Abstract
We present an algorithm to generate complete evolutionary profiles that represent the topology of the molecular phylogenetic tree of the homologous group. The method, based on the multidimensional QR factorization of numerically encoded multiple sequence alignments, removes redundancy from the alignments and orders the protein sequences by increasing linear dependence, resulting in the identification of a minimal basis set of sequences that spans the evolutionary space of the homologous group of proteins. We observe a general trend that these smaller, more evolutionarily balanced profiles have comparable and, in many cases, better performance in database searches than conventional profiles containing hundreds of sequences, constructed in an iterative and computationally intensive procedure. For more diverse families or superfamilies, with sequence identity <30%, structural alignments, based purely on the geometry of the protein structures, provide better alignments than pure sequence-based methods. Merging the structure and sequence information allows the construction of accurate profiles for distantly related groups. These structure-based profiles outperformed other sequence-based methods for finding distant homologs and were used to identify a putative class II cysteinyl-tRNA synthetase (CysRS) in several archaea that eluded previous annotation studies. Phylogenetic analysis showed the putative class II CysRSs to be a monophyletic group and homology modeling revealed a constellation of active site residues similar to that in the known class I CysRS.
Keywords: archaeal cysteinyl-tRNA synthetase, gene annotation, lipocalin superfamily, triosephosphate isomerase superfamily
Bioinformatics has developed as a data-driven science with a primary focus on storing and accessing the vast and exponentially growing amount of sequence and structure data. The rapid accumulation of data has led to an extraordinary problem of redundancy, which must be confronted in almost any type of statistical analysis. Attwood and Miller (1) observe that the non-redundant database (NRDB) of the National Center for Biotechnology Information “is not non-redundant, but non-identical, and is thus massively redundant.” Similarly, the current version of Swiss-Prot, a well curated sequence database and valuable research tool, is highly skewed toward the Bacteria and Eucarya (2).
An important goal of bioinformatics is to use the vast and heterogeneous biological data to extract patterns and make discoveries that bring to light the “unifying” principles in biology. Because these patterns can be obscured by bias in the data, we approach the problem of redundancy by appealing to a well known unifying principle in biology, evolution. Modern protein sequences and their three-dimensional structures are descendants of successful realizations of the evolutionary process. The entries in the sequence and structure databases are not merely an unconnected and seemingly endless array of biological novelty; rather, they can be clustered and treated as a smaller set of homologous groups. Hierarchical classifications of structures, such as SCOP (Structural Classification of Proteins) (3) and CATH (Class, Architecture, Topology, and Homologous superfamily) (4), and of sequences, such as Pfam (Protein Families Database of Alignments and Hidden Markov Models) (5), have made significant contributions in this direction, yet the problem of redundancy has not been addressed in an evolutionary context.
Here we present an algorithm based on the multidimensional QR factorization, which produces minimally redundant sets of protein sequences. This algorithm differs from traditional sequence identity threshold and sequence weighting approaches to the problem of redundancy, which we have recently reviewed in ref. 6, in two important ways. First, the QR algorithm has been designed to systematically choose a maximally linearly independent subset of sequences that best span the evolutionary space of the homologous group at any given level of diversity. In contrast, sequence identity cutoff algorithms arbitrarily remove sequences that contribute to pairwise identities above the given threshold, and sequence weighting schemes assign ad hoc weights to the sequences, giving more common sequences relatively less weight than rare ones. Second, the QR algorithm produces an ordering of the sequences in such a way that altering the desired level of diversity of the reduced set only requires adding or subtracting sequences from the precomputed order rather than launching a new calculation each time a different diversity threshold is applied.
Having introduced a structure-based analog of this procedure in which the QR factorization is computed over the cartesian space of the protein structures (6), here we detail the sequence-based algorithm and test its efficacy in forming evolutionarily well balanced profiles, termed evolutionary profiles (EPs), for homology searches over large sequence and genomic databases. In the case of distantly related homologous groups, we show that, by supplementing structure-based alignments with the appropriate sequences, single EPs can be built for diverse protein families or superfamilies (see Supporting Text, which is published as supporting information on the PNAS web site) and that these profiles perform as well in a single database search as the combined results from several database searches with profiles of the component subfamilies or families. Finally, we describe an application of this technology, in combination with homology modeling and phylogenetic analysis, to assign the putative function of a previously misannotated group of archaeal class II cysteinyl-tRNA synthetases (CysRSs) (7).
Theory and Methods
As the basis for the EPs, sequences and structures were selected and multiple alignments were generated by following the procedures outlined in Supporting Text. The QR factorization of an alignment matrix, a numerical encoding of a multiple sequence alignment, produces an ordering of the aligned proteins. The ordering can then be used to define a minimal basis set of spanning sequences to any desired level of redundancy, and the evolutionarily well balanced profiles computed from these minimal sets are termed EPs.
