Abstract
Safe, effective, orally delivered, live adenovirus vaccines have been in use for three decades. Recombinant derivatives of the live adenovirus vaccines may prove an economical alternative to current vaccines for a variety of diseases. To explore that possibility, we constructed a series of recombinants that express the major capsid protein (L1) of canine oral papillomavirus (COPV), a model for mucosal human papillomavirus (HPV) infection. Vaccination with virus-like particles (VLPs) composed of recombinant HPV L1 completely prevents persistent HPV infection [Koutsky, L. A., Ault, K. A., Wheeler, C. M., Brown, D. R., Barr, E., Alvarez, F. B., Chiacchierini, L. M. & Jansen, K. U. (2002) N. Engl. J. Med. 347, 1645–1651], suggesting that L1 expressed from recombinant adenoviruses might provide protective immunity. In our recombinants, COPV L1 is incorporated into adenovirus late region 5 (Ad L5) and is expressed as a member of the adenoviral major late transcriptional unit (MLTU). COPV L1 production by the most prolific recombinant is comparable to that of the most abundant adenoviral protein, hexon. COPV L1 production by recombinants is influenced by Ad L5 gene order, the specific mRNA processing signals associated with COPV L1, and the state of a putative splicing inhibitor in the COPV L1 gene. Recombinant COPV L1 protein assembles into VLPs that react with an antibody specific for conformational epitopes on native COPV L1 protein that correlate with protection in vivo. The designs of these recombinants can be applied directly to the production of recombinants appropriate for assessing immunogenicity and protective efficacy in animal models and in human trials.
Keywords: viable recombinant, virus-like particle, late-gene expression
Alive, orally administered vaccine has been used effectively for >30 years to protect United States military personnel from severe respiratory disease caused by adenovirus serotypes 4 (Ad4) and 7 (Ad7) (1). The adenovirus vaccine induces both cell-mediated and humoral immunity and confers >90% protection from respiratory adenovirus infection. In addition to efficacy, the adenovirus vaccines have economic properties that are desirable in settings where resources are limiting: Oral delivery eliminates the need for expensive equipment and extensively trained personnel, and the vaccine is effective in a single dose, making costly multiple immunizations unnecessary. Nonreplicating recombinant adenoviruses have been shown to induce cell-mediated and humoral immune responses and, in some cases, protection against a variety of pathogens (2). However, nonreplicating recombinant adenovirus vaccines generally are administered by repeated injection and frequently are most effective when combined with other methods of immunization, adding to their cost. In an effort to combine the economy, ease of administration, and efficacy of the military vaccine with the ability to efficiently induce immune responses against exogenous antigens, we have developed live recombinant adenoviruses that abundantly express foreign antigens by a mechanism that exploits the viral late-gene-expression machinery.
The eventual target of this work is human papillomavirus, the etiologic agent of cervical cancer. More than 500,000 cases of cervical cancer are diagnosed annually, and >200,000 women die each year from the disease (3). Vaccination with empty virus-like particles (VLPs), which spontaneously assemble from recombinant papillomavirus L1 capsid protein, confers immunity to papillomavirus-induced tumors in animal systems (4–6). VLPs induce strong immune responses in humans (7, 8), and a VLP vaccine was 100% effective in preventing persistent infection with HPV 16 (9). The success of the VLP vaccine is of great potential importance in the development of an effective public-health response to cervical cancer. However, because VLPs must be extensively purified and are injected, currently in three doses, VLP vaccines are likely to be expensive to manufacture and administer. Because most mortality due to cervical cancer occurs in developing countries (10) where limited public-health budgets impede vaccination programs, potentially less-expensive alternatives to VLP vaccines warrant exploration. In particular, a live recombinant adenovirus vaccine that expresses HPV L1 as it replicates in vaccinees and delivers a high, prolonged dose of antigen to both the humoral and cell-mediated immune systems is a potentially valuable complement to the VLP vaccine for use in such settings.
Canine oral papillomavirus (COPV) is a tumorigenic papillomavirus that mimics many of the features of HPV infection, including transmission at mucosal surfaces and progression of benign lesions to malignancy (5). COPV provides a papillomavirus-challenge model in one of the few experimental animals that is susceptible to human adenovirus infection (11), and immunization with recombinant COPV L1 is protective in the model. Therefore, we have focused initially on the expression of COPV L1 from viable adenovirus recombinants. Our best recombinants produce high levels of COPV L1, which assembles into VLPs and possesses conformational epitopes correlated with protection in the challenge model (12). The studies establish design principles for the construction of candidate COPV L1 vaccine strains that can be tested in the canine challenge model for HPV L1-producing strains that potentially are suitable for human clinical trials and for the construction of viable recombinant adenoviruses that express a wide variety of other antigens.
Materials and Methods
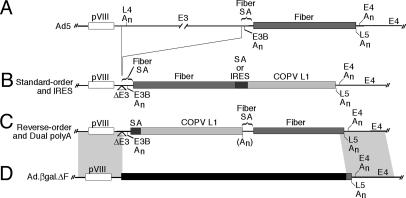
Construction and Characterization of Recombinants. FFIL (see Table 1 for nomenclature) was constructed in the adenovirus yeast artificial chromosome system (13). Other viruses were produced by recombination in transfected mammalian cells between the adenovirus fiber-deletion mutant Ad5.βgal.ΔF (14) and shuttle plasmids (Fig. 1). Construction details are presented in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Immunoprecipitations, immunoblots, and Northern blots were performed as described in ref. 15.
