Abstract
Our previous study revealed that microRNA (miR) −30c represents a potential tumor suppressor gene, the expression of which is associated with decreased oncogenic potential in prostate cancer (PCa) cell lines. However, the functional role and underlying mechanisms of miR-30c in PCa remain to be fully elucidated. Reverse transcription-quantitative polymerase chain reaction and immunohistochemical analysis were used to detect the expression levels of alternative splicing factor/splicing factor 2 (ASF/SF2) in PCa tissues. A luciferase reporter assay was used to investigate whether ASF/SF2 may be a direct target gene of miR-30c. In addition, the effects of miR-30c on the proliferation and apoptosis of PCa cell lines were examined, following transfection with miR-30c mimics. Furthermore, correlation analysis was performed to investigate the relationship between the expression of miR-30c and ASF/SF2 and various clinicopathological parameters of patients with PCa. The present results demonstrated that PCa tissues exhibited higher levels of alternative splicing factor/splicing factor 2 (ASF/SF2), compared with normal tissues. In addition, miR-30c was revealed to targete the 3′-untranslated region of the ASF/SF2 gene, causing a decrease in the mRNA and protein levels of ASF/SF2. Furthermore, miR-30c was reported to decrease cell proliferation, increase the percentage of cells in the G1 cell cycle phase, and promote apoptosis through the inhibition of ASF/SF2. Following correlation analysis using patient samples, the expression of ASF/SF2 was revealed to be tightly correlated with the pathological stage of PCa and biochemical recurrence (BCR). In addition, patients with PCa exhibiting low expression levels of miR-30c and high expression of ASF/SF2 had significantly lower rates of BCR-free survival. In conclusion, the present study suggested that the tumor suppressor miR-30c may be involved in PCa tumorigenesis, possibly via targeting ASF/SF2. The combined analysis of the expression of ASF/SF2 and miR-30c may be a valuable tool for early prediction of BCR in patients with PCa following radical prostatectomy.
Keywords: prostate cancer, microRNA-30c, alternative splicing factor/splicing factor 2, cell cycle, apoptosis, biochemical recurrence
Introduction
Prostate cancer (PCa) is the second most commonly diagnosed cancer in men worldwide (1). A number of factors are critical in the development of PCa, including genetic mutations and the tumor microenvironment (2,3). Several reports have indicated that genetic alterations are observed in advanced PCa; however, no single oncogenic event by itself is sufficient to determine a large proportion of the diseases (4–6). Accordingly, further understanding how oncogenic lesions may collaborate to augment the progression of PCa is likely to assist in clinical diagnosis and treatment.
MicroRNAs (miRNAs) are RNA polymerase II-transcribed RNAs, with a length of 18–24-nucleotides, which can negatively regulate gene expression (7). Altered levels of miRNAs have been observed in several solid tumors, including PCa (8–10). Dysfunctional miRNAs can mediate divergent cellular processes, including cellular proliferation, migration, differentiation and apoptosis (9). Several reports have indicated that specific miRNAs have been associated with PCa tumorigenesis and the clinical outcome of patients (10–12).
Our previous data demonstrated that that miR-30c represents a potential tumor suppressor gene, the expression of which is associated with decreased oncogenic potential in PC cell lines (13,14). Several genes, including UBC9, B-Myb and BCL9 have also been identified as miR-30c targets in tumor cells (15–17). As one miRNA can target multiple genes simultaneously, it is suggested that the functional role and mechanisms underlying the effect of miR-30c in PCa remain to be fully elucidated.
The serine- and arginine-rich (SR) proteins, and the heterogeneous nuclear ribonucleoproteins (hnRNPs) are two families of mRNA processing proteins, which serve, respectively, as splicing activators and repressors (18,19). These RNA-binding proteins can regulate the alternative splicing of several genes and have been associated with splicing dysregulation in various diseases (20,21).
The prototypical SR protein, of alternative splicing factor/splicing factor 2 (ASF/SF2; also termed SF2/ASF or SRSF1) functions in mRNA constitutive and alternative splicing, and is also incolved in stability, nuclear export and translation (22). SF2/ASF has been found to be involved in several malignancies, and can be oncogenic with effects on cellular transformation (20,21). Although ASF/SF2 is frequently upregulated in PCa (23), its roles in the cancer initiation and progression remain to be fully elucidated.
In the present study, it was demonstrated that the overexpression of ASF/SF2 was inhibited by miR-30c in PCa cells. A luciferase reporter assay further confirmed that ASF/SF2 was a direct target of miR-30c. The ectopic expression of miR-30c also resulted in the inhibition of proliferation, and induced cell cycle arrest and apoptosis by downregulating ASF/SF2. In tissue samples, ASF/SF2 was significantly associated with the pathological stage of PCa. Finally, the prognostic value of the combined analysis of ASF/SF2 and miR-30c was determined in biopsies from patients with PCa.
Materials and methods
Ethical approval
The clinical sample cohort used in the present study was approved by the Ethical Committee of Guangzhou First People's Hospital, Guangzhou Medical University (Guangzhou, China). All patients provided written informed consent prior to enrollment in the present study.
Patients and tissues samples
For RNA extraction, tumor samples were obtained from 111 patients with PCa, who underwent radical prostatectomy at Guangzhou First People's Hospital between January 2002 and August 2012. The detailed characteristics of the patients are presented in Table I. For immunohistochemistry, tumor tissues and adjacent normal tissues were obtained from 20 patients with PCa. Any patients who received either radiotherapy or hormonal treatment prior to surgery were not included in the investigation. Biochemical recurrence (BCR) was described as postoperative serum prostate-specific antigen (PSA) ≥0.2 ng/ml.
