Abstract
The spliceosome is an extraordinarily dynamic molecular machine in which significant changes in composition as well as protein and RNA conformation are required for carrying out pre-mRNA splicing. Single-molecule fluorescence resonance energy transfer (smFRET) can be used to elucidate these dynamics both in well-characterized model systems and in entire spliceosomes. These types of single-molecule data provide novel information about spliceosome components and can be used to identify sub-populations of molecules with unique behaviors. When smFRET is combined with single-molecule fluorescence colocalization, conformational dynamics can be further linked to the presence or absence of a given spliceosome component. Here, we provide a description of experimental considerations, approaches, and workflows for smFRET with an emphasis on applications for the splicing machinery.
Keywords: single-molecule, FRET, fluorescence, spliceosome, RNA, dynamics
1. Introduction
1.1. Pre-mRNA splicing
In biology, RNAs can broadly be classified into two functional groups—those that code for proteins (mRNAs) and those that do not (non-coding RNAs, ncRNAs). Eukaryotic mRNAs are first produced as precursors (pre-mRNAs) that undergo lengthy, regulated maturation processes between their transcription and translation [1]. ncRNAs can be used for a variety of cellular functions including catalytic activities, structural scaffolds, and cellular signals [2]. Both ncRNAs and pre-mRNAs come together in the spliceosome where five small nuclear RNAs (snRNAs) and dozens of proteins recognize and excise non-coding regions (introns) of a pre-mRNA and splice together the protein coding RNA (exons). Splicing is a highly dynamic process as the spliceosome is assembled anew onto each substrate pre-mRNA. While the structures of multiple spliceosome complexes have recently been recently determined [3-16], it is not yet clear how the spliceosome transitions between these complexes or how RNA and protein dynamics participate in these steps.
smFRET can be used to study the conformation and dynamics of biological factors, even when they are in low abundance or when the system is not kinetically synchronized [17]. In splicing, smFRET has been used to interrogate the dynamics and conformational landscape of the pre-mRNA and the snRNAs that participate in the reaction [18-27]. In combination with recently determined cryo-EM and crystal structures of the splicing machinery, these dynamics can now be linked to molecular structures to provide new insights into the spliceosome.
1.2. Single-molecule fluorescence microscopy
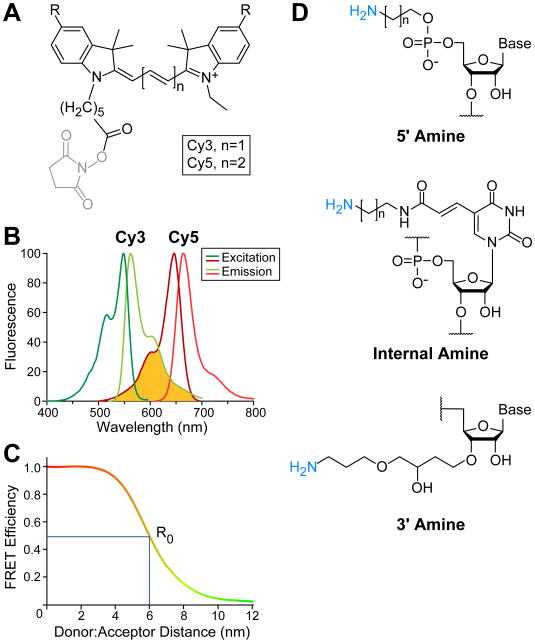
FRET is the transfer of energy from one fluorophore to another in a distance dependent manner [28]. For fluorophores typically used in biochemical experiments, FRET is observed when the fluorophores are within 2 to 10 nm [29]. Measuring FRET involves the excitation of a donor fluorophore and observation of the increased fluorescence emission from the acceptor fluorophore. Since the donor a n d acceptor fluorescence are anti-correlated with one another, the donor emission decreases as the acceptor fluorescence increases. Changes in FRET signal can be used to identify dynamic transitions as well as calculate relative distances between the fluorophores based on the efficiency of the energy transfer [28, 30]. When choosing fluorophores for a FRET experiment, consideration should be given to the photostability, extinction coefficient, quantum yield, size, and fluorescence excitation and emission wavelengths of the molecules. A common descriptor of fluorescent molecules is brightness which refers to the combined product of the extinction coefficient, number of photons able to be absorbed, and the quantum yield, how efficiently those photons are converted to fluorescence [31]. A host of small molecule dyes with unique properties are available and the properties of many fluorophores have been recently reviewed by Grimm et al. [32]. Commonly, cyanine (Cy) dyes are used for smFRET experiments since these fluorophores are bright and relatively photostable, which permits longer observation of FRET signals before irreversible photobleaching occurs. Two molecules from this family, Cy3 and Cy5, are a donor:acceptor pair widely used in biological FRET studies due to their well-matched spectral overlap and the ease by which Cy3- and Cy5-fluorescence emissions can be optically separated from one another (Fig. 1A,B); Cy3 has excitation maximum at 550 nm and emission maximum at 570 nm, while Cy5 has excitation and emission maxima at 649 and 670 nm, respectively. Because Cy5 is spectrally distinct from Cy3, excitation of Cy3 with a commonly-used 532 nm laser does not directly excite Cy5 to a significant degree but is sufficient to achieve robust FRET (Fig. 1B). These fluorophores can be easily linked to primary amino groups in nucleic acids or other biomolecules through amidation with N-hydroxysuccinimide (NHS) derivatized fluorophores (Fig. 1A).
Figure 1.
Cy3 and Cy5 fluorophore characteristics and attachment to RNA for FRET studies. (A) Chemical structures of Cy dyes frequently used for FRET with NHS leaving groups (grey) for fluorophore attachment. R represents sulfonate groups typically added to Cy dyes to increase solubility. (B) Excitation (Ex) and emission (Em) spectra for Cy3 and Cy5 fluorophores. The spectral overlap between Cy3 emission and Cy5 excitation is shown (yellow). Spectra obtained from GE Healthcare Life Sciences. (C) Example plot of the FRET between a Cy3:Cy5 pair. At 0.5 FRET efficiency, the fluorophore pair distance is equal to the Förster distance, R0, which is ∼6 nm for Cy3:Cy5 [32]. (D) Examples of commercially available options for fluorophore labeling RNAs either at the 5′ or 3′ ends or internally. Chemical structures shown are those commercially available from Integrative DNA Technologies.
