Abstract
Glucocorticoid receptor (GR) and androgen receptor (AR) are steroid-inducible transcription factors (TFs). The GR and the AR are central regulators of various metabolic, homeostatic and differentiation processes and hence important therapeutic targets, especially in inflammation and prostate cancer, respectively. Hormone binding to these steroid receptors (SRs) leads to DNA binding and activation or repression of their target genes with the aid of interacting proteins, coregulators. However, protein interactomes of these important drug targets have remained poorly defined. We used proximity-dependent biotin identification to map the protein interaction landscapes of GR and AR in the presence and absence of their cognate agonist (dexamethasone, 5α-dihydrotestosterone) and antagonist (RU486, enzalutamide) in intact human cells. We reproducibly identified more than 30 proteins that interacted with the GR in an agonist-specific manner and whose interactions were significantly influenced by the DNA-binding function of the receptor. Interestingly, the agonist-dependent interactome of the GR overlapped considerably with that of the AR. In addition to known coactivators, corepressors and components of BAF (SWI/SNF) chromatin-remodeling complex, we identified a number of proteins, including lysine methyltransferases and demethylases that have not been previously linked to glucocorticoid or androgen signaling. A substantial number of these novel agonist-dependent GR/AR-interacting proteins, e.g. BCOR, IRF2BP2, RCOR1, and TLE3, have previously been implicated in transcription repression. This together with our data on the effect of BCOR, IRF2BP2, and RCOR1 on GR target gene expression suggests multifaceted functions and roles for SR coregulators. These first high confidence SR interactomes will aid in therapeutic targeting of the GR and the AR.
The glucocorticoid receptor (GR)1 and androgen receptor (AR) are hormone-activated transcription factors that belong to the steroid receptor (SR) subfamily of nuclear receptors (NRs). GR mediates the effects of glucocorticoids in a plethora of fundamental biological processes in the human body, such as metabolism, cell proliferation, development, inflammation, and immune responses (1–3). Synthetic glucocorticoid agonists are widely used pharmaceuticals because of their potent anti-inflammatory and anti-immune effects (4). Androgens and the AR are imperative for the development, differentiation and function of male reproductive organs and they also regulate sexually dimorphic characteristics and processes in nongenital tissues, including development of muscle strength (5, 6). The AR is also an important drug target (7). Synthetic AR antagonists, antiandrogens, such as enzalutamide, are widely used in the treatment of metastatic prostate cancer (8).
The GR and the AR, like all NRs, consist of three main functional domains: The N-terminal transactivation domain (NTD), the central DNA-binding domain (DBD), and the C-terminal ligand-binding domain (LBD). Steroid binding to the LBD causes a conformational change, allowing the SR to homodimerize and translocate into the nucleus. In the nucleus, the SR binds to palindromic DNA motifs, steroid response elements (SREs). The SRE recognition is mediated by two zinc fingers of the DBD which is the most conserved domain among the NRs (9). The SREs reside often on distal enhancers where the SRs cooperate with other transcription factors to activate target gene transcription, whereas direct DNA-binding of SRs seems to be less frequently involved in repression of SR target genes (10). The DBDs of SRs are conserved to an extent allowing the AR and the GR but also other 3-keto SRs, mineralocorticoid receptor and progesterone receptor, to recognize the same canonical androgen/glucocorticoid/progesterone/mineralocorticoid response element (ARE/GRE/PRE/MRE), whereas estrogen receptor (ER) α and β bind to a different element (11). Moreover, the AR and the GR share a significant amount of chromatin binding sites (12, 13), and the shared binding sites are associated with genes that are regulated by both androgens and glucocorticoids (12). However, also AR-selective binding sites not recognized by the GR exist (14). The NTD of SRs contains the ligand-independent transcription activation function 1 (AF1) required for the maximal transcriptional activity of the SRs, and the LBDs contain the second transcription activation function (AF2) that is ligand-dependent (15).
Transcriptional regulation by NRs requires, in addition to RNA polymerase II and general TFs, several NR-interacting proteins, coregulators. The coregulators regulate transcription through a variety of functions, such as chromatin remodeling, histone-binding, and post-translational modification of histones and other proteins (16, 17). Initially, coactivators, such as NCOA1, NCOA2, and NCOA3 (SRC-1, SRC-2, and SRC-3), were thought to be recruited by hormone-bound nuclear receptors to enhance gene expression and corepressors, e.g. NCOR1 (N-CoR) and NCOR2 (SMRT), in turn by nonliganded or antagonist-bound nuclear receptors to repress gene expression (18–20). Most of the NR coregulators were originally identified through genetic screens with LBDs as baits in yeast (yeast two-hybrid systems), a milieu that does not normally express any NR or homologue. The early studies focused on coregulator interactions with the AF2 domain in the NR LBD (21). Interactions of full-length NRs with other proteins are still relatively ill-defined and, proteomics-based NR interactomes, including those of the AR and the GR, have remained surprisingly poorly defined (22). The latter may be because of transient nature of interactions and challenges in solubility and solubilization of chromatin-associated proteins when using traditional proteomic methods, such as affinity purification coupled to mass spectrometry (AP-MS) (23, 24).
In this work, we applied proximity-dependent biotin identification (BioID) to reveal the agonist-specific protein-protein interactome of the full-length GR and that of the full-length AR. We fused the GR and the AR with a mutated form of the E. coli biotin ligase (BirA*) that freely attaches biotin to primary amines within 10 nm of the ligase (25–27). Biotin labeling in intact human cells exposed to hormonal agonist, antagonist or vehicle was followed by affinity purification and detection of the biotinylated proteins by MS. Approximately one third of the identified high-confidence agonist-specific GR-interacting proteins (10 of 33) have been previously identified as GR-interacting proteins (GR interactors) by other means. Interestingly, practically all of these interactions were dependent on intact DNA-binding function of the receptor, suggesting that they take place on chromatin. Moreover, the interactome of agonist-bound AR was in qualitative terms highly like that of the GR, and the interactomes of antagonist RU486-bound GR and antiandrogen enzalutamide-bound AR were largely devoid of the proteins interacting with the agonist-bound receptors. Overall, these first unbiased high confidence steroid receptor interactomes provide novel insights into the molecular mechanisms of GR and AR action, which can be applied in the pharmaceutical targeting of glucocorticoid and androgen signaling.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
For generation of the expression vector for tetracycline-inducible expression of N-terminally BirA*-tagged GR, AR and enhanced green fluorescence protein (EGFP), cDNA of the human GR isoform alpha (GR), human AR (AR), or EGFP were transferred with Gateway-cloning (Invitrogen, Carlsbad, CA) to the destination vector pcDNA5-FRT-TO-HA-BirA-GW. The GR DNA-binding mutant R447A was generated by mutagenesis PCR using QuickChange II XL Site-Directed Mutagenesis Kit (forward primer, 5′-GCTGTAAAGTTTTCTTCAAAGCAGCAGTGGAAGGACAGCAC-3′; reverse primer, 5′-CTGTCCTTCCACTGCTGCTTTGAAGAAAACTTTACAGCTTC-3′) and transferred to the same vector.
Cell Line Generation, Culture, and Hormone Treatments
Flp-In 293 T-REx™ cells (Invitrogen) containing a single genomic FRT site and stably expressing the Tet repressor were grown in Dulbecco's modified Eagle medium (DMEM, Gibco, Invitrogen, 41965–039) supplemented with 10% (v/v) FBS, 25 U/ml penicillin, 25 μg/ml streptomycin, 100 μg/ml zeocin (Invitrogen), and 15 μg/ml blasticidin (Invitrogen) (antibiotics were excluded prior to transfection). For cell line generation, Flp-In 293 T-REx™ cells were cotransfected with pcDNA5-FRT-TO-HA-BirA-GR, pcDNA5-FRT-TO-HA-BirA-GR-R447A, pcDNA5-FRT-TO-HA-BirA-AR, or pcDNA5-FRT-TO-HA-BirA-EGFP expression plasmids together with pOG44 vector (Invitrogen) for coexpression of the Flp-recombinase using the TransIT-LT1 transfection reagent (Mirus Bio, Madison, WI). Two days after transfection, the cells were selected with 50 μg/ml hygromycin-B (Invitrogen) and 15 μg/ml blasticidin for 3 weeks. Cells were grown in DMEM supplemented with 2.5% (v/v) charcoal-treated FBS (steroid-depleted medium) before treatments. In all experiments, hormone/antagonist concentrations were as follows: 100 nm dexamethasone (dex, Sigma-Aldrich, St. Louis, MO), 1 μm RU486 (mifepristone, Sigma-Aldrich), 100 nm 5α-dihydrotestosterone (DHT, Steraloids Inc., Newport, RI), 10 μm enzalutamide (MDV3100, Medeia Therapeutics Ltd., Kuopio, Finland) or vehicle (etoh).