Table I.
Patients and clinicopathologic features.
| Characteristic | Patients with PCa (n=111) |
|---|---|
| Age range (years) | 47–83 |
| Median age (years) | 58 |
| Preoperative prostate-specific antigena | |
| <4 ng/ml | 17 |
| 4–10 ng/ml | 68 |
| ≥10 ng/ml | 24 |
| Gleason scorea | |
| 6 | 35 |
| 7 | 55 |
| 8 | 8 |
| 9 | 9 |
| Pathological stagea | |
| pT2 | 69 |
| pT3 | 30 |
| pT4 | 7 |
| Follow-up range (months) | 1–128 |
| Biochemical recurrence (n) | 25 |
| 1–12 months | 12 |
| 13–24 months | 6 |
| >24 months | 7 |
Data missing from some patients.
Transient transfection of mature miRNA
miR-30c mimics, negative control mimics, anti-miR-30c inhibitors and negative control inhibitors were purchased from GenPharma (Shanghai, China). The sequences of the oligoribonucleotides were as follows: miR-30c mimic, 5′-CCAGUGUAGGGUAAACACCUCUCUCAGCUUGG-3′; and anti-miR-30c inhibitor, 5′-GCUGAGAGUGUAGGAUGUUUACA-3′. The miR-30c mimics, anti-miR-30c inhibitors, and controls were transiently transfected into the human DU145 and 22RV1 prostatic carcinoma cell lines (American Type Culture Collection, Manassas, VA, USA) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. PCa cells were cultured in RPMI 1640 medium (Hyclone; GE Healthcare LifeSciences, Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientifc, Inc.) and 1% penicillin and streptomycin (Invitrogen; Thermo Fisher Scientifc, Inc.). For transient transfection, 0.5×105 cells were plated in 24-well plates 24 h prior to transfection and maintained at 37°C in an atmosphere of 5% CO2. Following transfection, ASF/SF2 mRNA and protein were extracted for further investigation.
Western blot analysis
Following standard protocol, total proteins were extracted from the cells. Cells and tumors containing at least 70% tumor cells were lysed using 200 µl M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, Inc.) and the supernatants were collected following centrifugation at 10,000 × g for 5 min at 4°C. Protein concentration was measured using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). Equal amounts of extracted protein samples (~20–30 µg) were boiled at 95°C for 5 min, separated by 10% SDS-PAGE (NuPAGE™ Novex 4–12% Bis-Tris Gel; cat. no. NP0322BOX; Thermo Fisher Scientific, Inc.), and transferred onto a nitrocellulose membrane (cat no. IPFL00010; EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% skim milk in PBS containing 0.1% Tween-20, and probed the following primary antibodies at 4°C overnight: Anti-ASF/SF2 (cat. no. HPA061301; dilution, 1:500; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and anti-GAPDH (cat. no. 2118; dilution, 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA). The membrane was then incubated with horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G secondary antibody (cat no. SAB3700852; 1:2,000; Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. Protein bands were visualized by enhanced chemiluminescence using the Super Signal West Pico Chemiluminescent Substrate kit (Pierce; Thermo Fisher Scientific, Inc.).
ASF/SF2 small interfering (si)RNA transfection and vector construction
PCa cells (0.5×105) were plated in 24-well plates and cultured in RPMI 1640 medium (Hyclone; GE Healthcare LifeSciences) at 37°C for 12 h prior to transfection. siRNA reverse transfection reactions were performed using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). The siRNA sequence targeting ASF/SF2 (Qiagen, Inc., Valencia, CA, USA) was 5′-CCAACAAGATAGAGTATAA-3′. AllStars Neg. Control siRNA (Qiagen, Inc.) was used as a negative control. The final concentration for control siRNA and ASF/SF2 siRNA transfection was 100 nM. The ASF/SF2 coding sequence, without the 3′-untranslated region (UTR) was cloned into the pGCMV/EGFP/Neo-Vector (GenePharma). A blank vector served as the negative control. The culture medium was replaced with fresh medium 48 h post-transfection, and the cell lysates were prepared at 72 h for western blot analysis.
In vitro luciferase assay
Using the databases TargetScan (http://targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and miRDB (http://www.mirdb.org/miRDB/), the putative target genes of miR-30c were investigated, and the ASF/SF2 gene was identified among them. To amplify the 3′UTR sequence of the human ASF/SF2 gene, primers were designed. Mlu1 and Spe1 sequences were cloned into the ends of primers (Promega Corporation, Madison, WI, USA). According to the manufacturer's protocol, 3′-UTR cloning was performed in a pMIR reporter vector (Promega Corporation). The pMIR-ASF/SF2 3′-UTR reporter vector (100 ng) was cotransfected with 50 ng of β-Gal and miRNA-30c mimic or anti-miR-30c using Lipofectamine 2000 transfection reagent. All Stars negative control siRNA (Qiagen, Inc.) and miScript inhibitor negative control (Qiagen, Inc.) were used as controls for mimic or inhibitor transfection, respectively. At 24 h post-transfection, luciferase reporter assays were performed, as described previously (9).