FRET efficiencies (EFRET) values can be calculated through bulk and single-molecule FRET experiments [33]. The relationship between EFRET and signal intensity is EFRET = 1- (IDA/ID), where ID is the intensity of the donor in the absence of acceptor and IDA is in the presence of acceptor. The distance between two fluorophores can be determined since FRET values scale with the inverse of the distance between the fluorophores to the 6th power and is normalized by the Förster distance, R0 (Fig 1C). The Förster distance is unique to each donor:acceptor pair and describes where the energy transfer efficiency is 50% by incorporating the quantum yield of the donor, the dipole-dipole orientation factor, and the spectral overlap between the donor and acceptor. Because many experiments assume the fluorophores exhibit free ranges of motion, the dipole-dipole orientation factor is frequently estimated to be giving the Cy3:Cy5 pair a R0 of ∼6 nm [34].
Observing fluorescence from a single fluorophore is challenging, in part due to high background from the excitation source. Fluorescence microscopes are designed to minimize this background, often by making use of total internal reflection fluorescence (TIRF) [35]. In TIRF, a laser light source used for fluorophore excitation is reflected from the interface between the slide and aqueous sample. This creates an evanescent field of limited depth (∼100 nm) that is used to excite the donor fluorophore for FRET and greatly reduces sample background fluorescence. TIRF microscopes can be constructed with either objective or prism-based excitation schemes to achieve the high incident angle required to generate the evanescent field. With either microscope design, the background fluorescence signal is greatly minimized.
Carrying out a smFRET experiment also requires producing fluorescently-labeled molecules. In Section 2, we discuss how to add fluorophores to commercially synthesized RNA oligonucleotides, a robust and frequently used approach. In Sections 3 and 4, we describe the experimental workflow for carrying out a smFRET experiment from sample preparation through data acquisition and analysis. This method has been applied in a number of ways to study the RNA dynamics of the spliceosome and several examples are discussed in detail in Section 5.
2. Preparing labeled RNA for smFRET
2.1. Design
The RNA locations for attaching fluorophores can be readily chosen to fit the experimental design since commercial oligonucleotide synthesis provides many options for dervitization. RNAs are commonly fluorescently labeled at either the 5′ or 3′ ends or internally on the non-Watson-Crick face of a nucleotide such as uracil (Fig. 1D). When selecting the location to incorporate a fluorescent dye, it should be noted that cyanine fluorophores are planar aromatic compounds and there is a possibility for them to π-stack on top of nucleic acid duplexes [36]. Adjacent guanosines have been reported to quench the fluorescence of labeled bases and may need to be avoided [37]. Linker length between the fluorophore and RNA can also be varied, with 3 to 12 carbon linkers being common. Multiple fluorophores are commercially available with the NHS moiety for simple attachment to primary amino groups incorporated into the RNA such as those shown in Fig. 1A. We prefer to incorporate our own fluorophores into the RNAs as this allows a single purchased RNA to be potentially labeled with many different fluorophores for different applications. However, it is also possible to order RNA oligonucleotides with fluorophores attached during synthesis. The RNA length is not limiting for fluorophore labeling but sequences longer than ∼90 nucleotides are not routinely available commercially and are often made by splinted ligation of two or more RNA fragments.
smFRET data are frequently acquired from surface-immobilized molecules since this enables many molecules to be observed simultaneously over long periods of time. In these cases, the RNA also needs to incorporate a “handle” for surface attachment. Often a biotin moiety is introduced into the sequence, and this permits immobilization on streptavidin-coated surfaces. The biotin can be attached directly to the RNA itself or the RNA can be annealed to a second, surface immobilized oligonucleotide and held in place by duplex formation.
2.2. Labeling and splinted ligation
Labeling of RNAs with NHS-fluorophores is relatively straightforward and many protocols are available from NHS-fluorophore manufacturers. We typically carry out these reactions protected from light, at room temperature, overnight, at slightly alkaline pH (e.g., in sodium bicarbonate or sodium tetraborate buffer), and under RNase-free conditions [38]. The fluorescently labeled RNA can then be purified away from free, unreacted fluorophore as well as unlabeled RNA via a desalting, spin column in combination with either denaturing PAGE or by HPLC [38]. The purified, labeled RNA can be stored in solution at -20°C for subsequent use.
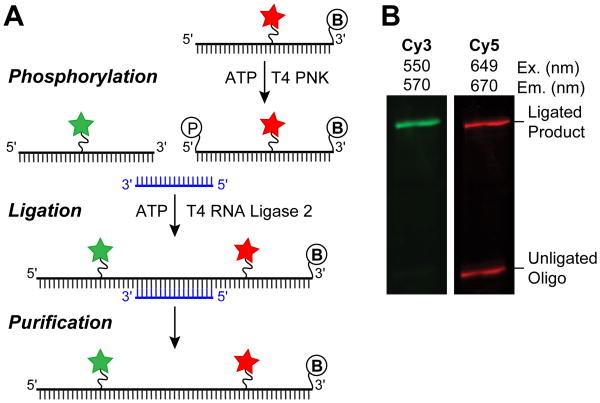
As mentioned in Section 2.1, splinted ligation gives the advantage of preparing the desired, full-length RNA from smaller building blocks [39]. The RNA oligonucleotide that donates the phosphoryl group in the ligation reaction must first be 5′ phosphorylated (Fig. 2A), which can either be accomplished during synthesis or by using T4 Polynucleotide kinase (PNK). Subsequently, the RNAs to be ligated are annealed to a complementary DNA splint prior to enzymatic ligation, often with T4 RNA ligase 2. We typically use a ratio of 1:1.2:2 of the 5′ RNA, DNA splint, and 3′ phosphodonor RNA, respectively. Denaturing PAGE is then performed to isolate the ligated, dual labeled RNA construct (Fig. 2B). The ligated RNA can be stored at -20°C and thawed several times before degradation is observed as long as RNase free conditions are maintained.
Figure 2.
Generating RNA constructs for FRET through splinted ligation. (A) Schematic overview of steps necessary for splinted ligation. The DNA splint (blue) basepairs to both the 5′ and 3′ oligos, bringing them in close proximity for ligation. The 5′ phosphoryl group and 3′ biotin are shown as circled “P” and “B”, respectively. (B) An example of the results from a splinted RNA ligation to prepare a smFRET reporter RNA, as analyzed by denaturing PAGE. The ligated product contains both the Cy3 and Cy5 RNAs and is resolved from excess, unligated Cy5 RNA. The ligated product can be excised and extracted from the gel for smFRET experiments.
