Abstract
Ever since early suggestions that instability at common fragile sites (CFSs) could be responsible for chromosome rearrangements in cancers, CFSs and associated genes have been the subject of numerous studies, leading to questions and controversies about their role and importance in cancer. It is now clear that CFSs are not frequently involved in translocations or other cancer-associated recurrent gross chromosome rearrangements. However, recent studies have provided new insights into the mechanisms of CFS instability, their impact on genome instability, and their role in generating focal copy number alterations that affect the genomic landscape of many cancers.
Introduction
CFSs were first described in 1984 as sites on human metaphase chromosomes that are particularly prone to forming cytogenetically-defined chromosomal gaps or breaks following partial inhibition of DNA synthesis1 (Figure 1). The same CFSs were seen in all individuals studied and thus were thought to represent a conserved component of chromosome structure. Interest in CFSs rapidly increased for two reasons. First, the agents and conditions leading to CFS formation (i.e., CFS expression) allowed the identification of factors involved in the cellular response to perturbed replication, or “replication stress”. Second, and more prominently, the association of CFSs with recurrent chromosome rearrangements in cancers, including those involving oncogenes, suggested that CFSs could be responsible for these events and be drivers of tumorigenesis or tumor progression2–6. Notably, these associations were made without the benefit of the human genome sequence or precise mapping of either the chromosome rearrangements or the CFSs. Most studies used classical cytogenetic techniques to identify and define CFSs, but as discussed below, advances in genomic technologies and new experimental findings have enabled the refinement of CFS locations and greatly expanded this experimental paradigm. It is now apparent that unlike the so-called early replicating fragile sites7, most CFSs are infrequently involved in translocations or other recurrent gross cancer chromosome rearrangements, and their locations do not coincide with oncogenes. Instead, genes underlying CFSs, such as fragile histidine triad (FHIT) and WW domain containing oxidoreductase (WWOX), were found to be sites of recurrent deletions in multiple tumor types, suggesting that they may function as tumor suppressor genes 8. The Cancer Genome Atlas (TCGA) and other studies have now revealed that de novo copy number variants (CNVs; synonymous with copy number alterations (CNAs) arise at a high frequency in most tumor types and that CFS loci are highly prone to their occurrence9. Recent studies have greatly advanced our understanding of the mechanistic basis for CFS instability and the resulting genomic effects, which have broad implications for CNVs and other genomic rearrangements arising in cancer and in normal cells. Our current view is that CFS gaps and breaks on metaphase chromosomes were just the first indicators of an unusually high instability of large, late replicating, actively transcribed genes that also manifests as many of the most frequently seen CNVs in cancer cells. This Perspective focuses on recent findings on the mechanisms of CFS instability and their biological consequences in cancer that support this view.
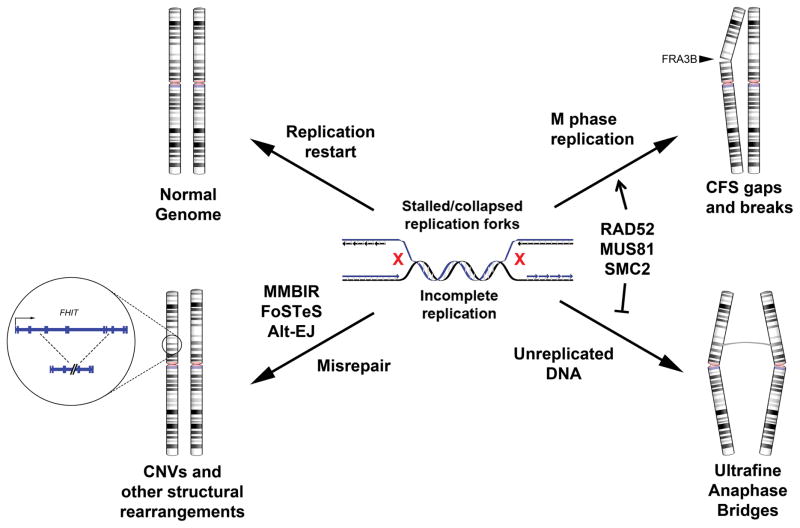
Figure 1. Possible genomic outcomes of replication stress.
DNA replication stress during S-phase leads to stalled or collapsed replication forks, which may be resolved in several ways. Clockwise, starting from top left: (a) Successful restart and completion of replication leads to an intact, normal genome. (b) If regions of unreplicated DNA persist through late-S phase, replication may be completed as late as M-phase, resulting in an apparent common fragile site (CFS) gap/break due to a lack of normal chromosome condensation. (c) If the unreplicated DNA is not resolved and persists to anaphase, ultrafine anaphase bridges can form at these sites. (d) If the stalled/collapsed forks are repaired and restarted through an error-prone mechanism, genome rearrangements, including copy number variants (CNVs) can occur. Alt-EJ, alternative end-joining; FoSTeS, fork stalling and template switching; MMBIR, microhomology-mediated break-induced replication; SMC2, structural maintenance of chromosomes protein 2.
Basic Properties of CFSs
CFSs are specific chromosomal loci that are prone to forming visible gaps and breaks on metaphase chromosomes under conditions that perturb normal DNA synthesis, thus causing replication stress1. Most commonly, cells are treated with low concentrations of the DNA polymerase inhibitor, aphidicolin (APH) to slow but not completely block DNA synthesis. Folate deficiency 1 and low doses of the ribonucleotide reductase inhibitor, hydroxyurea (HU)10, which reduces cellular dNTP pools, have lesser but similar effects. CFS gaps and breaks were initially defined and mapped at low resolution on metaphase chromosomes using accepted cytogenetic definitions of chromosome gaps and breaks11 without knowledge of the specific underlying DNA integrity or chromatin structure, including the presence or absence of DNA double-strand breaks (DSBs)1. The original report, which nominated a CFS if it was expressed in at least six of 50 cells, identified 17 CFS loci in cultured human lymphocytes with the most frequent being 3p14 (FRA3B), 16q23 (FRA16D) and 6q26 (FRA6E). Subsequent reports defined CFSs using similar criteria and led to 75 APH-type CFSs being listed in the NCBI Genome Database (GDB)12. However, there was no consensus about the statistical criteria needed to differentiate a ‘true’ CFS from a random or low frequency APH-induced gap or break13. Furthermore, most of these CFSs were mapped only at the chromosome band level with a resolution of >1Mb. Thus, while still a useful guide, the GDB CFS data are now largely outdated based on our current ability to precisely localize CFSs in different cell types and measure their relative instability at the genomic level. Nonetheless, it has been clear that in all cell types, the majority of CFS breaks occur at roughly 10–20 of the most sensitive and unstable sites in APH-treated cells. For example, in lymphocytes, gaps and breaks at just 20 CFSs represent over 80% of all cytogenetic lesions following low-dose APH treatment1. Experiments in cultured cells also showed that, in addition to metaphase gaps and breaks, CFSs are hotspots for sister chromatid exchanges (SCEs)14, 15, can lead to translocations and deletion breakpoints in somatic cell hybrid systems16, 17 and are preferred sites of integration for transfected plasmid DNA18. These and other properties of CFSs have been described in detail a number of past reviews19–21.
CFSs are late replicating
Late replication has long been known to be a key factor in CFS instability. Using fluorescence in situ hybridization (FISH) probes in FRA3B, LeBeau et al. demonstrated that sequences at FRA3B undergo DNA replication very late in S-phase of the cell cycle and that APH causes a further delay in replication, with many loci remaining unreplicated even in G2 22. Subsequent replication timing studies demonstrated that other CFSs experience similar difficulty in either the progression or completion of replication19. Importantly, late replication is not sufficient to fully define a CFS, as other, stable regions of the genome also replicate late in S-phase. Nonetheless, it is clear that late-replicating DNA at CFSs is particularly sensitive to replication inhibitors and plays a central role in their instability.
CFSs are enriched in large genes
The availability of FISH probes specific for sub-megabase genomic spans allowed for more precise mapping and molecular characterization of breakage across a number of CFSs, beginning with the most frequent CFSs in lymphocytes, FRA3B and FRA16D23–25. These studies revealed that CFS fragility extended over hundreds of kilobases, identifying CFSs as large, regional genomic features. Importantly, FRA3B mapped within the large 1.5 Mb FHIT gene at 3p14.2 and FRA16D mapped within the 1.1 Mb WWOX gene at 16q2326, 27. The association of CFSs with unusually large genes (the median human gene size is ~23 kb) was later extended to 13 other CFSs that only recently were characterized at the molecular level19, 28, 29. Reports of CFSs extending over several megabases may be influenced by two or more large CFS-associated genes mapping so close together as to be indistinguishable at the cytogenetic level, as was shown for CFSs in the AUTS2 and MAGI2 genes on chromosome 730. Recent findings, discussed below, offer mechanistic explanations for this interesting association between large gene size and the occurrence of a CFS and suggest that most, if not all, large, actively transcribed, late replicating genes have the potential to be CFSs.
CFSs show cell type specificity
An important factor in defining and studying CFSs is their cell type specificity. Most early CFS studies were performed using cultured lymphocytes and lymphoblastoid cell lines. Fibroblast-specific fragile sites were noted in early studies by Murano et al.31, 32 and others33, but these observations were not extended to other cell types. The importance of cell type differences in CFSs was recently brought into focus by Le Tallec et al.34, 35 and Hosseini et al.36, who studied CFSs in a variety of cell types, including fibroblast, epithelial and erythroid cell lines. While there were a number of CFSs found across all cell types, cell type-specific CFSs were also found in most cells, including fibroblasts and epithelial cell lines. For example, FRA3B, in FHIT, is highly expressed (i.e. exhibits chromosome gaps breaks) in lymphocytes, lymphoblasts, HCT116 and HeLa cells, but is not expressed in normal fibroblasts, which instead contain a novel CFS at 3q13.3. Other CFSs such as FRA16D, in WWOX, are expressed in most cell types, although at different frequencies. As described below, recent data have revealed that cell-type-specific CFS expression is influenced by the transcription of large genes with direct implications for genomic rearrangements that directly impact the mutational landscape of cancer cells.
