Significance
Cellular accumulation of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) is associated with stress responses as well as aging. The reactive nature of ROS marks these molecules as a serious threat to cell integrity. Consequently, eukaryotic cells deploy numerous antioxidant enzymes that detoxify ROS to protect them from ROS-induced damage to proteins. Although the importance of antioxidant enzymes is well understood, how these proteins avoid becoming damaged in the hostile, ROS-rich environments in which they function remains unknown. We show that in plant cells the oxidoreductase Nucleoredoxin 1 (NRX1) protects antioxidant enzymes such as catalase from ROS-induced oxidation. Importantly, this protective effect of NRX1 boosted the H2O2 detoxification capacity of catalase, thereby protecting the plant cell from oxidative stress.
Keywords: Nucleoredoxin, Thioredoxin, catalase, oxidative stress, reactive oxygen species
Abstract
Cellular accumulation of reactive oxygen species (ROS) is associated with a wide range of developmental and stress responses. Although cells have evolved to use ROS as signaling molecules, their chemically reactive nature also poses a threat. Antioxidant systems are required to detoxify ROS and prevent cellular damage, but little is known about how these systems manage to function in hostile, ROS-rich environments. Here we show that during oxidative stress in plant cells, the pathogen-inducible oxidoreductase Nucleoredoxin 1 (NRX1) targets enzymes of major hydrogen peroxide (H2O2)-scavenging pathways, including catalases. Mutant nrx1 plants displayed reduced catalase activity and were hypersensitive to oxidative stress. Remarkably, catalase was maintained in a reduced state by substrate-interaction with NRX1, a process necessary for its H2O2-scavenging activity. These data suggest that unexpectedly H2O2-scavenging enzymes experience oxidative distress in ROS-rich environments and require reductive protection from NRX1 for optimal activity.
In eukaryotic cells, production of reactive oxygen and nitrogen species (ROS/RNS) is associated with many different developmental processes and stress responses. ROS/RNS such as superoxide (O2−), hydrogen peroxide (H2O2), and nitric oxide (NO) are continuously produced throughout cellular compartments as a by-product of metabolic pathways and in case of H2O2 during periods of stress by a class of cell membrane-associated NADPH-dependent oxidases known as respiratory burst oxidase homologs (RBOHs) (1, 2). The inherent reactivity of these molecules and their ability to traverse membranes pose a significant danger to all cellular compartments (3), with common events including membrane lipid peroxidation, DNA damage, and irreversible hyperoxidation of proteins leading to loss of function (4). Consequently, ROS/RNS play key roles in cellular senescence and aging and have been linked to diseases such as cancer and the onset of neurodegenerative conditions (2, 5). Despite the dangers posed by ROS/RNS, they also function as indispensable short- and long-range signaling molecules in numerous cellular responses. Thus, ROS/RNS homeostasis needs to be carefully controlled to support signaling while preventing cellular damage.
To protect themselves against damage caused by oxidative stress and to redirect the actions of ROS/RNS into signaling pathways, plants and animals have evolved a plethora of antioxidant enzymes. The redoxin superfamily of oxidoreductases reverses the oxidative state of proteins modified by ROS/RNS. Allocation of Thioredoxin (TRX) activity is particularly important to protect and repair cell signaling proteins from undesired cysteine oxidation (6, 7). In addition, peroxidase family enzymes are up-regulated during periods of oxidative stress and directly metabolize ROS (1, 8). Although some peroxidases such as catalases use a haem center for dismutation of H2O2, other non–haem-containing peroxidases, including Ascorbate Peroxidases (APX) and Peroxiredoxins (PRX), often use thiol-based mechanisms to decompose H2O2 (9, 10). Notably, dysregulation of peroxidase enzymes is associated with cellular accumulation of H2O2 and redox imbalance, resulting in accelerated aging and various pathological disorders (11–14). Although these findings indicate that this class of antioxidant enzymes is indispensable for maintenance of cellular redox, how they do so without sustaining significant structural damage in the hostile oxidative environments in which they function remains unknown.
Although cooperation between peroxidase and redoxin antioxidant systems during oxidative stress is apparent, the extent of this cooperation beyond the requirement for reducing power is currently unclear. In this study, we present evidence that activity of the plant immune-inducible TRX superfamily member, Nucleoredoxin 1 (NRX1), is necessary for the integrity of antioxidant systems. Our data unexpectedly suggest that H2O2-scavenging enzymes such as catalase suffer oxidative suppression in hostile ROS-rich environments and require oxidoreductase activity of NRX1 for optimal function.
Results
A Pathogen-Inducible NRX Regulates Plant Immune Responses.
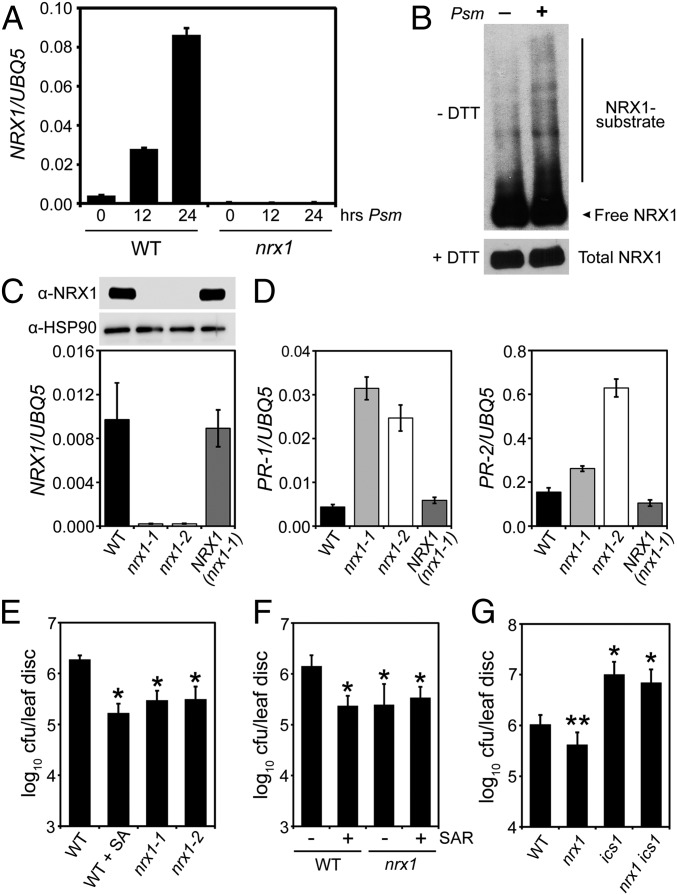
Pathogen infection and subsequent activation of plant immune responses is associated with an oxidative burst and expression of selected TRX gene family members. Infection of wild-type (WT) plants with the bacterial leaf pathogen Pseudomonas syringae pv. maculicola (Psm) ES4326 resulted in strong gene expression of a member of the NRX subfamily of TRX enzymes (Fig. 1A). Psm ES4326 infection also increased abundance of mixed disulfides between NRX1 and its substrates (Fig. 1B), indicating its oxidoreductase activity was pathogen-inducible. To investigate if NRX1 plays a functional role in plant immunity, we examined knockout lines (Fig. 1 A and C). Mutant nrx1 plants showed constitutive expression of pathogenesis-related (PR) genes but could be rescued by expression of Flag-tagged NRX1 from a constitutive 35S promoter (Fig. 1 C and D). Elevated PR gene expression in nrx1 was associated with enhanced disease resistance against Psm ES4326, which was comparable to WT plants immunized by the immune hormone salicylic acid (SA) or immunized by prior exposure to an avirulent pathogen (Fig. 1 E and F). This autoimmune phenotype of nrx1 mutants was dependent on SA, as mutation of the SA biosynthesis enzyme isochorismate synthase (ICS1) diminished immunity against Psm ES4326 (Fig. 1G).
Fig. 1.