3. smFRET experimental workflow
A common method for localizing molecules to a slide surface for TIRF microscopy is to passivate the slide with a coating of PEG and biotin-derivitized PEG [40]. The slide is then incubated with streptavidin, which binds the biotin-PEG molecules. Biotinylated RNAs or other molecules are attached to the surface by occupying the remaining biotin-binding sites on the immobilized streptavidin. The slide is mounted on the TIRF microscope and the fluorescence signals from the donor and acceptor fluorophore are recorded.
3.1. Slide and sample preparation
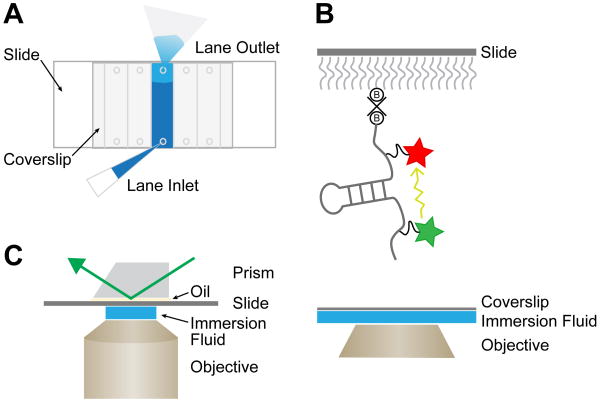
Carrying out smFRET experiments on reusable quartz slides (1 × 3 in) avoids background fluorescence from glass in the emission channel being used to image Cy5. A single slide can accommodate multiple individual compartments, or lanes, in which independent experiments can be run, and these lanes can be configured in many unique ways depending on the experiment. An example of a slide design with five lanes is shown in Fig. 3A and described here. Small holes have been drilled into the slide to allow for sample loading and buffer exchange into each lane. The spacing of the holes sets the dimensions for the lanes as well as whether the lanes run across the short or long axis of the slide. Before the lanes are created, the slide is first cleaned and silanized in preparation for passivation with PEG. The silanization procedure itself amounts to a brief incubation with aminosilane, which also provides the reactive amine groups necessary for PEG attachment.
Figure 3.
Single-molecule FRET data collection. (A) Illustration of an assembled flow-cell for smFRET assembled from a quartz slide, glass coverslip, double-sided tape, and vacuum grease. Holes drilled into the quartz slide provide inlets and outlets for sample application and buffer exchange. (B) 2D representation of an RNA molecule immobilized to a quartz slide for smFRET. Biotin-PEG molecules (B) are shown attached to a streptavidin tetramer (x). Energy transfer (yellow arrow) occurs with distance dependent efficiency between the two fluorophores. (C) A prism-based TIRF microscope setup for smFRET data collection. The slide is placed between the prism and the objective. A thin layer of oil creates even contact between the slide and the prism while the objective immersion fluid is located between the coverslip and the objective. The incoming laser excites the donor fluorophore by entering the prism from above (green arrow).
After silanization, thin strips of double-sided tape are laid across the slide between the holes to set the boundaries for each lane. Vacuum grease is then applied at the ends of each lane to create a seal at the top and bottom. A cover slip is carefully placed on top, and a tight seal is formed by the tape and grease to create each lane. We typically create lanes such that each compartment will hold ∼20 μL of liquid. Next, a mixture of amine-reactive PEG and biotin-PEG (50:1, w/w) is loaded into each lane and allowed to react for 2-16h. This results in passivation of the slide surface with PEG as well as attachment of biotin moieties that can be used for streptavidin attachment. Quartz slides can be cleaned and re-passivated using this procedure, typically 5-10 times before deterioration of the surface limits their usefulness for smFRET.
Immediately prior to the smFRET experiment, the PEG solution is removed and the slide lanes are thoroughly washed with buffer. The lane is then prepared by applying the streptavidin layer, attaching the biotinylated RNA or other molecule, and carrying out any other additions as per the specific experimental design. It is important to wash the lane sufficiently between additions to completely remove the previously loaded solution. Filter paper can be used to easily exchange solutions in each lane by wicking away the solution from one hole as solutions are being applied at the other end of the lane (Fig. 3A).
It is important that the biotinylated molecules sparsely populate the slide surface to ensure that single molecules are observed. As a result, very low concentrations of biotinylated RNAs (10-100 pM) are needed to achieve adequate densities of immobilized molecules for single-molecule experiments. Often the amount of usable smFRET data collected is limited by photobleaching of the fluorophores. To limit this, an imaging buffer containing an oxygen scavenging system (e.g., glucose oxidase/catalase) and triplet state quenchers (e.g., trolox) is often used [38, 41, 42]. The importance of these oxygen scavenging systems can become apparent during data analysis when fluorophore blinking or photobleaching due to oxygen may confound interpretation of the single molecule fluorescence signals. Defining experimental conditions in which these effects are minimized is often an important first step in a smFRET experiment.
If upon data analysis fluorophore blinking or photobleaching become more pronounced during the course of an experiment, it may be a sign that the oxygen scavenging system may need to be refreshed since these components have a limited lifetime in solution.
3.2. Data collection
On a prism-based TIRF microscope, the slide is mounted between the prism and the objective (typically 60-100× with a high numerical aperture (NA) to facilitate TIRF, Fig. 3C). The coverslip faces the objective and its thickness must be matched to the objective being used. It is important that the outside surfaces of the slide and cover slip are clean to allow light to pass through unhindered. Excitation lasers enter the prism from above, while fluorescence emission is collected through the bottom of the slide. Within the emission optical path, the donor and acceptor emission are separated by a dichroic mirror such that they are imaged on separate regions of the microscope's camera. The resulting images contains fluorescent signal recorded for both the donor and acceptor fluorescence channels. Additional information about prism-based TIRF can be found in Roy et al. [43].
Hundreds of single, fluorescent molecules can be imaged simultaneously using a conventional TIRF microscope. We often employ an alternating laser excitation scheme in which Cy5-labeled molecules are first directly excited with a 640 nm laser and imaged. This allows the locations of the Cy5-labeled molecules to be determined and confirms that these molecules contain a function Cy5 fluorophore. Next, FRET data are collected by turning off the 640 nm laser and turning on a 532 nm laser. The laser powers required to achieve sufficient signal-to-noise is often empirically determined and depends on fluorophore brightness and the camera frame rate among other factors.
