Stem cells hold enormous potential for application in regenerative medicine and tissue engineering.[1–5] It is hoped that the controlled expansion and differentiation of stem cells may allow physicians and scientists to augment, replace, or reconstruct damaged or diseased tissues and organs. While much progress has been achieved in stem cell research, additional improvements are needed to maximize their utility. Since cells typically discern and react to multiple cues rather than to a single signal, optimization of differentiation conditions can prove challenging. To address this problem, we have developed an integrated high-throughput (HT) polymer synthesis and rapid material/protein/cell-interaction assays to accelerate the optimization of the stem-cell microenvironment. Herein, an array of 496 different biomaterials was synthesized and studied to examine the attachment of human embryonic-body (hEB) cells, partially differentiated human embryonic stem (hES) cells. An optimized laser scanning cytometry system was developed to allow for rapid quantification of cell-material interaction. Additionally, fibronectin (Fn) adsorption on the polymer array was quantified. Analysis of the relationship between cell attachment and Fn adsorption indicated a significant difference in the cell adhesion activities between the adsorbed Fn on the polymer array, and we hypothesize that this difference can, in part, be explained by the capacity of the polymers to induce different conformations of the adsorbed Fn.[6] Finally, representative biological properties identified within the polymer array have been reproduced in large-scale polymeric films, and their effects on cell-behavior validated.
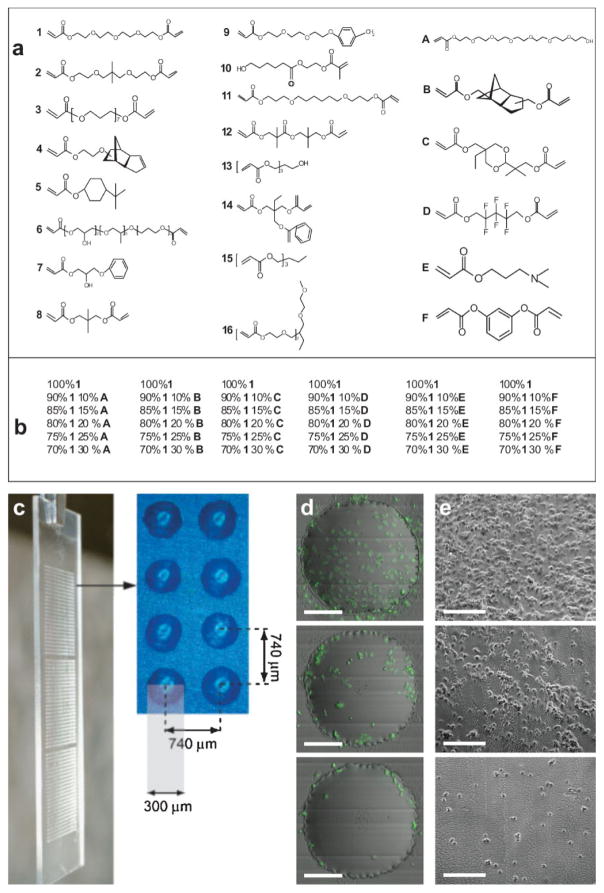
Previously, acrylate-based biomaterial arrays were developed for HT investigation of stem cell/materials interactions. These were fabricated using a robotic stage modified with a long-wave UV source.[7,8] In this study, 22 acrylate monomers were chosen to maximize the diversity of their hydrophobicity/hydrophilicity and crosslinking density. They were subsequently divided into 16 major constituent monomers and 6 minor constituent monomers. As shown in Figure 1a, the major monomers were named numerically, and the minor monomers were named in alphabetic order. The arrays were prepared by the copolymerization of each one of 16 major monomers with each one of 6 minor monomers at 6 different ratios (100:0, 90:10, 85:15, 80:20, 75:25, 70:30 (v/v)). In this way, arrays with 16 + 16 × 6 × 5 = 496 different combinations were created, primarily composed of the major monomer (70–100%), and to a lesser extent with the minor monomers (0–30%). These monomers were robotically deposited in triplicate on a layer of poly(hydroxyl ethyl methacrylate) (poly(HEMA))-covered conventional glass slides (75 mm × 25 mm). The systematic variation of the ratio between major and minor monomers permits a global understanding of the effects of each major/minor monomer on the cellular response.