Table 1. Structure of late region five in recombinants.
| Group | Code | First SA | First gene | Second SA | Second gene | L1 mRNA | L1 (Blot) | L1 (35S) |
|---|---|---|---|---|---|---|---|---|
| Dual SA | FFFL | Fiber† | Fiber | Fiber | L1 | - | - | - |
| FFHL | Fiber | Fiber | Hexon‡ | L1 | - | - | nd | |
| FFSL | Fiber | Fiber | SV40 (200) | L1 | (+) | + | + | |
| FLFF | Fiber | L1 | Fiber | Fiber | - | - | nd | |
| SLFF | SV40 (200)§ | L1 | Fiber | Fiber | - | - | nd | |
| S*LFF | SV40 (346)¶ | L1 | Fiber | Fiber | (+) | (+) | nd | |
| FFFO | Fiber | Fiber | Fiber | Opti L1 | + | ++ | ++ | |
| FFSO | Fiber | Fiber | SV40 (200) | Opti L1 | ++++ | ++++ | ++++ | |
| FOFF | Fiber | Opti L1∥ | Fiber | Fiber | - | - | nd | |
| Dual PolyA | FLAFF | Fiber | L1 | Fiber | Fiber | + | ++ | + |
| FOAFF | Fiber | Opti L1 | Fiber | Fiber | ++ | +++ | ++ | |
| IRES | FFIL | Fiber | Fiber | IRES†† | L1 | ++ | ++ | nd |
| — | H2d/810 | Fiber | Fiber | None | None | na | na | na |
The first (5′) and second (3′) genes and associated SAs are listed for each recombinant. Codes indicate the arrangement of elements in the recombinants. For example, FFFL contains a Fiber SA, the Fiber gene, a second Fiber SA, and COPV L1. O, optimized L1 gene; A, poly(A) site between the two genes; I, IRES. All recombinants carry a poly(A) site at the 3′ end of the second gene that is not explicitly indicated. The last three columns give an indication of the quantities of COPV L1 protein and RNA produced from undetectable (-) to extravagant (++++). (+), weak or irreproducible signal; na, not applicable; nd, not determined.
Fiber SA region; Ad5 nucleotides 30810-30979.
Hexon SA region; Ad5 nucleotides 18641-18841.
SV40 large T antigen intron SA region; SV40 nucleotides 4572-4770.
Complete SV40 large T antigen intron; SV40 nucleotides 4572-4917.
COPV L1 gene mutagenized in the region 91-133 to relieve inhibition of splicing.
Poliovirus internal ribosome entry site; poliovirus nucleotides 3186-3536. H2d/810 is an adenovirus 2 mutant that carries an E3 deletion mutation identical to that present in the COPV L1-containing recombinants.
Fig. 1.
Structure of Ad L5 in recombinants. The organization of Ad L5 in Ad5 (A), standard-order (B), and reverse-order (C) recombinants is diagrammed. Recombination between shuttle plasmids and Ad.βgal.ΔF (D) occurs in 1-kb regions of homology that flank Ad L5 (shaded). An, poly(A) site. The region deleted in Ad.βgal.ΔF is indicated by a black bar (14).
VLP Purification and Electron Microscopy. Adenovirus particles and COPV VLPs were recovered from recombinant-infected cells, concentrated by centrifugation on discontinuous CsCl gradients (16), and purified by centrifugation in successive equilibrium CsCl gradients of average density 1.315 g/ml and 1.29 g/ml. A prominent band that contains COPV L1 was visible in each gradient. Carbon-coated grids, negatively charged by glow discharge, were floated on fractions from the final gradient for 70 s. The grids were rinsed on drops of water and PBS. For gold labeling (17), the rinsed grids were transferred to drops of PBS containing COPV L1 conformation-specific rabbit antibody (1:500) (12) for 20 min. After three rinses with PBS, the grids were placed on drops containing gold-conjugated anti-rabbit IgG (6-nm particles 1:10, Jackson ImmunoResearch) for 6 min. The grids were washed three times in PBS and twice in water and were negatively stained by floating on a drop of 2% uranyl acetate for 1 min. Excess stain was wicked from the grid, and the grids were allowed to air dry. The grids were examined with a Philips CM12 electron microscope.
Results
Vector Design Rationale. Abundant antigen production probably will be required to achieve optimal protection by vaccination with recombinant adenoviruses. By far, the highest levels of expression in adenovirus-infected cells are of proteins of the major late transcriptional unit (MLTU) (18). The MLTU encodes ≈15 “late” proteins, including most of the proteins found in virus particles. Transcription of the MLTU is directed by the major late promoter (MLP), which lies near nucleotide 6,000 on the 36-kb adenoviral genome (Fig. 2A). The primary transcript of the MLTU is ≈30 kb long, extending from the MLP rightward, essentially to the end the genome. Individual mRNAs for each of the late proteins are produced from this transcript by alternative splicing and polyadenylation. MLTU-derived late mRNAs share an untranslated leader consisting of three short promoter-proximal exons that, in most late mRNAs, is spliced to a splice acceptor (SA) located immediately upstream of one of the late genes. The position of the acceptor for the final splice determines the protein produced from a particular RNA. Polyadenylation of late mRNAs occurs alternatively at five sites, and late genes are assigned to a late region (Ad L1–Ad L5; Fig. 2 A) based on the poly(A) site used in the genes' mRNAs. Four late regions produce multiple 3′-coterminal mRNA species that differ from one another in the position of the final SA used and encode different proteins (Fig. 2B). The fifth, Ad L5, encodes a single protein, fiber, in adenovirus type 5 (Ad5).
