Abstract
Mistranslation describes errors during protein synthesis that prevent the amino acid sequences specified in the genetic code from being reflected within proteins. For a long time, mistranslation has largely been considered an aberrant cellular process that cells actively avoid at all times. However, recent evidence has demonstrated that cells from all three domains of life not only tolerate certain levels and forms of mistranslation, but actively induce mistranslation under certain circumstances. To this end, dedicated biological mechanisms have recently been found to reduce translational fidelity, which indicates that mistranslation is not exclusively an erroneous process and can even benefit cells in particular cellular contexts. There currently exists a spectrum of mistranslational processes that differ not only in their origins, but also in their molecular and cellular effects. These findings suggest that the optimal degree of translational fidelity largely depends on a specific cellular context. This review aims to conceptualize the basis and functional consequence of the diverse types of mistranslation that have been described so far.
Keywords: Mistranslation, tRNA, ribosome, misacylation, miscoding, adaptation, stress response
Introduction
The accurate flow of information from nucleic acids to proteins is of principal importance to every biological system. The protein sequences contained within the genetic code have been refined through eons of evolution, but effective utilization of these optimized sequences is contingent on the accuracy of protein synthesis. Despite the need for accurate translation, ribosomes of higher accuracy can inhibit cell growth (Ruusala et al., 1984, Zaher and Green, 2010), indicating that translational fidelity is not the paramount objective of translation. Cells must balance translational accuracy with translational speed and there is a tradeoff between both factors (Drummond and Wilke, 2008, Johansson et al., 2008, Wohlgemuth et al., 2010, Wohlgemuth et al., 2011, Johansson et al., 2012). Therefore, biology must tolerate some errors in protein synthesis in order to maintain the rapidity of translation.
The mistranslational cost for high speed translation is approximately one error per 103 to 105 amino acids (Loftfield and Vanderjagt, 1972, Parker, 1989, Ibba and Soll, 1999, Ogle and Ramakrishnan, 2005, Kramer and Farabaugh, 2007, Zaher and Green, 2009, Kramer et al., 2010, Rodnina, 2012). Mistranslation can be engendered during any process that mediates the flow of information from the genetic code to proteins (Figure 1). Beginning with DNA replication, nucleotide misincorporations into the new genomic copy occurs about once every 108 nucleotides (Kunkel, 2004) and this event can lead to an enduring change in the amino acid identity of a codon. More commonly, nucleotide misincorporation errors during mRNA transcription occur once every 105 nucleotides (Blank et al., 1986) and can also alter the amino acid identity of a codon. However, inaccuracies are far more common during translation itself.
Figure 1. Cellular processes engendering mistranslation.
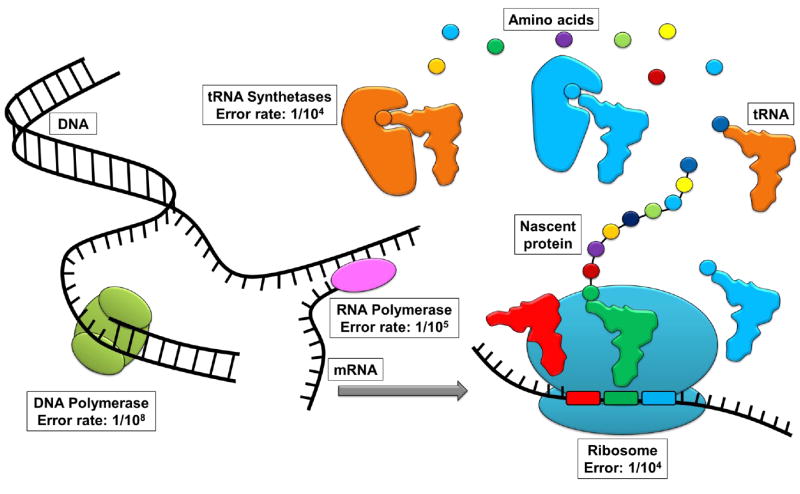
Mistranslation can result from any of the processes that mediate the flow of information from DNA to proteins.
Errors occurring during translation can occur when a tRNA is charged with a noncognate amino acid by an aminoacyl-tRNA synthetase (aaRS) or when the ribosome accepts a tRNA of the incorrect amino acid identity for a given codon. Each aaRS possesses a specific amino acid identity and is solely responsible for charging a single type of amino acid to specific, cognate tRNAs. The catalytic sites and editing domains of aaRS help ensure that only cognate amino acids are used in the aminoacylation reaction; tRNA synthetases have been shown to have in vitro fidelities greater than 10-4 (Ibba and Soll, 2000, Ling et al., 2009). After tRNAs have been ligated with amino acids, they must be correctly selected by the ribosome through accurate tRNA-mRNA base pairing (Dale and Uhlenbeck, 2005), which occurs with an in vitro accuracy of approximately 10-4 (Zaher and Green, 2009, Rodnina, 2012) (Figure 1).
Mistranslation in excess of the inherent inaccuracy required for rapid protein synthesis has long been considered an undesirable yet inexorable process that can exert deleterious consequences. However, the degree to which translational accuracy should be compromised for translational speed is probably not optimized in all conditions. Despite the need for translational fidelity, cells tolerate some types of mistranslation surprisingly well and specific mistranslational processes are even induced under certain conditions (Netzer et al., 2009, Meyerovich et al., 2010, Javid et al., 2014, Schwartz and Pan, 2016b, Schwartz et al., 2016). Mistranslation that is actively induced has been shown to serve beneficial functions in cells under the appropriate circumstances. These findings do not undermine the known and potential deleterious effects of mistranslation, but rather, they reveal that regulation of mistranslation is more multifaceted than previously expected. While many mistranslated proteins can be inactive or misfolded, there are also many ways by which mistranslation has the potential to alter the activity of proteins that are derived purely from the genetic code. Mistranslation describes innumerable types, locations, and degrees of amino acid substitutions within proteins—the vast effects of which will be largely dependent on cellular context. Since the fidelity of protein synthesis can be affected by a multitude of factors, the challenge going forward will be to deduce the basis and functional consequence of each distinct mistranslational process. This review explores the vast array of cellular conditions that prompt mistranslation and describes the different effects that have been shown for mistranslation in different cellular contexts.
Inherent mistranslation
Error is inherent to all biological systems and protein synthesis is no exception. The intrinsic translational error in biological systems is a function of a common design that compromises accuracy for speed (Johansson et al., 2008, Wohlgemuth et al., 2010, Wohlgemuth et al., 2011, Johansson et al., 2012). This general error rate of translation is independent of circumstance and appears to be common to all biological systems. However, all translational errors are not equally likely to occur in every organism and there are inherent predispositions for specific types of errors. For example, mischarging of tRNAs with noncognate amino acids by an aaRS is far more likely to occur with an amino acid of similar structure, which can differ by as little as a methyl or hydroxyl group. For example, the isoleucine tRNA synthetase (IleRS) mischarges Val to tRNAIle at ~1% of the time due to the structure of Val (Hale et al., 1997), which only differs from Ile by one methyl group. The vast majority of Val-tRNAIle can be eliminated by the editing activity of the IleRS (Hendrickson et al., 2002), but the IleRS does inherently charge Val to tRNAIle in appreciable levels. Similarly, mammalian tyrosine-tRNA synthetases (TyrRS) have been shown to charge Phe to tRNATyr at relatively high levels and this low specificity is further exacerbated during Tyr limitation (Raina et al., 2014). Additionally, the human alanine-tRNA (AlaRS) synthetase readily accepts noncognate tRNAs that have a G4:U69 base pair (e.g. tRNACys) in the acceptor stem (Sun et al., 2016). Despite the low specificity recently discovered for the mammalian TyrRS and AlaRS, the bacterial TyrRS and AlaRS still have high specificity for cognate amino acids and tRNAs, respectively, indicating that the inexplicable low specificity of these mammalian synthetases is not inherent to all biological systems (Raina et al., 2014, Sun et al., 2016).
In both E. coli and mammalian cells, Asn limitation results in Lys substitutions and His limitation results in Gln substitutions within proteins (Parker et al., 1978, Parker et al., 1980). It is well supported that the low levels of charged tRNAAsn and tRNAHis during starvation (reading AAC/AAU and CAU/CAC codons, respectively) allows charged tRNALys and tRNAGln (reading AAG/AAA and CAA/CAG codons, respectively) to read noncognate codons that differ only in the third position. However, some codons are misread in the third codon position more readily than others (Parker and Friesen, 1980, Parker et al., 1980). Surprisingly, Lys limitation does not elicit Asn mistranslation and Gln limitation does not elicit His mistranslation despite almost identical mismatches present in the third position (Parker et al., 1978, Parker and Friesen, 1980). However, it is now well known that the codon/anticodon pairing is not the only factor that controls tRNA acceptance by the ribosome (Cochella and Green, 2005, Ledoux et al., 2009, Murakami et al., 2009, Schmeing et al., 2011). These results indicate that the ribosome permits specific codon-anticodon mismatches more readily than others, which would make certain translational errors more prevalent. However, ribosome miscoding does not occur during all amino acid limitations despite similar violations of translational quality control. These results indicate that some miscodings are not derived from necessity, but may be exploiting specific vulnerabilities in tRNA acceptance by the ribosome. Relatedly, numerous other significant codon/anticodon mismatching preferences have been found for specific tRNA identities, type of mismatches, and corresponding mismatching positions in E. coli (Zhang et al., 2015, Zhang et al., 2016). Altogether, these results reveal numerous intrinsic tRNA-mRNA mismatching tendencies on the ribosome that have the potential to produce preferred amino acid substitutions within proteins.
