Abstract
Sco1 and Cox17 are accessory proteins required for the correct assembly of eukaryotic cytochrome c oxidase. At variance with Sco1, Cox17 orthologs are found only in eukaryotes. We browsed bacterial genomes to search proteins functionally equivalent to Cox17, and we identified a class of proteins of unknown function displaying a conserved gene neighborhood to bacterial Sco1 genes, all sharing a potential metal binding motif H(M)X10MX21HXM. Two members of this group, DR1885 from Deinococcus radiodurans and CC3502 from Caulobacter crescentus, were expressed, and their interaction with copper was investigated. The solution structure and extended x-ray absorption fine structure data on the former protein reveal that the protein binds copper(I) through a histidine and three Mets in a cupredoxin-like fold. The surface location of the copper-binding site as well as the type of coordination are well poised for metal transfer chemistry, suggesting that DR1885 might transfer copper, taking the role of Cox17 in bacteria. On the basis of our results, a possible pathway for copper delivery to the CuA center in bacteria is proposed.
Keywords: cytochrome c oxidase assembly protein, copper protein, cupredoxin-like fold
During the last few years, an increasing number of genes have been discovered to be implicated in many intracellular pathways of metal trafficking (1–4). Some of these genes are conserved in all organisms, whereas others are present in prokaryotic or eukaryotic organisms only. The repeated occurrence of genes in each other's neighborhood in bacterial genomes has been shown to indicate a functional association between the proteins they encode, i.e., the proteins are involved in the same biochemical pathway (5, 6). Following this approach, we have analyzed the gene neighborhood of a bacterial protein, Sco1, which is proposed to be involved in copper ion delivery to the CuA center of cytochrome c oxidase (CcO) complex (7), with the purpose to locate new proteins potentially involved in copper delivery and insertion in this enzyme. There is a large variability in proteins involved in copper delivery to CcO. As examples, the copper-binding protein Cox11, which is implicated in the assembly of the CuB site of CcO (8), is found only in eukaryotes and Gram-negative bacteria (9). No prokaryotic homologs are found for the eukaryotic copper chaperone Cox17, which has been recently shown to be implicated in the copper delivery to both CuA and CuB centers in eukaryotes (10).
The search of genes with conserved neighborhood to Sco1 identified, in eight bacteria, a protein having a conserved potential metal binding motif, H(M)X10MX21HXM. A blast search (www.ncbi.nlm.nih.gov/BLAST) then allows us to individuate further 42 conserved sequences having the same metal binding motif. Four sequences belonging to the above protein family were selected, cloned and expressed in a high-throughput approach. We solved the solution structure of the C-terminal domain of the gene product of this family from Deinococcus radiodurans (DR1885 hereafter) in its apoform and Cu(I) form. The whole analysis, including extended x-ray absorption fine structure (EXAFS) and NMR data, indicates that DR1885 is a copper protein, possibly involved in the assembly of CcO. In particular, we propose that it can take the role of the mitochondrial Cu(I) chaperone Cox17 in the extracytoplasmic environment of bacteria.
Materials and Methods
Sequence Analysis. The string program (Search Tool for the Retrieval of Interacting Genes/Proteins, www.bork.emblheidelberg.de/STRING) was used to identify the bacterial Sco1 neighboring genes. The blast program was used to search over all nonredundant GenBank database genomes for the DR1885 homolog sequences. Sequence alignments were performed with clustalw (11). Prediction of transmembrane helices and membrane topology of all sequences was obtained by using the hmmtop and tmpred programs (12, 13).
Protein Cloning and Purification. The genes from D. radiodurans, Campylobacter jejuni, Caulobacter crescentus, and Pseudomonas aeruginosa were amplified by PCR from the corresponding genomic DNA template. The PCR products were cloned into the His-fusion expression vector pET21a(+) (Novagen). The plasmids were sequenced with an ABI PRISM 310 Genetic Analyzer (Applied Biosystem).
The sequenced plasmids were used to transform Escherichia coli [DH5α for plasmid propagation and BL21 (DE3) pLysS (Novagen) for protein expression]. Because of its higher expression yields, only the protein from D. radiodurans was produced as labeled in the minimal medium with [13C]glucose and/or (15NH4)2SO4. The protein was purified through affinity chromatography, and the polyHis tag was cleaved with Factor Xa protease (Roche). The unlabeled protein from C. crescentus also was obtained by a similar procedure.
Protein Characterization. The concentration of DR1885 was determined by using an extinction coefficient of 6970 M–1·cm–1 at 280 nm, as estimated by protparam software (www.expasy.org/tools/protparam.html). Electrospray ionization (ESI)-MS spectra were taken with an Applied Biosystems ESI-TOF Mariner mass spectrometer. Circular dichroism (CD) spectra were collected on a Jasco J-500C spectropolarimeter (Easton, MD). The CD spectrum of apo- and Cu(I)DR1885 was analyzed, without applying any data smoothing routine, with the dicroprot v2.5 application package (14) to estimate secondary structure composition. Copper content was checked through atomic absorption measurements with a PerkinElmer 2380 instrument.
NMR samples were at 1–2 mM in protein concentration in 100 mM potassium phosphate buffer (pH 7) containing 10% D2O. To a 1 mM 15N apoDR1885 sample containing 2 mM d-isoascorbic acid, a stepwise addition up to 1.2 equivalents of Cu(I) was performed, and 2D 1H-15N heteronuclear single quantum correlation (HSQC) spectra were acquired for each addition. Copper(I) solution was obtained by reducing a CuSO4 solution with an excess of d-isoascorbic acid.