Immunohistochemical staining
Immunohistochemical assays were performed to detect the expression of ASF/SF2 in 4-cm thick formalin-fixed, paraffin-embedded sections of PCa and normal tissues. Using the DAKO EnVision system (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA), hematoxylin and eosin or immunohistochemical staining were performed on the formalin-fixed paraffin-embedded tissue sections. Endogenous peroxidase activity was quenched using 3% hydrogen peroxidase for 10 min at room temperature. Microwave antigen retrieval was performed with EDTA buffer for 30 min. The slides were then incubated overnight with primary antibody against ASF/SF2 protein (cat. no. HPA061301; Sigma-Aldrich; Merck KGaA) at a dilution of 1:200 at 4°C. Following washing with PBS, the sections were incubated with horseradish peroxidase-labeled polymer-conjugated secondary antibodies (DAKO EnVision system; Dako; Agilent Technologies, Inc.) for 1 h at room temperature and the proteins of interest were visualized following incubation with 3,3′-diaminobenzidine (Dako; Agilent Technologies, Inc.) at room temperature for 5 min, under an Olympus AX70 microscope (Olympus Corporation, Tokyo, Japan).
The extent and intensity of staining was evaluated independently by two experienced pathologists in a blinded manner. The extension was scored as the percentage of positive cells, and a score of 0–4 was assigned (0, 0%; 1, 1–25%; 2, 26–50%; 3, 51–75%; or 4, 76–100%). The percentage of positive cells was also calculated (0, no staining; 1, weak; 2, moderate; 3, strong staining). The sum of the two scores was used as the final staining score. Tumor specimens with a score of ≥3 were considered to be positive.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
RT-qPCR analysis was performed as previously described (13). miRNA was extracted from the 111 frozen PCa tissues and the two transfected PCa cell lines (22RV1 and DU145) following transfection using a miRNA extraction kit (BioTek China, Beijing, China). cDNA synthesis was performed according to the manufacturer's protocol using the All-in-One™ miRNA qRT-PCR Detection kit (GeneCopoeia, Inc., Rockville, MD, USA) with specific primers. The sequences of the primers, purchased from Ambio; Thermo Fisher Scientific, Inc. were as follows: miR-30c, forward 5′-TGTGTTTTTATTGTTTTTGTTGTCCCA-3′, reverse 5′-GGGACAGAACAGGTTAATGGGAA-3′; RNU6B, forward 5′-CTCGCTTCGGCAGCACA-3′, reverse 5′-AACGCTTCACGAATTTGCGT-3′; ASF/SF2, forward 5′-TTTAGATCTCACGAGGGAGAA-3′, reverse 5′-CGTGGTGATCCTCTGCTTC-3′; and β-actin, forward 5′-GGTGGCTTTTAGGATGGCAAG-3′ and reverse 5′-ACTGGAACGGTGAAGGTGACAG-3′. RNU6B and β-actin were used as the internal controls for miR-30c and ASF/SF2, respectively. qPCR was performed using TaqMan Gene Expression assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) and the MyiQ™2 Two-Color Real-Time PCR Detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Amplification conditions were as follows: Initial 1 step at 95°C for 5 min, followed by 40 cycles at 95°C for 10 sec and at 60°C for 20 sec, with a final extension step at 72°C for 20 sec. Gene expression was quantified using the comparative Cq method (24), with the IQ™5 Optical System software version 2.0 (Bio-Rad Laboratories, Inc.). The experiments were performed in triplicate.
Cell proliferation assay
For the cell proliferation assay, 2,000 cells/well were seeded into 96-well plates, cultured in RPMI 1640 medium (Hyclone; GE Healthcare Life Sciences) supplemented with 10% FBS for 24, 48 and 72 h, and then incubated with 20 µl CCK-8 solution (cat no. C0038; Beyotime Institute of Biotechnology, Haimen, China) for 4 h at 37°C. The absorbance of each sample was measured at 495 nm using a spectrophotometer.
Cell cycle analysis
Flow cytometric analysis was performed to assess the distribution of cells in different phases of the cell cycle. Briefly, at 24 h post-transfection, a total of 10,000 cells were harvested, fixed in ice-cold 70% ethanol at −20°C overnight, treated with 0.2 mg/ml RNase A (Sigma-Aldrich; Merck KGaA) for 4 h at 4°C, and then stained with 10 µg/ml propidium iodide (PI; Sigma-Aldrich; Merck KGaA) for 30 min at room temperature in the dark. The percentages of cells in the G1, S and G2 phases were analyzed using a FACScan flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Flow cytometric data were analyzed using the ModiFit LT software version 3.0 (Verity Software House, Inc., Topsham, ME, USA). The assays were performed in triplicate.
Apoptosis analysis
The percentages of apoptotic cells following SF2/ASF (or si-ASF/SF2) and miR-30c cotransfection were determined according to the protocol of the fluorescein isothiocyanate (FITC)/Annexin V Apoptosis Detection kit I (BD Pharmingen; BD Biosciences). Briefly, at 24 h post-transfection, the PCa cells were harvested, processed according to the manufacturer's protocol, and then analyzed on the FACScan flow cytometer (BD Biosciences). Flow cytometric data were analyzed using the BD FACSuite™ software (BD Biosciences). Apoptotic cells were defined as Annexin V/FITC- and PI-positive cells.
Statistical analysis
All statistical analyses were performed using SPSS software version 17.0. (SPSS, Inc., Chicago, IL, USA). The statistical significance of the differences between groups was assessed using Student's t-test for pair-wise comparisons or one-way analysis of variance followed by a post hoc Tukey-Kramer test for multiple comparisons. The patients were stratified in two groups according to the median values of the relative levels for miR-30c and SF2/ASF. The correlation between ASF/SF2 and clinicopathologic variables was assessed using Fisher's exact test or Pearson's χ2 test. Kaplan-Meier survival curves were generated to evaluate the significance of the expression of miR-30c and ASF/SF2 on the prognosis of the patients with PCa. Cox proportional hazards regression was used to analyze the independent factors for BCR. P<0.05 was considered to indicate a statistically significant difference.
