Abstract
The RNA-binding proteins that comprise the La-related protein (LARP) superfamily have been implicated in a wide range of cellular functions, from tRNA maturation to regulation of protein synthesis. To more expansively characterize the biological function of the LARP6 subfamily, we have recombinantly expressed the full-length LARP6 proteins from two teleost fish, platyfish (Xiphophorus maculatus) and zebrafish (Danio rerio). The yields of the recombinant proteins were enhanced to > 2 mg/L using a tandem approach of an N-terminal His6-SUMO tag and an iterative solubility screening assay to identify structurally stabilizing buffer components. The domain topologies of the purified fish proteins were probed with limited proteolysis. The fish proteins contain an internal, protease-resistant 40 kDa domain, which is considerably more stable than the comparable domain from the human LARP6 protein. The fish proteins are therefore a lucrative model system in which to study both the evolutionary divergence of this family of La-related proteins and the structure and conformational dynamics of the domains that comprise the LARP6 protein.
Keywords: RNA binding protein, La-related protein, bacterial expression, protein solubility
Introduction
The La-related superfamily of RNA-binding proteins are implicated in a wide range of eukaryotic cellular activities, from tRNA maturation to the regulation of protein synthesis (reviewed in [1]). Each member of this family binds to its respective RNA ligands using a bipartite RNA binding domain termed the “La Module”, which consists of a well-conserved La motif and an RNA Recognition Motif (Fig. 1). Of the five La-related protein subfamilies, the identification of the biochemical and cellular functions of LARP6 has been the most elusive. The Genuine La protein (now called LARP3) binds to the 3′ oligopyrimidine end of pre-tRNAs to assist maturation[2]; LARP7 similarly binds to the 3′ end of lncRNAs [3]. LARP1 and LARP4 bind to the 5′ and 3′ untranslated regions of mRNAs to regulate translation and degradation [4-7]. In contrast, there is no analogous cellular function yet identified for the LARP6 subfamily. Only one endogenous ligand has been identified: in mammals, LARP6 binds to a secondary structure element in the 5′ untranslated region (5′UTR) of type I collagen mRNAs [8,9]. Recently, the high-resolution structures of the individual La motif domain and the RRM domain of human LARP6 protein were solved by solution NMR [10]. The biochemical experiments that accompanied these structures demonstrated that the interdomain linker between the La motif and the RRM plays a critical role in the RNA binding activity [10].
Figure 1. Expression of fish LARP6 proteins and filter-based screening assay.
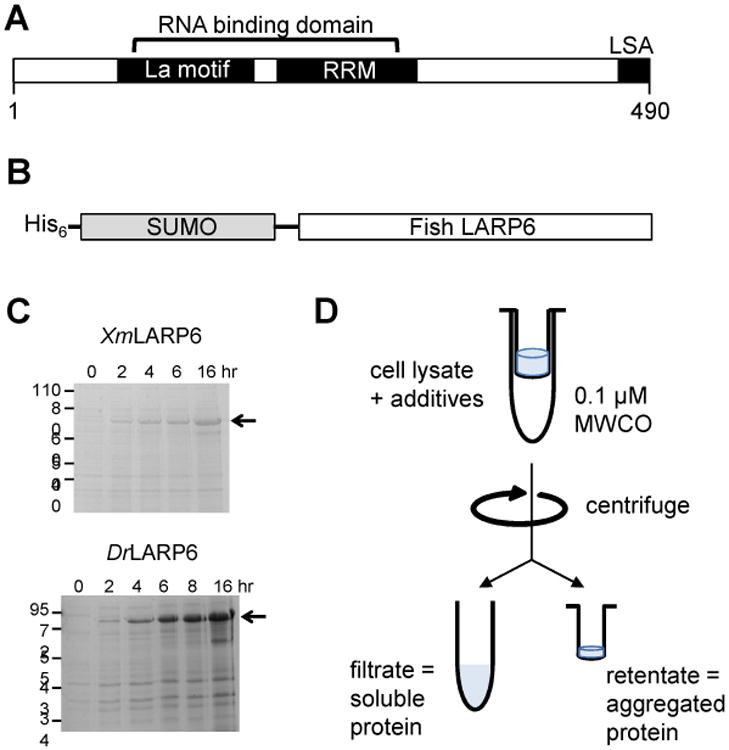
(A) The LARP6 protein superfamily shares a bipartite RNA binding domain, consisting of a La motif and an RNA recognition motif, and a C-terminal LSA motif. (B) The fish LARP6 proteins were cloned into a bacterial expression vector as a fusion with an N-terminal His6-SUMO solubility tag. (C) Both the Xiphophorus maculatus (Xm) LARP6 and Danio rerio (Dr) LARP6a proteins are robustly expressed in Rosetta (DE3) Escherichia coli cells. (D) The centrifugal filter assay for screening buffer additives that promote protein solubility.
