Abstract
In mammals, the cell surface receptors encoded by the leukocyte receptor complex (LRC) regulate the activity of T lymphocytes and B lymphocytes, as well as that of natural killer cells, and thus provide protection against pathogens and parasites. The chicken genome encodes many Ig-like receptors that are homologous to the LRC receptors. The chicken Ig-like receptor (CHIR) genes are members of a large monophyletic gene family and are organized into genomic clusters, which are in conserved synteny with the mammalian LRC. One-third of CHIR genes encode polypeptide molecules that contain both activating and inhibitory motifs. These genes are present in different phylogenetic groups, suggesting that the primordial CHIR gene could have encoded both types of motifs in a single molecule. In contrast to the mammalian LRC genes, the CHIR genes with similar function (inhibition or activation) are evolutionarily closely related. We propose that, in addition to recombination, single nucleotide substitutions played an important role in the generation of receptors with different functions. Structural models and amino acid analyses of the CHIR proteins reveal the presence of different types of Ig-like domains in the same phylogenetic groups, as well as sharing of conserved residues and conserved changes of residues between different CHIR groups and between CHIRs and LRCs. Our data support the notion that the CHIR gene clusters are regions homologous to the mammalian LRC gene cluster and favor a model of evolution by repeated processes of birth and death (expansion–contraction) of the Ig-like receptor genes.
Keywords: birth and death, conserved synteny, genomic organization, Ig-like receptors
Natural killer (NK) cells are a subpopulation of lymphocytes that function in innate immunity by recognizing and destroying virally infected and cancerous cells (1). NK cells are regulated by the interaction of inhibitory and activating signals emitted by cell surface receptors. The inhibitory receptors contain immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in their cytoplasmic (CYT) region, whereas the activating receptors contain a positively charged residue in their transmembrane (TM) region (2). The mammalian NK cell receptors fall into two categories, one belonging to the Ig superfamily and the other to the C-type lectin superfamily. The Ig-like receptors are encoded by a genomic region called the leukocyte receptor complex (LRC) (3). The LRC contains several gene families [e.g., the killer cell Ig-like receptors (KIRs), the leukocyte Ig-like receptors (LILRs), and the paired Ig-like receptors] and singleton genes. The presence of the LRC in all mammals studied thus far and the overall structural similarities of the LRC genes suggest that all of these genes have evolved from a common ancestral sequence and that the LRC region was formed before the mammalian radiation.
The common origin of the LRC genes is also supported by the existence of several homologous sequences in the chicken (4–7). It has been proposed that the Ig-like domains of the chicken Ig-like receptors (CHIR) are most closely related to the mammalian LRC domain sequences. These domains form two evolutionary groups that probably diverged before the separation of the mammalian and bird lineages (7). At present, however, the number and the overall organization of the CHIR genes are not known. Here, we describe the genomic organization of the CHIR genes and the evolutionary relationships between the members of the CHIR multigene family and those of the LRC.
Methods
Identification and Analysis of the CHIR Genomic Regions. We used the chicken sequences identified in a previous study (7) in blastn and tblastn similarity searches using default parameters (8) against the chicken whole genome shotgun database (9), the nucleotide database, the nonredundant database, and the high-throughput genomic sequences of the National Center for Biotechnology Information (NCBI). We also searched the EST database of NCBI and The Institute for Genomic Research (www.tigr.org/tdb/tgi). Overlapping genomic regions were identified by using blastn. CHIR genes were identified by using genomescan (10), and all gene predictions were manually corrected by taking into account pairwise alignments of the genomic DNA with full cDNAs and ESTs, as described in ref. 11. (The complete annotation is available from M.N. upon request.) Domain architecture analysis was performed by using the SMART (12) and PFAM (13) databases.
Sequence and Phylogenetic Analyses. The coding sequences were extracted exon by exon and were aligned by using the profile alignment option of clustalx 1.81 (14). Phylogenetic trees were constructed by using the neighbor-joining (NJ) method with p distances (proportion of differences) [mega 2.1 (15)]. The p distances are known to give a higher resolution of branching pattern because of the smaller standard errors (16). We also constructed maximum parsimony trees, but because they were essentially the same as the NJ trees with respect to the major branching patterns, they will not be presented here.