QR Factorization. The multidimensional QR factorization with pivoting algorithm (8), as applied to multiple structure alignments was presented in ref. 6. Here we provide only the salient points of the algorithm and its adaptation to multiple sequence alignments. Because the sequence databases are biased, any multiple sequence alignment encoded in matrix A will contain redundant information. The goal is to find a reduced set of sequences that well represent the major evolutionary changes in the alignment data. The problem is similar to a least-squares problem, Ax = b, of an overdetermined system, A. Although the problem of redundancy in multiple alignments cannot be fit into any single least-squares-like problem, redundancy in multiple alignments can be treated with the same methods used to remove redundancy in the least-squares problem. The QR factorization uses a combination of Householder transformations (9) and column pivoting (10) to establish an ordering of the columns (protein sequences) of A by increasing linear dependence (see Fig. 5, which is published as supporting information on the PNAS web site). The QR algorithm can be applied to an alignment matrix, A, if an information-preserving numerical encoding of the multiple sequence alignment is determined.
The sequence alignment data are encoded in the alignment matrix, A, which is of dimension m × n × d. m is the total length of the multiple alignment, and n is the number of proteins in the alignment. Each “column” in A is a (protein) matrix of dimension m × d that corresponds to a single protein sequence. The description dimension, d = 24, is used to encode the amino acids and gaps in the alignment. The first 23 components correspond to the 20 amino acids and three ambiguous amino acids (B, X, and Z) and the 24th component corresponds to the gapped positions. For example, the presence of an Ala at a particular position in the alignment, e.g., in the ith alignment position in the jth protein sequence, is encoded by aijk=1,24 = (1, 0, 0, 0,..., 0); a Cys at the same position would be encoded by aijk=1,24 = (0,0,1,0,...,0);a gapped position would be encoded by ãijk=1,24 = g × (0,0,0,0,...,1).Although it is logical to give gaps weight equal to the amino acids, i.e., g = 1, we have tested the performance by varying the gap scale parameter γ:
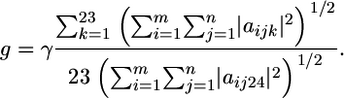 |
The range of allowed values is given in Fig. 6, which is published as supporting information on the PNAS web site.
The multidimensional QR factorization, for a matrix of dimension m × n × d, is the simultaneous QR factorization of d matrices each of size m × n. The algorithm is formally expressed as 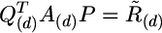 in which the matrix A encodes the multiple sequence alignment. The pivoting step, encoded by the permutation matrix, P, is applied to ensure that the transformation occurs such that the most linearly independent protein columns are segregated from the linearly dependent protein columns. The permutation matrix, thus, rearranges the column matrices of A such that the redundant column matrices (protein sequences) are moved to the right-hand side of the matrix. The permutation matrix, P, is independent of the d-dimension, so the amino acid components are not scrambled during pivoting operations. The choice of the first protein in the representative set is somewhat arbitrary, so we chose the protein with the smallest average percent identity with the whole set of proteins. In general, the quality of a profile appears to depend on the total composition of the profile, i.e., whether the profile represents the major evolutionarily distinct groups, and not on the specific choice of the first protein. At the kth step in the factorization before the application of the kth Householder transformation, the permutation P(k) is constructed to exchange the kth column of A over each d-dimension simultaneously, with the column of maximum Frobenius-like matrix p-norm, maxj=k,..., n(||aj||Fp), where
in which the matrix A encodes the multiple sequence alignment. The pivoting step, encoded by the permutation matrix, P, is applied to ensure that the transformation occurs such that the most linearly independent protein columns are segregated from the linearly dependent protein columns. The permutation matrix, thus, rearranges the column matrices of A such that the redundant column matrices (protein sequences) are moved to the right-hand side of the matrix. The permutation matrix, P, is independent of the d-dimension, so the amino acid components are not scrambled during pivoting operations. The choice of the first protein in the representative set is somewhat arbitrary, so we chose the protein with the smallest average percent identity with the whole set of proteins. In general, the quality of a profile appears to depend on the total composition of the profile, i.e., whether the profile represents the major evolutionarily distinct groups, and not on the specific choice of the first protein. At the kth step in the factorization before the application of the kth Householder transformation, the permutation P(k) is constructed to exchange the kth column of A over each d-dimension simultaneously, with the column of maximum Frobenius-like matrix p-norm, maxj=k,..., n(||aj||Fp), where
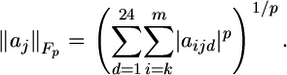 |
The pivoting step ensures that the kth protein is chosen based on its linear independence to the basis set formed by the first (k – 1) proteins. The value of P = 2 is determined numerically in Supporting Text. In addition, certain sequences, e.g., sequences with known structure, can be constrained to be a part of the representative set. These proteins are taken to be the first l members of the alignment, and no pivoting is performed for the first l steps of the QR factorization, which ensures that the structure-based alignment information is retained in the final profile.