Multiple sequence alignments of eukaryotic LARP6 sequences have identified significant regions of conservation, most notably in the La motif, the core of the RRM, and a C-terminal proline-rich domain called the LSA motif (Supp. Fig. 1) [6,11]. However, considerable evolutionary divergence exists between these conserved regions, including within the biochemically relevant interdomain linker (Supp. Fig. 1). Additionally, sequence divergence is found in the extended loops between beta strands of the RRM, which may form part or all of the RNA binding surface (Supp. Fig. 1). To test the effect of these sequence variations on the structure and function of the LARP6 protein, the LARP6 proteins from non-mammalian vertebrates should be studied closely.
Here, we report the successful expression and purification of recombinant zebrafish and platyfish LARP6 proteins. Recombinant expression in bacteria was significantly enhanced using the SUMO fusion system [12] (Fig. 1). We then identified optimal buffer conditions using an iterative filter-based screening assay for buffer additives that would enhance stability, which was originally developed for aggregation-prone intrinsically disordered proteins [13,14].
Materials and Methods
Plasmid constructs
The coding sequence for Xiphophorus maculatus LARP6 was directly amplified from X. maculatus cDNA, a kind gift of R. Walter (Texas State University). Apparent nonspecific binding of PCR primers to Danio rerio cDNA required that the coding sequence for D. rerio LARP6a be commercially synthesized as a pcDNA3.1 construct (GenScript, Piscataway, NJ). The pcDNA3.1-LARP6a construct was then used as the template for amplification of the LARP6a coding sequence. Both PCR products were digested with BamHI and XhoI, and ligated into the pET28a-SUMO expression vector (a kind gift of C. Lima, Rockefeller University, and Cornell University). The resulting plasmids were confirmed for correct fragment insertion by Sanger sequencing (Quintara Biosciences). The expression construct for human LARP6 protein, pET28a-HsLARP6, was a kind gift of M. Bayfield (York University).
Recombinant expression
The expression of human LARP6 was performed as described [15]. Expression constructs for the two fish LARP6 proteins were transformed into chemically-competent BL21(DE3) or Rosetta(DE3) expression lines of Escherichia coli (the former a kind gift of R. McLean, Texas State University). One liter cultures were grown at 37 °C until an optical density of 0.6 – 0.8, then cooled on ice before inducing expression with 1 mM IPTG for 8 hours at 18°C. Cells were harvested by centrifugation at 5000×g at 4°C for 5 min. The cell pellet was transferred to a conical vial and stored at -20°C until purification.
Purification of LARP6 proteins
The human LARP6 protein was performed as described [15]. The fish LARP6 proteins were purified essentially as described for other His6-SUMO-tagged proteins [16]. Briefly, cell pellets were thawed on ice before resuspension into lysis buffer (50 mM NaH2PO4/ Na2HPO4 [pH 8.0], 200 mM NaCl, 0.5 M glucose, 300 mM NaI and 10 mM imidazole [pH 8.0])containing protease inhibitor cocktail tablet (Pierce). Resuspended cells were sonicated and cell debris pelleted by centrifugation at 20,000×g at 4°C for 20 min. The cleared lysate was incubated with nickel-NTA resin for 1 – 2 hr at 4°C with mixing. Following a wash of the resin with buffer + 50 mM imidazole, and the tagged proteins eluted batchwise with lysis buffer + 300 mM imidazole. The eluate was analyzed by SDS-PAGE and Coomassie blue staining. LARP6-containing fractions were pooled and concentrated to < 3 mL in a 5000 MWCO centrifugal concentrator (Sartorius) and application to a Sephadex 200 gel filtration column pre-equilibrated in 50 mM NaH2PO4/ Na2HPO4 [pH 8.0], 200 mM NaCl, 0.5 M glucose, 300 mM NaI. Fractions containing His6-SUMO-LARP6 were identified by chromatogram and confirmed by SDS-PAGE and Coomassie blue staining, and then pooled.
