Abstract
p53, the tumor suppressor protein, functions as a dimer of dimers. However, how the tetramer binds to the DNA is still an open question. In the crystal structure, three copies of the p53 monomers (containing chains A, B, and C) were crystallized with the DNA-consensus element. Although the structure provides crucial data on the p53–DNA contacts, the active oligomeric state is unclear because the two dimeric (A–B and B–C) interfaces present in the crystal cannot both exist in the tetramer. Here, we address the question of which of these two dimeric interfaces may be more biologically relevant. We analyze the sequence and structural properties of the p53–p53 dimeric interfaces and carry out extensive molecular dynamics simulations of the crystal structures of the human and mouse p53 dimers. We find that the A–B interface residues are more conserved than those of the B–C. Molecular dynamics simulations show that the A–B interface can provide a stable DNA-binding motif in the dimeric state, unlike B–C. Our results indicate that the interface between chains A–B in the p53–DNA complex constitutes a better candidate for a stable biological interface, whereas the B–C interface is more likely to be due to crystal packing. Thus, they have significant implications toward our understanding of DNA binding by p53 as well as p53-mediated interactions with other proteins.
Keywords: p53 dimeric interface, p53 tetramer, hot spots, cancer, gene regulation
One of the most connected hubs in the cell is p53 (1). It regulates >160 genes (2), acting as a tumor suppressor and maintaining genome stability (2). Activated p53 either arrests the cell cycle to allow repair of damaged DNA or eliminates damaged cells through apoptosis (2). Because of its critical role, dysfunction of p53 disrupts basic cellular functions, particularly the DNA-damage response and tumor-predisposing stress (1). The structure of p53 contains several domains: the acidic, transactivation, SH3-binding, core-DNA-binding, tetramerization, and regulatory domains. DNA binding is critical for the biological functions of p53. Proper p53–DNA binding requires a well folded core-binding domain (CBD) and a p53 homotetramer.
The interdomain and intradomain interactions of p53 are critical for its association with other molecules (3–8). The tetramer domain provides a direct way for the p53 oligomerization, by forming a dimer of dimers (9). The tetramer domain is particularly important for the p53 binding of DNA loops (10, 11) or of supercoiled DNA (11). However, without the tetramer domain, the CBD binds DNA with a weaker affinity, also in a cooperative manner (12).
Structures of the individual isolated domains have been solved (13–17). The crystal structures of the CBD have been solved with (13) and without (14) DNA.
In the structure of the p53 CBD–DNA complex (Fig. 5, which is published as supporting information on the PNAS web site), the DNA mainly interacts with chain B. Chain C contacts both chain B and the DNA with a very similar orientation as chain B. Chain A is in contact with chain B; however, it has only limited interaction with the DNA. Thus, it has been believed that the interaction of chain A is caused by crystal packing (13). Interestingly, both protein–protein interfaces (A–B and B–C), which are involved in the CBD–DNA complex are also observed in crystal structures of the p53 core domain oligomers in the absence of DNA. The crystal structure of the mouse p53 core domain has a noncrystallographic trimer with three nearly identical dimer contacts. These contacts are similar to the chains A–B interface, with a 12° rotation (14). The crystal structure of a superstable quadruple mutant of the human p53 CBD was also solved in its DNA-free form. Interestingly, the crystal contact between the two core domains in the mutant structure is very similar to the chains B–C interface (18). Although a recurring pattern of the core domain interactions in the crystal may indicate a biological relevance, it is still possible that they reflect packing artifacts (12). To ascertain the interactions in solution, two NMR spectroscopy studies were directed at the CBD dimer interaction and their effect on DNA binding. The two studies differ in their explanation of the interactions. However, they both agree on the importance of the helix 1 region (V173–C182) (12, 19).
Although the available results provide crucial data relating to the p53–DNA contact, the information regarding the active biological oligomeric state is confusing. How the tetramer binds to the DNA is still an open question. Such information is crucial to understand the biological activity of p53. Here, we investigate the biological p53–p53 CBD dimeric interaction. We expect that knowledge of their mode of dimerization would serve as a key to figuring out the p53 oligomerization, CBD interactions with other domains, and ultimately p53–DNA interactions.
In this study, we analyzed the sequence and structural features of the protein–protein interfaces present in the p53–DNA structure (PDB ID code 1TSR) and performed extensive molecular dynamics simulations (≈50 ns in total) of the p53 dimers. Combined, these studies led us to conclude that chains A–B in the p53 trimer–DNA complex are more likely to constitute a stable biological interface than B–C.