NRX1 negatively regulates plant immune responses. (A) Plants were infected with Psm ES4326 (5 × 105 cells) for the indicated times. Expression of the NRX1 gene was analyzed and normalized against UBQ5. Error bars represent SD (n = 3). (B) WT plants were infiltrated with or without Psm ES4326 (5 × 105 cells). Total protein was denatured and alkylated to capture true mixed disulphide intermediates between NRX1 and its substrates, separated by SDS/PAGE in the presence or absence of DTT, and analyzed by Western blot against NRX1. Indicated are free NRX1 monomer, mixed disulfide intermediates (NRX1–substrate), and total NRX1. (C) Expression of NRX1 was analyzed in WT, nrx1-1, nrx1-2, and 35S::Flag-NRX1 (in nrx1-1) plants. Gene expression was normalized against UBQ5 (error bars represent SD; n = 3) and immunoblotting performed with anti-NRX1 and anti-HSP90 antibodies. (D) SA-dependent PR-1 and PR-2 gene expression. Error bars represent SD (n = 3). (E) Plants were mock-treated or sprayed with 0.5 mM SA 24 h before infection with Psm ES4326 (5 × 106 cells). Growth of Psm ES4326 was assessed after 3 d. Cfu, colony-forming units. Error bars represent statistical 95% confidence limits (n = 8). Asterisks indicate statistically significant differences compared with the WT (Tukey–Kramer ANOVA test; α = 0.05, n = 8). (F) Lower leaves were infiltrated with avirulent P. syringae pv. tomato DC3000/avrRpt2 (5 × 107) or 10 mM MgSO4. After 2 d, upper leaves were infected with Psm ES4326 (5 × 106 cells) and pathogen growth assessed after 3 d. Cfu, colony-forming units. Error bars represent statistical 95% confidence limits (n = 8). Asterisks indicate statistically significant differences compared with the WT (Tukey–Kramer ANOVA test; α = 0.05, n = 8). (G) Plants were infected with Psm ES4326 (5 × 106 cells) and pathogen growth assessed after 5 d. Cfu, colony-forming units. Error bars represent statistical 95% confidence limits (n = 8). *P < 0.05 for statistical differences with WT and nrx1; **P < 0.05 for statistical differences with WT, ics1, and nrx1 ics1 (Tukey–Kramer ANOVA test; α = 0.05, n = 8).
Two Active Sites Make Differential Contributions to NRX1 Disulfide Reductase Activity.
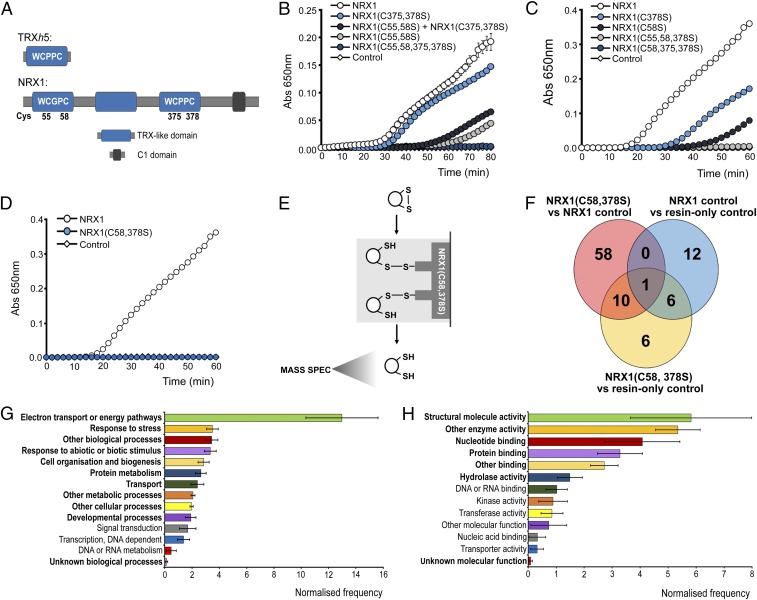
The NRX1 protein harbors three TRX-like domains, two of which contain an active site sequence consisting of WC(G/P)PC (Fig. 2A). Accordingly, NRX1 exhibits disulfide reductase activity (15), but the respective contributions of each active site have not yet been analyzed. In contrast to WT NRX1, mutation of either the first or third TRX-like domain in NRX1(C55,58S) and NRX1(C375,378S) proteins, respectively, resulted in reduced disulfide reduction activity (Fig. 2B and Fig. S1). However, NRX1(C375,378S) was much more efficient than NRX1(C55,58S), indicating that the majority of disulfide reduction activity occurred at the first TRX-like domain of NRX1. Simultaneous mutation of both the first and third TRX-like domains in the NRX1(C55,58,375,378S) protein completely abolished its ability to reduce insulin, demonstrating that these two domains contain all necessary active sites for disulfide reduction (Fig. 2B and Fig. S1). Because NRX1 proteins can dimerize (15), we investigated if active sites from different NRX1 proteins cooperate during disulfide reduction. Interestingly, when equal amounts of NRX1(C55,58S) and NRX1(C375,378S) were combined in an attempt to reconstitute the full enzymatic capacity of native NRX1 protein, we observed only a low level of activity that was comparable to NRX1(C55,58S) protein alone (Fig. 2B). This suggests that the two active sites do not function independent from one another. Moreover, a single cysteine in one active site was not sufficient to support the second active site, as the two single cysteine mutants, NRX1(C58S) and NRX1(C378S), both exhibited reduced activity (Fig. 2C and Fig. S1), indicating a requirement for both active-site resolving cysteines for full oxidoreductase activity. Accordingly, disulfide reduction activity of the mutant proteins NRX1(C55,58,378) and NRX1(C58,375,378S) (Fig. S1), in which only a single cysteine of one active site was left intact, was completely abolished (Fig. 2C). Taken together, these findings imply that NRX1 reduces disulfide bonds of insulin by a dicysteinic mechanism and that disulfide reduction may require cooperative or sequential interaction of a substrate with two different active sites of NRX1.
Fig. 2.
Identification of substrates of NRX1 oxidoreductase activity. (A) Unique domain structures of AtNRX1 compared with AtTRXh5, a conventional immune-inducible TRX. (B–D) Oxidized insulin (130 µM) was incubated with 0.3 mM DTT either alone (control) or together with 6 µM native NRX1 or indicated NRX1 active-site mutants. Formation of reduced insulin was measured at 650 nm. Error bars indicate SD (n = 3). (E) Schematic of NRX1 substrate capture experiment, using immobilized mutant NRX1(C58,378S). (F) Venn diagram illustrating proteins identified in the substrate capture experiment. Each circle represents proteins found to be enriched (P < 0.05, ratio > 1.5) in one of the following comparisons between columns: NRX1(C58,378S) column compared with WT NRX1 control column (pink); NRX1(C58,378S) column compared with resin-only control column (yellow); WT NRX1 control column compared with resin-only control column (blue). (G and H) GO term analysis was performed on the 74 targets enriched in the immobilized NRX1(C58,378S) mutant column for biological process (G) and molecular function (H) using Classification Superviewer on bar.utoronto.ca/. Normalized frequencies, SD, and P values were determined from absolute values as described in ref. 36. Error bars represent SD. P ≤ 0.05 are printed bold.
Fig. S1.
Purity of His-tagged recombinant NRX1 proteins. E. coli expressing WT and cysteine mutants of His6-NRX1 were treated with IPTG and total protein extracted. His6-NRX1 proteins were purified by incubation with His-binding cobalt resin, washed, and eluted with imidazole. Proteins were separated by SDS/PAGE and stained with Coomassie blue.
Identification of Stress-Induced Substrates of NRX1.