X-ray absorption spectroscopy data were collected at Deutsches Elektronen Synchrotron (Hamburg, Germany) on samples consisted of ascorbate-reduced 1.0 mM Cu(I)DR1885 in 100 mM phosphate buffer at pH 7.0 (see Supporting Text, which is published as supporting information on the PNAS web site, for details).
The titration of apoDR1885 with Cu(II), added as CuSO4, was followed through electronic, EPR, and NMR spectroscopy in 100 mM potassium phosphate buffer (pH 7).
NMR Spectroscopy and Structure Calculations. The NMR spectra were acquired at 298 K on Avance 800, 700, 600, and 500 Bruker spectrometers, the latter equipped with a triple resonance cryoprobe. The NMR experiments for resonance assignment and for obtaining structural restraints are summarized in Table 1, which is published as supporting information on the PNAS web site. The candid approach (15) was used to assign ambiguous nuclear Overhauser effect (NOE) cross-peaks and to have a preliminary protein structure. Structure calculations then were performed through iterative cycles of dyana (16), followed by restrained energy minimization with amber 5.0 (17) applied to each member of the final family. The copper ion was included in the calculations by adding new restraints obtained from EXAFS following a procedure described in ref. 18. The assessment of the structures was performed by using the program procheck-nmr (19, 20). Fold similarities in the Protein Data Bank (PDB) were searched by using the dali (21) program.
15N R1, R2, and steady-state heteronuclear NOEs were measured with pulse sequences as described by Farrow et al. (22), with the water signal suppressed with “water flipback” scheme (23). All spectra were processed with the nmrpipe program (24) and analyzed with sparky 3 software (T. D. Goddard and D. G. Kneller, University of California, San Francisco). The experimental relaxation rates were used to map the spectral density function values, J(ωH), J(ωN), and J(0), following a procedure described in ref. 25. The overall rotational correlation time τm value was estimated from the R2/R1 ratio with the program quadric_diffusion (26, 27).
Results and Discussion
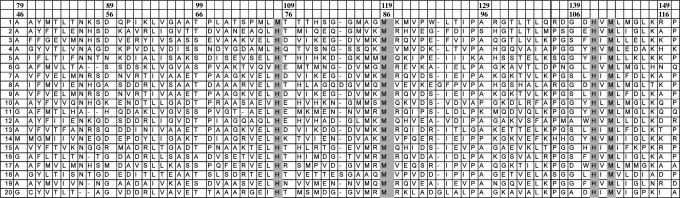
Genome Browsing. A search was performed starting from the YpmQ gene of Bacillus subtilis, codifying for Sco1 protein, through a string web server that retrieves for a given query gene all of the genes that repeatedly occur within potential operons (6). In this way, for eight bacteria, a Sco1 gene with a common neighboring gene of unknown function was found. The latter gene might be functionally associated with Sco1, which, in turn, is functionally associated with the clusters of genes encoding for the three subunits of bacterial CcO (see Fig. 6, which is published as supporting information on the PNAS web site). A blast search over all nonredundant GenBank genomes (as deposited by April, 2004) for proteins sharing >10% sequence identity with the amino acid sequences of any of the eight Sco1 neighboring proteins located 42 further protein sequences. The average residue identity within this protein class is 29 ± 8%, indicating a conserved protein group. In particular, the alignment of the 50 protein sequences shows a conserved potential metal binding motif: H(M)X10MX21HXM (Fig. 1). The 50 sequences belong to 43 Gram-negative and 7 Gram-positive bacterial organisms, and none belongs to eukaryotes.
Fig. 1.
Amino acid sequence alignment. At the top, amino acid numberings are reported according to the full-length and cloned sequence of DR1885, respectively. The alignment includes residues 46–117 of 20 selected protein sequences. The metal binding residues are shaded in gray. Row 1, Gene Info identifier (gi)|15806885|, D. radiodurans; row 2, gi|24375395|, Shewanella oneidensis; row 3, gi|15600808|, Vibrio cholerae; row 4, gi|22984054|, Burkholderia fungorum; row 5, gi|15792238|, C. jejuni; row 6, gi|26989104|, Pseudomonas putida; row 7, gi|27367620|, Vibrio vulnificus; row 8, gi|22959236|, Rhodobacter sphaeroides; row 9, gi|37675728|, Vibrio vulnificus YJ016; row 10, gi|15598980|, P. aeruginosa; row 11, gi|22979433|, Ralstonia metallidurans; row 12, gi|28871996|, Pseudomonas syringae pv. tomato str. DC3000; row 13, gi|28901482|, Vibrio parahaemolyticus; row 14, gi|15606478|, Aquifex aeolicus; row 15, gi|23104168|, Azotobacter vinelandii; row 16, gi|22965901|, Rhodospirillum rubrum; row 17, gi|23001688|, Magnetococcus sp. MC-1; row 18, gi|23017359|, Thermobifida fusca; row 19, gi|22972174|, Chloroflexus aurantiacus; and row 20, gi|16127732|, C. crescentus CB15.
A hydropathy and topological membrane analysis indicated that all proteins belonging to Gram-positive bacteria contain a single N-terminal transmembrane-spanning helix, and the main part of the protein is exposed to the outer leaflet of the cell membrane. In Gram-negative bacteria, these proteins are putative periplasmic soluble proteins or anchored to the inner membrane by a single N-terminal transmembrane helix where the water-soluble region faces the periplasm. Therefore, a water-soluble form can be produced in all cases if the N-terminal transmembrane segment is removed.