Figure 1.
Biomaterial array design: a) monomers used for array synthesis, b) 36 different combinations for the major monomer 1 with all six different minor monomers, c) photograph showing one polymer microarray in triplicate with eight polymer spots, to show dimension and separation. d) Merged images from fluorescence and scattered channel collected from iCys cytometry of three representative cell attachments (high, intermediate, low) on the polymer spots. The scale bar in the figure is 100 μm. e) Reproduced cell attachment on large-scale polymer films; the scale bar in the figure is 200 μm.
To generate polymer arrays with large diversity, monomers with a wide range of hydrophobicity/hydrophilicity were selected as major monomers. One way to quantify these properties is through analysis of the logarithm of the partition coefficient (log P). For reference, the log P values for hydrophilic methanol and hydrophobic hexamethylbenezene are −0.82 and 4.61, respectively. In this study, the log P values differ from −0.03 (monomer 6) to 3.74 (monomer 14).[9] It is important to note that the crosslink densities vary significantly within the array, which can affect the mechanical properties.[10,11] Minor monomers were chosen with diverse chemical structures (Fig. 1a, A–F). For example, minor monomer A has a PEO side chain that was anticipated to reduce the cell attachment, while minor monomer F was thought to increase the cell attachment, since it has a benzene ring structure that could increase the hydrophobicity of the materials.[12,13]
hES cells have the potential to differentiate into nearly every cell in the body.[14] In vitro differentiation is often initiated by the formation of embryonic bodies (EBs), and is believed to recapitulate early stages of primitive streak development and its derivative germ layers, mesoderm and endoderm.[15,16] Cell adhesion is essential for many complicated cellular activities, such as proliferation and differentiation. To better understand the effects of material composition on cell adhesion, the polymer arrays described here were used as substrates to examine hEB cells attachment.
To initiate the differentiation, hES cells were allowed to form embryonic bodies (EBs) for eight days, and were then trypsinized and cultured on polymer arrays for 16 h to test their initial attachment. An incubation time of 16 h was chosen because it allowed good cell attachment and spreading, and is significantly shorter than the doubling time of HESCs (36 h).[17] The cells were then fixed and stained for DNA/nucleus with SYTO 24, a fluorescent DNA-binding dye.
One bottleneck in the analytics of cellular microarrays is the lack of automated, quantitative data acquisition and analysis methods. In particular, the position of each spot in the polymer array has to be identified and coordinated with their chemical composition, so that the cellular response can be correlated with the materials properties. Here, we developed an automated cell analytical system based on laser scanning cytometry.[18,19] Two protocols were developed –with low and high resolutions. In the low-resolution scan, the distinct optical-scattering property of polymer spots permitted us to locate the spacial locations of the polymer spots, and then the index of each spot allowed us to identify its chemical composition (Supporting Information Fig. 1). During the high-resolution scan, only the polymer spots were scanned, and the intensities of fluorescently labeled cellular markers on the polymer spots enabled a quantitative measurement of cellular behavior. Equipped with three different lasers, as many as eight different cellular markers could be labeled and quantified in a high-throughput manner. Notably, the throughput of the laser scanning cytometer allows the quantification of an entire polymer array composed of 576 × 3 =1728 different polymers within 15 h. The methodology developed here is flexible enough to apply to other cellular microarray formats (see methods).
In this study, the number of cells on each polymer spot was quantified as the metric for cell attachment. A collage of the merged images of the cell attachment on the entire array has been shown in the Supporting Information, and Figure 1d shows three representative images of varying cell attachment in the polymer spots. To quantitatively assess the capacity of each major monomer to influence the cell adhesion, the number of cells on polymer spots composed of 100% major monomers was quantified and ranked in Table 1. The major monomers were then categorized into three different groups: high adhesion (60–100 cells/spot), intermediate adhesion (40–60 cells/spot), and low adhesion (10–40 cells/spot).