Immunoblotting
Cell harvesting, lysis and immunoblotting was performed as described (28). Anti-GR (sc-1003), anti-GAPDH (sc-25778), anti-Lamin B1 (sc-6216) and anti-CoREST (sc-376567) were from Santa Cruz Biotechnology (Dallas, TX), anti-IRF2BP2 (A303–190A), anti-BCOR (A301–673A), anti-RAI1 (A302–317A), and anti-NUP98 (A301–786A) were from Bethyl Laboratories (Montgomery, TX), anti-LSD1 (ab17721) was from Abcam (Cambridge, United Kingdom), and anti-AR as described (29). After primary antibody incubation and washes, membranes were incubated in blocking buffer containing horseradish peroxidase (HRP)-conjugated secondary antibody (Life Technologies, Carlsbad, CA) and detected with chemiluminescence reagent (Pierce, Thermo Fisher Scientific, Waltham, MA). For immunoblotting of the biotinylated proteins, membranes were blocked in 2.5% (w/v) BSA in PBS supplemented with 0.4% (v/v) Triton X-100 and incubated with streptavidin-HRP (Molecular Probes, Life Technologies).
Confocal Microscopy
Confocal microscopy was used to analyze the cellular localization of the biotinylated proteins. Flp-In T-Rex 293 cells expressing BirA*-fused GR, GR-R447A or AR were seeded to coverslips and grown for 24 h. Medium was replaced with steroid-depleted medium and cells were induced with 0.03 μg/ml tetracycline for 24 h before fixing. Cells were treated with 50 μm biotin for 6 h before fixing. In the case of BirA*-GR and BirA*-GR-R447A cell lines, cells were additionally treated with dex, RU486, or vehicle for 6 h before fixing. For the BirA*-AR cell line, cells were treated with DHT or vehicle. Cells were washed with PBS, fixed with 4% (w/v) formaldehyde-PBS for 20 min, and permeabilized with permeabilization buffer (PBS supplemented with 0.1% [v/v] Triton X-100 and 0.5% [w/v] BSA) for 20 min. Coverslips were incubated in primary antibody in permeabilization buffer for 1 h, washed three times for 5 min with PBS, incubated in secondary antibody in permeabilization buffer for 1 h, and washed three times for 5 min with PBS. BirA*-GR, BirA*-GR-R447A and BirA*-AR were detected with anti-HA (MMS-101P, Nordic BioSite AB, Taby, Sweden), biotin with fluorescence-coupled streptavidin DyLight 488 (SA-5488, Vector Laboratories Inc., Burlingame, CA), and lamin with anti-Lamin B1 (sc-6216, Santa Cruz Biotechnology). Rhodamine Red-X (715-295-150, Jackson ImmunoResearch Laboratories Inc., West Grove, PA) and Cy5 (705-175-003, Jackson ImmunoResearch Laboratories Inc.) were used as secondary antibodies. After immunolabeling, coverslips were mounted with ProLong Diamond antifade reagent (Thermo Fisher Scientific) and imaged on Zeiss LSM 800 confocal microscope.
RNAi
For the functional validation of the novel coregulators, A549 cells were seeded to 6-well plates in steroid-depleted medium and transfected with 20 nm ON-TARGETplus SMARTpool siRNAs (Dharmacon) against BCOR (l-004584–01-0005), RAI1 (l-012295–00-0005), RCOR1 (l-014076–00-0005), KDM1A (l-009223–00-0005), IRF2BP2 (l-007177–02-0005) for 72 h using Lipofectamine RNAiMAX (Invitrogen) reagent, and cells exposed to dex or vehicle for 6 h before collecting. ON-TARGETplus Non Targeting Pool (GE Dharmacon, Lafayette, LA) was used as the control siRNA. GR-expressing HEK293 cells were transfected as above, but seeded to their regular growth medium 1 day prior to change to steroid-depleted medium. RNA extraction, cDNA synthesis and fold change calculations were done as previously described for the GR (30) using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) messenger RNA levels for normalization. RT-qPCR primer sequences are available upon request.
Affinity Purification
GR, AR or EGFP -expressing Flp-In 293 T-REx™ cells were grown for 72 h, washed with PBS and the medium was replaced with steroid-depleted medium. After growing in serum-depleted medium for 24 h, cells were induced with tetracycline (0.03 μg/ml, Sigma-Aldrich) for the next 18 h, after which biotin (50 μm, Sigma-Aldrich) with either vehicle, dex, RU486, DHT or enzalutamide was added for 6 h. The cells were washed with ice-cold PBS supplemented with 0.1 mm MgCl2 and 0.1 mm CaCl2, harvested in PBS supplemented with 1 mm EDTA, snap frozen in liquid nitrogen and stored at −70 °C until purification. For affinity purification, ∼1 × 108 cells were lysed in 3 ml of lysis buffer (50 mm Hepes-NaOH [pH 8.0], 5 mm EDTA [pH 8.0], 150 mm NaCl, 50 mm NaF, 0.5% Nonidet P-40, 1 mm PMSF, 1.5 mm Na3VO4, 0.1% SDS, 83 U/ml benzonase, and 0.1× protease inhibitor mixture from Sigma-Aldrich) and sonicated with a probe sonicator (Branson Digital Sonifier) in ice-water bath using eighteen 30 s bursts with 60 s pauses with 30% amplitude. The lysates were then centrifuged at 16,000 × g for 15 min at 4 °C, transferred to new tubes and centrifuged for an additional 10 min at 16,000 × g at 4 °C. The cleared lysates were loaded on spin columns (Bio-Rad Laboratories, Hercules, CA) containing 400 μl Strep-Tactin beads (2-1201-010, IBA Lifesciences, Goettingen, Germany) prewashed in lysis buffer and the beads were washed three times with 1 ml of wash buffer 1 (lysis buffer without SDS and benzonase) and four times with 1 ml of wash buffer 2 (50 mm Hepes-NaOH [pH 8.0], 5 mm EDTA [pH 8.0], 150 mm NaCl, 50 mm NaF). Proteins were eluted twice with 300 μl of 0.5 mm biotin in wash buffer 2, and frozen at −20 °C until further processing. Eluates were neutralized with 100 mm NH4HCO3. Cysteines were reduced with 5 mm Tris(2-carboxyethyl)phosphine and alkylated with 10 mm iodoacetamide. The proteins were then trypsinized to peptides by adding 1 μg trypsin (Promega, Madison, WI). After overnight incubation at 37 °C, samples were quenched with 10% trifluoroacetic acid, purified with C18 Micro SpinColumns (The Nest Group Inc., Southborough, MA) according to the manufacturer's instructions and re-dissolved in 30 μl buffer A (0.1% trifluoroacetic acid and 1% acetonitrile in LC-MS grade water).
MS Analyses
The MS analysis was performed on Orbitrap Elite hybrid mass spectrometer coupled to EASY-nLC II -system using the Xcalibur version 2.7.0 SP1 (Thermo Fisher Scientific). 4 μl of the tryptic peptide mixture was loaded into a C18-packed pre-column (EASY-Column™ 2 cm x 100 μm, 5 μm, 120 Å, Thermo Fisher Scientific) in 10 μl volume of buffer A and then to C18-packed analytical column (EASY-Column™ 10 cm x 75 μm, 3 μm, 120 Å, Thermo Fisher Scientific). Sixty-minute linear gradient at the constant flow rate of 300 nl/minute from 5 to 35% of buffer B (98% acetonitrile and 0.1% formic acid in MS grade water) was used to separate the peptides. Analysis was performed in data-dependent acquisition: one high resolution (60,000) FTMS full scan (m/z 300–1700) was followed by top20 CID- MS2 scans in ion trap (energy 35). Maximum FTMS fill time was 200 ms (Full AGC target 1,000,000) and the maximum fill time for the ion trap was 200 ms (MSn AGC target of 50,000). Precursor ions with more than 500 ion counts were allowed for MSn. To enable the high resolution in FTMS scan preview mode was used.
Search Parameters and Acceptance Criteria
Proteins were identified using Proteome Discoverer™ software with SEQUEST search engine (version 1.4, Thermo Scientific). Thermo .raw files were searched against the human component of the UniProt-database (release 2014_11; 20130 entries) complemented with trypsin, BSA, GFP and tag sequences. Trypsin was used as the enzyme specificity. Search parameters specified a precursor ion tolerance of 15 ppm and fragment ion tolerance of 0.8 Da, with up to two missed cleavages allowed for trypsin. Carbamidomethylation (+57.021464 Da) of cysteine residues was used as static modification whereas oxidation (+15.994491 Da) of methionine and biotinylation (+226.078 Da) of lysine residues or N terminus were used as dynamic modification. Peptide false discovery rate (FDR) was calculated using Percolator node of software and set to <0.01. Spectral counting was used to produce semiquantitative data. Identification metrics for each sample are listed in supplemental Table S1.
RT-qPCR
For analyzing the effect of RU486 in GR target gene expression, GR-expressing HEK293 cells were seeded to 6-well plates and grown 24 h. Medium was replaced with steroid-depleted medium and the cells were grown for 48 h before collecting. Cells were treated with 100 nm dex, 1 μm RU486 or vehicle (ethanol) for 6 h before collecting. RT-qPCR analyses were otherwise performed as described for the siRNA experiments. RT-qPCR primer sequences are available upon request.