In summary, the QR factorization produces an ordering of the sequences from the original multiple alignment. The number of proteins retained in the representative set is easily determined by applying a sequence identity threshold or some other pairwise similarity threshold that indicates the retention of the first k proteins in the QR order such that protein (k + 1)th has an above-threshold pairwise similarity relationship with one of the first k proteins. The threshold selection can be aided by visualizing the alignment data with a phylogenetic tree (see Supporting Text).
EPs. The EPs tested in Results and Discussion are based on alignments of distantly related homologous groups at the SCOP family and superfamily (see Supporting Text) levels. Because sequence-based alignments are not always reliable in this regime of diversity, structure-based alignments of representative structures (except for the HisA–HisF family) were used as seeds to which supplemental sequences were added to completely represent the major evolutionary transitions of the homologous group up to a defined set of sequence identity thresholds. The structure-based, sequence-supplemented multiple alignments are the basis for the complete EPs. Unless otherwise noted, the structure-based QR factorization (6) was applied to the multiple structure alignment of all known protein structures for a particular homologous group with an upper limit threshold equivalent to the upper limit threshold applied for sequences. A series of threshold values were applied to produce different profiles for each homologous group as noted in Results and Discussion. Because proteins with pairwise sequence identity at <30% cannot be aligned well by using sequence-based methods, the sequence QR was applied to closely related subgroups with a lower limit of 30% sequence identity. Complete EPs for the distantly related groups discussed below were simply amalgamations of the representatives from the more closely related subgroups, as detailed in Supporting Text.
Results and Discussion
Profiles of the HisA–HisF Family. The HisA and HisF proteins form a family of enzymes involved in the fourth and sixth steps of His biosynthesis (11). These proteins belong to the triosephosphate isomerase (TIM) barrel fold, which consists of a (β/α)8-barrel with parallel β-strands. The TIM barrel is thought to be one of the most abundant folds in the cell, representing 8% of the yeast transcriptome and the most common fold therein (12). According to the SCOP database (3), the HisA–HisF family is a member of the ribulose-phosphate binding (RPB) barrel superfamily, which encompasses three additional enzyme families. All members of the superfamily bind a ribulose phosphate-like ligand at the C terminus of the barrel. These proteins are classified in the same superfamily in SCOP because their common function and clear structure similarity indicate a common evolutionary origin (3).
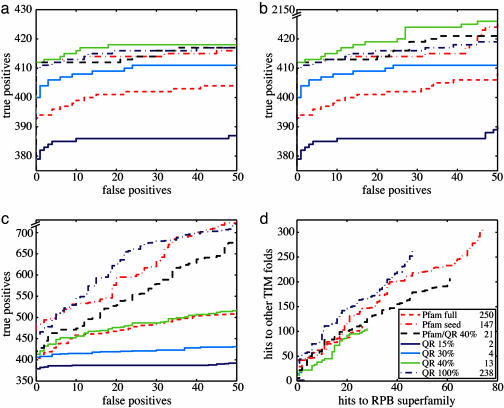
By using the QR factorization and the phylogenetic tree in Fig. 7, which is published as supporting information on the PNAS web site, we constructed EPs of the HisA–HisF family at various sequence identity thresholds from a multiple alignment of all of the HisA and HisF proteins in Swiss-Prot. The sensitivity of these profiles is measured by their ability to detect all members within the homologous group (true positives) in a database search. The results are shown as a receiver operating characteristic (ROC)50 plot in Fig. 1 in which the sensitivity is plotted against the specificity of a profile, here measured by the occurrence of the first 50 proteins outside the homologous group (false positives). The performance and reliability of a profile in such database searches is proportional to the area under each curve in the receiver operating characteristic plots. The database searches were performed with blast over the nonredundant database (NRDB) of the National Center for Biotechnology Information. Results for the database search using hmmer (see Fig. 8, which is published as supporting information on the PNAS web site) are strikingly similar to the blast results. Although hmmer requires an ≈100-fold increase in computation time with respect to blast, hmmer does outperform blast in searches with profiles of the more distantly related class II aminoacyl-RS (AARS) family discussed below or with profiles of the lipocalin superfamily presented in Fig. 9, which is published as supporting information on the PNAS web site (for a depiction of structural conservation, see Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 1.
Comparison of HisA–HisF family homology recognition. The key indicates the seven different profiles tested. Here and throughout, the number of sequences used to build each profile is listed to the right of the profile name in the key. (a–c) The definition of true positives is increasingly relaxed from the family level (a) to the superfamily level (b) and finally to the fold level (c). In family and superfamily recognition, the QR 40% EP, composed of just 13 sequences, outperforms all other profiles. In fold recognition, however, it is apparent that profiles containing the most sequences (QR 100%, Pfam seed, and Pfam full) performed best. (d) The surprising abundance of hits to TIM folds outside the RPB superfamily as compared with a relatively small number of hits to other RPB superfamily members.