Mechanisms of CFS Instability
CFS sequences
The agents and growth conditions that induce CFS breaks inhibit replication fork progression, increasing the frequency of stalled and/or collapsed forks genome-wide. But why are CFS loci uniquely sensitive to replication stress? Early studies proposed non-exclusive mechanisms underlying this observation1, 14, 22, 37–39, which included the presence of difficult-to-replicate sequences, late replication timing, and a paucity of replication origins within CFSs. With the molecular characterization and sequencing of the first CFS 26, 40–42, it was apparent that they are AT-rich and contain long stretches of perfect AT microsatellite sequences that have the potential to form complex secondary structures that could impede replication fork progression. This idea was furthered by the identification of increased numbers of DNA flexibility peaks, defined as areas of high local variation in the twist angle between stacked nucleotide base pairs43, suggesting that the formation of abnormal DNA structures could be a causal factor in mediating CFS expression40, 44, 45. Indeed, studies performed in yeast demonstrated that these AT-rich sequences lead to replication arrest and DNA breaks46. Questions regarding the necessity and sufficiency of DNA flexibility peaks on CFS instability arose from findings that cells containing FRA3B deletions that removed most DNA flexibility peaks still exhibited CFS breakage47 and the fact that the human genome contains numerous AT-rich regions of high flexibility that are not CFSs. Thus, AT-rich, flexible DNA is not sufficient to fully define most CFSs, but likely contributes to replication failure at these sites. The contributions of sequence composition and non-B forming DNA secondary structures to CFS instability are important questions that require further elucidation.
Genetic factors
Deficiency of a number of factors involved in DNA replication and the replication stress response influence CFS breakage. The first such study found that the ataxia-telangiectasia and Rad3-related (ATR) kinase plays a major role in maintaining CFS stability by activating a cell cycle checkpoint in response to the incomplete replication at these sites48. Cells lacking ATR showed a dramatic increase in CFS expression following APH treatment. Even untreated cells had a low frequency of spontaneous CFS breaks, demonstrating that ATR is required for CFS stability during normal DNA replication. Inhibition of several downstream effectors of ATR, including CHK1, HUS1, Claspin, and structural maintenance of chromosomes protein 1 (SMC1; also known as SMC1A), showed similar effects on CFS expression49–52. Other proteins involved in the resolution or repair of DSBs or stalled or collapsed replication forks resulting from incomplete replication also affect CFS stability. These include RAD51, RAD52, DNA-dependent protein kinase (DNA-PK), DNA Ligase IV, BRCA1, Fanconi anemia group D2 (FANCD2), the Bloom syndrome protein (BLM) and Werner syndrome ATP-dependent (WRN) helicases, the MUS81-EME1 nucleases as well as the specialized DNA polymerases Polκ, Polη, POLD3 and REV3 and the SNM1 homologue B (SNM1B; also known as APOLLO and DCLRE1B) 53–66 (Table 1); however, not all have been independently confirmed.
Table 1.
Proteins reported to play a role in CFS stability.
| Protein | Function | Role in CFS expression* | Refs |
|---|---|---|---|
| DNA Replication | |||
|
| |||
| MUS81 | Structure-specific endonuclease subunit | M-phase replication | 57–59 |
| EME1 | Structure-specific endonuclease subunit | M-phase replication | 58, 59 |
| RAD52 | Homologous recombination protein | M-phase replication | 55 |
| BLM | RecQ helicase | Suppresses inappropriate recombination | 63 |
| TOP1 | Type I topoisomerase | Relieves torsional stress downstream of replication and transcriptional machineries | 144, 145 |
|
| |||
| Cell Cycle Checkpoints | |||
|
| |||
| ATR | Serine/Threonine kinase | Cell cycle arrest in response to single stranded, unreplicated DNA | 48, 146 |
| ATM | Serine/Threonine kinase | Cell cycle arrest in response to DNA DSBs | 147 |
| CHK1 | Serine/Threonine kinase | Cell cycle arrest in response to DNA damage or unreplicated DNA | 49 |
| Claspin | BRCA1, CHK1 adapter protein | Facilitates the ATR-dependent phosphorylation of BRCA1 and CHK1; Replication fork sensor | 51 |
| HUS1 | Component of the 9-1-1 cell-cycle checkpoint response complex | p53-dependent checkpoint activation and apoptosis | 50 |
| SNM1b | Metallo-β-Lactamase super family protein | Promotes fork collapse, ATR activation | 56 |
|
| |||
| DNA Repair | |||
|
| |||
| RAD51 | Recombinase | Homologous Recombination | 66 |
| BRCA1 | E3 ubiquitin-protein ligase | Genome Surveillance | 65 |
| FANCD2 | Fanconi anemia-linked repair/replication protein | Facilitates Replication | 54, 64 |
| DNA-PK | Serine/Threonine protein kinase | Nonhomologous End-Joining | 66 |
| Ligase IV | ATP-dependent DNA ligase | Nonhomologous End-Joining | 66 |
| XLF | XRCC4-like factor | Nonhomologous End-Joining | 66, 148 |
| WRN | RecQ helicase | DSB repair | 62 |
|
| |||
| Translesion Synthesis | |||
|
| |||
| Pol η | Y family DNA polymerase | DNA synthesis at stalled forks in S phase | 60 |
| Pol κ | Y family DNA polymerase | Replication through repetitive elements | 61 |
| REV3 | Polymerase ζ subunit | G2/M-phase replication | 53 |
|
| |||
| Structural | |||
|
| |||
| SMC1(SMC1A) | Cohesin subunit | Prevents collapse of stalled replications forks | 52 |
Proteins have multiple roles in the cell; only roles relevant to CFSs are listed here.
CFS, Common Fragile Site; BLM, Bloom syndrome helicase; TOP1, DNA Topoisomerase 1; ATR, ataxia-telangiectasia and Rad3-related; ATM, Ataxia Telangiectasia Mutated; SNM1B, SNM1 homologue B, also known as APOLLO and DCLRE1B; FANCD2, Fanconi anemia group D2; DNA-PK, DNA-dependent protein kinase; LIG4, DNA Ligase 4; XLF, XRCC4-Like Factor, also known as Non-Homologous End Joining Factor 1 (NHEJ1); WRN, Werner syndrome ATP-dependent helicase; SMC1 (SMC1A), Structural Maintenance of Chromosomes 1A; XRCC4, X-Ray Repair Cross Complementing 4.
CFSs have a paucity of replication origins
The importance of replication origin organization and firing in CFS stability is now clear. In an early study of replication origins, Palakodeti et al.38 used a nascent strand abundance method to identify four putative origins within a 50kb region of FRA3B and concluded that CFS origins fire less efficiently than control regions upon APH treatment. In a series of elegant DNA combing experiments, Letessier et al.67 subsequently demonstrated that FRA3B does not show significant differences in replication fork speed or stalling relative to the bulk genome upon APH treatment in lymphoblasts. However, mapping of replication initiation events along FRA3B revealed an initiation-poor region that coincides with the center of the CFS. This paucity of active replication origins was apparent in lymphoblasts that express FRA3B instability, but not in fibroblasts, where FRA3B is not expressed. Conversely, fibroblasts, but not lymphoblasts, showed a paucity of active replication origins within a fibroblast-specific CFS at 3q13.3. These data are consistent with a reduced binding of the origin recognition complex (ORC; which seeds replication origin assembly) within CFSs68 and suggest a model wherein a paucity of replication origins in the center of CFSs requires converging replication forks from flanking genomic regions to cover long distances in S phase, greatly increasing the risk of incomplete replication under replication stress.
Consequnces of CFS Instability
It now appears clear that a paucity of active replication origins leads to incomplete DNA replication at CFSs during S phase and is a key factor in initiating CFS instability. Gaining insight into the downstream genomic consequences of incomplete replication and the mechanisms involved in resolving the associated lesions is fundamental to understanding the biologic importance of CFS instability in cancers and other disorders. In addition to error-free replication fork restart, there are at least four other major non-exclusive outcomes of incomplete replication at CFSs, each with different genomic and functional consequences, including mitotic replication and fragile site expression, persistence of unreplicated DNA and anaphase bridge formation, aberrant repair leading to CNV formation, and DSBs and chromosome rearrangements (Figure 1).
M-phase replication – CFS gaps and breaks on metaphase chromosomes
It has been known that CFSs complete replication late in S phase and even later in the presence of replication inhibitors, with unreplicated DNA persisting as late as G2/early M22. Minocherhomji and colleagues59 have extended these findings by exploring the events occurring at CFSs during the G2/M phases of the cell cycle. Remarkably, they found that after APH treatment, the passage of incompletely replicated DNA at CFSs into mitotic chromosomes serves as the trigger to activate a novel and distinct M-phase replication pathway. This mitotic DNA synthesis requires the SLX4 scaffold protein, MUS81-EME endonuclease and POLD3. The requirement of POLD3 suggested that replication could occur by break-induced replication (BIR), which, in yeast, requires the POLD3 orthologue. In addition, Bhowmick et al.,55 and Sotiriou et al.69 recently showed that RAD52 plays a key role in the repair of collapsed replication forks and is essential for CFS replication. Thus, replication at expressed CFSs is not completed until mitotic prophase, suggesting that the cytogenetic manifestation of CFSs results from extremely late replication and subsequent failure of these regions to undergo normal chromosome condensation. Indeed, these data can explain why CFSs typically appear as cytogenetically defined gaps and breaks on metaphase chromosomes, as opposed to acrocentric fragments or translocations that might be expected for chromosome DSBs.