Methods
The experimental B factors of the Cα atoms of each residue were averaged over the three chains of the p53 core domain–DNA complex (PDB ID code 1TSR). The residues were ranked according to their B-factor values and were divided equally into the following three categories: low-, medium-, and high-B-factor value regions. Residues in low B-factor regions were examined by their surface exposure. If a residue in the low B-factor region is surface-exposed, it is identified as potential binding site.
The surface area was calculated by using a rolling water ball with a radius of 1.4 Å. A surface residue was defined when its accessible surface area was >25% of the residue. Two residues are considered to be in contact across the interface if there is at least a pair of atoms, one from each residue, at a distance smaller than the sum of their van der Waals radii plus a threshold of 0.5 Å.
Molecular dynamics simulations were performed by using the charmm package (20) and the charmm 27 force field (21). Long-range electrostatic interactions were calculated with the particle mesh Ewald method (22). The systems were kept in constant pressure ensembles (NPTs) with the Hoover temperature control (23). The reference pressure was 1 atm (1 atm = 101.3 kPa), and the temperature was 300 K. The time step was 2 fs, with a shake constraint on all bonds with hydrogen atoms. Dimers were built by using the crystal structure conformations. Chloride atoms were added to make the overall system neutral. The overall systems contained 390 aa from the protein dimers and 11,457 water molecules.
The p53 dimer–DNA complexes were simulated by using the generalized Born method with molecular volume (GBMV) (24). To evaluate the energy as accurately as possible, no distance cutoff is used, and the grid-based GBMV module is used. In the GBMV calculation, the dielectric constant of water is set at 80, and the Debye–Huckel ionic term is 0.2 to reflect the salt effect.
Three Cys residues that are coordinated with Zn were deprotonated. The distances of the three Zn—S bonds and Zn—N between the Zn—His were fixed during the simulation. The charge and van der Waals parameters of the Zn and deprotonated Cys were taken from Maynard and Covell (25).
Results
Surface Analysis of the p53 CBD: Identification of Protein–Protein Interaction Hot Spots. The CBD has 193 residues. We picked 60 of them from the low B-factor region (see Methods). The first 24 residues are buried residues, with B factors ranging from 17.6 to 25.0. The remaining 36 residues are a mixture of surface exposed and other buried residues, with B factors ranging from 25.2 to 29.0. The 14 exposed residues are given in Table 1. Comparisons of these residues with known protein–protein interface contacts in the crystal structures show that nine residues appear in the binding sites. Five of these residues (His-178, Glu-171, Arg-174, Ala-138, and Leu-137) are at the A–B interface, two of these residues (His 233 and Lys-101) are in the B–C interface, and one of these residues (Asn-210) contributes to both the A–B and B–C interfaces. We analyzed the sequential conservation of these low-B-factor surface residues by using 10 divergent species. All highly conserved residues are located in the A–B interface (Table 1). Backbone hydrogen bonds across the interface have been shown to be important for protein–protein interactions (26). We identified five backbone hydrogen bonds between chains A and B (Fig. 6 and Table 2, which are published as supporting information on the PNAS web site). There are no backbone hydrogen bonds between chains B and C.
Table 1. Surface residues with low B factors, which are potentially important for binding.
| Residue | B factor | Exposed surface, % | Amino acid replacements in the homologous sequences† | Interface binding site | No. of known cancer mutations |
|---|---|---|---|---|---|
| His-178 | 25.2 | 59.9 | H (10) | A—B* | 12 (frameshift) |
| Glu-171 | 25.3 | 37.7 | D (3) E (7) | A*—B | |
| His-233 | 25.9 | 36.9 | H (5) L (5) | B—C* | 3 (His233Asp) |
| Tyr-107 | 26.5 | 25.7 | H (1) L (1) Y (8) | ||
| His-168 | 26.6 | 25.9 | D (1) H (7) F (1) Y (1) | DNA binding from A | |
| Asn-210 | 26.7 | 67.1 | N (5) Q (2) H (1) I (1) Y (1) | A*—B, B*—C | 10 (frameshift + Asn210Tyr) |
| Arg-174 | 27.6 | 46.1 | R (9) K (1) | A*—B | 6 (Arg174Ala) |
| Asn-131 | 28.1 | 37.1 | N (9) K (1) | ||
| Lys-101 | 28.5 | 78.9 | D (1) K (5) S (2) T (2) | B*—C (DNA—b—A) | |
| Ala-138 | 28.6 | 33.7 | A (10) | A—B* | 12 (Ala138Ser; Ala138Pro) |
| Leu-137 | 28.9 | 25.8 | L (10) | A—B* | |
| Trp-146 | 34.6 | 34.6 | R (2) L (1) W (5) V (2) | ||
| Phe-212 | 38.0 | 65.9 | G (1) L (2) K (2) F (5) | A*—B | 5 (frameshift) |
| Met-243 | 38.7 | 69.9 | M (10) |
Bold indicates absolute conservation.
