Abstract
Objectives
Drug transporters affect ART tissue disposition, but quantitative measures of drug transporter protein expression across pre-clinical species are not available. Our objective was to use proteomics to obtain absolute transporter concentrations and assess agreement with corresponding gene and immunometric protein data.
Design
In order to make interspecies comparisons, two humanized mouse (hu-HSC-Rag (n=41); BLT (n=13)) and one primate (rhesus macaque, (NHP, n=12)) models were dosed to steady-state with combination ART. Ileum and rectum were collected at necropsy and snap frozen for analysis.
Methods
Tissues were analyzed for gene (qPCR) and protein (LC-MS proteomics and Western blot) expression and localization (immunohistochemistry) of ART efflux and uptake transporters. Drug concentrations were measured by LC-MS/MS. Multivariable regression was used to determine the ability of transporter data to predict tissue ART penetration.
Results
Analytical methods did not agree, with different trends observed for gene and protein expression. For example, qPCR analysis showed a 2-fold increase in permeability glycoprotein (Pgp) expression in NHPs versus mice, however proteomics showed a 200-fold difference in the opposite direction. Proteomics results were supported by IHC staining showing extensive efflux transporter localization on the luminal surface of these tissues. ART tissue concentration was variable between species, and multivariable regression showed poor predictive power of transporter data.
Conclusions
Lack of agreement between analytical techniques suggests that resources should be focused on generating downstream measures of protein expression to predict drug exposure. Taken together, these data inform the use of pre-clinical models for studying ART distribution and the design of targeted therapies for HIV eradication.
Keywords: drug transporters, antiretrovirals, GALT, tissue reservoirs
Introduction
Despite the efficacy of combination antiretroviral therapy (cART), early studies of cART-treated patients showed persistent infection and rapid rebound viremia after drug removal even from patients with undetectable viral loads, necessitating lifelong therapy[1], [2]. There is evidence that HIV replication can persist within certain anatomic sites, or tissue reservoirs, including the central nervous system, lymphatic system, gut-associated lymphoid tissue (GALT), and genital tract[3]–[5]. HIV persistence in GALT is of particular concern given that ongoing replication at this site may result in prolong immune dysregulation and delayed T cell recovery even after cART initiation, suggesting that inadequate ART exposure may propagate HIV replication at this site[6]. It has been shown by our group and others that ART tissue penetration is highly variable between anatomic sites and between ART within a single tissue[7]. Further, Fletcher et al demonstrated that higher tissue ART concentrations were significantly associated with faster HIV decay within the lymph nodes and GI tract[8]. More recently, we have shown our ability to image ART within harvested tissue slices, and found heterogeneous efavirenz (EFV) distribution in several anatomic sites, particularly in the GI tract[9]. Further investigation into what factors govern these distribution patterns is critical for understanding how to increase ART exposure in GALT.
Drug transporters play an important role in the disposition of many antiretrovirals and are extensively expressed throughout the gut. Several groups, including our own, have published studies evaluating the expression and localization of drug transporters in tissues relevant for HIV prevention[10], [11] and, more recently, cure[12]. However, there has been no consensus in the field on the optimal way to measure transporter expression. There is little agreement between publications with regard to what is being measured (i.e. gene vs protein expression), and there has been no assessment of the extent of agreement between techniques (e.g. qPCR vs Western blot (WB) vs immunohistochemistry (IHC)). Further, although proteomics-based methods have been used to obtain absolute concentrations of specific proteins including drug transporters[13], [14], this technology has not been compared against other methods in the context of HIV infection. A comprehensive evaluation of transporter expression and localization using multiple techniques within the same study is greatly needed to inform the field as to the best way to measure transporter expression for their effect on ART concentration in tissues.
In addition to methodological considerations, another important variable to address is expression differences between species. Animal models are commonly used to study HIV infection, and any evaluation of the tissue exposure of a new or existing ART must first be performed in animals before moving into humans. While there are some data showing similarities in ART exposure between humans and animals[15], there is a paucity of data comparing important variables for drug distribution between animal models or between animals and humans. Further, the effect of HIV infection on these variables has not been elucidated. Identification and quantitation of these differences, if they exist, will help to prevent the inappropriate extrapolation of data from one species to another, determine whether pharmacokinetic information should be obtained during infection, and streamline the drug development process.
In the present study, we perform a comprehensive evaluation of drug transporter expression and localization in two tissues of the GI tract[4], [16] using multiple methodologies and three animal models from two species. These data help identify important variables for ART exposure into tissue reservoirs, while at the same time identifying the best way to measure drug transporter expression. Finally, the generation of novel inter-species data can help determine the applicability of animal models to future ART development.
Materials and Methods
Animal Dosing and Tissue Collection
Three commonly used animal models were employed in this study: the hu-HSC-Rag (n=41) and bone marrow-liver-thymus (BLT; n=13) humanized mouse models and a non-human primate model (n=12). Detailed dosing and infection information for all animals can be found in the Supplementary Methods. Doses for all drugs were chosen based on commonly used treatment doses for HIV infection in these models[15], [17]–[20]. ART dosing combinations were chosen based on the limited resources available (i.e. NHPs) or on toxicity (e.g. EFV in BLT mice). Two animals from each mouse model were not dosed with ART and used as controls. Ileum and rectum were collected at necropsy and snap frozen. All animal experiments were performed in accordance with locally approved IACUC protocols.