To express the protein, sequences from D. radiodurans, C. jejuni, C. crescentus, and P. aeruginosa organisms were selected. Protein cloning was successfully performed on all selected sequences by using pET21a (Novagen) as expression vector. The His-tag recombinant DR1885 protein showed the highest level of expression and was thus selected for the following structural studies.
Expression, Purification, and Characterization of DR1885 Protein. The DR1885 gene codes for a protein of 178 amino acids. The sequence possesses the potential metal binding motif MX10MX21HXM at positions 108, 119, 141, and 143. DR1885 is the only sequence of this class of proteins that has a Met instead of a His residue at position 108 in the potential metal-binding motif (Fig. 1). We cloned and expressed the soluble domain where the first 34 residues, including the N-terminal transmembrane helix (7–28), were omitted. In the expressed protein, four amino acids (IEGR), corresponding to the restriction enzyme recognition site, are further present at the C terminus as a consequence of the cloning, thus producing a final construct of 149 aa with a theoretical molecular mass of 15,473 Da (hereafter residue numbering is based on this construct). Within a week the protein transformed in a species with a molecular mass of 13,447 Da as determined by ESI-MS spectra, corresponding to a fragment of 129 aa, i.e., lacking the first 20 aa of the cloned construct. All of the characterization then was performed on this shorter construct (21–149).
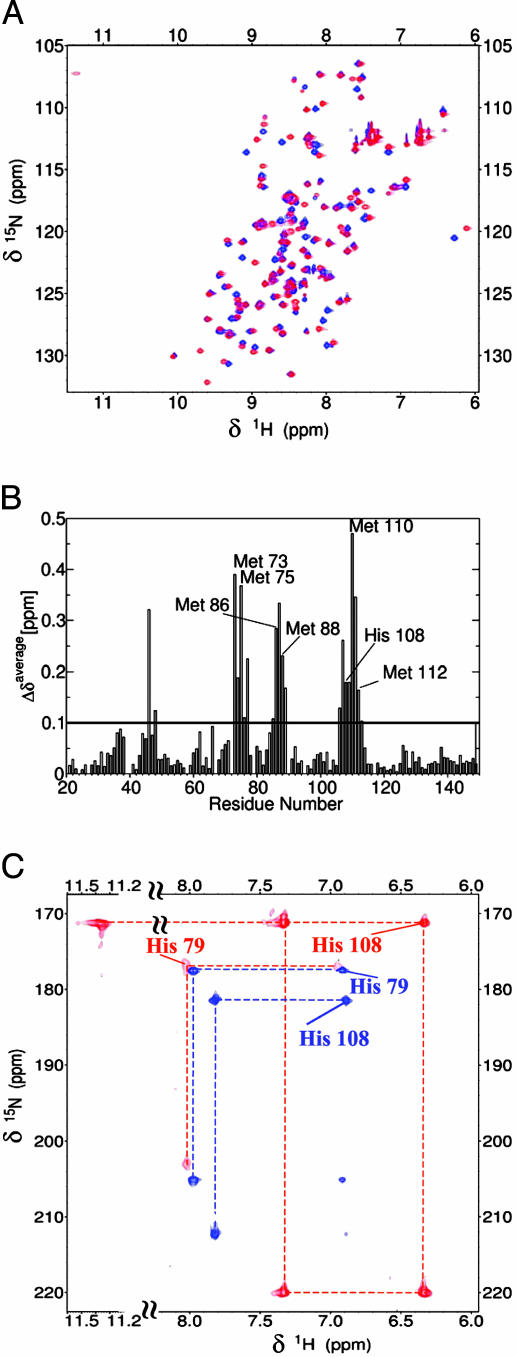
The CD spectrum, characteristic of a protein with a high content of β-sheet, indicates an increase in β-sheet content (from 45% to 60%) with a decrease of random coil (from 55% to 40%) upon addition of one equivalent of Cu(I) (see Fig. 7, which is published as supporting information on the PNAS web site). The amide resonances in the 1H-15N HSQC spectra indicate that Cu(I) binding occurs with a rate slower than the NMR time scale (i.e., <103 s–1). Indeed, upon addition of Cu(I), new amide resonances, corresponding to the holo form, appear with the concomitant disappearance of those of the apo form, their disappearance being completed with a 1:1 Cu(I)/protein ratio (Fig. 2A). Atomic absorption spectroscopy confirms the 1:1 Cu(I)/protein ratio. The charge-state distribution analysis of ESI-MS spectra indicates that one Cu(I) ion remains bound to the protein even after protein ionization and that Cu(I) binding produces a more compact conformational state with respect to the apoform. In fact, the apoform of DR1885 has four main peaks in the mass spectrum with charges +13, +12, +11, and +10, whereas the copper-loaded form has two main peaks with charges +9 and +8. A similar behavior is observed in the case of the copper chaperone Cox17 (28).
Fig. 2.
Effect of Cu(I) binding to DR1885 on NMR spectra. (A) Superposition of the 2D 1H–15N HSQC spectra (500 MHz, 298 K) of apoDR1885 (blue) and Cu(I)DR1885 (red). (B) The weighted average chemical shift differences between apoDR1885 and Cu(I)DR1885, Δavg(HN) = {[(ΔH)2 + (ΔN/5)2]/2}1/2, where ΔH and ΔN are chemical shift differences for 1H and 15N, respectively. (C) 1H-15N HSQC spectra optimized for the detection of 2JNH of His rings, recorded on apoDR1885 (blue) and Cu(I)DR1885 (red).