4. Data processing
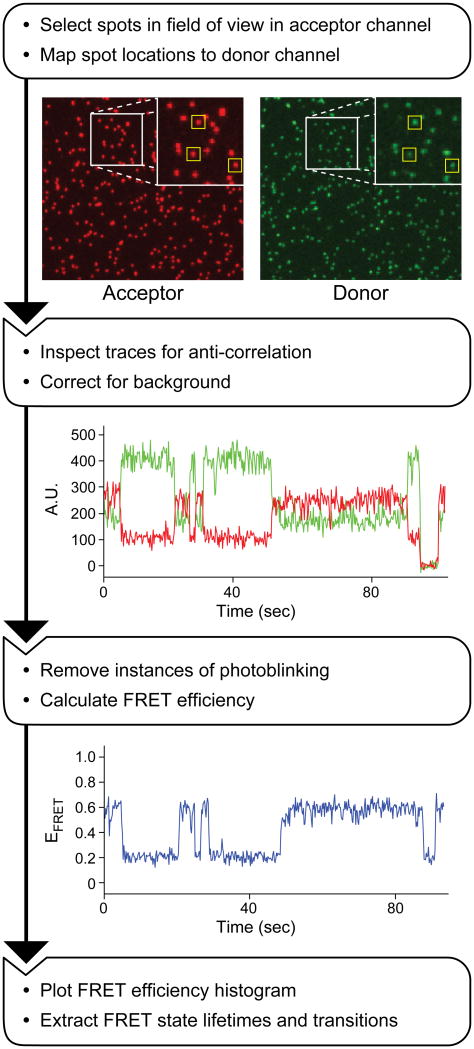
smFRET data contain information about molecular conformation as well as information about molecular heterogeneity in a population. A variety of approaches and software can be used to process smFRET data to extract the distribution of calculated FRET values, the dwell times of molecules in given FRET states, and the probability of molecules transitioning between different states [44]. A typical workflow diagram of smFRET data processing is shown in Fig. 4.
Figure 4.
Workflow of smFRET data processing. In the first step, the camera images from the donor (green) and acceptor (red) channels are used to select molecules present in both (yellow boxes). Next, integrated fluorescence trajectories of the fluorescent signal from both the donor (green) and acceptor (red) are plotted for each molecule to confirm anti-correlation and correct for background. Finally, the EFRET is calculated and plotted for the molecule for each time point (blue line).
4.1. Choosing molecules for analysis
In order to avoid bias towards molecules in low FRET states, we first pick molecules for analysis based on direct excitation of Cy5. Areas of interest (AOIs) are created around each fluorescent “spot” that define regions from which the fluorescent signal intensity will be integrated. It is important move or delete AOIs around spots if there is an indication of overlap of more than one molecule in that region. Once AOIs have been selected in the Cy5 channel, those AOIs need to be mapped onto the equivalent locations in the Cy3 channel. Mapping parameters should be determined with a control sample that fluoresces brightly in both the Cy3 and Cy5 channels such as streptavidin-coated, multi-wavelength fluorescent beads. After the mapping parameters have been defined, the Cy5-selected AOIs can then be mapped to the Cy3 channel such that each molecule now has two AOIs associated with it in the camera image: one in the Cy3 channel capturing donor fluorescence and one in the Cy5 channel capturing FRET acceptor fluorescence. The pixel intensities can then be integrated over time for the duration of the movie to create a fluorescence time trajectory for each AOI.
4.2. Selecting traces for analysis
Once the AOIs have been defined and the integrated time trajectories calculated, each trajectory is inspected to determine if the fluorescence from the associated single molecule is suitable for further analysis. For example, since we often choose AOIs based on Cy5 fluorescence alone, some molecules will not contain a fluorescent Cy3 fluorophore and should not be analyzed further. These molecules can be easily identified by comparing the paired fluorescence trajectories generated from the Cy3 and Cy5 channels for each AOI. In practice, it is also wise to remove molecules from further analysis that are in close proximity to others since this may interfere with assignment of FRET states. If changes in FRET are observed, the fluctuations in Cy3 and Cy5 fluorescence should be anti-correlated. Sometimes other changes in fluorescence are observed, such as multi-step blinking or photobleaching of the fluorescence spot. These molecules should not be analyzed since this is potentially indicative of the presence of more than one fluorescent molecule within that particular AOI.
Selected time trajectories should then be background corrected in each channel by subtracting measured intensities observed when both fluorophores are dark due to blinking or photobleaching. Depending on the type of analysis being used, it may also be necessary to remove instances of fluorophore blinking from fluorescence time trajectories and/or to “stitch” the trajectories into one file for subsequent analysis.
4.3. Calculating FRET efficiency
We typically calculate the apparent EFRET for each time point for each molecule using the integrated fluorescence intensities from both the donor and acceptor fluorophore according to the equation apparent EFRET= IA /((IA + IDA)) , in which IA is the acceptor fluorescence intensity upon excitation of the FRET donor. All of the apparent EFRET values observed in a given experiment are typically represented as a histogram that can be normalized by the total number of observations. The observed distribution can be fit mathematically, often as a sum of individual Gaussian distributions. For molecules with complex distributions of FRET states, statistical algorithms are employed to distinguish between states as discussed in Blanco et al., Rohlman et al. and briefly in Section 4.4 [44, 45]. EFRET values can be optimally distinguished from one another between 0.2 and 0.8 EFRET [29]. At regions outside this range, the signal-to-noise ratio is often poor for either the Cy3 or Cy5 emission, and this may obfuscate accurate identification of FRET values or conformational transitions.
4.4. Identifying FRET transitions and kinetics
Much insight into an RNA's structure:function relationship can be obtained from analysis of the time a molecule remains in a specific FRET state (dwell time) and the pathways by which FRET states are populated (transitions). For these types of analyses, the number of FRET states must first be determined in order to accurately assign each observed dwell time to a specific FRET state. For simple FRET distributions where there is no overlap between states, FRET efficiency thresholding can be used without biasing the analysis [44]. However, many molecules exhibit more complex behavior and to accurately determine the number of states present requires robust statistical analysis. The Hidden Markov Modeling (HMM) algorithm can be used to iteratively improve a set of probability matrices to generate a model for the number FRET states present that agrees with the smFRET data [46]. This analysis can be used to interpret complex histograms, by revealing hidden or overlapping states. An excellent review of HMM and software available for smFRET data analysis is given in Blanco and Walter [44]. The Walter lab has more recently developed a cluster-based analysis method, SiMCAn, with which they were able to quickly assign FRET states with improved accuracy [45, 47].