Results
ASF/SF2 is upregulated in PCa tissues
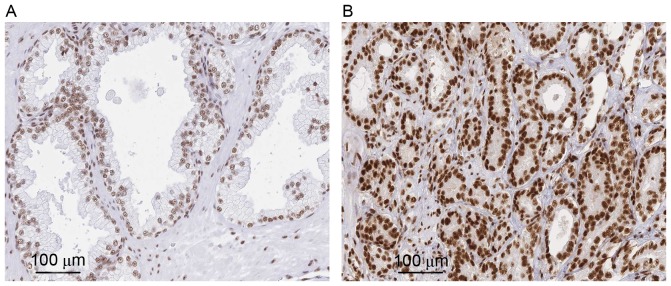
To evaluate the role of ASF/SF2 in PCa, the present study analyzed its expression in 20 tumor and normal tissues. In general, it was found the antibody staining was primarily in the nuclei of the PCa cells and exhibited an evenly distributed staining pattern. Compared with the PCa tissues, faint BCL9 staining was observed in certain sections of the normal tissues. Statistical analysis showed that the positive expression rate in normal tissue was significantly lower, compared with that in the tumor counterpart (P<0.001; Fig. 1A and B).
Figure 1.
ASF/SF2 is upregulated in human PCa tissues. An immunohistochemical assay was used to analyze the expression of ASF/SF2 in PCa and normal prostate tissues. Representative images of (A) normal and (B) tumor sections for the expression of ASF/SF2 are shown. Scale bar=100 µm. PCa, prostate cancer; ASF/SF2, of alternative splicing factor/splicing factor 2.
ASF/SF2 is a direct target of miR-30c in PCa cells
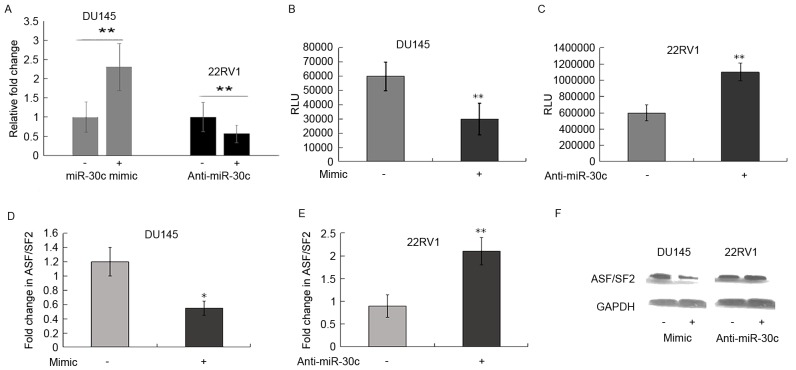
Using the TargetScan, PicTar and miRDB databases, the ASF/SF2 3′-UTR was found to contain one putative binding site for miR-30c. To further determine whether miR-30c interacts directly with the ASF/SF2 protein, a luciferase reporter assay was performed. The DU145 cells were transiently transfected cells with miR-30c mimic and its control, respectively. Following transfection, the increased expression level of miR-30c was confirmed using RT-qPCR analysis (P<0.01; Fig. 2A). However, decreased expression of miR-30c was observed following transfection with anti-miR-30c (P<0.01; Fig. 2A).
Figure 2.
ASF/SF2 is a direct target of miR-30c in PCa cells. (A) RT-qPCR verification of the induced ectopic expression of miR-30c in DU145 and 22RV1 cells following transfection with miR-30c or anti-miR-30c inhibitor. Luciferase assays were performed with the (B) miR-30c mimic or (C) anti-miR-30c inhibitor in PCa cells following cotransfection with pMIR-3′-UTR-ASF/SF2 and a β-gal expression construct. **P<0.01. DU145 and 22RV1 cells were transfected with (D) miR-30c (20 nM) or (E) anti-miR-30c inhibitor (50 and 100 nM) for 48 h *P<0.05, **P<0.01. RT-qPCR analysis was performed to detect the expression levels of ASF/SF2. (F) Cell lysates were used for western blot analysis to measure the levels of ASF/SF2 and GAPDH following miR-30c mimic or anti-miR-30c inhibitor transfection for 48 h. PCa, prostate cancer; ASF/SF2, of alternative splicing factor/splicing factor 2; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
To determine whether miR-30c binds to the ASF/SF2 3′-UTR, the DU145 cells were transfected with the pMIRreporter construct containing the ASF/SF2 3′-UTR together with miR-30c mimic, and a luciferase assay was performed. Luciferase activity was significantly downregulated in the pMIR-ASF/SF2 3′-UTR-transfected cells, compared with that in the scramble-transfected cells (P<0.01; Fig. 2B). By contrast, luciferase activity was significantly upregulated when 22Rv1 cells were transfected with the pMIR-ASF/SF2 3′-UTR construct together with anti-miR-30c inhibitor (P<0.01; Fig. 2C). These results indicated that miR-30c may modulate the expression of ASF/SF2by binding to its 3′-UTR.
