Abstract
The iron-regulated metastasis suppressor N-myc downstream-regulated gene 1 (NDRG1) has been shown to inhibit numerous oncogenic signaling pathways in cancer cells. Recent findings have demonstrated that NDRG1 inhibits the ErbB family of receptors, which function as key inducers of carcinogenesis. NDRG1 attenuates ErbB signaling by inhibiting formation of epidermal growth factor receptor (EGFR)/human epidermal growth factor receptor 2 (HER2) and HER2/HER3 heterodimers and by down-regulating EGFR via a mechanism involving its degradation. Understanding the complex interplay between NDRG1, iron, and ErbB signaling is vital for identifying novel, more effective targets for cancer therapy.
Keywords: cancer therapy, cell signaling, epidermal growth factor receptor (EGFR), iron, metastasis, NDRG1
Metastasis
Metastasis is a multistep process that remains the leading cause of cancer-related deaths worldwide (1). Beginning at the primary tumor site, cancer cells disseminate into the bloodstream and migrate to distant organs where they form secondary tumors (1). Metastasis of these cells can be triggered by a plethora of biochemical factors and molecules as well as the extracellular environment (1). Considering its key role in patient mortality, it is pertinent to understand how metastasis is regulated so as to design therapeutic modalities to inhibit cancer progression.
This review specifically addresses the regulation of the metastasis suppressor N-myc downstream-regulated gene 1 (NDRG1) by iron and its associated downstream signaling pathways that have also been linked to iron homeostasis. This is important, as iron is crucial for tumor cell proliferation and metastasis (2), and thus, it is vital to understand its role in oncogenic signaling.
Iron-regulated metastasis suppressor, NDRG1
The molecule NDRG1 is a well-known metastasis suppressor that results in decreased metastases and improved patient prognosis in several cancer types, including those of the breast, prostate, pancreas, and colon (3–5). Paradoxically, NDRG1 expression is demonstrated to be increased in tumors of the liver, esophagus and cervix, where it stimulates oncogenesis (6–8), although in the majority of studies, it is associated with metastasis suppression (3, 5). In fact, NDRG1 has been recognized for its involvement in cellular signaling, growth, differentiation, lipid biosynthesis, and the stress response (3, 5). Most importantly, in its role as a potent metastasis suppressor, NDRG1 has been of great interest due to its ability to inhibit tumor growth and to decrease cell proliferation, migration, invasion, and angiogenesis (9–19).
Recently, the roles of LSD1 and N-myc in the regulation of NDRG1 expression were investigated, and it was shown that LSD1 co-localizes with N-myc at the promoter region of the NDRG1 gene to inhibit its expression (20). Furthermore, it is well known that NDRG1 is a hypoxia-regulated gene and can be induced by hypoxic conditions via an HIF-1α (hypoxia-inducible factor-1α)-dependent mechanism (21). It has also been demonstrated that NDRG1 expression is regulated by intracellular iron in a wide variety of studies in vitro and in vivo (9, 13, 15, 16, 21–31). In fact, a decrease in cellular iron results in robust up-regulation of NDRG1 at the mRNA and protein level, whereas supplementation of iron leads to decreased NDRG1 expression (21, 22, 27). As an appropriate positive control, the same regulation was also reported for other classical iron-regulated molecules, e.g. the TfR14 (transferrin receptor 1), VEGF1 (vascular endothelial growth factor 1), etc., confirming the physiological nature of the iron depletion reported (21, 22, 27).
Critically, the up-regulation of NDRG1 occurs also in vivo in tumors after treatment of mice (intravenous) with the iron chelator, di-2-pyridylketone 4,4-dimethyl-3-thiosemicarbazone (Dp44mT) (26). This occurred concurrently in tumors with up-regulation of the TfR1 and also VEGF1 (26), both of which are classically known to be up-regulated by iron depletion (32). Dp44mT is a known iron chelator and results in cellular iron efflux and inhibits iron uptake from transferrin, leading to cellular iron depletion in vitro (33). Its in vivo effects were consistent with it binding iron in the tumor and forming an intracellular iron complex that sequestered or redistributed iron away from the cellular iron-sensing mechanisms, resulting in up-regulation of genes classically increased during iron depletion i.e. TfR1 and VEGF1 (25).
Furthermore, because of the treatment of mice with this novel chelator, there were higher iron levels in the liver, which corresponded with down-regulation of NDRG1 and also TfR1 and VEGF1 (26). This effect of increasing liver iron levels was a specific property of Dp44mT and also a related thiosemicarbazone, Triapine® (25), and it proved useful in demonstrating that NDRG1 was regulated by iron levels in vivo. Hence, the regulation of NDRG1 in vivo mirrors the regulation by iron observed in vitro in multiple studies performed by various independent investigators using many different protocols of iron depletion (9, 13, 15, 16, 21–31). Also consistent with this regulation are studies examining patients infected with hepatitis C virus, in which liver iron loading occurs, and NDRG1 is down-regulated (34). In summary, studies in vitro and in vivo clearly demonstrate that NDRG1 is regulated by cellular iron depletion (32). It is noteworthy that whereas iron has been shown to regulate the expression of NDRG1, further studies are required to confirm whether this regulation of NDRG1 levels occur due to a direct response to changes in physiological iron availability.
In terms of the iron-mediated regulation of NDRG1, studies by the authors first demonstrated that this occurred via both HIF-1α-independent and -dependent mechanisms (21). This is important to consider because iron plays an important role in oncogenesis, due to its role in cell cycle progression, DNA synthesis, and proliferation (35, 36). Of note, the iron chelators, desferrioxamine (DFO) and 2-hydroxyl-1-naphthaldehyde isonicotinoyl hydrazone (311), which cause cellular iron depletion and up-regulate HIF-1α, can then transactivate NDRG1 (21). Additionally, the eukaryotic initiation factor, eIF3a, was also shown to positively regulate NDRG1 expression during iron depletion (22). This occurs through the induction and recruitment of eIF3a-positive stress granules that, during stressful conditions, cause NDRG1 mRNA and protein levels to increase (22). This was demonstrated to occur in the absence of HIF-1α expression and may constitute the HIF-1α-independent pathway of NDRG1 up-regulation after iron depletion (22). An additional HIF-1α-independent pathway has also been reported by Zhang et al. (31) who, by using a series of NDRG1 promoter deletion constructs, identified a minimal cis-acting element conferring up-regulation by hypoxia and iron depletion that was localized to an Egr-1 (early growth response 1) and Sp1-overlapping binding site. This is of further interest especially as Egr-1 has been also demonstrated to be regulated by cellular iron levels in vitro (27). Further deletion studies examining the putative hypoxia-response element sites (i.e. upstream of the promoter at −1376 bp and −7503 bp) will be required to assess the importance of the HRE in the direct regulation of NDRG1 by iron (21).
Role of NDRG1 in oncogenic signaling
The role of NDRG1 as a metastasis suppressor has been of great interest over the past decade, with its ability to affect key molecules implicated in carcinogenesis (9, 13–17). It has been reported that NDRG1 can influence multiple signaling pathways responsible for cancer cell migration, proliferation, the epithelial to mesenchymal transition (EMT), and angiogenesis, all of which are hallmarks of metastasis (11, 15, 16, 23, 24), and will be further discussed below.
An important oncogene that is regulated by NDRG1 is cellular Src (c-Src (15)). In fact, c-Src is known to be involved in promoting tumorigenesis by facilitating cellular migration and invasion and has been linked to the EMT (37). This process occurs when cells lose their tight junctions and epithelial phenotype, instead adopting a mesenchymal appearance with associated aggressive characteristics leading to metastasis (37). Although it has been suggested that non-mutated c-Src may not be oncogenic by itself, studies have indicated that c-Src can interact and activate receptor tyrosine kinases and other growth factor receptors (38, 39). These proteins include epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor, and platelet-derived growth factor receptor (PDGFR) (38, 39).
In light of this, it was recently discovered that c-Src signaling can be suppressed via up-regulation of NDRG1 (Fig. 1) (15). This mechanism occurred via inhibition of c-Src activation, with NDRG1 preventing the phosphorylation of Src at its activating site, Tyr-416 (15). This led to inhibition of its downstream targets, Crk-associated substrate (p130Cas) and Abelson murine leukemia viral oncogene homolog 1 (ABL1, also known as c-Abl), both of which participate in modulating Ras-related C3 botulinum toxin substrate 1 (Rac1), a key regulator of cell migration (15). Additionally, NDRG1 was able to inhibit the binding of EGFR to c-Src (15), which is known to be an important means by which c-Src activity is increased (40).