The 1H-15N HSQC spectra of apoDR1885 obtained with increasing amounts of Cu(II) are completely superimposable to those observed for Cu(I) titration (see Fig. 8, which is published as supporting information on the PNAS web site). No evidence of Cu(II) complex is found from EPR and visible spectroscopy, suggesting that the protein has the ability to reduce Cu(II) in vitro. Other copper proteins, such as the prion and the β-amyloid precursor proteins, bear the capacity to reduce Cu(II) to Cu(I) in vitro (29, 30). In these human proteins, Met residues are susceptible to copper-stimulated oxidation, and it has been speculated that this Met oxidation may have a role in the neurotoxicity of these two proteins (30–32). Several Met residues, in addition to those belonging to the metal binding motif, are present in the amino acid sequences of these novel copper proteins, some of them even close to the conserved Cu(I) ligands (Fig. 1). Therefore, a Met residue might be responsible for reducing Cu(II) to Cu(I).
All these results indicate the presence of a single copper binding site within the protein scaffold, which is strongly specific for Cu(I).
The metal binding site in DR1885 has been characterized through x-ray absorption spectroscopy spectra recorded on a protein sample loaded with one equivalent of Cu(I). Fig. 9, which is published as supporting information on the PNAS web site, shows the edge region of the Cu(I)DR1885 sample compared with other Cu(I) binding proteins like Pseudomonas syringae CopC (33) and Sinorhizobium meliloti Cox11 (9) (see Fig. 9A), as well as the EXAFS (Fig. 9B) and its Fourier transform (see Fig. 9C). These spectra provide clear and definitive information on the oxidation state of the metal ion as well on its coordination number and nature of the ligands. The spectra on Cu(I)DR1885 indicate that Cu(I) is coordinated by 3S and 1N from a His. The EXAFS and the Fourier transform (see Fig. 9 B and C) of Cu(I)DR1885 are well reproduced by a coordination polyhedron constituted by 3 S atoms at an average distance of 2.30(1) Å and by 1 N atom from a His imidazole ring coordinating copper at 2.00(1) Å (see Table 2, which is published as supporting information on the PNAS web site). The 2.30-Å bond distance is larger than the usual 2.24–2.26 Å Cu-S distance observed in three-coordinated Cu(I) complexes (34, 35) and is consistent with a four-coordinated Cu(I) center. The x-ray absorption spectroscopy and EXAFS spectra are same as those observed in CopC, which presents the same set of copper ligands (33). The converging evidence consists of the following: (i) the large improvement of the fit quality upon adding the Cu-N-contribution; (ii) the multiple scattering contributions of the imidazole ring to the spectrum; and (iii) the edge and near-edge features allow us to confidently identify a Cu(I)-S3-N(His) metal site in the DR1885 protein.
Structure and Dynamics of Apo- and Cu(I)DR1885. NMR resonance assignment was obtained from the analysis of 3D triple-resonance experiments performed on a 13C/15N-labeled sample. In the apoform, all backbone NHs were assigned, whereas in the Cu(I) form the backbone NH of His-79 is missing. In total, the resonances of 90% carbon atoms, 98% nitrogen atoms, and 92% protons were assigned for both forms. The 1H, 13C, and 15N resonance assignments of the apo- and Cu(I)DR1885 protein are reported in Tables 3 and 4, which are published as supporting information on the PNAS web site. Several NH signals exhibit shift variations between the two forms, the most significant being localized in the stretches 73–77, 85–89, and 106–113 (Fig. 2B), which comprise the residues of the metal binding motif MX10MX21HXM. The nonexchangeable His-ring protons were assigned through a 1H-15N HSQC experiment tailored to the detection of 2J 1H-15N couplings and from the analysis of the 2D NOE spectroscopy and total correlation spectroscopy maps. From the pattern of 2J 1H-15N couplings, it appears that, in the apoform, both His-79 and His-108 are protonated on Nε2. In the Cu(I) form, His-79 has the same 2J 1H-15N coupling pattern, whereas His-108 drastically changes (Fig. 2C), with Nδ1 protonated and Nε2 coordinated to the Cu(I) ion, as indicated by its chemical shift of 220 ppm (Fig. 2C), typical of His-ring nitrogens bound to metal ions (36, 37). The exchangeable His-ring NH signal for both His residues are not detected in the apoform, whereas the Nδ1H signal of His-108 is observed in the Cu(I) form (Fig. 2C). This result indicates that the NH exchange with the solvent is slowed down by the coordination bond with Cu(I).
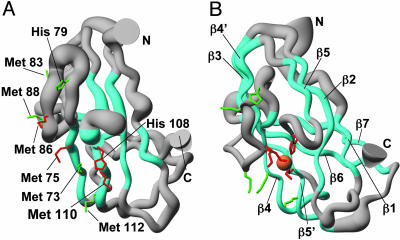
NOE-derived H-H upper limit distances and backbone dihedral angles restraints were used for structure calculations of apoDR1885 (Fig. 3A). The statistical analysis of apoDR1885 is reported in Table 5, which is published as supporting information on the PNAS web site. The N-terminal residues 21–32 do not show long- and medium-range NOEs, being so highly disordered. To identify which Met residues are involved in Cu(I) coordination, we have performed various structure calculations by linking the copper atom to the ring Nε2 of His-108 and to three sulfur atoms out of Met-73, -75, -83, -86, -88, -110, and -112 in all possible combinations. The calculation with the lowest penalty function is obtained when the three conserved Met-75, -86, and -110 are linked. On the contrary, when one or more of the nonconserved Mets are linked to the Cu ion substituting one or more of the conserved ones, a significant increase of the penalty function is observed. From all of the NMR and EXAFS data, it can be concluded that the Cu(I) ion of Cu(I)DR1885 is coordinated by Met-75, -86, and -110 and by the ring Nε2 of His-108 (Fig. 3B). The statistical analysis of Cu(I)DR1885 structure is reported in Table 6, which is published as supporting information on the PNAS web site.
