Abstract
The Ser/Thr kinase AKT, also known as protein kinase B (PKB), was discovered 25 years ago and has been the focus of tens of thousands of studies in diverse fields of biology and medicine. There have been many advances in our knowledge of the upstream regulatory inputs into AKT, key multifunctional downstream signaling nodes (GSK3, FoxO, mTORC1), which greatly expand the functional repertoire of Akt, and the complex circuitry of this dynamically branching and looping signaling network that is ubiquitous to nearly every cell in our body. Mouse and human genetic studies have also revealed physiological roles for the AKT network in nearly every organ system. Our comprehension of AKT regulation and functions is particularly important given the consequences of AKT dysfunction in diverse pathological settings, including developmental and overgrowth syndromes, cancer, cardiovascular disease, insulin resistance and type-2 diabetes, inflammatory and autoimmune disorders, and neurological disorders. There has also been much progress in developing AKT-selective small molecule inhibitors. Improved understanding of the molecular wiring of the AKT signaling network continues to make an impact that cuts across most disciplines of the biomedical sciences.
25 Years of AKT Signaling
Thirty years ago, Stephen Staal identified and cloned the v-Akt oncogene from the AKT8 transforming retrovirus (Staal, 1987). Four years later, three laboratories independently cloned and characterized the cellular homolog of v-AKT, a 57 Kd Ser/Thr protein kinase. Bellacosa and Tsichlis used cDNA hybridization with v-AKT to clone the protein kinase and termed it c-AKT (Bellacosa et al., 1991). The Hemmings group used degenerate PCR for sequences encoding protein kinase catalytic domains to identify the kinase, which they named Related to A- and C-kinase (RAC) (Jones et al., 1991). Woodgett and Coffer used library screening and identified a protein kinase they named protein kinase B (PKB), due to the similarity with PKA and PKC (Coffer and Woodgett, 1991). We now know there are three AKT/PKB isoforms conserved in mammalian genomes, AKT1 (PKBα), AKT2 (PKBβ) and AKT3 (PKBγ). AKT was propelled into the signal transduction limelight a few years later, when it was found that AKT activation occurs downstream of phosphoinositide 3-kinase (PI3K), a lipid kinase linked to cellular transformation and the insulin response (Cantley, 2004).
Class I PI3K phosphorylates the 3′ hydroxyl of the inositol head group of phosphoinositides, resulting in the production of the lipid second messengers PtdIns-3,4-P2 (PI3,4P2) and PtdIns-3,4,5-P3 (PIP3). However, downstream effectors of the PI3K products were unknown in the mid-90’s. Franke, Kaplan and Tsichlis working with PDGF receptor mutants developed by Kazlauskas showed that stimulation of cells with PDGF results in the activation of AKT in a manner that depends exclusively on the ability of PI3K to bind to the PDGF receptor (Franke et al., 1995). Burgering and Coffer (Burgering and Coffer, 1995) as well as the Roth laboratory (Kohn et al., 1995) used similar approaches to show that AKT is activated by growth factors in a PI3K-dependent manner. AKT was firmly established as the first bona-fide effector of PI3K in cells. What remained to be elucidated is the precise mechanism by which PI3K and its lipid products activate AKT. AKT possesses a Pleckstrin Homology (PH) domain at its amino-terminus, which Downes and Alessi initially showed can bind to PIP3 (James et al., 1996). Subsequently, both PI3,4P2 and PIP3 were shown to directly bind to the PH domain of AKT, and PI3,4P2 binding was found to induce partial activation of the protein kinase in vitro (Franke et al., 1997; Frech et al., 1997; Klippel et al., 1997).
Other landmark findings in the field were the mechanisms of termination of AKT activity and its first substrates. PI3K activity is opposed by the tumor suppressor PTEN, first cloned by the Parsons and Steck laboratories (Li et al., 1997; Steck et al., 1997), and characterized as a PIP3 phosphatase by Dixon (Maehama and Dixon, 1998). Concomitant with the identification of AKT as a PI3K effector, much work was being undertaken to uncover the role of PI3K in insulin signaling, leading to the discovery that GSK-3β is a substrate of AKT in insulin-stimulated cells (Cross et al., 1995). The identification of this first substrate of AKT was also instrumental in subsequent studies to define the optimal AKT consensus phosphorylation motif (Alessi et al., 1996b), which has since facilitated the discovery of over a hundred AKT substrates linked to cell physiology and disease.
In the past 25 years, the Akt signaling field has seen remarkable expansion and discoveries that have central relevance to health and human disease. Here, we provide an update and expansion to a review from 2007 (Manning and Cantley, 2007) that provides a more network view of Akt signaling. We highlight the detailed mechanisms that account for AKT regulation, the key downstream branches regulated by AKT, how the AKT network is wired and integrated with other cellular signals, how this in turn influences the physiology and pathobiology of AKT, and finally how therapeutic targeting of the AKT network can be used to treat human diseases.
Upstream Regulation of AKT
PI3K-Dependent AKT Activation
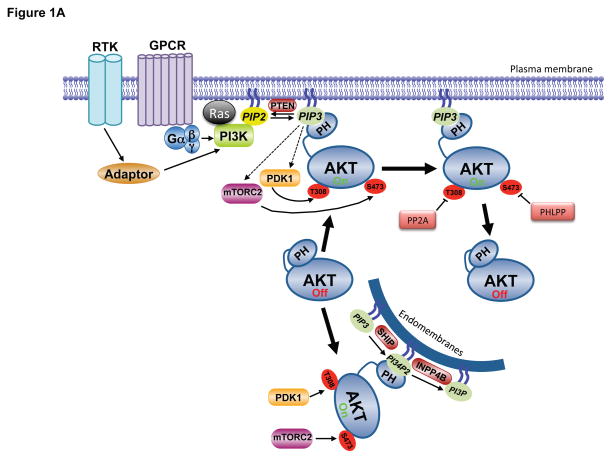
Activation of PI3K by extracellular stimuli results in activation of AKT in virtually all cells and tissues. As such, PI3K and its lipid products are generally considered to be obligate and rate-limiting for proper AKT activation. The canonical pathway leading to AKT activation is initiated by the stimulation of receptor tyrosine kinases (RTK) or G protein coupled receptors (GPCR) leading to plasma membrane recruitment and activation of one or more isoforms of the class I PI3K family (Figure 1A, for a detailed review on PI3K, see (Vanhaesebroeck et al., 2010)). Also important for regulation of PI3K is interaction with members of the Ras family of small GTPases (Rodriguez et al., 1994). Class I PI3Ks predominantly phosphorylate PI4, 5P2, thereby producing PIP3 (Vanhaesebroeck et al., 2010), whereas the synthesis of PI3,4P2 typically follows, perhaps resulting from the action of the 5′phosphatase SH2 domain-containing inositol 5′-phosphatase (SHIP) on PIP3 (Franke et al., 1997; Guilherme et al., 1996). PI3,4P2 can also be synthesized by class II PI3Ks using PI4P as substrate (reviewed in (Hawkins and Stephens, 2016)).. Although the specific PI3K isoform activated in a given cellular context may differ, the ultimate output is the same - relocalization of inactive AKT to membrane sites of PI3,4P2 or PIP3 accumulation via engagement of the AKT PH domain.
Figure 1. Molecular mechanisms of Akt regulation.
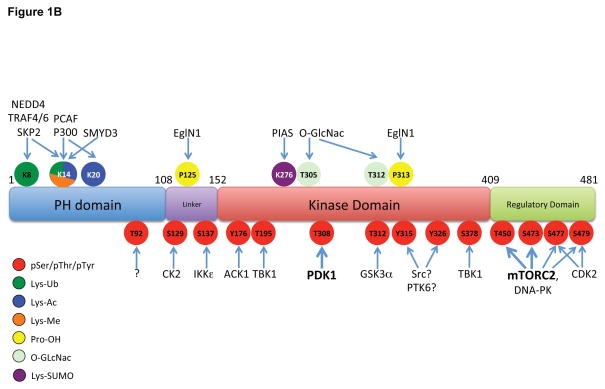
A. Stimulation of RTKs or GPCRs leads to activation of PI3K, leading to PIP3 production at the plasma membrane. Cytosolic inactive AKT is recruited to the membrane and engages PIP3 through PH domain binding. This leads to phosphorylation of T308 and S473 by PDK1 and mTORC2, respectively, resulting in full activation. Signal termination is achieved by the PIP3 phosphatase PTEN, and the PP2A and PHLPP protein phosphatases. A separate endomembrane pool of active AKT likely exists that is activated through engagement of PI3,4P2 through the action of the SHIP phosphatase, and terminated by INPP4B. B. The modular structure of AKT1 with position of PTMs color coded for phosphorylation (pSer/pThr/pTyr), acetylation (Lys-Ac), ubiquitylation (Lys-Ub), methylation (Lys-Me), hydroxylation (Pro-OH), glycosylation (O-GlcNac) and SUMOylation (Lys-SUMO).
Signal termination of PI3K/PIP3 signaling is primarily achieved by the phosphatase PTEN, which dephosphorylates PIP3 converting it back to PI4,5P2. The initial synthesis of PI3K lipid products is observed within seconds to minutes of growth factor stimulation, exhibits a peak, generally in the first hour, and is then downregulated with a timing that depends on the cell type and stimulus (Auger et al., 1989). The transient nature of this signal is largely achieved by PTEN action combined with temporal inactivation of PI3K. While AKT is the most widely-studied effector of PI3K signaling, and influences most phenotypes associated with PI3K pathway activation, it is worth noting that many other downstream effectors of PI3K exist that are activated in parallel to Akt and contribute to the subsequent cellular response (Vanhaesebroeck et al., 2010).
Major Regulatory Phosphorylation Events on Akt
Activation of PI3K results in the phosphorylation of two key residues on AKT1, T308 in the activation, or T-loop, of the catalytic protein kinase core, and S473 in a C-terminal hydrophobic motif (Alessi et al., 1996a) (Figure 1A,B). Phosphorylation of both residues is required for maximal activation of the kinase. Regulation also occurs on corresponding residues in AKT2 (T309 and S474) and AKT3 (T305 and S472). The phosphoinositide-dependent protein kinase 1 (PDK1) was discovered for its ability to phosphorylate AKT1 at T308, which is required for AKT activity (Alessi et al., 1997; Stokoe et al., 1997). Relocalization of both AKT and PDK1 to membrane sites of PIP3 or PI3,4P2 synthesis induces conformational changes providing access of PDK1 to AKT for phosphorylation of T308. In the inactive conformation, the AKT PH domain is inhibitory, and this ‘PH-in’ conformation is relieved by PH domain engagement of PI3K products, resulting in a ‘PH-out’ conformation that releases the kinase domain and allows its phosphorylation by PDK1 (Calleja et al., 2007; Calleja et al., 2009). PDK1 is also required for activation loop phosphorylation of other AGC family protein kinases (referring to those related to Protein Kinases A, G, and C), including all isoforms of the growth factor-stimulated kinases PKC, S6K, SGK, and RSK (Mora et al., 2004). However, none of these kinases possess a PIP3-binding domain, and AKT appears to be the only obligate PIP3-dependent PDK1 target amongst this group (Collins et al., 2003; McManus et al., 2004).
Maximal activation of AKT requires phosphorylation of S473 in the hydrophobic motif. The primary AKT S473 kinase is the mechanistic target of rapamcyin (mTOR) complex 2 (mTORC2) (Sarbassov et al., 2005) (Figure 1A,B)..While AKT lacking S473 phosphorylation has activity, it is greatly diminished, and phosphorylation of S473 stabilizes T308 phosphorylation and the activation state of AKT (Alessi et al., 1996a; Yang et al., 2002). By analogy with PDK1, mTORC2 also phosphorylates other AGC kinases at their corresponding hydrophobic motif residues, although the corresponding motif in S6K1 (T389) is targeted by mTORC1 (Saxton and Sabatini, 2017). Also like PDK1, some targets of mTORC2 are phosphorylated constitutively (e.g., PKC), whereas others are phosphorylated only in response to PI3K signaling (e.g., AKT, SGK). The protein kinase activity of mTORC2, assayed in immunopurifications, is stimulated by growth factors in a PI3K-dependent manner (Huang et al., 2008; Sarbassov et al., 2005). A recent study suggested a mechanism of this regulation, indicating that a PH domain within the SIN1 component of mTORC2 serves to bind to PIP3, leading to relief of autoinhibition of mTOR kinase activity within the complex (Liu et al., 2015). PIP3 binding would therefore have the dual function of relocalizing mTORC2 to membranes where AKT is being recruited as well as relieving conformational constraints on mTOR allowing AKT phosphorylation. However, a separate study using intracellular compartment-specific reporters concluded that PI3K activation is dispensable for mTORC2 activity on membranes (Ebner et al., 2017b). In this model, it is the relocalization of AKT to specific membranes through its PH domain that allows mTORC2 to gain access to S473. Although earlier studies suggested that PIP3 is exclusively localized to the plasma membrane, more recent reports have provided evidence for endomembrane pools of PIP3 and PI3,4P2 that directly contribute to AKT activation (Jethwa et al., 2015) (Figure 1A). Clearly, the molecular and spatial regulation of mTORC2 and its relationship to Akt activation remains an important and active area of investigation. Finally, the mTOR-related kinase DNA-dependent protein kinase (DNA-PK) can substitute for the activity of mTORC2 for AKT S473 phosphorylation in response to DNA damage (Bozulic et al., 2008), but the spatial nature of this regulation has not been defined.
The lifetime of active, fully phosphorylated AKT at the plasma membrane is relatively short (Calleja et al., 2007; Jethwa et al., 2015). Since phosphorylated AKT can be detected intracellularly and can phosphorylate substrates up to two hours post-stimulation (Kunkel et al., 2005), it is generally believed that AKT can dispense with PIP3-binding once phosphorylated and in an active conformation. However, a recent study found that PIP3 binding allosterically activates AKT and that dissociation from PIP3 is rate-limiting for Akt dephosphorylation and inactivation (Ebner et al., 2017a). In this provocative model, AKT-mediated substrate phosphorylation must be restricted to membranes containing PI3K lipid products, rather than through active AKT being released into cytosolic compartments. However, AKT phosphorylates a myriad of protein substrates with diverse subcellular localizations (Manning and Cantley, 2007). Some AKT targets localize to endomembrane surfaces (e.g., TSC2 (Menon et al., 2014; Roberts et al., 2004)), but others do not, such as nuclear-localized transcription factors (e.g., FoxO (Brunet et al., 1999)). This caveat notwithstanding, endomembranes contain PI3,4P2, and this mode of AKT regulation could represent a mechanism by which PI3,4P2 and PIP3 engage distinct pools of AKT (Braccini et al., 2015) (Figure 1A). This model is supported by studies on the PI3,4P2 phosphatase and tumor suppressor INPP4B,, the loss of which leads to elevated PI3,4P2 at endosomes and activation of AKT2 (Braccini et al., 2015; Fedele et al., 2010; Gewinner et al., 2009; Li Chew et al., 2015). Restricting AKT activity to membranes where PI3K lipid products are present could serve to provide specificity and fidelity in substrate selection as well as a mechanism of spatial segregation for specific signaling events.
Although a multitude of studies have demonstrated an obligate requirement for PI3K in activation of AKT, there are reports suggesting that AKT activation can proceed in a manner that is independent of PI3K and presumably phosphoinositide binding to AKT. However, whether functional AKT activation can occur in the absence of productive PI3K signaling has not been firmly established, and any such mechanism would have to be supported by clear cut evidence of PH domain release and exposure of the catalytic kinase core.
Other Regulatory Modifications of Akt
While T308 and S473 phosphorylation are considered rate-limiting and obligatory for maximal AKT activation downstream of PI3K, many other post-translational modifications have been detected and presumably serve to fine tune AKT activation, inactivation, cellular localization or, perhaps, substrate specificity (Figure 1B). Numerous phosphorylation sites on AKT have been mapped and some of these have been linked to AKT function (Guo et al., 2014). T450 in a region termed the turn-motif is constitutively phosphorylated by mTORC2, occurs co-translationally and is required for proper folding of the nascent AKT polypeptide (Facchinetti et al., 2008; Ikenoue et al., 2008).. S477 and T479 in the regulatory domain can be phosphorylated in a cell-cycle dependent-manner by the cyclin A-CDK2 complex but can also be targeted, along with S473, by mTORC2 to enhance AKT activity (Liu et al., 2014). CK2 has been shown to phosphorylate S129 and increase catalytic activity (Di Maira et al., 2005), whereas GSK-3α-mediated phosphorylation of T312 appears to attenuate AKT activity (Gulen et al., 2012). Many additional phosphorylation sites on AKT, including a number of tyrosine residues, have been mapped by phosphoproteomics, but the mechanisms that account for these modifications and their physiological significance are unknown (reviewed in (Risso et al., 2015), see http://www.phosphosite.org).
Beyond phosphorylation, various other post-translational modifications on Akt isoforms have been identified (Figure 1B). Acetylation of K14 in the AKT PH domain has been documented, and the histone deacetylase SIRT1 deacetylates this residue. K14 acetylation has been proposed to be required for AKT binding to PIP3 and therefore membrane translocation (Sundaresan et al., 2011). The K14 residue in the PH domain seems to be functionally important for regulation since it is modified by acetylation, ubiquitylation and methylation, depending on the cellular conditions used. Oxidation of Cys residues in the AKT2 linker region has been reported and may provide isoform-specific regulation, since these residues are not conserved in AKT1 or AKT3 (Wani et al., 2011). Glycosylation of AKT on T305 and T312 in the catalytic core has also been reported (Wang et al., 2012). AKT is hydroxylated by the prolyl hydroxylase EglN1/PHD2 on a number or Pro residues, particularly P125 and P313, thereby triggering interaction with pVHL (Guo et al., 2016).