Gene Expression Analysis
Transporter gene expression (including human genes in the humanized mouse samples) was analyzed by qPCR on five efflux and four uptake transporters (Supplementary Table 2), chosen based on their relevance to ART disposition and expression in the GI tract[21]. Approximately 30mg of tissue was homogenized in lysis buffer using a Precellys Tissue Homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) and RNA was extracted using a Qiagen RNAeasy Mini Kit (Qiagen, Valencia, CA) per manufacturer’s protocol. 200ng of RNA was reverse transcribed to cDNA using the VILO Superscript cDNA Synthesis Kit (Thermo Fisher). Forty cycles of qPCR were performed using Taqman primers and probes (Supplementary Table 3) on a QuantStudio6 (Life Technologies). Expression for all transporters was normalized to the housekeeping gene GAPDH using the 2−ΔCT method[22]. All samples were run in triplicate, and variability within and between reaction plates was low, with standard deviation less than 0.2 CT.
Protein Expression Analysis
Protein used for WB and LC-MS proteomics was isolated using a modified version of an extraction method optimized for proteomics as described previously[23], [24] and listed in detail in Supplementary Methods. For WB, up to 10μg of protein was loaded onto a 4–12% electrophoresis gel (NuPage) and run for 110 minutes at 180V. Transfer onto a PVDF membrane (NuPage) occurred over 90 minutes at 30V. After blocking in 5% milk, membranes were incubated in primary antibody for 1–3 hours (Supplementary Table 4), then rinsed and incubated in secondary antibody for 1–2 hours. Development occurred using Clarity ECL reagents (Bio-Rad, Hercules, CA) with a Chemi-Doc XRS+ Imager (Bio-Rad), and densitometry relative to GAPDH was calculated using ImageLab 5.2.1 (Bio-Rad). A combination of 15μg each of mouse brain extract, liver extract, and T98G cell lysate was used as the positive control sample. For proteomics analysis, up to 50ug of protein underwent 18 hour digestion with 1pmol stabile isotope labeled (SIL) peptide standards added (Supplementary Methods) and was analyzed using a nanoACQUITY system (Waters, Milford, MA) coupled to a QTRAP 5500 mass spectrometer (AB SCIEX, Framingham, MA) equipped with Nanospray III source.
Immunohistochemistry
Tissues were sliced frozen at 10μm thickness using a cryostat (Leica Biosystems, Wetzlar, Germany) and thaw mounted onto glass microscope slides. The frozen slides were then stained with primary antibody (Supplementary Table 3) for 15–60 minutes followed by pH antigen retrieval (Leica). DAB (3,3′-diaminobenzidine) was used as a substrate-chromagen for detection. All staining was performed on a Leica Bond automated tissue stainer (Leica). Samples were visually evaluated for transporter localization.
Statistical Analysis
Comparisons between dosing cohorts, species, and anatomic compartments were made using one-way Kruskal-Wallis ANOVA on ranks. Pearson correlation on log-transformed values was used to determine the relationship between gene expression and protein expression by each method for all combined samples. To determine which transporter evaluation method best predicted tissue ART penetration, univariate regression analysis was performed using log-transformed TPR values versus transporter expression results (as measured by qPCR, WB, QTAP [Quantitative Targeted Absolute Proteomics], or IHC). Those variables achieving p<0.05 in the univariate analysis were included in a multivariable analysis to identify combinations of variables significantly predicting TPR. Descriptive statistics and between-group comparisons were conducted using SigmaPlot 13.0 (Systat Software Inc., San Jose, CA), and the univariate and multivariable analyses were performed using SAS 9.3 (Cary, NC); p<0.05 was considered significant.
Results
Drug Transporter Gene Expression
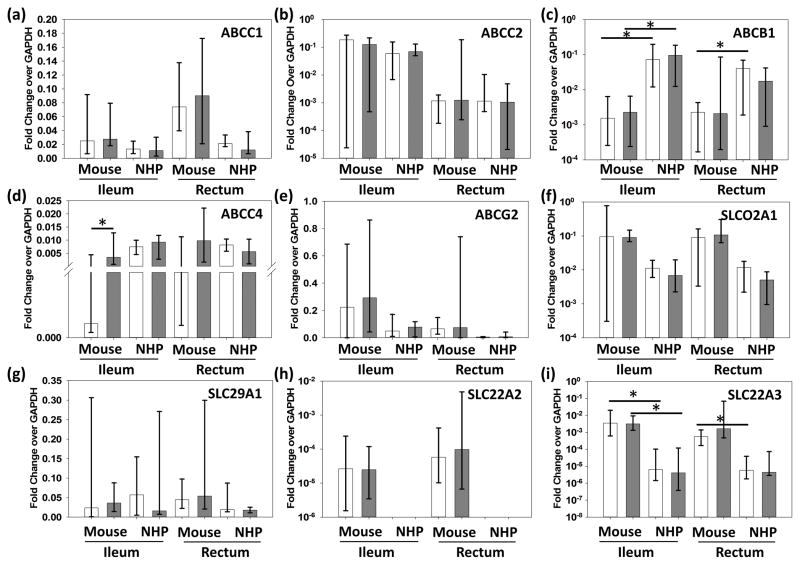
Comparisons between individual dosing cohorts for the ileum and rectum showed no significant differences between individual dosing cohorts, so these data were combined to assess total gene expression. Pooled ileum and rectum data from mice and macaques were compared in Figure 1. ABCB1 demonstrated a significant 2-fold increase in expression in macaques vs mice (Figure 1C), while the other efflux transporters did not differ significantly between species or tissue site. The uptake transporters SLCO2A1 and SLC29A1 did not differ between species. SLC22A2 was increased 0.5-fold in mouse rectum versus ileum and was not detected at all in macaque tissues (Figure 1H). SLC22A3 was 2–3 logs more highly expressed in mice ileum and rectum compared to macaques (Figure 1I).