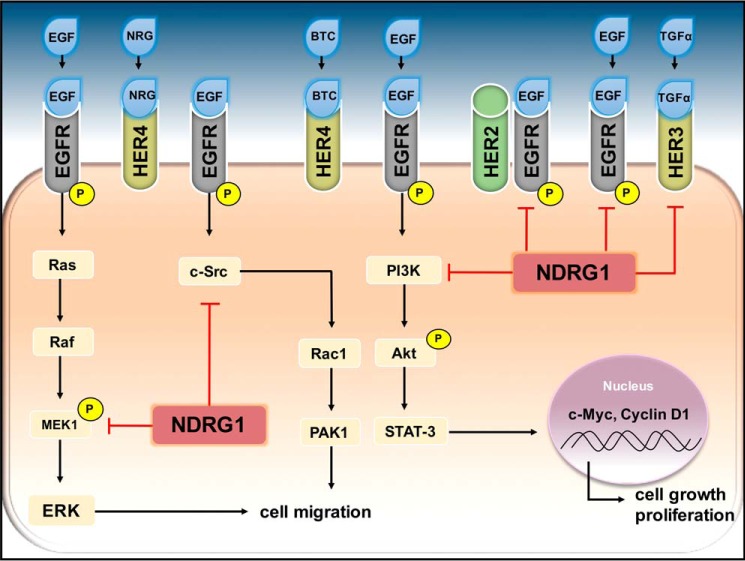
Figure 1.
NDRG1 inhibits oncogenic signaling pathways by down-regulating the ErbB family of receptors. Located on the cell membrane, the ErbB family of receptor tyrosine kinases consists of EGFR, HER2, HER3, and HER4. These receptors are stimulated by various ligands, including the epidermal growth factor (EGF), betacellulin (BTC), transforming growth factor α (TGFα), and neuregulin (NRG). Unlike its other family members, HER2 does not possess a ligand-binding site. The phosphorylation of the ErbB receptors leads to the activation of numerous signaling cascades, including c-Src, PI3K/Akt, and MAPK pathways. As shown, activated EGFR can stimulate Ras to recruit rapidly accelerated fibrosarcoma (Raf), which then activates mitogen-activated protein kinase kinase 1 (MEK1) and subsequently activates the extracellular signal-regulated kinase (ERK), leading to the promotion of cell migration. Interestingly, NDRG1 is able to halt this process by blocking MEK1 activation. Also contributing to the migratory capabilities of tumor cells is the c-Src pathway, which promotes Rac1 and PAK1 expression, contributing to cell migration. NDRG1 can inhibit this pathway by reducing c-Src levels and inhibit its activation. Additionally, NDRG1 is able to inhibit the PI3K/Akt pathway. These latter pathways can promote STAT3, leading to the transcription of c-Myc and cyclin D1, both of which promote cell growth and proliferation. In addition to acting on these key molecules in the above-mentioned oncogenic pathways, NDRG1 is able to inhibit EGFR, HER2, and HER3 levels and negatively regulate their activation and dimerization.
Another oncogenic signaling mechanism that is inhibited by NDRG1 is the phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt) pathway, which is involved in solid tumor growth and is able to further cancer progression and angiogenesis (13, 25). PI3K signaling is commonly activated in cancer, particularly in prostate tumors, where the PI3K/Akt pathway is known to be the most predominant growth factor-activated pathway (41).
The PI3K enzymes are divided into three main classes based on their structure, distribution, and function (42). Class I PI3Ks are of greatest interest, as they result in the biosynthesis of phosphatidylinositol 3,4,5-trisphosphate, resulting in the phosphorylation of Akt by its upstream activator, phosphoinositide-dependent kinase 1 (PDK1) (43). When active, Akt has been correlated with poor patient prognosis due to malignant transformation (44). Interestingly, the tumor suppressor gene, phosphatase and tensin homologue deleted on chromosome 10 (PTEN), can inhibit the PI3K/Akt pathway by dephosphorylating phosphatidylinositol 3,4,5-trisphosphate to phosphatidylinositol 4,5-bisphosphate (41). Acting through this pathway, PTEN can promote cell apoptosis and induce cell cycle arrest (45).
Of note, NDRG1 has been demonstrated to up-regulate PTEN expression leading to suppression of the Akt pathway (13, 46). This is important, as PTEN loses function when PI3K signaling is activated during oncogenesis (41). Interestingly, PTEN is also able to up-regulate NDRG1 expression (46). The role of NDRG1 in affecting the PI3K/Akt signaling cascade can also be mediated by its ability to decrease expression of the PI3K subunits, p-p85α and p-p55γ, as well as the levels of phosphorylated Akt (pAkt) (13). Notably, in glioma cells where PI3K/Akt signaling is linked to a poor prognosis, NDRG1 silencing increases pAkt levels (47). These findings are of significance, as the PI3K/Akt pathway has been implicated in many cancers by inducing the progression of the cell cycle and increasing the expression of pro-proliferative proteins such as cyclin D1 (48). These studies therefore further support the crucial role of NDRG1 in regulating molecules that are known to be major players in cancer metastasis.
The Ras/Raf/MEK/ERK signaling cascade is responsible for regulating the activity of various pro-oncogenic transcription factors (49) and is also targeted by NDRG1 (13, 25). Ras can become activated by growth factors leading to the recruitment of rapidly accelerated fibrosarcoma (Raf) to the cell membrane, where it becomes phosphorylated, activating mitogen-activated protein kinase kinase 1 (MAPKK1, also known as MEK1) and which subsequently activates extracellular signal-regulated kinase (ERK) (Fig. 1) (49). These kinases are then able to translocate to the nucleus to initiate the transcription of genes involved in proliferation and to further promote cell migration (49). A recent study demonstrated that NDRG1 plays a role in halting this cascade of signaling by inhibiting the phosphorylation of MEK1/2 at its activating sites (Ser-217/221) in pancreatic and colon cancer cells (17). Furthermore, NDRG1 was also found to inhibit ERK1/2 activation in prostate cancer cells (25).
ErbB family of receptor tyrosine kinases
The ability of NDRG1 to attenuate the plethora of oncogenic signaling pathways described above could be mediated by the ErbB family of receptors, which function as master regulators of cellular signaling (17, 50), and they were found to be inhibited by NDRG1 (17).
The ErbB family of receptors, which include EGFR, HER2 (human epidermal growth factor receptor 2), HER3 (human epidermal growth factor receptor 3), and HER4 (human epidermal growth factor receptor 4), are all receptor tyrosine kinases that are localized on the cell membrane (51). These receptors are known to be stimulated by numerous ligands, including the epidermal growth factor (EGF), betacellulin (BTC), transforming growth factor α (TGFα), and also several neuregulins (NRGs) (51, 52). Specifically, EGF has the ability to activate EGFR, HER2, and HER3; TGFα specifically activates HER3; NRG1–4 can bind HER3 and HER4; and BTC activates EGFR and HER4 (Fig. 1) (17, 51, 52). This binding of ligand to receptor occurs with high affinity and is rapidly followed by receptor homo- or heterodimerization (51). This event results in the activation of protein kinase activity, leading to phosphorylation of tyrosine residues (53). Notably, unlike the other ErbB receptors, HER2 does not have a ligand-binding site and instead relies on heterodimerization with other ErbB family members for subsequent activation (53). In fact, HER2 is the most preferred interacting partner of the ErbB family members in terms of heterodimerization (54).
The phosphorylation sites of the ErbB family serve as docking sites for different proteins that participate in the regulation and subsequent activation of various intracellular signaling cascades, such as c-Src, PI3K/Akt, and MAPK pathways (55). Specifically, upon heterodimer formation, the EGFR kinase can phosphorylate tyrosine sites of its dimerization partner (56). The site at which the ErbB receptor is phosphorylated is believed to be dependent on the dimerization partner, hence accounting for differences in cellular signaling (57). Although autophosphorylation of EGFR occurs at the carboxyl terminus to maintain an activated state, proteins such as Grb-2, Src, and Abl can directly dock to tyrosine residues at 1068, 891, and 1086, respectively, initiating various downstream signaling cascades (58). This is important, as the interaction between EGFR and Grb-2, for instance, is vital for the induction of the Ras/MAPK pathway (59).
Each of the oncogenic signaling pathways that can be inhibited by NDRG1 are also activated by the receptor tyrosine kinase, EGFR (38, 49, 60). Specifically, EGFR can interact with the regulatory subunit p85 to activate PI3K, as well as directly bind to c-Src to stimulate its activity (Fig. 1) (38, 60). Similarly, the Ras/Raf/MEK/ERK pathway is initiated by EGFR (49). These findings highlight the significance of EGFR as a crucial modulator of signaling pathways in cancer and point to its major role in the anti-oncogenic activity of NDRG1.
Of significance, NDRG1 was demonstrated to attenuate EGF-mediated activation of EGFR and its subsequent phosphorylation at Tyr-1068, Tyr-1086, and Tyr-1148 (17) (all of which participate in the activation of oncogenic pathways PI3K, RAS, and c-Src (Fig. 1) (38)).