To cleave the fusion tag from the fish proteins, the pooled fractions were incubated with 1000:1 molar ratio of target protein:Ulp1 protease at 16°C for 2 hours. The His6-SUMO tag was subsequently removed from the mixture by batchwise purification over fresh nickel-NTA resin. The flowthrough was concentrated and subjected to a final purification and buffer exchange over an S200 column pre-equilibrated as described above. Fractions were identified by chromatogram and confirmed by SDS-PAGE and Coomassie blue staining, pooled, concentrated as described above, and stored at -70°C.
Solubility screening assay
To identify buffer conditions that would promote stability of both XmLARP6 and DrLARP6a, a filter-based screening assay was performed essentially as described [13]. Briefly, the cleared cell lysate was treated with one of a series of additives, incubated on ice for 2 h, and filtered through a 0.1 μm microcentrifugal filter (EMD Millipore) at 4°C. The filter was rinsed with 1× SDS sample buffer (5×: 0.5 M Tris [pH 7.5 at 4 °C], 4 mM βME, 0.4 M SDS) to release aggregated proteins within the retentate from the filter surface. Both the filtrate and retentate were analyzed by SDS-PAGE and Western blotting using HisProbe-HRP conjugate (ThermoFisher) against the polyhistidine tag of the fusion protein. Buffer additives that increased the amount of either full-length protein or smaller His-reactive species in the filtrate were considered “hits”, and used to determine the next round of additives to screen.
Limited proteolysis
The purified human, platyfish, and zebrafish LARP6 proteins were diluted to ∼0.5 mg/mL into reaction mixes so that each contained equal final concentrations of salts and additives (150 mM NaI, 250 mM glucose, and 250 mM NaCl). After removing a sample for a zero timepoint, trypsin was added at a mass ratio of 60:1 LARP6:trypsin and the reaction incubated at 4°C. Aliquots were removed at the indicated timepoints and the reaction stopped with the addition of 5× SDS sample buffer. Samples were analyzed by gel electrophoresis on 10% polyacrylamide gel and either silver staining or Western blotting using a commercial antibody raised against the C-terminus of the human LARP6 protein (Abcam, cat. no. ab94640).
Results
Buffer additive screening
Initial expression of SUMO-tagged XmLARP6 and DrLARP6a in Rosetta (DE3) cells was robust (Fig. 1). However, the proteins exhibited aggregation and degradation during purification and storage in the same buffers used for the human LARP6 protein [15] (see size exclusion chromatogram below). To optimize the buffer conditions for the two recombinant fish proteins, an iterative buffer additive screen was performed to identify helpful solubility-promoting agents for XmLARP6 [13]. Briefly, cleared cell lysate was incubated with an additive for 2 h before separating soluble and insoluble proteins through a 0.1 μm centrifugal filter. The amount of His6-tagged SUMO-XmLARP6 fusion protein in both the flowthrough and retentate was measured by Western blot. We found that the vast majority of the His6-SUMO-XmLARP6 protein always flowed through the filter, presumably due to the stabilizing influence of the N-terminal His6-SUMO tag. As a result, we used two strategies to identify solubilizing agents: (1) the increased presence of smaller molecular weight His-reactive species, which represent C-terminal truncations of the fusion protein, and (2) the relative amount of His6-SUMO-XmLARP6 in the retentate fractions, which was often only visible in overexposed images.
The first round of screening identifies the category of additive that works best: kosmotrope, osmolyte, chaotrope, amino acid, and/or detergent. All five categories of additives improve the solubility of XmLARP6 degradation products, as shown by the increase in soluble degradation products in the filtrate and decrease in the amount of insoluble full-length protein in the retentate (Fig. 2A). Of the six additives tested, the largest improvement in His6-SUMO-XmLARP6 stability was induced by osmolytes (arginine, glycerol, and TMAO) and a chaotrope (urea). For the second round of screening, we tested the effect of several different osmolytes and weak chaotropes on protein solubility (Fig. 2B). Of the osmolytes, glycerol and glucose most significantly enhanced the fraction of soluble protein. Of the chaotropes, urea and sodium iodide were found to be equally effective at reducing the amount of aggregated protein in the retentate. In contrast, the chloride salts induced extensive precipitation in both solutions, preventing any protein from being recovered in the flowthrough nor from the filter membrane.