Logos of sequence conservation were generated by using weblogo (17). The multiple sequence alignments can be found in logo format in Figs. 5–7, which are published as supporting information on the PNAS web site. Theoretical models of representative CHIR Ig-like domains were predicted by using homology modeling as it is implemented in the Swiss-Model (18) and the 3DPSS servers (19), and figures were drawn by using pymol (http://pymol.sourceforge.net).
Results
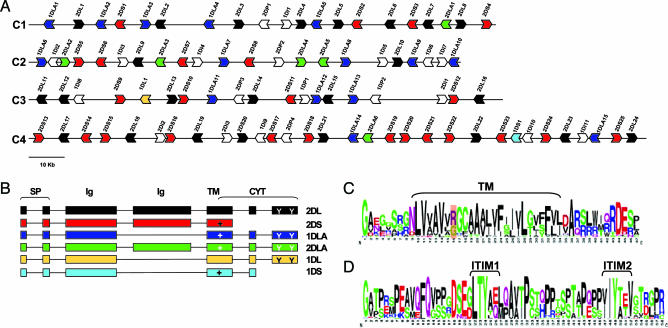
Characterization of the CHIR Genomic Regions. We have identified seven high-throughput genomic sequences of the chicken genome (phase 3) present in the nucleotide database of the NCBI (Aug. 29, 2004) that display significant alignments with the CHIR genes. These seven clones could be assembled into four nonoverlapping contigs (C1–C4 in Fig. 1A) ranging from 120 to 170 kb. The accession numbers of the clones were as follows: C1, BX663529; C2, BX663530 (containing the entire BX897752 clone); C3, BX663526 and BX663523; C4, BX663527 and BX663534. Three of the seven clones have recently been confirmed experimentally to encode at least four CHIR genes. It is not known, however, whether these clones reside on the same or different chromosomes (see ref. 6 and information for the clones at www.animalsciences.nl/chickfpc). The four contigs (C1–C4) contain, on average, one gene per 5 kb (including partial sequences that encode at least three exons). Comparisons with other randomly selected genomic regions of the chicken genome (from both micro- and macrochromosomes) showed that the average gene number ranges from one gene per 40 kb to one gene per 200 kb. The gene density of the CHIR regions is comparable to that of the major histocompatibility complex region in the chicken, which contains one gene per 4.4 kb (20).
Fig. 1.
The CHIR gene family. (A) Genomic organization of the CHIR genes. Each arrowhead represents a single gene, and direction of arrowhead shows transcriptional orientation. Genes with similar color have similar structure (see B). Truncated sequences and pseudogenes are shown as white arrowheads. Only sequences homologous to CHIR genes encoding more than three exons are shown. (B) Structure of the CHIR genes. Six types of different CHIR genes were identified. Boxes represent exons, and lines represent introns. Plus sign indicates the presence of the positive residue in the TM region, and Y indicates the ITIM motifs in the CYT region. Coloring of genes corresponds to that in A. (C) Conservation of the TM (in brackets) and CYT regions presented in a logo format. The position of the positive residue is highlighted. (D) Conservation of the CYT region presented in a logo format. The two ITIMs are shown in brackets. Note that alignment gaps were excluded.
Genes Identified and CHIR Nomenclature. The four genomic contigs (C1–C4) encode >120 genes and gene fragments (Fig. 1 A). The majority of the genes are homologous to the CHIR genes, and some of these had been reported previously (4, 6, 7). The remaining genes probably represent homologues of the mammalian G protein-coupled receptors 40–43 (GPR40–43), which, in humans and mice, reside in the “extended” LRC region (3).