Specific errors during tRNA charging and tRNA selection by the ribosome are not the only inherently erroneous processes that occur during translation. Many bacteria lack a specific aaRS for glutamine and asparagine (Sheppard et al., 2008). In these organisms, non-discriminatory glutamate and aspartate-tRNA synthetases intentionally mischarge tRNAGln and tRNAAsn, respectively, along with their cognate tRNAs. Glu/Asp are then transamidated directly on the mischarged Glu-tRNAGln to Gln or Asp-tRNAAsn to Asn by the GatCAB enzyme complex to generate correctly charged tRNAGln and tRNAAsn (Curnow et al., 1997). It was believed that the naturally misacylated tRNA intermediates would not be used for protein synthesis since EF-Tu was shown to discriminate against Glu-tRNAGln and Asp-tRNAAsn (Roy et al., 2007). However, expression of non-discriminatory AspRSs in trans results in significant Asn-to-Asp mistranslation in E. coli (Ruan et al., 2008, Nair et al., 2016), which is consistent with results showing that the ribosome can use certain misacylated tRNAs almost indistinguishably from correctly acylated tRNAs (Effraim et al., 2009). It has recently been shown that Mycobacterium tuberculosis, which makes charged tRNAGln and tRNAAsn through the tRNA dependent transamidation pathway, naturally uses the misacylated Glu-tRNAGln and Asp-tRNAAsn in translation (Su et al., 2016). These results indicate that the indirect tRNAGln and tRNAAsn aminoacylation pathway is an inherent source of mistranslation. Bacteria are typically assumed to have either a GlnRS/AsnRS or lack these synthetases and have non-discriminatory GluRS/AspRS with a GatCAB complex for generating charged tRNAGln and tRNAAsn. However, the AspRSs from several Bacillus species have retained their recognition of tRNAAsn despite the presence of an active AsnRS (Nair et al., 2016). Phylogenetic analysis of bacterial AspRSs reveals that the presence of a non-discriminatory AspRS may be pervasive in bacteria and cannot be discounted by the simple presence of an AsnRS. Like the indirect tRNAGln and tRNAAsn aminoacylation pathway, the retention of non-discriminatory AspRSs in bacteria is also likely an inherent source of Asn-to-Asp mistranslation.
Mistranslation due to stress or damage
The previous examples have focused on mistranslation that is inherent to various translational systems due to the fundamental pathway design. However, mistranslation can also be the direct result of stress or damage to the translation machinery. For example, the E. coli threonine-tRNA synthetase (ThrRS) has a conserved Cys residue in its editing domain. This residue is essential for the editing function of the ThrRS; Cys oxidation during oxidative stress can inactivate the editing domain and consequently generate significant amounts of mischarged Ser-tRNAThr (Ling and Soll, 2010, Wu et al., 2014). Although oxidative stress itself impedes growth of E. coli, the addition of Ser to the growth medium (to potentiate Ser mistranslation) further inhibits growth in the presence of hydrogen peroxide. Therefore, Thr-to-Ser mistranslation itself contributes to cellular damage during oxidative stress—an effect that is suppressed through the addition of Thr to the growth medium (to reduce Ser mistranslation). Oxidative stress has also been shown to facilitate mistranslation by damaging cellular amino acids (Bullwinkle et al., 2014). Phe can be oxidized by reactive oxygen species to form the non-proteinogenic amino acid meta-Tyr (Stadtman and Levine, 2003), which can be charged to tRNAPhe by the phenylalanine-tRNA synthetase (PheRS) in E. coli (Klipcan et al., 2009). Although most of the tRNAPhe mischarged with meta-Tyr is eliminated by PheRS editing, inactivating the editing domain of PheRS increases the meta-Tyr incorporated into proteins. During oxidative stress that facilitates the formation of meta-Tyr, the lack of effective meta-Tyr editing by PheRS results in growth deficiency, indicating that oxidative injury is exacerbated by translation with meta-Tyr despite normally being mitigated by PheRS editing (Bullwinkle et al., 2014). The danger of mistranslation with meta-Tyr is substantiated by its cytotoxicity in mammalian cells and its ability to inhibit the growth of a wide range of plant species when it is used in protein synthesis (Gurer-Orhan et al., 2006, Bertin et al., 2007).
The aminoglycoside class of antibiotics directly targets the ribosome and has been known to reduce the fidelity of translation for quite some time (Davies et al., 1964, Edelmann and Gallant, 1977, Kotra et al., 2000). Some aminoglycosides engender mistranslation by causing tRNA-mRNA mismatch on the ribosome (Weisblum and Davies, 1968, Davis, 1987) which has been shown to cause general protein misfolding (Kohanski et al., 2008). In addition, the antibiotic induced mistranslation can compromise the integrity of cellular membranes through the insertion of mistranslated membrane protein channels that are nonspecific (Anand and Davis, 1960, Anand et al., 1960, Eaton and Caffrey, 1961, Davis et al., 1986). These mistranslated, nonspecific protein channels have been proposed to contribute to cell death by allowing leakage and entry of molecules which are normally excluded from transport, such as additional antibiotic molecules (Davis et al., 1986). Many additional agents and conditions likely have the potential to compromise translation fidelity. However, in these instances, mistranslation is primarily due to dysfunction of the translational machinery and may not be adaptive in most circumstances.
Synthetic mistranslation
The difficulty in identifying cases of naturally high mistranslation to study its effects has necessitated the use of mistranslation systems that rely on mutated tRNAs, aaRSs, or ribosomes in order to engender high levels of mistranslation. A popular strategy has been to inactivate the editing domain of an aaRS to increase charging of noncognate amino acids. In mice, mutations in the editing domain of the alanine-tRNA synthetase causes significant mischarging of tRNAAla with Ser. Defective AlaRS editing in mice results in damage to cerebellar Purkinje cells, cardiac fibrosis and dysfunction, and even embryonic lethality (Lee et al., 2006, Liu et al., 2014). In Drosophila melanogaster, mutations in the editing domain of the phenylalanine-tRNA synthetase cause Tyr misacylation to tRNAPhe and the mutant flies have numerous defects including loss of neuronal cells, shortened lifespan, and smaller organ size (Lu et al., 2014). In mammalian cells, an editing deficient valine-tRNA synthetase disrupts cell morphology, membrane blebbing, and activates the apoptotic response (Nangle et al., 2006).
Yet, the expression of an editing defective PheRS in Saccharomyces cerevisiae and E. coli has only a small effect on growth despite the significant generation of Tyr-mischarged tRNAPhe (Reynolds et al., 2010). Furthermore, E. coli can tolerate an ambiguous genetic code that generates up to 10% amino acid substitutions in proteins by using multiple editing defective aaRSs (Ruan et al., 2008). Removing the editing domain from the isoleucine-tRNA synthetase in bacteria can increase the growth rate during isoleucine limitation despite being deleterious to growth under most conditions (Pezo et al., 2004, Bacher et al., 2005, Bacher et al., 2007). Relatedly, mistranslation engendered through editing defects in PheRS in E. coli has also been shown to increase tolerance to specific types of amino acid stress (Bullwinkle and Ibba, 2016). These results demonstrate that certain types of mistranslation can be beneficial in very specific contexts.
Candida albicans can survive a genetic code alteration that completely changes the amino acid identity of CUG codons, although the level of CUG miscoding can be controlled through endogenous tRNA knockout and foreign tRNA expression (Bezerra et al., 2013). Strains with increased CUG miscoding in C. albicans have diverse cell and colony morphologies, are more resistant to oxidative stress, antifungal agents, and phagocytotic killing by macrophages, despite growing slower in a rich medium compared to the wild-type strain (Miranda et al., 2007, Bezerra et al., 2013, Miranda et al., 2013). Although mistranslation can have diverse tangential effects on cellular physiology, specific miscoding of an adhesion and invasion protein in trans was sufficient to recapitulate phenotypes observed from increasing global CUG miscoding in C. albicans (Miranda et al., 2013).