AKT is ubiquitylated on multiple distinct Lys residues. With respect to degradative poly-ubiquitylation, multiple E3 ubiquitin-ligases have been shown to catalyze K48-linked ubiquitylation of AKT, thereby promoting proteasome-dependent degradation (reviewed in (Chan et al., 2014)). By contrast, distinct ubiquitin ligases that couple K63-linked ubiquitin to AKT serve to regulate AKT activation. For example, various growth factors elicit activation of the TRAF6 (Yang et al., 2009), Skp2 (Chan et al., 2012) and NEDD4-1 (Fan et al., 2013) E3 ligases that target Lys residues in the AKT PH domain, and these modifications enhance membrane localization. Termination of this signal is achieved by the deubiquitinating enzymes CYLD (Lim et al., 2012). Finally, SUMOylation of AKT at several Lys residues, including K276, mediated by the SUMO E3 ligase PIAS, has also been reported and suggested to be involved in AKT activation, (Li et al., 2013).
Signal Termination by AKT Phosphatases
In addition to signal termination by lipid phosphatases such as PTEN and INPP4B, two critical protein phosphatases function to directly inactivate AKT (Figure 1A). Protein phosphatase 2A (PP2A) dephosphorylates AKT T308, leading to kinase inactivation (Andjelkovic et al., 1996). The PP2A B55α regulatory subunit can directly bind to AKT in lymphoid cells (Kuo et al., 2008), whereas the B56β subunit directs PP2AC to AKT in adipocytes (Padmanabhan et al., 2009). The PH domain leucine-rich repeat protein phosphatases (PHLPP1 and PHLPP2) were discovered as the physiological AKT S473 phosphatases (Gao et al., 2005). PHLPP1 and PHLPP2 dephosphorylate S473 on specific AKT isoforms (Brognard et al., 2007). Since loss of PHLPP activity leads to hyperphosphorylation of AKT, it is not surprising that PHLPP1/2 expression is reduced or lost in many cancers (Chen et al., 2011).
AKT Substrates and Functions: Key Signaling Nodes
There are well over one hundred AKT substrates reported in the literature. While not all of these targets have been rigorously validated, as discussed previously (Manning and Cantley, 2007), the collective studies on AKT signaling over the years suggest a widely diverse repertoire of downstream effects in different settings stemming from its parallel regulation of multiple substrates (examples in Figure 2). In a sequence context-dependent manner, AKT directly phosphorylates protein targets of numerous functional classes, including protein and lipid kinases, transcription factors, regulators of small G proteins and vesicle trafficking, metabolic enzymes, E3 ubiquitin ligases, cell cycle regulators, and many others. AKT phosphorylates these targets on Ser/Thr residues primarily within a minimal consensus recognition motif of R-X-R-X-X-S/T-ϕ (where X is any amino acid and ϕ denotes a preference for large hydrophobic residues) to either activate or, more often, inhibit the function of the given protein. However, the mere existence of this motif, which can be found in thousands of proteins, does not render that protein a bona fide AKT substrate. Factors such as accessibility of the site on the substrate, secondary interactions with AKT, and subcellular compartmentalization are also likely to contribute. In addition, a few well-established AKT substrates are phosphorylated on modified versions of this motif, including the AMP-regulated protein kinase (AMPK) and ATP-citrate lyase (ACLY), both of which have a Pro residue at the -5 position, rather than the canonical Arg residue (Berwick et al., 2002; Horman et al., 2006). It is also now recognized that even the most well-established AKT substrates are not exclusively regulated by AKT, with context-dependent redundancy in substrate regulation being a prevalent feature of the signaling network (see below).
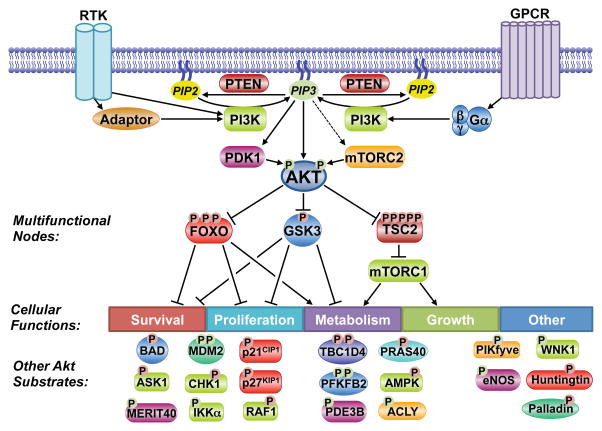
Figure 2. Substrates and functions of the Akt signaling network.
Akt phosphorylates downstream substrates involved in the regulation of diverse cellular functions, including multifunctional substrates. A partial list of known substrates is shown. P indicates phosphorylation, with red and green denoting inhibitory and activating regulation, respectively.
Key considerations for validating a candidate target of AKT as a bona fide substrate and the function of many established targets have been reviewed previously (Manning and Cantley, 2007). AKT isoforms have cell- and tissue-specific functions, but most prominently, AKT activation can promote cell survival, proliferation, growth, and changes in cellular metabolic pathways through its numerous downstream targets. While the AKT-mediated phosphorylation of many of its substrates has just one physiological consequence, AKT also controls key signaling nodes that subsequently regulate multiple cellular targets and functions (Figure 2). In the context of a network view of AKT signaling, we highlight the three best-established downstream targets of AKT, which are also key signaling nodes that integrate AKT signaling with additional cellular regulatory circuits.
Glycogen Synthase Kinase 3 (GSK3)
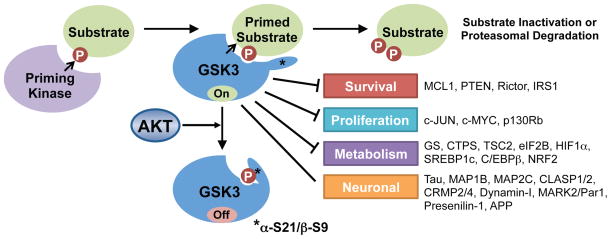
The multi-functional Ser/Thr protein kinase GSK3 was the first AKT substrate reported (Cross et al., 1995). The two isoforms, GSK3α and β, share 85% sequence homology and are functionally redundant in some contexts, but isoform-specific functions have been identified in specific tissues (Kaidanovich-Beilin and Woodgett, 2011). Through complex formation with distinct signaling components, GSK3 participates in different signaling pathways in cells, most notably the Wnt/β-catenin pathway. Importantly, GSK3 regulation in such pathways is believed to be independent of its regulation by growth factor signaling through PI3K and AKT. GSK3 is generally active in the absence of exogenous signals and is thus acutely inactivated upon stimulation of cells with growth factors. AKT exerts an inhibitory phosphorylation on an amino-terminal motif conserved in both GSK3α (S21) and GSK3β (S9). The molecular nature of this regulation stems from GSK3’s own substrate specificity, which includes a strong preference for phosphorylation of Ser/Thr residues that are four residues amino-terminal to a previously phosphorylated Ser/Thr (referred to as priming at the +4 position). The phosphate from the priming site on the substrate is recognized by a phosphate-binding pocket in the kinase domain of GSK3, which positions the target Ser/Thr residue for phosphorylation by the adjacent catalytic site of the kinase (Dajani et al., 2001; Frame et al., 2001; ter Haar et al., 2001). The AKT-mediated phosphorylation of GSK3 on the amino-terminus creates an intramolecular pseudosubstrate that occludes the phosphate-binding pocket and inhibits substrate accessibility to GSK3 (Figure 3).
Figure 3. GSK3 regulation and substrate phosphorylation.
GSK3 recognizes and phosphorylates substrates that are previously phosphorylated by a priming kinase. A partial list of known GSK3 substrates is shown. Akt’s phosphorylation of GSK3 inactivates it by blocking its access to primed substrates.
GSK3 regulates a large, functionally diverse set of direct downstream targets, most of which are inhibited or degraded upon GSK3-mediated phosphorylation (Kaidanovich-Beilin and Woodgett, 2011). As such, growth factor signaling through AKT positively regulates these targets through the inhibition of GSK3. The fact that distinct signaling pathways and protein kinases are responsible for the priming of individual GSK3 substrates adds an extra layer of complexity that allows the integrated regulation of these targets by multiple signaling inputs. The phosphorylation of some GSK3 targets involved in control of cell survival or proliferation creates a “phospho-degron” that is recognized by specific E3 ubiquitin ligases that subsequently target the substrate for proteasomal degradation. These include the prosurvival BCL-2 family member MCL-1 (Ding et al., 2007; Maurer et al., 2006; Morel et al., 2009) and the transcription factor c-Myc (Sears et al., 2000; Welcker et al., 2004), which are primed for GSK3 recognition following phosphorylation by JNK and ERK, respectively. Thus, AKT signaling can stabilize these proteins by inhibiting GSK3. GSK3 also regulates cellular metabolism, either directly, through the phosphorylation and inhibition of metabolic enzymes, such as its namesake substrate glycogen synthase (GS) (Parker et al., 1983; Rylatt et al., 1980), or indirectly, through the inhibitory regulation of transcription factors that globally regulate specific metabolic programs, including c-Myc, SREBP1c, HIF1α, and NRF2 (Kaidanovich-Beilin and Woodgett, 2011). Knock-in mice lacking the AKT phosphorylation sites on GSK3α and β (Gsk3αS21A; Gsk3βS9A) are impaired for insulin-stimulated glycogen synthesis in muscle (McManus et al., 2005). However, the degree to which the AKT-mediated regulation of GSK3 influences the functions of other GSK3 substrates is less clear.
Forkhead Box O (FoxO) Family Transcription Factors
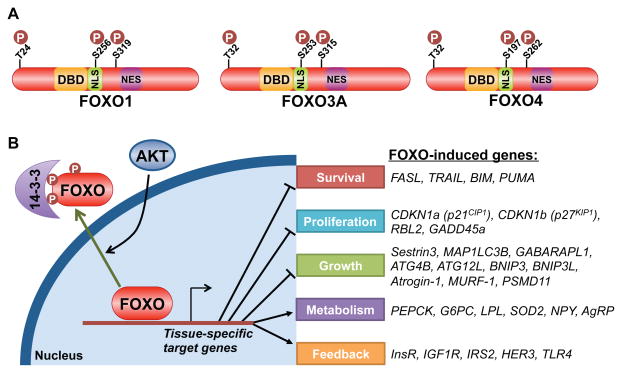
The FoxO transcription factors, comprised of FoxO1, 3, 4, and 6, control a diverse set of gene targets that are, among other responses, involved in adaptation to fasting and low insulin/IGF1 signaling (van der Vos and Coffer, 2011; Webb and Brunet, 2014). Activation of PI3K-AKT signaling leads to acute translocation of FoxO proteins out of the nucleus and attenuation of their transcriptional program (Brunet et al., 1999; Kops et al., 1999). AKT mediates this regulation through direct phosphorylation of three conserved residues on these factors (Figure 4A). Phosphorylation of the most amino-terminal site and a second site within a nuclear localization sequence (NLS) on the FoxO proteins (T24 and S256 on FoxO1) generates recognition motifs for the 14-3-3 family of phospho-binding proteins, which facilitate the export and sequestration of phosphorylated FoxO proteins in the cytosol (Figure 4B). In this manner, AKT signaling suppresses the expression of FoxO targets involved in the induction of apoptosis (e.g., BIM and PUMA), cell-cycle arrest (e.g., p21 and p27), catabolism and growth inhibition (e.g., Sestrin3, MAP1LC3B and BNIP3), and tissue-specific metabolic changes (e.g., PEPCK and G6PC) (van der Vos and Coffer, 2011; Webb and Brunet, 2014).
Figure 4. Akt-mediated regulation and transcriptional targets of FoxO family members.
A. Schematic of three FoxO family members, with the three conserved Akt phosphorylation sites denoted relative to the DNA-binding domain (DBD), nuclear localization sequence (NLS) and nuclear export sequence (NES). B. Akt-mediated phosphorylation of FoxO leads to its binding and cytosolic sequestration by 14-3-3 proteins, thereby attenuating the expression of its gene targets, a partial list of which is shown.
The highly conserved genetic relationship between AKT and FoxO family members provides definitive proof of the vital regulatory interaction between AKT signaling and suppression of the FoxO transcriptional program. This connection was first recognized in C. elegans, where dauer stage arrest induced by depletion of the two AKT isoforms (akt-1 and akt-2) was completely rescued by loss of the sole FoxO family member (daf-16) (Paradis and Ruvkun, 1998). In mice, liver-specific ablation of Akt1 and Akt2 gives rise to severe hepatic insulin resistance and hyperglycemia, phenotypes that are reversed by co-deletion of the primary isoform of FoxO in the liver, Foxo1 (Lu et al., 2012). Collectively, these studies demonstrate that the primary phenotypes caused by loss of AKT signaling in these settings are driven by sustained FoxO-mediated transcription. Thus, FoxO is the key downstream target of AKT signaling for many physiological processes (see below).
Tuberous Sclerosis Complex 2 (TSC2) and the Mechanistic Target of Rapamycin Complex 1 (mTORC1)
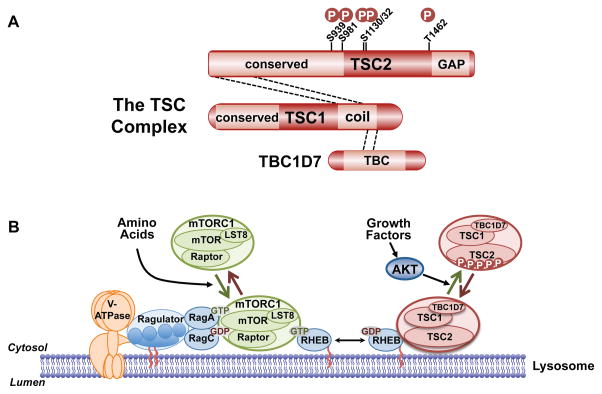
PI3K and AKT play an evolutionarily conserved role in promoting cell, tissue, and organismal growth downstream of growth factors such as IGF1. This regulation is primarily through the AKT-mediated activation of the protein kinase complex mTORC1, which stimulates the biosynthetic processes underlying cell growth (Saxton and Sabatini, 2017). The primary mechanism by which AKT activates mTORC1 is through the phosphorylation and inhibition of tuberous sclerosis complex 2 (TSC2, also known as tuberin) (Inoki et al., 2002; Manning et al., 2002; Potter et al., 2002), which functions within a protein complex also containing TSC1 and TBC1D7, collectively referred to as the TSC complex (Figure 5A). Through a carboxyl-terminal domain, TSC2 acts as a GAP specific for the Ras-related GTPase Rheb, thereby promoting the conversion of Rheb-GTP to Rheb-GDP (Saxton and Sabatini, 2017). In its GTP-bound form, Rheb is an essential activator of mTORC1. Thus, through its Rheb-GAP activity, the TSC complex is a potent inhibitor of mTORC1. The AKT-mediated phosphorylation of TSC2 relieves this inhibition to activate mTORC1.
Figure 5. Regulation of mTORC1 via the TSC complex and downstream functions of mTORC1.
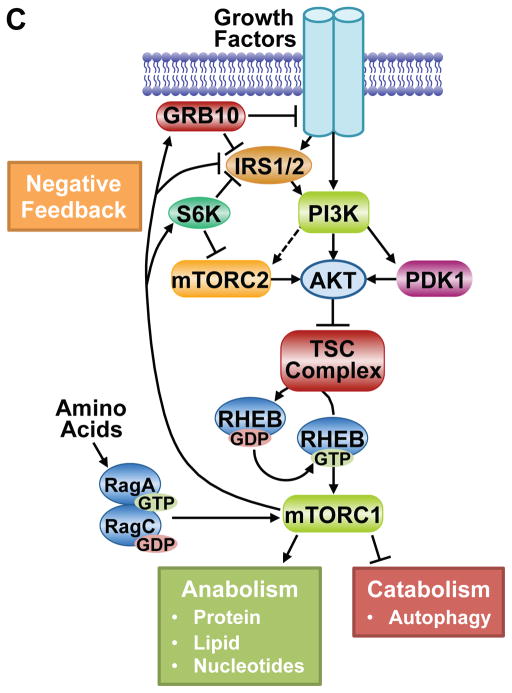
A. Schematic of the TSC complex components, their regions of association (dashed lines), and Akt phosphorylation sites on TSC2. Conserved domains of unknown function and GAP, coiled-coil, and TBC domains of the components are shown. B. Model of signal integration by growth factors and amino acids for regulation of mTORC1. The Rag heterodimer interacts with the Ragulator and V-ATPase at the lysosomal surface, and amino acids promote mTORC1 binding to this complex. The TSC complex maintains Rheb in the GDP-bound state. Growth factor-stimulated Akt phosphorylates TSC2, resulting in dissociation from the lysosomal surface, allowing Rheb to become GTP loaded and activate mTORC1. C. The PI3K-mTOR signaling pathway, depicting downstream functions and feedback regulation.
The activation state of mTORC1 is controlled in an integrated manner by largely independent signals impinging on Rheb and a second class of small GTPases, called the Rags (Saxton and Sabatini, 2017). A heterodimer of Rag isoforms localizes to the cytoplasmic face of the lysosome through an interaction with a protein complex referred to as the Ragulator, which itself interacts with the lysosomal V-ATPase. Through a variety of sensing mechanisms, the guanine nucleotide-binding state of the Rag proteins is altered by amino acid availability in a manner that influences its ability to interact with mTORC1. In the presence of amino acids, the Rag proteins recruit mTORC1 to the lysosomal surface where a sub-population of Rheb resides. In the absence of growth factors, the TSC complex associates with Rheb on the lysosome and maintains it in the GDP-bound state unable to activate mTORC1 (Menon et al., 2014). Growth factor stimulation causes immediate release of the TSC complex from Rheb at this location in a manner that is dependent on AKT and its five-phosphorylation sites on TSC2 (Figure 5A,B). Release of the TSC complex allows Rheb to become GTP loaded and locally activate mTORC1 recruited by the Rag proteins. This regulatory circuit serves as a spatial integrator of distinct signals, assuring that mTORC1 is only maximally activated when sufficient intracellular amino acids are sensed upstream of the Rag proteins and an exogenous signal from growth factors is propagated through AKT and the TSC complex.