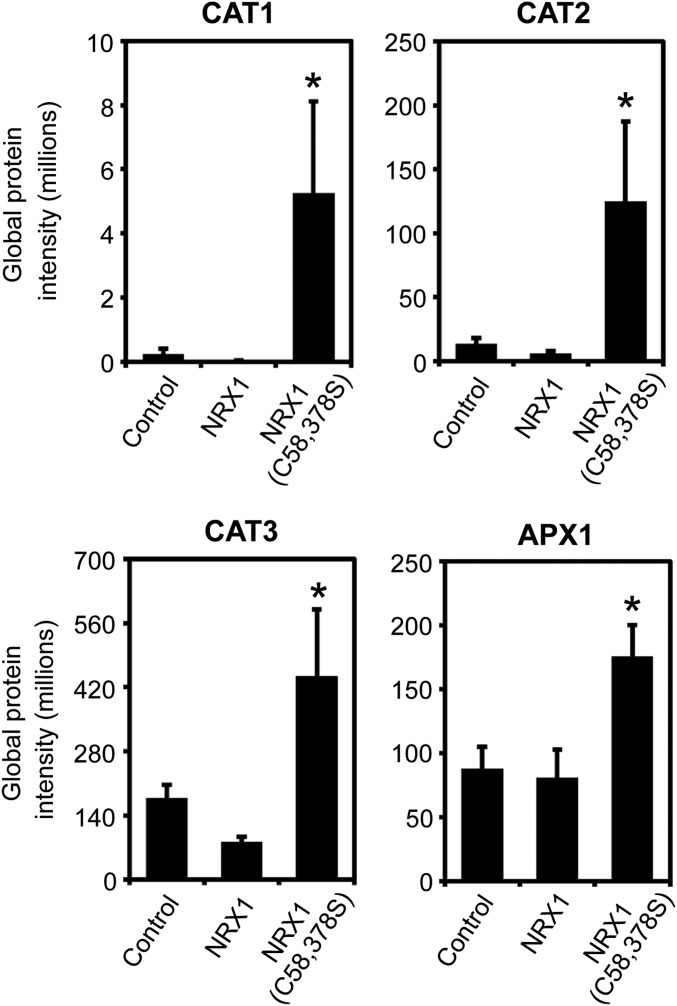
Next we sought to identify NRX1 substrates using a previously designed capture strategy that exploits the mechanism by which disulfides are reduced by TRX family members (16, 17). Mutation of the second active-site cysteine of TRX enzymes prevents complete resolution of the disulfide reduction reaction, resulting in trapping of the substrate via an intermediate mixed disulfide bond. Therefore, we replaced the second cysteine with serine in each active site of NRX1. The resulting NRX1(C58,378S) mutant protein was unable to reduce insulin, confirming its inability to complete disulfide reduction reactions (Fig. 2D and Fig. S1). The NRX1(C58,378S) protein was then immobilized on a column containing NHS-activated resin and incubated with total protein extracted from Psm ES4326-inoculated plants. After trapping substrates via the formation of mixed disulfide bonds, the column was rigorously washed, substrates eluted with DTT, and finally identified by mass spectrometry (Fig. 2E). Two control columns were also included and treated identically. Nonspecific binding to the column was monitored by inclusion of a control column containing only quenched resin without any NRX1 protein. A second control column contained immobilized WT NRX1, allowing us to distinguish between targets that interacted as substrate or nonsubstrate. Compared with the WT NRX1 control column, 69 proteins were found to be enriched in the NRX1(C58,378S) column (P < 0.05, ratio > 1.5) (Fig. 2F, pink circle). Additionally, 23 proteins were enriched in comparison with the resin-only control column (P < 0.05, ratio > 1.5) (Fig. 2F, yellow circle). To further eliminate nonspecific background binding, a comparison was run between the WT NRX1 control column and the resin-only control column (P < 0.05, ratio >1.5). This identified 19 proteins, 7 of which overlapped with those previously identified as potential NRX1(C58,378S) substrates (Fig. 2F, blue circle) and were therefore excluded from further analysis. Thus, a total of 74 proteins were uniquely identified as potential substrates on the NRX1(C58,378S) column (Fig. 2F, pink and yellow circles excluding overlap with blue circle; Dataset S1). A more stringent analysis still retained 45 potential NRX1 substrates with high confidence (Fig. S2A).
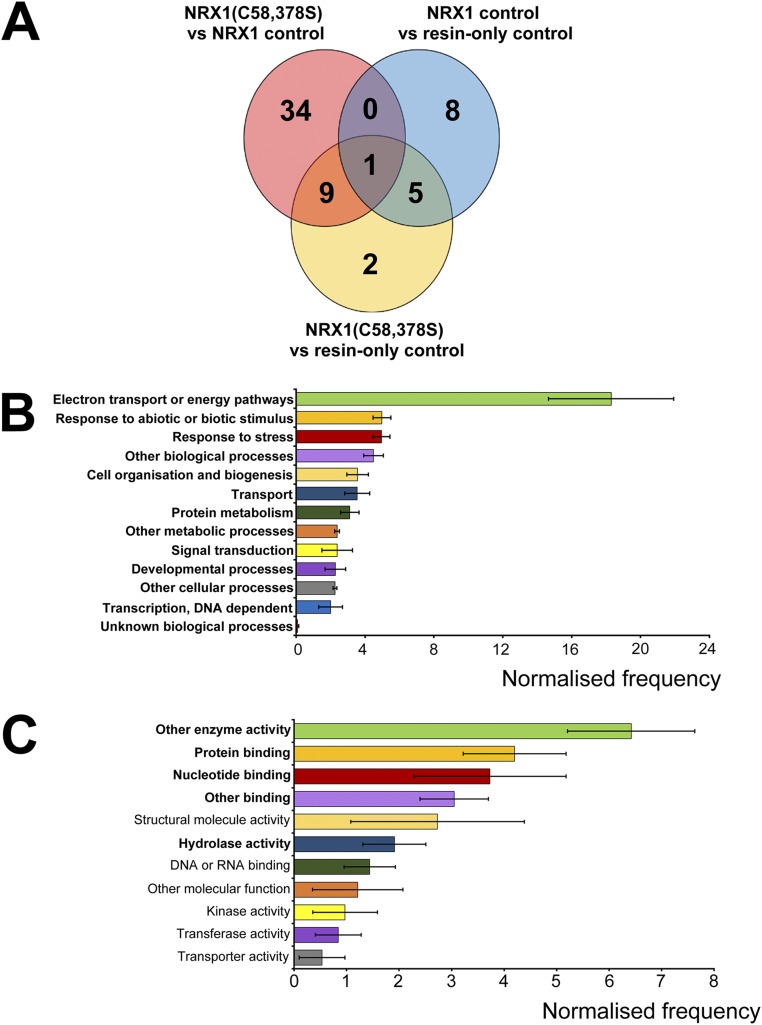
Fig. S2.
Identification and analysis of potential NRX1 substrates under stringent conditions. (A) Venn diagram illustrating proteins identified in the substrate capture experiment with an additional requirement that the number of peptides detected and used in quantification was equal to or more than two for each protein. Each circle represents proteins found to be enriched (P < 0.05, ratio > 1.5) in one of the following comparisons between columns: NRX1(C58,378S) column compared with WT NRX1 control column (pink); NRX1(C58,378S) column compared with resin-only control column (yellow); WT NRX1 control column compared with resin-only control column (blue). (B and C) GO term analysis was performed on the 45 targets (peptide count ≥ 2, see A) enriched in the immobilized NRX1(C58,378S) mutant column for biological process (B) and molecular function (C) using Classification Superviewer on bar.utoronto.ca/. Normalized frequencies, SD, and P values were determined from absolute values as described in ref. 36. Error bars represent SD. P values ≤ 0.05 are printed bold.
Gene ontology (GO) analysis of the identified targets revealed the most prominent biological processes were electron transport and energy pathways (Fig. 2G and Fig. S2B), consistent with the fact that these pathways often involve highly oxidative conditions. In agreement with our experimental approach, responses to stress as well as responses to abiotic or biotic stimuli were also found to be prominent biological processes (Fig. 2G and Fig. S2B). GO analyses for molecular function revealed that many potential NRX1 substrates exhibit structural molecule or enzymatic activity (Fig. 2H and Fig. S2C). As enzymes often contain active-site cysteines, these findings could suggest that NRX1 regulates their activity.
NRX1 Interacts with Enzymes of the Antioxidant H2O2-Scavenging Pathway.