No replication – Ultrafine anaphase bridges
A quite different outcome for CFSs is seen if DNA replication is not completed in M-phase. The persistence of a replication intermediate and, most likely, unreplicated DNA at CFSs into late mitosis can lead to the formation of ultrafine anaphase bridges (UFBs), which form a “thread” of DNA linking CFS loci on the separating sister chromatids. UFBs differ from typical anaphase bridges in that they lack histones and cannot be visualized with conventional DNA dyes such as DAPI. Instead, they are detected by visualizing bound proteins including PLK1-interacting checkpoint helicase (PICH; also known as ERCC6L), a member of the SNF2/SWI family of DNA-dependent ATPases, FANCD2, FANCI and BLM70–72, all of which are important for the resolution of the UFBs. While UFBs can be seen in cells after treatment with low-dose APH, their frequency is greatly increased after depletion of MUS81, SMC2 or other proteins involved in M-phase DNA synthesis59. These findings, coupled with the formation of large p53-binding protein 1 (53BP1; also known as TP53BP1) foci that colocalize with CFSs in the subsequent G1-phase indicate that the CFS-UFBs represent DNA that fails to complete replication during M phase59. The fate and biological importance of UFBs are not completely understood, but they are hypothesized to play a role in generating DNA breaks, genome rearrangements and chromosomal non-disjunction wherein chromsomes do not segregate properly during cell division59, 70.
Aberrant repair - CNV formation
A third consequence of unreplicated DNA at CFSs with major biological importance is CNV formation. CNVs are genomic deletions and duplications of tens of base pairs to over a megabase. They occur as normal genomic variants, with tens of thousands described in human populations73. Germline CNVs extensively to human genetic and developmental genomic disorders74, 75, and likely contribute to disease through somatic tissue mosaicism76, 77. Importantly, CNVs (a.k.a. CNAs) also arise frequently in cancers78, 79, with many containing tens to hundreds of de novo deletions and duplications.
Durkin et al. provided the first evidence that replication stress and CFS instability can lead to CNVs similar to those observed in both normal and tumor cells47. They tested the hypothesis that APH treatment of cultured mammalian cells would not only result in CFSs on metaphase chromosomes, but would also give rise to deletions at the CFS, FRA3B. Remarkably, up to 23% of treated cells contained deletions that mimicked the FRA3B deletions previously described in cancer cells in both size and breakpoint structure. Using emerging genomic array and sequencing technologies, Arlt and colleagues expanded these findings, showing that exogenous replication stress, in the form of low-dose APH, hydroxyurea, and ionizing radiation (IR), is potent inducer of CNVs genome-wide in cultured normal human cells80–82, and that some CNVs occurred within CFS-associated genes. These CNV breakpoint junctions primarily had short, 2–10 bp microhomologies or blunt ends, which are typical of almost all CNVs that arise in cancers as well as the majority of CNVs found in normal genomes and and de novo, often pathogenic CNVs arising by new mutations. These CNVs included complex rearrangements with multiple junctions that likely arose in a single mutational event. Importantly, CNV formation at CFSs does not rely on canonical DSB repair by the non-homologous end-joining (NHEJ) pathway, supporting the hypothesis that other DNA replication-associated repair mechanisms play critical roles in CNV formation83. These results support models that invoke fork stalling and template switching (FoSTeS), in which nascent DNA strands are proposed to switch replication templates.]. Microhomology-mediated break-induced replication (MMBIR), a related pathway in which fork cleavage and DSB resection expose the switching DNA strand, and/or alternative end joining likely lead to the genesis of CNVs arising at CFSs84.
The relationship between replication timing, experimentally-induced CNVs, CFSs, and the transcription of large CFS-associated genes was examined further by Wilson et al. 30. A striking feature of the CNVs was their genomic distribution. Although the majority of CNVs appeared random, occurring as isolated events at different genomic locations, a subset of events arose in focal clusters or hotspots, with over 40% of all CNVs arising in nine hotspots. CFSs were also examined, which revealed that CNV hotspots were also all CFSs in these normal human fibroblast cell lines, demonstrating that CFSs and CNVs are different manifestations of replication stress at the same cell-type-specific loci. Nascent transcript analysis, conducted with the same cell lines, revealed that all but one of the nine CNV hotspots were within large (>500kb) genes that were transcribed. Indeed, of the 12 genes that are >1Mb the long isoforms of which were transcribed in the cells studied, 11 contained CNVs. Conversely, large genes that were not transcribed in these cells did not have CNVs. Importantly, comparing two fibroblast lines with different transcription profiles for specific CFS/CNV hotspot genes demonstrated that locus instability was only found in the cell line transcribing a long isoform; expression of a short isoform of the same gene did not correlate with CNV or CFS formation. Finally, replication timing revealed that large CNV--CFS hotspot genes have consistently late replication timing despite being actively transcribed, consistent with earlier studies of CFSs22.
The above studies provide strong evidence that the most frequent and unstable CFSs and CNV hotspots occur at the same loci and that active transcription of associated large genes is a key factor in their extreme instability. The association with transcription supported the findings of Helmrich et al.85 who examined the correlation of CFS breaks, transcriptional activity, and R-loops at three CFSs in cells exhibiting different transcription profiles. They proposed that CFS instability was caused by replication-transcription conflicts resulting in R-loop formation and suggested that these conflicts are promoted at CFS genes because they are transcriptionally active in S-phase, since transcription of such large genes requires the entire duration of the cell cycle. Both studies demonstrated that differential expression of large genes can explain cell type differences in the expression patterns of CFSs. The findings of Wilson et al.30 further extend this mechanistic relationship to CNVs. An extrapolation of these data is that one could predict CFSs and CNV hotspot locations in any cell (or tumor) type for which the transcription pattern is known. Importantly, since most long genes have multiple isoforms, and shorter isoforms are often more prevalent in RNA-seq data, this predictive power is only possible if the complete nascent transcription profile is known. In addition, it requires that transcription be assessed in the same cells in which CFS and CNVs are analyzed, as highlighted by differences in transcription and CFS--CNV hotspots between two normal human fibroblast cell lines.
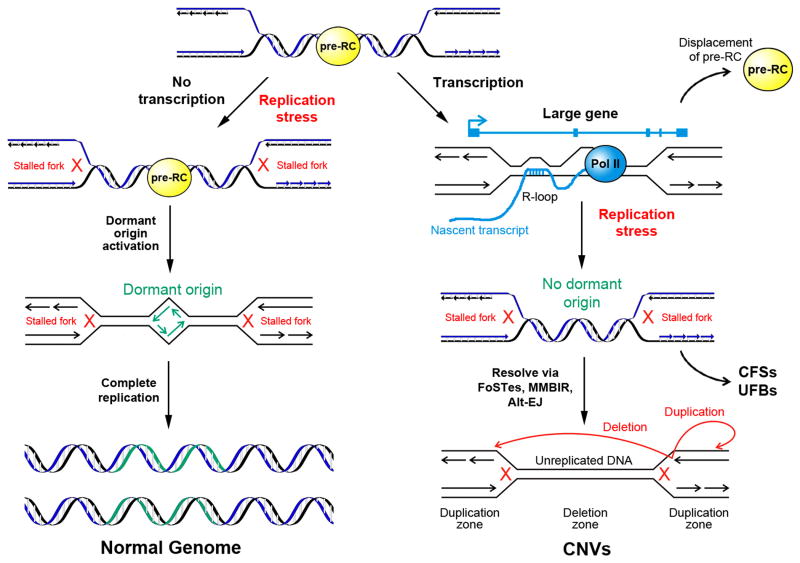
These collective findings have led to a model for the instability of CFSs and associated hotspot CNVs that invokes transcription-dependent double-fork failure30 (Figure 2). The model posits that large, late replicating genes are replicated via forks proceeding inward from the genomic flanks. Dormant origins within the large transcription units are displaced by RNA Pol II complex and are therefore not accessible for activation following replication stress. In this way, large active transcription units organize the locations of fork failures in a manner consistent with patterns of CNV formation. The associated nonlinear increase in CNV risk is consistent with mathematical models of the probability of double-fork failure as a function of inter-origin distance86. The model infers a paucity of usable origins within large active transcription units that is consistent with observations of reduced firing of late or dormant origins in CFSs that would normally fire under replication stress in other genomic regions67, 87, but further invokes transcription as a mechanistic feature. Importantly, pre-replication complexes (pre-RCs) are only licensed in G1 and must remain bound for firing to occur in S-phase. Movement of RNA pol II through an origin can displace a pre-RC, as demonstrated in yeast88, 89. Thus, Wilson et al. 30 suggested that dormant origins fail to rescue transcription-dependent double fork failure at CNV hotspots and CFSs because transcription has persisted throughout S phase and removed those pre-RCs before they could be utilized as replication origins. Consistent with findings of Helmrich et al.85, replication fork failure within transcribed genes could be further enhanced by transcription-dependent R-loop formation. Importantly, these transcription-driven phenomena are likely not the only mechanisms leading to a paucity of active replication origins and a high susceptibility to replication stress, but they appear to account for at least the majority of the most frequent and unstable CFS loci.
Figure 2. Model of genomic instability at active large transcription units.
Replication fork failures resulting from replication stress that occur at most genomic loci, including non-transcribed large genes, can be rescued by the firing of late, or “dormant” origins within the unreplicated region leading to complete replication (left). Large genes in which transcription persists into S-phase, are at high risk for incomplete replication leading to copy number variants (CNVs), common fragile sites (CFSs) and ultrafine anaphase bridges (UFBs; right). The Transcription-dependent Double-Fork Failure (TrDoFF) model for extreme locus instability under replication stress30 proposes that this results from the simultaneous failure of two converging forks, e.g., through the formation of R-loops, and that this creates large, late-replicating domains where displacement of pre-replication complexes (pre-RCs) by prolonged transcription into S-phase prevents dormant origin firing. CFS breaks and deletion CNVs arise within the resulting unreplicated DNA region while duplications arise on the flanks, (red arrows), likely due to fork stalling and template switching (FoSTeS), microhomology-mediated break-induced replication (MMBIR) or alternative end-joining (Alt-EJ), Modified with permission from Wilson et al.30.