It is also worth noting that AKT has been suggested to directly phosphorylate mTOR on S2448 (Nave et al., 1999; Sekulic et al., 2000), and this phosphorylation is frequently used as a marker of mTORC1 activation. However, subsequent studies demonstrated that S2448 is phosphorylated by S6K downstream of mTORC1 (Chiang and Abraham, 2005; Holz and Blenis, 2005), and this phosphorylation occurs on mTOR within both mTORC1 and mTORC2 (Rosner et al., 2010). Furthermore, mutation of S2448 does not appear to influence mTOR function, and its functional significance remains unknown. While mTOR phosphorylation on S2448 can correlate with mTORC1 signaling, these factors invalidate this phosphorylation as a specific marker for mTORC1 activity.
Proline-rich AKT substrate of 40 kDa (PRAS40; also known as AKT1S1) is another AKT target involved in mTORC1 regulation (Sancak et al., 2007; Vander Haar et al., 2007). PRAS40 is a protein of unknown function that is a non-essential component of mTORC1. AKT phosphorylates PRAS40 on T246 (Kovacina et al., 2003), and this substrate has become a reliable readout of AKT activity in cells and tissues. PRAS40 has inhibitory activity toward mTORC1 that is attenuated upon T246 phosphorylation (Sancak et al., 2007; Vander Haar et al., 2007). However, the role of PRAS40 phosphorylation in relaying the signal from AKT to mTORC1 activation remains unclear, with conflicting findings on whether loss of PRAS40 leads to growth factor-independent activation of mTORC1 (Sancak et al., 2007; Vander Haar et al., 2007). This is in contrast to components of the TSC complex, the loss of which lead to full AKT-independent activation of mTORC1, despite PRAS40 being in the dephosphorylated state in such settings (Sancak et al., 2007). Furthermore, insulin fails to activate mTORC1 signaling in cells lacking the AKT phosphorylation sites on TSC2, despite normal induction of PRAS40 phosphorylation (Menon et al., 2014). Thus, the TSC-Rheb circuit appears to be dominant over PRAS40 for mTORC1 regulation by Akt, at least in some settings. Interestingly, PRAS40 is also a substrate of mTORC1, and independent studies suggest that it might exert its negative regulatory effect through a substrate competition mechanism (Fonseca et al., 2007; Oshiro et al., 2007).
Through its regulation of the TSC-Rheb-mTORC1 circuit, AKT serves to link growth factor signals to a major signaling node controlling the metabolic changes that underlie cell growth (Saxton and Sabatini, 2017). mTORC1 activation serves to promote a variety of anabolic processes, such as protein, lipid, and nucleotide synthesis, while inhibiting the catabolic process of autophagy. In addition, mTORC1 is both a key downstream effector of PI3K-AKT signaling and a pathway inhibitor that exerts potent negative feedback effects on AKT activation by RTKs (Figure 5C, see below).
Central Features of the AKT Signaling Network
Studies of the AKT signaling network using increasingly sophisticated genetic, pharmacological, cell biological, and biochemical approaches have revealed the complex wiring of this branching and looping network and its intimate regulatory links to other cellular signaling networks. These features are key to understanding signal propagation within the network, context-dependent cellular responses, and the inherent challenges of targeting the network in human diseases.
Feedback mechanisms
Like all signaling pathways, the PI3K-AKT pathway is subjected to negative feedback regulation to assure that stimulatory signals are sensed and relayed in a transient manner. The shear number of distinct negative feedback mechanisms that have been identified for PI3K-AKT signaling underscore the importance of switch-like behavior for this pathway and that turning AKT signaling back off is as central to proper pathway function as turning it on. Thus, several downstream effectors of AKT signaling also exert negative regulatory inputs into AKT activation, thereby acting as rheostats that both perceive signaling through the pathway and regulate pathway activity.
Among the downstream targets of AKT signaling, mTORC1 appears to play a particularly important role in acute feedback inhibition of AKT through a variety of mechanisms (Figure 5C). It has been recognized for many years that short-term treatment with the mTORC1 inhibitor rapamycin enhances the responsiveness of AKT to RTK signaling, most notably to insulin and IGF1 (Manning, 2004). Much of this regulation has been attributed to mTORC1-dependent degradation of the insulin receptor substrates (IRS) IRS1 and IRS2, which serve as scaffolding adaptors linking the insulin and IGF1 receptors to PI3K-AKT activation. mTORC1 activation promotes IRS1/2 degradation through multiple serine phosphorylation events on these proteins mediated by mTORC1, S6K, or other unknown downstream protein kinases, thereby greatly dampening PI3K activation (Harrington et al., 2004; Shah and Hunter, 2006; Tzatsos and Kandror, 2006). Thus, mTORC1 inhibition increases IRS1/2 stability and allows more robust and sustained insulin/IGF1 signaling to PI3K and AKT. Another adaptor protein, growth factor receptor bound protein 10 (GRB10) has also been found to be a direct target of mTORC1 that negatively regulates RTK signaling (Hsu et al., 2011; Yu et al., 2011). The mTORC1-mediated phosphorylation of GRB10 stabilizes the protein and enhances its ability to attenuate signaling from the insulin/IGF1 receptors and IRS proteins, thus blocking PI3K-AKT activation. There are also points of cross-talk between mTORC1 and mTORC2 that influence the full activation of AKT. S6K1-mediated phosphorylation of the mTORC2 components Rictor (T1135) and Sin1 (T86, T398) decreases the mTORC2-dependent phosphorylation of AKT-S473 (Dibble et al., 2009; Julien et al., 2010; Liu et al., 2013b). Curiously, this regulation of mTORC2 does not appear to affect its activity toward other substrates, including AKT T450. In addition to insulin and IGF1 signaling, mTOR inhibitors also enhance the activation of other RTKs upstream of PI3K and AKT. This includes members of the EGF Receptor family (EGFR, ErbB2/Her2, Erb3/Her3, and Erb4/Her4), which are acutely activated upon mTOR inhibition, without immediate effects on receptor levels (Rodrik-Outmezguine et al., 2011). While the post-translational mechanisms underlying this regulation are not currently known, the effects are most prominently seen with mTOR kinase inhibitors, rather than the allosteric inhibitor rapamycin. As this class of compounds inhibits mTORC1 and mTORC2 equally, some of this feedback regulation could be mediated by mTORC2 signaling.
Other downstream branches of AKT signaling also exert feedback effects on AKT activation. In cancer research, the use of pharmacological compounds inhibiting PI3K isoforms, AKT, and/or mTOR have uncovered robust feedback regulation of RTK protein levels by the FoxO transcription factors (Chandarlapaty et al., 2011; Muranen et al., 2012; Zhuang et al., 2013). Prolonged inhibition of AKT signaling was found to enhance expression of the insulin and IGF1 receptors and HER3 through FoxO-dependent transcriptional induction, thereby rendering cells more prone to subsequent growth factor-mediated activation of PI3K-AKT signaling (Chandarlapaty et al., 2011).
Cross-talk with other major signaling pathways
Points of cross-regulation between the PI3K-AKT pathway and other major signaling pathways in are common. These integration points act to either directly regulate pathway components or converge on the regulation of downstream targets controlled by other pathways. While it is plausible that PI3K-AKT signaling intersects with all major signal transduction pathways in cells at one or more points, the diversity and complexity of cross-talk mechanisms are exemplified by those with the RAS-ERK and AMPK pathways (Figure 6A).
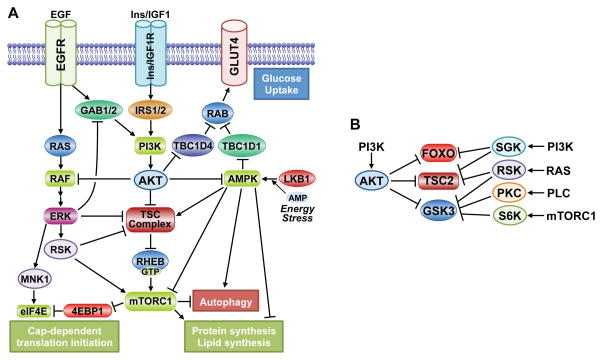
Figure 6. Signaling crosstalk and redundancy in the AKT network.
A. Several points of cross-regulation exist between the PI3K-Akt pathway and both the RAS-ERK and AMPK pathways, leading to both reciprocal pathway regulation and convergent regulation of downstream processes. B. Various AGC family kinases can redundantly phosphorylate overlapping sites on key downstream substrates of AKT, thereby altering the regulatory input into these targets.
There is a particularly intimate relationship between the PI3K-AKT and RAS-ERK pathways, where inhibitors of one pathway will often activate the other (Mendoza et al., 2011). One of the first substrates of AKT identified was the protein kinase c-Raf (or Raf1) (Rommel et al., 1999; Zimmermann and Moelling, 1999), which is activated by RAS and initiates a kinase cascade culminating in ERK activation. AKT phosphorylates c-RAF on S259 and an equivalent site on B-RAF (S364), which inhibits RAF activation or downstream signaling by promoting binding to 14-3-3 proteins, particularly when other regulatory sites on these proteins are also phosphorylated by PKA or AMPK (Dumaz and Marais, 2003; Guan et al., 2000; Shen et al., 2013; Zimmermann and Moelling, 1999). In addition to this point of direct crosstalk, many of the feedback mechanisms inherent to the AKT signaling network (discussed above) exert their effects through RTK signaling and scaffolding adaptors, which activate multiple mitogenic signaling pathways, including the RAS-ERK and PLCγ-PKC pathways. ERK activation can also suppress RTK-mediated induction of the PI3K-AKT pathway via phosphorylation of the scaffolding adaptors GAB1 and GAB2 (Mendoza et al., 2011). An unbiased kinome-wide siRNA screen revealed that attenuation of ERK signaling results in a general enhancement of AKT activation, indicating that there are likely to be additional inhibitory mechanisms between these pathways (Lu et al., 2011). The PI3K-AKT and RAS-ERK pathways also converge to regulate many of the same downstream effectors (e.g., TSC2, FOXO, GSK3) and cellular processes (Mendoza et al., 2011)..One clear example of this convergent regulation is in the control of cap-dependent translation. This occurs, in part, through mTORC1 regulation via ERK and RSK-mediated multi-site phosphorylation of both TSC2 and the mTORC1 component Raptor (Ma et al., 2005; Romeo et al., 2012; Roux et al., 2004). Like RSK, the MAPK-interacting kinases (MNK1 and MNK2) are activated by ERK signaling. MNK1/2 phosphorylate the 5′-mRNA cap-binding protein eIF4E, which appears to promote its ability to initiate cap-dependent translation (Siddiqui and Sonenberg, 2015). This regulatory input from ERK signaling occurs downstream of the mTORC1-mediated phosphorylation of the 4E-BP proteins, which stimulates their release from inhibitory binding of eIF4E. Thus, while the PI3K-AKT and RAS-ERK pathways can cross-inhibit one another, they can also act in a cooperative manner to robustly regulate key cellular processes involved in cell growth and proliferation.
AKT signaling has several points of cross-regulation with AMPK, a master cellular energy sensor that facilitates adaptation to ATP depletion (Figure 6A). As AKT signaling promotes glucose uptake and glycolysis, it stimulates ATP production and thereby indirectly prevents AMPK activation. However, AKT has been found to directly phosphorylate a carboxyl-terminal residue on AMPK (AMPKα1-S487), which hinders the activating phosphorylation of AMPK by LKB1 (Hawley et al., 2014; Horman et al., 2006). This regulation is interesting in light of the fact that AKT and AMPK have both redundant and counteracting functions in the regulation of cellular metabolism and growth. AMPK and AKT can both induce glucose uptake in metabolic tissues, such as skeletal muscle, through their respective phosphorylation and inhibition of the RAB-GAPs TBC1D1 and TBC1D4/AS160, resulting in RAB-mediated translocation of GLUT4 to the plasma membrane (Chavez et al., 2008; Eguez et al., 2005; Sano et al., 2003). While both kinases stimulate glucose uptake, they do so in response to distinct cues: AKT in response to insulin and AMPK in response to ATP depletion, such as occurs during muscle contraction. On the other hand, AKT signaling, largely through its activation of mTORC1, stimulates ATP-consuming anabolic processes, whereas AMPK activation blocks anabolic metabolism in favor of ATP-producing catabolic processes (Dibble and Manning, 2013; Mihaylova and Shaw, 2011). Through a variety of shared and distinct downstream targets, AKT and AMPK have opposing effects on mTORC1 signaling, protein, lipid, and glycogen synthesis, and the induction of autophagy. Thus, cross-talk between these ubiquitous pathways is a key control point for adaptive switching between catabolic and anabolic states in cells and tissues.
Modularity and redundancy
It is now well recognized that signaling is modular in nature and that substrates of AKT in one cell- and stimulus-specific context can be regulated by protein kinases related to AKT in another setting. This redundant regulation of substrates allows for key regulatory phosphorylation sites on a given protein to be responsive to a more diverse array of upstream inputs, which differentially regulate members of the AGC kinase family. This redundancy comes primarily through growth factor-regulated kinases that are stimulated in a manner that is parallel to (e.g., SGK, RSK, PKC) or downstream of (e.g., S6K) AKT signaling. While alternative regulation through the same sequence motif might be the case for all AKT substrates, the three signaling nodes detailed above are excellent examples of this feature of the AKT signaling network (Figure 6B). The N-terminal inhibitory site on GSK3 isoforms (GSK3α-S21, GSK3β-S9) can be phosphorylated by RSK or S6K, downstream of ERK or mTORC1 signaling, respectively (Kaidanovich-Beilin and Woodgett, 2011). The sequestration of FOXO3a in the cytosol can also be promoted by SGK-mediated phosphorylation of T32 (Brunet et al., 2001). Growth signals can activate mTORC1 signaling independent of AKT activity, including through the ERK-RSK pathway in a manner believed to involve RSK-mediated phosphorylation of at least two of the AKT sites on TSC2 (S939 and T1462), together with additional regulatory sites (Ma et al., 2005; Roux et al., 2004). Interestingly, SGK1 or SGK3 can also replace AKT for phosphorylation of TSC2 and induction of mTORC1 signaling, which was revealed in cancer cells upon prolonged exposure to pharmacological inhibitors of PI3K or AKT and provides a mechanism of cellular resistance to such inhibitors (Bago et al., 2016; Castel et al., 2016). In these examples, it appears that AKT is the dominant regulator, but this is unlikely to be the case for all AKT substrates and settings.
Selective signaling to downstream branches
Given the diverse functions of AKT substrates, the ability to regulate subsets of downstream branches under different conditions would seem to be an essential feature. However, outside of differential expression of AKT substrates in a given biological context, this is a poorly understood area. A few parameters are likely to contribute to selective signaling to specific targets. The strength or duration of AKT activation might influence which substrates get phosphorylated. One indication of this property is in settings where mTORC2 is genetically or pharmacologically inhibited, thereby blocking AKT S473 phosphorylation, an event required to stabilize T308 phosphorylation for maximal AKT activation (Alessi et al., 1996a). Upon loss of mTORC2, unknown mechanisms are induced to stabilize T308 phosphorylation resulting in sustained AKT signaling (Guertin et al., 2006; Jacinto et al., 2006). The one notable exception is the AKT-mediated phosphorylation of FOXO isoforms, which is attenuated in the absence of mTORC2, despite the T308 compensation. One interpretation of these observations is that phosphorylation of FOXO, and perhaps other substrates, requires a higher threshold of AKT activity relative to other AKT substrates. Another likely mechanism directing differential substrate phosphorylation could be through spatial effects influencing AKT trafficking to its substrates or vice versa, which is an important future area of research.
For many AKT substrates, properties intrinsic to the substrate itself and its control by other regulatory inputs will direct specific downstream outputs. Through specific downstream targets, AKT signaling often intersects with cellular stress response and nutrient sensing pathways. In such cases, signals from intracellular stress or nutrient depletion are generally dominant over exogenous signals from growth factors and cytokines propagated by the PI3K-AKT pathway. For instance, opposing the AKT-mediated sequestration of FoxO family members in the cytosol (Figure 4B) are a number of stress and nutrient responsive pathways (e.g., p38, JNK, AMPK, SIRT1) that also directly modify FoxO transcription factors and promote their nuclear translocation and activation of gene targets, many of which are involved in adaptions to cellular stress (Webb and Brunet, 2014). TSC2 and mTORC1 are also subjected to opposing regulation that senses the depletion of intracellular ATP or nutrients and dominantly inhibits the ability of AKT signaling to activate mTORC1 (Saxton and Sabatini, 2017). Integration of these distinct signals assures that, regardless of the activation state of AKT, mTORC1 will not be activated to promote energy- and nutrient-consuming anabolic processes when these resources are limited. This manner of signal integration at the level of specific substrates, especially those that also play a role in feedback regulation of AKT, is likely key to matching the appropriate cellular response to the dynamically changing cellular state. For instance, growth factor signals to PI3K and AKT received in a cell that is nutrient deprived will result in enhanced survival signaling from AKT without the promotion of cell growth due to attenuation of mTORC1 signaling and its feedback effects on AKT. It seems likely that other such mechanisms exist that are inherent to specific AKT substrates that direct AKT action toward certain targets and away from others, thereby tailoring the signal to compliment the cellular state.