For the purpose of this study, we annotated the CHIR homologues, and we further analyzed only those CHIR sequences that putatively encode functional proteins. More specifically, genes that code for a signal peptide, complete Ig-like domains, a TM region, and a CYT tail were considered as putative functional genes (see also refs. 4, 6, and 7). The remaining CHIR-related genes could either represent pseudogenes, because, for some of them, the coding region is interrupted by stop codons, or appear truncated because of assembly errors. The overall gene structure of the 70 putative functional CHIR genes identified resembles that of the mammalian LRC genes (Fig. 1B). In contrast to the mammalian KIR, LILR, and paired Ig-like receptor gene families, the members of the CHIR gene family do not have the same transcriptional orientation (Fig. 1 A and refs. 2 and 3).
In naming the CHIR genes, we followed the nomenclature proposed for the human KIR genes (21). Specifically, the genes are named 2DL if they encode two Ig-like domains (2D) and a long CYT tail (L) and are named 2DS if they encode two Ig-like domains (2D) and a short CYT tail (S). The 2DS genes code for a positively charged TM residue (Fig. 1C) and thus specify activating receptors. The 2DL genes encode one or two ITIMs in their CYT region (Fig. 1D) and can thus specify inhibitory receptors. Almost one-third of the CHIR proteins contain both a positive TM residue and a long CYT tail with ITIMs. Their function is not known. We call these genes 1DLA or 2DLA, because they encode one or two Ig-like domains, a long CYT tail (L), and a charged TM residue (A) (Fig. 1B).
To test the possibility that the contigs may contain allelic regions, we compared the genomic organization and the percentage similarity for the CHIR and the G protein-coupled receptor (GPR) sequences between and within the four contigs. Our analyses showed that: (i) the CHIR genes of the different contigs did not have the same transcription orientation, (ii) the average percentage of nucleotide identity was ≈75 (Fig. 1 A and data not shown), and (iii) the average identity between the GPR sequences was 77% at the amino acid level. Although these observations suggest that the four contigs may represent different genomic regions, we cannot exclude the possibility that they represent allelic genomic regions, because it has been shown in primates that different KIR haplotypes contain different numbers of genes in different combinations (22).
Homology Between the CHIR and the LRC Genes. Phylogenetic analysis of the CHIR, mammalian LRC, and mammalian Fc receptor sequences indicates that the three types of receptor genes form three monophyletic groups and that the CHIR group is closely related to the group of mammalian LRC genes (see Fig. 8, which is published as supporting information on the PNAS web site, and refs. 4, 6, and 7). These observations suggest that the CHIR and LRC sequences shared a common ancestor and that the CHIR gene family has been expanded by gene duplication.
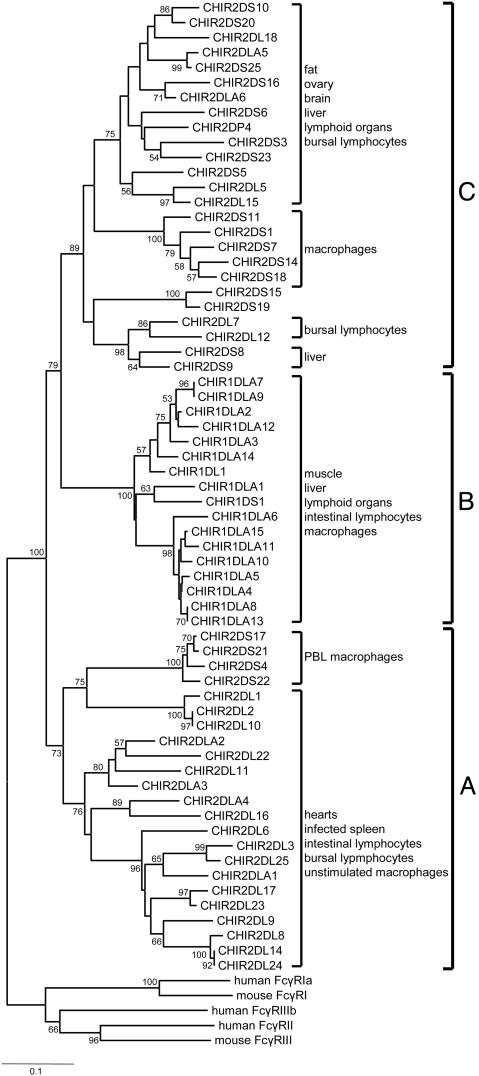
To gain insight into the expression patterns of the CHIR genes, 35 unique EST sequences were identified (Fig. 2). The results suggest that the CHIR genes, like the LRC genes, are expressed in tissues and cells involved in immunity (lymphoid organs, leukocytes, and macrophages) and provide an additional link between the LRC and CHIR genes.