To further substantiate these findings, the effects of miR-30c mimic and inhibitor on the mRNA and protein levels of ASF/SF2 were measured. The ectopic expression of miR-30c was associated with a reduction in the mRNA and protein expression of ASF/SF2; however, transfection of the 22Rv1 cells with anti-miR-30c inhibitor increased the expression of ASF/SF2 (Fig. 2D-F).
miR-30c inhibits cell proliferation and induces apoptosis by negatively regulating ASF/SF2
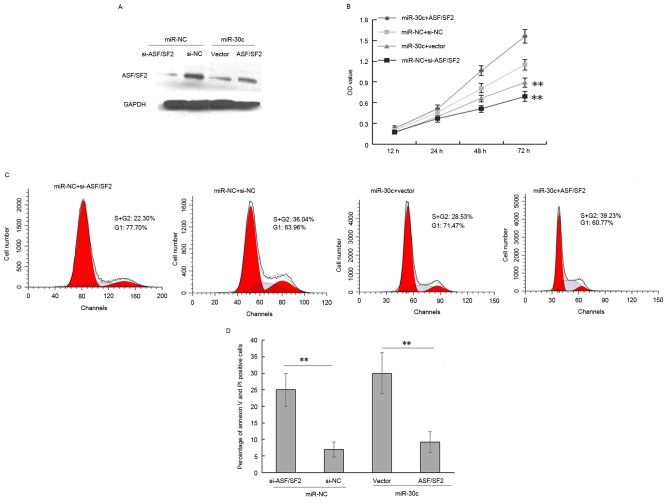
The data obtained in our previous study showed that the enforced expression of miR-30c inhibited tumorigenesis in PCa cells (13,14). Others have reported that the downregulation of SF2/ASF arrests cell proliferation and induces apoptosis in lung cancer and breast tumor (25,26). Therefore, the present study focused predominantly on whether miR-30c mediated ASF/SF2 has a similar effect on PCa cells. The expression of ASF/SF2 in DU145 cells was knocked down via siRNA (Fig. 3A). As shown in Fig. 3, the overexpression of ASF/SF2 eradicated the effects of miR-30c on proliferation of the DU145 cells (Fig. 3B; P<0.001). The cell cycle of the cells was analyzed using flow cytometry to determine whether the decrease in cell proliferation due to SF2/ASF-knockdown was associated with cell cycle arrest. The, cell cycle assay showed that ASF/SF2 promoted cell proliferation via an increase in the ratio of S+G2 phase cells in miR-30c-overexpressing cells (Fig. 3C; P<0.01). Therefore, the data indicated that miR-30c inhibited PCa cell proliferation and induced apoptosis, partially mediated by the targeting of ASF/SF2. Furthermore, the detection and quantification of apoptotic cells using flow cytometry showed a significant increase in the percentage of apoptotic cells in the miR-30c-overexpressing cells, whereas ASF/SF2 delayed the apoptotic effect induced by miR-30c (P<0.01; Fig. 3D).
Figure 3.
miR-30c inhibits cell proliferation and induces apoptosis by negatively regulating ASF/SF2. (A) Western blot analysis of the protein expression of ASF/SF2 in DU145 cells with or without miR-30c mimic transfection. GAPDH was used as an internal loading control. (B) Overexpression of ASF/SF2 eliminated the effects of miR-30c on the proliferation of DU145 cells. (C) Cell cycle analysis revealed that ASF/SF2 promoted cell proliferation by increasing the S+G2 phase ratio in miR-30c-overexpressing cells. (D) miR-30c-overexpressing cells had a significantly higher percentage of apoptotic cells, whereas ASF/SF2 delayed the apoptotic effect induced by miR-30c. **P<0.01. ASF/SF2, of alternative splicing factor/splicing factor 2; miR, microRNA; siRNA, small interfering RNA; si-NC, scramble siRNA negative control; si-ASF/SF2, siRNA for ASF/SF2; vector, blank vector negative control; ASF/SF2, ASF/SF2-overexpression vector; OD, optical density.
ASF/SF2 is associated with clinicopathological features
The present study examined the possible correlation between ASF/SF2 and clinicopathological features in the study cohort. Significantly increased expression levels of ASF/SF2 were observed in patients with PCa at an advanced pathological stage (P=0.038) and BCR (P=0.014), as showed in Table II. However, no significant associations were found between ASF/SF2 and other clinicopathological features, including preoperative PSA levels, Gleason score and surgical margin (all P>0.05).
Table II.
Expression levels of ASF/SF2 and clinicopathological features.
| Expression of ASF/SF2 | ||||
|---|---|---|---|---|
| Clinical feature | Low, n | High, n (%) | n (%) | P-value |
| Preoperative | 0.835 | |||
| PSA (ng/ml)a | ||||
| <10 | 85 | 41 (48.2) | 44 (51.8) | |
| ≥10 | 24 | 11 (45.8) | 13 (54.2) | |
| Gleason scorea | 0.450 | |||
| ≤6 | 32 | 16 (50.0) | 16 (50.0) | |
| 7 | 55 | 29 (52.7) | 26 (47.3) | |
| ≥8 | 17 | 6 (35.3) | 11 (64.7) | |
| Pathological stagea | 0.038 | |||
| pT2 | 69 | 35 (50.7) | 34 (49.3) | |
| pT3-pT4 | 37 | 11 (29.7) | 26 (54.1) | |
| Surgical margin statusa | 0.857 | |||
| Negative | 89 | 44 (49.4) | 45 (50.6) | |
| Positive | 17 | 8 (47.1) | 9 (52.9) | |
| Biochemical recurrencea | 0.014 | |||
| Negative | 80 | 45 (56.2) | 35 (43.8) | |
| Positive | 25 | 7 (28.0) | 18 (72.0) | |
Data missing from some patients. ASF/SF2, alternative splicing factor/splicing factor 2; PSA, prostate-specific antigen.
miR-30c and ASF/SF2 synergistically predict BCR
The data obtained in our previous study showed that miR-30c was correlated with the progression PCa and may be an independent predictor of BCR in PCa. As shown in Fig. 4A, the tumors with levels of ASF/SF2 above the median were significantly more likely to experience early recurrence (P=0.023; log-rank test; Fig. 4A). As ASF/SF2 was negatively regulated by miR-30c, the combined use of these two biomarkers merits further investigation. Of note, the group of PCa patients with low expression levels of miR-30c and high expression levels of ASF/SF2 had significantly lower rates of BCR-free survival compared with other groups of patients (P=0.017; log-rank test; Fig. 4B).