AKT Isoform-specific Functions and Substrates
The majority of AKT substrates are phosphorylated and functionally regulated by all three AKT isoforms. However, a number of substrates have been identified that are uniquely targeted by AKT1, AKT2 or AKT3. This substrate-selectivity likely accounts for some isoform-specific phenotypes observed in genetic studies, suggesting unique properties inherent to AKT isoforms or their regulation that account for such specificity. For example, independent breast cancer studies have shown that AKT1 suppresses migration and metastasis, whereas AKT2 promotes metastatic dissemination (Dillon et al., 2009; Hutchinson et al., 2004; Irie et al., 2005; Maroulakou et al., 2007). However, such opposing and non-redundant functions of AKT isoforms are likely to be context-dependent. For example, in prostate cancer cells with PTEN inactivation, AKT2 is exclusively required for cell-autonomous tumor maintenance (Chin et al., 2014), whereas in Pten+/− mice, the AKT1 isoform appears to suppresses prostate tumor development (Chen et al., 2006), while ablation of AKT2 has little impact (Xu et al., 2012). Global phospho-proteomic screening approaches have identified hundreds of novel phosphorylation sites that conform to the AKT consensus motif and have also provided new insights into potential AKT isoform-specific functions and substrates, many that await further validation (Lee et al., 2014; Moniz et al., 2017; Sanidas et al., 2014). Mechanisms that likely contribute to Akt isoform-specificity for downstream substrates include molecular features of individual isoforms, discrete subcellular localization, and relative expression levels in a given setting. Selective activation of specific AKT isoforms in certain settings might also contribute (Kim et al., 2011). Finally, the presence of oncogenic, activating somatic mutations in either AKT1, AKT2 or AKT3 will likely influence the pattern of specific substrate phosphorylation and signaling output (Lien et al., 2016). While the mechanistic aspects remain an interesting area of future investigation, clear evidence for AKT isoform-specific or -selective substrate phosphorylation has accumulated.
Physiology and Pathology of AKT Signaling
Given the broad spectrum of AKT substrates and functions discovered through genetic, biochemical, and cell biological studies, it is not surprising that AKT plays a central but diverse role in the response of various cell-types and tissues to hormones, growth factors, cytokines, and neurotrophic factors, among other stimuli. Mouse genetics indicate that AKT1, AKT2, and AKT3 have both redundant and specific functions in different tissues (Dummler and Hemmings, 2007). All tissues appear to express one or more AKT isoform, with AKT1 being the most widely expressed, AKT2 being enriched in insulin-responsive metabolic tissues, and AKT3 in the brain. Consistent with this tissue distribution, Akt1−/− mice display growth retardation and perinatal lethality (Chen et al., 2001; Cho et al., 2001b), Akt2−/− mice develop a diabetes-like syndrome (Cho et al., 2001a), Akt3−/− mice display decreased brain size (Easton et al., 2005; Tschopp et al., 2005), and compound knockouts develop a range of severe developmental abnormalities (Dummler and Hemmings, 2007). Here, we provide an overview of our current knowledge of AKT function in various organ systems and effects of AKT dysfunction in specific pathological states (Table 1).
Table 1.
Physiological functions of Akt and pathological implications from Akt dysregulation in different cell types and tissues.
| Cell Type/Tissue | Functions | Key targets | Potential Disease Implications* |
|---|---|---|---|
| Most cell types (epithelial or mesenchymal) | Survival, growth, migration, and invasion | Numerous targets | Overgrowth syndromes; adenoma; hamartoma; carcinoma; sarcoma |
| Endothelial cells and blood vessels | Survival and Angiogenesis | eNOS | Vascular anomalies; hemangioma; atherosclerosis. |
| Innate immune cells | Macrophage metabolism and polarization; Decreased inflammatory signals from dendritic cells; Neutrophil chemotaxis and respiratory burst | TSC-mTORC1, ACLY, FoxO1, GSK3 | Chronic inflammatory diseases; sepsis |
| Lymphocytes | Thymocyte survival; Th cell activation; Treg suppression; CTL activation?; memory T cell suppression; B cell maturation, survival, and Ig class switching; | TSC-mTORC1, FoxO1 | Autoimmune diseases |
| Neurons and CNS | Neuronal survival; polarity and axon specification; synaptic plasticity | TSC-mTORC1, GSK3, NFkB | Megalencephaly; epilepsy; autism; cognitive deficits; mood disorders; neurodegenerative diseases |
| Pancreas | Islet growth and insulin production | TSC-mTORC1 | ? |
| Liver | Suppress gluconeogenesis; lipid synthesis | FoxO, TSC-mTORC1 | Insulin resistance, type-2 diabetes, hepatosteatosis |
| Adipose | Glucose uptake; suppression of lipolysis | TBC1D4, PDE3B, TSC-mTORC1, FoxO | Insulin resistance; type-2 diabetes; lipodystrophy |
| Muscle | Glucose uptake; glycogen synthesis; protein synthesis; suppression of protein degradation | TBC1D4, GSK3, TSC-mTORC1, FoxO | Insulin resistance; type-2 diabetes; sarcopenia |
| Hypothalamus | Suppress feeding | FoxO, TSC-mTORC1 | Hyperphagia, obesity? |
| Heart | Physiological hypertrophy | TSC-mTORC1 | Pathological hypertrophy? |
Potential manifestations from aberrant activation or diminished Akt signaling in the given setting.
Overgrowth Syndromes
Multiple distinct overgrowth syndromes in humans have been associated with one or more genetic defects in the PI3K-AKT signaling network. The genetic lesions that are causally implicated in these syndromes all elicit a constitutive increase in AKT-TSC-mTORC1 signaling and include amplification or somatic mosaic mutations in PIK3CA, all three AKT isoforms, or mTOR, or germline inactivating mutations in PTEN or components of the TSC complex (Keppler-Noreuil et al., 2014). The clinical manifestations of these overgrowth disorders range from small skin lesions to extreme overgrowth in multiple tissues and increased tumor susceptibility. For instance, PIK3CA-Related Overgrowth Spectrum (PROS) is a congenital or early childhood overgrowth syndrome, where patients are at risk for development of malignancies, consistent with activating PIK3CA hotspot mutations at E545K and H1047R being frequent events in sporadic carcinomas (Samuels et al., 2004). Proteus syndrome (PS) is a distinct progressive overgrowth abnormality in which affected individuals have a propensity to develop a range of benign tumors. The genetic basis for PS is a gain of function mutation in AKT1 at E17K in the PH domain (Lindhurst et al., 2011). Affected individuals develop progressive overgrowth of most organs and tissues, in addition to severe vascular malformations. Patients with activating mutations in AKT2 at E17K do not develop PS, but instead display severe insulin-independent hypoglycemia, asymmetric overgrowth and obesity (Hussain et al., 2011). Patients with the equivalent AKT3 E17K mutation develop brain overgrowth and megalencephaly (Poduri et al., 2012). The downstream TSC-mTORC1 branch appears to be particularly important in driving overgrowth in these syndromes associated with activation of the PI3K-Akt pathway, with patients often showing improvement upon treatment with mTORC1 inhibitors.
Cancer
Multiple genetic lesions confer hyperactivation of AKT in human solid tumors and hematological malignancies. These include amplification or recurring oncogenic somatic mutations in EGFR, HER2, or other RTKs, PDK1, and PIK3CA. Similarly, inactivating mutations or loss-of-heterozygosity in tumor suppressor genes such as PTEN, INPP4B and PHLPP also lead to hyperactivation of AKT (reviewed in (Mayer and Arteaga, 2016)). However, AKT activation is not always concordant with PIK3CA mutation (Vasudevan et al., 2009), and other PI3K effectors are also likely to contribute to malignancy (Lien et al., 2017). While AKT is most frequently activated in human cancers by mutations affecting upstream regulators, the three AKT isoforms themselves are bona fide oncogenes.
Albeit at lower frequencies than core Akt regulators, such as PIK3CA, amplification or activating mutations in the AKT genes have been identified in multiple solid tumors (Altomare and Testa, 2005). The first-described somatic activating mutation of any AKT gene was the E17K mutation in the PH domain of AKT1 in breast cancer patients (Carpten et al., 2007). AKT1 E17K is a low frequency (1.5% −9%) recurring mutation in breast cancers (Rudolph et al., 2016). This mutation confers constitutive kinase activation due to a charge switch at Glu17 in the phosphoinositide-binding pocket of the PH domain that allows constitutive membrane localization of Akt (Carpten et al., 2007; Landgraf et al., 2008). However, AKT1 E17K is incapable of promoting tumorigenesis in the absence of other driver mutations (Lauring et al., 2010; Mancini et al., 2016). Like PIK3CA mutations, AKT E17K mutations are found in luminal, estrogen receptor (ER)-positive breast cancers, suggesting that PI3K-AKT pathway activation confers a selective advantage in this lineage (Salhia et al., 2012). It is also noteworthy that in addition to E17K, other activating mutations in AKT1 have been described in human cancers (Yi and Lauring, 2016). While the equivalent AKT2 E17K mutation has been identified in one breast cancer patient though a large scale sequencing effort (Stephens et al., 2012), AKT2 E17K does not appear to be a recurring event in breast cancer. By contrast, the equivalent AKT3 E17K has been identified as a recurring mutation, particularly in melanoma (Davies et al., 2008). In all of these cases, the array of downstream pro-survival and pro-growth effects of AKT signaling, including changes to cellular metabolism, are likely to contribute to its role in tumor growth and progression (Manning and Cantley, 2007).
Endothelial Cell Function, Angiogenesis, and Vascular Biology
The AKT signaling network plays a major functional role in cells that are often dysregulated in vascular abnormalities, including endothelial cells (ECs), vascular smooth muscle cells (VSMCs) and macrophages. AKT1 appears to be the major isoform that contributes to normal EC physiological functions, and activation of AKT1 by vascular endothelial growth factor (VEGF) stimulates EC proliferation, migration and survival (Chen et al., 2005). This is consistent with the finding that endothelial nitric oxide (NO) synthase (eNOS), which controls vascular tone, is an AKT1-specific substrate in endothelial cells (Lee et al., 2014). Thus, loss of AKT1, but not AKT2, in mouse ECs results in reduced NO release and impaired angiogenesis (Ackah et al., 2005). AKT1 has also been implicated in vascular remodeling. Expression of an activated AKT1 allele in ECs blocks the formation of neointimal lesions following arterial injury (Mukai et al., 2006) and induces pathological angiogenesis concomitant with increased vascular permeability (Phung et al., 2006).
AKT1 is the predominant isoform expressed in VSMCs and promotes VSMC cell proliferation, survival, and migration, with loss of Akt1 enhancing the development and severity of atherosclerosis in mouse models (Fernandez-Hernando et al., 2007; Fernandez-Hernando et al., 2009). Atherosclerosis is a widespread cardiovascular disorder that is dominated by the formation of localized vascular plaques comprised of lipid-laden macrophages, referred to as foam cells. AKT isoforms have prominent functions in macrophages related to innate immunity (see below), but they have also been found to influence foam cell formation. Mice lacking AKT2, but not AKT1, in hematopoietic cells display decreased atherosclerotic lesions (Babaev et al., 2014). By contrast, AKT3 knockout mice have enhanced development of atherosclerosis due to an increase in formation of foam cells, which appears to be an intrinsic property of Akt3-deficient macrophages resulting from enhanced uptake of lipoproteins (Ding et al., 2012). Taken together with the critical functions of Akt1 in ECs and VSMCs, the three Akt isoforms play a complex, yet critical, role in vascular health and disease.
Insulin response and systemic metabolism
In metazoans, AKT signaling has co-evolved with insulin and IGF1 signaling, with PI3K-dependent AKT activation being the primary effector pathway for these hormones in model organisms including worms, flies, and rodents. In mammals, the insulin-mediated activation of AKT is central to proper glucose disposal and other metabolic adaptations after feeding through differential actions in metabolic tissues. Evidence suggests that AKT1 activation in pancreatic islet cells increases β-cell mass and insulin production (Buzzi et al., 2010). The TSC-Rheb-mTORC1 circuit is the likely downstream effector for this function as islet cell-specific activation of mTORC1 results in a robust expansion of islets, hyperinsulinemia, and improved glucose tolerance (Howell and Manning, 2011). Post-prandial insulin secretion from the pancreas initiates the systemic metabolic response, which requires distinct functions of AKT in the major insulin-responsive tissues, including the liver, muscle, and fat. With some partial redundancy from AKT1, AKT2 appears to be the dominant functional isoform in these tissues for the response to insulin. As such, Akt2 knockout mice exhibit insulin resistance and glucose intolerance and have type-2 diabetes (Cho et al., 2001a; Garofalo et al., 2003). Interestingly, rare dominant negative mutations in AKT2 have been found to underlie the genetic development of severe diabetes in humans (George et al., 2004). Importantly, AKT signaling is attenuated in metabolic tissues in the insulin resistant state that underlies type-2 diabetes. Insulin resistance occurs, at least in part, through chronic activation of feedback and cross-talk mechanisms inherent to the PI3K-AKT signaling network (see above). The decreased ability to activate AKT disrupts the key metabolic actions of insulin.
Insulin signaling to AKT in the liver is essential for suppression of hepatic glucose production and for the stimulation of lipid synthesis (Dummler et al., 2006; Leavens et al., 2009; Ono et al., 2003). Insulin suppresses gluconeogenesis in hepatocytes via the AKT-mediated phosphorylation and inhibition of FoxO1, which in the fasted state resides in the nucleus and induces expression of the key gluconeogenic enzymes phosphoenolpyruvate carboxykinase (PEPCK) and glucose 6-phosphatase (G6Pase) (Matsumoto et al., 2007; Puigserver et al., 2003). Strikingly, the uncontrolled hepatic glucose production and insulin resistance in mice with liver-specific loss of Akt1 and Akt2 is reversed by co-deletion of Foxo1 (Lu et al., 2012), providing definitive genetic evidence that inhibition of FoxO1 is an essential function of AKT in the liver. A particularly surprising implication of this finding is that in the absence of FoxO1, hepatic AKT signaling appears to be dispensable for the control of glucose metabolism by fasting and feeding. AKT signaling can also decrease hepatic glucose release by channeling glucose 6-phosphate toward glycogen synthesis. However, the insulin-stimulated, AKT2-dependent promotion of glycogen synthesis in the liver has been found to occur through an unknown mechanism that is independent of GSK3 phosphorylation (Wan et al., 2013). Insulin and AKT signaling in the liver also enhances de novo lipid synthesis, at least in part, through activation of the SREBP1c transcription factor (Leavens et al., 2009). Downstream of AKT, mTORC1 plays a major role in the activation of SREBP isoforms to promote de novo lipid synthesis (Duvel et al., 2010; Porstmann et al., 2008). Induction of SREBP1c and lipid synthesis in the liver has been found to depend on both AKT2 and mTORC1, with FoxO1 suppression also contributing in this setting (Wang et al., 2015).
Another major effect of insulin on systemic metabolism is through the suppression of lipolysis and fatty acid release from adipose tissue, which likewise influences systemic insulin responsiveness (Czech et al., 2013). The inhibition of lipolysis is achieved, at least in part, through the suppression of catecholamine signaling to PKA, which induces lipolysis in the fasted state through the activation of acyl-glycerol lipases, including adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL). PKA signaling promotes ATGL and HSL access to the triglyceride-rich lipid droplet of adipocytes, and insulin signaling attenuates this effect.
However, an essential role for AKT in the ability of insulin to suppress lipolysis has not been fully established. Adipose-specific knockout of the mTORC2 component Rictor, which leads to attenuation of AKT signaling, results in a failure of insulin to block lipolysis (Kumar et al., 2010), but mice lacking AKT2 exhibit only a partial defect in insulin-mediated suppression of lipolysis (Koren et al., 2015). AKT has been found to directly phosphorylate phosphodiesterase 3B (PDE3B) on S273, which appears to enhance its ability to hydrolyze cAMP, thereby blocking PKA activation (Kitamura et al., 1999). Consistent with this being a key point of regulation, insulin fails to inhibit lipolysis in PDE3B knockout mice (Choi et al., 2006). However, the ability of insulin to block lipolysis is restored to PDE3B-null adipocytes with re-expression of either wild-type PDE3B or a phosphorylation-site mutant (S273A), suggesting that other regulatory sites on PDE3B or parallel mechanism are also required for this regulation (DiPilato et al., 2015). Rapamycin-mediated inhibition of mTORC1, downstream of Akt, leads to an increase in plasma lipids in humans (Morrisett et al., 2002), and S6K1 knockout mice show a similar phenotype with a corresponding decrease in adiposity (Um et al., 2004). These effects have been attributed to a role for mTORC1 in suppressing lipolysis, as mTORC1 inhibition induces lipolysis in cultured adipocytes, which correlates with increased ATGL expression and PKA-dependent activating phosphorylation of HSL (Chakrabarti et al., 2010; Soliman et al., 2010). Recent studies have also found that the process of autophagy, which mTORC1 acutely inhibits, is intimately linked to the induction of lipolysis (Cingolani and Czaja, 2016).
The clearance of circulating glucose via uptake into adipose tissue and, especially, skeletal muscle is induced by insulin through the activation of AKT, predominantly AKT2 (Cho et al., 2001a; Garofalo et al., 2003). Insulin-stimulated PI3K-AKT signaling leads to rapid translocation of the primary glucose transporter in these tissues, GLUT4, to the plasma membrane (Leto and Saltiel, 2012). AKT2 has been found to associate with GLUT4-containing vesicles and promotes their trafficking and plasma membrane fusion in response to insulin (Calera et al., 1998; Ng et al., 2008). AKT appears to phosphorylate several direct downstream targets that regulate exocytosis of these vesicles (Leto and Saltiel, 2012). The best characterized of these is the Rab-GAP TBC1D4 (or AS160), which acts to retain Glut4-containing vesicles in the cytosol, a function that is disrupted by AKT-mediated phosphorylation of multiple regulatory sites leading to TBC1D4 binding to 14-3-3 proteins (Eguez et al., 2005; Ramm et al., 2006; Sano et al., 2003). Subsequently, the newly acquired glucose from GLUT4-mediated uptake is stored as glycogen in skeletal muscle through the action of glycogen synthase, which is activated by AKT signaling via GSK3 phosphorylation and inhibition (McManus et al., 2005).