In terms of the precise molecular mechanism by which NDRG1 can inhibit EGFR, it has been demonstrated that NDRG1 could play a role in promoting EGFR degradation (17). A recent study has explored the interaction of HER3 with the ubiquitin ligase NEDD4 (neural precursor cell expressed developmentally down-regulated gene 4), which facilitated the receptor's degradation and showed that this binding was greater in the presence of NDRG1 (61). Interestingly, NDRG1 can up-regulate NEDD4 expression (13), which could promote the degradation of EGFR and would be consistent with the results of recent studies (17).
EGFR and iron
As cellular iron levels can regulate NDRG1 expression (21, 22), it is important to consider the regulation of EGFR by this metal ion. As discussed previously, it is well understood that iron is an essential requirement for cancer cell DNA synthesis and cell cycle progression (26). The TfR1 is responsible for the transport and internalization of iron bound to transferrin from the cell surface into the cell via receptor-mediated endocytosis (Fig. 2) (62).
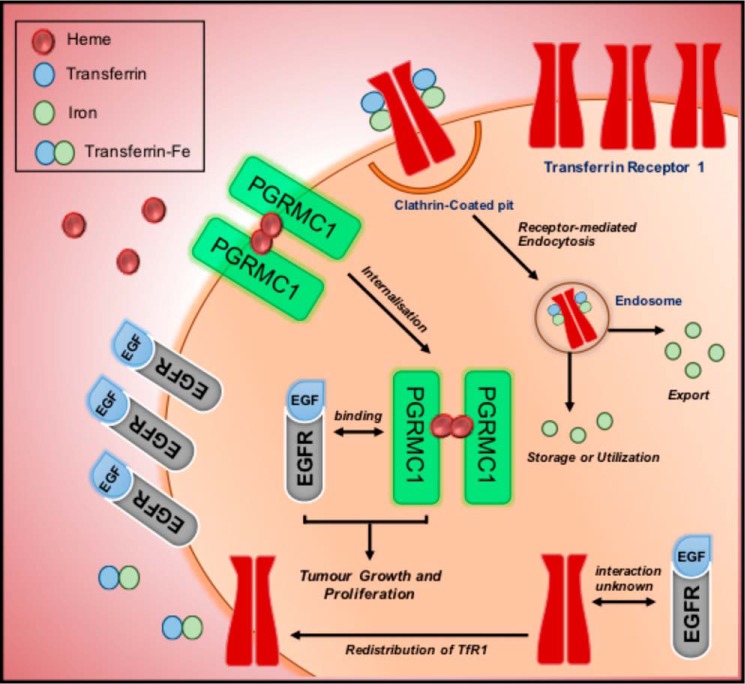
Figure 2.
Relationship between EGFR and iron. Transferrin receptor 1 (TfR1) is responsible for the transport and internalization of iron bound to transferrin (Tf) and is brought into the cell via receptor-mediated endocytosis. After being transported to the endosome, iron is released from Tf and either utilized, stored, or exported out by the cell. Although the relationship between iron-bound transferrin receptor and EGFR is still not fully understood, it has been shown that EGFR may play a role in regulating iron homeostasis by redistributing TfR1 to the membrane to increase iron import for use by the cancer cell. In addition, it is believed that EGFR may play a role in iron metabolism through its association with the progesterone-receptor membrane component 1 (PGRMC1). PGRMC1 is able to form a dimeric structure with heme, and can bind directly to EGFR to promote tumor growth and proliferation.
Although little is known regarding EGFR's role in iron metabolism, a recent study revealed that EGFR could regulate iron homeostasis by directly binding and re-distributing the TfR1 (Fig. 2) (63). In fact, in non-small cell lung carcinoma cells, it was demonstrated that inactivation of EGFR reduced cell surface TfR1 expression, causing a decrease in iron import, and cell cycle arrest (63).
The relationship between EGFR and iron metabolism is also further substantiated by recent studies examining PGRMC1 (progesterone-receptor membrane component 1) (64). PGRMC1 is highly expressed in a variety of tumor types where it can promote proliferation and survival (65).
Until recently, the crucial role of iron in the heme prosthetic groups of PGMRC1 in terms of its structure and function were unknown (66). A recent study has revealed that in the presence of heme, PGRMC1 is able to form a unique dimeric structure between the heme moieties (66). Study of its crystal structure demonstrated that PGRMC1 is able to undergo dimerization through stacking interactions of heme prosthetic groups from each monomer (Fig. 2) (66).
In addition to this, it is understood that PGRMC1 is able to associate with EGFR and cytochrome P450, to regulate proliferation, chemo-resistance, and also sensitivity to EGFR inhibitors such as Erlotinib (64, 66). In fact, silencing of PGRMC1 led to a reduction of EGFR phosphorylation after binding to the EGF ligand (66). Downstream targets of EGFR were also negatively correlated with PGRMC1 silencing, resulting in a down-regulation of phosphorylated Akt and ERK (66).
Ahmed et al. (64) demonstrated that PGRMC1 may directly bind to EGFR and co-localize in endosomes, where PGRMC1 promotes the endosomal recycling of EGFR back to the cell membrane, in preference to its lysosomal degradation. It has been proposed that the heme-mediated dimerization of PGRMC1 enables its interaction with EGFR (Fig. 2) (66). This was validated through studies with succinylacetone, a known inhibitor of δ-aminolevulinic acid dehydratase, which is an essential enzyme in the heme synthesis pathway (66). This study demonstrated that incubation with succinylacetone significantly decreased both PGRMC1 dimerization and its binding to EGFR (66).
The association of PGRMC1 with EGFR can also be further explained by a recent study that showed inactivation of EGFR signaling when PGRMC1 was not maintained on the cell membrane (67). It was suggested that PGRMC1 acts as an adaptor protein to regulate EGFR membrane expression (67). Additionally, PGRMC1 can also be regulated by phosphorylation at Tyr-113, whereby membrane trafficking by PGRMC1 is required for co-localization with EGFR (66). Interestingly, Kabe et al. (66) demonstrated that phosphorylation at this site could cause steric interference, thus discouraging heme binding. It was therefore suggested that heme binding and vesicular trafficking are mutually exclusive processes for PGMRC1 (68).
Although there is still no definitive evidence to suggest a direct link of iron regulation on EGFR, these findings are particularly pertinent considering the roles EGFR and PGRMC1 play in tumor and cancer progression (66).
Degradation mechanisms of EGFR
The link between the ErbB receptors and their signaling in cancer pathogenesis has been widely studied for decades. Considering their role in promoting cancer development, it is important to understand how ErbB family members may be degraded.
Upon ligand binding, EGFR is rapidly internalized into endosomes where it can either be recycled back to the cell surface or transported to lysosomes for degradation (Fig. 3) (69). Numerous studies have reported the role of c-Cbl (cellular casitas B-lineage lymphoma), an E3 ubiquitin ligase, in facilitating the endocytosis of EGFR (70). In fact, c-Cbl acts as a major substrate of tyrosine kinase phosphorylation, undergoing enhanced phosphorylation in response to ligand stimulation by EGF (70, 71).
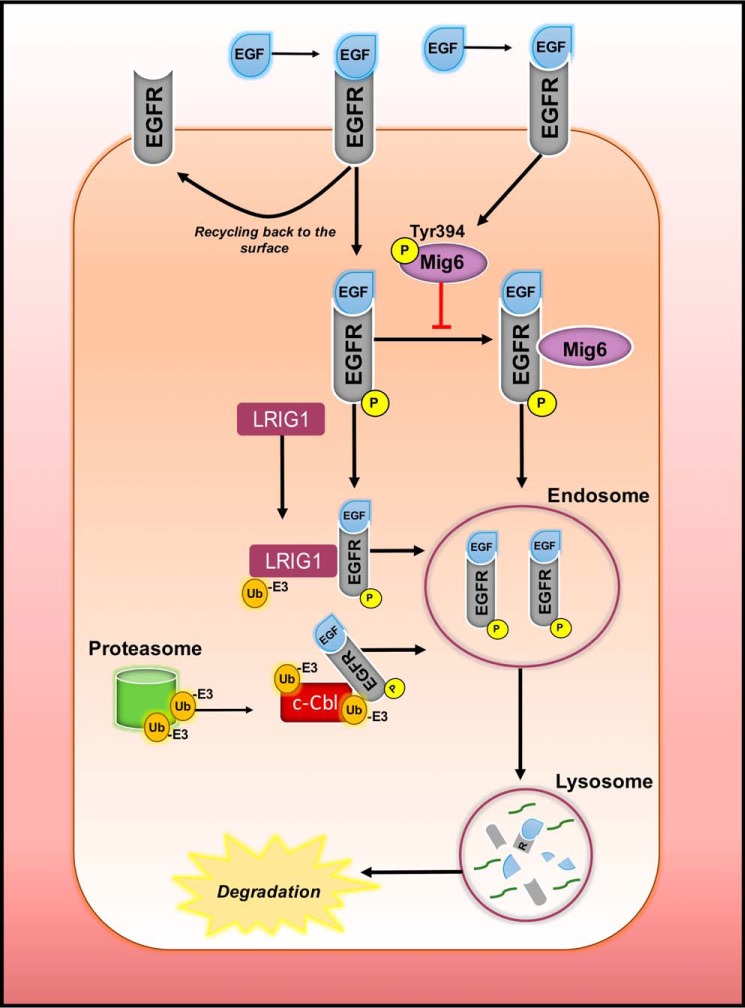
Figure 3.