Residue location on interface.
Amino acid replacements in the equivalent positions, obtained from the alignment of p53 sequences from the following sources: 1, Homo sapiens (human); 2, Platichthys flesus (European flounder); 3, Oryzias latipes (Medaka fish) (Japanese ricefish); 4, Xenopus laevis (African clawed frog); 5, Oncorhynchus mykiss (rainbow trout) (Salmo gairdneri); 6, Brachydanio rerio (zebrafish) (Danio rerio); 7, Mus musculus (mouse); 8, Cricetulus griseus (Chinese hamster); 9, Bos taurus (bovine); 10, Felis silvestris catus (cat).
The A–B interface has two strongly interacting regions (Fig. 6), which are also highly conserved. The helix 1 region in chain B interacts strongly with the Arg-175 region in chain A (Fig. 6, II, Arg cluster). B-chain Arg-181 forms strong hydrogen bonds with both its backbone carbonyl oxygen and side chain. The side chain of B-Arg-181 forms three hydrogen bonds with chain A residues (two with backbone carbonyl oxygen of Arg-175 and Gly-244, and one with Glu-180), whereas its backbone carbonyl oxygen forms hydrogen bond with Arg-174 in chain A. In another cluster (Fig. 6, III, Phe cluster), Chain A Val-172 and Chain B Cys-182 form backbone–backbone hydrogen bond, whereas the side chain of Thr-211 chain A forms a hydrogen bond with the backbone carbonyl oxygen of Ser-185 in chain B. In this region, there is also a potentially important Phe-211 in chain A, forming a hydrophobic patch with chain B. Phe is one of three interface hot spots (Trp, Phe, and Met) conserved only for interface binding (27).
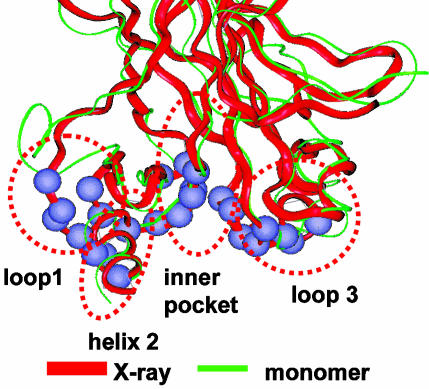
Conformational Changes of the p53 CBD Monomer. The conformational changes of the p53 CBD, especially those around the DNA-binding domain, will affect the p53–DNA interactions. We focused on the flexibility around the DNA-binding site, dividing it into four regions (Fig. 1). Region 1 is loop1, with six residues in contact with the DNA (G117, T118, A119, K120, S121, and V122); region 2 is helix 2 (binding residues: R280, D281, R283, and T284); region 3 has two loops in the inner pocket (Q136, L137, V272, R273, V274, C275, A276, and C277); and region 4 consists of loop3 (N239, S240, S241, M243, G244, N247, R248, and R249). Among these residues, the mutational hot spots R248 and R249 are in region 4, R273 is in the inner region 3, and R282 is within the helix region 2.
Fig. 1.
Definition of structure and dynamics of p53 CBD. The red ribbon represents the B chain in the crystal structure. The green ribbon is a snapshot from a monomer simulation.
The structure of the CBD is relatively stable in the 300-K simulation, with the overall rms deviation (rmsd) from the crystal structure reaching 3 Å. The core β-sheets do not change during the simulation. However, several loop regions are flexible, and they are responsible for the large rmsd of the monomer (data not shown).
The red ribbon in Fig. 1 represents the backbone crystal structure. The green ribbon represents the final snapshot from the molecular dynamics simulation of the CBD monomer. It shows that the loop1 is the most flexible part of the CBD. Interestingly, loop1 completely flipped away from the binding site, with an average deviation of 8–10 Å for the six residues binding DNA. Three other binding regions also fluctuate, with the helix 2 region fluctuating the most, from 2 to 4 Å. The results suggest that the binding site of the core domain monomer may need to be stabilized for DNA binding. Similar trends were observed in the exposed DNA binding sites in the dimer simulations.