Reporter Gene Assays
Reporter gene assay for the GR DNA-binding mutant characterization was done similarly as described for the androgen receptor (31, 32) with the following modifications. COS-1 cells on 12-well plates were cotransfected with pGRE4-tk-luc (Ikonen 1997; Tian 2002) reporter (100 ng/well) together with expression vectors encoding GR and GR-R447A (20 ng/well), and the cells were treated with 100 nm dexamethasone, 1 μm RU486 or vehicle (ethanol) before lysis in passive lysis buffer (Promega). pCMVβ (10 ng/well, Promega) was used for the transfection control. Luciferase and β-galactosidase activities were measured as described before (31).
FRAP
Fluorescence recovery after photobleaching (FRAP) was used to study the mobility of the GR DNA-binding mutant. Flp-In T-Rex 293 cells were seeded to μ-slide 8-well chambers (Ibidi GmbH, Munich, Germany) and transfected with constructs expressing EGFP -tagged GR and GR-R447A. Cells were induced with 100 nm dex, and the nucleus was scanned using 488 nm excitation at 500-ms intervals with Zeiss LSM 700 microscope. After 10 scans, a high intensity bleach pulse at 488 nm was applied to a 1-μm wide rectangular area spanning the nucleus, and scanning of the nucleus was continued until equilibrium in fluorescence distribution was reached. The fluorescence recovery was analyzed from the bleached area.
Experimental Design and Statistical Rationale
GR and AR-specific interactors from three biological replicates were discriminated from background contaminants by using 10 individual BirA*-EGFP control purifications as the control. Significance Analysis of INTeractome (SAINT) (33) V.2.5.0 with default settings was used to determine the statistical significance of the detected interactions. SAINT input and output files are in supplemental Table S2. Interactions with FDR < 0.05 were considered significant with the following exceptions: Acetyl-CoA carboxylase 2 (ACACB, endogenously biotinylated) (34), keratins (KRT2, KRT5, KRT14) and trypsin (unspecific interactors), and tubulin beta-4A chain (TUBB4A, nonspecific mapping of peptides to different tubulin isoforms).
RESULTS
The Protein Interaction Landscape of Agonist and Antagonist-bound GR
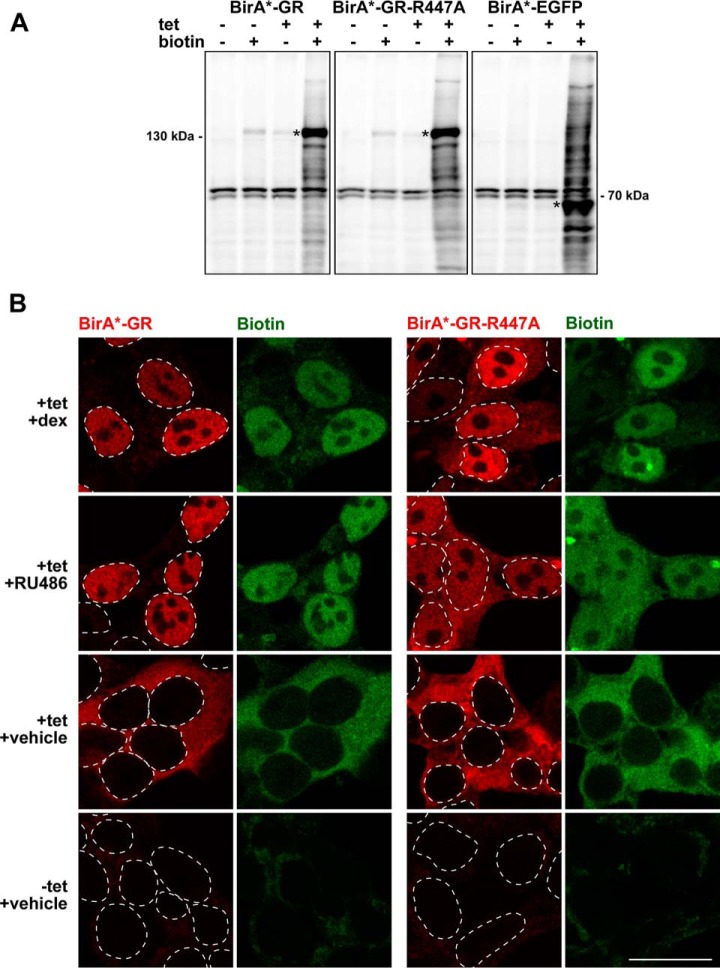
To detect GR-interacting proteins with proximity-dependent biotin identification, we generated HEK293 flp-in T-REx™ cell lines that express BirA*-fused wild-type GR, a DBD-mutated GR (GR-R447A), or enhanced green fluorescent protein (EGFP, a control) in a tetracycline (tet)-inducible fashion (supplemental Fig. S1). The tet-induced expression of BirA*-GR that exceeded the level of endogenous GR in the HEK293 cells by ∼3-fold was used for further experiments. Immunoblotting confirmed biotinylation of endogenous proteins of various sizes when the BirA* -fused bait proteins were expressed in the presence of excess biotin (Fig. 1A). In the absence of ligand, both the BirA*-GR and the BirA*-GR-R447A resided entirely in the cytoplasm. Exposure of the cells to GR-specific agonist dexamethasone (dex) or antagonist RU486, resulted in a complete or nearly complete transfer of the BirA*-GR and the BirA*-GR-R447A to the nucleus, respectively (Fig. 1B). Furthermore, confocal imaging showed that the cellular localization of the biotinylated proteins matched that of the BirA*-GR and the BirA*-GR-R447A (Fig. 1B).
Fig. 1.
Validation of the cell lines expressing BirA*-fused GR, GR-R447A and EGFP. A, Streptavidin-HRP immunoblots of cells with (+) or without (-) added biotin (50 μm) and tetracycline (tet, 0.03 μg/ml). Asterisks depict fusion protein positions. B, Confocal fluorescence microscopy images of BirA*-GR and BirA*-GR-R447A-expressing cells treated with dexamethasone (dex, 100 nm), RU486 (1 μm) or vehicle in the presence of biotin and with or without tetracycline (tet, 0.03 μg/ml). BirA*-fusion proteins were detected with anti-HA (red) and biotinylated proteins with fluorescently labeled streptavidin (green). Dashed lines indicate positions of cell nuclei as inferred from DAPI DNA staining. Scale bar: 20 μm.
Biotinylated proteins from three biological replicates were affinity purified with streptavidin and detected by mass spectrometry (MS). BirA*-tagged EGFP was used to control unspecific interactions. Confocal imaging showed that BirA*-EGFP localizes both to the nucleus and the cytosol (supplemental Fig. S2A). Moreover, analysis of the MS data showed that subcellular distribution of biotinylated proteins in BirA*-EGFP cells was comparable to that of BirA*-GR and BirA*-AR cells, confirming the suitability of BirA*-EGFP as a control (supplemental Fig. S2B). Significance Analysis of INTeractome (SAINT) (33) was utilized to determine the statistical significance of the detected interactions by setting BirA*-EGFP as the control. In total, 33 specific interactors were identified with high confidence (FDR < 0.05) for dex-exposed BirA*-GR (Fig. 2). Using the same statistical and background criteria, vehicle-exposed GR interacted only with four proteins, NUP210, CPVL, DBT, and DDX19A, of which NUP210 and CPVL were also identified as high-confidence interactors with dex. Interestingly, only about one third (10 of 33) of these high confidence interactors observed with dex have been previously found by other techniques for the GR. This demonstrates that BioID can identify well-established as well as novel GR interactors. The known interactors include NCOA1, NCOA2 and NCOA3, Mediator (MED) complex subunit 1 (TRAP220), and subunits, e.g. ARID1A and SMARCA4 (BRG1), of the BAF (Brahma-associated factor)/SWI/SNF (SWItch/Sucrose Non Fermentable) chromatin-remodeling complex. Lysine demethylases KDM1A (LSD1) and JMJD1C (TRIP8), methyltransferase KMT2D (MLL4), transcriptional coactivator TCF20 (SPBP) and its homolog RAI1, and putative corepressors SPEN (SHARP), BCOR, RCOR1 (CoREST), and IRF2BP2 are among the group of novel GR-interacting proteins.
Fig. 2.