Prospective mechanisms of degradation of EGFR. Once bound to the ligand EGF, EGFR is rapidly activated and subsequently internalized into endosomes, from which it can either be recycled back to the surface or proceed to lysosomal degradation. A mechanism that explains the degradation process is via E3 ubiquitin ligase (Ub−E3) that has been shown to facilitate the endocytosis of EGFR. The protein leucine-rich repeat and immunoglobulin-like domain 1 (LRIG1) may also be involved by simultaneously ubiquitinating itself and EGFR and recruiting c-Cbl to sort them into endosomes. There has also been evidence of participation of the proteasome through the release of Ub−E3 to further facilitate the degradation process. Additionally, a known inhibitor of EGFR is the mitogen-inducible gene 6 (MIG6) which binds to the carboxyl-terminal lobe of EGFR to deter its signaling. However, in tumor cells, this is unable to occur, as phosphorylation of MIG6 by EGFR at Tyr-394 stops the binding of MIG6 to EGFR, inhibiting receptor degradation.
In addition, c-Cbl was recently linked to the protein LRIG1 (leucine-rich repeat and immunoglobulin-like domain 1) in terms of regulating EGFR and its other family members (72). It was shown that following EGF stimulation, there is a recruitment of c-Cbl to simultaneously ubiquitinate EGFR and LRIG1, sorting them for degradation (Fig. 3) (72, 73). Furthermore, LRIG1 has also demonstrated the ability to suppress ErbB receptor levels by associating with EGFR, HER2, HER3, and HER4 to enhance ligand-stimulated ErbB receptor ubiquitination (73).
The MIG6 (mitogen-inducible factor 6) is also known as ERBB receptor feedback inhibitor 1, or the receptor-associated late transducer (74), and is involved in EGFR degradation (75). MIG6 is an immediate early response gene, encoding a non-kinase scaffolding adaptor protein that is induced by various stresses, hormones, and growth factors, such as EGF, NRG, and TGF-α (74). MIG6 acts to inhibit EGFR by associating with the activated receptor through a carboxyl-terminal binding domain (75). Specifically, a 25-residue epitope from MIG6 binds to the carboxyl-terminal lobe and blocks the formation of the activating dimer interface of EGFR that is required for its signaling (75). Therefore, although MIG6 has been reported to have several roles, including the regulation of stress responses and homeostasis (74), it has an important function as a tumor suppressor (76).
Interestingly, the ability of MIG6 to inhibit tumor growth has also been linked to cellular iron depletion (27). Indeed, the high affinity iron chelators DFO and 311 both induce cellular iron depletion (77) and up-regulate MIG6 (27). Furthermore, it has also been demonstrated that MIG6 is an HIF-1α target gene, with this latter transcription factor directly binding to the MIG6 promoter region, as confirmed by chromatin immunoprecipitation (ChIP) analysis (78, 79).
Considering its ability to promote EGFR degradation, MIG6 presents as a key anti-oncogenic molecule. In fact, MIG6 is able to increase EGFR internalization and trafficking to the lysosome (80). It is believed that EGFR endocytosis is initiated with the recruitment of c-Cbl by Grb-2 to further promote its lysosomal degradation (80). Furthermore, MIG6 can physically obstruct EGFR dimerization and bind to the proteins, syntaxin-8 and intersectin1/2, to foster lysosomal degradation (80). However, in tumor cells, the action of MIG6 to selectively target these receptors is hindered when it becomes phosphorylated at Tyr-394 (Fig. 3) (81). A recent study showed that phosphorylation of MIG6 by EGFR at Tyr-394 and by Src at Tyr-395 can inhibit MIG6 activity (82). Hence, the ability of MIG6 to promote EGFR degradation is prevented under these conditions.
In light of the evidence that EGFR is regulated by NDRG1 (17), it is important to consider whether this metastasis suppressor causes its inhibition of EGFR by promoting the aforementioned mechanisms of degradation. It was recently demonstrated that in the presence of NDRG1, levels of EGFR monomer and dimer were both rapidly reduced in response to the ligand, EGF, suggesting that internalization and subsequent degradation of the receptor may occur (17).
We speculate here that NDRG1 may assist in the endosomal trafficking of EGFR to foster its degradation due to evidence of its role in processing of the low-density lipoprotein (LDL) receptor to the endosome (83). This was identified by a clear reduction of the LDL receptor at the cell surface and decreased accumulation in early endosomes in the absence of NDRG1 (83). Additionally, in NDRG1-depleted cells it was found that LDL receptor degradation was slowed, and this could be explained by NDRG1's ability to interact with known regulators of degradation such as Rab GTPases (83). As such, the role of NDRG1 in LDL degradation could analogously be applied to EGFR, in ushering it to the endosome for eventual degradation.
Other metastasis suppressors
In addition to the metastasis suppressor, NDRG1, there are a host of other proteins that have been identified to perturb cancer progression by down-regulating molecules commonly implicated in metastasis (Fig. 4).
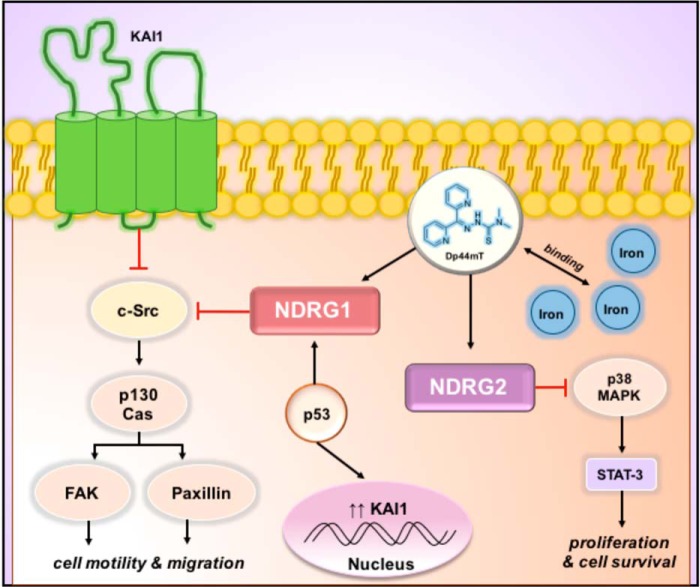
Figure 4.
Metastasis suppressors KAI1, NDRG2, and the tumor suppressor, p53, are regulated by cellular iron levels. KAI1 is a tetraspanin that resides on the cell membrane and has the ability to associate with c-Src to inhibit key molecules such as p130Cas, FAK, and paxillin that are involved in its regulation of cell motility and migration in cancer cells. It is known that NDRG1 is also able to down-regulate p130Cas in an Src-dependent manner, and in turn, it could possibly regulate KAI1. The established tumor suppressor p53 may also play a role in regulating KAI1, by increasing its transcription. The involvement of iron can be understood by evidence demonstrating that iron depletion by chelators such as Dp44mT increases NDRG1 levels in the presence of p53. NDRG2 is also up-regulated by cellular iron depletion and may inhibit cancer cell proliferation and survival by blocking STAT-3 activation in a p38 MAPK-dependent manner.
Although it is not as extensively studied as NDRG1, the Myc-repressed gene NDRG2, which also belongs to the NDRG family, has demonstrated tumor-suppressive functions in malignant carcinomas (84, 85). This was demonstrated by reduced overall survival of prostate cancer patients with low NDRG2 expression (84). The anti-tumorigenic activity of NDRG2 is mediated through its role in signal transduction pathways, whereby expression of NDRG2 inhibited STAT3 activation in a p38 MAPK-dependent manner, leading to decreased proliferation and survival of breast cancer cells (85). Similarly to NDRG1, NDRG2 is also regulated by iron levels, being up-regulated in response to iron depletion (86). In fact, studies examining hepatocellular carcinoma demonstrated that the iron chelator, Dp44mT (33), was able to up-regulate NDRG2, leading to reduced EMT and tumor metastasis via its effects on this latter molecule (86). Specifically, NDRG2 and Dp44mT reduced levels of the receptor gp130 and the activation of its downstream targets STAT3 and ERK1/2 (86).
