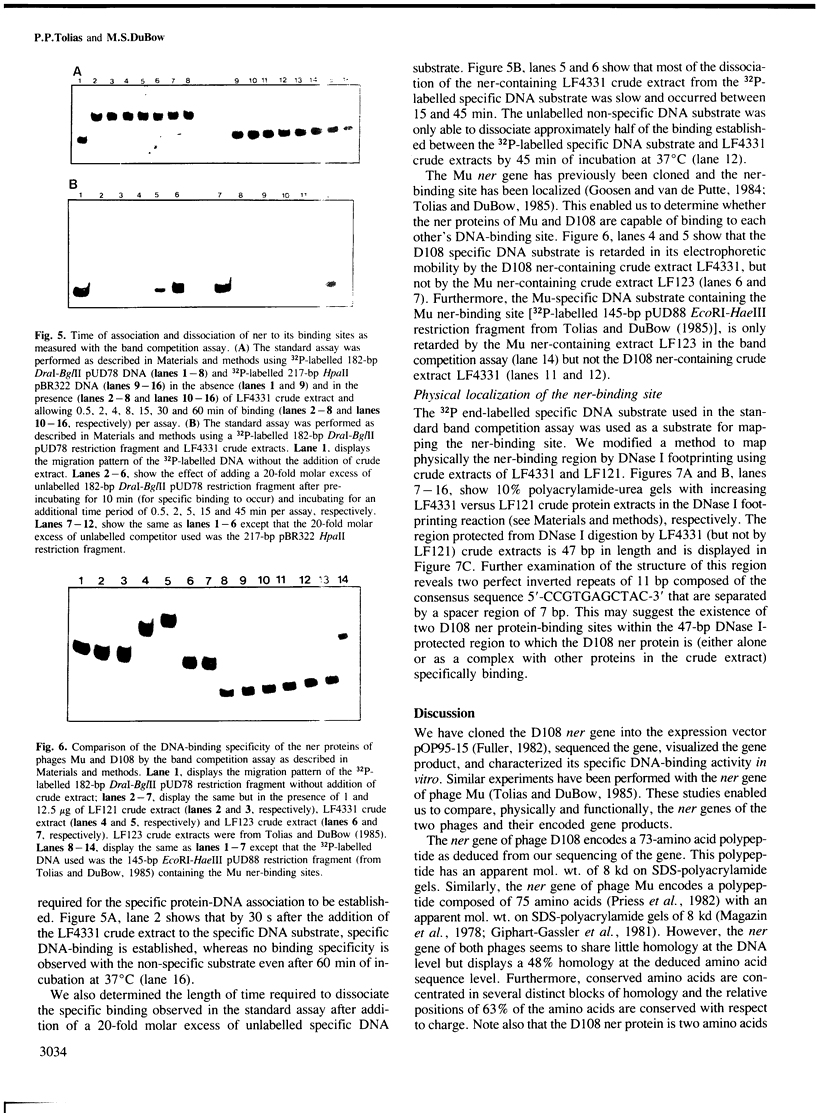
Abstract
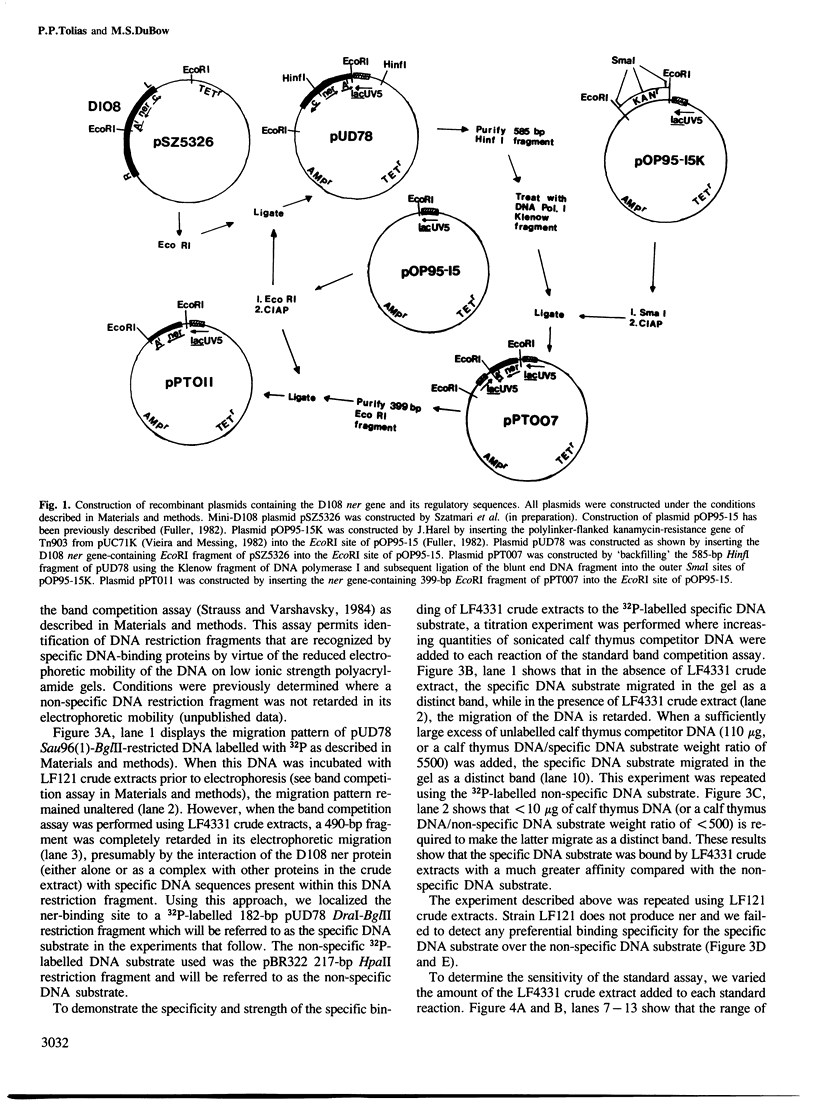
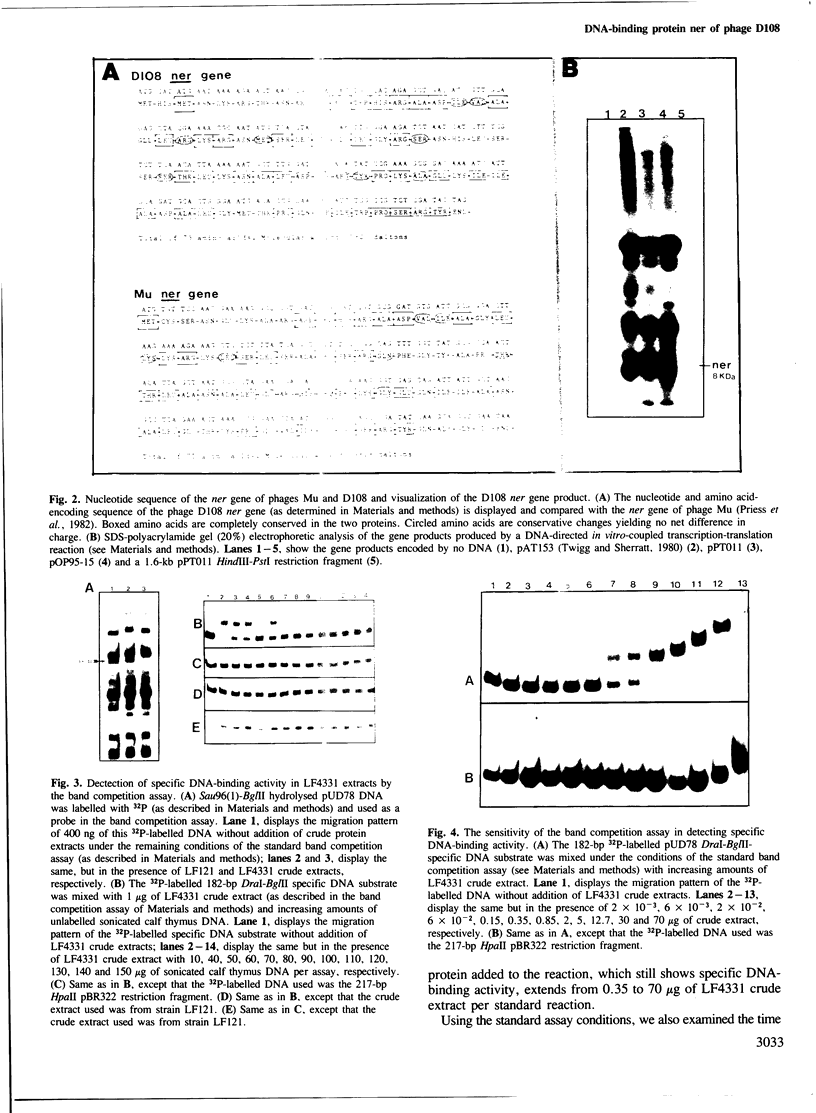
From the transposable Mu-like bacteriophage D108 we have cloned the ner gene under the control of the lac UV5 promoter in the expression vector pOP95-15. The recombinant plasmid, pPT011, overproduced the 8-kd D108 ner protein (visualized by in vitro-coupled transcription-translation) and served as a substrate for DNA sequencing of the D108 ner gene. The ner protein of D108 was found to be 48% homologous to the Mu ner protein, though the DNA sequences that encode these proteins are quite divergent. We used the retardation of migration of 32P-labelled DNA restriction fragments by ner-containing crude protein extracts in polyacrylamide gels (band competition assay) to determine which DNA restriction fragment(s) contained the ner-binding sites. DNA footprinting using crude extracts physically identified the 47-bp DNA sequence that the ner protein was interacting with in the D108 early gene regulatory region. This sequence is located 10 bp downstream from the presumed D108 early gene transcription initiation site. Therefore, by binding strongly to this 47-bp DNA sequence, the D108 ner protein can regulate D108 early gene transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Chaconas G., de Bruijn F. J., Casadaban M. J., Lupski J. R., Kwoh T. J., Harshey R. M., DuBow M. S., Bukhari A. I. In vitro and in vivo manipulations of bacteriophage Mu DNA: cloning of Mu ends and construction of mini-Mu's carrying selectable markers. Gene. 1981 Jan-Feb;13(1):37–46. doi: 10.1016/0378-1119(81)90041-x. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Fuller F. A family of cloning vectors containing the lacUV5 promoter. Gene. 1982 Jul-Aug;19(1):43–54. doi: 10.1016/0378-1119(82)90187-1. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. S., Hull R. C., Curtiss R., 3rd Mutator bacteriophage D108 and its DNA: an electron microscopic characterization. J Virol. 1981 Jan;37(1):420–430. doi: 10.1128/jvi.37.1.420-430.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giphart-Gassler M., Reeve J., van de Putte P. Polypeptides encoded by the early region of bacteriophage Mu synthesized in minicells of Escherichia coli. J Mol Biol. 1981 Jan 5;145(1):165–191. doi: 10.1016/0022-2836(81)90339-9. [DOI] [PubMed] [Google Scholar]
- Goosen N., van de Putte P. Regulation of Mu transposition. I. Localization of the presumed recognition sites for HimD and Ner functions controlling bacteriophage Mu transcription. Gene. 1984 Oct;30(1-3):41–46. doi: 10.1016/0378-1119(84)90103-3. [DOI] [PubMed] [Google Scholar]
- Hull R. A., Gill G. S., Curtiss R., 3rd Genetic characterization of Mu-like bacteriophage D108. J Virol. 1978 Sep;27(3):513–518. doi: 10.1128/jvi.27.3.513-518.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ljungquist E., Bukhari A. I. State of prophage Mu DNA upon induction. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3143–3147. doi: 10.1073/pnas.74.8.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mise K. Isolation and characterization of a new generalized transducing bacteriophage different from P1 in Escherichia coli. J Virol. 1971 Jan;7(1):168–175. doi: 10.1128/jvi.7.1.168-175.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess H., Kamp D., Kahmann R., Bräuer B., Delius H. Nucleotide sequence of the immunity region of bacteriophage Mu. Mol Gen Genet. 1982;186(3):315–321. doi: 10.1007/BF00729448. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Takeda Y., Ohlendorf D. H., Anderson W. F., Matthews B. W. DNA-binding proteins. Science. 1983 Sep 9;221(4615):1020–1026. doi: 10.1126/science.6308768. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Faelen M., Desmet L., Allet B. The products of gene A of the related phages Mu and D108 differ in their specificities. Mol Gen Genet. 1983;190(1):70–79. doi: 10.1007/BF00330326. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Faelen M., Résibois A. Chromosomal rearrangements induced by mini-Mu and mini-D108: mini review and new data. Gene. 1981 Jun-Jul;14(1-2):115–119. doi: 10.1016/0378-1119(81)90153-0. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Giphart-Gassler M., Goosen N., Goosen T., van Leerdam E. Regulation of integration and replication functions of bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):347–353. doi: 10.1101/sqb.1981.045.01.048. [DOI] [PubMed] [Google Scholar]