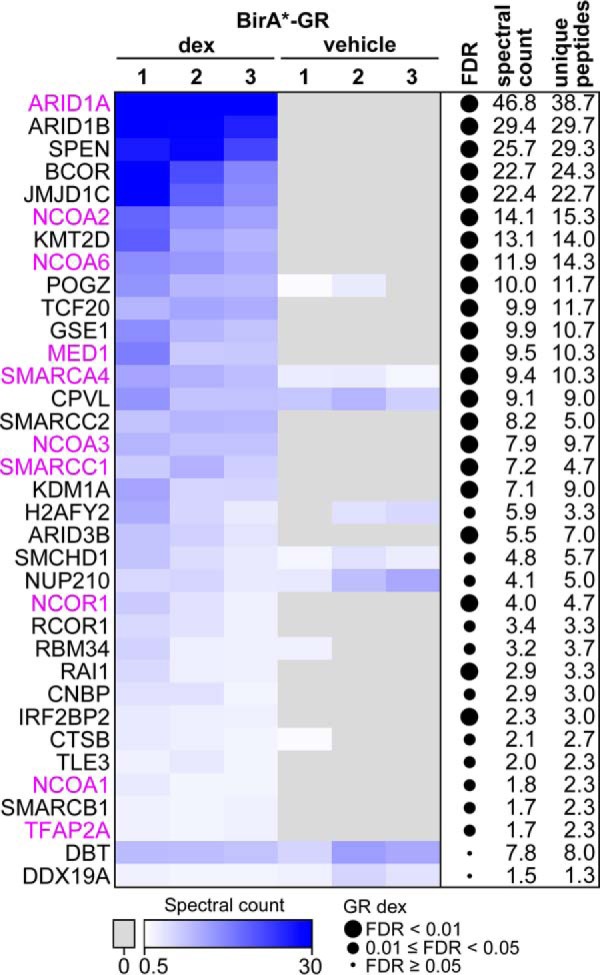
BioID-derived interactome of the GR. Heatmap showing the spectral counts of high confidence interactors (FDR < 0.05 after SAINT analysis) identified with BirA*-GR upon dex or vehicle exposure. Values for three biological replicates from dex-and vehicle-exposed samples are shown. DDX19A and DBT were the only interactors unique to vehicle treatment. Previously reported GR interactors in the BioGRID database (39, 40) and in individual publications (42) are shown in purple. On the right, FDRs, spectral count averages and unique peptide averages are shown for the dex-treated samples. Spectral counts have been normalized to that of GR in each sample.
Novel GR Interactors Coregulate the GR Activity in a Target Gene-selective Fashion
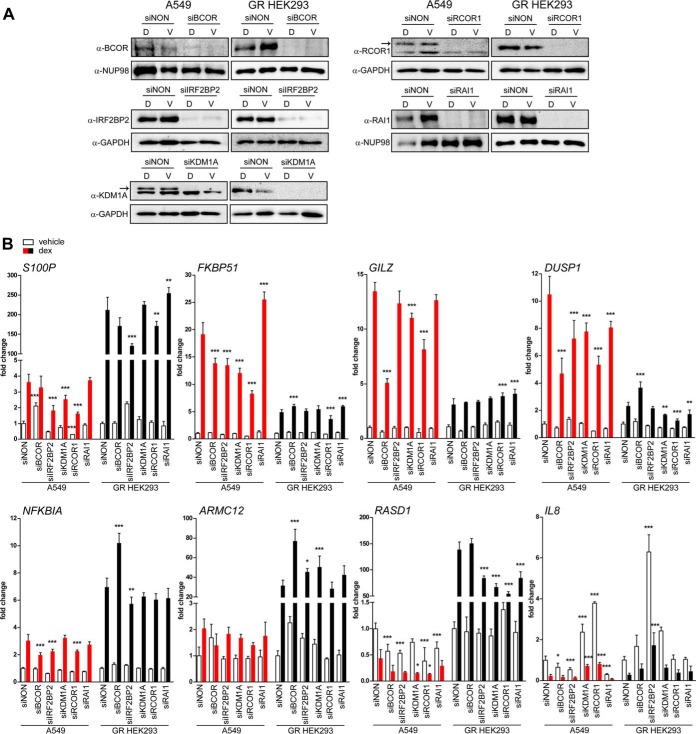
We next tested the gene regulatory effects of some of the novel agonist-bound GR-interacting proteins with a pattern of GR target genes. To that end, we silenced the BCOR, the RCOR1, the IRF2BP2, the RAI1, and the KDM1A in A549 and GR-expressing HEK293 cells by RNA interference (RNAi) (Fig. 3A). Silencing of these putative coregulators did not generally influence the GR protein levels in either cell line as assessed by immunoblotting, with the exception that silencing of BCOR in A549 cells resulted in a slight decrease in GR protein level (supplemental Fig. S3). After silencing, we monitored the effect of the depletion on selected dex-regulated GR target genes by RT-qPCR. As shown in Fig. 3B, the depletion of GR-interacting proteins resulted in gene and cell line specific change in dex-regulation of GR target genes. Interestingly, the depletion of the putative corepressors BCOR, RCOR1, and IRF2BP2 often blunted dex-induced gene expression (e.g. DUSP1 in A549 cells), implying their function as GR coactivators in the regulation of these genes. These results support the biological relevance of these novel GR-interacting proteins as GR coregulators as well as the notion that the activating or repressing effects of SR coregulators are multifaceted and dependent on the target gene context (35, 36).
Fig. 3.
Effect of selected novel GR-interacting proteins on GR target gene expression. A, Immunoblots from A549 and GR-expressing HEK293 cells after siRNA silencing of BCOR, IRF2BP2, KDM1A, RCOR1 or RAI1. Cells were transfected with specific siRNAs for 72 h and exposed for 6 h to 100 nm dexamethasone (D) or vehicle (V) before harvesting. Arrows depict specific bands for KDM1A and RCOR1 in A549 cells. B, Effect of the coregulator silencing on GR target gene expression. Gene expression levels were measured from vehicle- (white) or dexamethasone-treated (red, black) samples by RT-qPCR after silencing coregulator expression. Columns represent the mean ± S.D. of at least three biological replicates. Values are fold changes compared with siNON vehicle treatment in each cell line. Significances are for the difference between siNON and respective siRNA in the corresponding treatment in each cell line. ***, p < 0.001; **, p < 0.01; *, p < 0.05; ANOVA and Bonferroni.
RU486 Promotes Different GR-protein Interactions Than Dexamethasone
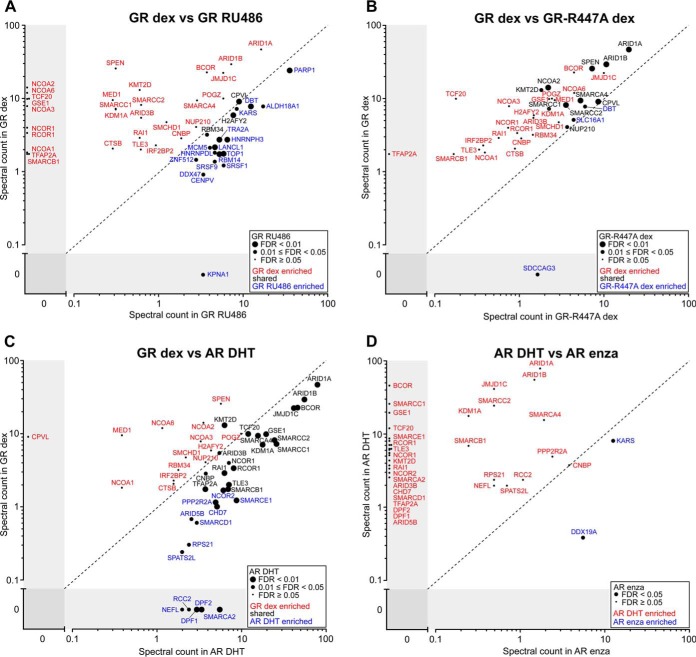
We next compared the interactome of the dex-bound GR to that of antagonist RU486-bound GR. RU486-bound GR showed 20 high confidence interactions (labeled with blue and black letters in Fig. 4A), of which about half are nuclear and half cytosolic proteins (supplemental Table S2). Many of these interactors contain RNA-binding motifs and are involved in RNA processing, such as RNA-binding proteins RBM14 and RBM34, splicing factors SRSF1 and SRSF9, and nuclear ribonucleoproteins HNRNPDL and HNRNPH3. Notably, the RU486-bound receptor was however largely devoid of the interactions seen with the dex-bound GR, including coactivators NCOA1, NCOA2, NCOA3, and NCOA6 (RAP250) (labeled with red letters in Fig. 4A). In keeping with the antagonistic nature of RU486, RT-qPCR analyses showed that RU486 has only a very limited effect on GR target gene expression, suggesting that on its own RU486 is unable to activate the GR (supplemental Fig. S4). However, although being below our selection criteria, many of the dex-bound GR-interacting proteins were detected at higher levels in RU486- than vehicle-exposed samples (supplemental Table S2), which together with GRE-driven reporter gene assay data (supplemental Fig. S5A) support the notion that RU486 possesses some residual agonistic activity.
Fig. 4.
Comparison of GR and AR interactomes induced with different ligands. Comparison of spectral counts in high confidence interactomes of (A) GR in dexamethasone (dex) and RU486 treatment, showing significances for the RU486 treatment, (B) GR and GR-R447A in dex treatment, showing significances for the GR-R447A, (C) GR in dex and AR in DHT treatment, showing significances for the AR DHT treatment, and (D) AR in DHT and enzalutamide (enza) treatment showing significances for the enza treatment. Red and blue letters depict proteins that were significantly enriched in only one of the two compared baits/treatments, whereas common interactors are in black letters.
