Bowie JU, Liu H, Heston W, Bander NH, Rajasekaran AK. A novel cytoplasmic tail MXXXL motif mediates the internalization of prostate-specific membrane antigen. Mol. Biol. Cell. 2003;14:4835–4845. doi: 10.1091/mbc.E02-11-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dassie JP, Liu X-y, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, II, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–846. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dassie JP, Giangrande PH. Current progress on aptamer-targeted oligonucleotide therapeutics. Ther. Deliv. 2013;4:1527–1546. doi: 10.4155/tde.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christiansen JJ, Rajasekaran SA, Moy P, Butch A, Goodglick L, Gu Z, Reiter RE, Bander NH, Rajasekaran AK. Polarity of prostate specific membrane antigen, prostate stem cell antigen, and prostate specific antigen in prostate tissue and in a cultured epithelial cell line. Prostate. 2003;55:9–19. doi: 10.1002/pros.10203. [DOI] [PubMed] [Google Scholar]
- 84.Tsai CH, Hong J-H, Hsieh K-F, Hsiao H-W, Chuang W-L, Lee C-C, McBride WH, Chiang C-S. Tetracycline-regulated intratumoral expression of interleukin-3 enhances the efficacy of radiation therapy for murine prostate cancer. Cancer Gene Ther. 2006;13:1082–1092. doi: 10.1038/sj.cgt.7700977. [DOI] [PubMed] [Google Scholar]
- 85.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. CRISPR knockout of PSMA in 22RV1 cells.
Fig. S2. CRISPR knockout of PSMA in TRAMP-C1 cells.
Fig. S3. PSMA knockdown increases the phosphorylation of ERK1/2 and decreases that of PDK in TRAMP-C1 cells.
Fig. S4. Cotransfection of TRAMP-C1 cells with FITC and PSMA blocking peptide.
Data file S1. R script document for analysis of GEO data set GSE32571.