Fig. 2.
Late transcription in Ad5 and recombinants. (A) The major late transcriptional region (MLTU) extends rightward from the major late promoter (MLP) and contains five late regions (Ad L1–Ad L5). Arrowheads mark polyadenylation sites. Black arrows, exons; gray lines, introns. (B) A typical multigenic adenoviral late region (Ad L4). The positions and sizes of the three Ad L4 genes are shown by black boxes; the corresponding mRNAs are indicated by arrows. The protein translated from each mRNA is indicated beside the arrow. Ad L4 mRNAs are spliced to the tripartite late leader (not shown) at their 5′ ends. L4 An, Ad L4 poly(A) site. (C) Recombinant Ad L5 regions and expected mRNAs. Dual SA, SAs precede each gene and the region possesses a single poly(A) site; Dual PolyA, each gene is individually associated with a SA and a poly(A) site; IRES, a poliovirus IRES separates the two genes. Single SA and poly(A) sites are located at the 5′ and 3′ ends of Ad L5, respectively.
The structures of the MLTU and late mRNAs suggest that an exogenous gene would be expressed as if it were a viral late gene if it were inserted with appropriate processing signals into the MLTU. To test that hypothesis, we constructed a series of 12 Ad5 recombinants containing the COPV L1 gene and a variety of regulatory signals inserted into Ad L5. Ad5 was chosen for these studies to permit analysis of the properties of diverse recombinants in an extensively characterized background. Recombinants fall into three structural categories (Fig. 2C and Table 1). (i) Dual-SA constructs carry the fiber and COPV L1 genes, each with an individual SA at its 5′ end. A single poly(A) site is located at the 3′ end of the distal gene. Dual-SA constructs simulate the organization of multigenic adenoviral late regions and should produce two 3′-coterminal RNAs by alternative splicing to the two SAs and polyadenylation at the 3′ end of Ad L5. Four different SAs were used in the construction of dual-SA vectors: the Ad5 fiber and hexon SAs, the complete SV40 large T antigen intron, and a 200-bp segment of the intron containing the SA. (ii) Dual-polyA constructs contain the COPV L1 and fiber genes each associated with both a SA and an individual poly(A) signal. This construction mimics the organization of the fiber regions in the enteric adenoviruses Ad40 and Ad41. Dual-polyA site constructs were expected to produce individual mRNAs for each gene by splicing to the SA immediately upstream of the gene and polyadenylation at the signal immediately downstream. (iii) An internal ribosome entry site (IRES) construct that contains the COPV L1 gene downstream of the fiber gene with an intervening IRES. This virus should produce a single bicistronic mRNA that can be translated to produce both products. Recently, this approach was used to express luciferase and p53 as part of Ad L5 in conditionally replicating vectors (19, 20).
COPV L1 Expression by Recombinants. The wild-type COPV L1 gene (21) initially was used to prepare two recombinants: FFFL and FFIL (see Table 1 for structures and nomenclature). The dual-SA recombinant FFFL (Fig. 2C1) contains a duplicate copy of the fiber SA and the COPV L1 gene located immediately downstream of the native fiber gene in Ad L5; FFIL contains a poliovirus IRES and the COPV L1 gene in the same position (Fig. 2C3). In both recombinants, the native Ad L5 poly(A) signal is ablated by mutation, and a new signal is positioned at the 3′ end of COPV L1.
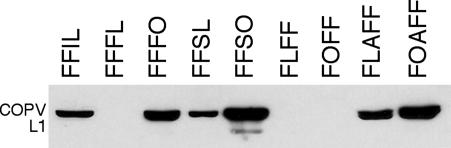
COPV L1 protein production by these recombinants was assessed by immunoblotting. FFIL produced COPV L1 but FFFL did not (Fig. 3). In Northern blots of RNA from FFIL-infected cells, both COPV L1 (Fig. 4A) and fiber (data not shown) probes detected an RNA of ≈4 kb, the size expected for a bicistronic message spliced at the SA at the 5′ end of the fiber gene and polyadenylated at the 3′ end of the COPV L1 gene. Because of the IRES, that mRNA should produce both proteins. In RNA from FFFL-infected cells, both probes detected the ≈4-kb band expected from splicing at the 5′ end of the fiber gene and polyadenylation at the 3′ end of COPV L1. That message should be translated to produce only fiber, encoded by the first ORF in the RNA. The COPV L1 probe did not detect the ≈2-kb mRNA that should arise by splicing to the duplicated fiber SA at the 5′ end of the COPV L1 gene and that should be translated to produce the COPV L1 protein. The failure of FFFL to produce COPV L1 protein, therefore, is a consequence of failure to accumulate the appropriate mRNA.
Fig. 3.
Expression of COPV L1 by recombinant adenoviruses. Infected cell lysates were examined by immunoblotting with anti-L1 antiserum. Three recombinants not shown (FFHL, SLFF, and S*LFF) produced no detectable COPV L1. For an explanation of the nomenclature, see Table 1.
Fig. 4.
L1- and fiber-containing RNAs in recombinant-infected cells. (A) RNAs detected with a COPV L1 probe. The approximate expected positions of RNAs containing both the fiber and L1 ORFs (≈4 kb) and L1 only (≈2 kb) are indicated on the left. Superimposed on each band is the protein(s) translated from that RNA (F, fiber; L1, COPV L1). RNAs marked with an asterisk are formed by splicing to an Ad L4 SA and polyadenylation at a site in Ad L5 and do not encode L1 or fiber protein. (B) RNAs detected with a fiber probe. The ≈2-kb RNA detected by this probe encodes fiber not COPV L1. Asterisks mark bands produced by polyadenylation at a duplicated E3B site located between L1 and fiber in some recombinants.