Akt signaling in response to insulin and IGF1 in skeletal muscle also enhances protein synthesis, while attenuating protein breakdown (Egerman and Glass, 2014). Through its induction of protein synthesis and inhibition of autophagy, mTORC1 signaling is a major driver of IGF1-stimulated muscle hypertrophy (Ohanna et al., 2005; Rommel et al., 2001). Skeletal muscle-specific knockout of mTORC1 results in muscle dystrophy (Bentzinger et al., 2008), and sustained mTORC1 signaling is sufficient to overcome atrophy during muscle immobilization (You et al., 2015). However, constitutive activation of mTORC1 in muscle can lead to organellar dysfunction and myopathy over time through chronic inhibition of autophagy (Castets et al., 2013). Interestingly, genetic activation of mTORC1 during denervation, which normally suppresses its activity, has been found to enhance muscle atrophy, at least in part through feedback suppression of AKT signaling to the FoxO transcription factors (Tang et al., 2014). AKT suppresses muscle protein breakdown through its inhibition of FoxO, thereby blocking expression of genes encoding the major muscle E3-ubiquitin ligases atrogin-1 and MuRF1 (Lee et al., 2004; Sandri et al., 2004; Stitt et al., 2004). By promoting an increase in muscle mass through such mechanisms, AKT signaling in skeletal muscle can improve systemic metabolism and overcome diet-induced obesity (Izumiya et al., 2008).
As a systemic negative feedback mechanism, insulin and leptin act on the hypothalamus to suppress food intake by stimulating AKT signaling to FoxO and mTORC1. Hypothalamic FoxO1 promotes food intake through the transcriptional induction of neuropeptide Y (NPY) and agouti-related peptide (AgRP), two major orexigenic peptides (Kim et al., 2006). AKT-mediated inhibition of FoxO1 in NPY/AgRP-producing neurons blocks the synthesis of these peptides in response to insulin or leptin. The production of NPY and AgRP is also blocked by hypothalamic mTORC1-S6K1 signaling, thereby providing an additional suppressive signal on food intake (Blouet et al., 2008; Cota et al., 2006). Additional studies are needed to understand the role of AKT signaling in other regions of the brain that influence feeding behavior and the potential pathophysiological functions of AKT dysregulation in feeding disorders.
Immunity and autoimmune diseases
The PI3K-AKT pathway plays a diverse role in both myeloid cells of the innate immune system and lymphoid cells of the adaptive immune system. While the immunological functions of the AKT signaling network is a rather broad and active area of research, the focus here is on established cell autonomous functions in these lineages.
A variety of different stimuli can activate AKT in myeloid cells, including specific growth factors, cytokines (e.g., IL-4), and ligands for toll-like receptors (TLRs; e.g., lipopolysaccharide (LPS)) and GPCRs (e.g., fMet-Leu-Phe (fMLP)). AKT activation stimulates a similar array of downstream effectors across the myeloid lineage, but how these signals impinge on the inflammatory action of each cell varies and is not well defined in most cases. AKT has emerged as mediator of macrophage polarization in response to different stimuli (Covarrubias et al., 2015; Weichhart et al., 2015). Polarization refers to specialized states adopted by activated macrophages (e.g., pro- or anti-inflammatory), which are dictated by the integration of diverse exogenous signals and alterations in the metabolic status of the cell. Along a continuum of functional states, the M1 state is the classical pro-inflammatory, anti-microbial macrophage promoted by LPS, whereas the M2 state mediates tissue repair, fibrosis, and response to parasitic infections and is induced by IL-4. While both M1 and M2 polarizing signals activate AKT, most genetic and pharmacological evidence suggests that AKT signaling promotes features of M2 macrophage polarization (Covarrubias et al., 2015; Weichhart et al., 2015). However, differential effects have been reported for AKT isoform-specific knockouts, with Akt1 null macrophages polarized toward the M1 state and Akt2 null macrophages toward M2 (Arranz et al., 2012; Babaev et al., 2014). What underlies this apparent functional difference is not well defined, but suggests that AKT2 might oppose the action of AKT1 in its promotion of M2 polarization. The TSC-mTORC1 circuit appears to play an important role in the control of this process. Mouse genetic models suggest that mTORC1 activation opposes the M2-polarizing effects of PI3K-AKT signaling, with negative feedback mechanisms dominating this regulation (Byles et al., 2013; Jiang et al., 2014; Zhu et al., 2014). Consistent with mTORC1 functioning downstream of AKT in the promotion of M2 polarization, rapamycin has been found to selectively kill human M2 macrophages, while enhancing M1 polarization (Mercalli et al., 2013). Alterations in cellular metabolism appear to underlie macrophage polarization (Covarrubias et al., 2015; Weichhart et al., 2015). Interestingly, AKT-induced glucose uptake, glycolysis, and production of cytosolic acetyl-CoA were recently found to promote M2 polarization by enhancing gene expression of specific targets in the M2 program, an effect largely attributed to epigenetic changes through directed histone acetylation (Covarrubias et al., 2016). An increase in ACLY-activating phosphorylation and protein levels downstream of AKT and mTORC1 were responsible for the increase in acetyl-CoA available for chromatin modifications. One mechanism by which AKT signaling might suppress M1 polarization in favor of M2 is through its inhibition of FoxO1, which promotes the expression of key M1 genes involved in inflammatory signaling, such as TLR4 (Fan et al., 2010). PI3K-AKT signaling in dendritic cells appears to dampen inflammatory signals, at least in part, by increasing expression of the anti-inflammatory cytokine IL-10 while decreasing expression of the pro-inflammatory cytokine IL-12 (Weichhart et al., 2015). Combined effects of the TSC-mTORC1, FoxO1, and GSK3 branches of AKT signaling likely contribute to this regulation. In addition, the mTORC1-inhibited process of autophagy has been suggested to play a key role in antigen processing and presentation by dendritic cells (Jagannath et al., 2009). In neutrophils, the control of cell polarity and chemotactic migration is key to their rapid response to infection or tissue damage. PI3Kγ, AKT2, and GSK3 are involved in the neutrophil response to GPCR agonists (e.g., fMLP), with mouse genetics demonstrating a prominent role in both neutrophil chemotaxis and induction of the respiratory burst involved in microbial killing (Chen et al., 2010b; Hirsch et al., 2000; Li et al., 2000; Liu et al., 2010; Tang et al., 2011). As in macrophage lineages, opposing functions for AKT isoforms have been observed in neutrophils, with Akt1-deficient neutrophils displaying enhanced migration and bacterial killing (Liu et al., 2013a). These genetic studies suggest that AKT2 promotes the mobilization and activation of neutrophils, while AKT1 has suppressive effects.
PI3K signaling plays a diverse and critical role in T-cell and B-cell functions in the adaptive immune system (So and Fruman, 2012). In lymphocytes, it is predominantly the PI3Kδ isoform that is activated by the T-cell receptor (TCR), B-cell receptor (BCR), and a variety of co-receptors and cytokine receptors, leading to activation of AKT and other effectors (Okkenhaug et al., 2002). The importance of properly linking transient PI3K-AKT activation to these coordinated immune signals is underscored by a variety of gain of function mouse models that give rise to severe lymphoproliferative and autoimmune disorders (Borlado et al., 2000; Parsons et al., 2001; Suzuki et al., 2001). AKT is important for the stimulated uptake of nutrients required for the survival and expansion of thymocyte populations, prior to differentiation into specific T cell lineages (Juntilla et al., 2007). Different classes of CD4+ T-helper (Th) cells exist that are defined by the cytokines they produce, which differentially influence the activity of other cells of both the innate and adaptive immune systems. The differentiation and clonal expansion of these populations involves AKT signaling to the TSC-mTORC1 circuit and FOXO family members, which serve to promote Th1, Th2 and Th17 cell differentiation while hindering the development of immunosuppressive Tregs (Delgoffe et al., 2009; Delgoffe et al., 2011; Gerriets et al., 2016; Kerdiles et al., 2010; Ouyang et al., 2010). While antigen and cytokine stimulation of CD8+ T-cells activate the PI3K-AKT pathway, the functional consequence of this signal is not entirely clear (Macintyre et al., 2011). However, mTORC1 plays an essential role in the differentiation and expansion of CD8+ cytotoxic T lymphocytes (CTLs), while attenuating the development of memory CD8+ T cells (Pollizzi et al., 2015). As in other cell types (Duvel et al., 2010), mTORC1 activation promotes glucose uptake, glycolysis and lipid synthesis to drive differentiation and anabolic cell proliferation in both Th cells and CTLs, while these metabolic changes appear to suppress Treg and memory cell functions (Gerriets et al., 2016; Kidani et al., 2013; Pollizzi et al., 2015; Shi et al., 2011). In B-cell development, PI3K-AKT signaling is required for transition of pro-B cells into IgM-expressing immature B cells (Okkenhaug et al., 2002). This process requires suppression of the recombination-activating gene (RAG) loci Rag1 and Rag2, which is mediated through AKT-dependent inhibition of FoxO1, a transcriptional activator of these genes (Amin and Schlissel, 2008). AKT signaling to FoxO1 also plays key roles in the maturation and survival of peripheral B cell populations and Ig class switching (Calamito et al., 2010; Chen et al., 2010a; Omori et al., 2006).
The opposing cell intrinsic functions of different AKT isoforms in distinct myeloid and lymphoid lineages, together with the critical communication between these populations of innate and adaptive immune cells that varies with the given challenge, makes interpretation of studies using whole body knockouts of AKT isoforms or systemic administration of PI3K, AKT, or mTOR inhibitors challenging at a mechanistic level.
Neuronal functions and neurological disorders
AKT signaling plays important roles in neuronal survival, growth, polarity, synaptic plasticity, and circuitry, thereby influencing brain development and function with implications in a diverse set of neurological disorders. In the developing brain, PI3K-AKT signaling is activated by a variety of growth and neurotrophic factors (e.g., IGF1, nerve-growth factor (NGF), brain-derived neurotrophic factor (BDNF)) and controls neuronal survival and morphology. The AKT-mediated inhibition of the proapoptotic protein Bad and Foxo transcription factors promotes neuronal cell survival (Brunet et al., 1999; Datta et al., 2000). Localized PI3K-AKT pathway activation and its inhibitory phosphorylation of GSK3 drives axon specification in neurites of developing neurons by activating collapsin response mediator protein 2 (CRMP-2), which associates with axonal growth cones in a manner that is inhibited by GSK3 (Jiang et al., 2005; Yoshimura et al., 2005). Through its activation of mTORC1, AKT signaling promotes the growth of multiple populations of brain cells, including neurons and astroglial cells, and plays a critical role in both developmental brain growth and pathological growth underlying megalencephaly and cortical malformations (Lipton and Sahin, 2014). The dynamic process of synapse formation (i.e., synaptic plasticity) is tightly regulated and required for the establishment of proper neuronal circuits and memory. Activation of AKT and mTORC1 signaling is important in the control of synaptic plasticity, affecting both long-term potentiation (LTP) and long-term depression (LTD) through its induction of localized protein synthesis (Hou and Klann, 2004; Li et al., 2010; Tang et al., 2002).
Dysregulation of the AKT signaling network underlies numerous neurodevelopmental, neurocognitive, neuropsychiatric, and neurodegenerative disorders. Germline or somatic mosaic mutations leading to increased activation of PI3K-AKT-mTORC1 signaling are frequently associated with epilepsy, autism spectrum disorders, and intellectual disabilities, a point that is exemplified by the multi-faceted TSC disease (Lipton and Sahin, 2014). Decreased AKT signaling has been associated with a number of mood disorders, including depression, schizophrenia, and bipolar disorder. Depression is believed to be caused by localized synaptic deficits and is associated with decreases in BDNF signaling to AKT and mTORC1, which can be reversed by anti-depressant drugs such as ketamine (Abdallah et al., 2015). Increased GSK3 activity upon attenuation of AKT signaling also contributes to depression, as well as schizophrenia and bipolar disorder (Emamian et al., 2004; Kaidanovich-Beilin and Woodgett, 2011). Gsk3αS21 or Gsk3β+S9 mice display anti-depression like behaviors that mimic those of animals on lithium, a GSK3 selective inhibitor used to treat mood disorders. While Gsk3αS21A; Gsk3βS9A mice are developmentally normal (McManus et al., 2005), they are sensitive to hyperactivity with phenotypes associated with anxiety in response to environmental changes and stress (Kaidanovich-Beilin and Woodgett, 2011; Polter et al., 2010). Various studies have also linked dysfunctional signaling of AKT and its downstream targets to neurodegenerative diseases, including Alzheimer’s. In the brains of Alzheimer’s patience, AKT signaling to GSK3 and mTORC1 appears elevated (An et al., 2003; Griffin et al., 2005). It has been suggested that mTORC1-mediated inhibition of autophagy might hinder the clearance of pathological protein aggregates in Alzheimer’s and other neurodegenerative diseases (Nixon, 2013). GSK3 has been implicated in Alzheimer’s disease and directly phosphorylates several proteins that contribute to the pathology of the disease, including amyloid precursor protein, presenilin, and tau (Kaidanovich-Beilin and Woodgett, 2011) (Figure 3). However, it is not clear whether this function of GSK3 is influenced by AKT signaling. Additional studies are needed to understand the role of FOXO family members in neurodegeneration, as there is increasing evidence that FOXO can promote the clearance of misfolded proteins (Webb and Brunet, 2014).
Therapeutic Targeting of AKT
While it is clear that PI3K-Akt signaling influences a wide variety of human diseases, therapeutic development of Akt inhibitors has been largely restricted to oncology. Over 50% of human tumors display hyperactivation of AKT. Over the last decade, numerous specific and potent PI3K pathway small molecule inhibitors have entered clinical trials. The in vitro use of two PI3K inhibitors, LY294002 and wortmannin, was instrumental in decoding many of the mechanisms by which PI3K initiates signaling, including AKT activation. However, subsequent studies showed that both compounds, in addition to blocking all PI3K isoforms, also directly inhibit mTOR, which is a PI3K-related protein kinase (Brunn et al., 1996). As more specific and potent PI3K and AKT inhibitors have since been developed, it is highly recommended that wortmannin and LY294002 no longer be used to make specific conclusions regarding the functions of PI3K or Akt. Some efficacy and therapeutic benefits have been observed in patients treated with new classes of PI3K inhibitors, but their use can be limited by significant toxicity (Mayer and Arteaga, 2016). Several AKT small molecule inhibitors have also been developed and are currently undergoing clinical evaluation.
A large number of ATP-competitive AKT inhibitors have been developed. Given the high degree of similarity of the ATP-binding pocket amongst AGC family kinases, it is not surprising that a lack of specificity is a major caveat of active site inhibitors. GSK690693 inhibits all AKT isoforms and showed efficacy in a number of preclinical studies, but dose-limiting toxicities that are likely on target, associated with hyperglycemia, led to termination of the clinical development of this drug (Crouthamel et al., 2009; Rhodes et al., 2008). AZD5363 is a nanomolar pan-AKT inhibitor that has shown efficacy in a number of hematological cancers and solid tumor models (Crabb et al., 2017; Davies et al., 2012). One of the more clinically advanced catalytic inhibitors is GDC0068 (Ipatasertib), a highly potent and specific orally-bioavailable inhibitor of all three AKT isoforms. Although GDC0068 has shown efficacy in preclinical models with PTEN inactivation or PIK3CA mutation (Lin et al., 2013), a phase I trial did not reveal significant antitumor activity (Saura et al., 2017). Afuresertib or GSK2110183, which inhibits AKT1 in the subnanomolar and AKT2 and AKT3 in the low nanomolar range, has also shown potent activity in preclinical models and in phase I trials appears to be well-tolerated (Dumble et al., 2014).
Catalytic AKT inhibitors block the phosphorylation of downstream substrates in a dose-dependent manner, but actually protect AKT from dephosphorylation at T308 and S473 (Lin et al., 2012; Okuzumi et al., 2009). Moreover, catalytic AKT inhibitors actually enhance PI3,4P2 and PIP3 binding, since the kinase adopts a conformation where the PH domain is released from its inhibitory interactions with the catalytic domain, leading to enhanced membrane binding (Vivanco et al., 2014). In a direct comparison, catalytic inhibitors show less efficacy in promoting cell death than allosteric inhibitors that target the PH domain (Vivanco et al., 2014) (see below).
MK2206 is a specific, potent and orally bioavailable allosteric AKT inhibitor that targets both the PH and catalytic domains (Hirai et al., 2010) and induces robust cancer cell death both alone and in combination with other compounds (Wisinski et al., 2016). As single agent therapy, however, MK2206 has not shown significant efficacy in the clinic (Ma et al., 2016). Unlike catalytic inhibitors, allosteric inhibitors target the closed, inactive conformation of AKT where the PH domain engages the kinase domain, thereby preventing phosphorylation and activation (Barnett et al., 2005; Hirai et al., 2010). Consistent with the notion that the AKT E17K mutation is constitutively membrane bound, this oncogenic AKT allele is less sensitive to MK2206 (Parikh et al., 2012). Finally, ARQ092 is a structurally distinct allosteric inhibitor that targets all three AKT isoforms, potently inhibits AKT signaling, and is currently under evaluation in phase I trails (Lapierre et al., 2016; Yu et al., 2015).
To date, both catalytic and allosteric inhibitors have shown limited efficacy in the clinic. A possible contributing factor to the lack of significant antitumor responses observed with all AKT inhibitors is a lack of patient stratification for those with activating AKT or PIK3CA mutations in the corresponding trials. In trials where patients were selected for the presence of pathway mutations, for example the AKT1 E17K mutation, a number of patients showed partial responses when treated with AZD5363 (Davies et al., 2012). Moreover, the relief of feedback inhibition in tumors treated with AKT inhibitors leads to a rebound effect resulting in robust PI3K pathway activation, thereby limiting the efficacy of such inhibitors, at least as single agents (Chandarlapaty et al., 2011). Importantly, PI3K pathway mutations, such as oncogenic PIK3CA, do not always coincide with elevated phosphorylation of AKT, with other PI3K effectors such as SGK isoforms driving downstream signaling in some settings (Vasudevan et al., 2009). The emergence of resistance to AKT inhibitors poses a major challenge to the effective implementation of small molecule inhibitors targeting the PI3K-AKT network in patients. These resistance mechanisms include increased expression of SGK isoforms (Bago et al., 2016; Sommer et al., 2013), further illustrating the challenge of overcoming regulatory redundancy within the network (see above).