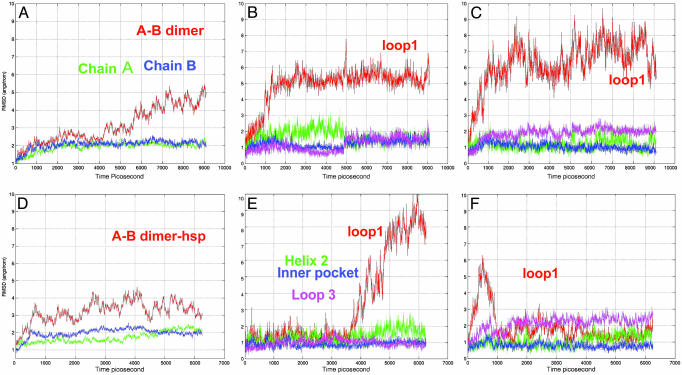
Conformational Changes of the p53 CBD A–B Chain Dimer. Conventional wisdom is that the A–B dimer derives from crystal packing (13). However, as indicated in our structural and sequential analysis, the A–B dimer interface has more binding hot spots than the B–C dimer. Molecular dynamics simulations reveal that the A–B dimer has a stable interface and a stable DNA-binding motif. The conformational changes for the overall structure and the binding motif are shown in Fig. 2. Fig. 2 A–C shows the A–B dimer in its neutral His form, and Fig. 2 D–F shows the dimer in the protonated His form (A–B dimer-hsp).
Fig. 2.
Molecular dynamics trajectories for the p53 A–B dimer with neutral and protonated His residues (A–B dimer-hsp). (A) The overall rmsd for the A–B dimer. (B) Binding-motif changes for chain A in the A–B dimer. (C) Binding-motif changes for chain B in the A–B dimer. (D) The overall rmsd for A–B dimer-hsp. (E) Binding-motif changes for chain A in the A–B dimer-hsp. (F) Binding-motif changes for chain B in the A–B dimer-hsp.
Compared with the isolated monomer, chains A and B have smaller rmsd values in the simulations, indicating a mutual stabilizing effect on the core domain. We used the overall rmsd for the whole dimer as a measure of the stability of the dimerization. As shown in Fig. 2 A, the fluctuation of the A–B dimer is <3 Å for ≈5 ns. Then, the rmsd gradually increases to 4–5 Å, and it fluctuates in this range. Compared with the monomeric state, the binding motif is stabilized by the A–B dimerization (Fig. 2 B and C). The loop1 is still flexible, although to a lesser extent than in the monomer. Other important binding sites, especially helix 1 and the inner pocket, are also much more stable than these in the monomeric state. Further, it is particularly interesting to note that there is an allosteric stabilization effect of the DNA-binding motif from the dimerization. In the A–B dimer, the DNA-binding motif in chain B is near the A–B interface, and thus, it is expected that the binding motif should be stabilized by the dimerization. However, the DNA-binding motif in chain A is exposed, and it is far away from the interface. As shown in Fig. 2B, the chain A DNA-binding motif is also allosterically stabilized.
If the A–B dimer is the biological free dimer in solution, the protonated protein should be closer to its final DNA-binding state because of the overall charged state of the DNA. His residues in the biologically active state should be protonated. We selectively protonated five His residues according to the suggestion of Wright et al. (28). The only two His residues left in the deprotonated state are His-179, which is coordinated to Zn, and His-214, which is close to R174. We term the A–B dimer with the protonated His residues A–B dimer-hsp. The dynamical behavior of A–B dimer-hsp is shown in Fig. 2 D–F.
Initially, the dynamics of the A–B dimer-hsp fluctuates more than that of the A–B dimer, with a slightly larger overall rmsd in the early stage of the simulation. However, it is greatly stabilized compared with the overall trajectory of the A–B dimer with neutral His residues. The most obvious effect of the protonation is the dynamics of the loop1 (Fig. 2F). Unlike its flexible nature in the neutral His form, loop1 is stabilized in the DNA-binding conformation in the protonated His form. Interestingly, the loop1 in chain B, which is close to the incoming DNA, fluctuates initially and then settles to its DNA-binding position and stays there for the rest of the simulation (Fig. 2F).
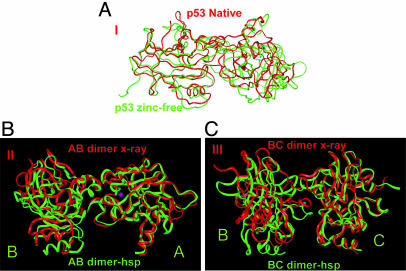
We also simulated the A–B dimers at the following different conditions (Table 3, which is published as supporting information on the PNAS web site). (i) A–B dimer-2: the A–B dimer from the crystal structure of the free mouse p53 CBD (without DNA, PDB: 1hu8). Here, the A–B dimer has a similar interface as in the DNA-coordinated p53 but with a twisted (12°) A–B orientation; (ii) A–B dimer-apo: Zn-free form of the A–B CBD dimer.
