Abstract
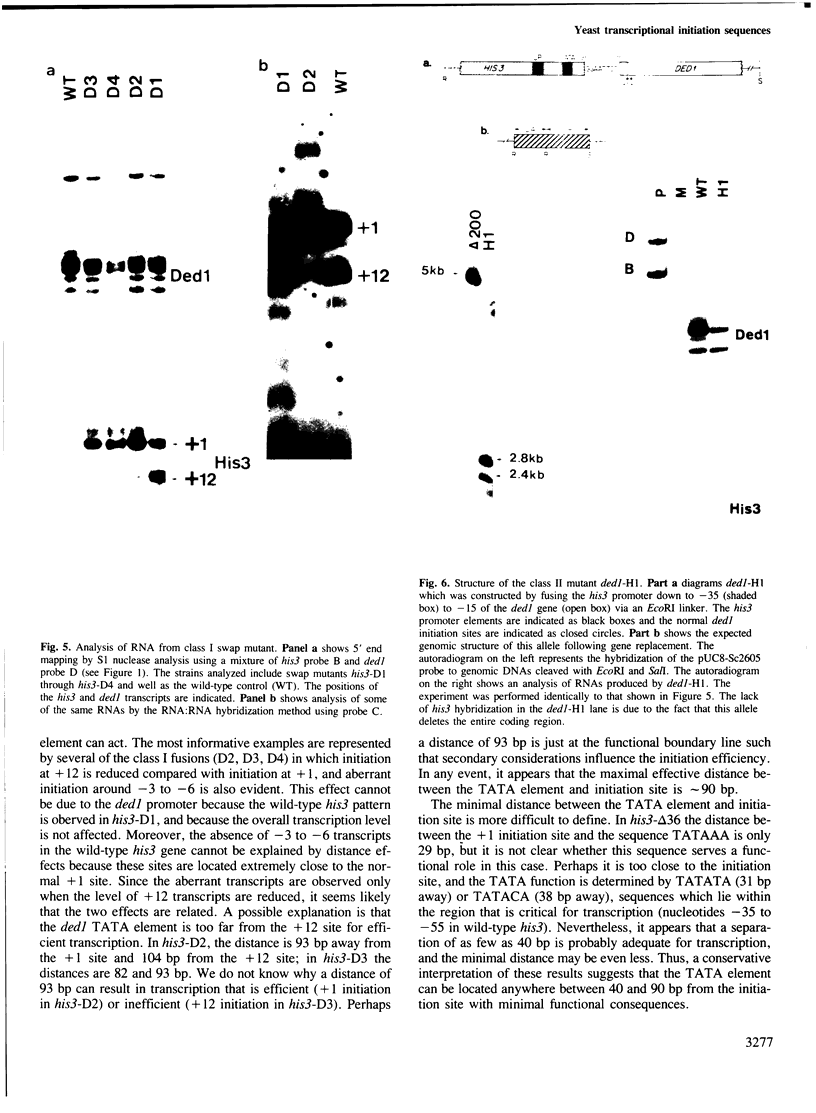
We present evidence suggesting that accurate mRNA initiation in yeast cells, unlike their higher eukaryotic counterparts, is determined primarily by specific sequences downstream from the TATA element. First, changing the distance between the his3 TATA element and the initiation region does not affect the sites of initiation or the level of RNA. Second, reciprocal his3-ded1 and ded1-his3 hybrid promoters containing the upstream and TATA elements of one gene fused to the mRNA coding region of the other gene initiate transcription at sites defined by wild-type mRNA coding sequences, not by the distance from the TATA element. Third, when the his3 or ded1 promoter region is fused to position +2 of the his3 gene, transcripts initiated from a position equivalent to +1 are not observed. The results also suggest that the spacing between the TATA element and initiation site is relatively flexible; distance ranging from 40 to 90 bp appear to be functionally acceptable.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker C. C., Herisse J., Courtois G., Galibert F., Ziff E. Messenger RNA for the Ad2 DNA binding protein: DNA sequences encoding the first leader and heterogenity at the mRNA 5' end. Cell. 1979 Oct;18(2):569–580. doi: 10.1016/0092-8674(79)90073-4. [DOI] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Will H., Hernandez N., Schaller H. Signals regulating hepatitis B surface antigen transcription. Nature. 1983 Sep 22;305(5932):336–338. doi: 10.1038/305336a0. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Mantei N., Weissmann C. DNA sequences preceding the rabbit beta-globin gene are required for formation in mouse L cells of beta-globin RNA with the correct 5' terminus. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1411–1415. doi: 10.1073/pnas.78.3.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P., Frisque R. J., Gluzman Y. Identification of a promoter component involved in positioning the 5' termini of simian virus 40 early mRNAs. Proc Natl Acad Sci U S A. 1981 Jan;78(1):100–104. doi: 10.1073/pnas.78.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S. Gel electrophoretic separation of the complementary strands of bacteriophage DNA. Virology. 1972 Jul;49(1):342–344. doi: 10.1016/s0042-6822(72)80042-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Jat P., Treisman R., Favaloro J., Folk W. R. 5' termini of polyoma virus early region transcripts synthesized in vivo by wild-type virus and viable deletion mutants. J Mol Biol. 1982 Aug 5;159(2):189–224. doi: 10.1016/0022-2836(82)90493-4. [DOI] [PubMed] [Google Scholar]
- Mathis D. J., Chambon P. The SV40 early region TATA box is required for accurate in vitro initiation of transcription. Nature. 1981 Mar 26;290(5804):310–315. doi: 10.1038/290310a0. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984 Jul;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. A physical, genetic and transcriptional map of the cloned his3 gene region of Saccharomyces cerevisiae. J Mol Biol. 1980 Jan 25;136(3):309–332. doi: 10.1016/0022-2836(80)90376-9. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Promotor mutants of the yeast his3 gene. J Mol Biol. 1981 Nov 5;152(3):553–568. doi: 10.1016/0022-2836(81)90268-0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Transcription of the his3 gene region in Saccharomyces cerevisiae. J Mol Biol. 1981 Nov 5;152(3):535–552. doi: 10.1016/0022-2836(81)90267-9. [DOI] [PubMed] [Google Scholar]
- Struhl K. Deletion mapping a eukaryotic promoter. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4461–4465. doi: 10.1073/pnas.78.7.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Direct selection for gene replacement events in yeast. Gene. 1983 Dec;26(2-3):231–241. doi: 10.1016/0378-1119(83)90193-2. [DOI] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington C. A., Leder P. Rescuing the in vitro function of a globin pseudogene promoter. Nature. 1982 Jul 8;298(5870):192–195. doi: 10.1038/298192a0. [DOI] [PubMed] [Google Scholar]