This model can explain the initial events leading to instability at CFS loci. The downstream consequences depend on the resolution mechanisms that follow. As discussed, CFSs result from M-phase replication and incomplete condensation, while UFBs appear to result from persistence of unreplicated DNA in anaphase. Exactly when or how CNVs arise remains an important unanswered question. They could form as a result of error-prone repair during the M-phase replication steps that create CFSs59. Alternatively, CNVs could form via BIR or template switching during late S or G2 phases, consistent with proposed mechanisms for CNV formation84. Finally, it is also possible that the unreplicated regions could persist into the next cell cycle, leading to CNV formation by erroneous repair, most likely in G1. Understanding the timing of CNV formation at CFS loci will be an important step in predicting and experimentally interrogating the repair mechanisms involved.
Double strand breaks and chromosome rearrangements
Wei et al.90 recently provided novel insight into the occurrence of DSBs at CFSs. Using a targeted sequencing assay to detect DSB-mediated translocations and other rearrangements, they characterized thousands of DSBs across the genome and identified 27 recurrent DSB clusters (RDCs) in neural stem and progenitor cells (NSPCs). In total, 26/27 RDCs mapped within genes larger than 400 kb, with half mapping in genes larger than 1Mb, including many known CFS-associated genes. Nearly all (24/27) of the RDC-associated genes in NSPCs have neural-specific functions and/or are linked to neurological disorders, including limbic system-associated membrane protein (Lsamp), neurexin 1 (Nrxn1) and deleted in colorectal carcinoma (Dcc). As with the CNVs reported by Wilson et al.30, all but one were transcribed in the NPSCs studied, and all but one were late replicating. The high DSB frequency may result from collapsed replication forks, leading to both CNVs and the translocations detected in this assay. Alternatively, DSBs leading to the detected translocations might represent outcomes distinct from the predominantly intrachromosomal template switching proposed to account for CNV formation. Understanding the disposition of transcription-dependent replicative lesions through different repair pathways and their coordination in the cell cycle are important challenges.
The fact that many of the largest mammalian genes have neuronal functions supports the potential for brain-specific genomic rearrangements affecting these genes. The potential biological importance of DSBs in these genes is underscored by the fact that NHEJ-mutant mice show impaired neuronal development, a motivating factor for the study of Wei et al.90. The studies described above and those of Wei et al.90 support a role of transcription in this process. It has long been known that the most frequently observed CFSs lie in large genes, many of which are involved in neuronal development and function and this relationship can now be better explained. It is unknown why many of these same genes, such as LSAMP and AUTS2 are also transcribed in human fibroblasts and presumably in some cancer cells, where they are CNV hotspots.
Cancer and Genomic Disorders
Viral integration sites
The finding that some viral integration sites are contained within certain CFSs has been known since early sequencing studies of a segment of the FRA3B locus revealed a previously identified human papillomavirus-16 (HPV-16) integration site in a primary cervical carcinoma91. Subsequent observations reported viral integrations at numerous CFSs in tumors and tumor cell lines92–97. Recent work with papillomaviruses demonstrated an association of viral E2 protein with chromatin at CFSs. This study suggested that HPV replication, which utilizes host DNA damage response proteins, occurs near regions of the genome prone to DNA damage (i.e. CFSs)98. In addition, Hu et al.99 found that HPV integration site junctions had short microhomology signatures between the host and viral genomes, similar to those found at CNV breakpoints. Based on these data, they proposed a model for viral integration in which HPV drives integration into the host genome by hijacking the FoSTeS and MMBIR repair pathways invoked in CNV formation, raising the intriguing possibility that viral integration, chromosome fragility, and CNV formation could be driven by the same molecular processes during replication stress.
Other studies failed to observe associations between viral integrations and CFSs. While a recent meta-analysis of HPV, hepatitis B virus (HBV), and Merkel cell polyomavirus (MCPyV) found integrations within CFSs in cervical and other carcinomas, these integrations were not at a higher frequency in CFSs than elsewhere in the genome100. It is noteworthy that integration hotspots have been described in large genes that are not previously-described CFSs, such as LDL receptor related protein 1B (LRP1B;1.9 Mb), CUB and Sushi multiple domains 1 (CSMD1; 2.1 Mb), and discs large MAGUK scaffold protein 2 (DLG2; 2.2 Mb)92, 93, 99 but are predicted to be fragile sites in cells in which they are transcribed30. A similar meta-analysis of viral integration site associations with large genes and transcription, which is highly dysregulated in cancer, could further inform the extent of integration at cell-type specific fragile sites.
Focal deletions at CFSs in cancer
Transcription of large genes in dividing cells leads to an increased risk for instability by creating a high probability for double fork failures, where two converging replication forks do not replicate the DNA between them, while simultaneously preventing the proper resolution of these unstable structures30. This model predicts that large genes will be CNV hotspots in the replicating cell types in which long isoforms are transcribed and has major implications for understanding CNV formation during tumorigenesis. Numerous reports have described frequent hemizygous or homozygous deletions of tens to hundreds of kilobases directly in CFS regions in cancer cell lines and primary tumors. Most studies have focused on FRA3B and FRA16D, which map within the large FHIT and WWOX genes, respectively8, 101, however similar deletion patterns were also shown for other CFSs, including FRA6E102, FRA9E103, and FRA7G104. These deletions can occur early and deletions in FHIT and other CFS genes are found in precancerous lesions including those in the colon, bladder and Barrett’s esophagus in association with activated DNA damage checkpoints 105–108.
At least four recent studies cataloged acquired CNVs in large cohorts of human tumors and concluded that many genomic loci repeatedly show focal deletions and amplifications in a cancer-specific manner, with most of the prevalent focal deletions targeting CFSs and large genes 9, 78, 79, 109. For example, a study of CNVs in 4,934 TCGA cancer samples found repeated focal CNVs in 140 regions (70 losses and 70 gains), 102 of which did not correspond to known cancer genes9. Among the 70 deletion regions, 22 localized to a subset of the 100 largest human genes, including FHIT, WWOX, and other CFS-associated genes.
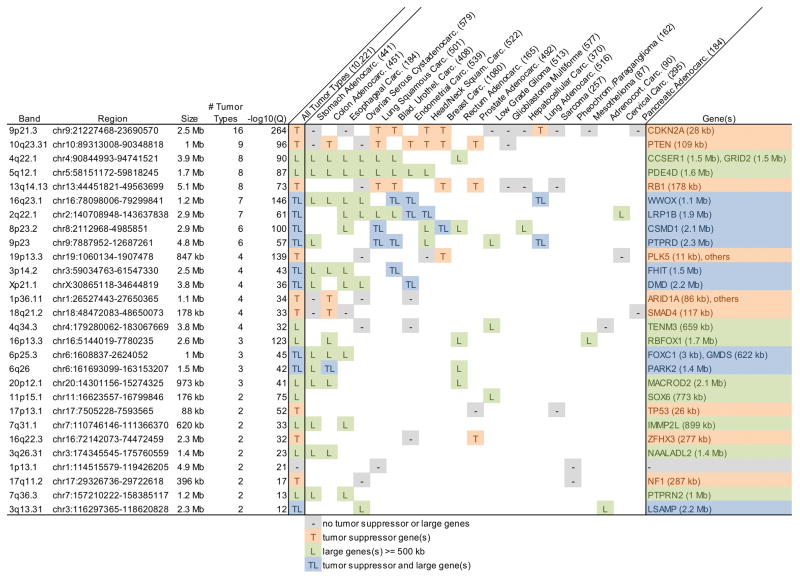
Our examination of the most recent TCGA data, encompassing 10,221 tumor specimens, reveals that many if not all of the CFSs and CNV hotspots described by Wilson et al.30 correspond to similar cancer focal deletions in one or more tumor types (Figure 3). Of the 28 strongest focal deletions in cancer, 19 were at genes > 500 kb in size, nine of which were listed in the Tumor Suppressor Gene Database (TSGene) 2.0 (https://bioinfo.uth.edu/TSGene/)110; the remaining nine all included smaller tumor suppressor genes. A number of the large genes are known CFSs, including WWOX, FHIT, PARK2 (also known as PRKN), inner mitochondrial membrane peptidase subunit 2 (IMMP2L) and LSAMP, and all are predicted to be CFSs and CNV hotspots in cells in which they are expressed30.
Figure 3. Gene content of pan-cancer focal deletions showing strong association with large genes.
Summary of The Cancer Genome Atlas (TCGA) somatic focal deletions in 30 tumor types representing 10,221 tumor specimens. GISTIC2-derived142 interstitial focal deletion calls with size <= 5 Mb and Q value <= 10−10 were merged into overlapping deletion regions. The maximum -log10(Q) (i.e. strongest focal deletion) in each region is reported. A summary of the properties of genes in each region is shown in column “All Tumor Types”, where “T” indicates the presence of a gene in the Tumor Suppressor Gene Database (TSGene) 2.0110 and “L” indicates the presence of a Large gene >= 500 kb. Similar entries are shown for each contributing tumor type, where genes had to lie within a focal deletion and have been identified as a down-regulated tumor suppressor in TSGene 2.0 for that tumor type (“-” indicates a focal deletion with no down-regulated tumor suppressor or large genes). Only tumor types with two or more focal deletions and regions with two or more contributing tumor types are shown. Of these 28 strongly recurrent focal deletions, 19 were at genes > 500 kb, nine of which were listed in TSGene 2.0, and the remaining nine all included smaller tumor suppressor gene loci. A number of the large genes are known CFSs, including WWOX, FHIT, PARK2, IMMP2L and LSAMP. Data summarized in this figure were generated in part by the TCGA Research Network: http://cancergenome.nih.gov/ and obtained from http://firebrowse.org/143.