Figure 4.
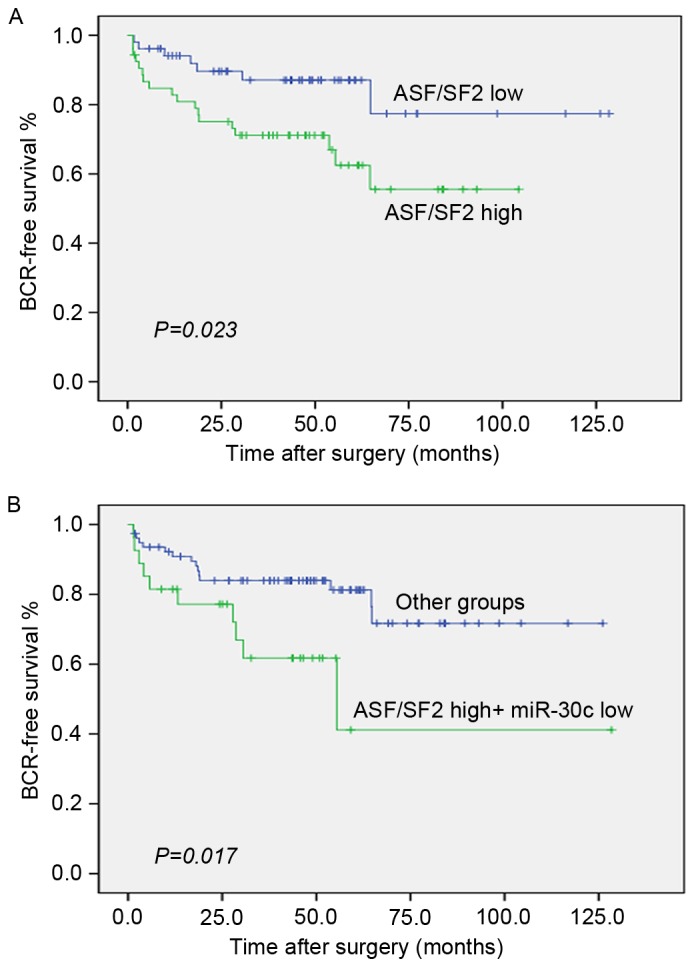
miR-30c and ASF/SF2 synergistically predict BCR. (A) Kaplan-Meier curves of BCR-free survival for patients with PCa according to the expression of ASF/SF. (B) Combined status of ASF/SF2 and miR-30c. Other groups comprised differential expression for miR-30c high and ASF/SF2 low; miR-30c high and ASF/SF2 high; and miR-30c low and ASF/SF2 low.
The Cox proportional hazards model was used to assess independent predictors of BCR-free survival, including preoperative PSA, Gleason score, surgical margin, pathological stage, and the coexpression of miR-30c and ASF/SF2. In the multivariate analysis, a low expression of miR-30c and high expression of ASF/SF2 (HR 2.89; P=0.024) were revealed to be independent prognostic factors for poor BCR-free survival (Table III).
Table III.
Univariate and multivariate analyses using Cox proportional hazard regression model for biochemical recurrence-free survival.
| Predictor | Hazard ratio (95% confidence interval) | P-value |
|---|---|---|
| Univariate | 0.022 | |
| miR-30c*ASF/SF2 status | 2.61 (1.15–5.90) | |
| Gleason score | 9.00 (4.30–18.84) | <0.001 |
| Preoperative PSA | 1.01 (1.00–1.01) | 0.010 |
| Pathological tumor stage | 5.83 (2.50–13.56) | <0.001 |
| Surgical margin status | 2.28 (0.98–5.30) | 0.056 |
| Multivariate | 0.024 | |
| miR-30c*ASF/SF2 status | 2.89 (1.15–7.22) | |
| Gleason score | 5.71 (2.46–13.29) | <0.001 |
| Preoperative PSA | 1.00 (1.00–1.03) | 0.001 |
| Pathological tumor stage | 2.84 (1.03–7.79) | 0.043 |
ASF/SF2, alternative splicing factor/splicing factor 2; miR, microRNA; PSA, PSA, prostate-specific antigen.
Discussion
Although miR-30c has been widely regarded as a tumor suppressor, its function in PCa remains to be elucidated as a result of relatively fewer investigations and known direct targets. Previous studies, including our own, have demonstrated that miR-30c is significantly reduced in tissue and plasma samples from patients with PCa, compared with their corresponding counterparts, and its expression predicts the risk of BCR and survival (13,27–29). Therefore, it is necessary to understand the role of miR-30c and its downstream effectors to determine its contribution to PCa tumorigenesis.
Verduci et al (30) demonstrated that two miRNAs (miR-28 and miR-505) affected mouse embryonic fibroblast senescence and apoptosis by modulating the expression of ASF/SF2. However, the possible functional link between ASF/SF2 and miR-30c remains to be elucidated. In the present study, it was observed that PCa tissues exhibited higher levels of ASF/SF2, compared with normal tissues. The results also showed that miR-30c targeted the 3′-UTR region of the ASF/SF2 gene, thereby decreasing the mRNA and protein levels of ASF/SF2. Of note, a negative correlation was observed between ASF/SF2 and miR-30c in PCa cells, wherein miR-30c was observed to decrease cell proliferation, and increase the percentage of cells in the G1 phase and apoptosis via the inhibition of ASF/SF2. The differences between the results of the present study and those of Verduci et al (30) indicated that the specific mechanisms underlying the induction of cell cycle arrest and apoptosis following a decrease in the expression of ASF/SF2 may be dependent on the cell type.