Table 1.
The ranking of the major monomers based on the cell density on the polymer spot [a].
| High-adhesion monomers | 15 (95 ± 8) | 8 (96 ± 12) | 14 (86 ± 14) | 13 (80 ± 14) | 4 (84 ± 13) | 12 (80 ± 14) | 9 (74 ± 9) | 1 (70 ± 13) |
| Intermediate-adhesion monomers | 2 (52 ± 12) | 11 (48 ± 14) | ||||||
| Low adhesion-monomers | 6 (40 ± 9) | 7 (31 ± 17) | 16 (16 ± 10) | 10 (23 ± 6) | 3 (14 ± 4) | 5 (12 ± 7) |
The first number is the ID of the major monomer, while the number in the bracket is the averaged cell density plus/minus standard deviation.
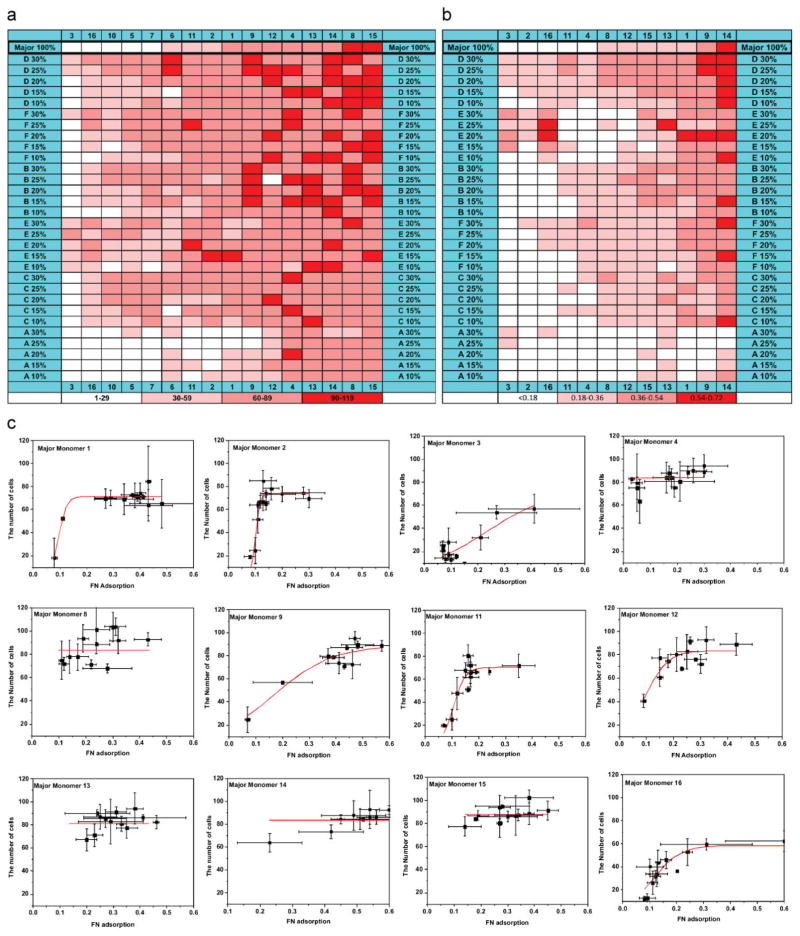
To better understand the effects of major and minor monomers, the number of cells on each polymer spot was mapped against their major monomer in the x-axis and minor monomer in the y-axis (Fig. 2a). The first row of the Figure 2a shows cell attachment on the homopolymers of the 16 major monomers. Ranking (the amount of cellular attachment) was used to define the position of each major monomer in the x-axis.
Figure 2.
a) Map of the relationship between cell attachment and polymer composition. Cell number per spot was grouped into four catagories 1–29, 30–59, 60–89, and 90–119 per spot. Cell numbers are mapped as a function of polymer composition. b) A map of the relationship between Fn adsorption and polymer composition, the major monomer 5, 6, 7, and 10 are excluded due to autofluorescence from the polymer spots. c) hEB cell attachment as a function of the Fn adsorption on the polymer array; the data has been grouped according to their major monomers.