Figure 1. Multispecies Comparison of Transporter Gene Expression.
Gene expression is represented as fold change of GAPDH for uninfected (white) and infected (gray) animals from multiple dosing cohorts. Data shown are median and range. SLC22A2 was observed in mouse tissues only. NHP=non-human primate; * represents p<0.05.
Interspecies Comparison of Transporter Protein Expression by Western Blot Analysis
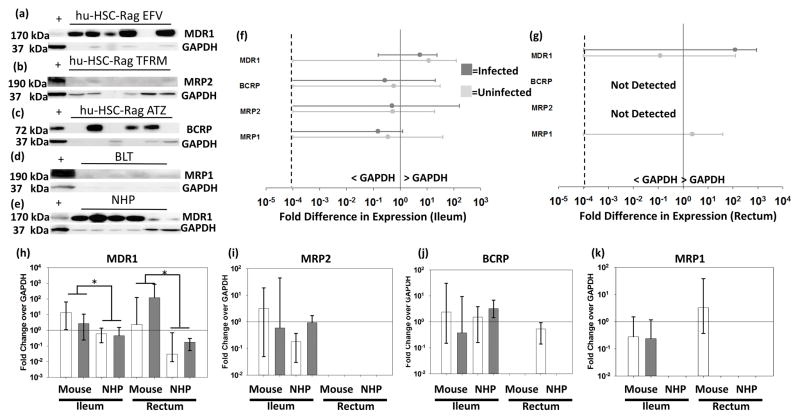
Figure 2 shows WB results, with representative blots shown in 2A–D. Densitometry data from all mouse samples are shown in Figure 2E and 3F, which demonstrates large variability in protein expression (0.2–100-fold GAPDH). Densitometry analysis of individual mouse dosing cohorts did not indicate significant differences between dosing cohorts or mouse models. Figure 2G–J compares mouse and NHP WB data. Relative protein expression trends were similar between mice and macaques for MRP1, MRP2, and BCRP. Interestingly, MDR1 protein expression showed a significant opposite trend compared to the ABCB1 gene expression between species, with relative MDR1 protein expression 1–2 logs higher than macaques in both the ileum and rectum.
Figure 2. Transporter Protein Expression Humanized Mice Ileum and Rectum.
Representative Western blots for four efflux transporters from each animal cohort (A–E). + represents the positive control sample. Samples with no detectable GAPDH were not included in subsequent analyses. Densitometry data from each blot was quantified for each transporter in mice (F[ileum]&G[rectum]), where protein expression is represented as a fold change over GAPDH for uninfected (light gray) and infected (dark gray) animals. Zero values were imputed at 10−4 (dashed line) for graphing purposes. Comparison of all mice and macaques is shown in H–K, where data are median and range. Solid line represents equal protein expression to GAPDH; * represents p<0.05.
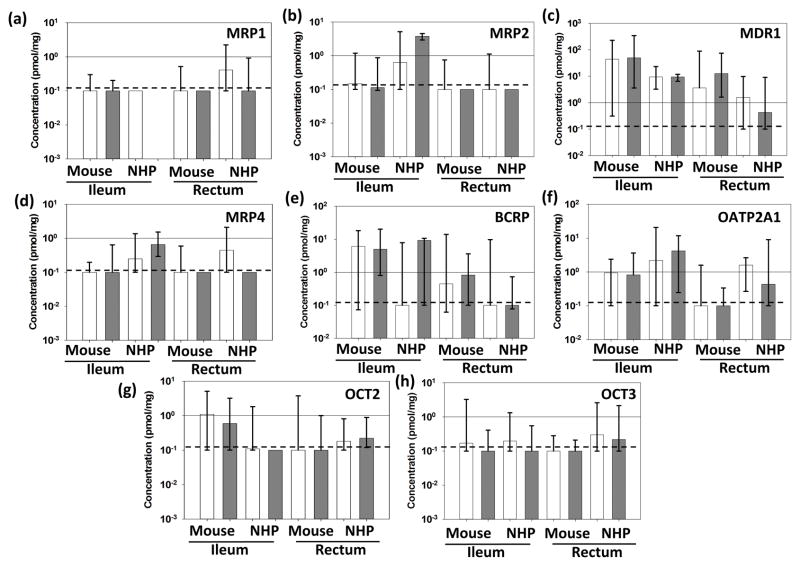
Figure 3. Multispecies Comparison of Transporter Protein Expression by QTAP.
Absolute protein concentrations are represented as pmol/mg protein for uninfected (white) and infected (gray) animals from multiple dosing cohorts. Solid lines represent 1pmol/mg; dashed lines represent the lower limit of quantitation. Data shown are median and range; * represents p<0.05.