The tetraspanin, KAI1, is another protein that acts as a metastasis suppressor through its ability to inhibit cancer cell motility and invasiveness (87). This is mediated by the ability of KAI1 to associate with proteins important for cell migration such as focal adhesion kinase (FAK) in its tetraspanin-enriched micro-domain, leading to the down-regulation of FAK function (87). This occurs by blocking the formation of the p130Cas–Crk complex, which is often described as a “molecular switch” for cell motility (88). In fact, as mentioned previously, NDRG1 is able to down-regulate p130Cas in a c-Src-dependent manner (15) and could therefore regulate KAI1. Notably, it has been demonstrated that KAI1 is a downstream target of NDRG1 (89). Specifically, NDRG1 targets the ATF3 (activating transcription factor 3) (89), which can directly bind to the KAI1 promoter. Therefore, these results establish a functional connection between these two metastasis suppressors (89). Furthermore, loss of KAI1 expression is common in metastatic cancers (87), further supporting its significant role as a suppressor of metastasis.
Interestingly, KAI1 has been reported to modulate the activity of several receptor tyrosine kinases, including EGFR (90). The molecular mechanism involved was demonstrated to involve association of KAI1 with the EGFR membrane complex, resulting in more rapid clearance of the ligand-bound receptor from the cell's surface (90). This event also led to a desensitization of the EGFR receptor, which is linked to an increased rate of receptor endocytosis (90).
Significantly, the classical tumor suppressor, p53, is able to increase the transcription of KAI1 (91). This occurs through a p53-responsive element and is of particular interest due to the known regulation of p53 by cellular iron depletion (91). In fact, HIF-1α, which is up-regulated upon iron depletion, leads to stabilization of transcriptionally active wild-type p53 (92). A link between p53, iron, and NDRG1 was also identified through the use of iron chelators (27). This latter study identified that p53 was necessary for the iron chelator-mediated up-regulation of NDRG1, with these agents failing to induce NDRG1 in p53-null H1299 cells (27). However, a more recent investigation has demonstrated that iron depletion markedly up-regulates NDRG1 irrespective of p53 status (93). This observation suggests that p53 may play some role in up-regulating NDRG1 under conditions of iron depletion depending on the cell type.
Iron-binding thiosemicarbazones that up-regulate NDRG1
Considering the aggressive nature of cancer, it is important to explore unique strategies for targeting molecules that function to inhibit key drivers of cancer progression and metastasis, such as the ErbB family of receptors. This can be achieved by agents such as the novel di-2-pyridylketone thiosemicarbazone (DpT) series, which bind cellular iron pools and potently up-regulate NDRG1 via HIF-1α-dependent and -independent mechanisms (21, 22).
These agents possess pronounced and selective anti-proliferative and anti-metastatic activity in vitro and in vivo (17, 18, 26, 28, 33, 86, 94, 95). The most active compounds of this series are Dp44mT and di-2-pyridylketone 4-cyclohexyl-4-methyl-3-thiosemicarbazone (DpC), both of which have shown potency and selectivity in vitro and in vivo against a broad spectrum of cancer types (26, 94–96). Although Dp44mT showed evidence of cardiotoxicity at high, non-optimal doses in mice (26), the recently developed DpC analog demonstrated potent anti-tumor activity in vivo, with no evidence of toxicity even at much higher doses (28, 94, 95). DpC was also demonstrated to be highly potent against the aggressive pancreatic cancer in vivo, almost completely inhibiting tumor growth and being more effective than both Dp44mT and the current gold standard for pancreatic cancer treatment, gemcitabine (17, 28).
These compounds elicit their anti-cancer activity by binding intracellular metal ions such iron and copper, leading to the generation of cytotoxic reactive oxygen species (97–99). These agents also markedly up-regulate the metastasis suppressor, NDRG1 (21, 26, 28), which has been shown to play a major role in their anti-cancer activity (15, 16, 18, 23, 24).
Because of their ability to up-regulate NDRG1, these agents can also potently inhibit the EGFR, Src, WNT, FAK, and PI3K/Akt pathways (14–17, 25). In particular, regarding the ErbB family of receptors, it was recently shown that Dp44mT and DpC significantly reduced EGFR levels and inhibited its activation in response to EGF (17). Additionally, DpC decreased the levels and activation of the oncogenes, HER2 and HER3 (17). This further supports the potential of these thiosemicarbazones as a novel strategy for the treatment of cancer.
Conclusions
It is clear that the iron-regulated metastasis suppressor NDRG1 plays a vital role in regulating oncogenic signaling. This was shown by NDRG1's inhibition of the ErbB family of receptor tyrosine kinases, especially considering that they promote metastasis, a major factor in the death of cancer patients (17). In elucidating the link between NDRG1 and EGFR with iron, a scaffold for controlling and inevitably deterring EGFR-dependent cancers can be identified. Through examination of this intricate and complex relationship, we have prompted further investigation into understanding the mechanisms by which NDRG1 exerts its potent anti-cancer activity.
Acknowledgments
We acknowledge Kyung Chan Park and Leyla Fouani (Molecular Pharmacology and Pathology Program, Department of Pathology, University of Sydney) for their critical evaluation of the manuscript prior to submission.
This work was supported in part by a University of Sydney Postgraduate Award (to S. V. M.), Project Grant 1021607 from the National Health and Medical Research Council of Australia (to D. R. R.), National Health and Medical Research Council of Australia Senior Principal Research Fellowship 1062607 (to D. R. R.), and a National Breast Cancer Foundation and Avner Pancreatic Cancer Foundation project grant (to D. R. R.). This is the sixth article in the Thematic Minireview series “Metals in Biology 2017: Iron transport, storage, and the ramifications.” D. R. R. is a stakeholder in Oncochel Therapeutics LLC and Pty. Ltd., which is developing the thiosemicarbazone, DpC, for the treatment of advanced and resistant cancer.
- TfR1
- transferrin receptor 1
- 311
- 2-hydroxyl-1-naphthaldehyde isonicotinoyl hydrazone
- BTC
- betacellulin
- DFO
- desferrioxamine
- Dp44mT
- 2-pyridylketone 4,4-dimethyl-3-thiosemicarbazone
- DpC
- di-2-pyridylketone 4-cyclohexyl-4-methyl-3-thiosemicarbazone
- EGFR
- epidermal growth factor receptor
- EMT
- epithelial to mesenchymal transition
- NRG
- neuregulin
- p130Cas
- Crk-associated substrate
- PTEN
- phosphatase and tensin homologue
- Raf
- rapidly accelerated fibrosarcoma
- Tf
- transferrin
- FAK
- focal adhesion kinase.