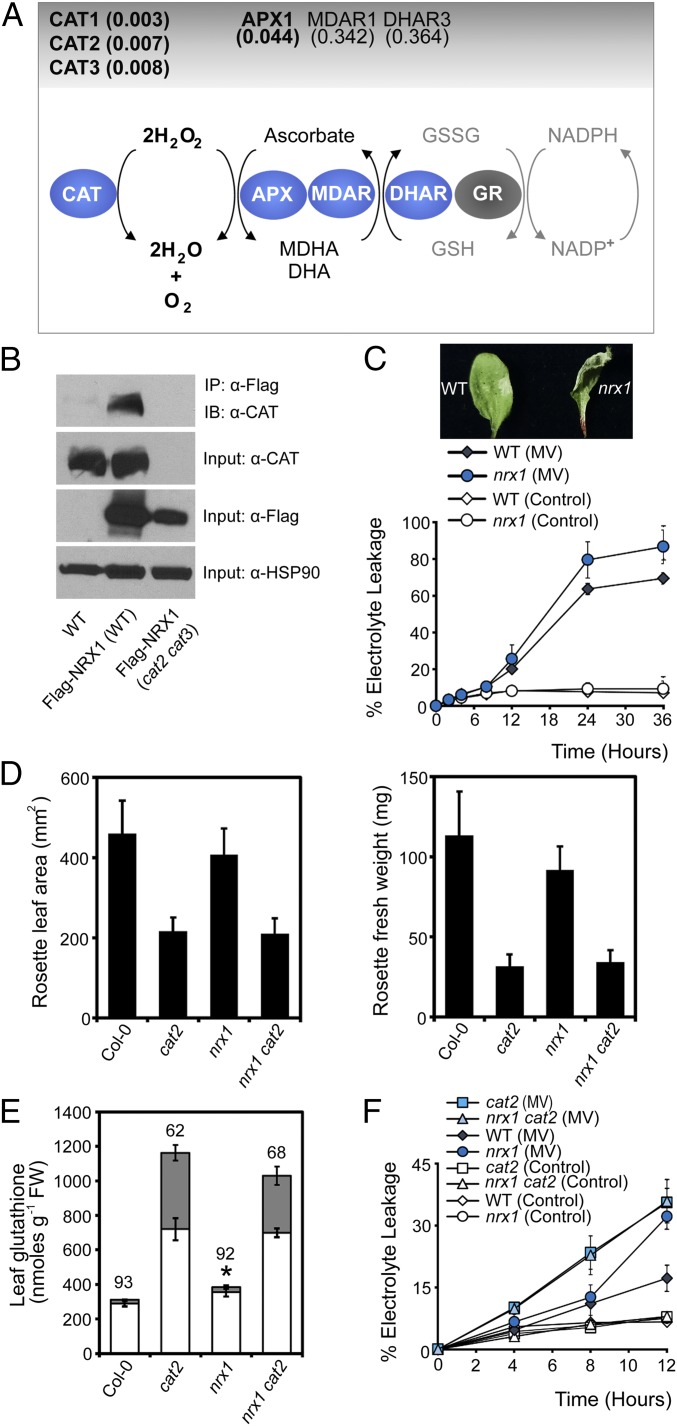
Upon further inspection of the entire list of identified potential NRX1 substrates, remarkably we identified many enzymes of the main cellular H2O2-scavenging pathway (Dataset S1). H2O2 is directly detoxified by both catalases and APX enzymes. Our quantitative mass spectrometry strategy identified as statistically significant several Arabidopsis catalases as well as the main cellular ascorbate peroxidase, APX1 (Fig. 3A, Fig. S3, and Dataset S1). Albeit outside the statistically significant range (P < 0.05), we also consistently found the ascorbate regenerating enzymes, monodehydroascorbate reductase (MDAR1) and dihydroascorbate reductase (DHAR3), that underpin APX1 activity (Fig. 3A and Dataset S1). The pattern emerging from these data implicates NRX1 in the regulation of antioxidant enzymes.
Fig. 3.
NRX1 protects plant cells from oxidative stress. (A) Diagram of the major H2O2-scavenging pathways. Enzymes enriched in the immobilized NRX1(C58, 378S) column are indicated in blue ovals and P values indicated above. (B) WT, 35S::Flag-NRX1 (Col-0), and 35S::Flag-NRX1 (cat2-2 cat3-1) plants were infected with Psm ES4326 (5 × 105 cells). Total protein was extracted under denaturing conditions and with the alkylating agent NEM to prevent nonspecific disulfide formation. Flag-NRX1 was immunoprecipitated and analyzed by Western blotting using an anti-CAT antibody. Anti-FLAG and anti-HSP90 were used as input controls. (C) Leaves were infiltrated with 5 µM MV or ddH2O (control). Electrolyte leakage was measured as a percentage of total electrolytes. Error bars represent SD (n = 3). Photographs were taken at 6 h. (D) Rosette leaf area (Left) and fresh weight (Right) were measured in 3-wk-old plants. Error bars represent SD (n = 20). (E) Total leaf glutathione of leaves. Asterisks indicate a significant difference (P ≤ 0.05) between total glutathione levels of WT and nrx1 plants. Reduced and oxidized glutathione levels are indicated by white and gray bars, respectively. The percentage reduction state of glutathione is indicated above bars. Error bars represent SD (n = 3). (F) Leaves were infiltrated with 5 µM MV or ddH2O (control). Electrolyte leakage was measured as a percentage of total electrolytes. Error bars represent SD of the mean (n = 3).
Fig. S3.
NRX1 targets enzymes of the major H2O2-scavenging pathways. Raw peptide abundance from mass spectrometry analysis of interaction between CAT1, CAT2, and CAT3 as well as APX1 with immobilized NRX1, NRX1(C58,378S), or resin-only (control) columns. Error bars represent SEM (n = 3). Asterisks indicate statistically significant differences compared with the NRX1 control column (*P < 0.05).
Because of their prominent role in H2O2 detoxification, we set out to validate the interaction of catalase enzymes with NRX1 in vivo. We performed a denaturing coimmunoprecipitation designed specifically to capture transient mixed disulfide interactions in vivo (SI Materials and Methods) and found that catalase readily coimmunoprecipitated with Flag-tagged NRX1 in the WT but not in the cat2 cat3 genetic background in which the two predominant catalase genes were knocked out (Fig. 3B) (18). These results confirm the mass spectrometry data and show that catalases are indeed substrates of NRX1 in vivo.
NRX1 Protects Cells Against Oxidative Stress.
The identification of ROS-scavenging enzymes as targets of NRX1 prompted us to investigate if NRX1 plays a role in oxidative stress. We first assessed the cell death response of mutant nrx1 plants treated with the ROS-generating chemical methyl viologen (MV). Compared with WT, mutant nrx1 plants displayed faster and more pronounced MV-induced cell death than the WT (Fig. 3C). This is reminiscent of the ROS intolerant phenotype of mutant cat2 plants (19), which, like nrx1 plants also display autoimmune phenotypes (20). We considered the possibility that NRX1 and catalases may act within the same ROS detoxification pathway. To investigate this further, we studied the phenotypes of an nrx1 cat2 double mutant. Mutation of NRX1 did not further aggravate the developmental phenotype of cat2 mutants (Fig. 3D). We then assessed the cellular redox status of these mutants. Compared with WT, mutant nrx1 plants accumulated significantly higher levels of total glutathione, but the ratio of oxidized versus reduced glutathione was normal (Fig. 3E). These data suggest that in absence of a stimulus, nrx1 mutants experience low levels of oxidative stress that are managed by increased glutathione levels. Mutation of NRX1, however, was not able to aggravate further the redox status of cat2 mutants, which as reported previously (19) contained very high levels of total and oxidized glutathione (Fig. 3E). These data suggest that NRX1 and CAT2 do not act additively to control the redox status of resting cells (Fig. 3E).
To assess if NRX1 and CAT2 function together or additively in stressed cells, we measured cell death in single and double mutants that had been treated with MV. As expected, both nrx1 and cat2 single mutants displayed increased levels of cell death in comparison with WT (Fig. 3F). However, the nrx1 cat2 double mutant was as susceptible to oxidative stress as cat2 single mutants, indicating that mutation of these genes also did not act additively during oxidative stress. Taken together, these data demonstrate that NRX1 is essential for protection against oxidative stress and suggest that it may act by modulating the activity of catalases.
Catalase Activity Is Controlled by NRX1-Mediated Reduction.