Fig. 2.
Neighbor-joining tree of the D1 Ig-like domain of the CHIR sequences. The tree was constructed with p distances for 90-aa sites after elimination of alignment gaps. The numbers on the interior branches represent bootstrap values (only values >50 are shown). The sequences of the mammalian Fc receptors were used as outgroups (see also figure 1 of ref. 7). The expression patterns (tissue or cell) of the different CHIRs according to the EST analysis are also shown. The cutoff values for assigning an EST sequence to a particular group of CHIR genes were >90% identity and >90% coverage of the EST sequence. The EST accession numbers are available from M.N. upon request.
Phylogenetic Relationships Between Different Domains of the CHIR Genes. We performed phylogenetic analysis separately for each domain (signal peptide, Ig-like, TM, and CYT), but we focused mainly on the membrane distal Ig-like (D1) and TM domains, because they are common to all CHIR genes. The phylogenetic analysis of the D1 domain sequences suggests the existence of three major groups (A–C in Fig. 2). Group A contains most of the 2DL and 2DLA sequences (putative inhibitory forms) and a small fraction of the 2DS sequences (putative activating forms). Group B contains the 1DLA sequences, as well as the unique 1DS1 and 1DL1 sequences. Group C consists primarily of type 2DS sequences, which intermingle with sequences of the 2DL-2DLA type.
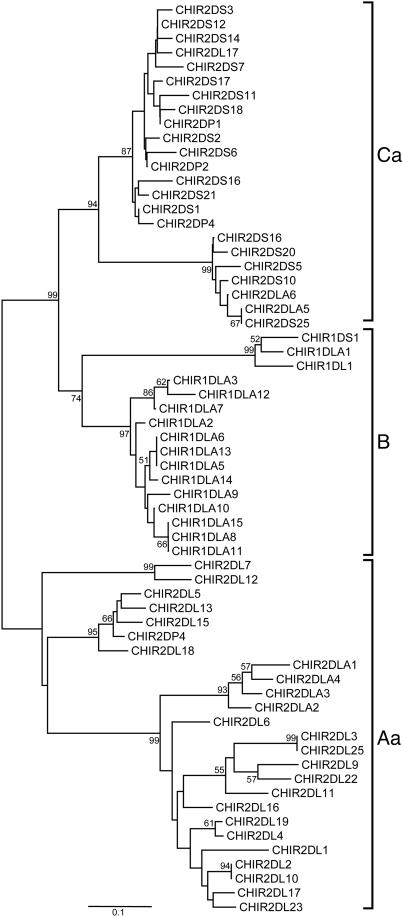
In the phylogenetic analysis of the TM region sequences, we present only representative CHIR sequences because the number of sites (47 aa) was limited in comparison to the Ig-like domains (90 aa). Hence, although the topology of all of the CHIR TM sequences was similar to the one presented in Fig. 3, the bootstrap values were very low. Fig. 3 shows the presence of three major groups (Aa, B, and Ca). Group Aa contains the 2DL and 2DLA sequences; group B contains the 1DLA, 1DS1, and 1DL1 sequences; and group Ca contains the 2DS sequences and three 2DLA sequences. The topology of the TM region tree (Fig. 3) shows that the three groups (Aa, B, and Ca) have similar gene content with the groups A–C in Fig. 2, with some exceptions. The exceptions are (i) the 2DS4, -17, -21, and -22 sequences, which in the Ig tree (Fig. 2) are clustered together in group A, in the TM tree (Fig. 3) are intermingled with the 2DS sequences of group Ca, and (ii) the 2DL5, -7, -12, -13, -15, and -18 sequences, which in the Ig tree (Fig. 2) intermingle with 2DS sequences of group C, in the TM tree are clustered together with sequences of group Aa (Fig. 3). These results suggest that CHIR genes with similar structure and function (inhibition or activation) are clustered together, with the exception of two cases, and that this grouping does not correlate with the genomic organization of the CHIR genes.