Figure 2. Initial buffer screening to promote protein solubility.
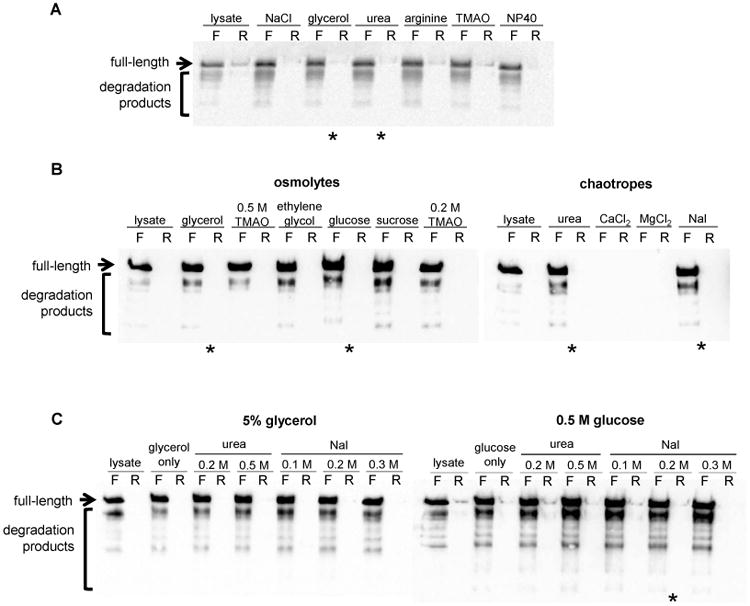
(A) The first round of additive screening evaluated several general chemical properties of additives, including chaotrope, kosmotrope, osmolyte, and amino acid. (B) The second round screened for specific chemicals within those categories.
The last set of screens was designed to screen optimal concentrations of the osmolytes and chaotropes, as well as assess the benefit of combinations thereof. Protein solubility was reduced with higher concentrations of both glycerol and glucose (Fig. 3). The combination of 0.5 M glucose and 0.3 M NaI increased solubility of both full-length and shorter fragments while minimizing the amount of aggregated protein in the retentate.
Figure 3. Final screening of buffer additive concentrations and conditions.
Multiple concentrations of the osmolytes glycerol and glucose were evaluated in combination with the indicated concentrations of the weak chaotropes urea and sodium iodide (NaI) to identify optimal conditions to promote the solubility of His6-SUMO-XmLARP6.
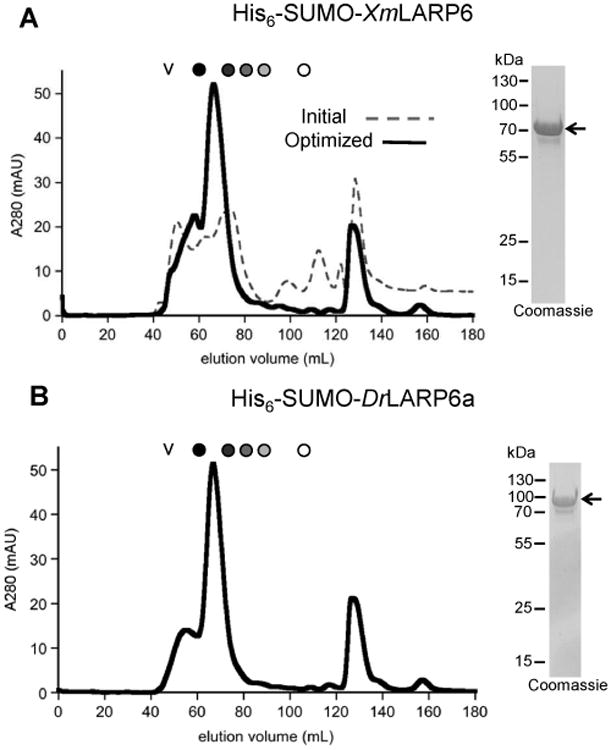
Purification of recombinant proteins with optimized buffer
Lysis, nickel affinity chromatography, and size exclusion chromatography of His6-SUMO-XmLARP6 was performed using the optimized buffer containing 0.5 M glucose and 0.3 M NaI. The improvement in protein solubility is clear when comparing the size exclusion chromatograms from the initial and optimized buffer conditions (Fig. 4). Conveniently, the same buffer conditions also enabled the purification of homogenous, soluble His6-SUMO-DrLARP6a protein (Fig. 4). These improved conditions produced yields of > 2 mg of tagged protein per liter of bacterial culture. To finish the purification, Ulp1 protease was used to cleave the His6-SUMO tag, and the resulting fish proteins polished by a final size exclusion column in the optimized buffer.