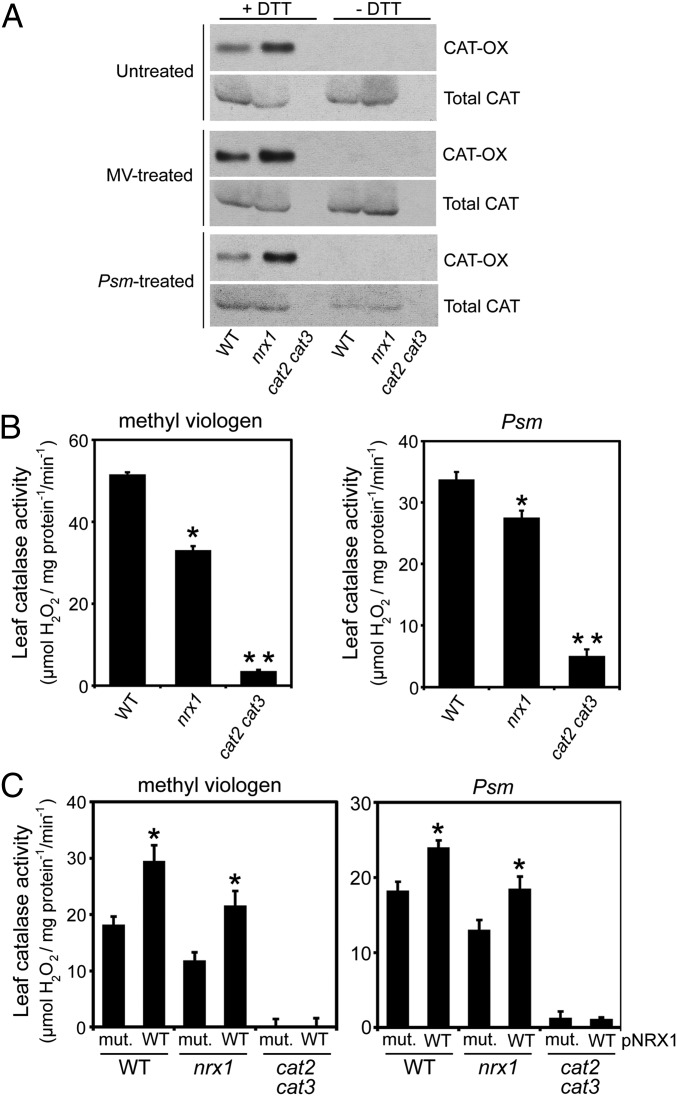
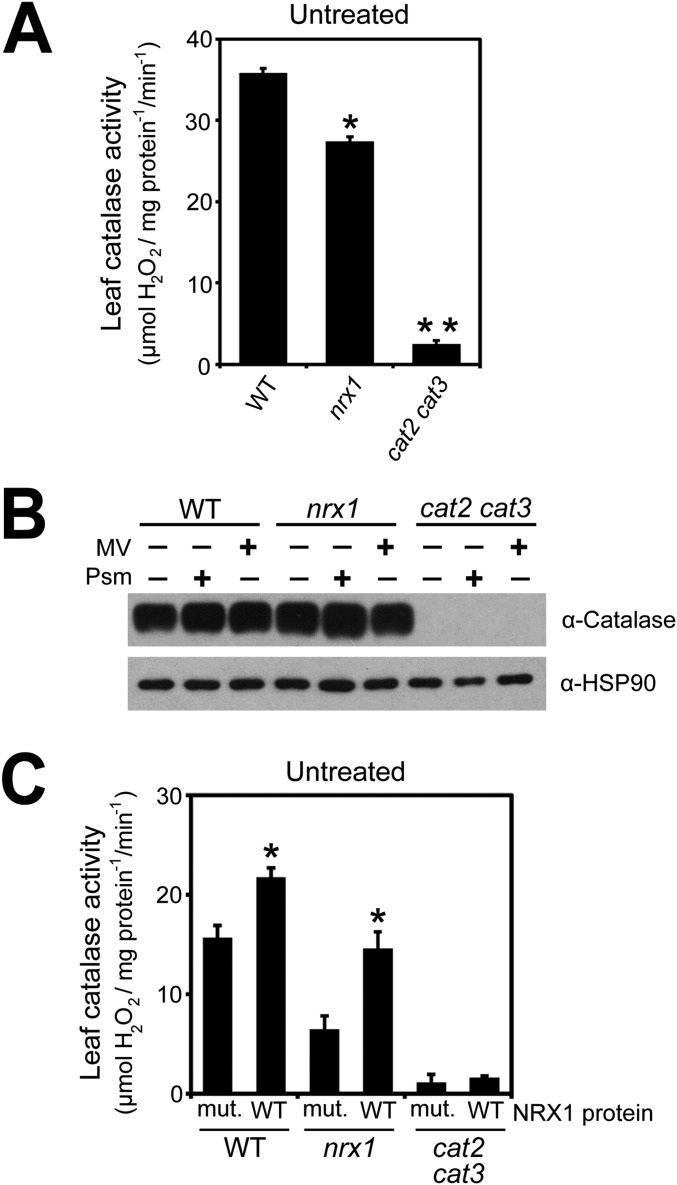
Our findings suggest that catalases may be recipients of oxidative posttranslational modifications and may require NRX1 to remain in the reduced state. To examine this possibility, we performed a reductive switch assay in which specifically oxidized cysteines undergo DTT-dependent labeling with biotin followed by pulldown with streptavidin. Indeed, catalase was subject to oxidative modification, and regardless of the treatment, the level of oxidized catalase was greatly enhanced in nrx1 mutants compared with WT, supporting the notion that NRX1 controls the oxidation state of catalase (Fig. 4A). To determine if NRX1 plays a role in modulating catalase activity, we examined catalase activity in protein extracts from untreated, MV-treated, and Psm ES4326-infected plants. Decomposition of H2O2 could be largely attributed to catalase activity, as cat2 cat3 double mutants displayed only residual activity (Fig. 4B and Fig. S4A). In striking contrast to WT, in all treatments nrx1 mutants displayed significantly decreased catalase activity (Fig. 4B and Fig. S4A), despite comparable catalase protein levels between genotypes (Fig. S4B). A direct link between NRX1 activity and the ability of catalase to decompose H2O2 was found by incubating plant extracts with recombinant WT NRX1 or mutant NRX1(C55,58,375,378S) protein. Addition of either recombinant protein to cat2 cat3 extracts did not increase H2O2 consumption, indicating NRX1 cannot detoxify H2O2 by itself (Fig. 4C and Fig. S4C). By contrast, addition of recombinant NRX1 but not mutant NRX1(C55,58,375,378S) to nrx1 extracts remarkably restored catalase activity to levels comparable to those found in WT extracts (Fig. 4C and Fig. S4C). Moreover, recombinant NRX1 significantly increased catalase activity in WT plant extracts, whereas mutant NRX1(C55,58,375,378S) protein was ineffective in this respect. In summary, these data demonstrate that NRX1 controls the oxidation status of catalase and thereby modulates its H2O2 dismutation activity.
Fig. 4.
NRX1 regulates redox modification and activity of catalase. (A) Protein extracts from untreated, MV- (5 µM), or Psm ES4326- (5 × 105 cells) treated plants were alkylated and incubated with or without DTT (2 mM). Free thiols were labeled with biotin and pulled down with Streptavidin. Oxidized catalase (OX-CAT) was visualized by immunoblotting with an anticatalase antibody and shown relative to total catalase. (B) The ability of protein extracts from untreated, MV- (5 µM), or Psm ES4326- (5 × 105 cells) treated plants to decompose H2O2 (5 mM) was measured at A240 and leaf catalase activity (mmol H2O2⋅mg⋅protein−1⋅min−1) calculated using the extinction coefficient of 0.036 cm2/µmol. Error bars represent SE (n = 3). *P < 0.05 for statistical differences with WT and cat2 cat3; **P < 0.05 for statistical differences with WT and nrx1 (Student’s t test). (C) As in B, but extracts were supplemented with 4 µM recombinant NRX1 or NRX1(C55,58,375,378S) and 0.33 mM DTT. pNRX1, recombinant NRX1 protein added to reaction mixture; mut., mutant recombinant NRX1 protein. Error bars represent SE (n = 3). *P < 0.05 for statistical differences compared to mutant recombinant NRX1 protein (Student’s t test).
Fig. S4.
