Abstract
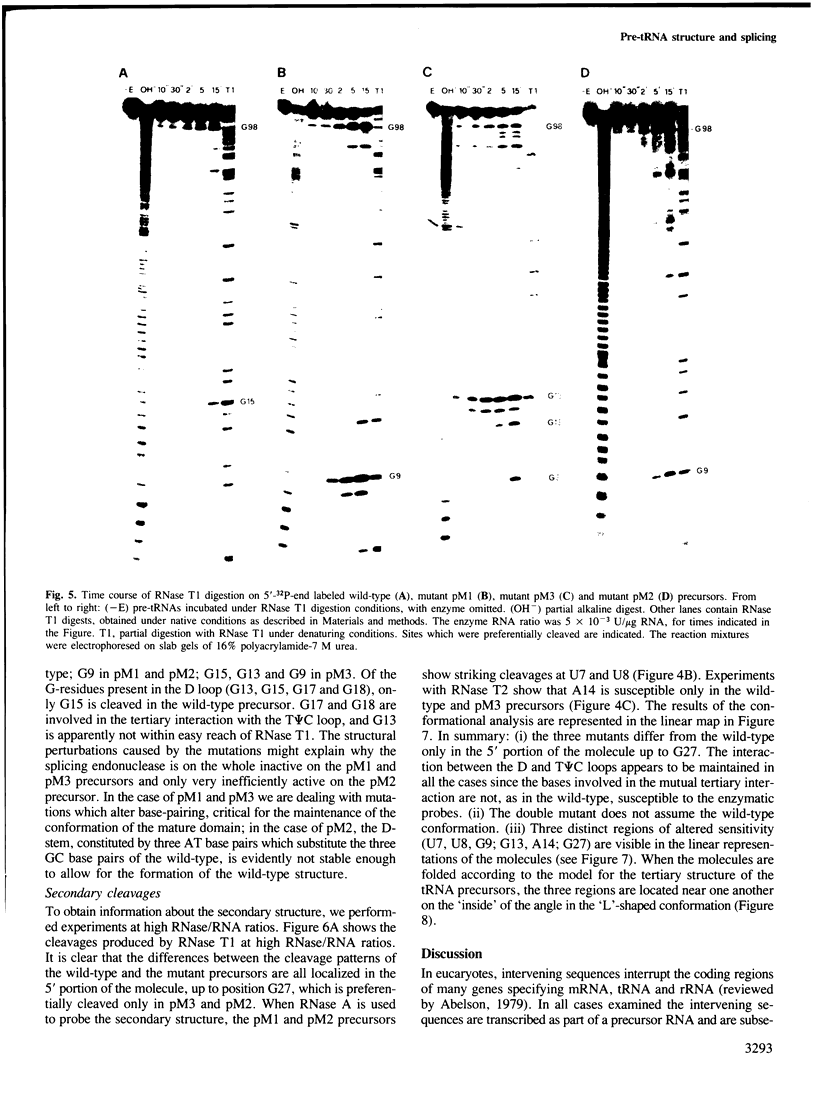
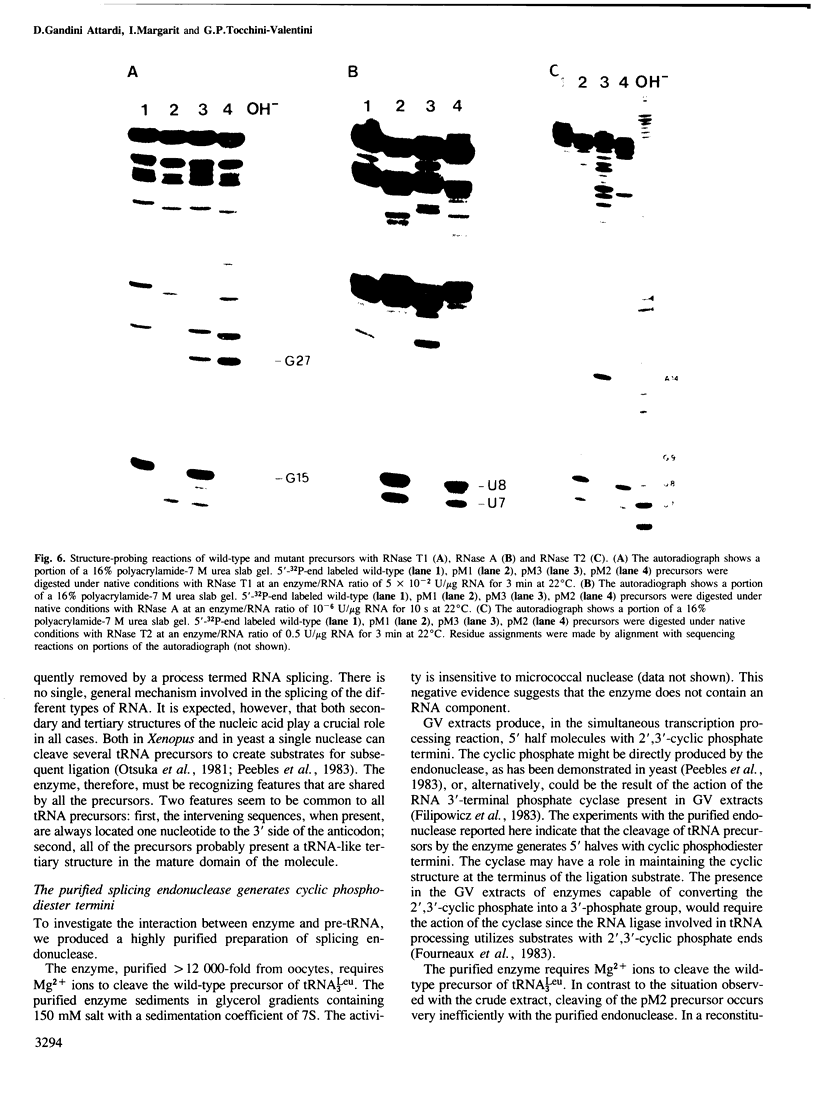
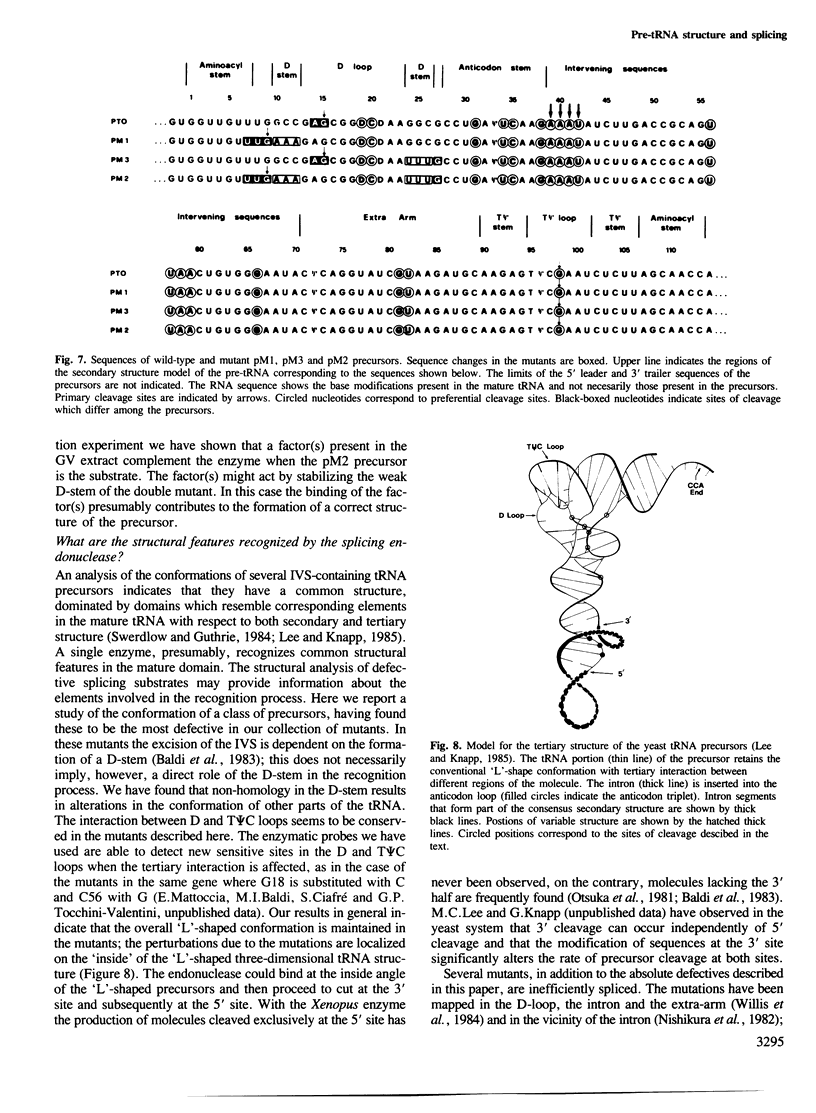
We have produced a highly purified preparation of the Xenopus laevis splicing endonuclease (XlaI RNase). The purified enzyme correctly cleaves tRNA precursors, creating substrates for subsequent ligation. The 5'-half molecules have a 2',3' cyclic phosphate at their 3' termini. Assuming that splicing enzymes recognize primarily structural elements in the 'mature domain', we have been studying the conformation of three splicing-defective precursors made from mutants of the yeast tRNALeu3 gene. The mutations alter base-pairing in the D-stem region and two of the mutants are absolute defectives. Enzymatic probing of the structures of the altered tRNA precursors shows that the structural perturbations in these mutants are localized on the 'inside' of the 'L'-shaped three-dimensional structure. The implications of this finding for the recognition process are discussed.
Full text
PDF
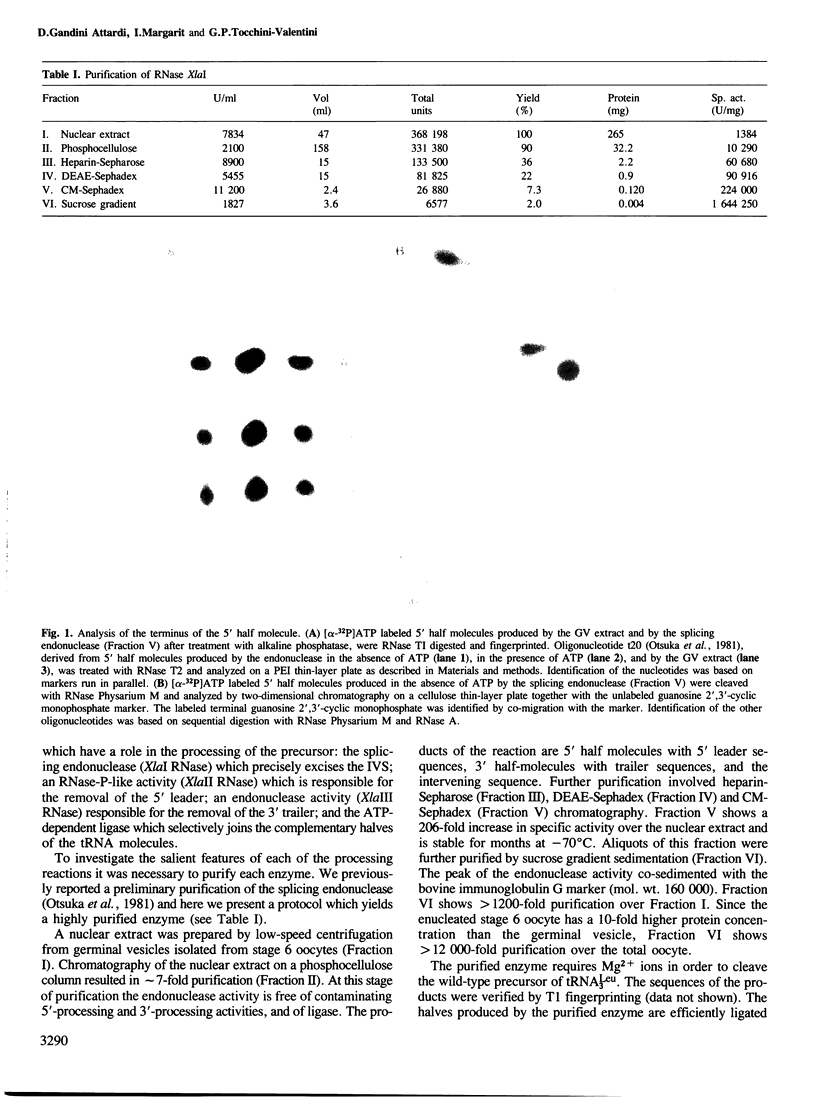

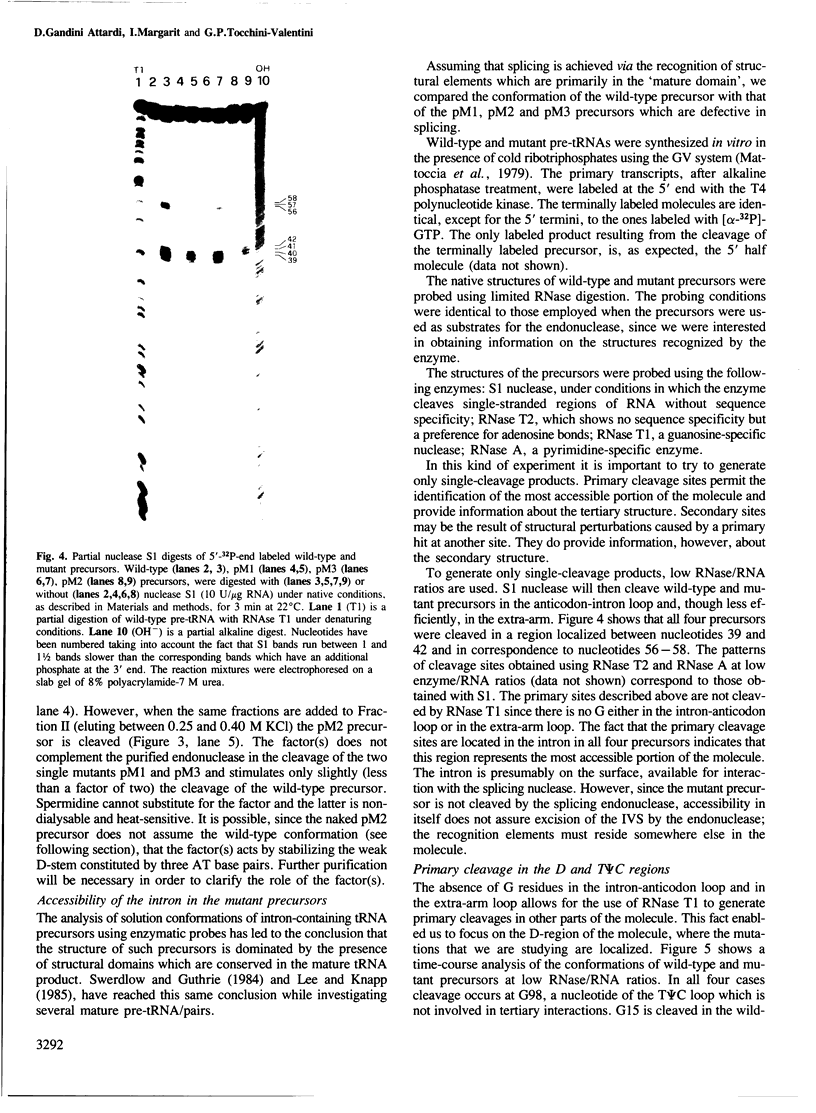