Synthetic mistranslation in Mycobacterium smegmatis has been shown to increase resistance to several types of antibiotics through the expression of mutator tRNAs, despite causing death if overexpressed in sufficient quantities (Javid et al., 2014). Mistranslated RNA polymerases from Mycobacteria are more resistant to inhibition by rifampicin demonstrating that mistranslation can synthesize proteins with altered susceptibilities to rifampicin that promote growth during prejudicial conditions. Furthermore, synthetic mistranslation activates transcriptional stress responses in Saccharomyces cerevisiae when CUG codon ambiguity is artificially imbued (Santos et al., 1999) and in E. coli through a ribosomal protein mutation (Battesti et al., 2011, Fan et al., 2015)—both of which broadly increase stress tolerance. However, mistranslation itself was not specifically shown to exert any specific effects in these cases and the increase in stress tolerance could also be derived from the general stress response that mistranslation induces.
The study of synthetic mistranslation has been indispensable for revealing the potential functional consequences of mistranslation. Numerous such studies have demonstrated that the functional consequences for mistranslation are diverse. This is not surprising considering the myriad of activity altering amino acid substitutions that can potentially be made through mistranslation. Only a small percentage of experimental measurements exist for the natural frequency of the 1,216 (64 × 19) possible codon-to-amino-acid events and a global functional consequence is lacking for the vast majority of amino acid substitutions (Drummond and Wilke, 2009, Ribas de Pouplana et al., 2014). It is evident that mistranslation can be deleterious, yet numerous studies show that mistranslation can be conditionally beneficial. Essentially all of the described benefits for synthetic mistranslation have been during periods of stress where the wild-type proteins may not be optimized for function, and thus may benefit from a mistranslational alteration of their standard activity. Therefore, the deterministic factor for the effect of a particular type of mistranslation seems to be the cellular context.
Natural and constitutive ambiguity
Although error is inevitable in translation, certain organisms have constitutively high levels of mistranslation. Mycoplasma parasites have been shown to have point mutations and deletions in the editing domains of their ThrRS, LeuRS and PheRS (Li et al., 2011). These mutations and deletions reduce the fidelity of tRNA aminoacylation at all times and result in constitutively high levels of mistranslation (Figure 2A) (Li et al., 2011, Yadavalli and Ibba, 2013). Mycoplasmas are obligate intracellular parasites and their survival therefore depends on the successful evasion of the host’s immune system. Constitutive mistranslation may help Mycoplasma create cell surface variability and evade the host’s immune system in a manner similar to antigenic variation that has been described for other pathogenic organisms (Finlay and McFadden, 2006).
Figure 2. Naturally ambiguous translation.
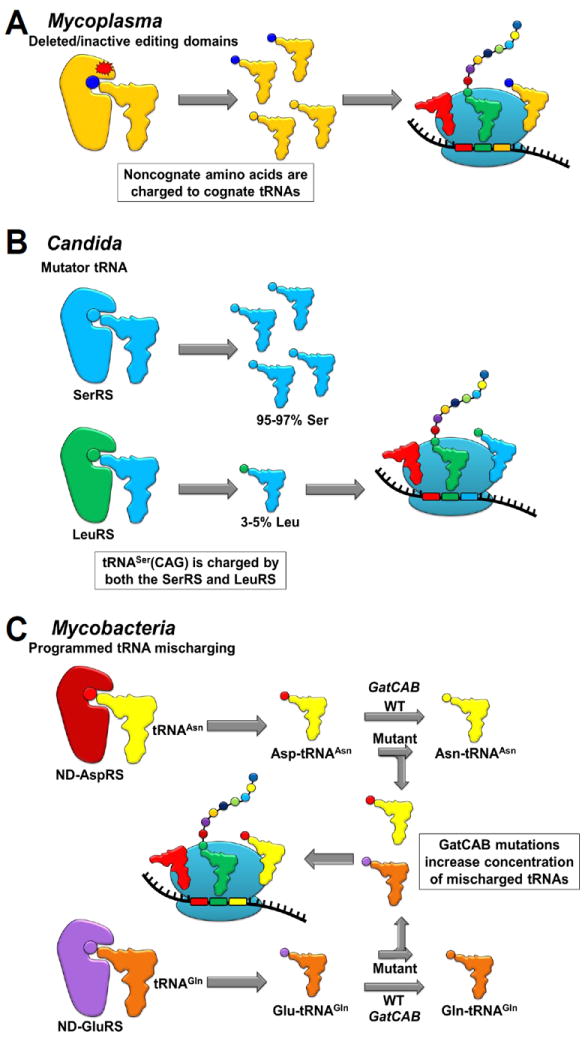
(A) Ambiguous translation in Mycoplasma species results from lost or inactive editing sites in several tRNA synthetases that allow noncognate amino acids to be charged to tRNAs. (B) Ambiguous translation occurs in Candida species due to tRNASerCAG being recognized by both SerRS and LeuRS and resulting in CUG codons being translated as both Ser and Leu. (C) Ambiguous translation in Mycobacteria results from an indirect tRNAAsn and tRNAGln synthesis that requires mischarged tRNA intermediates, which can be used in translation. Mutations in the GatCAB complex, which generates correctly acylated tRNAAsn and tRNAGln from the mischarged tRNA intermediates, increases Asn-to-Asp and Gln-to-Glu mistranslation.
The pathogenic fungus Candida albicans also performs constitutive mistranslation, but through CUG codon ambiguity (Gomes et al., 2007). Candida species have recoded their genomes so that CUG codes for Ser (rather than the standard Leu); the CUG reading tRNASer in Candida contains identity elements for both the serine and leucine-tRNA synthetases (Suzuki et al., 1997, Giege et al., 1998). Therefore, tRNASerCAG is charged with serine at 95% to 97% and with leucine at 3% to 5% in cells and this charging heterogeneity is reflected commensurately at CUG sites within the Candida proteome (Figure 2B) (Gomes et al., 2007). Artificially increasing the level of CUG codon ambiguity to misincorporate 28% Leu decreases phagocytotic killing by macrophages and increases adherence to host molecules relative to the wild-type cells (Miranda et al., 2013). C. albicans contains two copies of the tRNASerCAG that mediates CUG codon ambiguity. Knocking out one copy reduces Leu misincorporation at CUG codons to 0.6% and concurrently increases its growth rate in rich medium by 25% (Bezerra et al., 2013). This result indicates that CUG codon ambiguity is a mechanism for environmental survival since it impairs growth in a rich medium. Since Candida albicans is a pathogen, it likely uses mistranslation as a mechanism to create cell surface variability to evade the host’s immune system (Miranda et al., 2013).
As previously mentioned, Mycobacterium species lack AsnRSs and GlnRSs and generate these charged tRNAs through natural Glu-tRNAGln and Asp-tRNAAsn misacylation followed by amino acid transamidation on the tRNA by the GatCAB complex. Although these mischarged tRNA intermediates can naturally be used in translation (Ruan et al., 2008, Nair et al., 2016), mutations in the GatCAB complex can increase Gln-to-Glu and Asn-to-Asp mistranslation in M. tuberculosis by increasing the concentration of the Glu-tRNAGln and Asp-tRNAAsn mischarged tRNA intermediates (Figure 2C). An increase in mistranslation mediated by GatCAB mutations confers increased resistance to rifampicin, which is consistent with an increase in antibiotic resistance that was described for synthetic mistranslation in Mycobacteria (Javid et al., 2014, Su et al., 2016). Remarkably, clinical isolates of M. tuberculosis were found to contain natural mutations in GatCAB that increase mistranslation and concomitantly confer greater resistance to rifampicin. Although the inherent design of the indirect Asn and Gln pathway is conducive to mistranslation, clinical M. tuberculosis isolates have potentiated the inherent mistranslation in this system. The mutations in GatCAB facilitating high levels of constitutive mistranslation provide a mechanism that enhances survival of M. tuberculosis during infections since GatCAB smutations reduce growth during laboratory cultivation (Su et al., 2016).
In Candida, Mycoplasma, and M. tuberculosis, it is evident that cells employ mistranslation specifically for its physiological effects rather than tolerating it as a tangential process. Although Mycoplasma species have editing domains in their aaRS, some are rendered inactive though mutations and deletions, suggesting that mistranslation provides a benefit in their obligatory intracellular environment. In Candida species, CUG codon ambiguity must increase fitness in the environment since higher translational fidelity can increase growth under optimal conditions. Similarly, M. tuberculosis clinical isolates have evolved to have higher levels of mistranslation, indicating that this is likely a desirable process during infection despite hindering growth during laboratory cultivation. The purposeful retention of these natural mistranslational processes is supported by the short time scale required to mitigate the effects of deleterious mistranslation. When synthetic mistranslation is introduced into S. cerevisiae through a mutator tRNA and allowed to evolve in the laboratory for 250 generations, cells quickly adapt to and surmount the detriments of mistranslation (Kalapis et al., 2015). Numerous compensatory mutations are made and cells also reduce the expression of the mutator tRNA and the overall levels of mistranslation resulting in increased laboratory fitness (Kalapis et al., 2015). Therefore, cells have the potential to rapidly reduce genetic code ambiguity and mitigate the deleterious effects of mistranslation if they evolved in an environment where mistranslation is not advantageous. It is therefore not a coincidence that organisms that employ high levels of constitutive translational ambiguity are pathogens not solely grown in the laboratory. Mistranslation seems to function as a mechanism to generate cell surface protein variation and help these organisms endure the adversities of the immune response.