Fig. 3.
Solution structures of apoDR1885 (A) and Cu(I)DR1885 (B). The side chains of the Met and His residues belonging to the conserved metal binding motif MX10MX21HXM are shown in red. The other Met and His residues present in the sequence are in green. The Cu(I) ion is shown in orange, and β-strands are cyan. The radius of the tube is proportional to the backbone rmsd value of each residue.
Internal protein mobility was characterized for both apo and Cu(I)DR1885 through the analysis of 15N R1, R2, and 1H-15N NOEs (see Fig. 10, which is published as supporting information on the PNAS web site). Average 15N R1, R2, and 1H-15N NOE values are 1.73 ± 0.06 s–1, 10.2 ± 0.3 s–1, and 0.65 ± 0.03 for apoDR1885, and 1.67 ± 0.05 s–1, 10.9 ± 0.3 s–1, and 0.74 ± 0.04 for Cu(I)DR1885 at 500 MHz, respectively. The relaxation rates are essentially homogeneous along the entire polypeptide sequence, with the exception of some residues in loop regions and in the C terminus, the latter ones showing a large decrease in 1H-15N NOE and R2 values for both apoform and Cu(I) form. This behavior, as also indicated by the higher J(ωH) spectral density function values in these regions (see Fig. 11, which is published as supporting information on the PNAS web site), indicates the presence of local motions in the nanosecond–picosecond timescale, i.e., faster than the overall protein tumbling rate. The most relevant difference in terms of backbone mobility between apoform and Cu(I) form involves the disordered region comprising residues 79–84. In the apoform, fast local motions affect this region, as indicated by high J(ωH) values, whereas these motions are essentially not present in the Cu(I) form (see Fig. 11). It is important to point out that this region is upstream of the Cu(I) binding ligand Met-86.
The correlation times for molecule reorientation of apo- and Cu(I)DR1885 are 8.9 ± 0.5 and 9.2 ± 0.6 ns, respectively, as expected for a protein of this size in a monomeric state, thus indicating that Cu(I) binding does not induce aggregation phenomena.
The structure of the soluble domain of Cu(I)DR1885 reveals a well defined domain composed entirely by β-strands (Fig. 3B), organized in two β-sheets forming a Greek key β-barrel motif that involves nine strands, two of which are parallel, whereas the others are antiparallel. The core of the protein has a rough cylindrical shape formed by the various β-strands connected by loops mainly located at the two opposite sides of the cylinder. While maintaining the same fold topology of the Cu(I) form [rms deviation (rmsd) between the two average structures superimposing the β-strand regions is 1.5 Å for the backbone], the absence of copper produces a less tight and compact structure, with shorter β-strands and lacking strand β7. Accordingly, the CD spectra show a considerable increase of the β-strand content upon Cu(I) binding, and the ESI-MS data indicate a more compact structural organization. As a matter of fact, the apo-structure has a lower number of NOEs, consistently with being globally more disordered than the Cu(I) form.
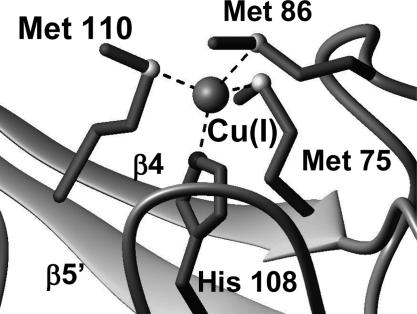
The β-sheet topology and the binding of Cu(I) bring all of the conser ved Met and His residues, belonging to the MX10MX21HXM copper-binding motif, close each other, whereas most of the remaining Met residues surround the metal-binding site, likely to protect and stabilize the Cu(I) state. In the Cu(I)DR1885 structure, Met-75, Met-110, and His-108 are located in the antiparallel strands β4 and β5′ facing each other, whereas Met-86 is lying above the plane defined by these two strands (Fig. 4). The four donor atoms are found in a roughly tetrahedral geometry around the metal ion (Fig. 4). This structural arrangement causes a low solvent accessibility of Cu(I), indicating that the three Met ligands and His-108 provide a hydrophobic pocket for the Cu(I) ion. Differently, in the apo-form, Met-86 is far from the copper-binding region (Fig. 3A), and the upstream region 79–84 is flexible and can easily adapt to structural changes induced by the presence of the Cu(I) ion.
Fig. 4.
The copper site of Cu(I)DR1885. The side chains of the Met and His ligands are shown in dark gray. The sulfur and nitrogen donor atoms of the ligands are shown as light gray and black spheres, respectively.