In a similar way, the position of each minor monomer in the y-axis is defined by its support for cellular attachment. Importantly, a range in the hEB cell attachment can be seen in the diagonal between bottom left to top right in Figure 2a, which can be attributed to a general relationship between the hEB cell attachment and the chemical composition of the polymeric materials. Simply put, the capacity of the copolymers to influence the cell attachment usually depends on both the major and minor monomers. In general, an enhanced cell attachment can be obtained by adding a “high-adhesion” minor monomer to a “low-adhesion” major monomer, while a decreased cell attachment can be achieved by adding a “low-adhesion” minor monomer to a “high-adhesion” major monomer. For example, a four-fold improvement (from ≈15 to ≈60 cells/spot) was found with the addition of 30% minor monomer F to the major monomer 3, one of the “lowest-adhesion” major monomers. The cell attachment on the homopolymer 8 can be reduced by 50% (from ≈100 to ≈50 cells/spot) with the addition of the 30% minor monomer A. By choosing the proper combination of the major and minor monomers, we are able to control the cell attachment on the polymeric materials. The capacity to precisely control the hEB cell adhesion may prove useful in the optimization of material supports for growth, propagation, and differentiation of hES cells.
After materials come in contact with serum-containing medium, serum proteins can adsorb onto the materials’ surfaces. This adsorbed protein layer plays an important role in the cellular response.[20,21] To control batch-to-batch variation in protein adsorption, a Fn-conditioning step was used to precoat the polymer array in this study.[6,22–25] To understand the effect of the polymer composition on Fn adsorption, fluorescently labeled Fn was adsorbed onto the polymer array, and the difference between the fluorescence intensity before and after Fn adsorption from each polymer spot was quantified in Figure 2b[26,27] (Supporting Information Fig. 3). Similar to the Figure 2a, the capacities of the major/minor monomers to influence Fn adsorption have been ranked, and the ranking was used to determine the position of each major/minor monomer in the x- and y-axis, respectively. The gradient in the diagonal direction from bottom left to top right clearly showed both major and minor monomer composition can have large effects on Fn adsorption, and that the certain combinations of the major and minor monomer can be used to control the amount of Fn adsorbed on a polymer spot. In general, a lesser amount of Fn has been found adsorbed on the “low-adhesion” monomers, such as 3, while a greater amount of Fn has been found on the “high-adhesion” monomers, such as 8. Compared to the hompolymers (the top row), minor monomer A has been found to reduce the Fn adsorption while the minor monomers B, D, and F can improve Fn adsorption. This correlates with the capacity of the monomers to modulate hEB cell attachment, and supports the conclusion that the adsorbed Fn plays a role in determining hEB cell attachment.
In an effort to understand the effect of adsorbed Fn on hEB cell attachment, the number of EB cells attached on each polymer spot was plotted against the amount of adsorbed Fn, and the results were further grouped according to their major monomers. (Fig. 2c) The relationship between cell adhesion and Fn adsorption on the substrate has been previously studied. For example, Keselowsky and coworkers studied the relationship between cell adhesion and Fn surface density on surfaces with a variety of properties.[6] For all substrates they examined, cell adhesion increased with Fn surface density in a sigmoidal fashion. In other words, saturated cell adhesion was found at high Fn surface density, a transition region of intermediate cell adhesion at intermediate Fn surface concentrations, and a background level of cell adhesion at low Fn surface coverage. The cell number versus Fn adsorption grouped by major monomer can be fit as part of a sigmoid curve in all cases (Fig. 2c), and the detailed fitting process can be found in the Supporting Information. Saturated cell attachment is observed on the “high-adhesion” major monomers, such as 4, 8, 13, 14, and 15, while a lower level of cell attachment can be found on “low-adhesion” spots containing monomers such as 3 and 16. For the “intermediate monomers”, such as 2 and 11, an incremental cell attachment was observed with the increase of Fn density, until a saturated cell attachment was reached.