DNA-binding Function of the GR Promotes Protein Interactions
To analyze if GR's protein interactions are influenced by the receptor's interaction with its DNA binding motif, we generated a GR-R447A mutant that is deficient in binding to GREs (37). As expected for severely compromised DNA-binding of the receptor, the GR-R447A was unable to activate GRE-driven transcription in reporter gene assays (supplemental Fig. S5A) and it (EGFP-GR-R447A) showed increased nuclear mobility compared with the wt receptor (EGFP-GR) in fluorescence recovery after photobleaching assay (supplemental Fig. S5B). Interactome analyses of the biotinylated proteins from the BirA*-GR-R447A-expressing HEK293 cells exposed to dex revealed only 13 high confidence interactions (labeled with black and blue letters in Fig. 4B). Essentially the same interacting proteins were identified as with the wtGR, but with considerably lower spectral counts compared with the wt receptor (labeled with red and black letters in Fig. 4B). These results suggest that GR's protein interactions are largely taking place on the chromatin and being stabilized by GRE-binding. The receptor's DBD may additionally act as a protein-protein interaction surface. However, it is less likely that the observed change in GR interactome is because of alteration(s) of the protein interaction interface of the receptor, as GR-R447A mutation does not directly alter the zinc coordination of the DBD and thus overall zinc finger structure is likely to be unaltered.
Protein Interactomes of the GR and the AR Are Similar but Not Identical
To compare the interactome of the GR to that of the AR, we generated a tetracycline-inducible HEK293 cell line expressing BirA*-AR (supplemental Fig. S6). Cells were treated with natural AR-specific agonist 5α-dihydrotestosterone (DHT), antagonist enzalutamide or vehicle and subjected to affinity purification, MS and statistical analyses as for the GR. Similarly to the situation with the GR, high confidence interactions (32) of the AR were detected only in the presence of DHT and not with vehicle (supplemental Fig. S7). More than half of the identified dex-enriched GR-interacting proteins (19 of 33) were detected with the DHT-bound AR (labeled with black letters in Fig. 4C), whereas the remaining dex-enriched GR-interacting proteins (14 of 33) were below SAINT detection criteria (FDR > 0.05) with the DHT-bound AR (labeled with red letters in Fig. 4C). However, except for CPVL, these interactors were DHT-enriched with the AR in the presence of DHT compared with the situation with vehicle (supplemental Table S2). Conversely, 13 AR-interacting proteins did not pass the SAINT detection criteria for the GR, and therefore were not high confidence interactors of the GR (labeled with blue letters in Fig. 4C). Only five AR-interacting proteins (SMARCA2, DPF2, DPF1, RCC2, and NEFL) were completely absent in the GR interactome. We next compared the interactome of the DHT-bound AR to that of the AR in the presence of antagonist enzalutamide. Interestingly, KARS and DDX19A were the only high confidence interactors identified in the presence of this second-generation antiandrogen (labeled with blue letters in Fig. 4D). In comparison to the situation with RU486 and GR, reduction in spectral counts of DHT-bound AR-interacting proteins was even more pronounced with enzalutamide. Our results indicate that these SR antagonists do not promote the same protein-protein interactions as the agonists. Our results are also in line with the “pure” AR antagonistic nature of enzalutamide (38).
Protein Networks of Agonist-bound GR and AR
A number of the GR and AR interactors found in the BioGRID database (39, 40), were not identified as high confidence interactors in our data (supplemental Fig. S8). For example, JUN, EP300 (Histone acetyltransferase p300), and TRIM24 (Transcription intermediary factor 1-alpha) that have been reported to interact with both the GR and the AR were actually detected as hormone-dependent interactors for both receptors in our data, but they did not pass the strict SAINT-filtering because of too low peptide abundancy. Heat shock proteins HSP90AA1 (heat shock 90 kDa protein 1A) and HSPA1A (heat shock 70 kDa protein 1A) and histone deacetylases HDAC1 and HDAC2 were in turn filtered out because of their presence in control BirA*-EGFP samples and were not therefore identified as high confidence interactors in our strict analysis. Out of the interactions submitted to the BioGRID and found in publications (41–43), 30% (47 of 157) of the previously reported GR interactors were detected with the BirA*-GR, and 27% (65 of 238) of the previously reported AR interactors were found with the BirA*-AR. It is of note that the selection and specificity criteria for a large portion of GR and AR interactors in the BioGRID database are highly variable and many of these interactions were detected in protein-specific studies under forced expression conditions, which hampers comparison of these interactions to our unbiased proteome-wide data.
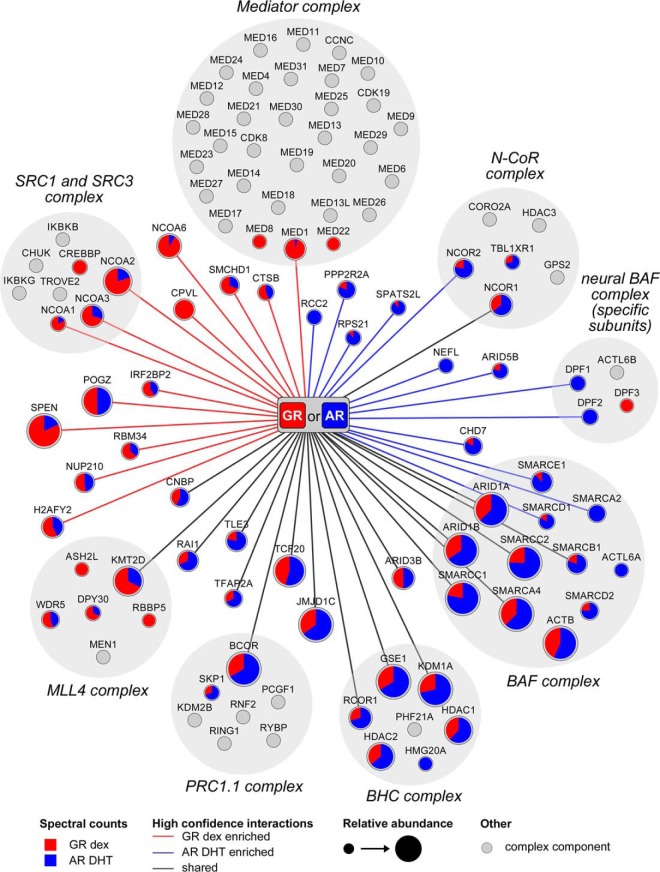
Fig. 5 shows the protein complexes and subunits therein that interact with the agonist-bound GR and the AR based on our unbiased BioID-derived data. In addition to several proteins classified as transcriptional coactivators and a number of chromatin remodeling complex subunits, the agonist-specific interactomes of the GR and the AR include corepressors, such as NCOR1 and NCOR2 reported to interact with both receptors (44–48). Moreover, BCOR (49), RCOR1 (50), SPEN (51), TLE3 (52), and IRF2BP2 (53), previously reported function as corepressors with other TFs, were identified as novel GR or AR-interacting proteins. Importantly, none of these putative corepressors was detected with the GR or the AR in the presence of antagonist or vehicle. Overall, the BioID-derived agonist-enriched interactomes of the GR and the AR reveal only a few receptor-specific interactions. However, the two SRs seem to possess some selectivity for coregulatory complexes: the GR favors MLL4, SRC and mediator complexes, whereas the AR favors BHC, PRC1.1, N-CoR, BAF, and neural BAF complexes.
Fig. 5.
Networks of the BioID-derived interactomes of the GR and the AR. Interactors of the GR with dex and the AR with DHT were associated with different coregulator complexes (gray background). Edges represent high confidence interactions for the GR (red edge), the AR (blue edge) and shared for both (black). Node size shows the relative abundance of each identified protein (spectral count average from the GR and the AR samples), with the color of the node showing the distribution of the detected spectral counts between the GR (red) and the AR (blue). Proteins commonly associated with a protein complex, but not detected in this study, are shown as gray nodes. Established complex compositions were acquired from the CORUM database (81) and for the neural BAF complex as described (82).
DISCUSSION
In addition to their DNA-binding motifs, NRs need to interact with their coregulatory proteins to exert their hormone-specific regulatory effects on the transcription machinery. Recent genome-wide approaches with chromatin immunoprecipitation-sequencing have provided us with comprehensive views of NRs' binding landscapes on chromatin (54), but there is still very scarce proteome-wide information of NRs' hormone-regulated interactomes in intact cells. This may at least be in part because of the challenges in the solubility of chromatin-bound proteins and transient nature of the interactions (23, 24). Indeed, a traditional AP-MS approach using strep-hemagglutinin-tagged GR showed a very limited success for identification of hormone-dependent interactions of the GR (Lempiäinen et al. unpublished). However, multimerized ER response element-containing DNA pull-down assays of nuclear extracts coupled to MS have revealed hormone-enhanced interaction of ERα with 17 proteins, including coactivators NCOA1, NCOA2, NCOA3, NCOA6, MED1, EP300, and CREBBP, and NRIP1 (55). A similar cell-free approach with a natural liver X receptor (LXR) target Abca1 promoter, led to identification of a similar number of proteins of which only one third however were pulled-down in an LXR-dependent manner, and NCOA5 was the only previously identified NR coregulator within the latter group (56). Immunoprecipitation of nuclear extracts of formaldehyde-crosslinked breast cancer cells coupled to MS in turn resulted in identification of a much larger number (108) of ERα-interacting proteins (57), but this group of proteins contained surprisingly few previously known NR coactivators or corepressors; merely NCOA2, EP300, NRIP1, and NCOR2.