CDKN2A, cyclin dependent kinase inhibitor 2A; PTEN, Phosphatase And Tensin Homolog; CCSER1, Coiled-Coil Serine Rich Protein 1; GRID2, Glutamate Ionotropic Receptor Delta Type Subunit 2; PDE4D, Phosphodiesterase 4D; RB1, RB Transcriptional Corepressor 1; WWOX, WW domain containing oxidoreductase; LRP1B, LDL Receptor Related Protein 1B; CSMD1, CUB And Sushi Multiple Domains 1; PTPRD, Protein Tyrosine Phosphatase, Receptor Type D; PLK5, Polo Like Kinase 5; FHIT, fragile histidine triad; DMD, Dystrophin; ARID1A, AT-Rich Interaction Domain 1A; SMAD4, SMAD Family Member 4; TENM3, Teneurin Transmembrane Protein 3; RBFOX1, RNA Binding Protein, Fox-1 Homolog 1; FOXC1, Forkhead Box C1; GMDS, GDP-Mannose 4,6-Dehydratase; PARK2, Parkin RBR E3 Ubiquitin Protein Ligase (also known as PRKN); MACROD2, MACRO Domain Containing 2; SOX6, SRY-Box 6; TP53, Tumor Protein P53; IMMP2L, inner mitochondrial membrane peptidase subunit 2; ZFHX3, Zinc Finger Homeobox 3; NAALADL2, N-Acetylated Alpha-Linked Acidic Dipeptidase Like 2; NF1, Neurofibromin 1; PTPRN2, Protein Tyrosine Phosphatase, Receptor Type N2; LSAMP, limbic system-associated membrane protein.
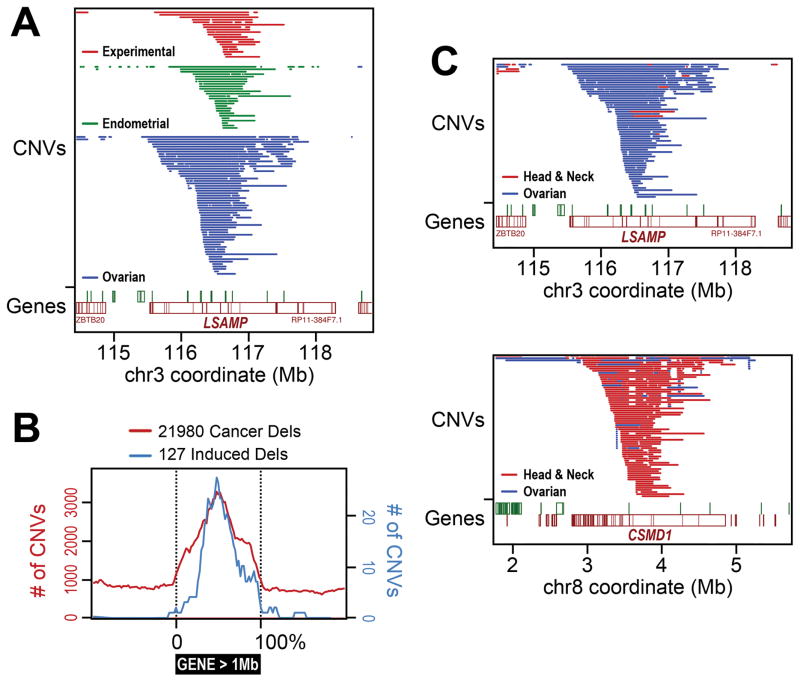
A higher resolution comparison of cancer focal deletions to CNVs experimentally induced by APH, HU and low-dose IR reveals a strikingly similar and highly specific pattern within these large genes (Figure 4). For example, both experimental and cancer deletions in LSAMP cluster around the center of the gene, with remarkably similar CNV sizes and breakpoint locations as illustrated in Uterine Corpus Endometrial Carcinomas (UCEC) and ovarian serous cystadenocarcinomas (Figure 4A). This same pattern is even more strongly evident in an aggregate analysis of all genes larger than 1 Mb (Figure 4B), strongly suggesting that CNVs are formed by the same mechanisms in both cell models and cancer. The consistent central location of large gene deletions is readily predicted by the replication- and transcription-dependent model described above (Figure 2) and suggests that this mechanism drives many large gene deletions, since less frequently observed terminal deletions would also disable gene function. Importantly, different tumor types show different focal deletion signatures. For example, the unstable experimentally induced CNV hotspot and CFS in fibroblasts within LSAMP is a strong focal deletion in ovarian serous cystadenocarcinoma and adrenocortical carcinoma, but not many other tumor types (Figures 3 and 4C). Other large genes show very different patterns, such as CSMD1, which is a focal deletion in six of the 21 tumor types (Figure 4C). The presence of cancer CNVs within large genes in only a subset of cancer types is consistent with the model that only cancers (or their precursor cells) that actively transcribe a long isoform of a large gene will exhibit a high risk for CNVs at that locus. In contrast, cyclin dependent kinase inhibitor 2A (CDKN2A) is a clear focal deletion hotspot in almost all cancer types despite not corresponding to a large gene as it is an important tumor suppressor gene that drives tumor formation 111.
Figure 4. Experimentally-induced CNVs and focal deletions in cancer arise in the centers of large genes.
(A) Example comparison of experimentally induced30 (in 090 human fibroblasts) and in vivo cancer focal deletion copy number variants (CNVs) at the limbic system-associated membrane protein (LSAMP) gene in in 539 endometrial carcinomas (focal deletion -log10(Q) value = 7.8, which is below the threshold used in Figure 3) and 579 ovarian serous cystadenocarcinomas143. Both sets cluster near the center of this large, 2.2 Mb gene. (B) Aggregate metagene analysis of all deletions <= 1Mb in or near genes >= 1Mb in 10,221 The Cancer Gene Atlas (TCGA) tumors representing 30 tumor types. Experimentally induced30 (blue) and a large proportion of cancer (red) deletion CNVs accumulate specifically and precisely at the centers of these large genes in a manner consistent with the model in Figure 2. (C) Examples of deletion CNV hotspot specificity by tumor type. Differences between ovarian serous cystadenocarcinoma (blue) and head and neck squamous carcinoma (red) are shown with respect to acquired deletion CNV occurrence in genes LSAMP and CUB and Sushi multiple domains 1 (CSMD1). Data summarized in this figure were generated in part by the TCGA Research Network, http://cancergenome.nih.gov/, and obtained from http://firebrowse.org/143.
Oncogene stress
The accumulation of CNVs in large CFS-associated genes in cancers suggests that the CNVs induced in cultured cells effectively mimic processes that occur in vivo, and that cancer cell CNVs also occur in response to replication stress. In cultured cells, APH and other inhibitors of DNA replication are used to induce replication stress, leading to CFSs and associated CNVs, but what creates this replication stress in cancer cells in vivo? A number of studies indicate that replication stress is a key component of the early tumorigenic process and that it can be induced by oncogene overexpression in vivo112, 113. Activated oncogenes are a key feature of early cancer development, with a major outcome being the induction of genome instability and DNA damage, including at CFSs 113–115.
Oncogene activation can deregulate replication in a number of ways including: by decreasing the number of licensed origins leading to under-replicated DNA, through unscheduled replication initiation leading to re-replication, by direct effects on replication fork progression, or by leading to accumulation of damaging reactive oxygen species116. Overexpression of Cyclin E and cell division cycle 25A (CDC25A) slows replication forks and induces fork reversal that activates the DNA damage response during G2/M in a MUS81-dependent manner, which is interpreted as evidence for increased topological stress during replication. These observations led to a model of nucleolytic processing of unusual replication intermediates during mitosis to limit oncogene-induced genotoxicity, which could modulate the anti-tumorigenic function of DNA damage checkpoints117. Similarly, Cyclin E deregulation causes cells to enter mitosis with incompletely replicated regions, including CFSs and late replicating domains 118. Interestingly, oncogene stimulation of transcription and associated R-loop formation, where RNA hybridizes to a complementary DNA strand, forming an RNA/DNA hybrid with displacement of the other DNA strand, is one important mechanism by which these replication outcomes arise, further supporting a link between transcription, replication and genomic instability in cancer119.
Consistent with these observations, genomic instability occurs preferentially at CFSs following experimentally-induced, aberrant oncogene expression114, 120. Recent studies demonstrated that overexpression of cyclin E or mutated HRAS in normal human fibroblasts leads to chromosomal breakage, with each oncogene creating a unique fragility landscape that overlaps with APH-induced CFSs as well as at a number of unique sites121. Oncogene-induced fragile sites share features with APH-induced CFSs, including colocalization with large genes and recurrent instability hotspots in cancer. This overlap between oncogene-induced fragile sites and large genes further supports the model that most large genes are actually fragile sites under specific replication stress conditions29, 30. If any CFS-CNV hotspots harbor tumor suppressor genes, genome instability driven by the overexpression of oncogenes could potentially preferentially inactivate these genes and drive the cell further down the tumorigenic pathway.
Functional role of CFS-associated genes in cancer
The observation that CFS genes are frequently deleted in cancers raises the important question of what function these deletions might play in tumorigenesis. FHIT, WWOX and a number of other CFS-associated genes have been proposed to be tumor suppressor genes and their loss may lead to cancer development122, 123. This hypothesis has been supported by studies indicating that deletion and/or reduced expression of these genes is a predictor of poor outcome in many different cancers. The largest body of data is on FHIT, with numerous reports of genomic alterations and loss of protein expression in preneoplasias, suggesting a tumor suppressive role beginning in the early stages of cancer development107, 124, 125. Loss of FHIT expression via promoter methylation and/or deletion have been associated with tumor progression and reduced survival in a number of cancer types126, 127. Homozygous and hemizygous deletions affecting the WWOX locus have been reported in several types of cancer123, 128 and similar but less extensive data are available for some other CFS-associated genes129.