Inducible mistranslation
Unlike the preceding forms of natural and constitutive mistranslation that are essentially immutable (because the causes of mistranslation are derived from DNA changes), inducible mistranslation can be actively regulated in response to growth conditions. The previous examples of mistranslation have demonstrated that the optimal levels of translational fidelity vary according to biological context. For example, the natural translational ambiguity of C. albicans increases fitness in natural contexts despite hindering growth in the laboratory (Bezerra et al., 2013). However, for C. albicans to optimize growth in all environments it would need to ablate mistranslation during laboratory growth (Bezerra et al., 2013), but also activate mistranslation during prejudicial conditions and infections (Bezerra et al., 2013, Miranda et al., 2013). The prospect of inducible mistranslation offers the benefits of mistranslation to be conferred when applicable, but also ensures that the inadvertent, deleterious consequences of mistranslation do not compromise growth during optimal conditions.
Although the potential benefits of inducible mistranslation suggest that this should be a pervasive biological mechanism, detecting conditional mistranslation can be difficult. Cells should not perform appreciable levels of mistranslation under optimal conditions in the laboratory since the genetically encoded proteome is optimized for function under these circumstances. Therefore, one would need to search for mistranslation in alternative conditions. Nonetheless, diverse organisms have now been shown to perform inducible mistranslation, although some instances are not yet well characterized. For example, Bacillus subtilis performs high levels of mistranslation through frameshifting and stop codon readthrough during low-temperature growth and stationary phase (Meyerovich et al., 2010). Similarly, Mycobacteria increase their rates of mistranslation during growth at low pH and stationary phase (Javid et al., 2014, Leng et al., 2015). Although it has not been unequivocally shown that these conditions induce a veritable mistranslational response rather than exploiting a natural frailty in translational fidelity, cells do make definitive responses to low pH (Cotter and Hill, 2003), low temperature (Weber and Marahiel, 2003), and most notably stationary phase (Battesti et al., 2011). Furthermore, none of the alternative conditions used to induce mistranslation should be severe enough to compromise translational fidelity (i.e. pH 6.1 and 23°C). Stationary phase growth in bacteria induces the general stress response which results in a broad increase in stress tolerance (Battesti et al., 2011) and is consistent with the effects described for synthetic mistranslation. The fact that both Mycobacteria and Bacillus species commonly activate mistranslation during stationary phase suggests that this is an intentional response. The challenge going forward will be to determine the mechanisms by which translational fidelity is reduced under these conditions and whether inducible mistranslation itself indeed has beneficial effects.
The best characterized case of veritable inducible mistranslation is the methionine mistranslation response (Netzer et al., 2009, Wiltrout et al., 2012, Schwartz and Pan, 2016b, Schwartz et al., 2016). Organisms from all three domains of life have now been shown to perform conditional mistranslation with Met under diverse conditions, and the attenuation of translational fidelity has been shown to be beneficial. Met mistranslation was first discovered in mammalian cells where ~1% of methionine is charged to nonmethionyl-tRNAs under standard tissue culturing conditions in the laboratory or in mouse dentric cells and mouse liver. Met misacylation can increase up to 10-fold during oxidative stress and in the presence of viruses and toll-like receptor ligands (Netzer et al., 2009). Remarkably, no tRNA misacylation is observed for Cys, Ile, Phe, Val, or Tyr, suggesting that mistranslation with Met is of particular significance. tRNA misacylation with Met is carried out by the human methionine-tRNA synthetase (MetRS) which is phosphorylated at two specific serine residues by the extracellular signal-related kinase (ERK1/2) during oxidative stress; the phosphorylated MetRS has significantly lower fidelity and readily charges noncognate tRNAs with Met (Figure 3) (Lee et al., 2014). Remarkably, constitutive expression of low-fidelity MetRSs that mimic the phosphorylated, low-fidelity form enhances oxidative stress response (Lee et al., 2014).
Figure 3. MetRS mechanisms mediating the conditional acceptance of noncognate tRNAs.
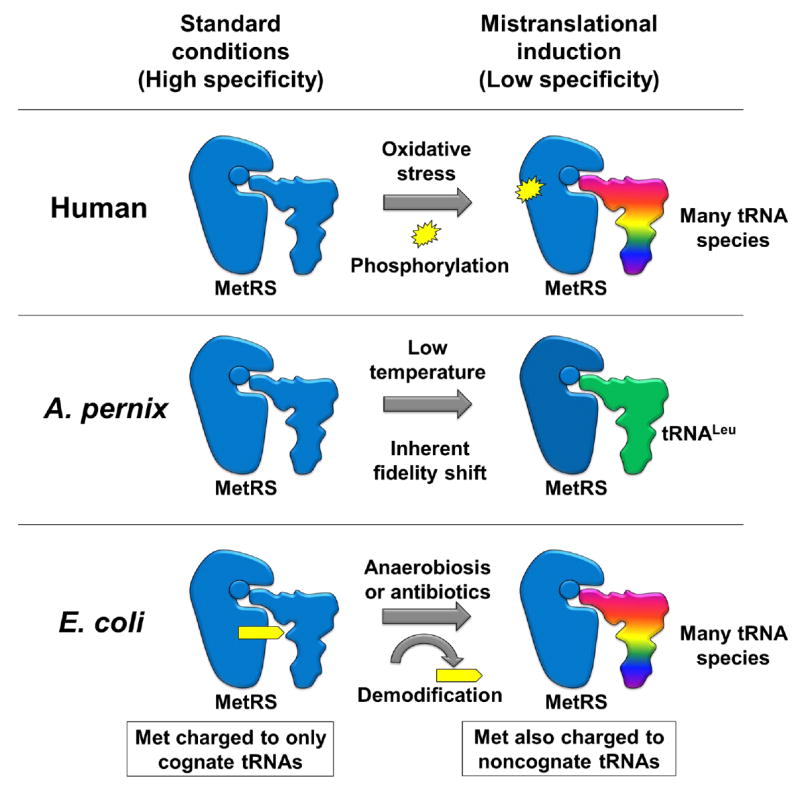
In humans, the unmodified MetRS has high specificity, but phosphorylation at two serine residues decreases the fidelity of the enzyme allowing it to charge noncognate tRNAs. In A. pernix, the MetRS is capable of undergoing a temperature dependent decrease in fidelity to specifically accept tRNALeu at lower physiological temperatures. In E. coli, the unmodified MetRS has low specificity and accepts noncognate tRNAs; a succinyllysine modification is required to impart high-tRNA charging fidelity. This modification is removed when Met mistranslation is induced in vivo.
Inducible tRNA misacylation with Met is also present in the archaeal domain of life. The archaeon Aeropyrum pernix is a natural inhabitant of deep sea hydrothermal vents and grows optimally at 90°C (Sako et al., 1996). Under standard laboratory growth at 90°C, no detectable tRNA misacylation with Met is observed. However, when A. pernix is grown at 75°C, Met is specifically misacylated to tRNALeu (Schwartz and Pan, 2016b). The fidelity of the A. pernix MetRS is regulated by temperature and it conditionally accepts tRNALeu at 75°C but not at 90°C in vitro (Figure 3). The activity of genetically encoded hyperthermophilic proteins from A. pernix may be significantly compromised at 75°C due to their extreme rigidity which precludes the degree of flexibly required for catalytic functions (Vihinen, 1987, Ladenstein and Antranikian, 1998). Accordingly, conditional Leu-to-Met mistranslation at 75°C can enhance enzymatic activity of hyperthermophilic proteins at lower temperatures (Schwartz and Pan, 2016b), likely by increasing conformational flexibility required for protein function (Vihinen, 1987, Gassner et al., 1996, Gassner et al., 2003). This destabilization mechanism for enhancing the low temperature activity of hyperthermophilic proteins is substantiated by the reduced activity of mistranslated A. pernix citrate synthase at higher temperatures compared to the wild-type counterparts (Schwartz and Pan, 2016b). The conditional low temperature mistranslation in A. pernix implicates a potential function for the low temperature mistranslation induced in Bacillus subtilis (Meyerovich et al., 2010).