NRX1 regulates catalase activity. (A) Protein extracts from untreated WT and nrx1-1 plants were desalted on NAP-5 columns and protein concentrations equalized. Decomposition of added H2O2 (5 mM) was measured at A240 and leaf catalase activity (mmol H2O2·mg·protein−1·min−1) calculated using the extinction coefficient of 0.036 cm2/µmol. Error bars represent SE (n = 3). Asterisks indicate statistically significant differences between samples (Student’s t test, P < 0.05). (B) Catalase protein abundance does not differ between WT and nrx1 mutant lines. Protein was extracted from untreated, MV-treated (5 µM), or Psm ES4326- (5 × 105 cells) treated WT, nrx1-1, and cat2 cat3 plants. Analysis was performed by Western blotting using an anti-CAT antibody and an anti-HSP90 antibody as loading control. (C) Protein extract from untreated WT and nrx1-1 plants was desalted on NAP-5 columns and protein concentrations equalized. Extracts were incubated with either 4 µM recombinant NRX1 or NRX1(C55,58,375,378S) and 0.33 mM DTT. Decomposition of added H2O2 (5 mM) was measured at A240 and leaf catalase activity (mmol H2O2·mg·protein−1·min−1) calculated using the extinction coefficient of 0.036 cm2/µmol. Error bars represent SE (n = 3). Asterisks indicate significant differences between WT and mutant recombinant NRX1 proteins (Student’s t test, P < 0.05).
SI Materials and Methods
Plant Material and Growth Conditions.
WT plants were of the Col-0 accession. Mutant lines used in this study include nrx1-1 (SALK_113401), nrx1-2 (SALK_100357), cat2-1 (SALK_076998), nrx1 cat2-1, cat2-2 cat3-1 (SALK_057998, SALK_092911), ics1 (37), and ics1 nrx1-1. Soil-grown plants were maintained at 21 °C and 100 μmoL⋅m−2⋅s−1 light on a “long day” 16 h light/8 h dark photoperiod at 65% day humidity and 55% night humidity. Rosette leaf area was measured using the ImageJ software.
Plant Transformation, Chemical Induction, and Pathogen Infection.
According to the manufacturer’s instructions, NRX1 (At1g60420) was cloned into pENTR/d-TOPO and recombined with Flag tag-containing pEarleyGate 202 (38) using LR clonase (Life Technologies) to generate the 35S::Flag-NRX1 transgene. The 35S::Flag-NRX1 construct was transformed into WT, nrx1-1, and cat2-2 cat3-1 lines by floral dipping in Agrobacterium tumefaciens (39). Transgenic plants were selected on soil by spraying glufosinate ammonium and confirmed by Western blotting against FLAG.
For pathogen infection, Psm ES4326 was grown overnight in liquid LB medium supplemented with streptomycin (100 µg/mL) and 10 mM MgSO4. Bacterial cells were diluted to 5 × 106 cells and pressure infiltrated via the abaxial side of leaves. In planta bacterial growth was determined 3 or 5 d after infection by spreading serial dilutions of leaf extracts on LB plates supplemented with streptomycin (100 µg/mL), 10 mM MgSO4, and 100 µM cycloheximide. To perform SAR tests, plants were first infiltrated with 2 × 107 cells of avirulent Pst DC3000 carrying the avrRpt2 gene [cultivated overnight in liquid LB medium supplemented with kanamycin (50 µg/mL) and 10 mM MgSO4]. After 2 d plants were infiltrated with virulent Psm ES4326 at 5 × 106 cells and subsequently processed as described above. For induction of the NRX1 gene, induction of NRX1 redox activity, and identification of NRX1 substrates, 5 × 105 cells of virulent Psm ES4326 were infiltrated.
For analysis of cell death, 4-wk-old soil-grown Arabidopsis plants were pressure infiltrated with 5 µM MV (Sigma) or a H2O control. Plants were photographed after 6 h under constant light. For electrolyte leakage assays, plants were first infiltrated with MV as above and then left for 1 h to ensure complete intake. Ten leaf discs (7 mm diameter) per genotype were then cut, washed extensively in H2O, and floated on 10 mL sterile H2O in fresh tubes. Samples were then put under constant light and electrolyte leakage measured at indicated time points using a conductivity meter. After the final time point, samples were boiled for 5 min and total electrolyte readings taken.
Site-Directed Mutagenesis and Protein Production.
NRX1 was cloned into the NdeI and EcoRI sites of vector pET28a (EMD Biosciences). Site-directed mutagenesis for NRX1 cysteines was performed using the QuikChange Lightning mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. Cys55/58/375/378 were replaced with Ser and sequences confirmed by DNA sequencing. NRX1 in pET28a and TRXh5 in pET24c (40) were used to produce recombinant proteins in Escherichia coli BL21 (DE3) cells by adding 1 mM IPTG after 2 h of culturing at 37 °C, followed by overnight growth at 16 °C or 5 h growth at 37 °C, respectively. Proteins were extracted in 1× Bugbuster (Novagen) and purified on HisPur Cobalt Resin (Pierce). Purified proteins were dialyzed against appropriate buffers.
Enzymatic Assays.
Disulfide reduction activities of NRX1 and TRXh5 proteins were measured using an insulin turbidity assay as described (41) with some minor alterations. Each 200 µL reaction volume consisted of 130 µM insulin, 100 mM potassium phosphate (pH 7.0), 2 mM EDTA, and indicated concentrations of purified redoxin. To initiate the assay, 0.33 mM DTT was added. Measurements at 650 nm were taken at 2-min intervals using an Infinite 200 NanoQuant (TECAN). Reactions for each experiment were performed simultaneously in triplicate.
Measurement of catalase activity was performed on plant protein extracted in 100 mM potassium phosphate buffer (pH 6.5) containing protease inhibitor mixture [50 µg/mL N-p-Tosyl-l-phenylalanine chloromethyl ketone (TPCK), 50 µg/mL Nα-Tosyl-l-lysine chloromethyl ketone hydrochloride (TLCK), 0.6 mM phenylmethylsulfonyl fluoride (PMSF)]. Following centrifugation, samples were desalted on NAP-5 columns (GE Healthcare) and protein concentration measured using the Bradford assay. Samples were diluted to 1.25 mg/mL protein, and 5 mM H2O2 was added either immediately or following a 1-h incubation with either 4 µM NRX1 or NRX1(C55,58,375,378S) recombinant protein and 0.33 mM DTT. Decomposition of H2O2 was measured at 240 nm and catalase activity calculated using the extinction coefficient of 0.036 cm2/µmol.
For measurement of the total glutathione, 3-wk-old rosette plants were collected individually and frozen in liquid nitrogen. Glutathione contents were determined using the recycling enzymatic assay described in ref. 42. Each measurement included three biological and three technical repetitions.
Protein Analysis and Identification of NRX1 Substrates.
Mixed disulfides between NRX1 and targets were assessed by extracting protein in HEN buffer [100 mM Hepes (pH 7.7), 1 mM EDTA, and 0.1 mM neocuproine] containing 2.5% SDS and 25 mM N-ethylmalemeide (NEM) and prepared directly for nonreducing or reducing SDS/PAGE gel electrophoresis and immunoblotting. Western blots were probed with an anti-NRX1 antibody generated against the full-length, recombinant NRX1 protein (ProteinTech).
For validation of in vivo interaction between NRX1 and Catalase, plant protein was extracted in fixation buffer [25 mM Hepes (pH 7.7), 1 mM EDTA, 0.1 mM neocuproine, 2.5% SDS, 25 mM NEM, and protease inhibitor mixture], incubated at 50 °C for 20 min, centrifuged, and acetone precipitated. Pellets were resuspended in diluted HEN buffer [25 mM Hepes (pH 7.7), 1 mM EDTA, 0.1 mM neocuproine] containing 0.5% SDS. Input aliquots were taken and the remaining samples mixed with 4 mL neutralization buffer (diluted HEN buffer containing 0.5% Triton X-100, 150 mM NaCl, and protease inhibitor mixture) containing 40 µL anti-FLAG M2 affinity resin (Sigma) prewashed with elution buffer [0.1 M glycine–HCl (pH 3.5)]. Samples were then incubated overnight at 4 °C with rotation. Next, samples were washed three times with neutralization buffer and incubated for 5 min in elution buffer with frequent mixing. Following centrifugation, supernatants were incubated for 5 min with 1/10 volume of StrataClean resin (Agilent Technologies). Protein was eluted from the resin by boiling for 5 min in neutralization buffer containing 2× SDS sample buffer and 200 mM DTT and analyzed by SDS/PAGE gel and Western blotting against anti-FLAG (Sigma), anti-Catalase (Agrisera), and anti-HSP90 (Santa Cruz Biotechnology).