Figure 4. Optimized buffer conditions produce higher yields of SUMO-tagged LARP6 proteins.
(A) The comparison of the size exclusion chromatograms of SUMO-tagged XmLARP6 in the initial buffer conditions (gray dashed line) and the optimized buffer with 0.5 M glucose and 0.3 M NaI (black solid line) show an increase in soluble protein. The resulting eluate is >98% pure as analyzed by SDS-PAGE and Coomassie stain (right panel). (B) The same buffer conditions are appropriate for the SUMO-tagged DrLARP6a protein, as analyzed by both size exclusion chromatogram (left) and Coomassie-stained SDS-PAGE (right panel).
Analysis of domain topology
The solubility stabilizing additives precluded a straightforward analysis of protein secondary structure by circular dichroism. Therefore, limited proteolysis was used to compare the domain topology of the purified human and fish proteins in the presence of the additives. To control for any common effects that the purification buffer additives may have on global protein structure, all three LARP6 proteins were diluted into a standard reaction buffer so that all salts and additives were at identical concentrations (150 mM NaI, 250 mM glucose, and 250 mM NaCl). Both fish proteins contain a highly protease-resistant domain of ∼ 40 kDa (Fig. 5). The human LARP6 protein also contains a similarly-sized domain that is slightly protease-resistant (Fig. 5). Despite the considerable sequence homology between the fish and human LARP6 proteins, the 40 kDa domain in the human protein is not formed as readily nor persists as long as those of the two fish proteins. Western blotting was then performed using an antibody raised against the C-terminus of the human protein, which cross-reacts with the highly conserved C-terminal sequence of the fish proteins. The antibody does not recognize the stable ∼40 kDa domain from any of the three LARP6 proteins, strongly suggesting that it is comprised of residues internal to the protein.
Figure 5. Fish LARP6 proteins contain a central protease-resistant domain.
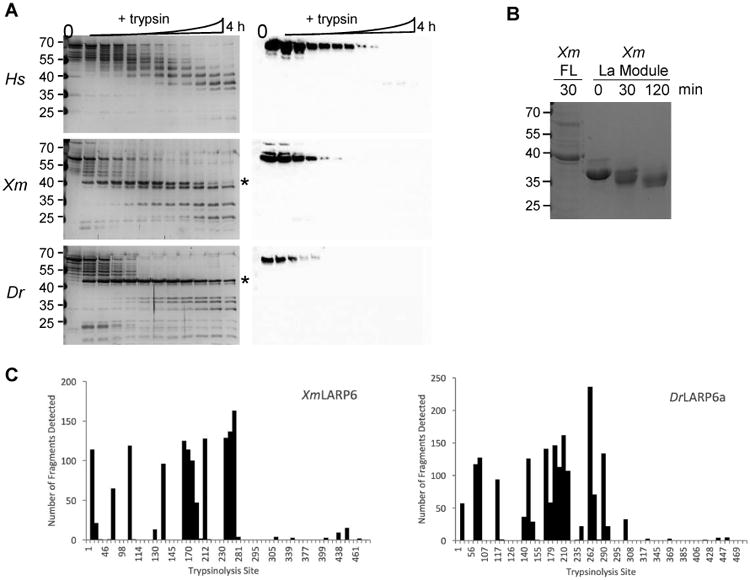
Limited trypsinolysis of the human (Hs), platyfish (Xm), and zebrafish (Dr) LARP6 proteins was analyzed by silver stain (left panels) and Western blot against the C-terminus of LARP6 (right panels). Timepoints (L-R) were: 0
Discussion
We report the novel expression and purification of two non-mammalian vertebrate LARP proteins, the LARP6 protein from Xiphophorus maculatus (Xm, platyfish) and the LARP6a protein from Danio rerio (Dr, zebrafish). Future biochemical and biophysical studies will be able to capitalize on the evolutionary divergence between these two fish proteins and the previously studied human LARP6 protein, to identify regions that are critical for protein structure and function.