Despite the specific functions and substrates attributed to the three AKT isoforms in pathobiology, highly selective and potent AKT isoform-specific inhibitors have yet to be developed. Based on our knowledge of isoform-specific Akt functions, optimizing Akt inhibitors to spare AKT2 might be particularly effective at relieving the major on-target toxicity of hyperglycemia, in cancer settings where AKT1 or 3 are the primary drivers.
Conclusions and Perspectives
The past 25 years of research on the AKT protein kinases has revealed an intricate molecular blueprint of AKT regulation and function and increasing insights into its role in cell, tissue, and organismal physiology. The diverse functional repertoire of AKT stems from the numerous downstream substrates that have evolved regulatory sequences that link the control of their cellular function to the myriad of upstream signals that stimulate AKT activity. The ever-expanding list of Akt substrates and their overlap and cross talk with other major cellular signaling networks creates a dynamic environment where the integration of distinct signals at the level of multiple downstream effectors is required for a cell to mount an appropriate response. It is now well recognized that signals from a combination of exogenous ligands and nutrients are perceived in a context of parallel cues monitored by pathways that sense intracellular nutrients, energy, and various stresses. Layered on top of this are feedback mechanisms, threshold effects, and spatial and temporal influences on signal propagation. Thus, the days of thinking about signal transduction in a binary, on-off fashion are long past. While the one-signal, one-substrate, one-function paradigm remains useful for reductionist research and will continue to play an essential role in discovering new components of signaling networks, we must also recognize and embrace the network view. The reality of the complexity of integrated signaling networks dictates that all signaling is contextual by nature, a fact that often leads to misinterpretation or overinterpretation of data. Now that the complexity of AKT signaling is glaringly evident, unraveling the detailed circuitry of the network and the strategies it employs to propagate cellular information will require novel biochemical, cell biological, genetic, computational, and systems approaches. Only with a more sophisticated understanding of the wiring and dynamic rewiring of the PI3K-AKT signaling network will we really grasp how to pharmacologically or biologically manipulate the network to treat the diverse set of human diseases exhibiting pathological features elicited by its dysregulation.
Acknowledgments
We apologize to the many colleagues and contributors over the last 25 years of research in the field whose work has added to our understanding of the AKT network, but could not be cited due to space limitations. This work was supported by grants from the National Institutes of Health: R35-CA197459 and P01-CA120964 (B.D.M.) and R01-CA200671 and R01-CA177910 (A.T.), and from the Ludwig Center at Harvard (A.T.)
Footnotes
Author Contributions B.D.M. and A.T. conceptualized, researched, and wrote this review.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdallah CG, Sanacora G, Duman RS, Krystal JH. Ketamine and rapid-acting antidepressants: a window into a new neurobiology for mood disorder therapeutics. Annu Rev Med. 2015;66:509–523. doi: 10.1146/annurev-med-053013-062946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackah E, Yu J, Zoellner S, Iwakiri Y, Skurk C, Shibata R, Ouchi N, Easton RM, Galasso G, Birnbaum MJ, et al. Akt1/protein kinase Balpha is critical for ischemic and VEGF-mediated angiogenesis. The Journal of clinical investigation. 2005;115:2119–2127. doi: 10.1172/JCI24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996a;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. Molecular basis for the substrate specificity of PKB; comparison with MAPKAP kinase-1 and p70S6kinase. FEBS Letts. 1996b;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An WL, Cowburn RF, Li L, Braak H, Alafuzoff I, Iqbal K, Iqbal IG, Winblad B, Pei JJ. Up-regulation of phosphorylated/activated p70 S6 kinase and its relationship to neurofibrillary pathology in Alzheimer’s disease. Am J Pathol. 2003;163:591–607. doi: 10.1016/S0002-9440(10)63687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andjelkovic M, Jakubowicz T, Cron P, Ming XF, Han JW, Hemmings BA. Activation and phosphorylation of a pleckstrin homology domain containing protein kinase (RAC-PK/PKB) promoted by serum and protein phosphatase inhibitors. Proc Natl Acad Sci USA. 1996;93:5699–5704. doi: 10.1073/pnas.93.12.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz A, Doxaki C, Vergadi E, Martinez de la Torre Y, Vaporidi K, Lagoudaki ED, Ieronymaki E, Androulidaki A, Venihaki M, Margioris AN, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci U S A. 2012;109:9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger KR, Serunian LA, Soltoff SP, Libby P, Cantley LC. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- Babaev VR, Hebron KE, Wiese CB, Toth CL, Ding L, Zhang Y, May JM, Fazio S, Vickers KC, Linton MF. Macrophage deficiency of Akt2 reduces atherosclerosis in Ldlr null mice. J Lipid Res. 2014;55:2296–2308. doi: 10.1194/jlr.M050633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago R, Sommer E, Castel P, Crafter C, Bailey FP, Shpiro N, Baselga J, Cross D, Eyers PA, Alessi DR. The hVps34-SGK3 pathway alleviates sustained PI3K/Akt inhibition by stimulating mTORC1 and tumour growth. EMBO J. 2016;35:2263. doi: 10.15252/embj.201670010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett SF, Defeo-Jones D, Fu S, Hancock PJ, Haskell KM, Jones RE, Kahana JA, Kral AM, Leander K, Lee LL, et al. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem J. 2005;385:399–408. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellacosa A, Testa JR, Staal SP, Tsichlis PN. A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- Blouet C, Ono H, Schwartz GJ. Mediobasal hypothalamic p70 S6 kinase 1 modulates the control of energy homeostasis. Cell Metab. 2008;8:459–467. doi: 10.1016/j.cmet.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlado LR, Redondo C, Alvarez B, Jimenez C, Criado LM, Flores J, Marcos MA, Martinez AC, Balomenos D, Carrera AC. Increased phosphoinositide 3-kinase activity induces a lymphoproliferative disorder and contributes to tumor generation in vivo. FASEB J. 2000;14:895–903. doi: 10.1096/fasebj.14.7.895. [DOI] [PubMed] [Google Scholar]
- Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBa/Akt1 Acts Downstream of DNA-PK in the DNA Double-Strand Break Response and Promotes Survival. Molecular Cell. 2008 doi: 10.1016/j.molcel.2008.02.024. In Press. [DOI] [PubMed] [Google Scholar]
- Braccini L, Ciraolo E, Campa CC, Perino A, Longo DL, Tibolla G, Pregnolato M, Cao Y, Tassone B, Damilano F, et al. PI3K-C2gamma is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling. Nat Commun. 2015;6:7400. doi: 10.1038/ncomms8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn GJ, Williams J, Sabers C, Wiederrecht G, Lawrence JC, Jr, Abraham RT. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 1996;15:5256–5267. [PMC free article] [PubMed] [Google Scholar]
- Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- Buzzi F, Xu L, Zuellig RA, Boller SB, Spinas GA, Hynx D, Chang Z, Yang Z, Hemmings BA, Tschopp O, et al. Differential effects of protein kinase B/Akt isoforms on glucose homeostasis and islet mass. Mol Cell Biol. 2010;30:601–612. doi: 10.1128/MCB.00719-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byles V, Covarrubias AJ, Ben-Sahra I, Lamming DW, Sabatini DM, Manning BD, Horng T. The TSC-mTOR pathway regulates macrophage polarization. Nat Commun. 2013;4:2834. doi: 10.1038/ncomms3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamito M, Juntilla MM, Thomas M, Northrup DL, Rathmell J, Birnbaum MJ, Koretzky G, Allman D. Akt1 and Akt2 promote peripheral B-cell maturation and survival. Blood. 2010;115:4043–4050. doi: 10.1182/blood-2009-09-241638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calera MR, Martinez C, Liu H, Jack AK, Birnbaum MJ, Pilch PF. Insulin increases the association of Akt-2 with Glut4-containing vesicles. J Biol Chem. 1998;273:7201–7204. doi: 10.1074/jbc.273.13.7201. [DOI] [PubMed] [Google Scholar]
- Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, Downward J, Parker PJ, Larijani B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja V, Laguerre M, Parker PJ, Larijani B. Role of a novel PH-kinase domain interface in PKB/Akt regulation: structural mechanism for allosteric inhibition. PLoS Biol. 2009;7:e17. doi: 10.1371/journal.pbio.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LC. The role of phosphoinositide 3-kinase in human disease. Harvey Lect. 2004;100:103–122. [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Castel P, Ellis H, Bago R, Toska E, Razavi P, Carmona FJ, Kannan S, Verma CS, Dickler M, Chandarlapaty S, et al. PDK1-SGK1 Signaling Sustains AKT-Independent mTORC1 Activation and Confers Resistance to PI3Kalpha Inhibition. Cancer Cell. 2016;30:229–242. doi: 10.1016/j.ccell.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castets P, Lin S, Rion N, Di Fulvio S, Romanino K, Guridi M, Frank S, Tintignac LA, Sinnreich M, Ruegg MA. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab. 2013;17:731–744. doi: 10.1016/j.cmet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Chakrabarti P, English T, Shi J, Smas CM, Kandror KV. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Jo U, Kohrman A, Rezaeian AH, Chou PC, Logothetis C, Lin HK. Posttranslational regulation of Akt in human cancer. Cell Biosci. 2014;4:59. doi: 10.1186/2045-3701-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez JA, Roach WG, Keller SR, Lane WS, Lienhard GE. Inhibition of GLUT4 translocation by Tbc1d1, a Rab GTPase-activating protein abundant in skeletal muscle, is partially relieved by AMP-activated protein kinase activation. J Biol Chem. 2008;283:9187–9195. doi: 10.1074/jbc.M708934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Limon JJ, Blanc C, Peng SL, Fruman DA. Foxo1 regulates marginal zone B-cell development. Eur J Immunol. 2010a;40:1890–1896. doi: 10.1002/eji.200939817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Somanath PR, Razorenova O, Chen WS, Hay N, Bornstein P, Byzova TV. Akt1 regulates pathological angiogenesis, vascular maturation and permeability in vivo. Nat Med. 2005;11:1188–1196. doi: 10.1038/nm1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tang H, Hay N, Xu J, Ye RD. Akt isoforms differentially regulate neutrophil functions. Blood. 2010b;115:4237–4246. doi: 10.1182/blood-2009-11-255323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O’Neill A, Castillo-Martin M, Nowak DG, Naguib A, Grace DM, et al. Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell. 2011;20:173–186. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Xu PZ, Peng XD, Chen WS, Guzman G, Yang X, Di Cristofano A, Pandolfi PP, Hay N. The deficiency of Akt1 is sufficient to suppress tumor development in Pten+/− mice. Genes Dev. 2006;20:1569–1574. doi: 10.1101/gad.1395006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15:2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280:25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- Chin YM, Yuan X, Balk SP, Toker A. Pten-deficient tumors depend on akt2 for maintenance and survival. Cancer discovery. 2014 doi: 10.1158/2159-8290.CD-13-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB, 3rd, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001a;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001b;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- Choi YH, Park S, Hockman S, Zmuda-Trzebiatowska E, Svennelid F, Haluzik M, Gavrilova O, Ahmad F, Pepin L, Napolitano M, et al. Alterations in regulation of energy homeostasis in cyclic nucleotide phosphodiesterase 3B-null mice. The Journal of clinical investigation. 2006;116:3240–3251. doi: 10.1172/JCI24867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani F, Czaja MJ. Regulation and Functions of Autophagic Lipolysis. Trends Endocrinol Metab. 2016;27:696–705. doi: 10.1016/j.tem.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffer PJ, Woodgett JR. Molecular cloning and characterisation of a novel putative protein- serine kinase related to the cAMP-dependent and protein kinase C families [published erratum appears in Eur J Biochem 1992 May 1;205(3):1217] Eur J Biochem. 1991;201:475–481. doi: 10.1111/j.1432-1033.1991.tb16305.x. [DOI] [PubMed] [Google Scholar]
- Collins BJ, Deak M, Arthur JS, Armit LJ, Alessi DR. In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J. 2003;22:4202–4211. doi: 10.1093/emboj/cdg407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, Seeley RJ. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- Covarrubias AJ, Aksoylar HI, Horng T. Control of macrophage metabolism and activation by mTOR and Akt signaling. Semin Immunol. 2015;27:286–296. doi: 10.1016/j.smim.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias AJ, Aksoylar HI, Yu J, Snyder NW, Worth AJ, Iyer SS, Wang J, Ben-Sahra I, Byles V, Polynne-Stapornkul T, et al. Akt-mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife. 2016;5 doi: 10.7554/eLife.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb SJ, Birtle AJ, Martin K, Downs N, Ratcliffe I, Maishman T, Ellis M, Griffiths G, Thompson S, Ksiazek L, et al. ProCAID: a phase I clinical trial to combine the AKT inhibitor AZD5363 with docetaxel and prednisolone chemotherapy for metastatic castration resistant prostate cancer. Invest New Drugs. 2017 doi: 10.1007/s10637-017-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Crouthamel MC, Kahana JA, Korenchuk S, Zhang SY, Sundaresan G, Eberwein DJ, Brown KK, Kumar R. Mechanism and management of AKT inhibitor-induced hyperglycemia. Clin Cancer Res. 2009;15:217–225. doi: 10.1158/1078-0432.CCR-08-1253. [DOI] [PubMed] [Google Scholar]
- Czech MP, Tencerova M, Pedersen DJ, Aouadi M. Insulin signalling mechanisms for triacylglycerol storage. Diabetologia. 2013;56:949–964. doi: 10.1007/s00125-013-2869-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, Greenberg ME. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- Davies BR, Greenwood H, Dudley P, Crafter C, Yu DH, Zhang J, Li J, Gao B, Ji Q, Maynard J, et al. Preclinical pharmacology of AZD5363, an inhibitor of AKT: pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol Cancer Ther. 2012;11:873–887. doi: 10.1158/1535-7163.MCT-11-0824-T. [DOI] [PubMed] [Google Scholar]
- Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, Lazar AJ, Gershenwald JE, Mills GB. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Maira G, Salvi M, Arrigoni G, Marin O, Sarno S, Brustolon F, Pinna LA, Ruzzene M. Protein kinase CK2 phosphorylates and upregulates Akt/PKB. Cell Death Differ. 2005;12:668–677. doi: 10.1038/sj.cdd.4401604. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol. 2009;29:5657–5670. doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RL, Marcotte R, Hennessy BT, Woodgett JR, Mills GB, Muller WJ. Akt1 and akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res. 2009;69:5057–5064. doi: 10.1158/0008-5472.CAN-08-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Biswas S, Morton RE, Smith JD, Hay N, Byzova TV, Febbraio M, Podrez EA. Akt3 deficiency in macrophages promotes foam cell formation and atherosclerosis in mice. Cell Metab. 2012;15:861–872. doi: 10.1016/j.cmet.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, Lee DF, Liu JC, Zhong Q, Wang X, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPilato LM, Ahmad F, Harms M, Seale P, Manganiello V, Birnbaum MJ. The Role of PDE3B Phosphorylation in the Inhibition of Lipolysis by Insulin. Mol Cell Biol. 2015;35:2752–2760. doi: 10.1128/MCB.00422-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N, Marais R. Protein kinase A blocks Raf-1 activity by stimulating 14-3-3 binding and blocking Raf-1 interaction with Ras. J Biol Chem. 2003;278:29819–29823. doi: 10.1074/jbc.C300182200. [DOI] [PubMed] [Google Scholar]
- Dumble M, Crouthamel MC, Zhang SY, Schaber M, Levy D, Robell K, Liu Q, Figueroa DJ, Minthorn EA, Seefeld MA, et al. Discovery of novel AKT inhibitors with enhanced anti-tumor effects in combination with the MEK inhibitor. PLoS One. 2014;9:e100880. doi: 10.1371/journal.pone.0100880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dummler B, Hemmings BA. Physiological roles of PKB/Akt isoforms in development and disease. Biochem Soc Trans. 2007;35:231–235. doi: 10.1042/BST0350231. [DOI] [PubMed] [Google Scholar]
- Dummler B, Tschopp O, Hynx D, Yang ZZ, Dirnhofer S, Hemmings BA. Life with a single isoform of Akt: mice lacking Akt2 and Akt3 are viable but display impaired glucose homeostasis and growth deficiencies. Mol Cell Biol. 2006;26:8042–8051. doi: 10.1128/MCB.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton RM, Cho H, Roovers K, Shineman DW, Mizrahi M, Forman MS, Lee VM, Szabolcs M, de Jong R, Oltersdorf T, et al. Role for Akt3/protein kinase Bgamma in attainment of normal brain size. Mol Cell Biol. 2005;25:1869–1878. doi: 10.1128/MCB.25.5.1869-1878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner M, Lucic I, Leonard TA, Yudushkin I. PI345P3 Engagement Restricts Akt Activity to Cellualr Membranes. Mol Cell. 2017a doi: 10.1016/j.molcel.2016.12.028. In press. [DOI] [PubMed] [Google Scholar]