To test whether other SA regions might mediate expression of the COPV L1 gene in constructs similar to FFFL and whether gene order critically affects Ad L5 gene expression, five additional dual-SA recombinants were prepared with the wild-type COPV L1 gene. Two recombinants retained the standard fiber–COPV L1 gene order with the hexon SA or a 200-bp segment of the SV40 large T antigen intron between fiber and COPV L1 (FFHL and FFSL). Three recombinants had the reverse gene order (COPV L1–fiber) with the fiber SA, the 200-bp SV40 large T intron SA, or the entire 346-bp SV40 large T intron upstream of COPV L1 (FLFF, SLFF, and S*LFF). A recombinant with the COPV L1–fiber gene order, a fiber SA upstream of each gene, and a newly created poly(A) signal between COPV L1 and the fiber gene (dual-polyA construct FLAFF; Fig. 2C2) also was prepared. Of these recombinants, two produced COPV L1: FFSL and FLAFF (Fig. 3). The remaining recombinants produced either no COPV L1 or small amounts detected irregularly (data not shown). All viruses examined produced fiber protein (see Fig. 7A, which is published as supporting information on the PNAS web site). Each COPV L1-positive virus tested made the protein with late-gene kinetics (see Fig. 8, which is published as supporting information on the PNAS web site).
Northern blots confirmed that each recombinant that produced COPV L1 protein also produced COPV L1 mRNA of the size and genetic content predicted from the recombinant's structure (Fig. 4). FFSL accumulates modest amounts of an mRNA of ≈2 kb that is detected only by the COPV L1 probe and larger quantities of an RNA of ≈4 kb that hybridizes to both probes, consistent with the expected pair of 3′-coterminal mRNAs. In RNA from FLAFF-infected cells, both probes detect RNAs of ≈2 kb, also as expected: FLAFF should generate two distinct mRNAs encoding fiber and COPV L1, respectively, each produced by splicing immediately upstream of the gene and polyadenylation immediately downstream and each ≈2 kb in length. COPV L1-negative recombinants fail to accumulate the mRNA species expected to encode COPV L1, again suggesting that a defect in mRNA synthesis or stability is responsible for the failure to produce COPV L1 protein.
Mutagenesis of the COPV L1 Gene Can Increase COPV L1 Expression. Collier et al. (22) identified regions in the 5′ end of the HPV-16 L1 gene that act to inhibit the appearance of HPV L1 mRNA in the cytoplasm and, more recently, have suggested that a conserved AU-rich element in that region specifically inhibits L1 splicing (Stefan Schwartz, personal communication, and ref. 45). A similar sequence in COPV L1 (base pairs 91–133) suggested that COPV L1 may also contain such an element. Therefore, an “optimized” COPV L1 gene containing mutations that alter nucleotide but not amino-acid sequence in that region was prepared (see Fig. 9, which is published as supporting information on the PNAS web site) and used to construct four new recombinants (FFFO, FFSO, FOAFF, and FOFF). Compared with analogous constructs containing wild-type COPV L1 genes, three of the optimized constructs (FFFO, FFSO, and FOAFF) produced substantially larger amounts of COPV L1 protein (Fig. 3). FFFO and FFSO also accumulated increased amounts of the ≈2-kb COPV L1-containing RNA that is predicted to arise from splicing immediately upstream of the COPV L1 gene (Fig. 4A). The properties of these recombinants are consistent with the hypothesis that the 5′ portion of the COPV L1 gene, like the HPV L1 gene, contains a sequence that inhibits mRNA accumulation, which was destroyed in constructing the optimized gene. Notably, these mutations do not enhance COPV L1 gene expression from a plasmid that contains a strong constitutive SA (pCI-neo, Promega) (data not shown). In FOAFF-infected cells, a 2-kb RNA species that hybridizes with the COPV L1 probe replaces a diffuse and less intense band from FLAFF-infected cells that may be a mixture of species with abnormal structures resulting from interference with splicing by the COPV L1inhibitory element. Increased COPV L1 protein production in FOAFF-infected cells may therefore reflect an increased level of properly spliced mRNA due to ablation of the inhibitory element. Like FLFF, FOFF does not produce COPV L1 protein or the expected COPV L1 mRNA. The sequence of the SA and the 5′ region of the COPV L1 gene in both recombinants was confirmed by sequencing viral DNA. Failure of FOFF, in particular, to produce COPV L1 must therefore reflect a property of the COPV L1 gene apart from the putative COPV L1-splicing inhibitor.
In addition to the expected fiber and COPV L1 mRNA species, all recombinants produced Ad L5 RNAs larger than those predicted from their structures (Fig. 4A and data not shown). The sizes of these RNAs are consistent with splicing to SAs in Ad L4 and polyadenylation at the first available site in Ad L5, and each large Ad L5 RNA also hybridizes to a probe from the distal Ad L4 gene pVIII (data not shown). The Ad L4 poly(A) site was removed in construction of the recombinants, and polyadenylation of Ad L4 mRNAs was expected to occur at the E3B site, which is retained immediately downstream of Ad L4 (Fig. 1). The expected Ad L4 mRNAs are made by recombinants in addition to the large species, indicating that both the E3B and Ad L5 poly(A) sites are used to form L4 messages. H2dl810, which carries an identical E3 deletion and also lacks the native Ad L4 poly(A) signal, produces Ad L4 mRNAs polyadenylated only at the E3B site, suggesting that the COPV L1 gene may influence poly(A) site selection in recombinants.