The reductive switch assay was performed on untreated, MV-treated, or Psm ES4326-treated plants. The assay was performed as in ref. 43, with the following exceptions: 20 mM NEM was used to alkylate free thiols, and thiols were reduced with 2 mM DTT instead of ascorbate. Oxidized and total CAT protein was detected using an anti-Catalase (Agrisera) antibody.
For the identification of NRX1 substrates, Psm ES4326-treated Col-0 plant tissue was extracted in 50 mM potassium phosphate buffer (pH 7.0) containing protease inhibitors. Following centrifugation, the supernatant was desalted on PD-10 columns (GE Healthcare) and then added to columns containing 8 mg/mL recombinant NRX1 or NRX1(C58/C378) that were immobilized on NHS-activated agarose (Pierce) according to the manufacturer’s instructions. Columns were then prereduced with 2 mM DTT and washed extensively in 50 mM potassium phosphate buffer (pH 7.0) before addition of samples. A column containing quenched NHS agarose only was included as a control. Following 1-h incubation with rotation at room temperature, columns were centrifuged for 2 min at 1,000 × g and then washed 10 times with 50 mM potassium phosphate buffer (pH 7.0) containing 0.5% Triton X-100, with the fifth wash containing 1 M NaCl. Samples were eluted from agarose by incubation with 2 mL of 50 mM DTT for 30 min at room temperature followed by 2 min of centrifugation at 1,000 × g.
Samples were concentrated by Speedvac, run on SDS/PAGE for 5 min, and then extracted and digested using the Shevchenko method (44). Peptide extracts were then cleaned on SPE reverse phase BondElut Omix tip C18 100 μL, (Agilent) and dried under low pressure (Speedvac). The dried peptide samples were resuspended in resuspension buffer (0.5% vol/vol trifluoroacetic acid in water). These samples were filtered using Millex filter and then analyzed by HPLC–MS. Nano-HPLC–MS/MS analysis was performed using an online system consisting of a nano-pump (Dionex Ultimate 3000, Thermo-Fisher) coupled to a QExactive instrument (Thermo-Fisher) with a precolumn of 300 µm × 5 mm (Acclaim Pepmap, 5 µm particle size) connected to a column of 75 µm × 50 cm (Acclaim Pepmap, 3 µm particle size). Samples were analyzed on a 90-min gradient in data-dependent analysis (1 survey scan at 70k resolution followed by the top 10 MS/MS). Data from MS/MS spectra were searched using MASCOT Versions 2.4 (Matrix Science Ltd.) against the Arabidopsis thaliana proteome downloaded from TAIR (https://www.arabidopsis.org/) with maximum missed-cut value set to 2. The following features were used in all searches: (i) variable methionine oxidation, (ii) fixed cysteine carbamidomethylation, (iii) precursor mass tolerance of 10 ppm, (iv) MS/MS tolerance of 0.05 amu, (v) significance threshold (p) below 0.05 (MudPIT scoring), and (vi) final peptide score of 20. Progenesis (version 4 Nonlinear Dynamics) was used for LC–MS label-free quantitation. Only MS/MS peaks with a charge of 2+, 3+, or 4+ were taken into account for the total number of “Feature” (signal at one particular retention time and m/z), and only the five most intense spectra per “Feature” were included. The associated unique peptide ion intensities for a specific protein were then summed to generate an abundance value. Based on the abundance values, within-group means were calculated, and from there, the fold changes between the different groups were evaluated. One-way ANOVA was used to calculate the P values based on the transformed values. Differentially expressed proteins were only considered significant in the current study if the following conditions were fulfilled: (i) P values (pair-wise) less than 0.05 and (ii) absolute fold change of at least 1.5 (i.e., ≥1.5-fold for up-regulated proteins, whereas ≤0.667-fold for down-regulated proteins), and (iii) higher abundance compared with control columns. Additionally, the more stringent analyses also included the condition that the number of peptides detected and used in quantification per protein was equal to or more than 2. Raw and processed data were uploaded to ProteoSAFe (massive.ucsd.edu/ProteoSAFe/) and have the massIVE accession no. MSV000080429 and proteomeXchange accession no. PXD005591.
Discussion
During periods of oxidative stress, ROS-scavenging by peroxidase enzymes is essential to maintaining cell integrity. However, how these enzymes withstand the toxic oxidizing conditions in which they function remains elusive. In this study, we demonstrate that in plants, NRX1, a member of the TRX superfamily of enzymes, maintains catalase enzymes in a reduced state, thereby protecting their H2O2-detoxifying activity and, importantly, ensuring efficient functioning of the antioxidant system during oxidative stress.
Using extracts from immune-induced plants, we identified 74 potential endogenous substrates of NRX1 (Fig. 2F). Although some of these are unlikely to be real substrates, strikingly we found that NRX1 engaged with enzymes of a major H2O2-scavenging pathway, including Arabidopsis catalases (Fig. 3A and Fig. S2). Indeed, we confirmed that catalase was a substrate of NRX1 in vivo, and epistasis experiments strongly suggested that NRX1 and the major leaf catalase CAT2 act together in the same H2O2-detoxification pathway (Fig. 3). The full extent to which catalase activity contributes to plant immune responses has only recently come to light. Catalases have previously been reported to bind SA, and a recent study showed that SA-induced suppression of CAT2 activity increased H2O2 levels upon pathogen infection (21). Here we show that additionally, during oxidative stress and pathogen infection, CAT2 activity is regulated by NRX1. Whether this is a conserved role of NRX1 in other eukaryotes remains to be seen, but in addition to this study, NRX1 has been implicated in immune responses in both a crop plant and mammals (22, 23).
Catalases are widely reported to localize to the peroxisomes where they remove toxic H2O2 generated in cellular metabolism. However, NRX1 lacks known peroxisomal targeting sequences (PTSs) and is localized to the cytoplasm and nucleus (15). Thus, it is unlikely that peroxisomal catalases are a substrate of NRX1. Nonetheless, using a coimmunoprecipitation trap, we found that NRX1 formed a mixed disulfide intermediate with catalase in vivo (Fig. 3D). In this respect, it should be noted that peroxisomal import of catalases is dependent on cytosolic interaction with the PTS import receptor, PEX5 (24, 25). PEX5 docks with the peroxisomal membrane where it releases its cargo into the peroxisome matrix. Under oxidizing conditions, however, PEX5 has been shown to undergo redox-dependent monoubiquitination at a cysteine residue in the N terminus, preventing it from being recycled back to the cytosol (26). Consequently, during periods of oxidative stress, PEX recycling is blocked, resulting in decreased peroxisomal import of PEX5 targets, including catalase (25). Accordingly, a recent report demonstrated that pathogen-mediated transient expression of CAT3 in tobacco leaves resulted in its localization to the peroxisome, cytoplasm, and plasma membrane (27). Our findings suggest that oxidation-induced retention of catalases in the cytoplasm may enable these critical detoxifying enzymes to become substrates of NRX1.
Catalases contain several surface-exposed cysteine residues that may be sensitive to oxidation (28–30). Therefore, it is surprising that these enzymes function efficiently in ROS-rich environments without incurring significant damage. Here we found that catalases are indeed subject to inhibitory oxidative modification and demonstrate that NRX1 may be dedicated to maintaining these enzymes in a reduced state to promote their H2O2-scavenging activity (Fig. 4). Studies on the green alga Chlamydomonas reinhardtii have demonstrated reversible, partial inactivation of catalase, which was attributed to redox regulation of a single cysteine residue that is conserved among Arabidopsis catalases (30). Oxidation of a single cysteine implies that rather than a disulfide, catalase may contain an S-nitrosothiol or sulfenic acid. Although the ability of NRX1 to reduce these modifications has not yet been demonstrated, they are targeted by conventional TRX family members (31–33). Regardless of these particulars, this study clearly demonstrates that NRX1-mediated reduction of catalase is an important mechanism for sustaining maximal catalase activity.