In Fig. 1a, only HisA–HisF family members are counted as true positives, whereas in Fig. 1b, which tests superfamily level recognition, all members of the RPB superfamily are counted as true positives. The EPs are compared to the results from the widely used Pfam profiles. Although in the family and superfamily recognition tests the EP constructed from two sequences with <15% sequence identity does not perform as well as the much larger seed and full Pfam profiles, the QR 40% profile based on only 13 sequences clearly performs better. This result is expected because the topology of the HisA–HisF phylogenetic tree is not adequately represented by the first two or four sequences in the QR ordering (see Fig. 7). In general, if the composition of the alignment reflects the evolutionary history of the homologous group, a profile made from such an alignment, with only a small number of sequences, can detect all of the proteins that belong to that group.
To estimate the amount of redundancy in the Pfam seed profile, we applied the QR factorization to the Pfam seed alignment with a 40% sequence identity threshold, giving a profile of 21 sequences referred to as Pfam/QR 40%. Interestingly, this profile performs comparably to the original Pfam seed profile, proving that the Pfam seed profile with 147 sequences is redundant with seven times more sequence information than required. Differences between the EP, QR 40%, and the Pfam seed profile are due to the presence of Swiss-Prot and TrEMBL sequences in the Pfam seed alignment. The Pfam seed profiles are constructed from sequences known to belong to the HisA–HisF family from the Swiss-Prot and TrEMBL databases, and then iterative database searches are performed until no additional family members are detected. The QR factorization, therefore, provides an efficient alternative to Pfam's computationally intensive profile construction procedure. A comparison of the performance of the HisA–HisF profiles on superfamily and fold recognition is also shown in Fig. 1 b and c. It was expected that once all of the proteins within the family were recognized, the profiles would hit sequences from other related families within the same RPB superfamily. We observed instead that all of the profiles found a larger number of hits to other TIM barrels, not within the RPB superfamily. In the fold recognition plot (Fig. 1c) the profiles with a larger number of sequences, including (in order of search accuracy) the QR 100% profile and Pfam seed and full profiles, exhibit better performance. Because it was surprising to find so few hits to RPB superfamily members (see Fig. 1d), we examined the structural alignment of a representative set of the TIM barrels found by the profile search.
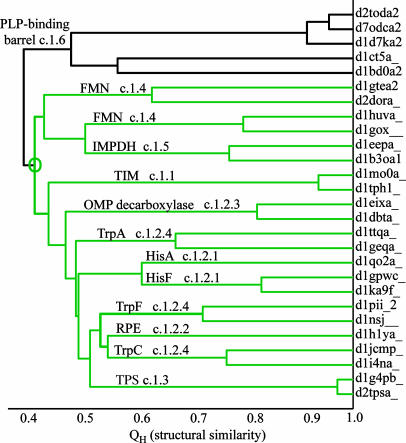
A careful investigation of evolutionary relatedness of the TIM barrel hits in this search reveals that the SCOP hierarchy does not reflect the correct evolutionary history of these distant relatives to the HisA–HisF family. The hits to TIM barrels outside the RPB superfamily are found to be members of four different TIM fold superfamilies; namely, TIMs, thiamin phosphate synthases, flavin mononucleotide-linked oxidoreductases, and the inosine monophosphate dehydrogenase superfamily. Representative structures of all five superfamilies were structurally overlapped with an “outgroup” superfamily, representatives of the pyridoxal-5′ phosphate-binding barrel superfamily, none of which were found by the HisA–HisF profile searches. The structure-based phylogenetic tree for these six superfamilies is shown in Fig. 2. Our recently developed structure-based phylogenetic methods are described in (6, 13). The five superfamilies found in the database search are evolutionarily related as a monophyletic group with respect to the pyridoxal-5′ phosphate-binding barrel superfamily. The members of the Trp biosynthesis family (c.1.2.4), however, display a polyphyletic distribution with respect to the SCOP classification, and, despite the fact that the proteins of this family are part of a common metabolic pathway, TrpC and TrpF are more closely related to d-ribulose-5-phosphate 3-epimerase (SCOP family code c.1.2.2) and even the thiamin phosphate synthases superfamily (SCOP superfamily code c.1.3) than either are to TrpA (SCOP family code c.1.2.4). The structure-based phylogeny along with the database search results reveal common structure and sequence features of the SCOP superfamilies (codes c.1.1, c.1.2, c.1.3, c.1.4, and c.1.5) and suggest their agglomeration into a single superfamily. A similar result was obtained by using a sequence-based psi-blast search with seed sequences from the flavin mononucleotide-linked oxidoreductases (14).