In general, synthesis of adenoviral late proteins is not strongly affected by the modifications made in the recombinants. However, fiber accumulation is reduced in cells infected by FFSO (Fig. 7A). The titer of FFSO stocks on 293 cells is reduced, compared with the other recombinants (≈20-fold), but is restored on fiber-producing C1B cells. We conclude that reduced fiber production limits growth of the FFSO recombinant. The significance of this impaired growth for the vaccine potential of FFSO is not clear. Reduction in growth may reduce activity of FFSO in inducing immunity but may also provide a measure of attenuation that increases safety.
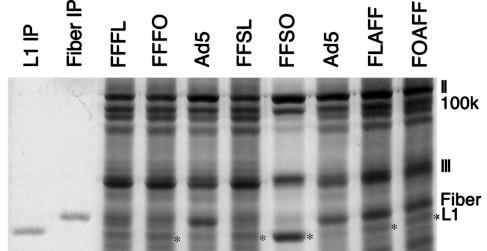
Immunoblotting gives little indication of the mass of protein produced. Therefore, we examined recombinant-infected cells after metabolic labeling with 35S cysteine and methionine to estimate production of COPV L1 relative to adenoviral late proteins. A COPV L1 band (identified by comparison to immunoprecipitated COPV L1 protein) is apparent in each lysate that contains COPV L1 detectable by immunoblotting (Fig. 5, asterisks). Strikingly, in FFSO lysates, COPV L1 is the most prominent single species after adenovirus hexon. We conclude that COPV L1 is produced in significant quantities by several adenovirus recombinants and in large quantities by FFSO.
Fig. 5.
35S metabolic labeling of proteins produced by recombinants. Proteins synthesized in infected cells were labeled, fractionated on a polyacrylamide/SDS gel, and autoradiographed. The positions of adenoviral proteins II (Hexon), 100k, III, Fiber, and COPV L1 are indicated on the right. The asterisks mark bands of COPV L1 protein. Lanes labeled L1 IP and Fiber IP contain labeled, immunoprecipitated proteins.
Recombinants Produce COPV L1 Protein with a Native Conformation. In animals, protective antibody is induced only by L1 protein with an appropriate native conformation (12). To determine whether the COPV L1 synthesized by adenovirus recombinants adopts the correct conformation, lysates of FOAFF-infected cells were examined by immunoprecipitation with a conformation-specific antibody (12). The antibody precipitated COPV L1 from extracts produced by gentle lysis of FOAFF-infected cells but not from FOAFF lysates boiled to denature proteins (see Fig. 10, which is published as supporting information on the PNAS web site). Antibody against a linear epitope detects COPV L1 protein in immunoblots of both native and boiled lysates. Therefore, COPV L1 expressed from adenovirus recombinants contains native conformational epitopes.
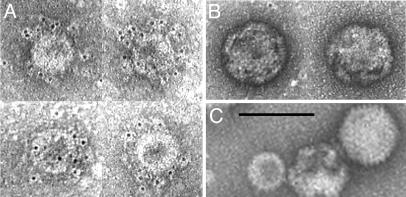
We also examined lysates of recombinant-infected cells for VLPs by electron microscopy. CsCl density-gradient centrifugation was used to isolate particles with the buoyant density of HPV 16 VLPs (1.29 g/cm3) from FFSO-infected cells. The particles were applied to carbon-coated grids, treated with the conformation-specific antibody, and then treated with gold-conjugated secondary antibody. The grids were negatively stained with uranyl acetate and photographed. The particle preparations contained both adenovirus particles (recognizable by size, shape, and some visible fibers) and numerous spherical particles of the size expected for VLPs, ≈50 nm (Fig. 6C). Most of the VLP-sized particles (Fig. 6A), but none of the adenoviral particles (Fig. 6B), were extensively labeled with the gold conjugate. We conclude that recombinants produce conformationally correct COPV L1 and direct the assembly of COPV VLPs in infected cells.
Fig. 6.
COPV VLP production by recombinants. (A) Purified VLPs from FFSO-infected cells reacted with COPV L1 conformation-specific antibody and labeled with gold-conjugated secondary antibody. The tiny black dots are the ImmunoGold stain. (B) Adenovirus particles from the same grid are not labeled. (C) Adenovirus particles and a VLP from a grid not treated with antibody. (Scale bar, 100 nm.)
Discussion
During papillomavirus infection, late-gene expression is restricted to terminally differentiated epithelial cells by transcriptional and posttranscriptional mechanisms. Posttranscriptional restrictions, in particular, also render papillomavirus late-gene expression inefficient in a variety of heterologous systems by inhibiting mRNA splicing and polyadenylation and by reducing mRNA stability (22–26). Among our recombinants, production of COPV L1 protein is correlated with accumulation of mRNA. Because adenoviral late mRNA levels are not generally affected by the inserted COPV L1 genes (data not shown), transcriptional regulation of COPV L1 expression is unlikely. However, COPV L1 mRNA levels are affected strongly by the nature and positions of associated SAs and poly(A) signals. Thus, it is likely that mRNA splicing and/or polyadenylation primarily influence production of COPV L1 by these recombinants.