Inducible mistranslation with Met also occurs in the bacterial domain. E. coli was shown to induce tRNA misacylation with Met during anaerobiosis and antibiotic stress, but exhibits high tRNA charging fidelity during standard aerobic growth (Schwartz et al., 2016). The E. coli MetRS has inherently low fidelity and accepts several nonmethionyl-tRNAs in vitro (Jones et al., 2011, Schwartz et al., 2016). However, a post-translational succinyl modification at either of two Lys residues along the tRNA binding interface of MetRS can confer high tRNA charging fidelity under optimal growth conditions. Succinyllysine demodification of MetRS allows E. coli to perform Met misacylation and subsequent mistranslation when prompted (Figure 3). Replacing either Lys residue with Glu imitates the terminal carboxyl-group of succinyllysine and results in MetRS enzymes of irreversibly high-fidelity both in vitro and in vivo (Lin et al., 2013, Schwartz et al., 2016). E. coli strains with chromosomal Lys-to-Glu MetRS mutations that produce endogenous, high fidelity MetRSs are less resistant to chemical stressors and antibiotics, which would induce tRNA misacylation in a wild-type strain. The broad increase in resistance to stressors and antibiotics is consistent with the merits of synthetic mistranslation that have been previously reported.
So far, every MetRS that has been assayed in eukaryotes (human, S. cerevisiae), bacteria (E. coli), and archaea (A. pernix) mischarges some noncognate tRNAs in vitro, which correlates with their propensity to perform Met mistranslation in vivo. The in vitro misacylation of noncognate tRNAs by MetRS can also be elevated by the addition of Ca2+ and it is likely that other conditions can also alter tRNA charging fidelity (Schwartz and Pan, 2016a). In the previous examples of tRNA mischarging described, an aaRS charges a noncognate amino acid to a cognate tRNA and the majority of mischarged tRNAs are eliminated by the editing domain of the same synthetase (Perona and Gruic-Sovulj, 2014). In contrast, during inducible Met mistranslation, the MetRS charges its cognate amino acid (Met) to noncognate tRNAs (Pan, 2013). Since no editing mechanisms have been identified that eliminate cognate amino acids charged to noncognate tRNAs, the MetRS cannot edit misacylated tRNAs with Met.
It is not a coincidence that nature has selected methionine to be the universal amino acid of choice for a conserved mistranslational response. Methionine residues can act as antioxidants within proteins through their ability to be reversibly oxidized and reduced (Levine et al., 1996, Levine et al., 2000, Delaye et al., 2007, Luo and Levine, 2009). In addition to being redox active, methionine has a side chain that is also both hydrophobic and polar and therefore can be utilized in both the interior and exterior of proteins (Brosnan and Brosnan, 2006). This chemical property of Met side chain reduces the potential for protein misfolding and aggregation that can result from mistranslation, especially in organisms that mistranslate Met for amino acids which reside on the surface of a protein (polar/charged) and in the core of a protein (hydrophobic).
The complete rubric that cells follow to incorporate tRNAs mischarged with Met into proteins is unclear. In E. coli, the utilization of mischarged tRNAs is thought to be partially dependent on their affinity for elongation factor Tu (EF-Tu) (Schrader et al., 2011), which could be either stronger or weaker than the affinity for the correctly acylated tRNA (LaRiviere et al., 2001). In theory, different tRNA species misacylated with Met could be used by the ribosome more or less readily depending on their respective EF-Tu affinities. However, many misacylated tRNAs are utilized almost identically as their correctly acylated counterparts by the ribosome and do not completely depend on EF-Tu affinities (Roy et al., 2007, Ruan et al., 2008, Effraim et al., 2009, Nair et al., 2016, Su et al., 2016), thus, ribosomal utilization of Met misacylated tRNAs is still difficult to predict. At this time, most of the methionine mistranslation events that have been identified substitute protein residues at the surface or at the oligomerization interface (Lee et al., 2014, Wang and Pan, 2015, Schwartz and Pan, 2016b).
The presence of methionine mistranslation in both prokaryotic domains indicates that this response likely evolved very early in evolution. In addition to arising early, Met mistranslation was perpetuated during the evolution from bacteria/archaea to eukaryotes (Gribaldo et al., 2010) and ostensibly found a suitable function in each domain of life. Therefore, despite the universal presence of Met mistranslation, this response is adapted for distinct functions in different organisms, and also occurs through distinct MetRS regulatory mechanisms. The examples of conditional mistranslation that have been described implicate many other potential benefits for mistranslation in diverse cellular contexts. Of particular significance, low temperature mistranslation in A. pernix demonstrates that inducible mistranslation can be used to specifically optimize a specific protein property through very selective amino acid substitutions to enhance activity in nonoptimal growth conditions (Schwartz and Pan, 2016b). In addition, this process demonstrates that the benefits of mistranslation can transcend the context of stress and subsequently implicates a fungible genetic code where protein sequences can be optimized via mistranslation for alternative growth conditions. Inducible mistranslation is an elegant method for acquiring the conditional benefits of mistranslation without compromising growth in all conditions—diverse examples of which are likely pervasive in the tree of life.
Benefits of mistranslation
The first and most obvious benefit for mistranslation is the acceleration of protein synthesis. There is an intrinsic trade-off between speed and accuracy during protein synthesis and the rapidity of translation is in part maintained by the permission of errors (Johansson et al., 2008, Wohlgemuth et al., 2010, Wohlgemuth et al., 2011). This potential is further exploited during certain types of amino acid limitations where relaxed translational fidelity can maintain or expedite protein synthesis via mistranslation (Parker et al., 1978, Parker and Friesen, 1980, Parker et al., 1980). However, this mistranslational benefit is a direct result of maintaining translational speed and is not employed to directly utilize the effects of amino acid substitutions within proteins. The second benefit for mistranslation is its potential to activate transcriptional stress responses. Both E. coli and S. cerevisiae have been shown to induce transcriptional stress responses during artificially generated protein mistranslation, which in turn confers a broad tolerance to stressors (Santos et al., 1999, Fan et al., 2015).
The most intriguing functions for mistranslation are the conditional benefits that mistranslation itself can confer. The basis for the phenotypic benefits of methionine mistranslation in both mammalian cells and the archaeon A. pernix is readily apparent. Methionine residues can act as antioxidants within proteins and increasing methionine incorporation in proteins can potentiate their antioxidant capacity, which may protect proteins sensitive to oxidative deactivation in mammalian cells (Levine et al., 1996, Temple et al., 2005, Luo and Levine, 2009). In A. pernix, the increased low temperature activity of mistranslated proteins is facilitated by the increase in protein flexibility conferred by Leu-to-Met substitutions (Gassner et al., 1996, Gassner et al., 2003).
The molecular basis for the increase in stress tolerance provided by mistranslation in other cases is less obvious, yet is likely the result of proteome diversification (Moura et al., 2009, Pan, 2013, Ribas de Pouplana et al., 2014). All genetically encoded or “wild-type” proteins have single optima for conditions such as temperature and pH, yet cells often occupy dynamic environments that venture outside the optimal functional range for protein activity. Proteome diversification could broaden the activity range of wild-type proteins (at the expense of function at the activity optimum) to potentially enhance protein function in nonstandard conditions (Figure 4). Obviously, this would not be advantageous in optimal growth conditions, where protein function is already at its pinnacle. Indeed, genetically engineered, mistranslating strains of different organisms universally have growth deficiencies when grown under optimal laboratory conditions. Furthermore, none of the inducible mistranslational processes are present at appreciable levels under optimal growth conditions. Yet, there is a potential benefit to protein mistranslation during conditions where the optimal function of the genetically encoded proteins is sufficiently hindered. Under these conditions, the activity optima of wild-type proteins cannot be utilized, so it can be advantageous to generate a collection of highly related protein variants with differential activity optima.
Figure 4. General effect of regulated and unregulated mistranslation on protein activity.
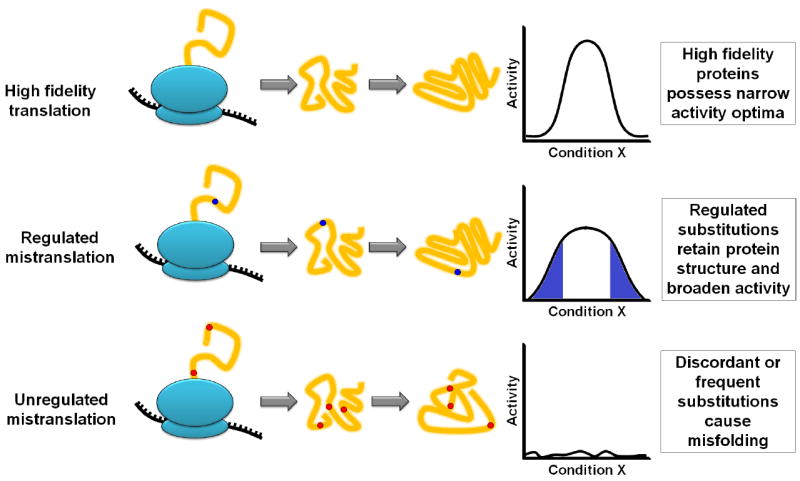
High fidelity translation produces a homogenous protein population that shares identical activity optima for all cellular conditions. Regulated mistranslation allows amino acid substitutions and their frequency to be controlled. This enables proteins to maintain their structures while diversifying their activities. The blue region in the graph indicates nonoptimal conditions where mistranslated proteins may have higher activity than their wild-type counterparts. Unregulated mistranslation can result in discordant amino acid substitutions that prevent proper protein folding resulting in protein inactivity.