Fig. 2.
A structure-based phylogeny of the TIM barrel proteins found in the HisA–HisF database searches with the pyridoxal-5′ phosphate (PLP)-binding barrel superfamily representatives used as an outgroup. In the upgma tree plots, structural similarity is measured by QH, and the branches are labeled by SCOP superfamily or family names and codes, with the SCOP/ASTRAL domain codes (3, 23) marking the leaves of the tree. The SCOP superfamilies (codes c.1.1, c.1.2, c.1.3, c.1.4, and c.1.5) form a monophyletic group, a result also supported by a neighbor-joining tree of the same group (data not shown). TPS, thiamin phosphate synthase; FMN, flavin mononucleotide; IMPDH, inosine monophosphate dehydrogenase; OMP, orotidine 5′-monophosphate.
A Single Profile for the Diverse Class II AARS Family. The class II AARS family, a group of enzymes that enforce the genetic code for 10 of the standard amino acids by catalyzing the correct aminoacylation of cognate tRNAs, is so diverse that it is not possible to produce a single, reliable multiple sequence alignment of the common catalytic domain of the group based on sequence information alone. Pfam, for example, represents this group by six separate subclass-level profiles (see Table 1). Multiple structure alignment of the entire group, however, is fairly straightforward (see also Fig. 11 a–d, which is published as supporting information on the PNAS web site) (13) so the known structures can be used as the basis for a single, sequence-supplemented multiple alignment and profile of the class II AARS family. The proteins in the single profile, which was developed from the QR factorization with a sequence identity threshold of 40% (sequence–structure QR 40%), have a sequence identity distribution with a mean of 11% and a range of 4–39%.
Table 1. Hits from subclass-level profiles of the class II AARS family.
| Pfam profile name | AARS specificities | Pfam hits | QR 40% hits |
|---|---|---|---|
| tRNA_Synt_2 | D K N | 299 (20) | 299 (7) |
| tRNA_Synt_2b | G(α2) H P T S, HisZ | 412 (129) | 420 (21) |
| tRNA_Synt_2c | A | 102 (25) | 102 (2) |
| tRNA_Synt_2d | F α-chain | 93 (48) | 92 (5) |
| tRNA_Synt_2e | G(αβ2) α-chain | 61 (9) | 61 (1) |
| N/A | F β-chain | N/A | 110 (16) |
| AsnA | AsnA | 15 (7) | 15 (2) |
| Total | — | 982 (238) | 1,099 (54) |
Shown are the seven subclass-level profiles of the class II AARS family along with the name of the corresponding Pfam profile. The Pfam hits and QR 40% hits columns give the number of true positives found by the Pfam profile and EP, respectively, at the subclass level. The values in parenthesis indicate the number of sequences used to build the profile. AsnA and HisZ catalyze similar enzymatic reactions and, according to sequence and structure, are clearly members of the class II AARS family. These pseudosynthetases, although not represented in the single class-level profiles, are counted as true positives in Fig. 3. —, not applicable; N/A, not available.
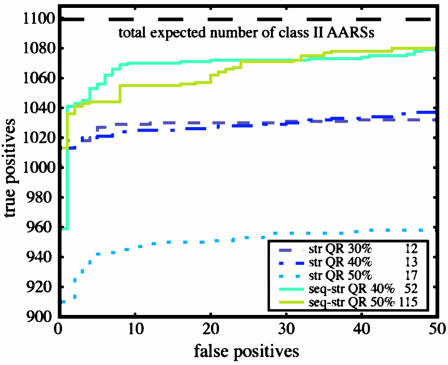
To test the accuracy of the single EP for the class II AARSs, database searches over Swiss-Prot (2) with hmmer and blast (see Fig. 12a, which is published as supporting information on the PNAS web site) were performed. The results from the hmmer search, shown in Fig. 3 and Table 1, were compared with those from the summed results of the seven subclass-level EPs and six Pfam profiles. The sum of these seven separate searches with the EPs should give the total number of class II AARSs in the database, and this value sets an upper limit on the number of true positives that the single class-level profiles are expected to find. The performance of the subclass-level EP for the PheRS α-chain group is compared to the Pfam profile of the same group in Fig. 12b. The combined subclass-level Pfam profiles, including 231 sequences, find 982 class II AARSs, whereas our subclass-level EPs, composed of 54 sequences total, find 1,099 class II AARSs. Pfam does not have a profile for one of the subgroups of the class II AARSs, i.e., PheRS β-chain, and that accounts for most of the discrepancy in performance between the subclass-level EPs and Pfam profiles.
Fig. 3.
AARS class II profile database search results. str, structure; seq, sequence.