The conformation of the A–B dimer-2 (Fig. 7 A–C, which is published as supporting information on the PNAS web site) is much less stable than the A–B dimer (Fig. 2). The individual domains themselves have a similarly small rmsd compared with the A–B dimer. However, the overall rmsd for the A–B dimer-2 jumps by as much as 6 Å. The motion of the A–B dimer-2 does not relax to the conformation of the A–B dimer. The dimeric contact of the A–B dimer-2 is held by two strong interactions, as observed in the A–B dimer itself. One is the hydrophobic interaction of A-Phe-212 with the B chain loop; another is the cross-interface backbone hydrogen bonds of B-Arg-181 to A-Arg-175 and A-Glu-180 (Fig. 6). Even though the orientation of the two core domains fluctuates during the simulation, these two key interaction patches are very stable. Protonation of His does not stabilize the A–B dimer-2 hsp (data not shown).
The Zn-free A–B dimer-apo partially unfolds during the simulation (Fig. 3A). Even though the two core domains are initially stable, the dimer is not. The rmsd for the Zn-free A–B dimer-apo quickly reaches 4 Å at ≈2 ns and gradually increases to 6 Å. Chain B partially unfolds during the simulation, with the N-terminal residues (Ser-96–Gln-104) flipping away.
Fig. 3.
Structure changes after simulation. (A) Superimposition of partially unfolded p53 A–B dimer-apo with the conformation in the crystal structure. (B) Superimposition of p53 A–B dimer-hsp with the conformation in the crystal structure. (C) Superimposition of p53 B–C dimer-hsp with the conformation in the crystal structure.
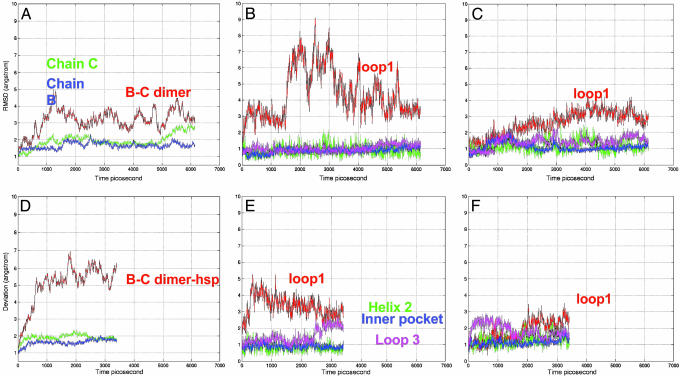
Conformational Changes of the p53 CBD B–C Chain Dimer. The B–C interface is another candidate for the p53 dimer–DNA interaction (13, 29). Chain B binds to the consensus-binding site. C is also in contact with the DNA, in a way that is similar to chain B. In our simulation, the B–C dimer initially has a very large fluctuation, with the rmsd of the dimer reaching 5 Å. Then, the two chains adjust their positions and reach a stable conformation that is close to the crystal structure, with the rmsd fluctuating at ≈3–4 Å (Fig. 4A). The individual domain rmsd values are similar to those in the A–B dimer, with chain C having a slightly higher rmsd. In the B–C dimer, loop1 of chain B is exposed. Loop1 of chain C is within the B–C interface. Thus, the dynamic behavior of loop1 in chains B and C is different, with the exposed loop1 in chain B fluctuating as usual, whereas loop1 in chain C is stabilized by the interface interaction. Still, the rmsd of the chain C loop1 is increasing. Then, the question is whether the His protonation can stabilize loop1 in the B–C dimer.
Fig. 4.
Molecular dynamics trajectories for the p53 B–C dimer with neutral and protonated His residues (B–C dimer-hsp). (A) Overall rmsd for the B–C dimer. (B) Binding-motif changes for chain B in the B–C dimer. (C) Binding-motif changes for chain C in the B–C dimer. (D) Overall rmsd for B–C dimer-hsp. (E) Binding-motif changes for chain B in the B–C dimer-hsp. (F) Binding-motif changes for chain C in the B–C dimer-hsp.
Simulations of the B–C dimer-hsp indicate that instead of stabilization of the B–C dimer, the His protonation greatly destabilizes the B–C dimer-hsp. Even though loop1 fluctuates a little less in the B–C dimer-hsp, the dimer itself is not stable. The rmsd quickly reaches 7 Å and fluctuates at ≈6 Å (Fig. 4 D–F). Thus, unlike the small structural variations observed in the simulations of the A–B dimer-hsp (Fig. 2B), the B–C dimer-hsp interface does not retain its stability (Fig. 4D).