To improve the yield of the fish protein preparations, we used two complementary strategies. First, the well-characterized SUMO fusion system was used to provide an affinity purification tag in the N-terminal His6 moiety as well as a stabilizing, well-folded N-terminal domain. Next, we employed a filter-based iterative screen to identify chemical additives to the purification buffers that enhanced protein stability. This screen was originally designed to promote the solubility of aggregation-prone natively-disordered proteins [13]. While the bipartite RNA binding domain of LARP6 is globular, very little is known about the structure outside of that domain. With three recombinant LARP6 proteins from evolutionarily diverged species in hand, we are now able to carry out detailed biochemical and structural analyses of the conserved and specialized functions of these proteins.
Analysis of the primary protein sequences by PONDR-FIT predicts that nearly two-thirds of the protein may be disordered (Supp. Fig. 2) [17]. Although the buffer components precluded analysis of secondary structure by CD, all three LARP6 proteins were shown to be folded by limited proteolysis. Those data demonstrate that there are structured, protease-resistant regions that comprise at least half of the apparent molecular mass of the full-length protein (Fig. 5). Even so, the C-terminal portion of LARP6 from all three species is consistently predicted to be disordered (Supp. Fig. 2). Therefore, we have demonstrated that the filter-based aggregation screen can be successfully applied to proteins that contain both a stably-folded domain and potentially disordered regions [13].
The robust production of recombinant fish LARP6 proteins demonstrates that comparative phylogenetics can be readily extended into biochemical studies of La-related protein structure and function. These reagents will allow the field to directly address critical questions about the function of evolutionary sequence divergence within loops and linker regions of the LARP6 family of proteins. By comparing and contrasting the biophysical and biochemical properties of these regions from three evolutionarily divergent LARP6 proteins, the field will be able to identify the physiological function of the C-terminal portion of LARP6 as well as critical conformational features of the RNA binding domain that contribute to function. Ultimately, the biochemical data that will be produced using these proteins will also provide a strong foundation for future genetic studies in the widely used model organisms of zebrafish and platyfish, to further characterize the role of LARP6 in development and physiology.
Supplementary Material
Supplemental Figure 1. Multiple sequence alignment of vertebrate LARP6 RNA binding domains (La motif + RRM). The secondary structures as solved by solution NMR are depicted as diagram above the sequence. The elongated loops between beta strands of the RRM are highlighted (L1, red; L3, blue; L5, green).
Supplemental Figure 2. The full-length sequences of HsLARP6, XmLARP6, and DrLARP6a were analyzed by four disorder prediction algorithms: VSL2B (blue), VL3 (red), VLXT (gray), and PONDR-FIT (green). The low predicted disorder across residues ∼80 - 270 in all three proteins represents the structured, bipartite RNA binding domain.
Highlights.
- Novel expression and purification of uncharacterized, post-transcriptional regulatory LARP6 proteins from fish
- Tandem solubility tag and buffer composition screens significantly enhanced quality and amount of recovered protein
- Well-folded subdomain in fish protein encompasses both N-terminus and RNA binding module, indicating intramolecular domain interactions
Acknowledgments
This work was supported by the National Institutes of Health – National Institute of General Medical Sciences through an Institutional Bridges to Doctorate grant (GM102783, supporting J.M.C.) and an Academic Research Enhancement Award (GM119096 to K.A.L.). J.M.C. was additionally supported by scholarships from the United States Department of Agriculture through the Food Safety and Agroterrorism Training: Educating our Future Workforce (USDA 2011-38422-30832) and Southwest Agriculture and Food Security Education (USDA 2014-38422-22084) programs.