Most functional studies on CFS genes have also focused on FHIT, WWOX and, more recently, PARK2, linking them to the DNA damage response in cultured cells and mouse cancer models122. Loss of FHIT has been reported to result in dNTP imbalance and spontaneous replication stress130 and WWOX has been reported to function in activation of the ATR-mediated DNA damage checkpoint response activation131. Consistent with these hypotheses, both Fhit- and Wwox-deficient mice exhibit increased cancer incidence and susceptibility to carcinogen-induced cancers132–134. Ectopic expression of WWOX in WWOX-deficient cancer cells suppresses cell and tumor growth in immune-compromised mice and ablation of WWOX in mice resulted in higher incidence of lesions resembling osteosarcomas and lung and mammary tumors135. The mechanisms by which FHIT, a dinucleosidetriphosphatase and WWOX, an oxidoreductase, might function in the DNA damage response are not entirely clear. A better understood mechanistic path to genome instability has been reported by Gong et al.136, who showed that the PARK2 E3 ubiquitin ligase coordinately controls the stability of both cyclin D and cyclin E. Inactivation of PARK2 results in the accumulation of cyclin D and acceleration of cell cycle progression. Thus, the PARK2 CFS regulates cyclin-CDK complexes, as does the CDK inhibitor p16 (which is encoded by CDKN2A), and acts as a major regulator of the stability of G1/S cyclins, critical factors in genome stability.
Others have argued that CFS-associated gene deletions in cancer cells are simply passenger mutations that result from the inherent instability of the CFSs79, 137. This viewpoint is supported by the predominance of hemizygous versus homozygous deletions, the paucity of inactivating point mutations in the genes in tumors and the frequent failure of deletions to affect RNA or protein expression of associated genes109, all of which are common features of tumor suppressor genes. Given the instability of CFS-associated genes and their high risk for CNV formation at these loci, focal intragenic deletions in CFS-associated genes are not sufficient evidence of tumor suppressor function. However, high inherent instability and functional importance are not necessarily mutually exclusive. CFS-associated genes with different functional properties should not be lumped together when considering their possible roles in cancer. Clearly, additional functional studies, verified in independent laboratories, are required to reach firm conclusions about a mechanistic role of specific CFS-associated genes in tumorigenesis.
CFS genes in genomic disorders
Finally, the potential important biological implications of instability of CFS-associated genes extend beyond cancer. Many of these genes play important roles in neurodevelopment and many CFS-associated CNV hotspots in cultured cells correspond to a subclass of clinically relevant human CNVs implicated in developmental disorders28, 30. For example, constitutional deletions within AUTS2, IMMP2L, and NRXN1 have been associated with autism spectrum disorder, intellectual disability, and psychiatric disorders138–140. Other large genes, such as CNTNAP2, in which intragenic CNVs are found in several neurodevelopmental disorders141 were not expressed in the cell types used by Wilson et al.30, but are predicted to be CFSs and CNV hotspots in the replicating cell types in which they are expressed, which could include neural progenitor cells, germ cell precursors, and early post-zygotic cells. Emerging genomic approaches in single cell genomics should soon allow tests of this prediction.
Conclusions and Perspective
CFS gaps and breaks on metaphase chromosomes were discovered over 35 years ago as the first indicators of the unusually high instability of large, late replicating genes that also manifests as frequent and, in many cases, unexplained focal CNVs in cancer. We now know that the most frequent and unstable CFSs are enriched within large genes with a paucity of replication origins and that active transcription greatly increases their instability, leading to late or incomplete replication under conditions of replication stress. These perturbations of DNA replication can lead to a number of outcomes depending on the resolution of these unreplicated regions including, most importantly for cancer and perhaps normal somatic tissues, CNV formation. In cancer, this relationship is particularly exacerbated because early stages of oncogenic transformation represent a form of transcription-associated replication stress that can potentiate further genomic instability and CNV formation. Data from our laboratory have shown that CFSs and CNV hotspots are different manifestations of the same mechanistic process driven by large, active transcription units. Transcription of large genes in dividing cells is an important factor in the mechanisms leading to CFSs and associated CNV hotspots, setting up a “perfect storm” of instability driven by replication stress and double fork failure. These data predict that large genes will be CFSs and CNV hotspots in the replicating cell types in which their long isoforms are expressed, enabling the identification of potential CFS and CNV hotspots from any cells or tissue in which the nascent transcription profile is known. They also suggest that transcriptional differences among individual tumors or the cells from which they arose likely explain differences in CNV hotpots observed in different cancers. It is important to note that not all genomic regions with a paucity of replication origins reside within large transcribed genes, and further attention should be also given to transcription-independent mechanisms of origin suppression and other mechanisms leading to incomplete replication that may function in less frequent CFSs and CNVs. Finally, the opposing views on whether CFS genes such as FHIT, WWOX and PARK2 are important players in cancer progression or simply passenger lesions that result from CFS hypermutability will only be resolved with continued rigorous and independently verified gene-specific functional studies.
Acknowledgments
The authors would like to thank John Moran for critical reading of this manuscript, and Rameen Beroukhim for sharing early versions of the TCGA CNV data analyses. This work was supported in part by grants ES020875 from the National Institute of Environmental Health Sciences and CA 200731 from the National Cancer Institute.
Biographies
Thomas W. Glover, Ph.D. is a Professor in the Departments of Human Genetics, Pathology, and Pediatrics, University of Michigan and a member of the University of Michigan Comprehensive Cancer Center. He first described common fragile sites in the early 1980s and has continued to study the mechanisms of their instability and associated genomic consequences. His current focus is on understating the relationship of CFSs to genomic structural alterations in cancer and genetic disorders.
Thomas E. Wilson holds a M.D. Ph.D. from Washington University. He is Professor of Pathology and Human Genetics at the University of Michigan Medical School, where he serves as Associate Director of the Molecular Diagnostics Laboratory. Dr. Wilson’s research laboratory uses in vitro and bioinformatics approaches to study mechanisms of DNA repair, replication and transcription and how they contribute to formation of chromosomal rearrangements in health and disease.
Martin F. Arlt earned his Ph.D. from Washington University. As a postdoctoral fellow and research faculty in the Department of Human Genetics at the University of Michigan Medical School, his published body of work has expanded the understanding of the molecular mechanisms underlying common fragile site instability. His current work focuses on understanding the genetic and environmental risk factors of chromosome structural rearrangements.
Footnotes
Competing interests statement
There is NO Competing Interest.
Contributor Information
Thomas W. Glover, Department of Human Genetics, University of Michigan Medical School, Ann Arbor, MI 48109 USA. Department of Pathology, University of Michigan Medical School, Ann Arbor, MI 48109 USA. Department of Pediatrics and Communicable Diseases, University of Michigan Medical School, Ann Arbor, MI 48109 USA
Thomas E. Wilson, Department of Human Genetics, University of Michigan Medical School, Ann Arbor, MI 48109 USA. Department of Pathology, University of Michigan Medical School, Ann Arbor, MI 48109 USA
Martin F. Arlt, Department of Human Genetics, University of Michigan Medical School, Ann Arbor, MI 48109 USA
References
- 1.Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67:136–42. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 2.Yunis JJ. Fragile sites and predisposition to leukemia and lymphoma. Cancer Genet Cytogenet. 1984;12:85–8. doi: 10.1016/0165-4608(84)90011-6. [DOI] [PubMed] [Google Scholar]
- 3.de Braekeleer M. Fragile sites and chromosomal structural rearrangements in human leukemia and cancer. Anticancer Res. 1987;7:417–22. [PubMed] [Google Scholar]
- 4.Yunis JJ. Fragile sites, mutagens and genomic rearrangements in cancer. Basic Life Sci. 1988;43:11–21. doi: 10.1007/978-1-4684-5460-4_3. [DOI] [PubMed] [Google Scholar]
- 5.Le Beau MM. Chromosomal fragile sites and cancer-specific rearrangements. Blood. 1986;67:849–58. [PubMed] [Google Scholar]
- 6.De Braekeleer M, Smith B, Lin CC. Fragile sites and structural rearrangements in cancer. Hum Genet. 1985;69:112–6. doi: 10.1007/BF00293279. [DOI] [PubMed] [Google Scholar]
- 7.Barlow JH, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–32. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huebner K, Hadaczek P, Siprashvili Z, Druck T, Croce CM. The FHIT gene, a multiple tumor suppressor gene encompassing the carcinogen sensitive chromosome fragile site, FRA3B. Biochim Biophys Acta. 1997;1332:M65–70. doi: 10.1016/s0304-419x(97)00009-7. [DOI] [PubMed] [Google Scholar]
- 9.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan ZA, Li XZ, Zhou XT. The effect of hydroxyurea on the expression of the common fragile site at 3p14. J Med Genet. 1987;24:593–6. doi: 10.1136/jmg.24.10.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An international system for human cytogenetic nomenclature--high-resolution banding (1981); ISCN. Report of the Standing Committee on Human Cytogenetic Nomenclature. Cytogenet Cell Genet. 1981;31:5–23. doi: 10.1159/000131621. [DOI] [PubMed] [Google Scholar]
- 12.GDB. ([Internet] Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004–2017. Available from: https://www.ncbi.nlm.nih.gov/gene/) [Google Scholar]
- 13.Mariani T. Fragile sites and statistics. Hum Genet. 1989;81:319–22. doi: 10.1007/BF00283683. [DOI] [PubMed] [Google Scholar]
- 14.Glover TW, Stein CK. Induction of sister chromatid exchanges at common fragile sites. Am J Hum Genet. 1987;41:882–90. [PMC free article] [PubMed] [Google Scholar]
- 15.Wenger SL. Chemical induction of sister chromatid exchange at fragile sites. Cancer Genet Cytogenet. 1995;85:72–4. doi: 10.1016/0165-4608(95)00137-9. [DOI] [PubMed] [Google Scholar]
- 16.Glover TW, Stein CK. Chromosome breakage and recombination at fragile sites. Am J Hum Genet. 1988;43:265–73. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang ND, Testa JR, Smith DI. Determination of the specificity of aphidicolin-induced breakage of the human 3p14.2 fragile site. Genomics. 1993;17:341–7. doi: 10.1006/geno.1993.1330. [DOI] [PubMed] [Google Scholar]