- Ebner M, Sinkovics B, Szczgiel M, Ribeiro DW, Yudushkin I. Localization of mTORC2 activity inside cells. J Cell Biol. 2017b doi: 10.1083/jcb.201610060. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol. 2014;49:59–68. doi: 10.3109/10409238.2013.857291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguez L, Lee A, Chavez JA, Miinea CP, Kane S, Lienhard GE, McGraw TE. Full intracellular retention of GLUT4 requires AS160 Rab GTPase activating protein. Cell Metab. 2005;2:263–272. doi: 10.1016/j.cmet.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36:131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, Newton AC, Mao Y, Miao RQ, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. Embo J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CD, Lum MA, Xu C, Black JD, Wang X. Ubiquitin-dependent regulation of phospho-AKT dynamics by the ubiquitin E3 ligase, NEDD4-1, in the insulin-like growth factor-1 response. J Biol Chem. 2013;288:1674–1684. doi: 10.1074/jbc.M112.416339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Morinaga H, Kim JJ, Bae E, Spann NJ, Heinz S, Glass CK, Olefsky JM. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 2010;29:4223–4236. doi: 10.1038/emboj.2010.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele CG, Ooms LM, Ho M, Vieusseux J, O’Toole SA, Millar EK, Lopez-Knowles E, Sriratana A, Gurung R, Baglietto L, et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc Natl Acad Sci U S A. 2010;107:22231–22236. doi: 10.1073/pnas.1015245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Hernando C, Ackah E, Yu J, Suarez Y, Murata T, Iwakiri Y, Prendergast J, Miao RQ, Birnbaum MJ, Sessa WC. Loss of Akt1 leads to severe atherosclerosis and occlusive coronary artery disease. Cell Metab. 2007;6:446–457. doi: 10.1016/j.cmet.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Hernando C, Jozsef L, Jenkins D, Di Lorenzo A, Sessa WC. Absence of Akt1 reduces vascular smooth muscle cell migration and survival and induces features of plaque vulnerability and cardiac dysfunction during atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29:2033–2040. doi: 10.1161/ATVBAHA.109.196394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem. 2007;282:24514–24524. doi: 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- Frame S, Cohen P, Biondi RM. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell. 2001;7:1321–1327. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- Franke TF, Yang SI, Chan TO, Datta K, Kazlauskas A, Morrison DK, Kaplan DR, Tsichlis PN. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- Frech M, Andjelkovic M, Ingley E, KRK, Falck JR, Hemmings BA. High Affinity Binding of Inositol Phosphates and Phosphoinositides to the Pleckstrin Homology Domain of RAC/Protein Kinase B and Their Influence on Kinase Activity. J Biol Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Garofalo RS, Orena SJ, Rafidi K, Torchia AJ, Stock JL, Hildebrandt AL, Coskran T, Black SC, Brees DJ, Wicks JR, et al. Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB beta. The Journal of clinical investigation. 2003;112:197–208. doi: 10.1172/JCI16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S, Rochford JJ, Wolfrum C, Gray SL, Schinner S, Wilson JC, Soos MA, Murgatroyd PR, Williams RM, Acerini CL, et al. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science. 2004;304:1325–1328. doi: 10.1126/science.1096706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, Warmoes MO, de Cubas AA, MacIver NJ, Locasale JW, et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat Immunol. 2016;17:1459–1466. doi: 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, Barretina J, Lin WM, Rameh L, Salmena L, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin RJ, Moloney A, Kelliher M, Johnston JA, Ravid R, Dockery P, O’Connor R, O’Neill C. Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer’s disease pathology. J Neurochem. 2005;93:105–117. doi: 10.1111/j.1471-4159.2004.02949.x. [DOI] [PubMed] [Google Scholar]
- Guan KL, Figueroa C, Brtva TR, Zhu T, Taylor J, Barber TD, Vojtek AB. Negative regulation of the serine/threonine kinase B-Raf by Akt. J Biol Chem. 2000;275:27354–27359. doi: 10.1074/jbc.M004371200. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Guilherme A, Klarlund JK, Krystal G, Czech MP. Regulation of PIP3 5-phosphatase activity by insulin. J Biol Chem. 1996;271:29533–29536. doi: 10.1074/jbc.271.47.29533. [DOI] [PubMed] [Google Scholar]
- Gulen MF, Bulek K, Xiao H, Yu M, Gao J, Sun L, Beurel E, Kaidanovich-Beilin O, Fox PL, DiCorleto PE, et al. Inactivation of the enzyme GSK3alpha by the kinase IKKi promotes AKT-mTOR signaling pathway that mediates interleukin-1-induced Th17 cell maintenance. Immunity. 2012;37:800–812. doi: 10.1016/j.immuni.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Gao M, Lu Y, Liang J, Lorenzi PL, Bai S, Hawke DH, Li J, Dogruluk T, Scott KL, et al. Coordinate phosphorylation of multiple residues on single AKT1 and AKT2 molecules. Oncogene. 2014;33:3463–3472. doi: 10.1038/onc.2013.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Chakraborty AA, Liu P, Gan W, Zheng X, Inuzuka H, Wang B, Zhang J, Zhang L, Yuan M, et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science. 2016;353:929–932. doi: 10.1126/science.aad5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PT, Stephens LR. Emerging evidence of signalling roles for PI(3,4)P2 in Class I and II PI3K-regulated pathways. Biochem Soc Trans. 2016;44:307–314. doi: 10.1042/BST20150248. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Ross FA, Gowans GJ, Tibarewal P, Leslie NR, Hardie DG. Phosphorylation by Akt within the ST loop of AMPK-alpha1 down-regulates its activation in tumour cells. Biochem J. 2014;459:275–287. doi: 10.1042/BJ20131344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Sootome H, Nakatsuru Y, Miyama K, Taguchi S, Tsujioka K, Ueno Y, Hatch H, Majumder PK, Pan BS, et al. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Ther. 2010;9:1956–1967. doi: 10.1158/1535-7163.MCT-09-1012. [DOI] [PubMed] [Google Scholar]
- Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, Wymann MP. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- Holz MK, Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem. 2005;280:26089–26093. doi: 10.1074/jbc.M504045200. [DOI] [PubMed] [Google Scholar]
- Horman S, Vertommen D, Heath R, Neumann D, Mouton V, Woods A, Schlattner U, Wallimann T, Carling D, Hue L, et al. Insulin antagonizes ischemia-induced Thr172 phosphorylation of AMP-activated protein kinase alpha-subunits in heart via hierarchical phosphorylation of Ser485/491. J Biol Chem. 2006;281:5335–5340. doi: 10.1074/jbc.M506850200. [DOI] [PubMed] [Google Scholar]
- Hou L, Klann E. Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2004;24:6352–6361. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol Metab. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008 doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain K, Challis B, Rocha N, Payne F, Minic M, Thompson A, Daly A, Scott C, Harris J, Smillie BJ, et al. An activating mutation of AKT2 and human hypoglycemia. Science. 2011;334:474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Jin J, Cardiff RD, Woodgett JR, Muller WJ. Activation of Akt-1 (PKB-alpha) can accelerate ErbB-2-mediated mammary tumorigenesis but suppresses tumor invasion. Cancer Res. 2004;64:3171–3178. doi: 10.1158/0008-5472.can-03-3465. [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, Brugge JS. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya Y, Hopkins T, Morris C, Sato K, Zeng L, Viereck J, Hamilton JA, Ouchi N, LeBrasseur NK, Walsh K. Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7:159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–276. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- James SR, Downes CP, Gigg R, Grove SJA, Holmes AB, Alessi DR. Specific Binding of the Akt-1 protein kinase to PIP3 without subsequent activation. Biochem J. 1996;315:709–713. doi: 10.1042/bj3150709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jethwa N, Chung GH, Lete MG, Alonso A, Byrne RD, Calleja V, Larijani B. Endomembrane PtdIns(3,4,5)P3 activates the PI3K-Akt pathway. J Cell Sci. 2015;128:3456–3465. doi: 10.1242/jcs.172775. [DOI] [PubMed] [Google Scholar]
- Jiang H, Guo W, Liang X, Rao Y. Both the establishment and the maintenance of neuronal polarity require active mechanisms: critical roles of GSK-3beta and its upstream regulators. Cell. 2005;120:123–135. doi: 10.1016/j.cell.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Jiang H, Westerterp M, Wang C, Zhu Y, Ai D. Macrophage mTORC1 disruption reduces inflammation and insulin resistance in obese mice. Diabetologia. 2014;57:2393–2404. doi: 10.1007/s00125-014-3350-5. [DOI] [PubMed] [Google Scholar]
- Jones PF, Jakubowicz T, Pitossi FJ, Maurer F, Hemmings BA. Molecular cloning and identification of a serine/threonine protein kinase of the second-messenger subfamily. Proc Natl Acad Sci U S A. 1991;88:4171–4175. doi: 10.1073/pnas.88.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien LA, Carriere A, Moreau J, Roux PP. mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol Cell Biol. 2010;30:908–921. doi: 10.1128/MCB.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntilla MM, Wofford JA, Birnbaum MJ, Rathmell JC, Koretzky GA. Akt1 and Akt2 are required for alphabeta thymocyte survival and differentiation. Proc Natl Acad Sci U S A. 2007;104:12105–12110. doi: 10.1073/pnas.0705285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidanovich-Beilin O, Woodgett JR. GSK-3: Functional Insights from Cell Biology and Animal Models. Front Mol Neurosci. 2011;4:40. doi: 10.3389/fnmol.2011.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler-Noreuil KM, Sapp JC, Lindhurst MJ, Parker VE, Blumhorst C, Darling T, Tosi LL, Huson SM, Whitehouse RW, Jakkula E, et al. Clinical delineation and natural history of the PIK3CA-related overgrowth spectrum. Am J Med Genet A. 2014;164A:1713–1733. doi: 10.1002/ajmg.a.36552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch’en IL, Stockmann C, Katayama CD, Hedrick SM. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EK, Yun SJ, Ha JM, Kim YW, Jin IH, Yun J, Shin HK, Song SH, Kim JH, Lee JS, et al. Selective activation of Akt1 by mammalian target of rapamycin complex 2 regulates cancer cell migration, invasion, and metastasis. Oncogene. 2011;30:2954–2963. doi: 10.1038/onc.2011.22. [DOI] [PubMed] [Google Scholar]
- Kim MS, Pak YK, Jang PG, Namkoong C, Choi YS, Won JC, Kim KS, Kim SW, Kim HS, Park JY, et al. Role of hypothalamic Foxo1 in the regulation of food intake and energy homeostasis. Nat Neurosci. 2006;9:901–906. doi: 10.1038/nn1731. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Kitamura Y, Kuroda S, Hino Y, Ando M, Kotani K, Konishi H, Matsuzaki H, Kikkawa U, Ogawa W, et al. Insulin-induced phosphorylation and activation of cyclic nucleotide phosphodiesterase 3B by the serine-threonine kinase Akt. Mol Cell Biol. 1999;19:6286–6296. doi: 10.1128/mcb.19.9.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A, Kavanaugh WM, Pot D, Williams LT. A specific product of PI 3-K directly activates the protein kinase Akt through its pleckstrin homology domain. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn AD, Kovacina KS, Roth RA. Insulin stimulates the kinase activity of RAC-PK, a pleckstrin homology domain containing ser/thr kinase. Embo J. 1995;14:4288–4295. doi: 10.1002/j.1460-2075.1995.tb00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- Koren S, DiPilato LM, Emmett MJ, Shearin AL, Chu Q, Monks B, Birnbaum MJ. The role of mouse Akt2 in insulin-dependent suppression of adipocyte lipolysis in vivo. Diabetologia. 2015;58:1063–1070. doi: 10.1007/s00125-015-3532-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, Roth RA. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- Kumar A, Lawrence JC, Jr, Jung DY, Ko HJ, Keller SR, Kim JK, Magnuson MA, Harris TE. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes. 2010;59:1397–1406. doi: 10.2337/db09-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J Biol Chem. 2005;280:5581–5587. doi: 10.1074/jbc.M411534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, Chiang CW. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem. 2008;283:1882–1892. doi: 10.1074/jbc.M709585200. [DOI] [PubMed] [Google Scholar]
- Landgraf KE, Pilling C, Falke JJ. Molecular mechanism of an oncogenic mutation that alters membrane targeting: Glu17Lys modifies the PIP lipid specificity of the AKT1 PH domain. Biochemistry. 2008;47:12260–12269. doi: 10.1021/bi801683k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre JM, Eathiraj S, Vensel D, Liu Y, Bull CO, Cornell-Kennon S, Iimura S, Kelleher EW, Kizer DE, Koerner S, et al. Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin -2-amine (ARQ 092): An Orally Bioavailable, Selective, and Potent Allosteric AKT Inhibitor. J Med Chem. 2016;59:6455–6469. doi: 10.1021/acs.jmedchem.6b00619. [DOI] [PubMed] [Google Scholar]
- Lauring J, Cosgrove DP, Fontana S, Gustin JP, Konishi H, Abukhdeir AM, Garay JP, Mohseni M, Wang GM, Higgins MJ, et al. Knock in of the AKT1 E17K mutation in human breast epithelial cells does not recapitulate oncogenic PIK3CA mutations. Oncogene. 2010;29:2337–2345. doi: 10.1038/onc.2009.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavens KF, Easton RM, Shulman GI, Previs SF, Birnbaum MJ. Akt2 is required for hepatic lipid accumulation in models of insulin resistance. Cell Metab. 2009;10:405–418. doi: 10.1016/j.cmet.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Luciano AK, Ackah E, Rodriguez-Vita J, Bancroft TA, Eichmann A, Simons M, Kyriakides TR, Morales-Ruiz M, Sessa WC. Endothelial Akt1 mediates angiogenesis by phosphorylating multiple angiogenic substrates. Proc Natl Acad Sci U S A. 2014;111:12865–12870. doi: 10.1073/pnas.1408472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Dai G, Hu Z, Wang X, Du J, Mitch WE. Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol. 2004;15:1537–1545. doi: 10.1097/01.asn.0000127211.86206.e1. [DOI] [PubMed] [Google Scholar]
- Leto D, Saltiel AR. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol. 2012;13:383–396. doi: 10.1038/nrm3351. [DOI] [PubMed] [Google Scholar]
- Li Chew C, Lunardi A, Gulluni F, Ruan DT, Chen M, Salmena L, Nishino M, Papa A, Ng C, Fung J, et al. In Vivo Role of INPP4B in Tumor and Metastasis Suppression through Regulation of PI3K-AKT Signaling at Endosomes. Cancer discovery. 2015;5:740–751. doi: 10.1158/2159-8290.CD-14-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer [see comments] Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, Li XY, Aghajanian G, Duman RS. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–964. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K, Wang P. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 2013;73:5742–5753. doi: 10.1158/0008-5472.CAN-13-0538. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science. 2000;287:1046–1049. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- Lien EC, Dibble CC, Toker A. PI3K Signaling in Cancer: Beyond AKT. Current Opinion in Cell Biology. 2017 doi: 10.1016/j.ceb.2017.02.007. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien EC, Lyssiotis CA, Juvekar A, Hu H, Asara JM, Cantley LC, Toker A. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat Cell Biol. 2016;18:572–578. doi: 10.1038/ncb3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Jono H, Komatsu K, Woo CH, Lee J, Miyata M, Matsuno T, Xu X, Huang Y, Zhang W, et al. CYLD negatively regulates transforming growth factor-beta-signalling via deubiquitinating Akt. Nat Commun. 2012;3:771. doi: 10.1038/ncomms1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Sampath D, Nannini MA, Lee BB, Degtyarev M, Oeh J, Savage H, Guan Z, Hong R, Kassees R, et al. Targeting activated Akt with GDC-0068, a novel selective Akt inhibitor that is efficacious in multiple tumor models. Clin Cancer Res. 2013;19:1760–1772. doi: 10.1158/1078-0432.CCR-12-3072. [DOI] [PubMed] [Google Scholar]
- Lin K, Lin J, Wu WI, Ballard J, Lee BB, Gloor SL, Vigers GP, Morales TH, Friedman LS, Skelton N, et al. An ATP-site on-off switch that restricts phosphatase accessibility of Akt. Sci Signal. 2012;5:ra37. doi: 10.1126/scisignal.2002618. [DOI] [PubMed] [Google Scholar]
- Lindhurst MJ, Sapp JC, Teer JK, Johnston JJ, Finn EM, Peters K, Turner J, Cannons JL, Bick D, Blakemore L, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Bi Y, Wang R, Shen B, Zhang Y, Yang H, Wang X, Liu H, Lu Y, Han F. Kinase AKT1 negatively controls neutrophil recruitment and function in mice. J Immunol. 2013a;191:2680–2690. doi: 10.4049/jimmunol.1300736. [DOI] [PubMed] [Google Scholar]
- Liu L, Das S, Losert W, Parent CA. mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev Cell. 2010;19:845–857. doi: 10.1016/j.devcel.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Begley M, Michowski W, Inuzuka H, Ginzberg M, Gao D, Tsou P, Gan W, Papa A, Kim BM, et al. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature. 2014;508:541–545. doi: 10.1038/nature13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Gan W, Chin YR, Ogura K, Guo J, Zhang J, Wang B, Blenis J, Cantley LC, Toker A, et al. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer discovery. 2015;5:1194–1209. doi: 10.1158/2159-8290.CD-15-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Gan W, Inuzuka H, Lazorchak AS, Gao D, Arojo O, Liu D, Wan L, Zhai B, Yu Y, et al. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat Cell Biol. 2013b;15:1340–1350. doi: 10.1038/ncb2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Wan M, Leavens KF, Chu Q, Monks BR, Fernandez S, Ahima RS, Ueki K, Kahn CR, Birnbaum MJ. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat Med. 2012;18:388–395. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Muller M, Smith D, Dutta B, Komurov K, Iadevaia S, Ruths D, Tseng JT, Yu S, Yu Q, et al. Kinome siRNA-phosphoproteomic screen identifies networks regulating AKT signaling. Oncogene. 2011;30:4567–4577. doi: 10.1038/onc.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CX, Sanchez C, Gao F, Crowder R, Naughton M, Pluard T, Creekmore A, Guo Z, Hoog J, Lockhart AC, et al. A Phase I Study of the AKT Inhibitor MK-2206 in Combination with Hormonal Therapy in Postmenopausal Women with Estrogen Receptor-Positive Metastatic Breast Cancer. Clin Cancer Res. 2016;22:2650–2658. doi: 10.1158/1078-0432.CCR-15-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Macintyre AN, Finlay D, Preston G, Sinclair LV, Waugh CM, Tamas P, Feijoo C, Okkenhaug K, Cantrell DA. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Mancini ML, Lien EC, Toker A. Oncogenic AKT1(E17K) mutation induces mammary hyperplasia but prevents HER2-driven tumorigenesis. Oncotarget. 2016;7:17301–17313. doi: 10.18632/oncotarget.8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/Akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Oemler W, Naber SP, Tsichlis PN. Akt1 ablation inhibits, whereas Akt2 ablation accelerates, the development of mammary adenocarcinomas in mouse mammary tumor virus (MMTV)-ErbB2/neu and MMTV-polyoma middle T transgenic mice. Cancer Res. 2007;67:167–177. doi: 10.1158/0008-5472.CAN-06-3782. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Mayer IA, Arteaga CL. The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu Rev Med. 2016;67:11–28. doi: 10.1146/annurev-med-062913-051343. [DOI] [PubMed] [Google Scholar]