Abbreviations
- RRM
RNA recognition motif
- LARP
La-related protein
- b-ME
beta-mercaptoethanol
- SDS
sodium dodecyl sulfate
- lncRNA
long non-coding RNA
- PAGE
polyacrylamide gel electrophoresis
- Ni-NTA
nickel nitrilotriacetic acid agarose
- HRP
horseradish peroxidase
- CD
circular dichroism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stavraka C, Blagden S. The La-Related Proteins, a Family with Connections to Cancer. Biomolecules. 2015;5:2701–2722. doi: 10.3390/biom5042701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Y, Bayfield MA, Intine RV, Maraia RJ. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol. 2006;13:611–618. doi: 10.1038/nsmb1110. [DOI] [PubMed] [Google Scholar]
- 3.Eichhorn CD, Chug R, Feigon J. hLARP7 C-terminal domain contains an xRRM that binds the 3′ hairpin of 7SK RNA. Nucleic Acids Res. 2016:gkw833. doi: 10.1093/nar/gkw833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki K, Adachi S, Homoto M, Kusano H, Koike K, Natsume T. LARP1 specifically recognizes the 3′ terminus of poly(A) mRNA. FEBS Lett. 2013;587:2173–2178. doi: 10.1016/j.febslet.2013.05.035. [DOI] [PubMed] [Google Scholar]
- 5.Yang R, Gaidamakov SA, Xie J, Lee J, Martino L, Kozlov G, et al. La-Related Protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w motif, and can promote mRNA stability. Mol Cell Biol. 2011;31:542–556. doi: 10.1128/MCB.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merret R, Martino L, Bousquet-Antonelli C, Fneich S, Descombin J, Billey É, et al. The association of a La module with the PABP-interacting motif PAM2 is a recurrent evolutionary process that led to the neofunctionalization of La-related proteins. Rna. 2012;19:36–50. doi: 10.1261/rna.035469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahr RM, Mack SM, Héroux A, Blagden SP, Bousquet-Antonelli C, Deragon JM, et al. The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5′TOP sequence. Nucleic Acids Res. 2015;43:8077–8088. doi: 10.1093/nar/gkv748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schartl M. Beyond the zebrafish: diverse fish species for modeling human disease. Dis Models Mech. 2014;7:181–192. doi: 10.1242/dmm.012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai L, Fritz D, Stefanovic L, Stefanovic B. Binding of LARP6 to the Conserved 5′ Stem–Loop Regulates Translation of mRNAs Encoding Type I Collagen. J Mol Biol. 2010;395:309–326. doi: 10.1016/j.jmb.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martino L, Pennell S, Kelly G, Busi B, Brown P, Atkinson RA, et al. Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module. Nucleic Acids Res. 2015;43:645–660. doi: 10.1093/nar/gku1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousquet-Antonelli C, Deragon JM. A comprehensive analysis of the La-motif protein superfamily. Rna. 2009;15:750–764. doi: 10.1261/rna.1478709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mossessova E, Lima CD. Ulp1-SUMO Crystal Structure and Genetic Analysis Reveal Conserved Interactions and a Regulatory Element Essential for Cell Growth in Yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/S1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 13.Churion KA, Bondos SE. Identifying solubility-promoting buffers for intrinsically disordered proteins prior to purification. Methods in Molecular Biology. 2012;896:415–427. doi: 10.1007/978-1-4614-3704-8_28. [DOI] [PubMed] [Google Scholar]
- 14.Bondos SE, Bicknell A. Detection and prevention of protein aggregation before, during, and after purification. Analytical Biochemistry. 2003;316:223–231. doi: 10.1016/s0003-2697(03)00059-9. [DOI] [PubMed] [Google Scholar]
- 15.Hussain RH, Zawawi M, Bayfield MA. Conservation of RNA chaperone activity of the human La-related proteins 4, 6 and 7. Nucleic Acids Res. 2013;41:8715–8725. doi: 10.1093/nar/gkt649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao T, Lubin JW, Armstrong GS, Tucey TM, Lundblad V, Wuttke DS. Structure of Est3 reveals a bimodal surface with differential roles in telomere replication. Proceedings of the National Academy of Sciences. 2014;111:214–218. doi: 10.1073/pnas.1316453111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Multiple sequence alignment of vertebrate LARP6 RNA binding domains (La motif + RRM). The secondary structures as solved by solution NMR are depicted as diagram above the sequence. The elongated loops between beta strands of the RRM are highlighted (L1, red; L3, blue; L5, green).
Supplemental Figure 2. The full-length sequences of HsLARP6, XmLARP6, and DrLARP6a were analyzed by four disorder prediction algorithms: VSL2B (blue), VL3 (red), VLXT (gray), and PONDR-FIT (green). The low predicted disorder across residues ∼80 - 270 in all three proteins represents the structured, bipartite RNA binding domain.