The splicing of L1 mRNAs is regulated by exonic splicing enhancers (ESEs) and exonic splicing suppressors (ESSs) in bovine papillomavirus 1 and HPV 16 (22, 23). ESS elements generally regulate splicing at SAs lacking good consensus branch-point sequences and/or a long uninterrupted polypyrimidine tract (27). Like most SAs in the MLTU (28), each of the SAs used here is suboptimal by these criteria and potentially subject to regulation by sequences in COPV L1. Consistent with regulation of splicing by an ESS in COPV L1, mutagenesis of a putative inhibitory element strikingly increased mRNA accumulation from recombinants. The sequence mutagenized in COPV L1 contains a potential binding site for the cellular polypyrimidine-tract-binding protein (PTB), a potent modulator of splicing (29). The site is destroyed by the mutations that enhance COPV L1 expression (Fig. 9), suggesting that PTB may inhibit the function of SAs located at the 5′ end of COPV L1. The web utility esefinder (30) revealed that the mutations introduced six potential SR-protein-binding sites in COPV L1 and eliminated one (Fig. 9); any of these alterations also might contribute to increased expression of the optimized gene.
None of the reverse-order (COPV L1–fiber) dual-SA recombinants expressed COPV L1. In particular, the combination of the fiber SA and the optimized COPV L1 gene, which is functional when located at the 3′ end of Ad L5 in FFFO, fails to mediate COPV L1 mRNA production when it lies at the 5′ end of Ad L5 in FOFF. Gene order in dual-SA constructs strongly affects the distance between the COPV L1-associated SA and the Ad L5 poly(A) site, which are separated by ≈1.5 kb in COPV L1-positive FFFO and by ≈3.4 kb in COPV L1-negative FOFF. All genes in the MLTU lie within long (up to 4 kb) 3′-terminal exons. 3′-terminal exons are believed to be defined by interactions between factors bound at poly(A) sites and SAs, and natural members of the MLTU may possess features that stimulate exon definition over long distances that other genes do not. The expression phenotypes of our recombinants, therefore, might be explained by failure of COPV L1 to support terminal exon definition over long distances.
In FLAFF and FOAFF, the insertion of a single nucleotide immediately 3′ of COPV L1 to create a new poly(A) signal dramatically improves COPV L1 mRNA accumulation compared with the otherwise identical FLFF and FOFF recombinants. This finding is consistent with the suggestion that a poly(A) signal close to COPV L1 enhances terminal-exon definition by a COPV L1-associated SA. Alternatively, addition of the single nucleotide might inactivate an element that suppresses COPV L1 expression. Sequences that destabilize L1 transcripts (25, 26) or inhibit polyadenylation (24) have been identified in the 3′ untranslated regions (UTRs) of L1 messages from other papillomaviruses. The COPV L1 DNA segment used here contains ≈20 bp of the COPV L1 3′ UTR but has no obvious homology to previously identified inhibitory sequences. It seems likely that the spacing of the polyadenylation and SA sites is the critical difference between FLFF/FOFF and FLAFF/FOAFF.
The military adenovirus vaccine was developed to prevent prostrating upper-respiratory disease induced by Ad4 and Ad7 (1, 31, 32). The vaccine consists of lyophilized stocks of live Ad4 and Ad7 strains originally isolated from affected individuals and incorporated into enteric-coated tablets given by mouth. Vaccination results in an asymptomatic intestinal-tract infection that induces neutralizing antibody in 80–95% of vaccinees, and a single dose of the oral vaccine protected vaccinees against subsequent adenoviral respiratory disease with an efficacy of 70–100% (average >90%) in controlled trials involving >42,000 soldiers (1). Serious adverse reactions to the Ad4 and Ad7 vaccines have not been reported despite administration to hundreds of thousands of recruits (1), including 21 with early HIV infection (mean CD4+ T cell count of 523 per mm3) (33). These properties of the military vaccines suggest that viable recombinant adenovirus vaccines could provide economical, safe, effective protection against additional pathogens.
Defective recombinant adenovirus vaccines have been extensively characterized. Such recombinants induce protective immunity in animals to infectious diseases ranging from malaria (34) to Ebola virus (35) and induce humoral and cellular responses to HPV 16 L1 in primates (36). However, defective adenoviruses generally are repeatedly injected, immunization requires substantial doses of purified virus [108 plaque-forming units (pfu) per mouse or 1010 pfu per monkey vs. 104.5 TCID50 for the Ad4/7 vaccines in humans], and most regimens combine vaccination by adenoviruses and another agent (e.g., DNA) (35). Thus, defective adenovirus vaccines have the disadvantage of expense of manufacture and administration. Live recombinant adenoviral vaccines that use a free-standing MLP-based expression system distinct from that developed here also have been constructed (11, 37, 38). Derivatives expressing simian immunodeficiency virus (SIV) proteins have been exhaustively examined in a SIV/macaque challenge model, where both humoral and cell-mediated responses to the SIV proteins were induced and were partially protective against mucosal challenge with SIV (39, 40). These vectors lack E3, as do our recombinants, indicating that E3 is not required for sufficient viral growth to induce immunity and that E3 deletions do not increase the pathogenicity of recombinants (41).