Once the number of FRET states has been determined and dwell times properly assigned, kinetic parameters for the molecule in each FRET state can be determined. The measured dwell times can be fit directly by maximum likelihood methods or the data can be compiled into a histogram and fit with exponential rate equations [48]. Transition density plots (TDPs) are used to graphically display observed transitions by plotting the FRET efficiencies of all transitioning molecules before and after each transition. TDPs indicate the number of and preference for a particular transition [49]. Transition occupancy density plots (TODP) weight observed transitions by the dwell time of the FRET state and can provide additional insights, particularly into the behavior of low populated states [44].
5. Examples of smFRET analysis of spliceosome dynamics
Saccharomyces cerevisiae (yeast) has been a key model organism for understanding spliceosome structure and mechanism. The smFRET experiments described in this section have been used to elucidate RNA dynamics within the yeast splicing machinery. Notably, these experiments have been carried out on a variety of RNA substrates and with a variety of methods to illustrate three types of insights obtainable by smFRET: visualization of dynamic transitions, kinetic characterization of transitions, and correlation of transitions with the presence or absence of particular factors. With these representative examples, we briefly review both the methods employed, the single-molecule results, and discuss the new insights into the spliceosome that were obtained.
5.1. Single snRNA dynamics visualized by smFRET
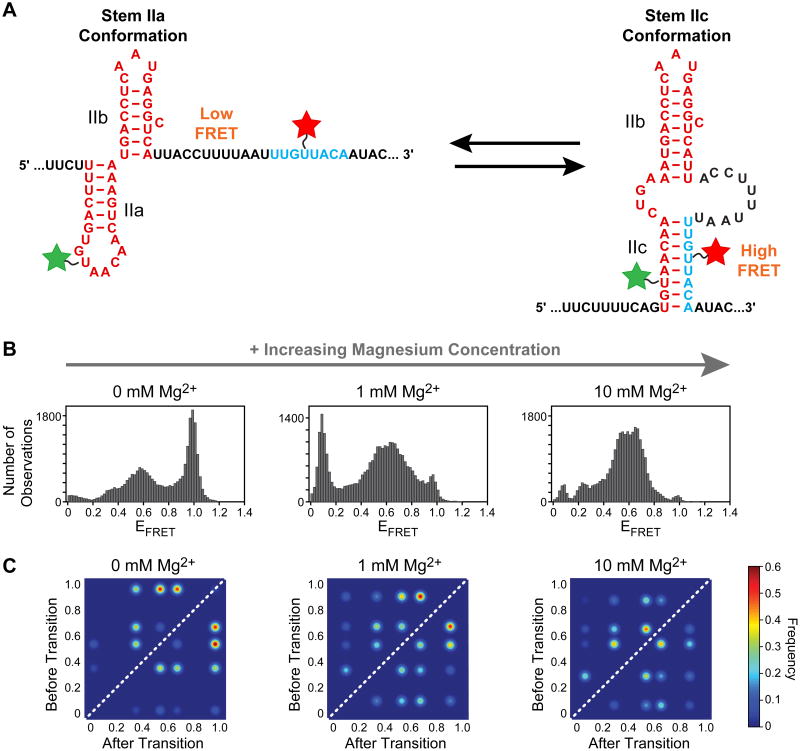
The U2 snRNA recognizes the branch site in pre-mRNA through direct basepairing to the snRNA's GUAGUA motif [50]. Both upstream and downstream of this region are RNA structural elements that impact U2's ability to recognize correct branchsite sequences. The stem II region lies downstream of the GUAGUA motif and folds into two mutually exclusive structures between which the snRNA alternates during splicing (Fig. 5A) [51]. The stem IIa structure is essential for spliceosome assembly and participates in rearrangement of the spliceosome in between 5′ splice site cleavage and exon ligation [52-54]. U2 adopts the stem IIc conformation just before or during both catalytic steps [51, 52].
Figure 5.
U2 snRNA dynamics analyzed by smFRET by Rodgers et al. [20]. (A) Two mutually exclusive RNA structures can be adopted U2 stem II and are described by different FRET states. The smFRET reporter RNA was labeled with Cy3 (green) and Cy5 (red). (B) Histograms plotting EFRET for the smFRET reporter RNA in response to Mg2+ (C) TODPs of EFRET for stem II molecular transitions. The structural transitions of the RNA increased and changed in EFRETin the presence of Mg2+ The figure shown was adapted from reference [20] and used with permission.
Rodgers et al. utilized smFRET to study stem II conformation in vitro in the presence of Mg2+ and Cus2, a protein known to genetically interact with stem II mutations in vivo [20, 55]. Since the stem II region is ∼100 nucleotides, a smFRET reporter RNA was designed from two smaller fragments that were labeled with fluorophores and joined via splinted ligation as described in Section 2. The reporter was designed such that a lower FRET state would correspond to stem IIa folding, while a higher FRET state would correspond to stem IIc formation (Fig. 5A).
smFRET experiments revealed that the wild type stem II RNA to be highly dynamic in vitro as a wide variety of FRET values were observed. To facilitate interpretation, smFRET experiments were repeated with variant stem II sequences designed to favor formation of either a IIa or IIc state. The stem IIa stabilize d construct had a broad EFRET distribution centered around 0.54, while the stem IIc stabilized construct possessed a high EFRET of 0.96. Together the data suggested that the wild type stem IIa RNA transiently sampled a stem IIc conformation in addition to range of conformations similar to those that appear upon stem IIa stabilization.
The authors probed how the EFRET distribution was affected by Mg2+. With increasing magnesium ion concentration, the population shifted from a predominantly IIc, high FRET state to a broader, lower FRET distribution more indicative of stem IIa (Fig. 5B). HMM analysis was performed on the complex FRET distributions to generate TODPs to describe the transitions at each Mg2+ concentration (Fig. 5C). The TODP analysis revealed how Mg2+ facilitates conformational sampling of stem II, properties partly shared by the Cus2 protein.
5.2. Kinetic characterization of snRNA dynamics
The U6 snRNA recognizes the 5′ splice site and is a key component of the catalytic active site of the spliceosome [56]. In the cell, U6 snRNA can basepair with itself or with the U4 or U2 snRNAs and intron [57-60]. U6 structure is heavily regulated throughout the splicing cycle, and much effort has gone towards understanding how U6 transitions between its different basepairing partners.