- 18.Rassool FV, et al. Preferential integration of marker DNA into the chromosomal fragile site at 3p14: an approach to cloning fragile sites. Proc Natl Acad Sci U S A. 1991;88:6657–61. doi: 10.1073/pnas.88.15.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–92. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 20.Sarni D, Kerem B. The complex nature of fragile site plasticity and its importance in cancer. Curr Opin Cell Biol. 2016;40:131–6. doi: 10.1016/j.ceb.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Arlt MF, Casper AM, Glover TW. Common fragile sites. Cytogenet Genome Res. 2003;100:92–100. doi: 10.1159/000072843. [DOI] [PubMed] [Google Scholar]
- 22.Le Beau MM, et al. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum Mol Genet. 1998;7:755–61. doi: 10.1093/hmg/7.4.755. [DOI] [PubMed] [Google Scholar]
- 23.Wilke CM, et al. Multicolor FISH mapping of YAC clones in 3p14 and identification of a YAC spanning both FRA3B and the t(3;8) associated with hereditary renal cell carcinoma. Genomics. 1994;22:319–26. doi: 10.1006/geno.1994.1390. [DOI] [PubMed] [Google Scholar]
- 24.Paradee W, et al. Precise localization of aphidicolin-induced breakpoints on the short arm of human chromosome 3. Genomics. 1995;27:358–61. doi: 10.1006/geno.1995.1057. [DOI] [PubMed] [Google Scholar]
- 25.Mangelsdorf M, et al. Chromosomal fragile site FRA16D and DNA instability in cancer. Cancer Res. 2000;60:1683–9. [PubMed] [Google Scholar]
- 26.Ohta M, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–97. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 27.Bednarek AK, et al. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–5. [PubMed] [Google Scholar]
- 28.Smith DI, Zhu Y, McAvoy S, Kuhn R. Common fragile sites, extremely large genes, neural development and cancer. Cancer Lett. 2006;232:48–57. doi: 10.1016/j.canlet.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 29.Gao G, Smith DI. Very large common fragile site genes and their potential role in cancer development. Cell Mol Life Sci. 2014;71:4601–15. doi: 10.1007/s00018-014-1753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson TE, et al. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 2015;25:189–200. doi: 10.1101/gr.177121.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murano I, Kuwano A, Kajii T. Cell type-dependent difference in the distribution and frequency of aphidicolin-induced fragile sites: T and B lymphocytes and bone marrow cells. Hum Genet. 1989;84:71–4. doi: 10.1007/BF00210675. [DOI] [PubMed] [Google Scholar]
- 32.Murano I, Kuwano A, Kajii T. Fibroblast-specific common fragile sites induced by aphidicolin. Hum Genet. 1989;83:45–8. doi: 10.1007/BF00274145. [DOI] [PubMed] [Google Scholar]
- 33.Kuwano A, Murano I, Kajii T. Cell type-dependent difference in the distribution and frequency of excess thymidine-induced common fragile sites: T lymphocytes and skin fibroblasts. Hum Genet. 1990;84:527–31. doi: 10.1007/BF00210803. [DOI] [PubMed] [Google Scholar]
- 34.Le Tallec B, et al. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol. 2011;18:1421–3. doi: 10.1038/nsmb.2155. [DOI] [PubMed] [Google Scholar]
- 35.Le Tallec B, et al. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Rep. 2013;4:420–8. doi: 10.1016/j.celrep.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Hosseini SA, et al. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Genes Chromosomes Cancer. 2013;52:1017–29. doi: 10.1002/gcc.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover TW. In: Genetic Instabilities and Neurological Diseases. Wells RDW, ST, editors. Academic Press; San Diego: 1998. pp. 75–83. [Google Scholar]
- 38.Palakodeti A, et al. Impaired replication dynamics at the FRA3B common fragile site. Hum Mol Genet. 2010;19:99–110. doi: 10.1093/hmg/ddp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover TW, Arlt MF, Casper AM, Durkin SG. Mechanisms of common fragile site instability. Hum Mol Genet. 2005;14(Spec No 2):R197–205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- 40.Boldog F, et al. Chromosome 3p14 homozygous deletions and sequence analysis of FRA3B. Hum Mol Genet. 1997;6:193–203. doi: 10.1093/hmg/6.2.193. [DOI] [PubMed] [Google Scholar]
- 41.Ried K, et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9:1651–63. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 42.Arlt MF, Miller DE, Beer DG, Glover TW. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes Cancer. 2002;33:82–92. doi: 10.1002/gcc.10000. [DOI] [PubMed] [Google Scholar]
- 43.Zlotorynski E, et al. Molecular basis for expression of common and rare fragile sites. Mol Cell Biol. 2003;23:7143–51. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishmar D, et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci U S A. 1998;95:8141–6. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Freudenreich CH. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol Cell. 2007;27:367–79. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durkin SG, et al. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc Natl Acad Sci U S A. 2008;105:246–51. doi: 10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–89. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 49.Durkin SG, Arlt MF, Howlett NG, Glover TW. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene. 2006;25:4381–8. doi: 10.1038/sj.onc.1209466. [DOI] [PubMed] [Google Scholar]
- 50.Zhu M, Weiss RS. Increased common fragile site expression, cell proliferation defects, and apoptosis following conditional inactivation of mouse Hus1 in primary cultured cells. Mol Biol Cell. 2007;18:1044–55. doi: 10.1091/mbc.E06-10-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Focarelli ML, et al. Claspin inhibition leads to fragile site expression. Genes Chromosomes Cancer. 2009;48:1083–90. doi: 10.1002/gcc.20710. [DOI] [PubMed] [Google Scholar]
- 52.Musio A, et al. SMC1 involvement in fragile site expression. Hum Mol Genet. 2005;14:525–33. doi: 10.1093/hmg/ddi049. [DOI] [PubMed] [Google Scholar]
- 53.Bhat A, Andersen PL, Qin Z, Xiao W. Rev3, the catalytic subunit of Polzeta, is required for maintaining fragile site stability in human cells. Nucleic Acids Res. 2013;41:2328–39. doi: 10.1093/nar/gks1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madireddy A, et al. FANCD2 Facilitates Replication through Common Fragile Sites. Mol Cell. 2016;64:388–404. doi: 10.1016/j.molcel.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhowmick R, Minocherhomji S, Hickson ID. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell. 2016;64:1117–1126. doi: 10.1016/j.molcel.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 56.Mason JM, et al. The SNM1B/APOLLO DNA nuclease functions in resolution of replication stress and maintenance of common fragile site stability. Hum Mol Genet. 2013;22:4901–13. doi: 10.1093/hmg/ddt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ying S, et al. MUS81 promotes common fragile site expression. Nat Cell Biol. 2013;15:1001–7. doi: 10.1038/ncb2773. [DOI] [PubMed] [Google Scholar]
- 58.Naim V, Wilhelm T, Debatisse M, Rosselli F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat Cell Biol. 2013;15:1008–15. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- 59.Minocherhomji S, et al. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528:286–90. doi: 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- 60.Bergoglio V, et al. DNA synthesis by Pol eta promotes fragile site stability by preventing under-replicated DNA in mitosis. J Cell Biol. 2013;201:395–408. doi: 10.1083/jcb.201207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walsh E, Wang X, Lee MY, Eckert KA. Mechanism of replicative DNA polymerase delta pausing and a potential role for DNA polymerase kappa in common fragile site replication. J Mol Biol. 2013;425:232–43. doi: 10.1016/j.jmb.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J Cell Biol. 2008;180:305–14. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fundia A, Gorla N, Larripa I. Non-random distribution of spontaneous chromosome aberrations in two Bloom Syndrome patients. Hereditas. 1995;122:239–43. doi: 10.1111/j.1601-5223.1995.00239.x. [DOI] [PubMed] [Google Scholar]
- 64.Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 65.Arlt MF, et al. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol Cell Biol. 2004;24:6701–9. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz M, et al. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005;19:2715–26. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Letessier A, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–3. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 68.Miotto B, Ji Z, Struhl K. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci U S A. 2016;113:E4810–9. doi: 10.1073/pnas.1609060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sotiriou SK, et al. Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol Cell. 2016;64:1127–1134. doi: 10.1016/j.molcel.2016.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–14. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 71.Chan KL, Hickson ID. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin Cell Dev Biol. 2011;22:906–12. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 72.Biebricher A, et al. PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol Cell. 2013;51:691–701. doi: 10.1016/j.molcel.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–81. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee C, Scherer SW. The clinical context of copy number variation in the human genome. Expert Rev Mol Med. 2010;12:e8. doi: 10.1017/S1462399410001390. [DOI] [PubMed] [Google Scholar]
- 76.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–7. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Campbell IM, et al. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am J Hum Genet. 2014;95:173–82. doi: 10.1016/j.ajhg.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–8. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arlt MF, et al. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet. 2009;84:339–50. doi: 10.1016/j.ajhg.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, Glover TW. Hydroxyurea induces de novo copy number variants in human cells. Proc Natl Acad Sci U S A. 2011;108:17360–5. doi: 10.1073/pnas.1109272108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. Copy number variants are produced in response to low-dose ionizing radiation in cultured cells. Environ Mol Mutagen. 2014;55:103–13. doi: 10.1002/em.21840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 2012;8:e1002981. doi: 10.1371/journal.pgen.1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22:211–20. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–77. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 86.Newman TJ, Mamun MA, Nieduszynski CA, Blow JJ. Replisome stall events have shaped the distribution of replication origins in the genomes of yeasts. Nucleic Acids Res. 2013;41:9705–18. doi: 10.1093/nar/gkt728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ozeri-Galai E, et al. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell. 2011;43:122–31. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 88.Snyder M, Sapolsky RJ, Davis RW. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2184–94. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Looke M, et al. Relicensing of transcriptionally inactivated replication origins in budding yeast. J Biol Chem. 2010;285:40004–11. doi: 10.1074/jbc.M110.148924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei PC, et al. Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells. Cell. 2016;164:644–55. doi: 10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wilke CM, et al. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integration sites and fragile sites. Hum Mol Genet. 1996;5:187–95. doi: 10.1093/hmg/5.2.187. [DOI] [PubMed] [Google Scholar]