Detailed information regarding the mechanism of transformation modulated by ASF/SF2 has been already described, primarily through inducing alternative splicing isoforms of certain cancer-associated genes (23,31,32). For example, Liu et al (31) found that RNA splicing of the ASF/SF2 protein was critical in the splicing of AR pre-mRNA into AR splice variant 7 in PCa cells. Olshavsky et al (23) also identified ASF/SF2 as a disease relevant effector of CCND1 alternative splicing, which can promote the allele-specific production of cyclin D1b in PCa. In other tumors, it has been found that induction of the mammalian target of rapamycin (mTOR) pathway by ASF/SF2 is necessary for ASF/SF2-mediated transformation (25,26,33). It is known that the mTOR pathway is constitutively activated in several tumors, which affects the transformed phenotype (34). However, whether the upregulation of mTOR signaling via the induction of ASF/SF2 in PCa cells by miR-30c requires further investigation.
Although the functions of ASF/SF2 in the maintenance of tumor cells have been widely described, few reports mention its role as a novel prognostic factor. In lung cancer, high expression levels of ASF/SF2 tend to be associated with poor prognosis (25). In breast cancer, the genomic region (17q23) of ASF/SF2 is commonly amplified, and this amplification is associated with poor prognosis (35). In the present study, it was found that the upregulation of ASF/SF2 was associated with PCa progression. Furthermore, the expression of ASF/SF2 was significantly correlated with BCR in patients with PCa. The results of the univariate and multivariate analyses indicated that patients with high levels of ASF/SF2 and low levels of miR-30c showed earlier BCR following radical prostatectomy in patients with PCa. These results support a hypothesis that ASF/SF2 has potential as a biomarker for PCa, and therapeutic approaches targeting aberrant levels of ASF/SF2 may be investigated as a potential approach to improve clinical outcomes. The fact that the combined expression of ASF/SF2 and miR-30c was more informative, compared with that of AS/SF2 alone indicated that ASF/SF2 is modulated by other factors in addition to miR-30c, and that ASF/SF2 is an important, but not exclusive, mediator of the tumor-suppressive effects of miR-30c, as it has been described.
In conclusion, the results of the present study showed that the tumor suppressor miR-30c was involved in PCa tumorigenesis, possibly by targeting ASF/SF2. Of note, the combined analysis of the expression of ASF/SF2 and miR-30c may be a valuable tool for the early prediction of BCR in PCa following radical prostatectomy. Further investigations are required to validate the clinical utility of ASF/SF2 and miR-30c as prognostic biomarkers comprising large and multicentre PCa patient cohorts.
Acknowledgements
This study was supported by grants from the Science and Technology Project of Guangdong (grant no. 2014A020212035), the China Postdoctoral Science Foundation (grant no. 2016M590842) and the Medical Scientific Research Foun-dation of Guangdong Province, China (grant no. A2016052).
References
- 1.Kakehi Y. Watchful waiting as a treatment option for localized prostate cancer in the PSA era. Jpn J Clin Oncol. 2003;33:1–5. doi: 10.1093/jjco/hyg011. [DOI] [PubMed] [Google Scholar]
- 2.Santanam U, Banach-Petrosky W, Abate-Shen C, Shen MM, White E, DiPaola RS. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev. 2016;30:399–407. doi: 10.1101/gad.274134.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miftakhova R, Hedblom A, Semenas J, Robinson B, Simoulis A, Malm J, Rizvanov A, Heery DM, Mongan NP, Maitland NJ, et al. Cyclin A1 and P450 aromatase promote metastatic homing and growth of stem-like prostate cancer cells in the bone marrow. Cancer Res. 2016;76:2453–2464. doi: 10.1158/0008-5472.CAN-15-2340. [DOI] [PubMed] [Google Scholar]
- 4.Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu G, Ye CJ. Genetic and epigenetic heterogeneity in cancer: A genome-centric perspective. J Cell Physiol. 2009;220:538–547. doi: 10.1002/jcp.21799. [DOI] [PubMed] [Google Scholar]
- 5.Ulaganathan VK, Sperl B, Rapp UR, Ullrich A. Germline variant FGFR4 p.G388R exposes a membrane-proximal STAT3 binding site. Nature. 2015;528:570–574. doi: 10.1038/nature16449. [DOI] [PubMed] [Google Scholar]
- 6.Blattner M, Liu D, Robinson BD, Huang D, Poliakov A, Gao D, Nataraj S, Deonarine LD, Augello MA, Sailer V, et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell. 2017;31:436–451. doi: 10.1016/j.ccell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Q, Song R, Ortogero N, Zheng H, Evanoff R, Small CL, Griswold MD, Namekawa SH, Royo H, Turner JM, Yan W. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J Biol Chem. 2012;287:25173–25190. doi: 10.1074/jbc.M112.362053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 9.Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, Lee DH, Borin JF, Naslund MJ, Alexander RB, et al. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 2012;40:3689–3703. doi: 10.1093/nar/gkr1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rane JK, Scaravilli M, Ylipää A, Pellacani D, Mann VM, Simms MS, Nykter M, Collins AT, Visakorpi T, Maitland NJ. MicroRNA expression profile of primary prostate cancer stem cells as a source of biomarkers and therapeutic targets. Eur Urol. 2015;67:7–10. doi: 10.1016/j.eururo.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Alhasan AH, Scott AW, Wu JJ, Feng G, Meeks JJ, Thaxton CS, Mirkin CA. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc Natl Acad Sci USA. 2016;113:10655–10660. doi: 10.1073/pnas.1611596113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Wan X, Chen H, Yang S, Liu Y, Mo W, Meng D, Du W, Huang Y, Wu H, et al. Identification of miR-133b and RB1CC1 as independent predictors for biochemical recurrence and potential therapeutic targets for prostate cancer. Clin Cancer Res. 2014;20:2312–2325. doi: 10.1158/1078-0432.CCR-13-1588. [DOI] [PubMed] [Google Scholar]