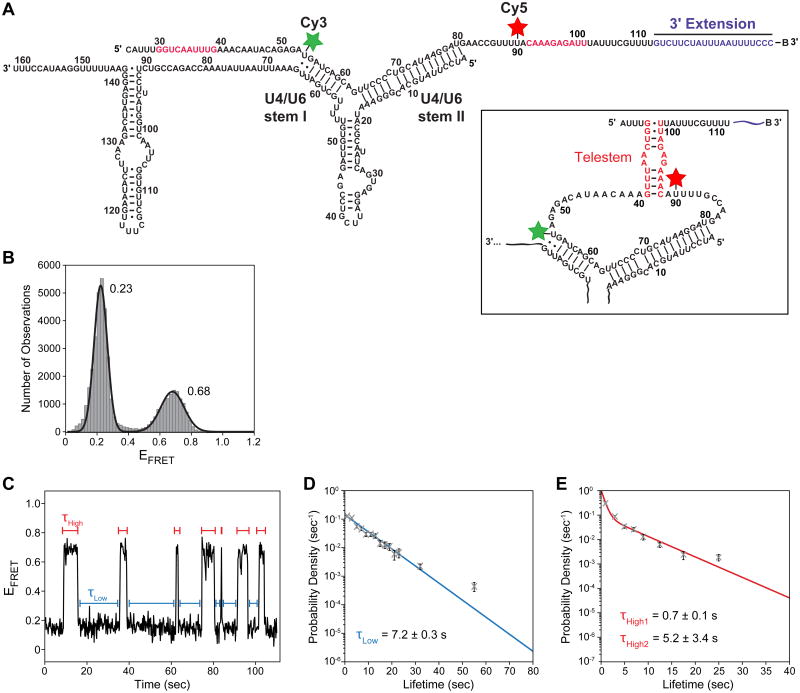
Rodgers et al. used smFRET in combination with biochemical and genetic assays to study U6 conformation when it is paired to U4 [21]. The authors designed a U6 snRNA smFRET construct that could report on potential intramolecular basepairing, leading towards telestem formation (Fig. 6A). With this construct, sw itching was observed between two conformational states of U6 only when U6 was annealed to U4. Importantly, they observed a 0.68 FRET state that corresponded to the formation of the telestem. When U6 was in a more extended conformation, the lower 0.23 FRET state was observed (Fig. 6B). In addition to FRET distributions, Rodgers et al. also analyzed the kinetics of the conformational switching [21]. This was achieved by measuring the dwell times, τ, for U6 in each FRET state followed by fitting the observed distribution of dwell times to a kinetic model using maximum likelihood methods (Fig. 6C-E). Together these data revealed that U6 is structurally dynamic when basepaired with U4, and these single molecule results were complemented by biochemical experiments showing that telestem formation leads to U4/U6 instability and unwinding.
Figure 6.
U4/U6 snRNA dynamics analyzed by smFRET by Rodgers et al. [21]. (A) 2D representation of the proposed structural states of U6 while basepaired to U4 snRNA. U6 was labeled with Cy3 (green) and Cy5 (red) fluorophores. (B) Histogram of EFRET values observed for the U4/U6 smFRET reporter shown in (A). Two states were observed, one centered at EFRET = 0.23 and one at EFRET = 0.68. (C) EFRET from a single U4/U6 molecule shows multiple and frequent transitions between the two states indicated by blue and red bars. (D and E) Measured dwell times τLow (blue) and τHigh (red) were combined and plotted as probability density histograms. Calculated lifetimes of the FRET states were obtained by fitting these distributions to maximum likelihood functions with either one (D) or two (E) kinetic parameters. The figure shown was adapted from reference [21] and used with permission.
5.3. Correlation of pre-mRNA dynamics with splicing factor binding
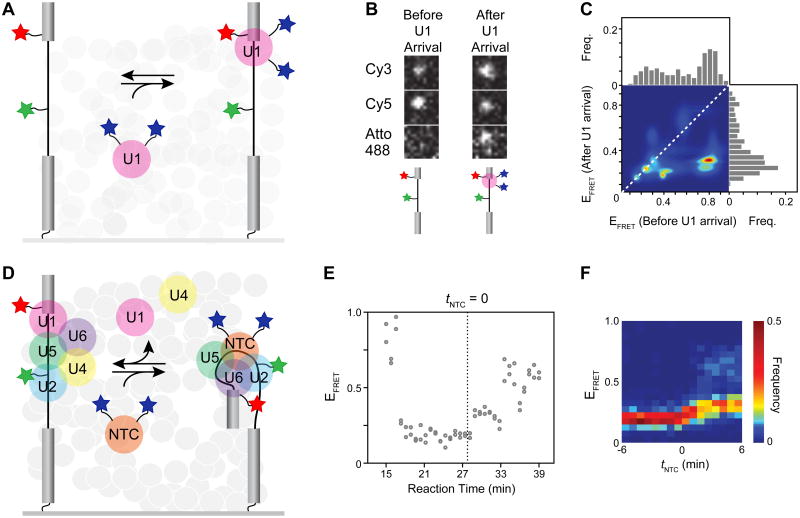
smFRET can be combined with a number of other single-molecule techniques. As an example, Crawford et al. incorporated smFRET with colocalization fluorescence microscopy to correlate changes in pre-mRNA conformation with distinct stages in spliceosome assembly [24]. The authors prepared a full-length pre-mRNA substrate for the spliceosome that included a Cy3/Cy5 FRET pair located seven nucleotides upstream of the 5′ splice site and branchsite. They monitored FRET of the immobilized RNA during splicing in yeast whole cell extract. The extract also contained fluorescent spliceosome components that had been labeled with Atto488 fluorophores (max ex/em 500/520) using the SNAP tag. In the case of labeled U1 snRNP (Fig. 7A), a 0.5 FRET state was observed which transitioned to a 0.2 state upon U1 snRNP arrival (Fig 7B). Histogram transition analysis confirmed that many molecules were indeed transitioning to a dominant state with FRET at 0.2 upon U1 binding (Fig. 7C). Surprisingly, this low FRET state persisted until after spliceosome activation had begun and the Nineteen Complex (NTC) arrived (Fig. 7D). NTC arrival was correlated with a subsequent shift to a FRET state of 0.3 (Fig. 7E-F). These results could be explained by a process occurring after NTC addition that results in bringing the branch site near the 5′ splice site in the catalytic core of the spliceosome. The smFRET experiments show NTC arrives ∼ 2 min prior to activation (Fig. 7F). This result agrees well with data reported by Blanco et al. as well as recent cryo-EM structures of activated yeast spliceosomes that were determined subsequently [10, 11, 13, 47]. In these NTC-containing structures, the branchsite and 5′ splice site are separated from one another by ∼50 Å [10, 11, 13]. These sequences only become juxtaposed following catalytic activation of the spliceosome by the Prp2 ATPase [47, 61-63].