Energetic Comparison of p53(A–B)–DNA Complex and p53(B–C)–DNA Complex. In our molecular dynamics simulations, the A–B dimer is more stable than the B–C dimer. By using the recently released dfire potentials (31), we calculated the binding energies. We found that the strength of the A–B interaction is almost twice (–10.4 kcal/mol) that of the B–C interaction (–5.5 kcal/mol). However, this protein–protein interaction might change upon DNA binding. To address the question of a possible change of the stability preference of the A–B versus the B–C dimer upon DNA binding, we simulated the p53(A–B)–DNA complex and p53(B–C)–DNA complex briefly with GBMV (24). The initial structures are taken from the p53–DNA complex by removing one copy of the monomer from the crystal structure [chain C for p53(A–B)–DNA complex, and chain A for p53(B–C)–DNA complex]. Ser-94 was removed from chain A to have the same number of amino acids as chain C. For each complex, 50 structures from the 50-ps molecular dynamics simulations were saved. The structures were minimized with 500 steps, and the total energies (including internal energy, van der Waals energy, coulombic interaction, and electrostatic-solvation energies) are evaluated with the grid-based GBMV. The average energy from the 50 p53(A–B)–DNA structures is –21,047 kcal/mol, and the average energy from the 50 p53(B–C)–DNA structures is –20,983 kcal/mol. Thus, the p53(A–B)–DNA complex is more stable than the p53(B–C)–DNA complex.
Discussion and Conclusions
The nature of the interactions between the p53 core domains is expected to be important for the mode of DNA binding, for modeling the molecular oligomer and for assembling the entire interaction network. Even though the exact conformation of the A–B dimer may change, this region is apparently important for the protein–protein interactions of the p53 core domain.
Surface Analysis of p53 CBD. Structural and sequential analysis of protein–protein interfaces revealed that structurally conserved residues correspond to energy hot spots, with packing playing a critical role. B factors are largely determined by local packing (30). Surface residues with low B-factor values correspond to highly packed regions. Here, we simply use the B factors of surface residues to locate potential protein–protein interaction hot spots. The surface analysis indicates that the A–B interface has more residues with low B factors than the B–C interface. A–B interface residues with low B factors are highly conserved. Their mutations cause cancer (Table 1). Thus, according to this analysis, the A–B interface is more important for CBD–CBD interactions.
A–B Dimer or B–C Dimer: Comparison with Experimental Results. Klein et al. (19) investigated the solution dimerization interface of the p53 CBD bound to the consensus DNA. Their results revealed the essential role of the short H1 helix (Pro-177–Cys-182). This helix contains mutational hot spots, particularly the three exposed residues Pro-177, His-178, and Arg-181. They also found that a change in the conformation of the helix 1 region may modify the p53 dimerization behavior and prevent cooperative DNA binding. Helix 1 forms part of the A–B interface. In our binding hot-spot analysis, we identified His-178, which has the smallest B factor among the surface residues. Rippin et al. (12) provided further insight into the intermolecular contacts of the p53 CBD by NMR analysis. They found that isolated CBDs dimerize at low salt concentration with a dissociation constant of 24 μM. Similar to the NMR results of Klein et al., Rippin et al. also identified helix 1 (V173–C182) as interface region. In addition, they also found that G244 and the loop1 region L114–T118 are also affected by the dimerization. Loop1 is within the B–C interface and may support a B–C interface. However, loop1 is also in the vicinity of the A–B interface. Also, A-G244 forms a backbone hydrogen bond with B-Arg-181, consistent with the observed NMR shifts.
In addition to the direct interface interaction, the chemical-shift difference observed experimentally could be the consequence of the DNA binding (12). The change in the helix 1 region may be translated from the DNA interface via the bound Zn. As to the loop1 region, because DNA binds the Arg at the tip of the loop, it is expected to change the loop dynamics. Further, DNA binding will protonate the His residue in the CBD, and we have demonstrated that the most significant effect of the His protonation is the stabilization of the loop1.
In either of these cases, combining the simulation results and available NMR studies, it is most likely that, of A–B and B–C, the functional form of the CBD dimer is A–B. The A–B dimer is stable in the neutral pH range and is highly stable in the protonated His form, which is crucial for DNA binding. In addition, only the A–B dimer can provide a critical loop1 conformation in a protonated environment. The B–C dimer might have some stability in the neutral pH form. However, it is not a functional (DNA binding) dimer. The B–C dimer interface most likely derives from crystal packing. Consistently, the energetic comparisons confirm that p53(A–B)–DNA complex is more stable than p53(B–C)–DNA complex.
The A–B Versus the B–C Dimer: DNA Binding, Dimerization of the CBD, and Tetramerization of p53. Our results suggest that chain A may lead to a stable DNA-binding motif in chain B, which binds DNA directly. Chain A has a few interactions with the DNA backbone. Thus, the A–B dimer still binds a half site with 10 base pairs. For the full-length p53, the N-terminal and, in particular, the C-terminal domains may lead to an altered tighter contact between chain A and the DNA, like the symmetrical form suggested in ref. 13.