References
- 1. Weigelt B., Peterse J. L., and van't Veer L. J. (2005) Breast cancer metastasis: markers and models. Nat. Rev. Cancer 5, 591–602 [DOI] [PubMed] [Google Scholar]
- 2. Fouani L., Menezes S. V., Paulson M., Richardson D. R., and Kovacevic Z. (2017) Metals and metastasis: exploiting the role of metals in cancer metastasis to develop novel anti-metastatic agents. Pharmacol. Res. 115, 275–287 [DOI] [PubMed] [Google Scholar]
- 3. Fang B. A., Kovačevic Ž., Park K. C., Kalinowski D. S., Jansson P. J., Lane D. J., Sahni S., and Richardson D. R. (2014) Molecular functions of the iron-regulated metastasis suppressor, NDRG1, and its potential as a molecular target for cancer therapy. Biochim. Biophys. Acta 1845, 1–19 [DOI] [PubMed] [Google Scholar]
- 4. Ellen T. P., Ke Q., Zhang P., and Costa M. (2008) NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis 29, 2–8 [DOI] [PubMed] [Google Scholar]
- 5. Kovacevic Z., and Richardson D. R. (2006) The metastasis suppressor, Ndrg-1: a new ally in the fight against cancer. Carcinogenesis 27, 2355–2366 [DOI] [PubMed] [Google Scholar]
- 6. Chua M. S., Sun H., Cheung S. T., Mason V., Higgins J., Ross D. T., Fan S. T., and So S. (2007) Overexpression of NDRG1 is an indicator of poor prognosis in hepatocellular carcinoma. Mod. Pathol. 20, 76–83 [DOI] [PubMed] [Google Scholar]
- 7. Nishio S., Ushijima K., Tsuda N., Takemoto S., Kawano K., Yamaguchi T., Nishida N., Kakuma T., Tsuda H., Kasamatsu T., Sasajima Y., Kage M., Kuwano M., and Kamura T. (2008) Cap43/NDRG1/Drg-1 is a molecular target for angiogenesis and a prognostic indicator in cervical adenocarcinoma. Cancer Lett. 264, 36–43 [DOI] [PubMed] [Google Scholar]
- 8. Ai R., Sun Y., Guo Z., Wei W., Zhou L., Liu F., Hendricks D. T., Xu Y., and Zhao X. (2016) NDRG1 overexpression promotes the progression of esophageal squamous cell carcinoma through modulating Wnt signaling pathway. Cancer Biol. Ther. 17, 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kovacevic Z., Fu D., and Richardson D. R. (2008) The iron-regulated metastasis suppressor, Ndrg-1: Identification of novel molecular targets. Biochim. Biophys. Acta 1783, 1981–1992 [DOI] [PubMed] [Google Scholar]
- 10. Kovacevic Z., Sivagurunathan S., Mangs H., Chikhani S., Zhang D., and Richardson D. R. (2011) The metastasis suppressor, N-myc downstream-regulated gene 1 (NDRG1), upregulates p21 via p53-independent mechanisms. Carcinogenesis 32, 732–740 [DOI] [PubMed] [Google Scholar]
- 11. Chen J., Chen J. K., Nagai K., Plieth D., Tan M., Lee T. C., Threadgill D. W., Neilson E. G., and Harris R. C. (2012) EGFR signaling promotes TGFβ-dependent renal fibrosis. J. Am. Soc. Nephrol. 23, 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun J., Zhang D., Bae D. H., Sahni S., Jansson P., Zheng Y., Zhao Q., Yue F., Zheng M., Kovacevic Z., and Richardson D. R. (2013) Metastasis supressor, NDRG1, mediates its activity through signaling pathways and molecular motors. Carcinogenesis 34, 1943–1954 [DOI] [PubMed] [Google Scholar]
- 13. Kovacevic Z., Chikhani S., Lui G. Y., Sivagurunathan S., and Richardson D. R. (2013) The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. Antioxid. Redox Signal. 18, 874–887 [DOI] [PubMed] [Google Scholar]
- 14. Jin R., Liu W., Menezes S., Yue F., Zheng M., Kovacevic Z., and Richardson D. R. (2014) The metastasis suppressor, NDRG1, modulates β-catenin phosphorylation and nuclear translocation by mechanisms involving FRAT1 and PAK4. J. Cell Sci. 127, 3116–3130 [DOI] [PubMed] [Google Scholar]
- 15. Liu W., Yue F., Zheng M., Merlot A., Bae D. H., Huang M., Lane D., Jansson P., Lui G. Y., Richardson V., Sahni S., Kalinowski D., Kovacevic Z., and Richardson D. R. (2015) The proto-oncogene c-Src and its downstream signaling pathways are inhibited by the metastasis suppressor, NDRG1. Oncotarget 6, 8851–8874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wangpu X., Lu J., Xi R., Yue F., Sahni S., Park K. C., Menezes S., Huang M. L., Zheng M., Kovacevic Z., and Richardson D. R. (2016) Targeting the metastasis suppressor, N-Myc Downstream Regulated Gene-1, with novel di-2-pyridylketone thiosemicarbazones: suppression of tumor cell migration and cell-collagen adhesion by inhibiting focal adhesion kinase/paxillin signaling. Mol. Pharmacol. 89, 521–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kovacevic Z., Menezes S. V., Sahni S., Kalinowski D. S., Bae D. H., Lane D. J., and Richardson D. R. (2016) The metastasis suppressor, N-MYC downstream-regulated gene-1 (NDRG1), down-regulates the ErbB family of receptors to inhibit downstream oncogenic signaling pathways. J. Biol. Chem. 291, 1029–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu W., Xing F., Iiizumi-Gairani M., Okuda H., Watabe M., Pai S. K., Pandey P. R., Hirota S., Kobayashi A., Mo Y. Y., Fukuda K., Li Y., and Watabe K. (2012) N-myc downstream regulated gene 1 modulates Wnt-β-catenin signalling and pleiotropically suppresses metastasis. EMBO Mol. Med. 4, 93–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hosoi F., Izumi H., Kawahara A., Murakami Y., Kinoshita H., Kage M., Nishio K., Kohno K., Kuwano M., and Ono M. (2009) N-myc downstream regulated gene 1/Cap43 suppresses tumor growth and angiogenesis of pancreatic cancer through attenuation of inhibitor of κB kinase β expression. Cancer Res. 69, 4983–4991 [DOI] [PubMed] [Google Scholar]
- 20. Ambrosio S., Amente S., Saccà C. D., Capasso M., Calogero R. A., Lania L., and Majello B. (2017) LSD1 mediates MYCN control of epithelial-mesenchymal transition through silencing of metastatic suppressor NDRG1 gene. Oncotarget 8, 3854–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le N. T., and Richardson D. R. (2004) Iron chelators with high antiproliferative activity up-regulate the expression of a growth inhibitory and metastasis suppressor gene: a link between iron metabolism and proliferation. Blood 104, 2967–2975 [DOI] [PubMed] [Google Scholar]
- 22. Lane D. J., Saletta F., Suryo Rahmanto Y., Kovacevic Z., and Richardson D. R. (2013) N-myc downstream regulated 1 (NDRG1) is regulated by eukaryotic initiation factor 3a (eIF3a) during cellular stress caused by iron depletion. PLoS ONE 8, e57273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Z., Zhang D., Yue F., Zheng M., Kovacevic Z., and Richardson D. R. (2012) The iron chelators Dp44mT and DFO inhibit TGF-β-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J. Biol. Chem. 287, 17016–17028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun J., Zhang D., Zheng Y., Zhao Q., Zheng M., Kovacevic Z., and Richardson D. R. (2013) Targeting the metastasis suppressor, NDRG1, using novel iron chelators: regulation of stress fiber-mediated tumor cell migration via modulation of the ROCK1/pMLC2 signaling pathway. Mol. Pharmacol. 83, 454–469 [DOI] [PubMed] [Google Scholar]
- 25. Dixon K. M., Lui G. Y., Kovacevic Z., Zhang D., Yao M., Chen Z., Dong Q., Assinder S. J., and Richardson D. R. (2013) Dp44mT targets the AKT, TGF-β and ERK pathways via the metastasis suppressor NDRG1 in normal prostate epithelial cells and prostate cancer cells. Br. J. Cancer 108, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whitnall M., Howard J., Ponka P., and Richardson D. R. (2006) A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc. Natl. Acad. Sci. U.S.A. 103, 14901–14906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saletta F., Suryo Rahmanto Y., Noulsri E., and Richardson D. R. (2010) Iron chelator-mediated alterations in gene expression: identification of novel iron-regulated molecules that are molecular targets of hypoxia-inducible factor-1α and p53. Mol. Pharmacol. 77, 443–458 [DOI] [PubMed] [Google Scholar]
- 28. Kovacevic Z., Chikhani S., Lovejoy D. B., and Richardson D. R. (2011) Novel thiosemicarbazone iron chelators induce up-regulation and phosphorylation of the metastasis suppressor N-myc downstream regulated gene 1: a new strategy for the treatment of pancreatic cancer. Mol. Pharmacol. 80, 598–609 [DOI] [PubMed] [Google Scholar]
- 29. Hickok J. R., Sahni S., Mikhed Y., Bonini M. G., and Thomas D. D. (2011) Nitric oxide suppresses tumor cell migration through N-Myc Downstream-regulated Gene-1 (NDRG1) expression: role of chelatable iron. J. Biol. Chem. 286, 41413–41424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salnikow K., Li X., and Lippmann M. (2004) Effect of nickel and iron co-exposure on human lung cells. Toxicol. Appl. Pharmacol. 196, 258–265 [DOI] [PubMed] [Google Scholar]
- 31. Zhang P., Tchou-Wong K. M., and Costa M. (2007) Egr-1 mediates hypoxia-inducible transcription of the NDRG1 gene through an overlapping Egr-1/Sp1 binding site in the promoter. Cancer Res. 67, 9125–9133 [DOI] [PubMed] [Google Scholar]