Figure 7.
Pre-mRNA dynamics analyzed by smFRET and colocalization fluorescence microscopy by Crawford et al. [24]. (A) Experimental design used by Crawford et al. The pre-mRNA was labeled just upstream of the 5′ splice site with Cy5 (red star) and the branchsite with Cy3 (green star). The U1 snRNP was doubly labeled with Atto488 (blue stars). (B) Colocalization of U1 with the pre-mRNA (Atto488 spot) coincides with a decrease in Cy5 acceptor FRET and an increase in Cy3 donor FRET. (C) Heat map representation of EFRET transitions observed before and after U1 binding. (D) Experimental design as in panel (A) except that the NTC was doubly labeled with Atto488 (blue stars) for monitoring later stages in spliceosome assembly. (E) EFRET plotted as a function of reaction time for a single pre-mRNA molecule shows a shift from low to mid EFRET occurring after NTC arrival (dotted line). (F) Heat map representation for a population of 80 pre-mRNA molecules shows that the EFRET transition shown in panel (E) is a common feature of spliceosome assembly. Shaded circles in (A) and (D) represent the yeast whole cell extract in which the experiments were performed. The figure shown was adapted from reference [24] and used with permission.
6. Conclusion
In addition to the studies described above, many other experiments have used smFRET to interrogate the splicing machinery. These studies have encompassed a wide variety of experimental questions from U2/U6 and U4/U6 di-snRNA structure and dynamics to characterizing spliceosome remodeling by DEAH-Box ATPases [19, 22, 23, 27, 64]. While we have not discussed each of these studies in detail, it is apparent that smFRET has become an increasingly important tool for studying spliceosome structure and mechanism.
smFRET and other single-molecule methods can provide unique insights into complex macromolecular machines such as the spliceosome that undergo intricate structural rearrangements. These methods can uniquely identify short-lived or rare conformations and reaction pathways that can be obscured in bulk e xperiments. Moreover, the small sample requirements for single-molecule experiments are compatible with studies of many cellular machines which are lowly abundant or difficult to isolate in large quantities. With the recent high resolution structures of spliceosomes at different stages in the splicing reaction, it is likely that future smFRET analysis of splicing will take advantage of this additional structural information to design new experiments further probing RNA conformation during splicing. Many of the RNA and protein dynamics necessary for transitioning the spliceosome between the different stages captured by cryo-EM are still waiting to be observed.
Highlights.
Single-molecule FRET is useful for studying spliceosome RNA dynamics.
This review summarizes steps involved in a typical single-molecule FRET experiment.
These methods have been used to study snRNA and pre-mRNA dynamics during splicing.
Acknowledgments
We thank Margaret Rodgers, Tucker Carocci, and Josh Larson for feedback on the manuscript and comments and Laura Vanderploeg for assistance with figure artwork. This work was supported by the National Institutes of Health (R01 GM112735), Shaw Scientist and Beckman Young Investigator Awards, and startup funding from the University of Wisconsin-Madison, Wisconsin Alumni Research Foundation (WARF), and the Department of Biochemistry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Pomeranz Krummel DA, Oubridge C, Leung AKW, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature. 2009;458(7237):475–80. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349(6253):1182–91. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 5.Kondo Y, Oubridge C, van Roon AM, Nagai K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife. 2015;4 doi: 10.7554/eLife.04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y. The 3.8 A structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351(6272):466–75. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen TH, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SH, Nagai K. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature. 2016;530(7590):298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Lührmann R, Stark H. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016;351(6280):1416–20. doi: 10.1126/science.aad2085. [DOI] [PubMed] [Google Scholar]
- 10.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science. 2016;353(6302):904–11. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 11.Wan R, Yan C, Bai R, Huang G, Shi Y. Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science. 2016;353(6302):895–904. doi: 10.1126/science.aag2235. [DOI] [PubMed] [Google Scholar]
- 12.Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537(7619):197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, Stark H, Luhrmann R. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353(6306):1399–1405. doi: 10.1126/science.aag1906. [DOI] [PubMed] [Google Scholar]
- 14.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast step II catalytically activated spliceosome. Science. 2017;355(6321):149–155. doi: 10.1126/science.aak9979. [DOI] [PubMed] [Google Scholar]
- 15.Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai XC, Newman AJ, Nagai K. Structure of a spliceosome remodelled for exon ligation. Nature. 2017;524(7641):377–80. doi: 10.1038/nature21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertram K, Agafonov DE, Liu WT, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature advance online publication. 2017 doi: 10.1038/nature21079. [DOI] [PubMed] [Google Scholar]
- 17.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods. 2008;5(6):475–89. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wozniak AK, Nottrott S, Kuhn-Holsken E, Schroder GF, Grubmuller H, Luhrmann R, Seidel CA, Oesterhelt F. Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA. 2005;11(10):1545–54. doi: 10.1261/rna.2950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karunatilaka KS, Rueda D. Post-transcriptional modifications modulate conformational dynamics in human U2-U6 snRNA complex. RNA. 2014;20(1):16–23. doi: 10.1261/rna.041806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodgers ML, Tretbar US, Dehaven A, Alwan AA, Luo G, Mast HM, Hoskins AA. Conformational dynamics of stem II of the U2 snRNA. RNA. 2016;22(2):225–36. doi: 10.1261/rna.052233.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodgers ML, Didychuk AL, Butcher SE, Brow DA, Hoskins AA. A multi-step model for facilitated unwinding of the yeast U4/U6 RNA duplex. Nucleic Acids Res. 2016;44(22):10912–10928. doi: 10.1093/nar/gkw686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardin JW, Warnasooriya C, Kondo Y, Nagai K, Rueda D. Assembly and dynamics of the U4/U6 di-snRNP by single-molecule FRET. Nucleic Acids Res. 2015;43(22):10963–74. doi: 10.1093/nar/gkv1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornilescu G, Didychuk AL, Rodgers ML, Michael LA, Burke JE, Montemayor EJ, Hoskins AA, Butcher SE. Structural Analysis of Multi-Helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA. J Mol Biol. 2016;428(5 Pt A):777–89. doi: 10.1016/j.jmb.2015.