The Fn density for the saturated cell attachment (Fnsat, defined as the value at which the cell number reaches a plateau) can be used to examine the cell-adhesion activity of the adsorbed Fn, and represents an inverse of the cell-adhesion activity.[6] Notably, Fnsat is characteristic for each major monomer, and it reflects the difference in the ability of each major monomer to modulate cell-adhesion activity of the adsorbed Fn. The semi-quantitative analysis of Fnsat in Figure 2c is summarized in Table 2. It reveals a lower Fnsat for high-cell-adhesion monomers (Fnsat < 0.2) and a higher Fnsat for the low-cell-adhesion ones (Fnsat > 0.3). The difference in the Fnsat reflects that the high-cell-adhesion major monomers tend to induce “active” Fn, while the low-cell-adhesion monomers tend to induce “inactive” Fn. Notably, the most “active” Fn was found for major monomer 4 (Fnsat < 0.05), while the most “inactive” Fn was found for major monomer 3 (Fnsat > 0.4).
Table 2.
Relationship between major monomers and Fnsat [a].
| High-adhesion monomers | 15 (0.1) | 8 (<0.1) | 13 (<0.1) | 4 (<0.05) | 12 (0.15) | 14 (0.15) | 9 (0.35) |
| Intermediate-adhesion monomers | 2 (0.15) | 11 (0.15) | |||||
| Low-adhesion monomers | 16 (0.3) | 3 (>0.4) |
The first number is the ID of the major monomer, while the number in the bracket is Fnsat..
The difference in Fnsat showed in the Figure 2c and Table 2 indicates that the diversity of the polymer array can not only generate a wide range of different amounts of adsorbed Fn, but is also capable of inducing adsorbed Fn to present different activities. Importantly, both the amount and the activities of the adsorbed Fn can affect cell attachment. For example, the capacities of ethylene oxide and propylene oxide moieties to reduce cell attachment have often been ascribed to their abilities to resist protein adsorption; the data presented here indicated the decreased cell attachment can also be attributed to a “deactivated” state of the adsorbed Fn, for example, monomer 3 and 16 in Figure 2c. We also hypothesize that the difference in Fn adhesion activities observed here could be attributed to the conformational change moderated by the materials.[6,24] It is important to note that it would be ideal to perform cell-adhesion experiments in a serum-free medium in order to avoid the possible exchange between serum proteins and preadsorbed Fn. However, serum-containing media are usually essential to maintain the normal function of cells, and have been widely used to investigate the interactions between adsorbed proteins and cells.[6,24,28] In addition, it is possible for serum protein to replace the preadsorbed Fn during a prolonged cell culture. However, our data and the existing literature showed the preadsorbed Fn is critical for the initial attachment of cells.[6,24,28]
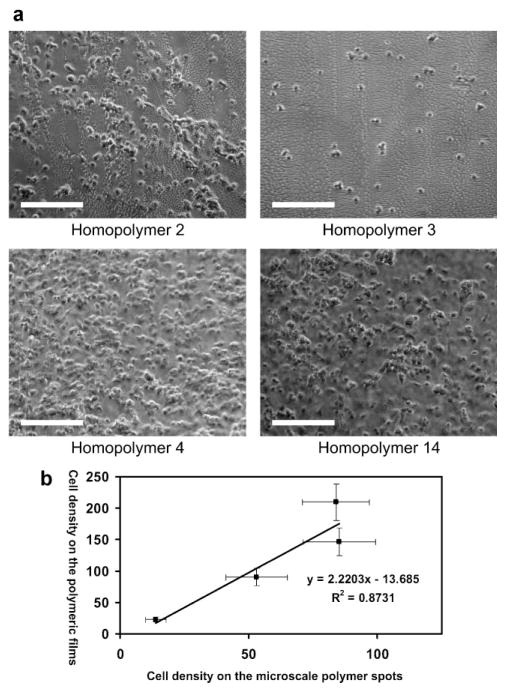
To investigate whether the same range of hEB cell attachment can be reproduced on polymeric films, four 1 cm × 1 cm polymeric films were prepared from two “high-adhesion” monomers (4,13), one “intermediate-adhesion” monomer (2), and one “low-adhesion” monomer (3) on poly(HEMA)-coated glass slides. Phase-contrast images of cell attachment on the polymeric films are shown in Figure 3a. The linear correlation (R2 =0.87) between the cell densities on the polymeric films and polymer spots (Fig. 3b) indicated that controllable cell attachment found on the polymer spots can be reproduced in the large scale films. Similar biological properties of the large films and microscale spots strongly indicate that potential “hit” properties identified via HTscreening could be reproduced in an industrially applicable scale. It is also important to note that the poly(HEMA) layer seems critically important to the scale up. Large films prepared on the acrylate silane instead of poly(HEMA) failed to recapitulate the biological properties found in the microarray format (data not shown).