The subclass database searches were more accurate in detecting all members of the same subclass than the single profile for the entire family was in detecting all members of each subgroup. As shown in Fig. 3, although the performance for the EP of the single class II profile has deteriorated in comparison to the subclass profiles, the single class II AARS profile (sequence–structure QR 50%) succeeded in finding 98% of all true positives in the database and 92% of those before, including a single false positive. Naturally, if the goal of the database search is to find all relatives of the class II AARSs, then a single profile is n times less as computationally expensive as n separate subclass-level profiles. Although the profiles based on representative sets of only the known structures do perform quite well, the best of which, structure QR 40%, detects 94% of all true positives in the database, the sequence supplemented EPs perform significantly better. In ref. 6, a similar profile for the class I AARSs performed comparably to the six Pfam profiles corresponding to class I AARSs. The slight decrease in the performance of the class II profile as compared with the class I profile is a result of the sparse structural data for some of the members in this family. Performance of the EP for the class II AARSs will be better if structures for the archaeal versions of AlaRS, PheRS α-chain and β-chain, and SerRS are determined experimentally.
Identification of a Putative Archaeal Class II CysRS. In all known examples to date, CysRS is a class I AARS of the Rossmann-fold type, and, being the only known route for charging tRNACys with Cys, this protein is essential to all cellular life. It was surprising, therefore, that CysRS has yet to be found in the completely sequenced genomes of Methanocaldococcus jannaschii (15), Methanothermobacter thermoautotrophicus, or Methanopyrus kandleri (16), despite the fact that these archaea are indeed capable of forming Cys-tRNACys and incorporating Cys into proteins (7). A large body of literature (for a review, see ref. 13) supports the notion that all amino acids are ligated to their cognate tRNA by a direct charging mechanism involving either a class I or class II AARS or by an indirect charging mechanism, as in the case Glu and Asp in some organisms. Even in the more complicated indirect mechanism, all aminoacylation reactions of the tRNA are catalyzed by a class I or class II AARS. We hypothesized, therefore, that the missing archaeal CysRS must be either a class I or class II AARS common to these three methanogens. This hypothesis prompted two searches of these genomes with the EPs for the complete class I AARSs (6) and the complete class II AARSs developed above. If the missing CysRS is a class II AARS, the subfamily-level profiles might fail to find this elusive group because it may form a distinct phylogenetic subgroup within the family.
By using hmmer, a genomic database search of these methanogenic archaea with the class I AARS profile revealed the nine known class I AARSs, and, as expected, no class I CysRS was identified. Also, there were no other class I AARS-like proteins found to be common to these organisms. In light of the existence of a class I and class II LysRS (17), it is possible that the missing CysRS is not a class I enzyme but rather belongs to class II. A similar homology search with the EP of class II AARSs found, in addition to the nine expected class II AARSs, a putative α-chain PheRS common to all three organisms. This putative PheRS is found among the hits to the class II AARS family and has the sequence motifs common to the class II AARS family. The multiple sequence alignment in Fig. 4a shows that these putative PheRSs lack residues that are known to recognize the substrate Phe. Furthermore, biochemical analysis (18) has indicated the absence of PheRS activity in this group, yet this misannotation still persists in the database. According to our homology model in Fig. 4d, these putative AARS-like proteins do have a constellation of conserved Cys residues and a His typical of the CysRS catalytic site as shown in Fig. 4c. For this reason, we now refer to this model as a putative class II CysRS. By using the numbering from PheRS [Protein Data Bank ID code 1PYS], R204 interacts with the anhydride bond in the aminoacyl-adenylate intermediate, whereas R321 forms a cation-π interaction with the adenosine base of the intermediate. The putative class II CysRS, in addition to the conserved Arg motifs corresponding to the class II AARSs, has a KMSK-like motif (gray highlighted residues in Fig. 4a) similar to that found in the class I AARSs. The role of this motif in the putative class II CysRS is unclear.
Fig. 4.
Bioinformatic identification of the elusive archaeal CysRSs. (a) The multiple alignment of archaeal, eukaryotic, and bacterial representatives of the α-chain PheRSs with three representative putative class II CysRSs. Although the putative class II CysRSs have conserved residues important for interaction with the aminoacyl-adenylate intermediate (highlighted in blue), residues important for substrate Phe recognition (highlighted in red) have not been conserved. The putative CysRS group displays three completely conserved Cys residues (highlighted in yellow) and a completely conserved His (highlighted in green). As shown, in class I CysRS, two completely conserved Cys residues and a His residue are critical for forming a zinc-binding site that recognizes the substrate Cys. (b) A percent accepted mutation (PAM) substitution matrix distance-based neighbor-joining tree (δPAM is the distance scale in PAM units) shows the confirmed canonical phylogenetic distribution of the α-chain PheRSs (20) and the monophyletic outgroup of the putative class II CysRSs. *, Uncultured archaeon GZfos26D8 (National Center for Biotechnology Information accession no. AAU43713). (c) A structure of the class I CysRS from E. coli (Protein Data Bank ID code 1LI7) is shown with substrate Cys recognition residues highlighted. (d) The modeled structure of a putative class II CysRS from M. jannaschii shows candidate Cys recognition residues in the class II active site region. Note the cluster of two Cys residues (yellow spheres are the sulfur atoms) and a His residue (green) in the active site suggests a possible zinc-binding motif. The AARS class II, completely conserved Arg residues, which are responsible for generic aminoacyl-adenylate and tRNA acceptor stem binding, are shown in blue.