Interspecies Comparison of Transporter Protein Expression by Targeted Quantitative Proteomic Analysis
No significant differences were observed between infected and uninfected animals or individual dosing cohorts, so ileac and rectal QTAP data from all cohorts of mice were combined and compared to those generated in macaques (Figure 3). MDR1 protein concentrations were 2 logs higher in the mouse ileum compared to macaques (Figure 3C), which is similar to the WB analysis, and contrary to observed ABCB1 gene expression. Further, the significant differences in ABCC4 expression between infected and uninfected mice were not replicated in the protein analysis (Figure 3D). The 3-log increase in SLC22A3 gene expression in mice over macaques was also not replicated here (Figure 3H). There were also several significant differences in protein concentrations that were not present in the qPCR analysis. SLCO2A1, for example, was not significantly different between species in gene expression, however a significant increase in OATP2A1 concentrations was observed in macaques compared to mice (Figure 3F).
Transporter Localization in the Ileum and Rectum
IHC staining revealed distinct localization of several drug transporters within the GI tract (representative images in (Supplementary Figure 2). In both the ileum and rectum, MDR1 was found to localize on the luminal surface of the gut mucosa in tissues from all three animal models, and was readily expressed. Conversely, MRP2 was not detected in any tissue from any animal, though protein was sporadically detected with WB (Figure 2) and QTAP (Figure 3). BCRP was detected in the ileum of both mouse models, and showed a similar localization profile to MDR1. MRP1 localized to the luminal surface in a similar fashion to MDR1 and was expressed in all tissues. MRP4 was localized to the lamina propria in all three species, with extensive positive staining on the basolateral surface of mucosal cells.
Human Transporter Expression in Humanized Mice
Supplementary Figure 3 provides an overview of human transporter gene expression as it relates to mouse gene expression in the same tissues. We observed human gene expression for more than half of the transporters evaluated. Expression was in general 1–5 logs lower than the mouse genes, with the notable exceptions of ABCB1 and ABCC4, which in some samples was 2 logs higher than mouse expression. An analysis of the relationship between the extent of humanization and the amount of human transporter gene expression did not show any significant relationship (data not shown). Western blot analysis using human-specific antibodies showed detectable bands for MDR1 only, which had fold-GAPDH values that were within 50% of mouse protein expression (data not shown). However, QTAP analysis using human-specific SILs did not detect any MDR1 in any humanized mouse sample, nor did it detect human protein from any other transporter (data not shown). To demonstrate that earlier interspecies comparisons for ABCB1 and ABCC4 were not confounded by a lack of accounting for human gene expression, we re-analyzed these data after accounting for the contribution of human gene expression of these transporters (Supplementary Figure 3c&d). Our ABCB1 results were not significantly altered, however median ABCC4 expression in the uninfected mouse rectum greatly increased (p<0.01) over corresponding data in NHPs.
Methodology Comparison for Drug Transporter Evaluations
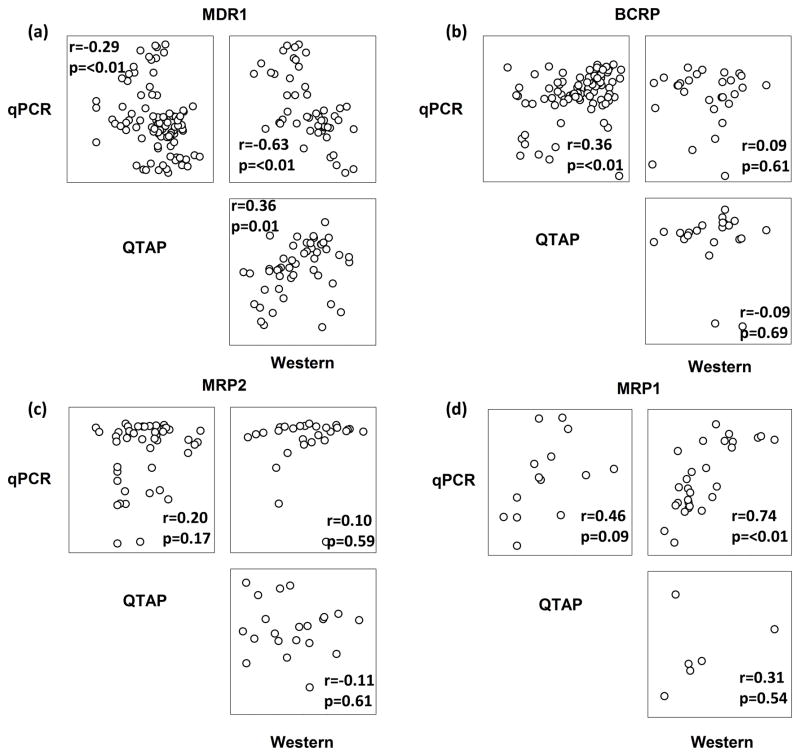
Correlation matrices were generated for the four efflux transporters evaluated by all three methods using combined data from all animals (Figure 4). Correlation coefficients were low for most comparisons, showcasing the lack of agreement between techniques. Comparison of QTAP and Western for MDR1and MRP1 showed the strongest correlation and reached statistical significance (p<0.01), however the large amount of variability in the data does not provide convincing evidence that these methods are in high agreement. Correlation between qPCR and QTAP for the uptake transporters was also poor, with no comparison reaching statistical significance (data not shown).