Figure 3.
a). Phase-contrast images of cell attachments on four different homopolymer films 2, 3, 4, 13; the scale bar in the figure is 200 μm. b) Cell density on the microscale polymer spots as a function of cell density on the polymer films
In summary, new analytical methods for high throughput analysis of cell-materials interaction have been developed. A diverse set of 496 arrayed materials have been designed and prepared with 16 major monomers and 6 minor monomers. Combining high throughput polymer synthesis and rapid quantification of material/protein/cell interactions, we have shown that it is possible to quickly map out the interactions among hEB cell attachment, Fn adsorption, and the chemical structures of the substrates. Both the major and minor monomers have been shown to affect Fn adsorption and hEB cell attachment, and certain combinations of major and minor monomers can lead to a controllable Fn adsorption and hEB cell attachment. Further analysis revealed that the chemical diversity created here can generate a diverse collection of materials with varying amounts of adsorbed Fn and cell adhesion. Controllable hEB cell attachment was shown to be reproducible on four polymer films with three distinct cell-attachment capacities, and the biological properties of the scaled up films were found to be similar to their microscale counterparts. We believe that the integrated high-throughput synthesis and rapid quantification of materials/protein/cell interactions may accelerate the development of biomaterials for various applications, such as materials-directed stem cell differentiations.
Experimental
Combinatorial Array Preparation
Polymers were printed in a humid Ar atmosphere on epoxy monolayer-coated glass slides (Xenopore XENO-SLIDE E, Hawthorne, NJ) that were first dip-coated in 4 vol% pHEMA (pHEMA =poly(hydroxyethyl methacrylate)), using modifications of robotic fluid-handling technology as described previously. Spots were polymerized via 10 s exposure to long-wave UV light, and dried at <50 mTorr (1 Torr =133.32 Pa) for at least seven days. The chips are sterilized for 30 min for each side, and then washed with PBS twice for 15 min to remove the residue monomer or solvent. After that, the chips were coated with 25 μg mL−1 Fn (Sigma) for 1 h, and then washed with PBS and medium before cell seeding.
hESC Culture and EB Formation
Undifferentiated hESCs (H13, WiCell, Wisconsin) were grown on an inactivated mouse embryonic fibroblast (MEF) feeder layer, as previously described. To induce the formation of EBs, undifferentiated hESCs were treated with 1 mg mL−1 type-IV collagenase for 40 min, and then transferred (2:1) to low-attachment plates (10 cm, Ref:3262, Corning) containing 10 mL of differentiation medium [80% knockout-DMEM, supplemented with 20% fetal bovine serum (FBS, Hyclone), 0.5% L-glutamine, 0.2% β-mercaptoethanol, and 1% nonessential amino acids (all from Invitrogen)]. EBs were cultured for eight days at 37 °C and 5% CO2, in a humidified atmosphere, with changes of media every two days.