- 13.Ling XH, Han ZD, Xia D, He HC, Jiang FN, Lin ZY, Fu X, Deng YH, Dai QS, Cai C, et al. MicroRNA-30c serves as an independent biochemical recurrence predictor and potential tumor suppressor for prostate cancer. Mol Biol Rep. 2014;41:2779–2788. doi: 10.1007/s11033-014-3132-7. [DOI] [PubMed] [Google Scholar]
- 14.Ling XH, Chen ZY, Luo HW, Liu ZZ, Liang YK, Chen GX, Jiang FN, Zhong WD. BCL9, a coactivator for Wnt/β-catenin transcription, is targeted by miR-30c and is associated with prostate cancer progression. Oncol Lett. 2016;11:2001–2008. doi: 10.3892/ol.2016.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- 16.Martinez I, Cazalla D, Almstead LL, Steitz JA, DiMaio D. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc Natl Acad Sci USA. 2011;108:522–527. doi: 10.1073/pnas.1017346108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao JJ, Lin J, Zhu D, Wang X, Brooks D, Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, et al. miR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res. 2014;74:1801–1813. doi: 10.1158/0008-5472.CAN-13-3311-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pino I, Pío R, Toledo G, Zabalegui N, Vicent S, Rey N, Lozano MD, Torre W, García-Foncillas J, Montuenga LM. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer. 2003;41:131–143. doi: 10.1016/S0169-5002(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 19.Zerbe LK, Pino I, Pio R, Cosper PF, Dwyer-Nield LD, Meyer AM, Port JD, Montuenga LM, Malkinson AM. Relative amounts of antagonistic splicing factors, hnRNP A1 and ASF/SF2, change during neoplastic lung growth: Implications for pre-mRNA processing. Mol Carcinog. 2004;41:187–196. doi: 10.1002/mc.20053. [DOI] [PubMed] [Google Scholar]
- 20.de Miguel FJ, Sharma RD, Pajares MJ, Montuenga LM, Rubio A, Pio R. Identification of alternative splicing events regulated by the oncogenic factor SRSF1 in lung cancer. Cancer Res. 2014;74:1105–1115. doi: 10.1158/0008-5472.CAN-13-1481. [DOI] [PubMed] [Google Scholar]
- 21.Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, et al. SRSF1-regulated alternative splicing in breast cancer. Mol Cell. 2015;60:105–117. doi: 10.1016/j.molcel.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olshavsky NA, Comstock CE, Schiewer MJ, Augello MA, Hyslop T, Sette C, Zhang J, Parysek LM, Knudsen KE. Identification of ASF/SF2 as a critical, allele-specific effector of the cyclin D1b oncogene. Cancer Res. 2010;70:3975–3984. doi: 10.1158/0008-5472.CAN-09-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Ezponda T, Pajares MJ, Agorreta J, Echeveste JI, López-Picazo JM, Torre W, Pio R, Montuenga LM. The oncoprotein SF2/ASF promotes non-small cell lung cancer survival by enhancing survivin expression. Clin Cancer Res. 2010;16:4113–4125. doi: 10.1158/1078-0432.CCR-10-0076. [DOI] [PubMed] [Google Scholar]
- 26.Anczuków O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy SK, Krainer AR. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Z, Zhang L, Yi X, Yu X. Diagnostic and prognostic values of tissue hsa-miR-30c and hsa-miR-203 in prostate carcinoma. Tumour Biol. 2016;37:4359–4365. doi: 10.1007/s13277-015-4262-9. [DOI] [PubMed] [Google Scholar]
- 28.Kachakova D, Mitkova A, Popov E, Popov I, Vlahova A, Dikov T, Christova S, Mitev V, Slavov C, Kaneva R. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015;34:189–200. doi: 10.1089/dna.2014.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren Q, Liang J, Wei J, Basturk O, Wang J, Daniels G, Gellert LL, Li Y, Shen Y, Osman I, et al. Epithelial and stromal expression of miRNAs during prostate cancer progression. Am J Transl Res. 2014;6:329–339. [PMC free article] [PubMed] [Google Scholar]
- 30.Verduci L, Simili M, Rizzo M, Mercatanti A, Evangelista M, Mariani L, Rainaldi G, Pitto L. microRNA (miRNA)-mediated interaction between leukemia/lymphoma-related factor (LRF) and alternative splicing factor/splicing factor 2 (ASF/SF2) affects mouse embryonic fibroblast senescence and apoptosis. J Biol Chem. 2010;285:39551–39563. doi: 10.1074/jbc.M110.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busà R, Geremia R, Sette C. Genotoxic stress causes the accumulation of the splicing regulator Sam68 in nuclear foci of transcriptionally active chromatin. Nucleic Acids Res. 2010;38:3005–3018. doi: 10.1093/nar/gkq004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karni R, Hippo Y, Lowe SW, Krainer AR. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proc Natl Acad Sci USA. 2008;105:15323–15327. doi: 10.1073/pnas.0801376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res Treat. 2003;78:313–322. doi: 10.1023/A:1023081624133. [DOI] [PubMed] [Google Scholar]