Figure 4. Lack of Agreement Between Transporter Evaluation Methods.
Correlation matrices are shown for the four efflux transporters evaluated by all three methods.
Results from the multivariable analysis are shown in Table 1. The ability to predict ART tissue penetration (shown in Supplementary Table 5) was generally low, with R2 values ranging from 0.09 (TFV TPR predicted by qPCR) to 0.51 (FTC TPR predicted by WB). qPCR data were able to generate significantly predictive regression models for each drug evaluated, though the resulting R2 values tended to be lower than those generated by WB. QTAP data generated significant models for TFV, FTC, and RAL only, and R2 values were lower than qPCR or WB in every case. There was little agreement between methods regarding which specific efflux transporters were found to significantly contribute to each model, though BCRP and MRP1 expression were most commonly implicated.
Table 1. Multivariable Regression Analysis of Drug Transporter Expression Methods on TPR.
Forward stepwise regression was performed using TPR for each drug as the dependent variable, with plasma concentration and efflux transporter expression as independent variables for the all methods used. Only the variables that were found to be significant in the univariate analysis were included. P values for each independent variable are shown in the table, with statistically significant variables bolded. R2 values for each regression model are shown on the far right. Models that did not identify any of the included variables as statistically significant in the univariate analysis do not have R2 values and are shown as “n/a”
| Drug | Method | MDR1 | BCRP | MRP2 | MRP1 | MRP4 | R2 |
|---|---|---|---|---|---|---|---|
| TFV | qPCR | - | - | 0.01 | - | - | 0.09 |
| WB | - | 0.01 | - | 0.05 | - | 0.38 | |
| QTAP | - | 0.01 | - | - | - | 0.11 | |
| IHC | - | - | - | - | - | n/a | |
| FTC | qPCR | - | 0.007 | - | 0.008 | - | 0.41 |
| WB | - | 0.01 | - | 0.002 | - | 0.51 | |
| QTAP | 0.04 | - | 0.05 | - | - | 0.15 | |
| IHC | - | - | - | - | - | n/a | |
| RAL | qPCR | 0.56 | 0.002 | - | 0.002 | - | 0.43 |
| WB | - | 0.03 | - | 0.01 | - | 0.51 | |
| QTAP | - | - | - | 0.58 | 0.008 | 0.16 | |
| IHC | - | - | - | - | - | n/a | |
| EFV | qPCR | - | 0.004 | - | 0.07 | - | 0.39 |
| WB | - | 0.12 | - | - | - | 0.22 | |
| QTAP | - | - | - | - | - | n/a | |
| IHC | - | - | - | - | - | n/a | |
| ATZ | qPCR | - | - | - | - | 0.06 | 0.17 |
| WB | - | - | - | - | - | n/a | |
| QTAP | - | - | - | - | - | n/a | |
| IHC | - | - | - | - | - | n/a | |
| MVC | qPCR | - | - | - | 0.002 | - | 0.23 |
| WB | - | - | - | - | - | n/a | |
| QTAP | - | - | - | - | - | n/a | |
| IHC | - | - | - | - | - | n/a |
Discussion
This is the first study comprehensively comparing drug transporter expression in the GI tract across animal models, and has demonstrated several novel findings with important implications for drug development and HIV eradication research. When gene and protein expression data were pooled to investigate differences between anatomic compartments and between species, several important differences were observed. The multiple log differences observed between mice and macaques for ABCB1/MDR1 and ABCC4/MRP4 indicate ART PK data generated in one model may not easily be extrapolated to the other. Given that several of these transporters have been shown to efflux numerous antiretrovirals, the species used in investigations of ART disposition into tissues, whether for prevention or eradication, is a critically important variable. RAL, for example, has been shown to reach rectal concentrations that are 35-fold greater than plasma in macaques[25]. We have also recently shown that RAL distributes readily throughout the macaque rectum, but that distribution in humanized mouse rectum is lacking[26]. In this study, RAL NHP ileum and rectum concentrations were increased over mice by 16- and 376-fold, respectively. Given that RAL is known to be effluxed by MDR1, it may be the case that RAL distributes into the intestinal mucosa in both species, but is effluxed back into the intestinal lumen by MDR1 to a greater extent in mice versus macaques, helping to explain the decreased tissue concentrations in this model. The distinct differences observed here provide support that transporter expression may also differ between animal models and humans.
Not only does the current study provide important information on transporter expression between animal models, we also are the first to formally compare methodologies for measuring transporter gene and protein expression in tissues relevant for HIV research. The extent of agreement between methods was generally poor, with ABCG2/BCRP showing the only significant relationship. There are several possible explanations for this lack of correlation, including the fact that, compared to the robust qPCR data, protein expression was highly variable and was not observed in all samples. Conditions for these experiments have been optimized by our lab, however lot-to-lot antibody variability and lack of an accepted standard for quantifying densitometry data are persistent challenges with the Western blot technique[27]. Further, mRNA inhibition by native micro-RNAs or post-translational protein modifications can affect the relationship between gene and protein expression and may be influencing the observed results. Additionally, differential rates of mRNA degradation between GAPDH and transporters may alter observed expression despite correction for this housekeeping gene.