The potential for mistranslational adaptability during nonoptimal growth conditions is mediated by the fact that amino acid substitutions can affect the function of a protein more than they affect the overall structure of a protein (Guo et al., 2004, Aharoni et al., 2005, Bloom et al., 2005). In Met mistranslation, single amino acid substitutions can have dramatic effects on protein function, activation, and intracellular localization (Wang and Pan, 2015). Furthermore, although the detriments of mistranslation have a fairly definitive molecular consequence (i.e. protein inactivation and misfolding), the potential means by which protein activity can be altered through amino acid substitutions is vastly more expansive. The benefits conferred by protein mistranslation can also be independent of direct changes in a protein’s activity. For example, mistranslation of surface proteins could generate cell surface variation to aid evasion of host immune responses in the Mycoplasma, Candida, and Mycobacterial pathogens that employ constitutive mistranslation.
In addition to general protein diversification, mistranslation has the potential to alter a specific property of genetically encoded proteins though specific amino acid substitutions that favor the mistranslated amino acid residues to be integrated at advantageous locations. This possibility was validated in A. pernix, which only mischarges methionine to tRNALeu, thus ensuring that Met will only be mistranslated for Leu residues. Conceivably, countless combinations of amino acid substitutions could be used to specifically alter genetically encoded proteins in different manners to optimize their function in diverse conditions without the burden of additional genes. This strategy would also minimize the effects of discordant amino acid substitutions which would facilitate protein misfolding and aggregation.
In addition to genetically encoded proteins having single optima for activity and function, they also share identical susceptilities. Protein diversification via mistranslation could allow a subset of proteins to continue their function in the presence of an agent that completely inactivates the wild-type protein. For example, the Mycobacterial RNA polymerase can gain resistance to its inactivating agent rifampicin via mistranslation (Javid et al., 2014). This principle is conceptually similar to the organismal diversification within a population, except on a molecular scale. Most individuals or proteins function perfectly well in conditions to which they are well adapted, yet the population of individuals or proteins will perish if a subset is not better adapted to pernicious environmental changes. Some protein variations will have decreased activity or may be inactive, yet this effect may be inconsequential if cellular conditions have already sufficiently inhibited the wild-type protein (as during intense rifampicin treatment). Although it is yet to be determined how many persisting functional copies of a given essential protein would be required for continued growth and survival during a detrimental stress, mistranslation may generate a sufficient number of crucial protein variants capable of maintaining their function in conditions which may fully inactivate the wild-type protein. The mistranslational response can also function on a much quicker time scale than transcriptional responses and may serve as a solution to conditions for which an effective transcriptional response does not exist or would be too costly to maintain for an infrequent demand.
The evolution of intentional mistranslation ultimately contributes to the biological strategy of perpetuating life through the generation of diversity on many scales. Diversity in the tree of life permeates all levels of the biological hierarchy and regulated mistranslation provides the means to generate diversity on the protein molecular scale. In an entirely static environment that never ventures outside optimal conditions, diversity on any scale would be undesirable since a single modality would be optimal and deviations from this pinnacle would likely be deleterious. However, all environments contain variations, and in many cases they cannot be anticipated or avoided. In the absence of diversity during unfavorable conditions, all members of a population—organismal or proteinaceous—may be impaired under a change that is sufficiently detrimental. Therefore, life employs biological diversity at the organismal or protein molecule scale to generate biological entities with different abilities and susceptibilities as a preemptive mechanism for enduring unanticipated changes. We are just now beginning to appreciate that this principle may be just as applicable at the molecular level.
Conclusions
Mistranslation is a complex process that can be engendered in the many steps required to synthesize proteins (Figure 1). The process of mistranslation describes 1,216 (64 × 19) possible codon-to-amino-acid events that can have a vast array of effects on protein function depending on the type, location, and degree of substitution. Mistranslation has generally been assumed to be an inexorable consequence of rapid protein synthesis that exerts deleterious effects. Indeed, unregulated mistranslation can obviously make discordant amino acid substitutions that promote protein misfolding and aggregation. However, regulated mistranslation allows cells to control the potential detriments associated with mistranslation. There are now numerous examples of cells performing mistranslation in regulated fashions. Furthermore, various benefits have been demonstrated for mistranslation, revealing that mistranslation can be advantageous under certain conditions. This review aimed to emphasize that mistranslation can have both diverse origins and effects, yet the overall functional consequence of a specific mistranslational process largely depends on the cellular context. Mistranslation is neither universally beneficial nor deleterious: it is a multifaceted process that has innumerable variations and degrees which have the potential to exert diverse effects at both the molecular and cellular levels. We are now beginning to appreciate that some of these effects are advantageous under specific conditions, which are mostly nonoptimal for the function of the genetically encoded, “wild-type” proteins (Figure 5). Like most aspects of biology, regulation of translational fidelity is far more complex than we initially expected. However, the intricacies of intentional mistranslation are revealing exciting new methods of cellular adaption that have the potential to reshape our views on the major objectives of protein synthesis.
Figure 5. Specific roles for mistranslation.
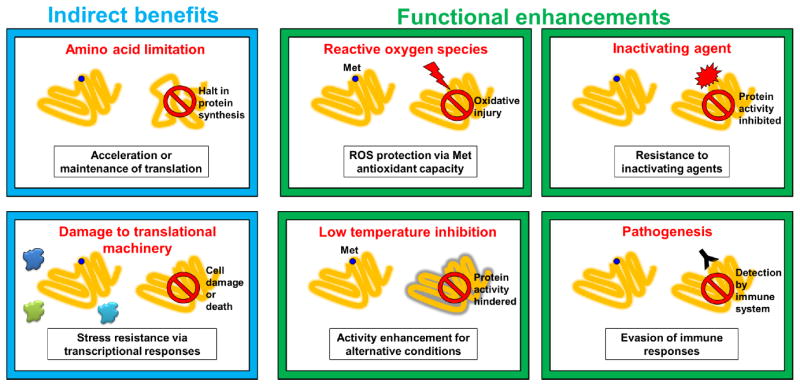
Mistranslation can have benefits independent of the direct effects of substituting amino acids within proteins such as expediting/maintaining translation during amino acid limitation and activating transcriptional stress responses (blue). Substituting amino acids within proteins can result in novel protein phenotypes that have shown to be beneficial in several ways (green). An increase in Met residues within proteins can aid the oxidative stress response in mammalian cells, likely by protecting proteins from oxidative inactivation. Mistranslation can render proteins resistant to a specific inactivating agent like rifampicin in bacteria. Mistranslation can alter a specific protein property such as rigidity to increase function in lower temperatures in archaea. Lastly, mistranslation can mediate variability of surface proteins to aid evasion of immune responses in pathogens.
Acknowledgments
This work is supported by the NIH MCB Training Grant (T32 GM007183) to M.S. and the NIH Director’s Pioneer Award (DP1GM105386) to T.P.
Footnotes
Disclosure statement
The authors declare no conflicts of interest.