The M. jannaschii putative class II CysRS in Swiss-Prot (Swiss-Prot accession code YG60_METJA) was used as a representative to search the nonredundant database NRDB of the National Center for Biotechnology Information, revealing the presence of this gene in 10 archaeal genomes (see Fig. 4b). Of these organisms, Methanococcus maripaludis does have the standard class I CysRS, but, in an experiment in which the class I CysRS was knocked out, the deletion strain was able to survive, indicating the existence of an alternative pathway for the formation of Cys-tRNACys (19). The α-chain PheRS orthologs display a full canonical phylogenetic distribution, with only a minor amount of horizontal gene transfer from the Archaea to some bacterial organisms (20). This three-domains-of-life pattern is also clear in a sequence similarity, distance-based neighbor-joining tree for a representative set of archaeal, eukaryotic, and bacterial proteins (Fig. 4b). The tree also reveals that the putative class II CysRSs form a distinct monophyletic outgroup with respect to the PheRS group, suggesting a functional divergence between the PheRS and putative class II CysRS groups rather than a speciation-related divergence. This observation is supported in the multiple alignment in Fig. 4a, where there are clear distinctions in the sequences of the PheRS group versus the putative class II CysRS group, which has idiosyncratic insertions and sequence signatures.
Although the class I and class II LysRSs show no global similarity in sequence or structure (13), their active sites contain a similar constellation of residues (21). Because the α-chain PheRS group is the most closely related of any other AARS group to the putative class II CysRSs, we used the structure of PheRS from Thermus thermophilus (Protein Data Bank ID code 1PYS) as a template for building a homology model of the putative class II CysRS from M. jannaschii. For comparison, the structure of the class I CysRS from Escherichia coli (Fig. 4c) is juxtaposed with our model of the putative class II CysRS (Fig. 4d) with the putative Cys recognition residues highlighted. In the class I CysRS, a zinc-binding pocket is formed by two strictly conserved Cys residues and a His residue, and structural analysis has shown that the substrate Cys is involved in a direct thiol coordination to the bound zinc (22). The model in Fig. 4d clearly shows the placement of these conserved residues (Cys residues 104 and 246 and His residue 198) within the active site region of this class II catalytic domain. Interestingly, there is a third highly conserved Cys residue in our model that appears to be too far from the putative active site to play a role in the zinc-binding pocket. Although our bioinformatic results strongly indicate that YG60_METJA and its orthologs are candidates for the missing archaeal CysRS, a high-resolution crystal structure and biochemical studies are required to confirm this suggestion. Although this enzyme appears to specifically bind a cysteinyl-adenylate and a tRNA acceptor stem, perhaps it is involved in some kind of indirect aminoacylation pathway, as in the nondiscriminating AspRS, or it is a pseudosynthetase enzyme involved in Cys biosynthesis, similar to the class II homolog AsnA, which is involved in Asp biosynthesis (for a review, see ref. 20).
Conclusion
The QR factorization allows an economy of information in constructing profiles at the family and superfamily (see Supporting Text) levels. Our studies indicate that it is better to build EPs from a minimal set of sequences that span the evolutionary space, rather than to iteratively add sequences to a profile until the “best” database search results are found. The appropriate size of the minimal EP depends on the level of diversity and the number of major evolutionarily related subgroups in the family. Before superfamily profiles can be developed systematically, the groupings of superfamily in the SCOP database need to be checked. In all cases, proteins exhibited such extreme diversity that sequence information had to be augmented by structural information to obtain a single, complete EP that could be used for sensitive database searches. The single EP of the diverse, full class II AARS family contained general signals of the class II AARS that allowed us to predict the presence of an elusive archaeal class II CysRS. In addition, the groupings observed from the database search also imply that the putative CysRS should group with the PheRS, GlyRS, and AlaRS tetramers.
Our approach to the problem of redundancy in protein sequence and structure data is based on the observation that the organizational structure created by the evolutionary process is the very structure that we seek to define and understand with the tools of bioinformatics. The molecular components of organisms are not a bewildering array of nearly endless novelty; rather, their evolutionary relationships indicate a smaller set of primordial forms that have left their imprint throughout the molecular characters of all cellular life. In this light, we have presented the notion that one way to deal with the massive amount of biological data is to make use of evolutionary relationships to motivate a reduction of the data to a smaller subset or basis set that well represents or characterizes the evolutionary space.