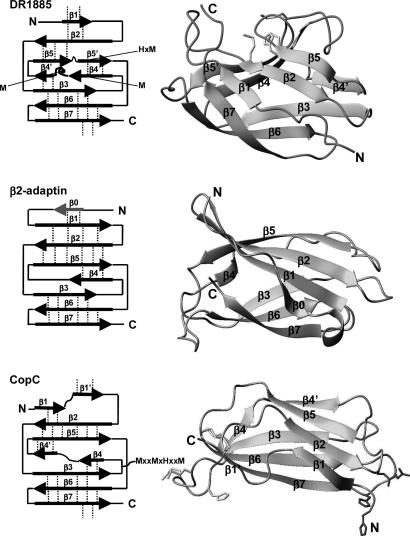
A Bacterial Cu(I) Protein. The Greek key topology of Cu(I)DR1885 resembles the so-called cupredoxin fold (38). Although this structure is the first structure for this previously undescribed class of copper proteins, a similar fold is shared by other proteins. The closest matches have been detected with some Ig plasma membrane adaptin proteins [PDB ID codes 1E42, β2-adaptin, Z-score of 4.6, rmsd on Cα atoms of 3.1 Å with Cu(I)DR1885; and 1GYU, γ-adaptin, Z-score of 3.7, rmsd on Cα atoms of 3.1 Å with Cu(I)DR1885]. A lower match was found with copper proteins (Z-score of 2.3) such as blue oxidases (PDB ID codes 1KV7 and 1AOZ) and a bacterial periplasmic copper trafficking protein CopC (PDB ID codes 1M42, rmsd on Cα atoms of 3.1 Å with Cu(I)DR1885, and 1LYQ), all possessing a cupredoxin-like fold. CopC is able to bind both Cu(I) and Cu(II) in two different sites (33). None of the above-mentioned copper proteins shares high sequence similarity with DR1885 protein.
Cu(I)DR1885 structurally differs with the closest structural homolog β2-adaptin (39) in the lack of the N-terminal β-strand (β0) and in the presence of an additional strand (β4′) and of a long Met-rich loop between strands β4 and β4′ (Fig. 5). Moreover, differently from the β2-adaptin structure, which has a long β-strand (β5) entirely antiparallel to strand β2, strand β5 of Cu(I)DR1885 is interrupted by a short loop. Its presence produces a shift of the subsequent strand β5′ where the H(V)M copper binding residues are located. All these structural variations, predominantly located close to the metal binding region, are important to arrange the copper-binding site in one face of the β-barrel. Cu(I)DR1885 structure maintains the same global Greek key topology of CopC, but the β-sheet association as well as the location of the Cu(I) binding site are different (Fig. 5). The Cu(I) site is still constituted by one His and three Mets, but is located in one extreme of the β-barrel (40, 41).
Fig. 5.
Comparison among the structures of Cu(I)DR1885, β2-adaptin (PDB ID code 1E42), and apoCopC (PDB ID code 1M42). (Right) The topology of each protein is shown. The side chains of the Cu(I) and Cu(II) binding residues are shown in light gray and black, respectively. The dashed lines indicate sheet formation between two β-strands.
Cu(I)DR1885 adopts a fold reminiscent of that of other bacterial extracytoplasmic copper proteins like CopC (1), constituting an emerging class of proteins that use Met-rich motifs to bind Cu(I). Interestingly, also in eukaryotes copper transport and import inside cells occur through a series of clustered Met residues arranged as [M(X)nM]m motifs (42), reminiscent of those present in this class of Cu(I) bacterial transporters. The ability of binding Cu(I) is also shared by the other protein members of this family, which all have a His instead of a Met at position 108 in the metal-binding motif, at variance of DR1885. Indeed, we have experimentally found that the protein from C. crescentus also binds Cu(I) with a 1:1 stoichiometry.
The Cu(I) binding site of DR1885 is sterically hindered but located on the protein surface. It can still allow a partner protein to access the metal site by means of conformational changes and coordination of an incoming side chain, probably a Cys of the metal acceptor, because the structurally flexible metal binding environment of DR1885 and the displacement of Met-86 can facilitate the interaction with the metal ligands of the receiving protein. DR1885 might also have a role in stabilizing the Cu(I) redox state in an extracytoplasmic oxidizing environment through a protected, hydrophobic Met-rich region. DR1885 thus might be considered an extracytoplasmic chaperone specific for Cu(I).
The functional role of DR1885 as Cu(I) chaperone for Sco1 finds an experimental support from the recently reported evidence that a DR1885 homolog from Vibrio cholerae bacterium (VCA0037 gene) (Fig. 1) has a coregulated expression with the Sco homolog (VCA0038 gene) (see http://string.embl.de).
The proposed metallochaperone function of DR1885 allows us to compare the eukaryotic and prokaryotic pathways for providing Cu(I) to the CuA center of CcO. In eukaryotes, the insertion of copper into the binuclear CuA center is known to require at least two proteins, Sco1 and Cox17 (43). The first is a Cu(I) binding protein that is known to interact with subunit 2 of CcO (44), and it is proposed to donate Cu(I) to the CuA center, whereas the second, exclusively present in eukaryotes, is a mitochondrial chaperone that provides copper to the CuA center of CcO (10, 45). DR1885 might take the role of Cox17 in the extracytoplasmic environment of bacteria and may donate one Cu(I) ion to Sco1, which binds only one Cu(I) ion per molecule (46). However, the CuA center is a binuclear copper center, where, in the oxidized state, the electron is totally delocalized over the two copper ions in a mixed-valence [Cu(1.5)... Cu(1.5)] state (47). Therefore, once DR1885 has provided one Cu(I) to the CuA site through Sco1, it may be possible in bacteria that Cu(II), which is available in the extracytoplasmic environment, enters the second site of the CuA center, forming the binuclear mixed-valence units. In accordance with this scheme of the copper “recruitment” for bacterial CuA center, kinetic and copper binding studies on a CuA center engineered into P. aeruginosa azurin showed that the formation of a Cu(I) monoderivative was observed and that the following addition of Cu(II) converts the intermediate to the final purple CuA center (48, 49). Apparently, this hypothesis on a sequential copper insertion mechanism for the formation of the binuclear CuA center in bacteria could be extended to eukaryotes with difficulty, because Cox17 binds up to four Cu(I) ions (28, 50) and therefore can simultaneously transfer more than one copper ion. However, it has been shown recently that only a subset of the bound Cu(I) ions of Cox17 is available for metal transfer (10). The recently solved solution structure of the yeast Cox17 indicates that Cox17 might also bind one Cu(I) ion (51), similarly to DR1885. Therefore, all these data suggest that a sequential copper insertion in the CuA center can also occur in eukaryotes similarly to what is here suggested for bacteria, even if a different kind of metallochaperone was selected by genome evolution (52).