Protein Labeling and Adsorption
Human fibronectin (1 mg mL−1) was prepared in a 0.1 M sodium carbonate buffer (pH 9). Rhodamine B isothiocyanate was dissolved in Dimethyl sulfoxide (1 mg mL−1), and 50 μL added to the Fn solution. This Fn-Rhodamine B solution was incubated for 8 h at 4 °C. Ammonium chloride was then added to a final concentration of 50 mM, and incubated for a further 2 h at 4 °C. The unbound Rhodamine ITC was then removed by dialysis.
A polymer microarray was incubated in a 0.03 mg mL−1 solution of the Fn-Rhodamine B for 2 h at 37 °C, after which it was removed and rinsed with fresh PBS. A Genepix 4000B scanner (laser wavelength 532 nm) was used to measure the fluorescence of the polymer spots before and after immersion. The intensity before immersion was then subtracted from the intensity after to account for background fluorescence of the polymers. The intensities were then normalized by dividing all values by the maximum intensity.
Immunohistochemsitry
Chips were washed with PBS, fixed with Accustain (Sigma) solution for 30 min, permeabilized with 1% Triton X-100 in PBS for 10 min, and then stained with Cyto 24 (invetrogen) for 1 h. The chips were washed with PBS and water to remove the salts, and air dried. The chips were imaged with iCys laser-scanning Cytometry.
Polymer Film
The polymerization solution was prepared by mixing 3 mL monomer with 40 mg 2,2-dimethoxy-2-phenyl-acetophenone dissolved in 1 mL DMF. 0.2 μL polymerization solution was spread into a 1 cm × 1 cm liquid thin film on the poly(HEMA)-coated glass slide, and polymerized via 6 min exposure to long-wave UV light in a humid Ar atmosphere, and dried at <50 mTorr (1 Torr =133.32 Pa) for at least seven days.
Supplementary Material
Acknowledgments
We gratefully acknowledge E. Luther for the help with the laser scanning cytometry. Funding was kindly provided by Mitsubishi Chemical Company, NIH grant R01 (DE016516-03), and BBSRC (project grant BBC5163791) and studentship to M. T. Supporting Information is available online from Wiley InterScience or from the author.
Contributor Information
Dr. Ying Mei, Department of Chemical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139 (USA)
Prof. Sharon Gerecht, Department of Chemical and Bimolecular Engineering, Johns Hopkins University, Baltimore, MD 21218 (USA)
Dr. Michael Taylor, Laboratory of Biophysics and Surface Analysis, School of Pharmacy, The University of Nottingham, Nottingham, NG7 2RD (UK)
Dr. Andrew J. Urquhart, Laboratory of Biophysics and Surface Analysis, School of Pharmacy, The University of Nottingham, Nottingham, NG7 2RD (UK)
Said R. Bogatyrev, Department of Chemical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139 (USA)
Dr. Seung-Woo Cho, Department of Chemical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139 (USA)
Prof. Martyn C. Davies, Laboratory of Biophysics and Surface Analysis, School of Pharmacy, The University of Nottingham, Nottingham, NG7 2RD (UK)
Prof. Morgan R. Alexander, Laboratory of Biophysics and Surface Analysis, School of Pharmacy, The University of Nottingham, Nottingham, NG7 2RD (UK)
Prof. Robert S. Langer, Department of Chemical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139 (USA)
Dr. Daniel G. Anderson, David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, 45 Carleton Street, Building E25-342, Cambridge, MA 02142 (USA)
References
- 1.Tutter AV, Baltus GA, Kadam S. Curr Opin Drug Discovery Dev. 2006;9:169. [PubMed] [Google Scholar]
- 2.Lindvall O, Kokaia Z. Nature. 2006;441:1094. doi: 10.1038/nature04960. [DOI] [PubMed] [Google Scholar]