A dimer of A–B dimers could represent a key feature of the p53 tetramer. The large conformational change of loop1 observed in our simulations provides insight into p53–DNA binding, which has been difficult to observe from the crystal structure alone. Mutations in the loop1 region enhance specific p53–DNA binding (32), probably because of the stabilization of loop1. The recently solved crystal structure of Cep-1, the human p53 Caenorhabditis elegans ortholog (33) also highlights the need to understand the function of the loop1. The conformation of loop1 in Cep-1 differs greatly from that of human p53. Huyen et al. (33) considered the possibility of DNA-binding-induced conformational change for loop1; however, they disfavored the conformational change and proposed a different DNA-binding orientation.
Based on the possible core-domain dimers, we propose p53–DNA binding mechanisms. Schemes 1 and 2 in Fig. 8 (which is published as supporting information on the PNAS web site) are based on the A–B dimer. In Scheme 1 (in Fig. 8), two A–B dimers are bound nonspecifically to the DNA at different sites, sliding along the DNA to form a tetramer at specific binding site. In Scheme 2, one dimer binds the DNA at a specific site, with lower affinity, and the second dimer joins to form a tetramer. In the less likely scenario in which B–C is a biological interface (Scheme 3 in Fig. 9, which is published as supporting information on the PNAS web site), the dimer binds the DNA occupying a half site. The second B–C dimer leads to a tetramer.
Protein sliding along the DNA chain is an efficient way to locate a specific binding site (34). Atomic-force microscopy has shown two modes of p53–DNA recognition, (i) direct binding, and (ii) an initial nonspecific binding with subsequent sliding to the specific site (35). A large protein–DNA interface interferes with the sliding (34). In this regard, again, an A–B dimer in which DNA-binding interactions center in chain B is better than a B–C dimer. Schemes 1 and 2 (in Fig. 8) are consistent with experimental observations for p53–DNA binding (36), which showed that, although one dimer within the tetramer is sufficient for binding to the DNA, concurrent interaction of the second dimer greatly enhances binding affinity.
The p53–p53 interface in the trimeric state without DNA is affected by crystal packing. Considering both A–B and B–C dimers, we propose possible mechanisms for the p53 core domain–DNA interaction. Overall, the dimer interactions between the p53 core domains and the resulting DNA interactions are better explained with the A–B dimer interactions as compared with the B–C. Last, the exposed binding site on chain A for the A–B dimer could provide a binding site for the DNA loop binding under suitable conditions. To elucidate the biological interface, the next step involves a comparison of the A–B interface with those in other models, like the symmetry one.
Supplementary Material
Acknowledgments
We thank Drs. O. Keskin, D. Zanuy, C.-J. Tsai, and H.-H. Tsai for discussion. The central processing unit times were provided by the National Cancer Institute Advanced Biomedical Computing Center (Frederick, MD) and the National Institutes of Health Biowulf. This work was supported by the National Cancer Institute under Contract No. NO1-CO-12400.
Author contributions: B.M., B.V., A.J.L., and R.N. designed research; B.M., Y.P., and K.G. performed research; B.M. analyzed data; and B.M. and R.N. wrote the paper.
Abbreviations: rmsd, rms deviation; CBD, core-binding domain; GBMV, generalized Born method with molecular volume.
References
- 1.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 2.Zhao, R., Gish, K., Murphy, M., Yin, Y., Notterman, D., Hoffman, W. H., Tom, E., Mack, D. H. & Levine, A. J. (2000) Cold Spring Harbor Symp. Quant. Biol. 65, 475–482. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, J. & Prives, C. (2001) Nat. Struct. Biol. 8, 730–732. [DOI] [PubMed] [Google Scholar]
- 4.Kussie, P. H., Gorina, S., Marechal, V., Elenbaas, B., Moreau, J., Levine, A. J. & Pavletich, N. P. (1996) Science 274, 948–953. [DOI] [PubMed] [Google Scholar]
- 5.Liu, W. L., Midgley, C., Stephen, C., Saville, M. & Lane, D. P. (2001) J. Mol. Biol. 313, 711–731. [DOI] [PubMed] [Google Scholar]