In this study, by using the BioID instead, we detected ∼30 high confidence interactions for the GR and the AR in intact human cells. Notably, about one third of these strictly agonist-dependent interactions have previously been implicated in glucocorticoid or androgen signaling. The previously reported interactors include proteins such as NCOA1, NCOA2, NCOA3, NCOR1, and NCOR2 and the novel interactors, for example, histone lysine methyltransferase KMT2D (MLL4), BCOR, and IRF2BP2. KMT2D acts as major mammalian H3K4 mono- and di-methyltransferase required for enhancer activation during cell differentiation (58), whereas BCOR and IRF2BP2 have been mainly reported to be involved in repressing functions (49, 53, 59–61). Some of the novel interactors of the GR and the AR have been previously reported only for one of them. For instance, the lysine demethylase JMJD1C has been reported to coactivate AR (62) and interact with ERβ (63), and the lysine demethylase KDM1A to coactivate both AR and ERα (41, 64), but neither of them has been identified as a GR coregulator before. Our interactome data with a GRE-binding deficient GR mutant provides further mechanistic insight into protein interactions of the agonist-bound GR, strongly suggesting that DNA binding significantly promotes the GR's protein interactions in the nucleus. It is likely that many of the SR-coregulator complexes are assembled on chromatin or the complex formation is enhanced therein. In keeping with this notion, the GR interactions that were also detected in the absence of hormone (CPVL, NUP210, DBT, and DDX19A) and therefore occurred in the cytoplasm were the ones least affected by the R447A mutation.
Numerous NR-interacting proteins have been classified as NR coactivators or corepressors based on the transcriptional outcome in overexpression/reporter gene-based assays. Initially, coactivators, such as NCOA1, NCOA2, and NCOA3, were postulated to be recruited by hormone-bound NRs to enhance target gene transcription, whereas nonliganded NRs excluding 3-keto SRs or antagonist-bound NRs were thought to interact with corepressors, such as NCOR1 and NCOR2, to repress transcription (18–20). Later however, many of the NR-interacting proteins were shown to have opposite effects on transcription depending on cellular environment and target gene. For example, the KDM1A was shown to harbor a dual function as an AR coregulator (65, 66), and the NCOA1 and the NCOR1 and the NCOR2 were also shown to have opposite effects in transcription in certain situations (67–70). Therefore, it has been suggested that coregulators would be classified based on their mechanism of action (e.g. histone acetylase/deacetylase or methyltransferase/demethylase) rather than transcriptional outcome (16). Our data support this notion. For example, depletion of the putative corepressors BCOR, RCOR1, and IRF2BP2 blunted dex-induced gene expression of FKBP51, DUSP1, and NFKBIA in A549 cells, implying their function as GR coactivators in the regulation of these genes. The target gene- and cell-specific effects may derive from differences in coregulator expression profiles and occupancy of other TFs on the enhancers and promoters of target genes. Others have reported similar results with other GR coregulators (35).
Notably, BioID approach did not show evidence for corepressor interactions with antagonist-bound GR or AR (RU486 or enzalutamide, respectively). It seems that the mechanism behind their antagonistic effect is on the reduction of coregulator interactions rather than on the recruitment of corepressors that would inhibit the GR or the AR function. Binding of antagonist to the LBD of the receptor most likely causes a deformed AF2 conformation, preventing the interactions that would occur by the binding of an agonist. Conformation of the enzalutamide-bound AR may diverge from that of the agonist-bound receptor more than that of the RU486-bound GR, leading to a more profound reduction in protein interactions.
SRs are known to share several coregulators (71), which agrees with the large overlap between the AR and the GR interactomes: more than half of their high confidence interactions were shared ones. This is likely to derive from the structural and functional similarities of the two SRs as well as similarities in their chromatin binding patterns (12, 13). In the HEK293 cell background used in this study, most (>90%) of the GR chromatin-binding sites are also bound by the AR (30, 72). Residence of the receptors in comparable chromatin environments may allow them to be exposed to a similar set of coregulators. However, complex-selective differences were also seen between the GR and AR interactomes: the AR seems to favor BHC, PRC1.1, N-CoR, BAF, and neural BAF complexes, whereas the GR appears to prefer MLL4, SRC, and mediator complexes. Dissimilarities in the GR and the AR interactomes may largely derive from differences in their N-terminal domains harboring the AF1 (15). For example, ZMIZ2 (hZimp7) binds to the AR AF1 and coactivates the AR but not the GR (73). Interestingly, the ZMIZ2 additionally interacts with the BAF complex subunits SMARCA4 and SMARCE1 (73), and it was detected as an interactor with the AR, but not with the GR (supplemental Fig. 8). This differential interaction of the ZMIZ2 may have promoted the preferential interaction of the AR with the BAF complex.
Comparison of the BioID-derived interactomes of the GR and the AR to their interactions in the BioGRID database showed that many of the previously reported interactions are not among our list of high confidence interactors. In some cases, this could be because of cell specific expression of the interactors. Furthermore, topology of protein complexes may explain the lack of some interactors. For example, visualization of the full-length ERα-coregulator complex (74) revealed that P300 does not interact directly with the receptor, but it is attached to the complex via interactions with two SRC3 proteins that are directly bound to the ERα. Some of the previously reported AR and GR interactors that were detected with too small peptide abundancies to pass the SAINT filtering, including EP300, may thus represent subunits of protein complexes which because of steric hindrances were not efficiently biotinylated by the BirA* ligase. Moreover, it is of note that the selection and specificity criteria for a large portion of GR and AR interactors in the BioGRID database are highly variable and several of these interactions were detected in protein-specific studies under forced expression conditions, which hampers their comparison to our unbiased proteome-wide data. In this study, BirA*-EGFP combined with SAINT filtering was used for robust and unbiased removal of nonspecific interactors from the data. The caveat of this approach is that it also removes some known SR-interacting proteins, including the heat shock proteins HSP90 and HSP70 (75), and the deacetylases HDAC1 and HDAC2 (76–78) because of their presence in the control BirA*-EGFP samples. As EGFP has also been reported to interfere with the ubiquitination pathway (79), its use as a control in this study may have led to removal of some biologically relevant ubiquitination pathway components from the GR and AR interactomes.
Taken together, our unbiased approach yielded the first high confidence interactomes of the AR and the GR from intact human cells. Only about one third of these strictly agonist-dependent interactions have previously been implicated in glucocorticoid or androgen signaling. The two closely related SRs have highly similar, albeit not identical, interactomes. Because the SRs do not function in isolation of each other, competition of the SRs for common coregulatory/interacting proteins or “coregulator squelching” may take place under conditions where the nuclear concentrations of these interacting proteins are limiting. Coregulator squelching caused by the simultaneous activation of both receptors could lead them to modulate each other's functions (13, 80). Overall, these interactome data provide important insights into the androgen and glucocorticoid signaling milieu, which can be applied in testing of novel steroid receptor ligands and targeting of the GR and the AR.
Data Availability
The mass spectrometry proteomics data have been deposited to PeptideAtlas (www.peptideatlas.org) with the dataset identifier PASS00971.
Supplementary Material
Acknowledgments
We thank Merja Räsänen for assistance in cell culturing, plasmid preparations and for performing the reporter gene assays and Sini Miettinen for assisting in MS sample preparation and MS runs. Dr. Maria Vartiainen is thanked for providing the gateway compatible pcDNA5-FRT-TO-BirA-GW construct.
Footnotes
Author contributions: E.A.N. and J.J.P. designed research; J.K.L., K.V., and R.E.L. performed research; K.V. and M.V. contributed new reagents or analytic tools; J.K.L., E.A.N., H.G., M.V., and J.J.P. analyzed data; J.K.L., E.A.N., and J.J.P. wrote the paper.
* This work was supported by the Academy of Finland, the Finnish Cancer Organisations, UEF Doctoral Programme in Molecular Medicine, and the Sigrid Jusélius Foundation.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- GR
- glucocorticoid receptor
- AP
- affinity purification
- AR
- androgen receptor
- ARE
- androgen response element
- BioID
- proximity-dependent biotin identification
- DBD
- DNA-binding domain
- dex
- dexamethasone
- DHT
- 5α-dihydrotestosterone
- ER
- estrogen receptor
- GRE
- glucocorticoid response element
- NR
- nuclear receptor
- RU486
- mifepristone
- SR
- steroid receptor
- TF
- transcription factor.