- McManus EJ, Collins BJ, Ashby PR, Prescott AR, Murray-Tait V, Armit LJ, Arthur JS, Alessi DR. The in vivo role of PtdIns(3,4,5)P3 binding to PDK1 PH domain defined by knockin mutation. EMBO J. 2004;23:2071–2082. doi: 10.1038/sj.emboj.7600218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus EJ, Sakamoto K, Armit LJ, Ronaldson L, Shpiro N, Marquez R, Alessi DR. Role that phosphorylation of GSK3 plays in insulin and Wnt signalling defined by knockin analysis. Embo J. 2005;24:1571–1583. doi: 10.1038/sj.emboj.7600633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercalli A, Calavita I, Dugnani E, Citro A, Cantarelli E, Nano R, Melzi R, Maffi P, Secchi A, Sordi V, et al. Rapamycin unbalances the polarization of human macrophages to M1. Immunology. 2013;140:179–190. doi: 10.1111/imm.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz LS, Surinova S, Ghazaly E, Velasco LG, Haider S, Rodriguez-Prados JC, Berenjeno IM, Chelala C, Vanhaesebroeck B. Phosphoproteomic comparison of Pik3ca and Pten signalling identifies the nucleotidase NT5C as a novel AKT substrate. Sci Rep. 2017;7:39985. doi: 10.1038/srep39985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Seminars in cell & developmental biology. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Morel C, Carlson SM, White FM, Davis RJ. Mcl-1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol Cell Biol. 2009;29:3845–3852. doi: 10.1128/MCB.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisett JD, Abdel-Fattah G, Hoogeveen R, Mitchell E, Ballantyne CM, Pownall HJ, Opekun AR, Jaffe JS, Oppermann S, Kahan BD. Effects of sirolimus on plasma lipids, lipoprotein levels, and fatty acid metabolism in renal transplant patients. J Lipid Res. 2002;43:1170–1180. [PubMed] [Google Scholar]
- Mukai Y, Rikitake Y, Shiojima I, Wolfrum S, Satoh M, Takeshita K, Hiroi Y, Salomone S, Kim HH, Benjamin LE, et al. Decreased vascular lesion formation in mice with inducible endothelial-specific expression of protein kinase Akt. The Journal of clinical investigation. 2006;116:334–343. doi: 10.1172/JCI26223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, Gao S, Mills GB, Brugge JS. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave BT, Ouwens DM, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J. 1999;344:427–431. [PMC free article] [PubMed] [Google Scholar]
- Ng Y, Ramm G, Lopez JA, James DE. Rapid activation of Akt2 is sufficient to stimulate GLUT4 translocation in 3T3-L1 adipocytes. Cell Metab. 2008;7:348–356. doi: 10.1016/j.cmet.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Ohanna M, Sobering AK, Lapointe T, Lorenzo L, Praud C, Petroulakis E, Sonenberg N, Kelly PA, Sotiropoulos A, Pende M. Atrophy of S6K1(−/−) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat Cell Biol. 2005;7:286–294. doi: 10.1038/ncb1231. [DOI] [PubMed] [Google Scholar]
- Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, Shokat KM. Inhibitor hijacking of Akt activation. Nat Chem Biol. 2009;5:484–493. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori SA, Cato MH, Anzelon-Mills A, Puri KD, Shapiro-Shelef M, Calame K, Rickert RC. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 2006;25:545–557. doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Ono H, Shimano H, Katagiri H, Yahagi N, Sakoda H, Onishi Y, Anai M, Ogihara T, Fujishiro M, Viana AY, et al. Hepatic Akt activation induces marked hypoglycemia, hepatomegaly, and hypertriglyceridemia with sterol regulatory element binding protein involvement. Diabetes. 2003;52:2905–2913. doi: 10.2337/diabetes.52.12.2905. [DOI] [PubMed] [Google Scholar]
- Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S, Mukhopadhyay A, Narasimhan SD, Tesz G, Czech MP, Tissenbaum HA. A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell. 2009;136:939–951. doi: 10.1016/j.cell.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh C, Janakiraman V, Wu WI, Foo CK, Kljavin NM, Chaudhuri S, Stawiski E, Lee B, Lin J, Li H, et al. Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc Natl Acad Sci U S A. 2012;109:19368–19373. doi: 10.1073/pnas.1204384109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker PJ, Caudwell FB, Cohen P. Glycogen synthase from rabbit skeletal muscle; effect of insulin on the state of phosphorylation of the seven phosphoserine residues in vivo. Eur J Biochem. 1983;130:227–234. doi: 10.1111/j.1432-1033.1983.tb07140.x. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Jones RG, Tsao MS, Odermatt B, Ohashi PS, Woodgett JR. Expression of active protein kinase B in T cells perturbs both T and B cell homeostasis and promotes inflammation. J Immunol. 2001;167:42–48. doi: 10.4049/jimmunol.167.1.42. [DOI] [PubMed] [Google Scholar]
- Phung TL, Ziv K, Dabydeen D, Eyiah-Mensah G, Riveros M, Perruzzi C, Sun J, Monahan-Earley RA, Shiojima I, Nagy JA, et al. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell. 2006;10:159–170. doi: 10.1016/j.ccr.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi KN, Patel CH, Sun IH, Oh MH, Waickman AT, Wen J, Delgoffe GM, Powell JD. mTORC1 and mTORC2 selectively regulate CD8(+) T cell differentiation. The Journal of clinical investigation. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polter A, Beurel E, Yang S, Garner R, Song L, Miller CA, Sweatt JD, McMahon L, Bartolucci AA, Li X, et al. Deficiency in the inhibitory serine-phosphorylation of glycogen synthase kinase-3 increases sensitivity to mood disturbances. Neuropsychopharmacology. 2010;35:1761–1774. doi: 10.1038/npp.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Ramm G, Larance M, Guilhaus M, James DE. A role for 14-3-3 in insulin-stimulated GLUT4 translocation through its interaction with the RabGAP AS160. J Biol Chem. 2006;281:29174–29180. doi: 10.1074/jbc.M603274200. [DOI] [PubMed] [Google Scholar]
- Rhodes N, Heerding DA, Duckett DR, Eberwein DJ, Knick VB, Lansing TJ, McConnell RT, Gilmer TM, Zhang SY, Robell K, et al. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res. 2008;68:2366–2374. doi: 10.1158/0008-5472.CAN-07-5783. [DOI] [PubMed] [Google Scholar]
- Risso G, Blaustein M, Pozzi B, Mammi P, Srebrow A. Akt/PKB: one kinase, many modifications. Biochem J. 2015;468:203–214. doi: 10.1042/BJ20150041. [DOI] [PubMed] [Google Scholar]
- Roberts MS, Woods AJ, Dale TC, Van Der Sluijs P, Norman JC. Protein kinase B/Akt acts via glycogen synthase kinase 3 to regulate recycling of alpha v beta 3 and alpha 5 beta 1 integrins. Mol Cell Biol. 2004;24:1505–1515. doi: 10.1128/MCB.24.4.1505-1515.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez VP, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer discovery. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo Y, Zhang X, Roux PP. Regulation and function of the RSK family of protein kinases. Biochem J. 2012;441:553–569. doi: 10.1042/BJ20110289. [DOI] [PubMed] [Google Scholar]
- Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- Rommel C, Clarke BA, Zimmermann S, Nunez L, Rossman R, Reid K, Moelling K, Yancopoulos GD, Glass DJ. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science. 1999;286:1738–1741. doi: 10.1126/science.286.5445.1738. [DOI] [PubMed] [Google Scholar]
- Rosner M, Siegel N, Valli A, Fuchs C, Hengstschlager M. mTOR phosphorylated at S2448 binds to raptor and rictor. Amino Acids. 2010;38:223–228. doi: 10.1007/s00726-008-0230-7. [DOI] [PubMed] [Google Scholar]
- Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M, Anzeneder T, Schulz A, Beckmann G, Byrne AT, Jeffers M, Pena C, Politz O, Kochert K, Vonk R, et al. AKT1 (E17K) mutation profiling in breast cancer: prevalence, concurrent oncogenic alterations, and blood-based detection. BMC Cancer. 2016;16:622. doi: 10.1186/s12885-016-2626-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylatt DB, Aitken A, Bilham T, Condon GD, Embi N, Cohen P. Glycogen synthase from rabbit skeletal muscle. Amino acid sequence at the sites phosphorylated by glycogen synthase kinase-3, and extension of the N-terminal sequence containing the site phosphorylated by phosphorylase kinase. Eur J Biochem. 1980;107:529–537. [PubMed] [Google Scholar]
- Salhia B, Van Cott C, Tegeler T, Polpitiya A, Duquette RA, Gale M, Hostteter G, Petritis K, Carpten J. Differential effects of AKT1(p.E17K) expression on human mammary luminal epithelial and myoepithelial cells. Hum Mutat. 2012;33:1216–1227. doi: 10.1002/humu.22100. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 Is an Insulin-Regulated Inhibitor of the mTORC1 Protein Kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanidas I, Polytarchou C, Hatziapostolou M, Ezell SA, Kottakis F, Hu L, Guo A, Xie J, Comb MJ, Iliopoulos D, et al. Phosphoproteomics screen reveals akt isoform-specific signals linking RNA processing to lung cancer. Mol Cell. 2014;53:577–590. doi: 10.1016/j.molcel.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Kane S, Sano E, Miinea CP, Asara JM, Lane WS, Garner CW, Lienhard GE. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Saura C, Roda D, Rosello S, Oliveira M, Macarulla T, Perez-Fidalgo JA, Morales-Barrera R, Sanchis-Garcia JM, Musib L, Budha N, et al. A First-in-Human Phase I Study of the ATP-Competitive AKT Inhibitor Ipatasertib Demonstrates Robust and Safe Targeting of AKT in Patients with Solid Tumors. Cancer discovery. 2017;7:102–113. doi: 10.1158/2159-8290.CD-16-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic A, Hudson CC, Homme JL, Yin P, Otterness DM, Karnitz LM, Abraham RT. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 2000;60:3504–3513. [PubMed] [Google Scholar]
- Shah OJ, Hunter T. Turnover of the Active Fraction of IRS1 Involves Raptor-mTOR- and S6K1-Dependent Serine Phosphorylation in Cell Culture Models of Tuberous Sclerosis. Mol Cell Biol. 2006;26:6425–6434. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CH, Yuan P, Perez-Lorenzo R, Zhang Y, Lee SX, Ou Y, Asara JM, Cantley LC, Zheng B. Phosphorylation of BRAF by AMPK impairs BRAF-KSR1 association and cell proliferation. Mol Cell. 2013;52:161–172. doi: 10.1016/j.molcel.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui N, Sonenberg N. Signalling to eIF4E in cancer. Biochem Soc Trans. 2015;43:763–772. doi: 10.1042/BST20150126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So L, Fruman DA. PI3K signalling in B- and T-lymphocytes: new developments and therapeutic advances. Biochem J. 2012;442:465–481. doi: 10.1042/BJ20112092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman GA, Acosta-Jaquez HA, Fingar DC. mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids. 2010;45:1089–1100. doi: 10.1007/s11745-010-3488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer EM, Dry H, Cross D, Guichard S, Davies BR, Alessi DR. Elevated SGK1 predicts resistance of breast cancer cells to Akt inhibitors. Biochem J. 2013;452:499–508. doi: 10.1042/BJ20130342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal SP. Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: amplification of AKT1 in a primary human gastric adenocarcinoma. Proc Natl Acad Sci U S A. 1987;84:5034–5037. doi: 10.1073/pnas.84.14.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- Stokoe D, Stephens LR, Copeland T, Gaffney PR, Reese CB, Painter GF, Holmes AB, McCormick F, Hawkins PT. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, Raghuraman H, Cunningham JM, Gupta M, Gupta MP. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal. 2011;4:ra46. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yamaguchi MT, Ohteki T, Sasaki T, Kaisho T, Kimura Y, Yoshida R, Wakeham A, Higuchi T, Fukumoto M, et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 2001;14:523–534. doi: 10.1016/s1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- Tang H, Inoki K, Lee M, Wright E, Khuong A, Khuong A, Sugiarto S, Garner M, Paik J, DePinho RA, et al. mTORC1 promotes denervation-induced muscle atrophy through a mechanism involving the activation of FoxO and E3 ubiquitin ligases. Sci Signal. 2014;7:ra18. doi: 10.1126/scisignal.2004809. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci U S A. 2002;99:467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zhang Y, Xu W, Harden TK, Sondek J, Sun L, Li L, Wu D. A PLCbeta/PI3Kgamma-GSK3 signaling pathway regulates cofilin phosphatase slingshot2 and neutrophil polarization and chemotaxis. Dev Cell. 2011;21:1038–1050. doi: 10.1016/j.devcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Coll JT, Austen DA, Hsiao HM, Swenson L, Jain J. Structure of GSK3beta reveals a primed phosphorylation mechanism. Nat Struct Biol. 2001;8:593–596. doi: 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- Tschopp O, Yang ZZ, Brodbeck D, Dummler BA, Hemmings-Mieszczak M, Watanabe T, Michaelis T, Frahm J, Hemmings BA. Essential role of protein kinase B gamma (PKB gamma/Akt3) in postnatal brain development but not in glucose homeostasis. Development. 2005;132:2943–2954. doi: 10.1242/dev.01864. [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Kandror KV. Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation. Mol Cell Biol. 2006;26:63–76. doi: 10.1128/MCB.26.1.63-76.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- van der Vos KE, Coffer PJ. The extending network of FOXO transcriptional target genes. Antioxid Redox Signal. 2011;14:579–592. doi: 10.1089/ars.2010.3419. [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, Chen ZC, Tanos B, Oldrini B, Hsieh WY, Yannuzzi N, Campos C, Mellinghoff IK. A kinase-independent function of AKT promotes cancer cell survival. Elife. 2014;3 doi: 10.7554/eLife.03751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Leavens KF, Hunter RW, Koren S, von Wilamowitz-Moellendorff A, Lu M, Satapati S, Chu Q, Sakamoto K, Burgess SC, et al. A noncanonical, GSK3-independent pathway controls postprandial hepatic glycogen deposition. Cell Metab. 2013;18:99–105. doi: 10.1016/j.cmet.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Huang X, Sun D, Xin X, Pan Q, Peng S, Liang Z, Luo C, Yang Y, Jiang H, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates Akt signaling. PLoS One. 2012;7:e37427. doi: 10.1371/journal.pone.0037427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Viscarra J, Kim SJ, Sul HS. Transcriptional regulation of hepatic lipogenesis. Nat Rev Mol Cell Biol. 2015;16:678–689. doi: 10.1038/nrm4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani R, Qian J, Yin L, Bechtold E, King SB, Poole LB, Paek E, Tsang AW, Furdui CM. Isoform-specific regulation of Akt by PDGF-induced reactive oxygen species. Proc Natl Acad Sci U S A. 2011;108:10550–10555. doi: 10.1073/pnas.1011665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014;39:159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Hengstschlager M, Linke M. Regulation of innate immune cell function by mTOR. Nat Rev Immunol. 2015;15:599–614. doi: 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JA, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisinski KB, Tevaarwerk AJ, Burkard ME, Rampurwala M, Eickhoff J, Bell MC, Kolesar JM, Flynn C, Liu G. Phase I Study of an AKT Inhibitor (MK-2206) Combined with Lapatinib in Adult Solid Tumors Followed by Dose Expansion in Advanced HER2+ Breast Cancer. Clin Cancer Res. 2016;22:2659–2667. doi: 10.1158/1078-0432.CCR-15-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PZ, Chen ML, Jeon SM, Peng XD, Hay N. The effect Akt2 deletion on tumor development in Pten(+/−) mice. Oncogene. 2012;31:518–526. doi: 10.1038/onc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat Struct Biol. 2002;9:940–944. doi: 10.1038/nsb870. [DOI] [PubMed] [Google Scholar]
- Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi KH, Lauring J. Recurrent AKT mutations in human cancers: functional consequences and effects on drug sensitivity. Oncotarget. 2016;7:4241–4251. doi: 10.18632/oncotarget.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- You JS, Anderson GB, Dooley MS, Hornberger TA. The role of mTOR signaling in the regulation of protein synthesis and muscle mass during immobilization in mice. Dis Model Mech. 2015;8:1059–1069. doi: 10.1242/dmm.019414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Savage RE, Eathiraj S, Meade J, Wick MJ, Hall T, Abbadessa G, Schwartz B. Targeting AKT1-E17K and the PI3K/AKT Pathway with an Allosteric AKT Inhibitor, ARQ 092. PLoS One. 2015;10:e0140479. doi: 10.1371/journal.pone.0140479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Yang T, Li L, Sun L, Hou Y, Hu X, Zhang L, Tian H, Zhao Q, Peng J, et al. TSC1 controls macrophage polarization to prevent inflammatory disease. Nat Commun. 2014;5:4696. doi: 10.1038/ncomms5696. [DOI] [PubMed] [Google Scholar]
- Zhuang G, Yu K, Jiang Z, Chung A, Yao J, Ha C, Toy K, Soriano R, Haley B, Blackwood E, et al. Phosphoproteomic analysis implicates the mTORC2-FoxO1 axis in VEGF signaling and feedback activation of receptor tyrosine kinases. Sci Signal. 2013;6:ra25. doi: 10.1126/scisignal.2003572. [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Moelling K. Phosphorylation and regulation of Raf by Akt (protein kinase B) Science. 1999;286:1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]