- 3.Levenberg S, Huang NF, Lavik E, Rogers AB, Itskovitz-Eldor J, Langer R. Proc Natl Acad Sci USA. 2003;100:12741. doi: 10.1073/pnas.1735463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levenberg S, Golub JS, Amit M, Itskovitz-Eldor J, Langer R. Proc Natl Acad Sci USA. 2002;99:4391. doi: 10.1073/pnas.032074999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langer R, Vacanti JP. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 6.Keselowsky BG, Collard DM, Garcia AJ. J Biomed Mater Res Part A. 2003;66:247. doi: 10.1002/jbm.a.10537. [DOI] [PubMed] [Google Scholar]
- 7.Anderson DG, Levenberg S, Langer R. Nat Biotechnol. 2004;22:863. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 8.Anderson DG, Putnam D, Lavik EB, Mahmood TA, Langer R. Biomaterials. 2005;26:4892. doi: 10.1016/j.biomaterials.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 9.Rawsterne RE, Todd SJ, Gough JE, Farrar D, Rutten FJ, Alexander MR, Ulijn RV. Acta Biomater. 2007;3:715. doi: 10.1016/j.actbio.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 11.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 12.Gombotz WR, Wang GH, Horbett TA, Hoffman AS. J Biomed Mater Res. 1991;25:1547. doi: 10.1002/jbm.820251211. [DOI] [PubMed] [Google Scholar]
- 13.Lee JH, Kopecek J, Andrade JD. J Biomed Mater Res. 1989;23:351. doi: 10.1002/jbm.820230306. [DOI] [PubMed] [Google Scholar]
- 14.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science. 1998;282:1145. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 15.Gadue P, Huber TL, Nostro MC, Kattman S, Keller GM. Exp Hematol. 2005;33:955. doi: 10.1016/j.exphem.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Keller GM. Curr Opin Cell Biol. 1995;7:862. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 17.Park SP, Lee YJ, Lee KS, Ah Shin H, Cho HY, Chung KS, Kim EY, Lim JH. Hum Reprod. 2004;19:676. doi: 10.1093/humrep/deh102. [DOI] [PubMed] [Google Scholar]
- 18.Luther E, Kamentsky L, Henriksen M, Holden E. In: Cytometry New Developments. 4. Darzynkiewicz Z, Roederer M, Tanke HJ, editors. Vol. 75. Elsevier; USA: 2004. p. 185. [Google Scholar]
- 19.Luther E, Kamentsky L, Henriksen M, Holden E. Methods Cell Biol. 2004;75:185. doi: 10.1016/s0091-679x(04)75008-6. [DOI] [PubMed] [Google Scholar]
- 20.Ratner BD, Bryant SJ. Annu Rev Biomed Eng. 2004;6:41. doi: 10.1146/annurev.bioeng.6.040803.140027. [DOI] [PubMed] [Google Scholar]
- 21.Castner DG, Ratner BD. Surf Sci. 2002;500:28. [Google Scholar]
- 22.Pankov R, Yamada KM. J Cell Sci. 2002;115:3861. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 23.Garcia AJ, Ducheyne P, Boettiger D. Biomaterials. 1997;18:1091. doi: 10.1016/s0142-9612(97)00042-2. [DOI] [PubMed] [Google Scholar]
- 24.Keselowsky BG, Collard DM, Garcia AJ. Biomaterials. 2004;25:5947. doi: 10.1016/j.biomaterials.2004.01.062. [DOI] [PubMed] [Google Scholar]
- 25.McClary KB, Ugarova T, Grainger DW. J Biomed Mater Res. 2000;50:428. doi: 10.1002/(sici)1097-4636(20000605)50:3<428::aid-jbm18>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 26.Urquhart AJ, Taylor M, Anderson DG, Williams PM, Langer RS, Alexander MR, Davies MC. Macromol Rapid Commun. 2008;29:1298. [Google Scholar]
- 27.Chernousov MA, Metsis ML, Koteliansky VE. FEBS Lett. 1985;183:365. doi: 10.1016/0014-5793(85)80811-5. [DOI] [PubMed] [Google Scholar]
- 28.Mei Y, Elliott JT, Smith JR, Langenbach KJ, Wu T, Xu C, Beers KL, Amis EJ, Henderson L. J Biomed Mater Res Part A. 2006;79:974. doi: 10.1002/jbm.a.30883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.