REFERENCES
- 1. Ramamoorthy S., and Cidlowski J. A. (2016) Corticosteroids: Mechanisms of action in health and disease. Rheum. Dis. Clin. North Am. 42, 15–31, vii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vandevyver S., Dejager L., and Libert C. (2014) Comprehensive overview of the structure and regulation of the glucocorticoid receptor. Endocr. Rev. 35, 671–693 [DOI] [PubMed] [Google Scholar]
- 3. Weikum E. R., Knuesel M. T., Ortlund E. A., and Yamamoto K. R. (2017) Glucocorticoid receptor control of transcription: Precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 18, 159–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kadmiel M., and Cidlowski J. A. (2013) Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 34, 518–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bardin C. W., and Catterall J. F. (1981) Testosterone: A major determinant of extragenital sexual dimorphism. Science 211, 1285–1294 [DOI] [PubMed] [Google Scholar]
- 6. Gao W., Bohl C. E., and Dalton J. T. (2005) Chemistry and structural biology of androgen receptor. Chem. Rev. 105, 3352–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Watson P. A., Arora V. K., and Sawyers C. L. (2015) Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scher H. I., Fizazi K., Saad F., Taplin M. E., Sternberg C. N., Miller K., de Wit R., Mulders P., Chi K. N., Shore N. D., Armstrong A. J., Flaig T. W., Flechon A., Mainwaring P., Fleming M., Hainsworth J. D., Hirmand M., Selby B., Seely L., de Bono J. S., and AFFIRM Investigators. (2012) Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 367, 1187–1197 [DOI] [PubMed] [Google Scholar]
- 9. Helsen C., and Claessens F. (2014) Looking at nuclear receptors from a new angle. Mol. Cell. Endocrinol. 382, 97–106 [DOI] [PubMed] [Google Scholar]
- 10. Kassel O., and Herrlich P. (2007) Crosstalk between the glucocorticoid receptor and other transcription factors: Molecular aspects. Mol. Cell. Endocrinol. 275, 13–29 [DOI] [PubMed] [Google Scholar]
- 11. Cotnoir-White D., Laperriere D., and Mader S. (2011) Evolution of the repertoire of nuclear receptor binding sites in genomes. Mol. Cell. Endocrinol. 334, 76–82 [DOI] [PubMed] [Google Scholar]
- 12. Sahu B., Laakso M., Pihlajamaa P., Ovaska K., Sinielnikov I., Hautaniemi S., and Janne O. A. (2013) FoxA1 specifies unique androgen and glucocorticoid receptor binding events in prostate cancer cells. Cancer Res. 73, 1570–1580 [DOI] [PubMed] [Google Scholar]
- 13. Pihlajamaa P., Sahu B., and Janne O. A. (2015) Determinants of receptor- and tissue-specific actions in androgen signaling. Endocr. Rev. 36, 357–384 [DOI] [PubMed] [Google Scholar]
- 14. Denayer S., Helsen C., Thorrez L., Haelens A., and Claessens F. (2010) The rules of DNA recognition by the androgen receptor. Mol. Endocrinol. 24, 898–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Warnmark A., Treuter E., Wright A. P., and Gustafsson J. A. (2003) Activation functions 1 and 2 of nuclear receptors: Molecular strategies for transcriptional activation. Mol. Endocrinol. 17, 1901–1909 [DOI] [PubMed] [Google Scholar]
- 16. Millard C. J., Watson P. J., Fairall L., and Schwabe J. W. (2013) An evolving understanding of nuclear receptor coregulator proteins. J. Mol. Endocrinol. 51, T23–T36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meier K., and Brehm A. (2014) Chromatin regulation: How complex does it get? Epigenetics 9, 1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collingwood T. N., Urnov F. D., and Wolffe A. P. (1999) Nuclear receptors: Coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23, 255–275 [DOI] [PubMed] [Google Scholar]
- 19. Perissi V., and Rosenfeld M. G. (2005) Controlling nuclear receptors: The circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 6, 542–554 [DOI] [PubMed] [Google Scholar]
- 20. Dasgupta S., and O'Malley B. W. (2014) Transcriptional coregulators: Emerging roles of SRC family of coactivators in disease pathology. J. Mol. Endocrinol. 53, R47–R59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., and Milburn M. V. (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395, 137–143 [DOI] [PubMed] [Google Scholar]
- 22. Khorasanizadeh S., and Rastinejad F. (2016) Visualizing the architectures and interactions of nuclear receptors. Endocrinology 157, 4212–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lambert J. P., Pawson T., and Gingras A. C. (2012) Mapping physical interactions within chromatin by proteomic approaches. Proteomics 12, 1609–1622 [DOI] [PubMed] [Google Scholar]
- 24. Lambert J. P., Tucholska M., Pawson T., and Gingras A. C. (2014) Incorporating DNA shearing in standard affinity purification allows simultaneous identification of both soluble and chromatin-bound interaction partners. J. Proteomics 100, 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choi-Rhee E., Schulman H., and Cronan J. E. (2004) Promiscuous protein biotinylation by escherichia coli biotin protein ligase. Protein Sci. 13, 3043–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim D. I., Birendra K. C., Zhu W., Motamedchaboki K., Doye V., and Roux K. J. (2014) Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U.S.A. 111, E2453–E2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rytinki M. M., Kaikkonen S., Sutinen P., and Palvimo J. J. (2011) Analysis of androgen receptor SUMOylation. Methods Mol. Biol. 776, 183–197 [DOI] [PubMed] [Google Scholar]
- 29. Karvonen U., Kallio P. J., Janne O. A., and Palvimo J. J. (1997) Interaction of androgen receptors with androgen response element in intact cells. roles of amino- and carboxyl-terminal regions and the ligand. J. Biol. Chem. 272, 15973–15979 [DOI] [PubMed] [Google Scholar]
- 30. Paakinaho V., Kaikkonen S., Makkonen H., Benes V., and Palvimo J. J. (2014) SUMOylation regulates the chromatin occupancy and anti-proliferative gene programs of glucocorticoid receptor. Nucleic Acids Res. 42, 1575–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Makkonen H., Jaaskelainen T., Rytinki M. M., and Palvimo J. J. (2011) Analysis of androgen receptor activity by reporter gene assays. Methods Mol. Biol. 776, 71–80 [DOI] [PubMed] [Google Scholar]
- 32. Kaikkonen S., Paakinaho V., Sutinen P., Levonen A. L., and Palvimo J. J. (2013) Prostaglandin 15d-PGJ(2) inhibits androgen receptor signaling in prostate cancer cells. Mol. Endocrinol. 27, 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2011) SAINT: Probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chandler C. S., and Ballard F. J. (1985) Distribution and degradation of biotin-containing carboxylases in human cell lines. Biochem. J. 232, 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu D. Y., Ou C. Y., Chodankar R., Siegmund K. D., and Stallcup M. R. (2014) Distinct, genome-wide, gene-specific selectivity patterns of four glucocorticoid receptor coregulators. Nucl. Recept. Signal. 12, e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Toropainen S., Malinen M., Kaikkonen S., Rytinki M., Jaaskelainen T., Sahu B., Janne O. A., and Palvimo J. J. (2015) SUMO ligase PIAS1 functions as a target gene selective androgen receptor coregulator on prostate cancer cell chromatin. Nucleic Acids Res. 43, 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hua G., Paulen L., and Chambon P. (2016) GR SUMOylation and formation of an SUMO-SMRT/NCoR1-HDAC3 repressing complex is mandatory for GC-induced IR nGRE-mediated transrepression. Proc. Natl. Acad. Sci. U.S.A. 113, E626–E634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tran C., Ouk S., Clegg N. J., Chen Y., Watson P. A., Arora V., Wongvipat J., Smith-Jones P. M., Yoo D., Kwon A., Wasielewska T., Welsbie D., Chen C. D., Higano C. S., Beer T. M., Hung D. T., Scher H. I., Jung M. E., and Sawyers C. L. (2009) Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stark C., Breitkreutz B. J., Reguly T., Boucher L., Breitkreutz A., and Tyers M. (2006) BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N. K., O'Donnell L., Oster S., Theesfeld C., Sellam A., Stark C., Breitkreutz B. J., Dolinski K., and Tyers M. (2017) The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45, D369–D379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Metzger E., Wissmann M., Yin N., Muller J. M., Schneider R., Peters A. H., Gunther T., Buettner R., and Schule R. (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 [DOI] [PubMed] [Google Scholar]
- 42. Ebert S. N., Ficklin M. B., Her S., Siddall B. J., Bell R. A., Ganguly K., Morita K., and Wong D. L. (1998) Glucocorticoid-dependent action of neural crest factor AP-2: Stimulation of phenylethanolamine N-methyltransferase gene expression. J. Neurochem. 70, 2286–2295 [DOI] [PubMed] [Google Scholar]
- 43. Mukhopadhyay N. K., Ferdinand A. S., Mukhopadhyay L., Cinar B., Lutchman M., Richie J. P., Freeman M. R., and Liu B. C. (2006) Unraveling androgen receptor interactomes by an array-based method: Discovery of proto-oncoprotein c-rel as a negative regulator of androgen receptor. Exp. Cell Res. 312, 3782–3795 [DOI] [PubMed] [Google Scholar]
- 44. Szapary D., Huang Y., and Simons S. S. Jr. (1999) Opposing effects of corepressor and coactivators in determining the dose-response curve of agonists, and residual agonist activity of antagonists, for glucocorticoid receptor-regulated gene expression. Mol. Endocrinol. 13, 2108–2121 [DOI] [PubMed] [Google Scholar]