- 92.Gao G, et al. Common fragile sites (CFS) and extremely large CFS genes are targets for human papillomavirus integrations and chromosome rearrangements in oropharyngeal squamous cell carcinoma. Genes Chromosomes Cancer. 2017;56:59–74. doi: 10.1002/gcc.22415. [DOI] [PubMed] [Google Scholar]
- 93.Kraus I, et al. The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res. 2008;68:2514–22. doi: 10.1158/0008-5472.CAN-07-2776. [DOI] [PubMed] [Google Scholar]
- 94.Thorland EC, et al. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 2000;60:5916–21. [PubMed] [Google Scholar]
- 95.Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004;64:3878–84. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 96.Matovina M, Sabol I, Grubisic G, Gasperov NM, Grce M. Identification of human papillomavirus type 16 integration sites in high-grade precancerous cervical lesions. Gynecol Oncol. 2009;113:120–7. doi: 10.1016/j.ygyno.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 97.Walline HM, et al. Integration of high-risk human papillomavirus into cellular cancer-related genes in head and neck cancer cell lines. Head Neck. 2017 doi: 10.1002/hed.24729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jang MK, Shen K, McBride AA. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog. 2014;10:e1004117. doi: 10.1371/journal.ppat.1004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu Z, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47:158–63. doi: 10.1038/ng.3178. [DOI] [PubMed] [Google Scholar]
- 100.Doolittle-Hall JM, Cunningham Glasspoole DL, Seaman WT, Webster-Cyriaque J. Meta-Analysis of DNA Tumor-Viral Integration Site Selection Indicates a Role for Repeats, Gene Expression and Epigenetics. Cancers (Basel) 2015;7:2217–35. doi: 10.3390/cancers7040887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Finnis M, et al. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum Mol Genet. 2005;14:1341–9. doi: 10.1093/hmg/ddi144. [DOI] [PubMed] [Google Scholar]
- 102.Denison SR, Callahan G, Becker NA, Phillips LA, Smith DI. Characterization of FRA6E and its potential role in autosomal recessive juvenile parkinsonism and ovarian cancer. Genes Chromosomes Cancer. 2003;38:40–52. doi: 10.1002/gcc.10236. [DOI] [PubMed] [Google Scholar]
- 103.Callahan G, Denison SR, Phillips LA, Shridhar V, Smith DI. Characterization of the common fragile site FRA9E and its potential role in ovarian cancer. Oncogene. 2003;22:590–601. doi: 10.1038/sj.onc.1206171. [DOI] [PubMed] [Google Scholar]
- 104.Huang H, Qian C, Jenkins RB, Smith DI. Fish mapping of YAC clones at human chromosomal band 7q31.2: identification of YACS spanning FRA7G within the common region of LOH in breast and prostate cancer. Genes Chromosomes Cancer. 1998;21:152–9. doi: 10.1002/(sici)1098-2264(199802)21:2<152::aid-gcc11>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 105.Lai LA, et al. Deletion at fragile sites is a common and early event in Barrett’s esophagus. Mol Cancer Res. 2010;8:1084–94. doi: 10.1158/1541-7786.MCR-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Michael D, Beer DG, Wilke CW, Miller DE, Glover TW. Frequent deletions of FHIT and FRA3B in Barrett’s metaplasia and esophageal adenocarcinomas. Oncogene. 1997;15:1653–9. doi: 10.1038/sj.onc.1201330. [DOI] [PubMed] [Google Scholar]
- 107.Gu J, et al. Genome-wide catalogue of chromosomal aberrations in barrett’s esophagus and esophageal adenocarcinoma: a high-density single nucleotide polymorphism array analysis. Cancer Prev Res (Phila) 2010;3:1176–86. doi: 10.1158/1940-6207.CAPR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 109.Rajaram M, et al. Two Distinct Categories of Focal Deletions in Cancer Genomes. PLoS One. 2013;8:e66264. doi: 10.1371/journal.pone.0066264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44:D1023–31. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Romagosa C, et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011;30:2087–97. doi: 10.1038/onc.2010.614. [DOI] [PubMed] [Google Scholar]
- 112.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 113.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 114.Georgakilas AG, et al. Are common fragile sites merely structural domains or highly organized “functional” units susceptible to oncogenic stress? Cell Mol Life Sci. 2014;71:4519–44. doi: 10.1007/s00018-014-1717-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dereli-Oz A, Versini G, Halazonetis TD. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol Oncol. 2011;5:308–14. doi: 10.1016/j.molonc.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gaillard H, Garcia-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–89. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 117.Neelsen KJ, Zanini IM, Herrador R, Lopes M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol. 2013;200:699–708. doi: 10.1083/jcb.201212058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Teixeira LK, et al. Cyclin E deregulation promotes loss of specific genomic regions. Curr Biol. 2015;25:1327–33. doi: 10.1016/j.cub.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kotsantis P, et al. Increased global transcription activity as a mechanism of replication stress in cancer. Nat Commun. 2016;7:13087. doi: 10.1038/ncomms13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsantoulis PK, et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27:3256–64. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 121.Miron K, Golan-Lev T, Dvir R, Ben-David E, Kerem B. Oncogenes create a unique landscape of fragile sites. Nat Commun. 2015;6:7094. doi: 10.1038/ncomms8094. [DOI] [PubMed] [Google Scholar]
- 122.Karras JR, Schrock MS, Batar B, Huebner K. Fragile Genes That Are Frequently Altered in Cancer: Players Not Passengers. Cytogenet Genome Res. 2017 doi: 10.1159/000455753. [DOI] [PubMed] [Google Scholar]
- 123.Schrock MS, Huebner K. WWOX: a fragile tumor suppressor. Exp Biol Med (Maywood) 2015;240:296–304. doi: 10.1177/1535370214561590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guler G, et al. Concordant loss of fragile gene expression early in breast cancer development. Pathol Int. 2005;55:471–8. doi: 10.1111/j.1440-1827.2005.01855.x. [DOI] [PubMed] [Google Scholar]
- 125.Sozzi G, et al. Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res. 1998;58:5032–7. [PubMed] [Google Scholar]
- 126.Wu X, Wu G, Yao X, Hou G, Jiang F. The clinicopathological significance and ethnic difference of FHIT hypermethylation in non-small-cell lung carcinoma: a meta-analysis and literature review. Drug Des Devel Ther. 2016;10:699–709. doi: 10.2147/DDDT.S85253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kandimalla R, van Tilborg AA, Zwarthoff EC. DNA methylation-based biomarkers in bladder cancer. Nat Rev Urol. 2013;10:327–35. doi: 10.1038/nrurol.2013.89. [DOI] [PubMed] [Google Scholar]
- 128.Baryla I, Styczen-Binkowska E, Bednarek AK. Alteration of WWOX in human cancer: a clinical view. Exp Biol Med (Maywood) 2015;240:305–14. doi: 10.1177/1535370214561953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gao G, Smith DI. WWOX, large common fragile site genes, and cancer. Exp Biol Med (Maywood) 2015;240:285–95. doi: 10.1177/1535370214565992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saldivar JC, et al. Initiation of genome instability and preneoplastic processes through loss of Fhit expression. PLoS Genet. 2012;8:e1003077. doi: 10.1371/journal.pgen.1003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Abu-Odeh M, Hereema NA, Aqeilan RI. WWOX modulates the ATR-mediated DNA damage checkpoint response. Oncotarget. 2016;7:4344–55. doi: 10.18632/oncotarget.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Drusco A, et al. Common fragile site tumor suppressor genes and corresponding mouse models of cancer. J Biomed Biotechnol. 2011;2011:984505. doi: 10.1155/2011/984505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aqeilan RI, et al. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 2007;67:5606–10. doi: 10.1158/0008-5472.CAN-07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zanesi N, et al. The tumor spectrum in FHIT-deficient mice. Proc Natl Acad Sci U S A. 2001;98:10250–5. doi: 10.1073/pnas.191345898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aqeilan RI, et al. Targeted deletion of Wwox reveals a tumor suppressor function. Proc Natl Acad Sci U S A. 2007;104:3949–54. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gong Y, et al. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nat Genet. 2014;46:588–94. doi: 10.1038/ng.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Le Beau MM, et al. An FHIT tumor suppressor gene? Genes Chromosomes Cancer. 1998;21:281–9. doi: 10.1002/(sici)1098-2264(199804)21:4<281::aid-gcc1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 138.Nagamani SC, et al. Detection of copy-number variation in AUTS2 gene by targeted exonic array CGH in patients with developmental delay and autistic spectrum disorders. Eur J Hum Genet. 2013;21:343–6. doi: 10.1038/ejhg.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Swaminathan S, et al. Analysis of copy number variation in Alzheimer’s disease: the NIALOAD/ NCRAD Family Study. Curr Alzheimer Res. 2012;9:801–14. doi: 10.2174/156720512802455331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gregor A, et al. Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC Med Genet. 2011;12:106. doi: 10.1186/1471-2350-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mikhail FM, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A. 2011;155A:2386–96. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- 142.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Broad. Broad Institute TCGA Genome Data Analysis Center. SNP6 Copy number analysis (GISTIC2) Broad Institute of MIT and Harvard; 2016. [DOI] [Google Scholar]
- 144.Arlt MF, Glover TW. Inhibition of topoisomerase I prevents chromosome breakage at common fragile sites. DNA Repair (Amst) 2010;9:678–89. doi: 10.1016/j.dnarep.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tuduri S, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–24. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Casper AM, Durkin SG, Arlt MF, Glover TW. Chromosomal instability at common fragile sites in Seckel syndrome. Am J Hum Genet. 2004;75:654–60. doi: 10.1086/422701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ozeri-Galai E, Schwartz M, Rahat A, Kerem B. Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene. 2008;27:2109–17. doi: 10.1038/sj.onc.1210849. [DOI] [PubMed] [Google Scholar]
- 148.Schwartz M, et al. Impaired replication stress response in cells from immunodeficiency patients carrying Cernunnos/XLF mutations. PLoS One. 2009;4:e4516. doi: 10.1371/journal.pone.0004516. [DOI] [PMC free article] [PubMed] [Google Scholar]