So does the advantageous effect of NRX1 on H2O2 scavenging extend beyond regulation of catalases? Extraordinarily our proteomic capture experiments recovered nearly all major enzymes of the cytosolic glutathione/ascorbate-dependent H2O2-scavenging pathway, including APX, MDAR, and DHAR (Fig. 3 and Dataset S1), some of which have been identified individually in other proteomic screens for substrates of conventional TRX enzymes (see ref. 34 and references herein). The importance of the cytosolic glutathione/ascorbate cycle in H2O2 scavenging is exemplified by reports that showed reduced levels of APX protein or ascorbate deficiency resulted in high levels of oxidative stress, spontaneous cell death, and hyperresponsiveness to plant pathogens (11, 35). Given that cysteine oxidation appears prevalent in controlling activities of enzymes in this H2O2-scavenging pathway, it is highly likely that the interactions found here between NRX1 and these enzymes are of functional significance to cellular redox homeostasis. Thus, NRX1 may play an important novel role in directly regulating the cellular capacity for H2O2 detoxification, thereby protecting plant cells from oxidative stress triggered by environmental challenges.
Materials and Methods
Detailed procedures for plant materials and treatments, protein assays, and identification of NRX1 substrates by mass spectrometry are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Royal Society University Research Fellowships (UF090321 and UF140600) and Research Grant Rg110495, Wellcome Trust–University of Edinburgh Institutional Strategic Support Fund (ISSF), and European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme Grant No 678511. Further, S.K. was supported by a studentship from the Biotechnology and Biological Sciences Research Council (BBSRC), R.K. by a studentship from the Higher Education Commission Pakistan, G.J.L. by BBSRC Grant BB/D011809/1, T.L.B. by European Union’s Seventh Framework Programme 602470, and V.D.-H. and J.-P.R. by the Centre National de la Recherche Scientifique and the Agence Nationale de la Recherche ANR-Blanc Cynthiol 12-BSV6-0011. Liquid chromatography–mass spectrometry (LC–MS) QExactive equipment was purchased by an ISSF award and Wellcome Trust for the Centre for Immunity, Infection and Evolution Award 095831/Z/11/Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw and processed proteomic data were uploaded to ProteoSAFe, massive.ucsd.edu/ProteoSAFe/ (massIVE accession no. MSV000080429 and proteomeXchange accession no. PXD005591).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703344114/-/DCSupplemental.
References
- 1.Mittler R, et al. ROS signaling: The new wave? Trends Plant Sci. 2011;16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrov VD, Van Breusegem F. Hydrogen peroxide—A central hub for information flow in plant cells. AoB Plants. 2012;2012:pls014. doi: 10.1093/aobpla/pls014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 5.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 6.Day AM, et al. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol Cell. 2012;45:398–408. doi: 10.1016/j.molcel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 7.Meyer Y, Buchanan BB, Vignols F, Reichheld J-P. Thioredoxins and glutaredoxins: Unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 8.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins A, Nelson KJ, Parsonage D, Poole LB, Karplus PA. Peroxiredoxins: Guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci. 2015;40:435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zamocky M, Furtmüller PG, Obinger C. Evolution of catalases from bacteria to humans. Antioxid Redox Signal. 2008;10:1527–1548. doi: 10.1089/ars.2008.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittler R, et al. Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc Natl Acad Sci USA. 1999;96:14165–14170. doi: 10.1073/pnas.96.24.14165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willekens H, et al. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 1997;16:4806–4816. doi: 10.1093/emboj/16.16.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koepke JI, Wood CS, Terlecky LJ, Walton PA, Terlecky SR. Progeric effects of catalase inactivation in human cells. Toxicol Appl Pharmacol. 2008;232:99–108. doi: 10.1016/j.taap.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Terlecky SR, Koepke JI, Walton PA. Peroxisomes and aging. Biochim Biophys Acta. 2006;1763:1749–1754. doi: 10.1016/j.bbamcr.2006.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchal C, et al. NTR/NRX define a new thioredoxin system in the nucleus of Arabidopsis thaliana cells. Mol Plant. 2014;7:30–44. doi: 10.1093/mp/sst162. [DOI] [PubMed] [Google Scholar]
- 16.Motohashi K, Kondoh A, Stumpp MT, Hisabori T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA. 2001;98:11224–11229. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T. Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:18–27. doi: 10.1093/pcp/pch019. [DOI] [PubMed] [Google Scholar]
- 18.Mhamdi A, et al. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J Exp Bot. 2010;61:4197–4220. doi: 10.1093/jxb/erq282. [DOI] [PubMed] [Google Scholar]
- 19.Queval G, et al. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 2007;52:640–657. doi: 10.1111/j.1365-313X.2007.03263.x. [DOI] [PubMed] [Google Scholar]
- 20.Chaouch S, et al. Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 2010;153:1692–1705. doi: 10.1104/pp.110.153957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan H-M, Liu W-C, Lu Y-T. CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe. 2017;21:143–155. doi: 10.1016/j.chom.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi T, et al. Nucleoredoxin negatively regulates Toll-like receptor 4 signaling via recruitment of flightless-I to myeloid differentiation primary response gene (88) J Biol Chem. 2010;285:18586–18593. doi: 10.1074/jbc.M110.106468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y-B, et al. The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 2016;170:2392–2406. doi: 10.1104/pp.15.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshima Y, et al. Plant catalase is imported into peroxisomes by Pex5p but is distinct from typical PTS1 import. Plant Cell Physiol. 2008;49:671–677. doi: 10.1093/pcp/pcn038. [DOI] [PubMed] [Google Scholar]
- 25.Legakis JE, et al. Peroxisome senescence in human fibroblasts. Mol Biol Cell. 2002;13:4243–4255. doi: 10.1091/mbc.E02-06-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apanasets O, et al. PEX5, the shuttling import receptor for peroxisomal matrix proteins, is a redox-sensitive protein. Traffic. 2014;15:94–103. doi: 10.1111/tra.12129. [DOI] [PubMed] [Google Scholar]
- 27.Zou J-J, et al. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell. 2015;27:1445–1460. doi: 10.1105/tpc.15.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown GC. Reversible binding and inhibition of catalase by nitric oxide. Eur J Biochem. 1995;232:188–191. doi: 10.1111/j.1432-1033.1995.tb20798.x. [DOI] [PubMed] [Google Scholar]
- 29.Clark D, Durner J, Navarre DA, Klessig DF. Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol Plant Microbe Interact. 2000;13:1380–1384. doi: 10.1094/MPMI.2000.13.12.1380. [DOI] [PubMed] [Google Scholar]
- 30.Michelet L, et al. Down-regulation of catalase activity allows transient accumulation of a hydrogen peroxide signal in Chlamydomonas reinhardtii. Plant Cell Environ. 2013;36:1204–1213. doi: 10.1111/pce.12053. [DOI] [PubMed] [Google Scholar]
- 31.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chae HZ, Chung SJ, Rhee SG. Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem. 1994;269:27670–27678. [PubMed] [Google Scholar]
- 33.Kneeshaw S, Gelineau S, Tada Y, Loake GJ, Spoel SH. Selective protein denitrosylation activity of Thioredoxin-h5 modulates plant immunity. Mol Cell. 2014;56:153–162. doi: 10.1016/j.molcel.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Montrichard F, et al. Thioredoxin targets in plants: The first 30 years. J Proteomics. 2009;72:452–474. doi: 10.1016/j.jprot.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Pavet V, et al. Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol. 2005;139:1291–1303. doi: 10.1104/pp.105.067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Provart N, Zhu T. A browser-based functional Classification SuperViewer for Arabidopsis genomics. Curr Comput Mol Biol. 2003;2003:271–272. [Google Scholar]
- 37.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 38.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 39.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 40.Tada Y, et al. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 42.Bashandy T, et al. Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell. 2010;22:376–391. doi: 10.1105/tpc.109.071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.