- 32. Torti S. V., and Torti F. M. (2013) Iron and cancer: more ore to be mined. Nat. Rev. Cancer 13, 342–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yuan J., Lovejoy D. B., and Richardson D. R. (2004) Novel di-2-pyridyl-derived iron chelators with marked and selective antitumor activity: in vitro and in vivo assessment. Blood 104, 1450–1458 [DOI] [PubMed] [Google Scholar]
- 34. Hagist S., Sültmann H., Millonig G., Hebling U., Kieslich D., Kuner R., Balaguer S., Seitz H. K., Poustka A., and Mueller S. (2009) In vitro-targeted gene identification in patients with hepatitis C using a genome-wide microarray technology. Hepatology 49, 378–386 [DOI] [PubMed] [Google Scholar]
- 35. Nyholm S., Mann G. J., Johansson A. G., Bergeron R. J., Gräslund A., and Thelander L. (1993) Role of ribonucleotide reductase in inhibition of mammalian-cell growth by potent iron chelators. J. Biol. Chem. 268, 26200–26205 [PubMed] [Google Scholar]
- 36. Kalinowski D. S., and Richardson D. R. (2005) The evolution of iron chelators for the treatment of iron overload disease and cancer. Pharmacol. Rev. 57, 547–583 [DOI] [PubMed] [Google Scholar]
- 37. Nagathihalli N. S., and Merchant N. B. (2012) Src-mediated regulation of E-cadherin and EMT in pancreatic cancer. Front. Biosci. 17, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 38. Bromann P. A., Korkaya H., and Courtneidge S. A. (2004) The interplay between Src family kinases and receptor tyrosine kinases. Oncogene 23, 7957–7968 [DOI] [PubMed] [Google Scholar]
- 39. Amanchy R., Zhong J., Hong R., Kim J. H., Gucek M., Cole R. N., Molina H., and Pandey A. (2009) Identification of c-Src tyrosine kinase substrates in platelet-derived growth factor receptor signaling. Mol. Oncol. 3, 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leu T. H., and Maa M. C. (2003) Functional implication of the interaction between EGF receptor and c-Src. Front. Biosci. 8, S28–S38 [DOI] [PubMed] [Google Scholar]
- 41. Cantley L. C., and Neel B. G. (1999) New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase AKT pathway. Proc. Natl. Acad. Sci. U.S.A. 96, 4240–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Domin J., and Waterfield M. D. (1997) Using structure to define the function of phosphoinositide 3-kinase family members. FEBS Lett. 410, 91–95 [DOI] [PubMed] [Google Scholar]
- 43. Song G., Ouyang G., and Bao S. (2005) The activation of Akt/PKB signaling pathway and cell survival. J. Cell. Mol. Med. 9, 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wegiel B., Bjartell A., Culig Z., and Persson J. L. (2008) Interleukin-6 activates PI3K/Akt pathway and regulates cyclin A1 to promote prostate cancer cell survival. Int. J. Cancer 122, 1521–1529 [DOI] [PubMed] [Google Scholar]
- 45. Lu X. X., Cao L. Y., Chen X., Xiao J., Zou Y., and Chen Q. (2016) PTEN Inhibits cell proliferation, promotes cell apoptosis, and induces cell cycle arrest via downregulating the PI3K/AKT/hTERT pathway in lung adenocarcinoma A549 cells. Biomed. Res. Int. 2016, 2476842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bandyopadhyay S., Pai S. K., Hirota S., Hosobe S., Tsukada T., Miura K., Takano Y., Saito K., Commes T., Piquemal D., Watabe M., Gross S., Wang Y., Huggenvik J., and Watabe K. (2004) PTEN up-regulates the tumor metastasis suppressor gene Drg-1 in prostate and breast cancer. Cancer Res. 64, 7655–7660 [DOI] [PubMed] [Google Scholar]
- 47. Ma W., Na M., Tang C., Wang H., and Lin Z. (2015) Overexpression of N-myc downstream-regulated gene 1 inhibits human glioma proliferation and invasion via phosphoinositide 3-kinase/AKT pathways. Mol. Med. Rep. 12, 1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Diehl J. A., Cheng M., Roussel M. F., and Sherr C. J. (1998) Glycogen synthase kinase 3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12, 3499–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McCubrey J. A., Steelman L. S., Chappell W. H., Abrams S. L., Wong E. W., Chang F., Lehmann B., Terrian D. M., Milella M., Tafuri A., Stivala F., Libra M., Basecke J., Evangelisti C., Martelli A. M., and Franklin R. A. (2007) Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 1773, 1263–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Citri A., and Yarden Y. (2006) EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 [DOI] [PubMed] [Google Scholar]
- 51. Roskoski R. (2014) The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 79, 34–74 [DOI] [PubMed] [Google Scholar]
- 52. Pinkas-Kramarski R., Shelly M., Guarino B. C., Wang L. M., Lyass L., Alroy I., Alimandi M., Kuo A., Moyer J. D., Lavi S., Eisenstein M., Ratzkin B. J., Seger R., Bacus S. S., Pierce J. H., et al. (1998) ErbB tyrosine kinases and the two neuregulin families constitute a ligand-receptor network. Mol. Cell. Biol. 18, 7602–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Appert-Collin A., Hubert P., Crémel G., and Bennasroune A. (2015) Role of ErbB receptors in cancer cell migration and invasion. Front. Pharmacol. 6, 283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Graus-Porta D., Beerli R. R., Daly J. M., and Hynes N. E. (1997) ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 16, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morandell S., Stasyk T., Skvortsov S., Ascher S., and Huber L. A. (2008) Quantitative proteomics and phosphoproteomics reveal novel insights into complexity and dynamics of the EGFR signaling network. Proteomics 8, 4383–4401 [DOI] [PubMed] [Google Scholar]
- 56. Fan Y. X., Wong L., and Johnson G. R. (2005) EGFR kinase possesses a broad specificity for ErbB phosphorylation sites, and ligand increases catalytic-centre activity without affecting substrate binding affinity. Biochem. J. 392, 417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Olayioye M. A., Graus-Porta D., Beerli R. R., Rohrer J., Gay B., and Hynes N. E. (1998) ErbB-1 and ErbB-2 acquire distinct signaling properties dependent upon their dimerization partner. Mol. Cell. Biol. 18, 5042–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jorissen R. N., Walker F., Pouliot N., Garrett T. P., Ward C. W., and Burgess A. W. (2003) Epidermal growth factor receptor: mechanisms of activation and signalling. Exp. Cell Res. 284, 31–53 [DOI] [PubMed] [Google Scholar]
- 59. Sakaguchi K., Okabayashi Y., Kido Y., Kimura S., Matsumura Y., Inushima K., and Kasuga M. (1998) Shc phosphotyrosine-binding domain dominantly interacts with epidermal growth factor receptors and mediates Ras activation in intact cells. Mol. Endocrinol. 12, 536–543 [DOI] [PubMed] [Google Scholar]
- 60. Zhu G., Decker S. J., Mayer B. J., and Saltiel A. R. (1993) Direct analysis of the binding of the abl src homology-2 domain to the activated epidermal growth-factor receptor. J. Biol. Chem. 268, 1775–1779 [PubMed] [Google Scholar]
- 61. Verma N., Müller A. K., Kothari C., Panayotopoulou E., Kedan A., Selitrennik M., Mills G. B., Nguyen L. K., Shin S., Karn T., Holtrich U., and Lev S. (2017) Targeting of PYK2 synergizes with EGFR antagonists in basal-like TNBC and circumvents HER3-associated resistance via the NEDD4-NDRG1 axis. Cancer Res. 77, 86–99 [DOI] [PubMed] [Google Scholar]
- 62. Morgan E. H. (1981) Transferrin, biochemistry, physiology and clinical significance. Mol. Aspects Med. 4, 1–123 [Google Scholar]
- 63. Wang B., Zhang J., Song F., Tian M., Shi B., Jiang H., Xu W., Wang H., Zhou M., Pan X., Gu J., Yang S., Jiang L., and Li Z. (2016) EGFR regulates iron homeostasis to promote cancer growth through redistribution of transferrin receptor 1. Cancer Lett. 381, 331–340 [DOI] [PubMed] [Google Scholar]
- 64. Ahmed I. S., Rohe H. J., Twist K. E., and Craven R. J. (2010) Pgrmc1 (Progesterone Receptor Membrane Component 1) associates with epidermal growth factor receptor and regulates erlotinib sensitivity. J. Biol. Chem. 285, 24775–24782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cahill M. A., Jazayeri J. A., Catalano S. M., Toyokuni S., Kovacevic Z., and Richardson D. R. (2016) The emerging role of progesterone receptor membrane component 1 (PGRMC1) in cancer biology. Biochim. Biophys. Acta 1866, 339–349 [DOI] [PubMed] [Google Scholar]
- 66. Kabe Y., Nakane T., Koike I., Yamamoto T., Sugiura Y., Harada E., Sugase K., Shimamura T., Ohmura M., Muraoka K., Yamamoto A., Uchida T., Iwata S., Yamaguchi Y., Krayukhina E., et al. (2016) Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat. Commun. 7, 11030–11042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aizen J., and Thomas P. (2015) Role of Pgrmc1 in estrogen maintenance of meiotic arrest in zebrafish oocytes through Gper/Egfr. J. Endocrinol. 225, 59–68 [DOI] [PubMed] [Google Scholar]