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5′ splice site and branch site. Proc Natl Acad Sci U S A. 2013;110(17):6783–8. doi: 10.1073/pnas.1219305110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, Wood M, Villa T, Ryan DE, Pleiss JA, Maeder C, Guthrie C, Walter NG. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol. 2010;17(4):504–12. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnan R, Blanco MR, Kahlscheuer ML, Abelson J, Guthrie C, Walter NG. Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing. Nat Struct Mol Biol. 2013;20(12):1450–7. doi: 10.1038/nsmb.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semlow DR, Blanco MR, Walter NG, Staley JP. Spliceosomal DEAH-Box ATPases Remodel Pre-mRNA to Activate Alternative Splice Sites. Cell. 2016;164(5):985–98. doi: 10.1016/j.cell.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25(1):78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 29.Stephenson JD, Kenyon JC, Symmons MF, Lever AM. Characterizing 3D RNA structure by single molecule FRET. Methods. 2016;103:57–67. doi: 10.1016/j.ymeth.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Kalinin S, Peulen T, Sindbert S, Rothwell PJ, Berger S, Restle T, Goody RS, Gohlke H, Seidel CA. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat Methods. 2012;9(12):1218–25. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- 31.Lavis LD, Raines RT. Bright ideas for chemical biology. ACS chemical biology. 2008;3:142–55. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimm JB, Heckman LM, Lavis LD. The chemistry of small-molecule fluorogenic probes. Progress in molecular biology and translational science. 2013;113:1–34. doi: 10.1016/B978-0-12-386932-6.00001-6. [DOI] [PubMed] [Google Scholar]
- 33.Lakowicz JR. Springer US. 3. New York, NY: 2006. Priniciples of Fluorescence Spectroscopy. [Google Scholar]
- 34.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86(4):2530–7. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Axelrod D, Thompson NL, Burghardt TP. Total internal inflection fluorescent microscopy. Journal of microscopy. 1983;129(Pt 1):19–28. doi: 10.1111/j.1365-2818.1983.tb04158.x. [DOI] [PubMed] [Google Scholar]
- 36.Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DM. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci U S A. 2008;105(32):11176–81. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behlke M, Huang L, Bogh L, Rose S, Devor EJ. Fluorescence Quenching by Proximal G-bases. Integrated DNA Technologies. 2005 http://biophysics.idtdna.com/pdf/Fluorescence_quenching_by_proximal_G_bases.pdf.
- 38.Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. Visualizing the splicing of single pre-mRNA molecules in whole cell extract. RNA. 2008;14(1):170–9. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akiyama BM, Stone MD. Assembly of complex RNAs by splinted ligation. Methods in Enzymology. 2009;469:27–46. doi: 10.1016/S0076-6879(09)69002-9. [DOI] [PubMed] [Google Scholar]
- 40.Lamichhane R, Solem A, Black W, Rueda D. Single-molecule FRET of protein-nucleic acid and protein-protein complexes: surface passivation and immobilization. Methods. 2010;52(2):192–200. doi: 10.1016/j.ymeth.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96(6):2371–81. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3(11):891–3. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 43.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–16. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanco M, Walter NG. Analysis of complex single-molecule FRET time trajectories. Methods in enzymology. 2010;472:153–78. doi: 10.1016/S0076-6879(10)72011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohlman CE, Blanco MR, Walter NG. Putting Humpty-Dumpty Together: Clustering the Functional Dynamics of Single Biomolecular Machines Such as the Spliceosome. Methods in enzymology. 2016;581:257–283. doi: 10.1016/bs.mie.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 46.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91(5):1941–51. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanco MR, Martin JS, Kahlscheuer ML, Krishnan R, Abelson J, Laederach A, Walter NG. Single Molecule Cluster Analysis dissects splicing pathway conformational dynamics. Nat Methods. 2015;12(11):1077–84. doi: 10.1038/nmeth.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colquhoun D, Sigworth FJ. Fitting and Statistical Analysis of Single-Channel Records. In: Bert Sakmann EN, editor. Single-Channel Recording. Springer US, New York; New York: 1995. pp. 483–585. [Google Scholar]
- 49.Ghosh S, Garcia-Blanco MA. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA. 2000;6(9):1325–1334. doi: 10.1017/s1355838200992537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49(2):229–39. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 51.Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21(7):821–34. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perriman R, Ares M. Invariant U2 snRNA Nucleotides Form a Stem Loop to Recognize the Intron Early in Splicing. Mol Cell. 2010;38(3):416–427. doi: 10.1016/j.molcel.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zavanelli MI, Ares M., Jr Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes Dev. 1991;5(12b):2521–33. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]
- 54.Zavanelli MI, Britton JS, Igel AH, Ares M., Jr Mutations in an essential U2 small nuclear RNA structure cause cold-sensitive U2 small nuclear ribonucleoprotein function by favoring competing alternative U2 RNA structures. Mol Cell Biol. 1994;14(3):1689–97. doi: 10.1128/mcb.14.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18(9):5000–9. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kandels-Lewis S, Seraphin B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262(5142):2035–9. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 57.Montemayor EJ, Curran EC, Liao HH, Andrews KL, Treba CN, Butcher SE, Brow DA. Core structure of the U6 small nuclear ribonucleoprotein at 1.7-Å resolution. Nat Struct Mol Biol. 2014;21(6):544–51. doi: 10.1038/nsmb.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bringmann P, Appel B, Rinke J, Reuter R, Theissen H, Lührmann R. Evidence for the existence of snRNAs U4 and U6 in a single ribonucleoprotein complex and for their association by intermolecular base pairing. EMBO J. 1984;3(6):1357–63. doi: 10.1002/j.1460-2075.1984.tb01977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334(6179):213–8. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- 60.Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71(5):803–17. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 61.Ohrt T, Prior M, Dannenberg J, Odenwalder P, Dybkov O, Rasche N, Schmitzova J, Gregor I, Fabrizio P, Enderlein J, Luhrmann R. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA. 2012;18(6):1244–56. doi: 10.1261/rna.033316.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lardelli RM, Thompson JX, Yates JR, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010;16(3):516–528. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu HL, Cheng SC. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol Cell Biol. 2012;32(24):5056–66. doi: 10.1128/MCB.01109-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo Z, Karunatilaka KS, Rueda D. Single-molecule analysis of protein-free U2-U6 snRNAs. Nat Struct Mol Biol. 2009;16(11):1154–9. doi: 10.1038/nsmb.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]