Supplementary Material
Acknowledgments
We thank Carl Woese for many helpful discussions and for communicating this work. This work was supported by National Science Foundation Grants MCB04-46227 and MCB02-35144. P.O. was supported in part by National Institutes of Health National Research Service Award 5T32GM08276 in Molecular Biophysics.
Author contributions: Z.L.-S. designed research; A.S., P.O., and Z.L.-S. performed research; A.S., P.O., and Z.L.-S. contributed new reagents/analytic tools; A.S., P.O., and Z.L.-S. analyzed data; and A.S., P.O., and Z.L.-S. wrote the paper.
Abbreviations: EP, evolutionary profile; TIM, triosephosphate isomerase; RPB, ribulose-phosphate binding; RS, tRNA synthetase; AARS, aminoacyl-RS.
References
- 1.Attwood, T. K. & Miller, C. J. (2001) Comput. Chem. 25, 329–339. [DOI] [PubMed] [Google Scholar]
- 2.Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M., Estreicher, A., Gasteiger, E., Martin, M. J., Michoud, K., O'Donovan, C., Phan, I., et al. (2003) Nucleic Acids Res. 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 4.Orengo, C. A., Jones, A. D. M. S., Jones, D. T., Swindells, M. B. & Thornton, J. M. (1997) Structure (Cambridge, MA, U. S.) 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- 5.Sonnhammer, E. L. L., Eddy, S. R. & Durbin, R. (1997) Proteins Struc. Funct. Genet. 28, 405–420. [DOI] [PubMed] [Google Scholar]
- 6.O'Donoghue, P. & Luthey-Schulten, Z. (2005) J. Mol. Biol. 346, 875–894. [DOI] [PubMed] [Google Scholar]
- 7.Ruan, B., Nakano, H., Tanaka, M., Mills, J. A., DeVito, J. A., Min, B., Low, K. B., Battista, J. R. & Söll, D. (2004) J. Bacteriol. 186, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heck, L. P., Olkin, J. A. & Naghshineh, K. (1998) J. Vib. Acoust. 120, 663–670. [Google Scholar]
- 9.Householder, A. S. (1958) J. Assoc. Comput. Mach. 5, 339–342. [Google Scholar]
- 10.Golub, G. (1965) Numer. Math. 7, 206–216. [Google Scholar]
- 11.O'Donoghue, P., Amaro, R. E. & Luthey-Schulten, Z. (2001) J. Struct. Biol. 134, 257–268. [DOI] [PubMed] [Google Scholar]
- 12.Jansen, R. & Gerstein, M. (2000) Nucleic Acids Res. 28, 1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Donoghue, P. & Luthey-Schulten, Z. (2003) Microbiol. Mol. Biol. Rev. 67, 550–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bork, P., Gellerich, J., Groth, H., Hoofth, R. & Martin, F. (1995) Protein Sci. 4, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bult, C. J., White, O., Olsen, G. J., Zhou, L., Fleischmann, R. D., Sutton, G. G., Blake, J. A., FitzGerald, L. M., Clayton, R. A., Gocayne, J. D., et al. (1996) Science 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 16.Jacquin-Becker, C., Ahel, I., Ambrogelly, A., Ruan, B., Söll, D. & Stathopoulos, C. (2002) FEBS Lett. 514, 34–36. [DOI] [PubMed] [Google Scholar]
- 17.Ibba, M., Morgan, S., Curnow, A. W., Pridmore, D. R., Vothknecht, U. C., Gardner, W., Lin, W., Woese, C. R. & Söll, D. (1997) Science 278, 1119–1122. [DOI] [PubMed] [Google Scholar]
- 18.Das, R. & Vothknect, U. C. (1999) Biochimie 81, 1037–1039. [DOI] [PubMed] [Google Scholar]
- 19.Stathopoulos, C., Kim, W., Li, T., Anderson, I., Deutsch, B., Palioura, S., Whitman, W. & Söll, D. (2001) Proc. Natl. Acad. Sci. USA 98, 14292–14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woese, C. R., Olsen, G., Ibba, M. & Söll, D. (2000) Microbiol. Mol. Biol. Rev. 64, 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terada, T., Nureki, O., Ishitani, R., Ambrogelly, A., Ibba, M., Söll, D. & Yokoyama, S. (2002) Nat. Struct. Biol. 9, 257–262. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, C. M., Christian, T., Newberry, K. J., Perona, J. J. & Hou, Y. M. (2003) J. Mol. Biol. 327, 911–917. [DOI] [PubMed] [Google Scholar]
- 23.Chandonia, J. M., Hon, G., Walker, N. S., Lo Conte, L., Koehl, P., Levitt, M. & Brenner, S. E. (2004) Nucleic Acids Res. 32, D189–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.