Supplementary Material
Acknowledgments
We thank Prof. R. Udisti for atomic absorption spectroscopy and the Centro Italiano Spettrometria di Massa facility for ESI-MS experiments. The work was supported by European Commission SPINE Contract No. QLG2-CT-2002–00988, Structural Proteomics in Europe, and by Ente Cassa Risparmio di Firenze.
Author contributions: L.B., I.B., and S.C.-B. designed research; S.C.-B., E.K., K.K., and S.M. performed research; S.C.-B., E.K., K.K., and S.M. analyzed data; and L.B., I.B., S.C.-B., N.K., K.K., and S.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CcO, cytochrome c oxidase; ESI, electrospray ionization; EXAFS, extended x-ray absorption fine structure; HSQC, heteronuclear single quantum correlation; NOE, nuclear Overhauser effect; PDB, Protein Data Bank; rmsd, rms deviation.
Data deposition: The derived atomic coordinates for a family of acceptable structures and for the average minimized structures of apoDR1885 and Cu(I)DR1885 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1X7L and 1X9L).
References
- 1.Finney, L. A. & O'Halloran, T. V. (2003) Science 300, 931–936. [DOI] [PubMed] [Google Scholar]
- 2.Banci, L. & Rosato, A. (2003) Acc. Chem. Res. 36, 215–221. [DOI] [PubMed] [Google Scholar]
- 3.Arnesano, F., Banci, L., Bertini, I. & Ciofi-Baffoni, S. (2004) Eur. J. Inorg. Chem. 2004, 1583–1593. [Google Scholar]
- 4.Cavet, J. S., Borrelly, G. P. & Robinson, N. J. (2003) FEMS Microbiol. Rev. 27, 165–181. [DOI] [PubMed] [Google Scholar]
- 5.Snel, B., Bork, P. & Huynen, M. A. (2002) Proc. Natl. Acad. Sci. USA 99, 5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Mering, C., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P. & Snel, B. (2003) Nucleic Acids Res. 31, 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattatall, N. R., Jazairi, J. & Hill, B. C. (2000) J. Biol. Chem. 275, 28802–28809. [DOI] [PubMed] [Google Scholar]
- 8.Hiser, L., Di Valentin, M., Hamer, A. G. & Hosler, J. P. (2000) J. Biol. Chem. 275, 619–623. [DOI] [PubMed] [Google Scholar]
- 9.Banci, L., Bertini, I., Cantini, F., Ciofi-Baffoni, S., Gonnelli, L. & Mangani, S. (2004) J. Biol. Chem. 279, 34833–34839. [DOI] [PubMed] [Google Scholar]
- 10.Horng, Y. C., Cobine, P. A., Maxfield, A. B., Carr, H. S. & Winge, D. R. (2004) J. Biol. Chem. 279, 35334–35340. [DOI] [PubMed] [Google Scholar]
- 11.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann, K. & Stoffel, W. (1993) Biol. Chem. Hoppe-Seyler 347, 166. [DOI] [PubMed] [Google Scholar]
- 13.Tusnady, G. E. & Simon, I. (1998) J. Mol. Biol. 283, 489–506. [DOI] [PubMed] [Google Scholar]
- 14.Deleage, G. & Geourjon, C. (1993) Comp. Appl. Biosci. 9, 197–199. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann, T., Güntert, P. & Wüthrich, K. (2002) J. Mol. Biol. 319, 209–227. [DOI] [PubMed] [Google Scholar]
- 16.Güntert, P., Mumenthaler, C. & Wüthrich, K. (1997) J. Mol. Biol. 273, 283–298. [DOI] [PubMed] [Google Scholar]
- 17.Pearlman, D. A., Case, D. A., Caldwell, J. W., Ross, W. S., Cheatham, T. E., Ferguson, D. M., Seibel, G. L., Singh, U. C., Weiner, P. K. & Kollman, P. A. (1997) amber 5.0 (University of California, San Francisco).