- 45. Cheng S., Brzostek S., Lee S. R., Hollenberg A. N., and Balk S. P. (2002) Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol. Endocrinol. 16, 1492–1501 [DOI] [PubMed] [Google Scholar]
- 46. Schulz M., Eggert M., Baniahmad A., Dostert A., Heinzel T., and Renkawitz R. (2002) RU486-induced glucocorticoid receptor agonism is controlled by the receptor N terminus and by corepressor binding. J. Biol. Chem. 277, 26238–26243 [DOI] [PubMed] [Google Scholar]
- 47. Dotzlaw H., Moehren U., Mink S., Cato A. C., Iniguez Lluhi J. A., and Baniahmad A. (2002) The amino terminus of the human AR is target for corepressor action and antihormone agonism. Mol. Endocrinol. 16, 661–673 [DOI] [PubMed] [Google Scholar]
- 48. Liao G., Chen L. Y., Zhang A., Godavarthy A., Xia F., Ghosh J. C., Li H., and Chen J. D. (2003) Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J. Biol. Chem. 278, 5052–5061 [DOI] [PubMed] [Google Scholar]
- 49. Huynh K. D., Fischle W., Verdin E., and Bardwell V. J. (2000) BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 14, 1810–1823 [PMC free article] [PubMed] [Google Scholar]
- 50. Andres M. E., Burger C., Peral-Rubio M. J., Battaglioli E., Anderson M. E., Grimes J., Dallman J., Ballas N., and Mandel G. (1999) CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. U.S.A. 96, 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shi Y., Downes M., Xie W., Kao H. Y., Ordentlich P., Tsai C. C., Hon M., and Evans R. M. (2001) Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 15, 1140–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen G., and Courey A. J. (2000) Groucho/TLE family proteins and transcriptional repression. Gene 249, 1–16 [DOI] [PubMed] [Google Scholar]
- 53. Childs K. S., and Goodbourn S. (2003) Identification of novel co-repressor molecules for interferon regulatory factor-2. Nucleic Acids Res. 31, 3016–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mundade R., Ozer H. G., Wei H., Prabhu L., and Lu T. (2014) Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell. Cycle 13, 2847–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Foulds C. E., Feng Q., Ding C., Bailey S., Hunsaker T. L., Malovannaya A., Hamilton R. A., Gates L. A., Zhang Z., Li C., Chan D., Bajaj A., Callaway C. G., Edwards D. P., Lonard D. M., Tsai S. Y., Tsai M. J., Qin J., and O'Malley B. W. (2013) Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Mol. Cell 51, 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gillespie M. A., Gold E. S., Ramsey S. A., Podolsky I., Aderem A., and Ranish J. A. (2015) An LXR-NCOA5 gene regulatory complex directs inflammatory crosstalk-dependent repression of macrophage cholesterol efflux. EMBO J. 34, 1244–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mohammed H., D'Santos C., Serandour A. A., Ali H. R., Brown G. D., Atkins A., Rueda O. M., Holmes K. A., Theodorou V., Robinson J. L., Zwart W., Saadi A., Ross-Innes C. S., Chin S. F., Menon S., Stingl J., Palmieri C., Caldas C., and Carroll J. S. (2013) Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell. Rep. 3, 342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee J. E., Wang C., Xu S., Cho Y. W., Wang L., Feng X., Baldridge A., Sartorelli V., Zhuang L., Peng W., and Ge K. (2013) H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2, e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gearhart M. D., Corcoran C. M., Wamstad J. A., and Bardwell V. J. (2006) Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 26, 6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cao Q., Gearhart M. D., Gery S., Shojaee S., Yang H., Sun H., Lin D. C., Bai J. W., Mead M., Zhao Z., Chen Q., Chien W. W., Alkan S., Alpermann T., Haferlach T., Muschen M., Bardwell V. J., and Koeffler H. P. (2016) BCOR regulates myeloid cell proliferation and differentiation. Leukemia 30, 1155–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stadhouders R., Cico A., Stephen T., Thongjuea S., Kolovos P., Baymaz H. I., Yu X., Demmers J., Bezstarosti K., Maas A., Barroca V., Kockx C., Ozgur Z., van Ijcken W., Arcangeli M. L., Andrieu-Soler C., Lenhard B., Grosveld F., and Soler E. (2015) Control of developmentally primed erythroid genes by combinatorial co-repressor actions. Nat. Commun. 6, 8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolf S. S., Patchev V. K., and Obendorf M. (2007) A novel variant of the putative demethylase gene, s-JMJD1C, is a coactivator of the AR. Arch. Biochem. Biophys. 460, 56–66 [DOI] [PubMed] [Google Scholar]
- 63. Iannone M. A., Consler T. G., Pearce K. H., Stimmel J. B., Parks D. J., and Gray J. G. (2001) Multiplexed molecular interactions of nuclear receptors using fluorescent microspheres. Cytometry 44, 326–337 [DOI] [PubMed] [Google Scholar]
- 64. Garcia-Bassets I., Kwon Y. S., Telese F., Prefontaine G. G., Hutt K. R., Cheng C. S., Ju B. G., Ohgi K. A., Wang J., Escoubet-Lozach L., Rose D. W., Glass C. K., Fu X. D., and Rosenfeld M. G. (2007) Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cai C., He H. H., Chen S., Coleman I., Wang H., Fang Z., Chen S., Nelson P. S., Liu X. S., Brown M., and Balk S. P. (2011) Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer. Cell. 20, 457–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cai C., He H. H., Gao S., Chen S., Yu Z., Gao Y., Chen S., Chen M. W., Zhang J., Ahmed M., Wang Y., Metzger E., Schule R., Liu X. S., Brown M., and Balk S. P. (2014) Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell. Rep. 9, 1618–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jeyakumar M., Tanen M. R., and Bagchi M. K. (1997) Analysis of the functional role of steroid receptor coactivator-1 in ligand-induced transactivation by thyroid hormone receptor. Mol. Endocrinol. 11, 755–767 [DOI] [PubMed] [Google Scholar]
- 68. Weiss R. E., Xu J., Ning G., Pohlenz J., O'Malley B. W., and Refetoff S. (1999) Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO J. 18, 1900–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tagami T., Madison L. D., Nagaya T., and Jameson J. L. (1997) Nuclear receptor corepressors activate rather than suppress basal transcription of genes that are negatively regulated by thyroid hormone. Mol. Cell. Biol. 17, 2642–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Berghagen H., Ragnhildstveit E., Krogsrud K., Thuestad G., Apriletti J., and Saatcioglu F. (2002) Corepressor SMRT functions as a coactivator for thyroid hormone receptor T3Ralpha from a negative hormone response element. J. Biol. Chem. 277, 49517–49522 [DOI] [PubMed] [Google Scholar]
- 71. McKenna N. J., and O'Malley B. W. (2010) SnapShot: NR coregulators. Cell 143, 172–172.e1 [DOI] [PubMed] [Google Scholar]
- 72. Sutinen P., Malinen M., Heikkinen S., and Palvimo J. J. (2014) SUMOylation modulates the transcriptional activity of androgen receptor in a target gene and pathway selective manner. Nucleic Acids Res. 42, 8310–8319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Huang C. Y., Beliakoff J., Li X., Lee J., Li X., Sharma M., Lim B., and Sun Z. (2005) hZimp7, a novel PIAS-like protein, enhances androgen receptor-mediated transcription and interacts with SWI/SNF-like BAF complexes. Mol. Endocrinol. 19, 2915–2929 [DOI] [PubMed] [Google Scholar]
- 74. Yi P., Wang Z., Feng Q., Pintilie G. D., Foulds C. E., Lanz R. B., Ludtke S. J., Schmid M. F., Chiu W., and O'Malley B. W. (2015) Structure of a biologically active estrogen receptor-coactivator complex on DNA. Mol. Cell 57, 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pratt W. B., Galigniana M. D., Morishima Y., and Murphy P. J. (2004) Role of molecular chaperones in steroid receptor action. Essays Biochem. 40, 41–58 [DOI] [PubMed] [Google Scholar]
- 76. Jee Y. K., Gilmour J., Kelly A., Bowen H., Richards D., Soh C., Smith P., Hawrylowicz C., Cousins D., Lee T., and Lavender P. (2005) Repression of interleukin-5 transcription by the glucocorticoid receptor targets GATA3 signaling and involves histone deacetylase recruitment. J. Biol. Chem. 280, 23243–23250 [DOI] [PubMed] [Google Scholar]
- 77. Kristensen A. R., Gsponer J., and Foster L. J. (2012) A high-throughput approach for measuring temporal changes in the interactome. Nat. Methods 9, 907–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gaughan L., Logan I. R., Cook S., Neal D. E., and Robson C. N. (2002) Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 277, 25904–25913 [DOI] [PubMed] [Google Scholar]
- 79. Baens M., Noels H., Broeckx V., Hagens S., Fevery S., Billiau A. D., Vankelecom H., and Marynen P. (2006) The dark side of EGFP: Defective polyubiquitination. PLoS ONE 1, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schmidt S. F., Larsen B. D., Loft A., and Mandrup S. (2016) Cofactor squelching: Artifact or fact? Bioessays 38, 618–626 [DOI] [PubMed] [Google Scholar]
- 81. Ruepp A., Waegele B., Lechner M., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., and Mewes H. W. (2010) CORUM: The comprehensive resource of mammalian protein complexes–2009. Nucleic Acids Res. 38, D497–D501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., Wu H., Aebersold R., Graef I. A., and Crabtree G. R. (2007) An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to PeptideAtlas (www.peptideatlas.org) with the dataset identifier PASS00971.