The lack of agreement between qPCR and Western blot data even after correction for GAPDH expression is concerning, as there is currently no accepted standard in the field for measurement of transporter expression. Several groups have published data generated using both methods[28],[12], however the utility of Western blot data is limited due to narrow dynamic range and often possible antibody cross reactivity. Further, relative gene expression data should be interpreted with caution, as the high sensitivity may lead to false positives. Using DNA standards, we have determined that 8–10,000 copies of GAPDH were present in each mouse sample, with 60–100,000 GAPDH copies present in each NHP sample (data not shown). Based on these values, relative transporter expression values of 10−4 or greater represent biologically plausible expression of these genes in our samples, however lower relative expression values may simply mean that the gene is not expressed at all.
Proteomics analysis of the same tissues showed much more robust data compared to those generated by Western blot in terms of overall frequency of detection (80% for QTAP vs 71% for WB). However even these data showed little agreement when compared to qPCR data. Despite the lack of agreement, QTAP tended to agree with transporter localization data determined by IHC, where MDR1 and BCRP were the most highly expressed throughout the ileum and rectum, with decreased expression of MRP1 and almost no expression of MRP2. The ability of QTAP to provide robust, downstream protein expression data with high sensitivity and specificity for multiple transporters from a single sample make this an appealing technology. The lack of agreement between QTAP and WB is inconsistent with previous reports showing good correlation between these methods[29]. However, those results have been generated using recombinant enzymes, which do not represent the complex biological tissue matrices studied here. To our knowledge, ours is the first study to compare these data using tissue homogenates. While this technology requires specialized equipment and expertise its use has become more widespread in recent years[30]–[32].
Despite these advantages, the multivariable regression analysis found that QTAP data did not provide a significant increase in predicting observed TPR values over data derived from other techniques (Table 1). These results may be explained in part by the large amount of undetectable samples for Western blot and QTAP, which may have reduced the ability to detect significant relationships due to limited statistical power. Further, the possibility of drug-drug interactions affecting tissue ART exposure, particularly in the NHPs receiving EFV, must be considered. In addition, we were limited by the use of whole tissue homogenates to determine TPR. Although this has been the current standard in the field, TPR cannot distinguish between ART penetration directly from the lumen versus penetration from peripheral blood. Measuring ART concentrations from luminal washes in addition to tissue concentrations can highlight the effects of drug efflux and may have provided a more discreet variable upon which to base our regression model.
The negative results of this regression analysis may indicate that drug transporters alone do not govern ART tissue concentrations in a significant way, but must be measured in the context of additional variables such as drug metabolizing enzyme expression or drug PK properties, which were not examined here. Understanding the contribution of drug metabolizing enzymes would be of particular interest, as it has been shown that the expression, regulation, and magnitude of inhibition/induction can vary drastically between rodent and non-rodent species.[33] Interspecies differences in other factors such as target cell expression and the makeup of intestinal microbiota are less well characterized, but may have affected our results. Further, although these humanized mouse models have shown success in reconstituting systemic human lymphocyte populations, it may be that these murine models do not fully recapitulate the human gut microenvironment. It is possible that consideration of these variables in tissue accumulation would have improved the predictive ability of the model. Given the high sensitivity and low variability of qPCR data compared to other methods, it is surprising that this method did not identify more expected transporters; however, qPCR was able to identify at least one significant variable for every drug. The fact that gene expression is not always reflected by protein expression could be seen as an issue with the qPCR technique.
One of the most notable findings of this analysis is the characterization of human drug transporter expression in the tissues of both humanized mouse models. The extent of peripheral immune humanization observed here was consistent with previous studies using these models[34], but this is the first study to quantify human transporter expression in these animals. The detection of some human transporter gene expression should not be surprising given that many of these transporters are expressed on the surface of human lymphocytes, which are abundant in the humanized mouse GI tract, particularly in the ileum. We observed human gene expression from nearly every transporter evaluated, and found that ABCB1 and ABCC4 were expressed at an extent equal to or greater than mouse transporters in tissues from five mice (Supplementary Figure 4). This implies some amount of underestimation of the size of the total transporter pool, and when human transporter isoforms were accounted for, ABCC4 results significantly changed. However, ABCB1 results remained consistent and the use of species-specific antibodies and SILs for Western blot and QTAP, respectively, preserved the validity of our protein results. Future studies of drug transporters and/or drug metabolizing enzymes in these mouse models should include the analysis of both mouse and human-derived proteins.
Conclusion
As the body of evidence for persistent HIV replication within tissue reservoirs continues to grow, so does the need to define and quantify the factors influencing ART disposition within these tissues. To that end, this analysis compares drug transporter expression between commonly used animal models, and assesses the effect of HIV infection on transporter expression. We also demonstrate that the methods commonly used to evaluate transporter expression have little agreement with each other, and that robust downstream measures may have the most utility. Finally, we are the first to quantify the contribution of human transporters to the overall transporter pool in the GI tissue of these humanized mouse models.
These data have important implications for future studies of ART disposition in relation to HIV persistence. Our finding that transporter expression methods are not in agreement, and that transporter data alone are insufficient to predict ART tissue penetration highlights the need for future studies on alternative endpoints. The contribution of drug metabolizing enzymes, physicochemical properties, and gut microbiota to the tissue and cellular disposition of ART and their metabolites remains undefined, but will be necessary to know toinform the development of future therapies targeted toward HIV reservoirs.