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Akaboshi E., Guerrier-Takada C., Altman S. Veal heart ribonuclease P has an essential RNA component. Biochem Biophys Res Commun. 1980 Sep 30;96(2):831–837. doi: 10.1016/0006-291x(80)91430-8. [DOI] [PubMed] [Google Scholar]
- Baldi M. I., Mattoccia E., Tocchini-Valentini G. P. Role of RNA structure in splicing: excision of the intervening sequence in yeast tRNA3leu is dependent on the formation of a D stem. Cell. 1983 Nov;35(1):109–115. doi: 10.1016/0092-8674(83)90213-1. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Konarska M., Gross H. J., Shatkin A. J. RNA 3'-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res. 1983 Mar 11;11(5):1405–1418. doi: 10.1093/nar/11.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Shatkin A. J. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983 Feb;32(2):547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- Furneaux H., Pick L., Hurwitz J. Isolation and characterization of RNA ligase from wheat germ. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3933–3937. doi: 10.1073/pnas.80.13.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. D., Ogden R., Johnson P., Abelson J., Dembeck P., Itakura K. Transcription and processing of a yeast tRNA gene containing a modified intervening sequence. Proc Natl Acad Sci U S A. 1980 May;77(5):2564–2568. doi: 10.1073/pnas.77.5.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Grabowski P. J., Padgett R. A., Sharp P. A. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature. 1985 Feb 14;313(6003):552–557. doi: 10.1038/313552a0. [DOI] [PubMed] [Google Scholar]
- Koontz S. W., Schimmel P. R. Aminoacyl-tRNA synthetase-catalyzed cleavage of the glycosidic bond of 5-halogenated uridines. J Biol Chem. 1979 Dec 25;254(24):12277–12280. [PubMed] [Google Scholar]
- Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J Biol Chem. 1985 Mar 10;260(5):3108–3115. [PubMed] [Google Scholar]
- Mattoccia E., Baldi M. I., Carrara G., Fruscoloni P., Benedetti P., Tocchini-Valentini G. P. Separation of RNA transcription and processing activities from X. laevis germinal vesicles. Cell. 1979 Nov;18(3):643–648. doi: 10.1016/0092-8674(79)90119-3. [DOI] [PubMed] [Google Scholar]