Animal tests of COPV L1 recombinants will be required to assess efficacy, the effects of such variables as preexisting immunity to the vector (42), and possible requirements for boosting. Because our recombinants express COPV L1 only in cells that are permissive for replication, most laboratory animals, including mice, are unsuitable for that purpose. Therefore, animal tests of COPV L1 recombinants probably will be conducted with derivatives of Ad7 in dogs, which are permissive for that serotype (11). Human trials of HPV vaccine strains also would be conducted with Ad7, which is less seroprevalent than Ad5 (43) and is licensed for human vaccine use. All the genes and regulatory elements used in the construction of our Ad5 recombinants are readily identifiable in the Ad7 sequence (44), confirming the feasibility of using our expression strategy in Ad7. A primate model for tests of viable adenovirus vaccines is also available in an Ad5 background for recombinants targeting appropriate pathogens (39).
The efficacy, safety, relative economy, and ease of administration of the oral adenovirus vaccines make it reasonable to envision recombinant adenovirus-based vaccines that are well suited for mass vaccination campaigns, even in locales where a lack of infrastructure and funding present obstacles to the use of other vaccine types. The general applicability of recombinant expression vectors and the high-level expression achieved here suggest that a wide variety of pathogens are potential targets of viable adenovirus recombinants. In the context of papillomaviruses, recombinants that abundantly express L1 protein capable of assembly into VLPs, combined with the advantages of the existing adenoviral vaccines, make the development of a viable adenovirus-based HPV vaccine a promising component of efforts to reduce the incidence of cervical cancer in those parts of the world where it imposes its greatest burden.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants 5R01CA82127, 5T32AI07417, and R01CA53371.
Author contributions: M.B. and G.K. designed research; M.B., J.D., E.H., and G.K. performed research; R.S. contributed new reagents/analytic tools; M.B., J.D., E.H., R.S., and G.K. analyzed data; and M.B. and G.K. wrote the paper.
Abbreviations: Ad4, 5, and 7, adenovirus serotypes 4, 5, and 7; Ad L5, adenovirus late region 5; COPV, canine oral papilloma virus; HPV, human papilloma virus; IRES, internal ribosome entry site; MLP, major late promoter; MLTU, major late transcriptional unit; SA, splice acceptor; VLP, virus-like particle.
References
- 1.Gaydos, C. A. & Gaydos, J. C. (2004) in Vaccines, eds. Plotkin, S. A. & Orenstein, M. D. (Saunders, Philadelphia), pp. 863–885.
- 2.Tatsis, N. & Ertl, H. C. (2004) Mol. Ther. 10, 616–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, F. X., Lorincz, A., Munoz, N., Meijer, C. J. & Shah, K. V. (2002) J. Clin. Pathol. 55, 244–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitburd, F., Kirnbauer, R., Hubbert, N. L., Nonnenmacher, B., Trin-Dinh-Desmarquet, C., Orth, G., Schiller, J. T. & Lowy, D. R. (1995) J. Virol. 69, 3959–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzich, J. A., Ghim, S. J., Palmer-Hill, F. J., White, W. I., Tamura, J. K., Bell, J. A., Newsome, J. A., Jenson, A. B. & Schlegel, R. (1995) Proc. Natl. Acad. Sci. USA 92, 11553–11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirnbauer, R., Chandrachud, L. M., O'Neil, B. W., Wagner, E. R., Grindlay, G. J., Armstrong, A., McGarvie, G. M., Schiller, J. T., Lowy, D. R. & Campo, M. S. (1996) Virology 219, 37–44. [DOI] [PubMed] [Google Scholar]
- 7.Harro, C. D., Pang, Y. Y., Roden, R. B., Hildesheim, A., Wang, Z., Reynolds, M. J., Mast, T. C., Robinson, R., Murphy, B. R., Karron, R. A., et al. (2001) J. Natl. Cancer Inst. 93, 284–292. [DOI] [PubMed] [Google Scholar]
- 8.Pinto, L. A., Edwards, J., Castle, P. E., Harro, C. D., Lowy, D. R., Schiller, J. T., Wallace, D., Kopp, W., Adelsberger, J. W., Baseler, M. W., et al. (2003) J. Infect. Dis. 188, 327–338. [DOI] [PubMed] [Google Scholar]
- 9.Koutsky, L. A., Ault, K. A., Wheeler, C. M., Brown, D. R., Barr, E., Alvarez, F. B., Chiacchierini, L. M. & Jansen, K. U. (2002) N. Engl. J. Med. 347, 1645–1651. [DOI] [PubMed] [Google Scholar]
- 10.Bosch, F. X. & de Sanjose, S. (2003) J. Natl. Cancer Inst. Monogr., 3–13. [DOI] [PubMed]
- 11.Chengalvala, M., Lubeck, M. D., Davis, A. R., Mizutani, S., Molnar-Kimber, K., Morin, J. & Hung, P. P. (1991) Vaccine 9, 485–490. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Y., Ghim, S. J., Jenson, A. B. & Schlegel, R. (1998) J. Gen. Virol. 79, 2137–2146. [DOI] [PubMed] [Google Scholar]
- 13.Ketner, G., Spencer, F., Tugendreich, S., Connelly, C. & Hieter, P. (1994) Proc. Natl. Acad. Sci. USA 91, 6186–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Von Seggern, D. J., Chiu, C. Y., Fleck, S. K., Stewart, P. L. & Nemerow, G. R. (1999) J. Virol. 73, 1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 16.Ketner, G. & Boyer, J. (1998) in Adenovirus Methods and Protocols, ed. Wold, W. S. M. (Humana, Totowa, NJ).
- 17.Hultgren, S. J., Porter, T. N., Schaeffer, A. J. & Duncan, J. L. (1985) Infect. Immun. 50, 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shenk, T. (2002) in Fundamental Virology, eds. Knipe, D. M. & Howlet, P. M. (Lippincott, Philadelphia), pp. 1053–1088.