References
- Aharoni A, Gaidukov L, Khersonsky O, Mc QGS, Roodveldt C, Tawfik DS. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005;37:73–6. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- Anand N, Davis BD. Damage by streptomycin to the cell membrane of Escherichia coli. Nature. 1960;185:22–3. doi: 10.1038/185022a0. [DOI] [PubMed] [Google Scholar]
- Anand N, Davis BD, Armitage AK. Uptake of streptomycin by Escherichia coli. Nature. 1960;185:23–4. doi: 10.1038/185023a0. [DOI] [PubMed] [Google Scholar]
- Bacher JM, De Crecy-Lagard V, Schimmel PR. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc Natl Acad Sci U S A. 2005;102:1697–701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher JM, Waas WF, Metzgar D, De Crecy-Lagard V, Schimmel P. Genetic code ambiguity confers a selective advantage on Acinetobacter baylyi. J Bacteriol. 2007;189:6494–6. doi: 10.1128/JB.00622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin C, Weston LA, Huang T, Jander G, Owens T, Meinwald J, Schroeder FC. Grass roots chemistry: meta-tyrosine, an herbicidal nonprotein amino acid. Proc Natl Acad Sci U S A. 2007;104:16964–9. doi: 10.1073/pnas.0707198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra AR, Simoes J, Lee W, Rung J, Weil T, Gut IG, Gut M, Bayes M, Rizzetto L, Cavalieri D, Giovannini G, Bozza S, Romani L, Kapushesky M, Moura GR, Santos MA. Reversion of a fungal genetic code alteration links proteome instability with genomic and phenotypic diversification. Proc Natl Acad Sci U S A. 2013;110:11079–84. doi: 10.1073/pnas.1302094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank A, Gallant JA, Burgess RR, Loeb LA. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986;25:5920–8. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- Bloom JD, Silberg JJ, Wilke CO, Drummond DA, Adami C, Arnold FH. Thermodynamic prediction of protein neutrality. Proc Natl Acad Sci U S A. 2005;102:606–11. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan JT, Brosnan ME. The sulfur-containing amino acids: an overview. J Nutr. 2006;136:1636S–1640S. doi: 10.1093/jn/136.6.1636S. [DOI] [PubMed] [Google Scholar]
- Bullwinkle TJ, Ibba M. Translation quality control is critical for bacterial responses to amino acid stress. Proc Natl Acad Sci U S A. 2016;113:2252–7. doi: 10.1073/pnas.1525206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullwinkle TJ, Reynolds NM, Raina M, Moghal A, Matsa E, Rajkovic A, Kayadibi H, Fazlollahi F, Ryan C, Howitz N, Faull KF, Lazazzera BA, Ibba M. Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. Elife. 2014:3. doi: 10.7554/eLife.02501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–80. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Hill C. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol Rev. 2003;67:429–53. doi: 10.1128/MMBR.67.3.429-453.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Soll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci U S A. 1997;94:11819–26. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale T, Uhlenbeck OC. Amino acid specificity in translation. Trends Biochem Sci. 2005;30:659–65. doi: 10.1016/j.tibs.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Davies J, Gilbert W, Gorini L. Streptomycin, Suppression, and the Code. Proc Natl Acad Sci U S A. 1964;51:883–90. doi: 10.1073/pnas.51.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BD. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev. 1987;51:341–50. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BD, Chen LL, Tai PC. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc Natl Acad Sci U S A. 1986;83:6164–8. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaye L, Becerra A, Orgel L, Lazcano A. Molecular evolution of peptide methionine sulfoxide reductases (MsrA and MsrB): on the early development of a mechanism that protects against oxidative damage. J Mol Evol. 2007;64:15–32. doi: 10.1007/s00239-005-0281-2. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–52. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–24. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton NR, Caffrey R. Effect of dihydrostreptomycin on Escherichia coli. J Bacteriol. 1961;81:918–23. doi: 10.1128/jb.81.6.918-923.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann P, Gallant J. Mistranslation in E. coli. Cell. 1977;10:131–7. doi: 10.1016/0092-8674(77)90147-7. [DOI] [PubMed] [Google Scholar]
- Effraim PR, Wang J, Englander MT, Avins J, Leyh TS, Gonzalez RL, Jr, Cornish VW. Natural amino acids do not require their native tRNAs for efficient selection by the ribosome. Nat Chem Biol. 2009;5:947–53. doi: 10.1038/nchembio.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Wu J, Ung MH, De Lay N, Cheng C, Ling J. Protein mistranslation protects bacteria against oxidative stress. Nucleic Acids Res. 2015;43:1740–8. doi: 10.1093/nar/gku1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Mcfadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–82. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Gassner NC, Baase WA, Matthews BW. A test of the “jigsaw puzzle” model for protein folding by multiple methionine substitutions within the core of T4 lysozyme. Proc Natl Acad Sci U S A. 1996;93:12155–8. doi: 10.1073/pnas.93.22.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassner NC, Baase WA, Mooers BH, Busam RD, Weaver LH, Lindstrom JD, Quillin ML, Matthews BW. Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability. Biophys Chem. 2003;100:325–40. doi: 10.1016/s0301-4622(02)00290-9. [DOI] [PubMed] [Google Scholar]
- Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–35. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AC, Miranda I, Silva RM, Moura GR, Thomas B, Akoulitchev A, Santos MA. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 2007;8:R206. doi: 10.1186/gb-2007-8-10-r206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat Rev Microbiol. 2010;8:743–52. doi: 10.1038/nrmicro2426. [DOI] [PubMed] [Google Scholar]
- Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc Natl Acad Sci U S A. 2004;101:9205–10. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurer-Orhan H, Ercal N, Mare S, Pennathur S, Orhan H, Heinecke JW. Misincorporation of free m-tyrosine into cellular proteins: a potential cytotoxic mechanism for oxidized amino acids. Biochem J. 2006;395:277–84. doi: 10.1042/BJ20051964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale SP, Auld DS, Schmidt E, Schimmel P. Discrete determinants in transfer RNA for editing and aminoacylation. Science. 1997;276:1250–2. doi: 10.1126/science.276.5316.1250. [DOI] [PubMed] [Google Scholar]
- Hendrickson TL, Nomanbhoy TK, De Crecy-Lagard V, Fukai S, Nureki O, Yokoyama S, Schimmel P. Mutational separation of two pathways for editing by a class I tRNA synthetase. Mol Cell. 2002;9:353–62. doi: 10.1016/s1097-2765(02)00449-5. [DOI] [PubMed] [Google Scholar]
- Ibba M, Soll D. Quality control mechanisms during translation. Science. 1999;286:1893–7. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- Javid B, Sorrentino F, Toosky M, Zheng W, Pinkham JT, Jain N, Pan M, Deighan P, Rubin EJ. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc Natl Acad Sci U S A. 2014;111:1132–7. doi: 10.1073/pnas.1317580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Lovmar M, Ehrenberg M. Rate and accuracy of bacterial protein synthesis revisited. Curr Opin Microbiol. 2008;11:141–7. doi: 10.1016/j.mib.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Johansson M, Zhang J, Ehrenberg M. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc Natl Acad Sci U S A. 2012;109:131–6. doi: 10.1073/pnas.1116480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TE, Alexander RW, Pan T. Misacylation of specific nonmethionyl tRNAs by a bacterial methionyl-tRNA synthetase. Proc Natl Acad Sci U S A. 2011;108:6933–8. doi: 10.1073/pnas.1019033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalapis D, Bezerra AR, Farkas Z, Horvath P, Bodi Z, Daraba A, Szamecz B, Gut I, Bayes M, Santos MA, Pal C. Evolution of Robustness to Protein Mistranslation by Accelerated Protein Turnover. PLoS Biol. 2015;13:e1002291. doi: 10.1371/journal.pbio.1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipcan L, Moor N, Kessler N, Safro MG. Eukaryotic cytosolic and mitochondrial phenylalanyl-tRNA synthetases catalyze the charging of tRNA with the meta-tyrosine. Proc Natl Acad Sci U S A. 2009;106:11045–8. doi: 10.1073/pnas.0905212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–90. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotra LP, Haddad J, Mobashery S. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob Agents Chemother. 2000;44:3249–56. doi: 10.1128/aac.44.12.3249-3256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010;16:1797–808. doi: 10.1261/rna.2201210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–8. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- Ladenstein R, Antranikian G. Proteins from hyperthermophiles: stability and enzymatic catalysis close to the boiling point of water. Adv Biochem Eng Biotechnol. 1998;61:37–85. doi: 10.1007/BFb0102289. [DOI] [PubMed] [Google Scholar]
- Lariviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–8. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- Ledoux S, Olejniczak M, Uhlenbeck OC. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat Struct Mol Biol. 2009;16:359–64. doi: 10.1038/nsmb.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–5. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Lee JY, Kim DG, Kim BG, Yang WS, Hong J, Kang T, Oh YS, Kim KR, Han BW, Hwang BJ, Kang BS, Kang MS, Kim MH, Kwon NH, Kim S. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J Cell Sci. 2014;127:4234–45. doi: 10.1242/jcs.152470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng T, Pan M, Xu X, Javid B. Translational misreading in Mycobacterium smegmatis increases in stationary phase. Tuberculosis (Edinb) 2015 doi: 10.1016/j.tube.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Levine RL, Moskovitz J, Stadtman ER. Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. IUBMB Life. 2000;50:301–7. doi: 10.1080/713803735. [DOI] [PubMed] [Google Scholar]
- Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci U S A. 1996;93:15036–40. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Boniecki MT, Jaffe JD, Imai BS, Yau PM, Luthey-Schulten ZA, Martinis SA. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci U S A. 2011;108:9378–83. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB, Jin W, Huang HH, Chen X. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun. 2013;441:191–5. doi: 10.1016/j.bbrc.2013.10.033. [DOI] [PubMed] [Google Scholar]
- Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- Ling J, Soll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci U S A. 2010;107:4028–33. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Satz JS, Vo MN, Nangle LA, Schimmel P, Ackerman SL. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci U S A. 2014;111:17570–5. doi: 10.1073/pnas.1420196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972;128:1353–6. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Bergert M, Walther A, Suter B. Double-sieving-defective aminoacyl-tRNA synthetase causes protein mistranslation and affects cellular physiology and development. Nat Commun. 2014;5:5650. doi: 10.1038/ncomms6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J. 2009;23:464–72. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerovich M, Mamou G, Ben-Yehuda S. Visualizing high error levels during gene expression in living bacterial cells. Proc Natl Acad Sci U S A. 2010;107:11543–8. doi: 10.1073/pnas.0912989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda I, Rocha R, Santos MC, Mateus DD, Moura GR, Carreto L, Santos MA. A genetic code alteration is a phenotype diversity generator in the human pathogen Candida albicans. PLoS One. 2007;2:e996. doi: 10.1371/journal.pone.0000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda I, Silva-Dias A, Rocha R, Teixeira-Santos R, Coelho C, Goncalves T, Santos MA, Pina-Vaz C, Solis NV, Filler SG, Rodrigues AG. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. MBio. 2013:4. doi: 10.1128/mBio.00285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura GR, Carreto LC, Santos MA. Genetic code ambiguity: an unexpected source of proteome innovation and phenotypic diversity. Curr Opin Microbiol. 2009;12:631–7. doi: 10.1016/j.mib.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Murakami H, Ohta A, Suga H. Bases in the anticodon loop of tRNA(Ala)(GGC) prevent misreading. Nat Struct Mol Biol. 2009;16:353–8. doi: 10.1038/nsmb.1580. [DOI] [PubMed] [Google Scholar]
- Nair N, Raff H, Islam MT, Feen M, Garofalo DM, Sheppard K. The Bacillus subtilis and Bacillus halodurans Aspartyl-tRNA Synthetases Retain Recognition of tRNA(Asn) J Mol Biol. 2016;428:618–30. doi: 10.1016/j.jmb.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13:1091–100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, Embry A, Dolan B, Das S, Hickman HD, Berglund P, Bennink JR, Yewdell JW, Pan T. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–6. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–77. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu Rev Genet. 2013;47:121–37. doi: 10.1146/annurev-genet-111212-133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989;53:273–98. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Friesen JD. “Two out of three” codon reading leading to mistranslation in vivo. Mol Gen Genet. 1980;177:439–45. doi: 10.1007/BF00271482. [DOI] [PubMed] [Google Scholar]
- Parker J, Johnston TC, Borgia PT. Mistranslation in cells infected with the bacteriophage MS2: direct evidence of Lys for Asn substitution. Mol Gen Genet. 1980;180:275–81. doi: 10.1007/BF00425839. [DOI] [PubMed] [Google Scholar]
- Parker J, Pollard JW, Friesen JD, Stanners CP. Stuttering: high-level mistranslation in animal and bacterial cells. Proc Natl Acad Sci U S A. 1978;75:1091–5. doi: 10.1073/pnas.75.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona JJ, Gruic-Sovulj I. Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. Top Curr Chem. 2014;344:1–41. doi: 10.1007/128_2013_456. [DOI] [PubMed] [Google Scholar]
- Pezo V, Metzgar D, Hendrickson TL, Waas WF, Hazebrouck S, Doring V, Marliere P, Schimmel P, De Crecy-Lagard V. Artificially ambiguous genetic code confers growth yield advantage. Proc Natl Acad Sci U S A. 2004;101:8593–7. doi: 10.1073/pnas.0402893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina M, Moghal A, Kano A, Jerums M, Schnier PD, Luo S, Deshpande R, Bondarenko PV, Lin H, Ibba M. Reduced amino acid specificity of mammalian tyrosyl-tRNA synthetase is associated with elevated mistranslation of Tyr codons. J Biol Chem. 2014;289:17780–90. doi: 10.1074/jbc.M114.564609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds NM, Ling J, Roy H, Banerjee R, Repasky SE, Hamel P, Ibba M. Cell-specific differences in the requirements for translation quality control. Proc Natl Acad Sci U S A. 2010;107:4063–8. doi: 10.1073/pnas.0909640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas De Pouplana L, Santos MA, Zhu JH, Farabaugh PJ, Javid B. Protein mistranslation: friend or foe? Trends Biochem Sci. 2014;39:355–62. doi: 10.1016/j.tibs.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Rodnina MV. Quality control of mRNA decoding on the bacterial ribosome. Adv Protein Chem Struct Biol. 2012;86:95–128. doi: 10.1016/B978-0-12-386497-0.00003-7. [DOI] [PubMed] [Google Scholar]
- Roy H, Becker HD, Mazauric MH, Kern D. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res. 2007;35:3420–30. doi: 10.1093/nar/gkm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan B, Palioura S, Sabina J, Marvin-Guy L, Kochhar S, Larossa RA, Soll D. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci U S A. 2008;105:16502–7. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusala T, Andersson D, Ehrenberg M, Kurland CG. Hyper-accurate ribosomes inhibit growth. EMBO J. 1984;3:2575–80. doi: 10.1002/j.1460-2075.1984.tb02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Nomura N, Uchida A, Ishida Y, Morii H, Koga Y, Hoaki T, Maruyama T. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 degrees C. Int J Syst Bacteriol. 1996;46:1070–7. doi: 10.1099/00207713-46-4-1070. [DOI] [PubMed] [Google Scholar]
- Santos MA, Cheesman C, Costa V, Moradas-Ferreira P, Tuite MF. Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp. Mol Microbiol. 1999;31:937–47. doi: 10.1046/j.1365-2958.1999.01233.x. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Ramakrishnan V. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat Struct Mol Biol. 2011;18:432–6. doi: 10.1038/nsmb.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader JM, Chapman SJ, Uhlenbeck OC. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc Natl Acad Sci U S A. 2011;108:5215–20. doi: 10.1073/pnas.1102128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MH, Pan T. Determining the fidelity of tRNA aminoacylation via microarrays. Methods. 2016a doi: 10.1016/j.ymeth.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MH, Pan T. Temperature dependent mistranslation in a hyperthermophile adapts proteins to lower temperatures. Nucleic Acids Res. 2016b;44:294–303. doi: 10.1093/nar/gkv1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MH, Waldbauer JR, Z L, Pan T. Global tRNA misacylation induced by anaerobiosis and antibiotic exposure broadly increases stress resistance in Escherichia coli. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Soll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–25. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–18. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- Su HW, Zhu JH, Li H, Cai RJ, Ealand C, Wang X, Chen YX, Kayani MU, Zhu TF, Moradigaravand D, Huang H, Kana BD, Javid B. The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat Microbiol. 2016;1:16147. doi: 10.1038/nmicrobiol.2016.147. [DOI] [PubMed] [Google Scholar]
- Sun L, Gomes AC, He W, Zhou H, Wang X, Pan DW, Schimmel P, Pan T, Yang XL. Evolutionary Gain of Alanine Mischarging to Noncognate tRNAs with a G4:U69 Base Pair. J Am Chem Soc. 2016;138:12948–12955. doi: 10.1021/jacs.6b07121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ueda T, Watanabe K. The ‘polysemous’ codon--a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 1997;16:1122–34. doi: 10.1093/emboj/16.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple MD, Perrone GG, Dawes IW. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005;15:319–26. doi: 10.1016/j.tcb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Vihinen M. Relationship of protein flexibility to thermostability. Protein Eng. 1987;1:477–80. doi: 10.1093/protein/1.6.477. [DOI] [PubMed] [Google Scholar]
- Wang X, Pan T. Methionine Mistranslation Bypasses the Restraint of the Genetic Code to Generate Mutant Proteins with Distinct Activities. PLoS Genet. 2015;11:e1005745. doi: 10.1371/journal.pgen.1005745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MH, Marahiel MA. Bacterial cold shock responses. Sci Prog. 2003;86:9–75. doi: 10.3184/003685003783238707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B, Davies J. Antibiotic inhibitors of the bacterial ribosome. Bacteriol Rev. 1968;32:493–528. [PMC free article] [PubMed] [Google Scholar]
- Wiltrout E, Goodenbour JM, Frechin M, Pan T. Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:10494–506. doi: 10.1093/nar/gks805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Pohl C, Mittelstaet J, Konevega AL, Rodnina MV. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos Trans R Soc Lond B Biol Sci. 2011;366:2979–86. doi: 10.1098/rstb.2011.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Pohl C, Rodnina MV. Optimization of speed and accuracy of decoding in translation. EMBO J. 2010;29:3701–9. doi: 10.1038/emboj.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Fan Y, Ling J. Mechanism of oxidant-induced mistranslation by threonyl-tRNA synthetase. Nucleic Acids Res. 2014;42:6523–31. doi: 10.1093/nar/gku271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadavalli SS, Ibba M. Selection of tRNA charging quality control mechanisms that increase mistranslation of the genetic code. Nucleic Acids Res. 2013;41:1104–12. doi: 10.1093/nar/gks1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–62. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol Cell. 2010;39:110–20. doi: 10.1016/j.molcel.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ieong KW, Johansson M, Ehrenberg M. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. Proc Natl Acad Sci U S A. 2015;112:9602–7. doi: 10.1073/pnas.1506823112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ieong KW, Mellenius H, Ehrenberg M. Proofreading neutralizes potential error hotspots in genetic code translation by transfer RNAs. RNA. 2016;22:896–904. doi: 10.1261/rna.055632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]