Supplementary Material
Acknowledgments
This project was funded by a National Institutes of Health (NIH) grant (R01 AI111891), and in part by an NIH instrument grant (S10 RR024595). C.T. is a recipient of the American Foundation for Pharmaceutical Education Pre-Doctoral Fellowship. Nonhuman primate studies were supported in part by the Base Grant to the California National Primate Research Center P51 OD011107. The authors have no conflicts of interest to declare.
Parts of this work have been presented previously as posters at the Seventeenth International Workshop on Clinical Pharmacology of HIV & Hepatitis Therapy, Washington, DC, USA, 2016 (Poster P_28), and at the 2016 Conference on Retroviruses and Opportunistic Infections Seattle, WA, USA (Abstract 450). C.T., P.L., J.G., R.A., P.S., and A.K. contributed to the design of the study. P.C., M.K., and L.A dosed the animals and collected tissues. C.T., N.S., and J.F. contributed to collection of transporter expression data. A.S and C.S performed LC-MS/MS analyses. M.M. performed the IHC staining and Y.F contributed to the analyses of these data. C.T and A.K wrote the manuscript. This project was funded by a National Institutes of Health (NIH) grant (R01 AI111891), and in part by an NIH instrument grant (S10 RR024595). C.T. is a recipient of the American Foundation for Pharmaceutical Education Pre-Doctoral Fellowship. Nonhuman primate studies were supported in part by the Base Grant to the California National Primate Research Center P51 OD011107.
Footnotes
Conflicts of Interest
None to declare
References
- 1.Chun T, Engel D, Berrey M, Shea T, Corey L, Fauci T. Early establishment of a pool of latently infected, resting CD4 T cells during primary HIV-1 infection. Proc Natl Acad Sci. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chun TW, Davey RT, Engel D, Lane HC, Fauci AS. Re-emergence of HIV after stopping therapy. Nature. 1999;401:874–875. doi: 10.1038/44755. [DOI] [PubMed] [Google Scholar]
- 3.Palmer S, Josefsson L, Coffin JM. HIV reservoirs and the possibility of a cure for HIV infection. J Intern Med. 2011;270:550–60. doi: 10.1111/j.1365-2796.2011.02457.x. [DOI] [PubMed] [Google Scholar]
- 4.Smith MZ, Wightman F, Lewin SR. HIV reservoirs and strategies for eradication. Curr HIV/AIDS Rep. 2012;9:5–15. doi: 10.1007/s11904-011-0108-2. [DOI] [PubMed] [Google Scholar]
- 5.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Kosakovsky Pond SL, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;513:51–6. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–17. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson CG, Cohen MS, Kashuba ADM. Antiretroviral pharmacology in mucosal tissues. J Acquir Immune Defic Syndr. 2013;63(Suppl 2):S240–7. doi: 10.1097/QAI.0b013e3182986ff8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A. 2014;111:2307–12. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson CG, Bokhart MT, Sykes C, Adamson L, Fedoriw Y, Luciw PA, et al. Mass spectrometry imaging reveals heterogeneous efavirenz distribution within putative HIV reservoirs. Antimicrob Agents Chemother. 2015;59:2944–8. doi: 10.1128/AAC.04952-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu M, Cost M, Poloyac S. Expression of Transporters and Metabolizing Enzymes in the Female Lower Genital Tract: Implications for Microbicide Research. AIDS Res Hum Retroviruses. 2013;29:1496–503. doi: 10.1089/aid.2013.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicol MR, Fedoriw Y, Mathews M, Prince HM, Patterson KB, Geller E, et al. Expression of six drug transporters in vaginal, cervical, and colorectal tissues: Implications for drug disposition in HIV prevention. J Clin Pharmacol. 2014;54:574–83. doi: 10.1002/jcph.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Hoque MT, Jenabian MA, Vyboh K, Whyte SK, Sheehan N, et al. Antiretroviral drug transporters and metabolic enzymes in human testicular tissue – potential contribution to HIV-1 sanctuary site. J Antimicrob Chemother. 2016;71:1954–65. doi: 10.1093/jac/dkw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson CG, Fallon JK, Nicol MR, Smith PC, Kashuba ADM. Quantification of Drug Transporters in Vaginal and Cervical Tissue Using a Novel Targeted Proteomics Approach: Implications for Small Molecule Disposition in Viral Reservoirs.[Abstract]. 20th International AIDS Conference; Melbourne, Australia. 2014. [Google Scholar]
- 14.Uchida Y, Zhang Z, Tachikawa M, Terasaki T. Quantitative targeted absolute proteomics of rat blood-cerebrospinal fluid barrier transporters: comparison with a human specimen. J Neurochem. 2015;134:1104–15. doi: 10.1111/jnc.13147. [DOI] [PubMed] [Google Scholar]