- 19.Rivera, A. A., Wang, M., Suzuki, K., Uil, T. G., Krasnykh, V., Curiel, D. T. & Nettelbeck, D. M. (2004) Virology 320, 121–134. [DOI] [PubMed] [Google Scholar]
- 20.Sauthoff, H., Pipiya, T., Heitner, S., Chen, S., Norman, R. G., Rom, W. N. & Hay, J. G. (2002) Hum. Gene Ther. 13, 1859–1871. [DOI] [PubMed] [Google Scholar]
- 21.Ghim, S. J., Jenson, A. B. & Schlegel, R. (1992) Virology 190, 548–552. [DOI] [PubMed] [Google Scholar]
- 22.Collier, B., Oberg, D., Zhao, X. & Schwartz, S. (2002) J. Virol. 76, 2739–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng, Z. M., He, P. & Baker, C. C. (1996) J. Virol. 70, 4691–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furth, P. A., Choe, W. T., Rex, J. H., Byrne, J. C. & Baker, C. C. (1994) Mol. Cell. Biol. 14, 5278–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennedy, I. M., Haddow, J. K. & Clements, J. B. (1991) J. Virol. 65, 2093–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokolowski, M., Zhao, C., Tan, W. & Schwartz, S. (1997) Oncogene 15, 2303–2319. [DOI] [PubMed] [Google Scholar]
- 27.Zheng, Z. M., He, P. J. & Baker, C. C. (1999) J. Virol. 73, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhlemann, O., Kreivi, J. P. & Akusjarvi, G. (1995) J. Virol. 69, 7324–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez, I., Lin, C. H., McAfee, J. G. & Patton, J. G. (1997) RNA 3, 764–778. [PMC free article] [PubMed] [Google Scholar]
- 30.Cartegni, L., Wang, J., Zhu, Z., Zhang, M. Q. & Krainer, A. R. (2003) Nucleic Acids Res. 31, 3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Top, F. H., Jr., Grossman, R. A., Bartelloni, P. J., Segal, H. E., Dudding, B. A., Russell, P. K. & Buescher, E. L. (1971) J. Infect. Dis. 124, 148–154. [DOI] [PubMed] [Google Scholar]
- 32.Dudding, B. A., Bartelloni, P. J., Scott, R. M., Top, F. H., Jr., Russell, P. K. & Buescher, E. L. (1972) Infect. Immun. 5, 295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhoads, J. L., Birx, D. L., Wright, D. C., Brundage, J. F., Brandt, B. L., Redfield, R. R. & Burke, D. S. (1991) J. Acquired Immune Defic. Syndr. 4, 724–731. [PubMed] [Google Scholar]
- 34.Rodrigues, E. G., Zavala, F., Eichinger, D., Wilson, J. M. & Tsuji, M. (1997) J. Immunol. 158, 1268–1274. [PubMed] [Google Scholar]
- 35.Sullivan, N. J., Sanchez, A., Rollin, P. E., Yang, Z. Y. & Nabel, G. J. (2000) Nature 408, 605–609. [DOI] [PubMed] [Google Scholar]
- 36.Tobery, T. W., Smith, J. F., Kuklin, N., Skulsky, D., Ackerson, C., Huang, L., Chen, L., Cook, J. C., McClements, W. L. & Jansen, K. U. (2003) Vaccine 21, 1539–1547. [DOI] [PubMed] [Google Scholar]
- 37.Natuk, R. J., Chanda, P. K., Lubeck, M. D., Davis, A. R., Wilhelm, J., Hjorth, R., Wade, M. S., Bhat, B. M., Mizutani, S., Lee, S., et al. (1992) Proc. Natl. Acad. Sci. USA 89, 7777–7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lubeck, M. D., Natuk, R., Myagkikh, M., Kalyan, N., Aldrich, K., Sinangil, F., Alipanah, S., Murthy, S. C., Chanda, P. K., Nigida, S. M., Jr., et al. (1997) Nat. Med. 3, 651–658. [DOI] [PubMed] [Google Scholar]
- 39.Buge, S. L., Richardson, E., Alipanah, S., Markham, P., Cheng, S., Kalyan, N., Miller, C. J., Lubeck, M., Udem, S., Eldridge, J. & Robert-Guroff, M. (1997) J. Virol. 71, 8531–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson, L. J., Malkevitch, N., Venzon, D., Pinczewski, J., Gomez-Roman, V. R., Wang, L., Kalyanaraman, V. S., Markham, P. D., Robey, F. A. & Robert-Guroff, M. (2004) J. Virol. 78, 2212–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patterson, L. J., Prince, G. A., Richardson, E., Alvord, W. G., Kalyan, N. & Robert-Guroff, M. (2002) Virology 292, 107–113. [DOI] [PubMed] [Google Scholar]
- 42.Xiang, Z. Q., Gao, G. P., Reyes-Sandoval, A., Li, Y., Wilson, J. M. & Ertl, H. C. (2003) J. Virol. 77, 10780–10789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogels, R., Zuijdgeest, D., van Rijnsoever, R., Hartkoorn, E., Damen, I., de Bethune, M. P., Kostense, S., Penders, G., Helmus, N., Koudstaal, W., et al. (2003) J. Virol. 77, 8263–8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kovacs, G. M., Davison, A. J., Zakhartchouk, A. N. & Harrach, B. (2004) J. Gen. Virol. 85, 2799–2807. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, X., Rush, M. & Schwartz, S. (2004) J. Virol. 78, 10888–10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.