- 68. Cahill M. A., Jazayeri J. A., Kovacevic Z., and Richardson D. R. (2016) PGRMC1 regulation by phosphorylation: potential new insights in controlling biological activity! Oncotarget 7, 50822–50827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mizuno E., Iura T., Mukai A., Yoshimori T., Kitamura N., and Komada M. (2005) Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell 16, 5163–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., and Yarden Y. (1998) c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12, 3663–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Galisteo M. L., Dikic I., Batzer A. G., Langdon W. Y., and Schlessinger J. (1995) Tyrosine phosphorylation of the c-Cbl protooncogene protein product and association with epidermal growth-factor (Egf) receptor upon Egf stimulation. J. Biol. Chem. 270, 20242–20245 [DOI] [PubMed] [Google Scholar]
- 72. Gur G., Rubin C., Katz M., Amit I., Citri A., Nilsson J., Amariglio N., Henriksson R., Rechavi G., Hedman H., Wides R., and Yarden Y. (2004) LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 23, 3270–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Laederich M. B., Funes-Duran M., Yen L., Ingalla E., Wu X., Carraway K. L. 3rd, and Sweeney C. (2004) The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J. Biol. Chem. 279, 47050–47056 [DOI] [PubMed] [Google Scholar]
- 74. Anastasi S., Sala G., Huiping C., Caprini E., Russo G., Iacovelli S., Lucini F., Ingvarsson S., and Segatto O. (2005) Loss of RALT/MIG-6 expression in ERBB2-amplified breast carcinomas enhances ErbB-2 oncogenic potency and favors resistance to herceptin. Oncogene 24, 4540–4548 [DOI] [PubMed] [Google Scholar]
- 75. Zhang X., Pickin K. A., Bose R., Jura N., Cole P. A., and Kuriyan J. (2007) Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 450, 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yu X. D., Yang R., and Leng C. J. (2016) Truncation, modification, and optimization of MIG6(segment) (2) peptide to target lung cancer-related EGFR. Comput. Biol. Chem. 61, 251–257 [DOI] [PubMed] [Google Scholar]
- 77. Chaston T. B., Lovejoy D. B., Watts R. N., and Richardson D. R. (2003) Examination of the antiproliferative activity of iron chelators: multiple cellular targets and the different mechanism of action of triapine compared with desferrioxamine and the potent pyridoxal isonicotinoyl hydrazone analogue 311. Clin. Cancer Res. 9, 402–414 [PubMed] [Google Scholar]
- 78. Saarikoski S. T., Rivera S. P., and Hankinson O. (2002) Mitogen-inducible gene 6 (MIG-6), adipophilin and tuftelin are inducible by hypoxia. FEBS Lett. 530, 186–190 [DOI] [PubMed] [Google Scholar]
- 79. Schödel J., Oikonomopoulos S., Ragoussis J., Pugh C. W., Ratcliffe P. J., and Mole D. R. (2011) High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 117, e207–e217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Frosi Y., Anastasi S., Ballarò C., Varsano G., Castellani L., Maspero E., Polo S., Alemà S., and Segatto O. (2010) A two-tiered mechanism of EGFR inhibition by RALT/MIG6 via kinase suppression and receptor degradation. J. Cell Biol. 189, 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang Z., Raines L. L., Hooy R. M., Roberson H., Leahy D. J., and Cole P. A. (2013) Tyrosine phosphorylation of mig6 reduces its inhibition of the epidermal growth factor receptor. ACS Chem. Biol. 8, 2372–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Park E., Kim N., Ficarro S. B., Zhang Y., Lee B. I., Cho A., Kim K., Park A. K., Park W. Y., Murray B., Meyerson M., Beroukhim R., Marto J. A., Cho J., and Eck M. J. (2015) Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6. Nat. Struct. Mol. Biol. 22, 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pietiäinen V., Vassilev B., Blom T., Wang W., Nelson J., Bittman R., Bäck N., Zelcer N., and Ikonen E. (2013) NDRG1 functions in LDL receptor trafficking by regulating endosomal recycling and degradation. J. Cell Sci. 126, 3961–3971 [DOI] [PubMed] [Google Scholar]
- 84. Ren G. F., Tang L., Yang A. Q., Jiang W. W., and Huang Y. M. (2014) Prognostic impact of NDRG2 and NDRG3 in prostate cancer patients undergoing radical prostatectomy. Histol. Histopathol. 29, 535–542 [DOI] [PubMed] [Google Scholar]
- 85. Park Y., Shon S. K., Kim A., Kim K. I., Yang Y., Cho D. H., Lee M. S., and Lim J. S. (2007) SOCS1 induced by NDRG2 expression negatively regulates STAT3 activation in breast cancer cells. Biochem. Biophys. Res. Commun. 363, 361–367 [DOI] [PubMed] [Google Scholar]
- 86. Wang J., Yin D., Xie C., Zheng T., Liang Y., Hong X., Lu Z., Song X., Song R., Yang H., Sun B., Bhatta N., Meng X., Pan S., Jiang H., and Liu L. (2014) The iron chelator Dp44mT inhibits hepatocellular carcinoma metastasis via N-Myc downstream-regulated gene 2 (NDRG2)/gp130/STAT3 pathway. Oncotarget 5, 8478–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liu W. M., and Zhang X. A. (2006) KAI1/CD82, a tumor metastasis suppressor. Cancer Lett. 240, 183–194 [DOI] [PubMed] [Google Scholar]
- 88. Zhang X. A., He B., Zhou B., and Liu L. (2003) Requirement of the p130CAS-Crk coupling for metastasis suppressor KAI1/CD82-mediated inhibition of cell migration. J. Biol. Chem. 278, 27319–27328 [DOI] [PubMed] [Google Scholar]
- 89. Liu W., Iiizumi-Gairani M., Okuda H., Kobayashi A., Watabe M., Pai S. K., Pandey P. R., Xing F., Fukuda K., Modur V., Hirota S., Suzuki K., Chiba T., Endo M., Sugai T., and Watabe K. (2011) KAI1 gene is engaged in NDRG1 gene-mediated metastasis suppression through the ATF3-NFκB complex in human prostate cancer. J. Biol. Chem. 286, 18949–18959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Odintsova E., Sugiura T., and Berditchevski F. (2000) Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr. Biol. 10, 1009–1012 [DOI] [PubMed] [Google Scholar]
- 91. Mashimo T., Watabe M., Hirota S., Hosobe S., Miura K., Tegtmeyer P. J., Rinker-Shaeffer C. W., and Watabe K. (1998) The expression of the KAI1 gene, a tumor metastasis suppressor, is directly activated by p53. Proc. Natl. Acad. Sci. U.S.A. 95, 11307–11311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. An W. G., Kanekal M., Simon M. C., Maltepe E., Blagosklonny M. V., and Neckers L. M. (1998) Stabilization of wild-type p53 by hypoxia-inducible factor 1α. Nature 392, 405–408 [DOI] [PubMed] [Google Scholar]
- 93. Moussa R. S., Kovacevic Z., and Richardson D. R. (2015) Differential targeting of the cyclin-dependent kinase inhibitor, p21(CIP1/WAF1), by chelators with anti-proliferative activity in a range of tumor cell-types. Oncotarget 6, 29694–29711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lovejoy D. B., Sharp D. M., Seebacher N., Obeidy P., Prichard T., Stefani C., Basha M. T., Sharpe P. C., Jansson P. J., Kalinowski D. S., Bernhardt P. V., and Richardson D. R. (2012) Novel second-generation di-2-pyridylketone thiosemicarbazones show synergism with standard chemotherapeutics and demonstrate potent activity against lung cancer xenografts after oral and intravenous administration in vivo. J. Med. Chem. 55, 7230–7244 [DOI] [PubMed] [Google Scholar]
- 95. Guo Z. L., Richardson D. R., Kalinowski D. S., Kovacevic Z., Tan-Un K. C., and Chan G. C. (2016) The novel thiosemicarbazone, di-2-pyridylketone 4-cyclohexyl-4-methyl-3-thiosemicarbazone (DpC), inhibits neuroblastoma growth in vitro and in vivo via multiple mechanisms. J. Hematol. Oncol. 9, 98–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kalinowski D. S., and Richardson D. R. (2007) Future of toxicology–iron chelators and differing modes of action and toxicity: the changing face of iron chelation therapy. Chem. Res. Toxicol. 20, 715–720 [DOI] [PubMed] [Google Scholar]
- 97. Richardson D. R., Sharpe P. C., Lovejoy D. B., Senaratne D., Kalinowski D. S., Islam M., and Bernhardt P. V. (2006) Dipyridyl thiosemicarbazone chelators with potent and selective antitumor activity form iron complexes with redox activity. J. Med. Chem. 49, 6510–6521 [DOI] [PubMed] [Google Scholar]
- 98. Kalinowski D. S., Sharpe P. C., Bernhardt P. V., and Richardson D. R. (2007) Design, synthesis, and characterization of new iron chelators with anti-proliferative activity: structure-activity relationships of novel thiohydrazone analogues. J. Med. Chem. 50, 6212–6225 [DOI] [PubMed] [Google Scholar]
- 99. Lovejoy D. B., Jansson P. J., Brunk U. T., Wong J., Ponka P., and Richardson D. R. (2011) Antitumor activity of metal-chelating compound Dp44mT is mediated by formation of a redox-active copper complex that accumulates in lysosomes. Cancer Res. 71, 5871–5880 [DOI] [PubMed] [Google Scholar]