- 15.Veselinovic M, Yang KH, LeCureux J, Sykes C, Remling-Mulder L, Kashuba ADM, et al. HIV Pre-Exposure Prophylaxis: Mucosal Tissue Drug Distribution of RT Inhibitor Tenofovir and Entry Inhibitor Maraviroc in a Humanized Mouse Model. Virology. 2014;0:253–263. doi: 10.1016/j.virol.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yukl S, Gianella S, Sinclair E, Epling L, Li W, Choi ALM, et al. Differences in HIV Burden and Immune Activation within the Gut of HIV+ Patients on Suppressive Antiretroviral Therapy. J Infect Dis. 2010;202:1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massud I, Aung W, Martin A, Bachman S, Mitchell J, Aubert R, et al. Lack of prophylactic efficacy of oral maraviroc in macaques despite high drug concentrations in rectal tissues. J Virol. 2013;87:8952–61. doi: 10.1128/JVI.01204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shytaj IL, Norelli S, Chirullo B, Della Corte A, Collins M, Yalley-Ogunro J, et al. A highly intensified ART regimen induces long-term viral suppression and restriction of the viral reservoir in a simian AIDS model. PLoS Pathog. 2012;8:e1002774. doi: 10.1371/journal.ppat.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neff CP, Ndolo T, Tandon A, Habu Y, Akkina R. Oral pre-exposure prophylaxis by anti-retrovirals raltegravir and maraviroc protects against HIV-1 vaginal transmission in a humanized mouse model. PLoS One. 2010;5:e15257. doi: 10.1371/journal.pone.0015257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denton PW, Krisko JF, Powell DA, Mathias M, Kwak YT, Martinez-Torres F, et al. Systemic administration of antiretrovirals prior to exposure prevents rectal and intravenous HIV-1 transmission in humanized BLT mice. PLoS One. 2010;5:e8829. doi: 10.1371/journal.pone.0008829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kis O, Robillard K, Chan GNY, Bendayan R. The complexities of antiretroviral drug-drug interactions: role of ABC and SLC transporters. Trends Pharmacol Sci. 2010;31:22–35. doi: 10.1016/j.tips.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 23.Higgins JW, Bao JQ, Ke AB, Manro JR, Fallon JK, Smith PC, et al. Utility of oatp1a/1b-knockout and OATP1B1/3-humanized mice in the study of OATP-mediated pharmacokinetics and tissue distribution: Case studies with pravastatin, atorvastatin, simvastatin, and carboxydichlorofluorescein. Drug Metab Dispos. 2014;42:182–192. doi: 10.1124/dmd.113.054783. [DOI] [PubMed] [Google Scholar]
- 24.Fallon JK, Smith PC, Xia CQ, Kim MS. Quantification of Four Efflux Drug Transporters in Liver and Kidney Across Species Using Targeted Quantitative Proteomics by Isotope Dilution NanoLC-MS/MS. Pharm Res. 2016;33:2280–2288. doi: 10.1007/s11095-016-1966-5. [DOI] [PubMed] [Google Scholar]
- 25.Massud I, Martin A, Dinh C, Mitchell J, Jenkins L, Heneine W, et al. Pharmacokinetic profile of raltegravir, elvitegravir and dolutegravir in plasma and mucosal secretions in rhesus macaques. J Antimicrob Chemother. 2015;70:1473–81. doi: 10.1093/jac/dku556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosen EP, Thompson CG, Sykes C, Adamson L, Charlins P, Remling-Mulder L, et al. Quantifying Intra- and Inter-species Variability of Antiretroviral (ARV) Distribution in GALT by MSI.[Abstract]. 23rd Conference on Retroviruses and Opportunistic Infections; Boston, MA. 2016. [Google Scholar]
- 27.MacPhee DJ. Methodological considerations for improving Western blot analysis. J Pharmacol Toxicol Methods. 2010;61:171–177. doi: 10.1016/j.vascn.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 28.De Rosa MF, Robillard KR, Kim CJ, Hoque MT, Kandel G, Kovacs C, et al. Expression of membrane drug efflux transporters in the sigmoid colon of hiv-infected and uninfected men. J Clin Pharmacol. 2013;53:934–945. doi: 10.1002/jcph.132. [DOI] [PubMed] [Google Scholar]
- 29.Fallon JK, Harbourt DE, Maleki SH, Kessler FK, Ritter JK, Smith PC. Absolute quantification of human uridine-diphosphate glucuronosyl transferase (UGT) enzyme isoforms 1A1 and 1A6 by tandem LC-MS. Drug Metab Lett. 2008;2:210–222. doi: 10.2174/187231208785425764. [DOI] [PubMed] [Google Scholar]
- 30.Asher GN, Fallon JK, Smith PC. UGT concentrations in human rectal tissue after multidose, oral curcumin. Pharmacol Res Perspect. 2016;4:e00222. doi: 10.1002/prp2.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchida Y, Toyohara T, Ohtsuki S, Moriyama Y, Abe T, Terasaki T. Quantitative Targeted Absolute Proteomics for 28 Transporters in Brush-Border and Basolateral Membrane Fractions of Rat Kidney. J Pharm Sci. 2016;105:1011–1016. doi: 10.1002/jps.24645. [DOI] [PubMed] [Google Scholar]
- 32.Aebersold R, Burlingame AL, Bradshaw RA. Western Blots vs. SRM Assays: Time to turn the tables? Mol Cell Proteomics 2013. 2013;12:2381–2382. doi: 10.1074/mcp.E113.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martignoni M, Groothuis GM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol. 2006;2:875–94. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- 34.Berges BK, Wheat WH, Palmer BE, Connick E, Akkina R. HIV-1 infection and CD4 T cell depletion in the humanized Rag2−/−gamma c−/− (RAG-hu) mouse model. Retrovirology. 2006:3. doi: 10.1186/1742-4690-3-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.