- Mattoccia E., Baldi M. I., Pande G., Ogden R., Tocchini-Valentini G. P. Mutation in the a block of the yeast tRNAleu3 gene that allows transcription but abolishes splicing and 5'-end maturation. Cell. 1983 Jan;32(1):67–76. doi: 10.1016/0092-8674(83)90497-x. [DOI] [PubMed] [Google Scholar]
- Nishikura K., Kurjan J., Hall B. D., De Robertis E. M. Genetic analysis of the processing of a spliced tRNA. EMBO J. 1982;1(2):263–268. doi: 10.1002/j.1460-2075.1982.tb01157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka A., de Paolis A., Tocchini-Valentini G. P. Ribonuclease "XlaI," an activity from Xenopus laevis oocytes that excises intervening sequences from yeast transfer ribonucleic acid precursors. Mol Cell Biol. 1981 Mar;1(3):269–280. doi: 10.1128/mcb.1.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Gegenheimer P., Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983 Feb;32(2):525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- Schoemaker H. J., Schimmel P. R. Effect of aminoacyl transfer RNA synthetases on H-5 exchange of specific pyrimidines in transfer RNAs. Biochemistry. 1977 Dec 13;16(25):5454–5460. doi: 10.1021/bi00644a009. [DOI] [PubMed] [Google Scholar]
- Starzyk R. M., Koontz S. W., Schimmel P. A covalent adduct between the uracil ring and the active site of an aminoacyl tRNA synthetase. Nature. 1982 Jul 8;298(5870):136–140. doi: 10.1038/298136a0. [DOI] [PubMed] [Google Scholar]
- Starzyk R., Schoemaker H., Schimmel P. Covalent enzyme-RNA complex: a tRNA modification that prevents a covalent enzyme interaction also prevents aminoacylation. Proc Natl Acad Sci U S A. 1985 Jan;82(2):339–342. doi: 10.1073/pnas.82.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow H., Guthrie C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J Biol Chem. 1984 Apr 25;259(8):5197–5207. [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Willis I., Hottinger H., Pearson D., Chisholm V., Leupold U., Söll D. Mutations affecting excision of the intron from a eukaryotic dimeric tRNA precursor. EMBO J. 1984 Jul;3(7):1573–1580. doi: 10.1002/j.1460-2075.1984.tb02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Wurst R., Vournakis J., Rich A. Conformational changes of yeast tRNAPhe and E. coli tRNA2Glu as indicated by different nuclease digestion patterns. J Biol Chem. 1979 Oct 10;254(19):9608–9616. [PubMed] [Google Scholar]