- 18.Arnesano, F., Banci, L., Bertini, I., Huffman, D. L. & O'Halloran, T. V. (2001) Biochemistry 40, 1528–1539. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R. & Thornton, J. M. (1996) J. Biomol. NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski, R. A., MacArthur, M. W. & Thornton, J. M. (1998) Curr. Opin. Struct. Biol. 8, 631–639. [DOI] [PubMed] [Google Scholar]
- 21.Holm, L. & Sander, C. (1996) Science 273, 595–603. [DOI] [PubMed] [Google Scholar]
- 22.Farrow, N. A., Muhandiram, R., Singer, A. U., Pascal, S. M., Kay, C. M., Gish, G., Shoelson, S. E., Pawson, T., Forman-Kay, J. D. & Kay, L. E. (1994) Biochemistry 33, 5984–6003. [DOI] [PubMed] [Google Scholar]
- 23.Grzesiek, S. & Bax, A. (1993) J. Am. Chem. Soc. 115, 12593–12594. [Google Scholar]
- 24.Delaglio, F., Grzesiek, S., Vuister, G., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 25.Peng, J. W. & Wagner, G. (1992) J. Magn. Reson. 98, 308–332. [Google Scholar]
- 26.Brüschweiler, R., Liao, X. & Wright, P. E. (1995) Science 268, 886–889. [DOI] [PubMed] [Google Scholar]
- 27.Lee, L. K., Rance, M., Chazin, W. J. & Palmer, A. G., III. (1997) J. Biomol. NMR 9, 287–298. [DOI] [PubMed] [Google Scholar]
- 28.Palumaa, P., Kangur, L., Voronova, A. & Sillard, R. (2004) Biochem. J. 382, 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Multhaup, G., Schlicksupp, A., Hesse, L., Beher, D., Ruppert, T., Masters, C. L. & Beyreuther, K. (1996) Science 271, 1406–1409. [DOI] [PubMed] [Google Scholar]
- 30.Wong, B. S., Wang, H., Brown, D. R. & Jones, I. M. (1999) Biochem. Biophys. Res. Commun. 259, 352–355. [DOI] [PubMed] [Google Scholar]
- 31.Barnham, K. J., Ciccotosto, G. D., Tickler, A. K., Ali, F. E., Smith, D. G., Williamson, N. A., Lam, Y. H., Carrington, D., Tew, D., Kocak, G., et al. (2003) J. Biol. Chem. 278, 42959–42965. [DOI] [PubMed] [Google Scholar]
- 32.Nishino, S. & Nishida, Y. (2001) Inorg. Chem. Commun. 4, 86–89. [Google Scholar]
- 33.Arnesano, F., Banci, L., Bertini, I., Mangani, S. & Thompsett, A. R. (2003) Proc. Natl. Acad. Sci. USA 100, 3814–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kau, L. S., Spira-Solomon, D. J., Penner-Hahn, J. E., Hodgson, K. O. & Solomon, E. I. (1987) J. Am. Chem. Soc. 109, 6433–6442. [Google Scholar]
- 35.Pickering, I. J., George, G. N., Dameron, C. T., Kurz, B., Winge, D. R. & Dance, I. G. (1993) J. Am. Chem. Soc. 115, 9498–9505. [Google Scholar]
- 36.Chen, Y.-L., Park, S., Thornburg, R. W., Tabatabai, L. B. & Kintanar, A. (1995) Biochemistry 34, 12265–12275. [DOI] [PubMed] [Google Scholar]
- 37.Eijkelenboom, A. P., Van den Ent, F. M., Vos, A., Doreleijers, J. F., Hard, K., Tullius, T. D., Plasterk, R. H., Kaptein, R. & Boelens, R. (1997) Curr. Biol. 7, 739–746. [DOI] [PubMed] [Google Scholar]
- 38.Adman, E. T. (1991) Adv. Prot. Chem. 42, 144–197. [DOI] [PubMed] [Google Scholar]
- 39.Owen, D. J., Vallis, Y., Pearse, B. M., McMahon, H. T. & Evans, P. R. (2000) EMBO J. 19, 4216–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnesano, F., Banci, L., Bertini, I. & Thompsett, A. R. (2002) Structure (London) 10, 1337–1347. [DOI] [PubMed] [Google Scholar]
- 41.Wernimont, A. K., Huffman, D. L., Finney, L. A., Demeler, B., O'Halloran, T. V. & Rosenzweig, A. C. (2003) J. Biol. Inorg. Chem. 8, 185–194. [DOI] [PubMed] [Google Scholar]
- 42.Puig, S., Lee, J., Lau, M. & Thiele, D. J. (2002) J. Biol. Chem. 277, 26021–26030. [DOI] [PubMed] [Google Scholar]
- 43.Carr, H. S. & Winge, D. R. (2003) Acc. Chem. Res. 36, 309–316. [DOI] [PubMed] [Google Scholar]
- 44.Lode, A., Kuschel, M., Paret, C. & Rodel, G. (2000) FEBS Lett. 485, 19–24. [DOI] [PubMed] [Google Scholar]
- 45.Punter, F. A. & Glerum, D. M. (2003) J. Biol. Chem. 278, 30875–30880. [DOI] [PubMed] [Google Scholar]
- 46.Balatri, E., Banci, L., Bertini, I., Cantini, F. & Ciofi-Baffoni, S. (2003) Structure (London) 11, 1431–1443. [DOI] [PubMed] [Google Scholar]
- 47.Farrar, J. A., Neese, F., Lappalainen, P., Kroneck, P. M. H., Saraste, M., Zumft, W. G. & Thomson, A. J. (1996) J. Am. Chem. Soc. 118, 11501–11514. [Google Scholar]
- 48.Hay, M. T. & Lu, Y. (2000) J. Biol. Inorg. Chem. 5, 699–712. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X., Ang, M. C. & Lu, Y. (1999) J. Am. Chem. Soc. 121, 2947–2948. [Google Scholar]
- 50.Heaton, D., Nittis, T., Srinivasan, C. & Winge, D. R. (2000) J. Biol. Chem. 275, 37582–37587. [DOI] [PubMed] [Google Scholar]
- 51.Abajian, C., Yatsunyk, L. A., Ramirez, B. E. & Rosenzweig, A. C. (2004) J. Biol. Chem. 279, 53584–53592. [DOI] [PubMed] [Google Scholar]
- 52.Boussau, B., Karlberg, E. O., Frank, A. C., Legault, B. A. & Andersson, S. G. (2004) Proc. Natl. Acad. Sci. USA 101, 9722–9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.