- 6.Bayle, J. H., Elenbaas, B. & Levine, A. J. (1995) Proc. Natl. Acad. Sci. USA 92, 5729–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller-Tiemann, B. F., Halazonetis, T. D. & Elting, J. J. (1998) Proc. Natl. Acad. Sci. USA 95, 6079–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, A. L., Raffo, A. J., Brandt-Rauf, P. W., Pincus, M. R., Monaco, R., Abarzua, P. & Fine, R. L. (1999) J. Biol. Chem. 274, 34924–34931. [DOI] [PubMed] [Google Scholar]
- 9.Jeffrey, P. D., Gorina, S. & Pavletich, N. P. (1995) Science 267, 1498–1502. [DOI] [PubMed] [Google Scholar]
- 10.Stenger, J. E., Tegtmeyer, P., Mayr, G. A., Reed, M., Wang, Y., Wang, P., Hough, P. V. & Mastrangelo, I. A. (1994) EMBO J. 13, 6011–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brazdova, M., Palecek, J., Cherny, D. I., Billova, S., Fojta, M., Pecinka, P., Vojtesek, B., Jovin, T. M. & Palecek, E. (2002) Nucleic Acids Res. 30, 4966–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rippin, T. M., Freund, S. M., Veprintsev, D. B. & Fersht, A. R. (2002) J. Mol. Biol. 319, 351–358. [DOI] [PubMed] [Google Scholar]
- 13.Cho, Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265, 346–355. [DOI] [PubMed] [Google Scholar]
- 14.Zhao, K., Chai, X., Johnston, K., Clements, A. & Marmorstein, R. (2001) J. Biol. Chem. 276, 12120–12127. [DOI] [PubMed] [Google Scholar]
- 15.Clore, G. M., Omichinski, J. G., Sakaguchi, K., Zambrano, N., Sakamoto, H., Appella, E. & Gronenborn, A. M. (1994) Science 265, 386–391. [DOI] [PubMed] [Google Scholar]
- 16.Clore, G. M., Ernst, J., Clubb, R., Omichinski, J. G., Kennedy, W. M., Sakaguchi, K., Appella, E. & Gronenborn, A. M. (1995) Nat. Struct. Biol. 2, 321–333. [DOI] [PubMed] [Google Scholar]
- 17.Rustandi, R. R., Baldisseri, D. M. & Weber, D. J. (2000) Nat. Struct. Biol. 7, 570–574. [DOI] [PubMed] [Google Scholar]
- 18.Joerger, A. C., Allen, M. D. & Fersht, A. R. (2004) J. Biol. Chem. 279, 1291–1296. [DOI] [PubMed] [Google Scholar]
- 19.Klein, C., Planker, E., Diercks, T., Kessler, H., Kunkele, K. P., Lang, K., Hansen, S. & Schwaiger, M. (2001) J. Biol. Chem. 276, 49020–49027. [DOI] [PubMed] [Google Scholar]
- 20.Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S. & Karplus, M. (1983) J. Comput. Chem. 4, 187–217. [Google Scholar]
- 21.MacKerell, A. D., Bashford, D., Bellott, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., et al. (1998) J. Phys. Chem. B 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- 22.Darden, T., York, D. & Pedersen, L. (1993) J. Chem. Phys. 98, 10089–10092. [Google Scholar]
- 23.Hoover, W. G. (1985) Phys. Rev. A 31, 1695–1697. [DOI] [PubMed] [Google Scholar]
- 24.Lee, M. S., Feig, M., Salsbury, F. R., Jr. & Brooks, C. L., III, (2003) J. Comput. Chem. 24, 1348–1356. [DOI] [PubMed] [Google Scholar]
- 25.Maynard, A. T. & Covell, D. G. (2001) J. Am. Chem. Soc. 123, 1047–1058. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez, A. & Scheraga, H. A. (2003) Proc. Natl. Acad. Sci. USA 100, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, B., Elkayam, T., Wolfson, H. & Nussinov, R. (2003) Proc. Natl. Acad. Sci. USA 100, 5772–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright, J. D., Noskov, S. Y. & Lim, C. (2002) Nucleic Acids Res. 30, 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagaich, A. K., Zhurkin, V. B., Durell, S. R., Jernigan, R. L., Appella, E. & Harrington, R. E. (1999) Proc. Natl. Acad. Sci. USA 96, 1875–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halle, B. (2002) Proc. Natl. Acad. Sci. USA 99, 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, C., Liu, S., Zhou, H. & Zhou, Y. (2004) Protein Sci. 13, 400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inga, A., Monti, P., Fronza, G., Darden, T. & Resnick, M. A. (2001) Oncogene 20, 501–513. [DOI] [PubMed] [Google Scholar]
- 33.Huyen, Y., Jeffrey, P. D., Derry, W. B., Rothman, J. H., Pavletich, N. P., Stavridi, E. S. & Halazonetis, T. D. (2004) Structure (Cambridge) 12, 1237–1243. [DOI] [PubMed] [Google Scholar]
- 34.Breyer, W. A. & Matthews, B. W. (2001) Protein Sci. 10, 1699–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao, Y., Cherny, D. I., Heim, G., Jovin, T. M. & Schaffer, T. E. (2001) J. Mol. Biol. 314, 233–243. [DOI] [PubMed] [Google Scholar]
- 36.McLure, K. G. & Lee, P. W. (1998) EMBO J. 17, 3342–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.