Fig. 3.
Neighbor-joining tree of the TM-CYT region of the CHIR sequences. The tree was constructed with p distances for 43-aa sites after elimination of alignment gaps.
The genes that show the different branching pattern between the Ig and TM trees could be the result of recombination between the TM-CYT encoding segments of different genes and/or the result of nucleotide substitutions. In an attempt to distinguish between these two possibilities, we produced pairwise alignments using the sequences of the introns that are located between the exons encoding the second Ig domain (D2) and the TM region of all CHIR genes (intron 4 in Fig. 1B). The analyses of these alignments showed that the intronic sequences from the 2DS genes belonging to group A (Fig. 2) are 93–100% identical with introns of 2DS genes from group C (Fig. 2 and data not shown); the introns of the 2DL genes (group C in Fig. 2), however, have significant similarity only to each other. These results suggest that homologous recombination between activating and inhibitory receptor genes could be responsible for shuffling the TM and CYT sequences of the 2DS genes of group A, whereas for the case of the 2DL sequences of group C, there is not enough evidence to support recombination unambiguously.
Phylogenetic analyses using all of the different domains of CHIR genes (Figs. 2 and 3; see also Figs. 9 and 10, which are published as supporting information on the PNAS web site) revealed that CHIR genes encoding both TM regions with a positively charged residue (putative activating receptors) and long CYT tails with ITIMs (putative inhibitory receptors) in the same molecule exist in all major groups. To explain these observations, we hypothesize that the primordial CHIR sequences could also have encoded both types of motifs (positive TM residues and ITIMs at the CYT tail) in a single molecule. This assumption is further supported by the observation that the nucleotide sequence downstream of the stop codon of one-third of the 2DS genes might encode degenerated ITIMs (data not shown). This finding suggests that the 2DS type of sequences could have evolved from 2DLAs by nucleotide substitutions that resulted in premature stop codons. In addition, the assumption that the common ancestor of CHIRs encoded both motifs (LA type) could explain the presence of 1DS and 1DL genes in the 1DLA subgroup and the presence of the 2DL genes in group C (Fig. 2), because these sequences might have evolved from an LA sequence by nucleotide substitutions.
By contrast, phylogenetic analysis using the extracellular part of the KIR, LILR, and Ly49 genes from humans, mice, and rats has revealed that inhibitory and activating forms intermingle in the tree, whereas analysis using only the TM regions has shown that activating and inhibitory types form two separate groups. To explain this difference in the branching pattern, several researchers have postulated that recombination is the main mechanism of exchange between the activating and inhibitory forms (2, 23–25). Contrary to these suggestions, analysis of the CHIR genes indicates that for most cases, there is not enough evidence to support the recombination hypothesis and that nucleotide substitutions might have played an important role in the evolution of the different forms of CHIR genes.
Evolution and Fold Recognition of the CHIR Ig-Like Domains. Similarity searches against the Protein Data Bank (PDB) showed that the CHIR Ig-like domain sequences produce significant alignment scores of E values <10–5 with the LRC Ig-like domains whose tertiary structures have been resolved. The best hits were scored by the KIRs, LILRs, and the NCR1 Ig-like domain sequences. These domains belong to the constant Ig-like domains and can be classified as s- or h-type. The s-type domains contain two β-sheets composed of three and four β-strands each; the h-type contains two β-sheets composed of four β-strands each (26). To predict which type of Ig-like domains the CHIR genes encode, we used homology modeling and structure-based alignments. The predicted models indicate that (i) the D1 domain of the CHIR sequences with two extracellular Ig-like domains resembles the membrane distal (D1) domain of LILRs (E values ranging from 10–5 to 10–9), (ii) the single domain of the 1DLA proteins resembles the second (D1) domain of KIRs (E value of 10–5), and (iii) the D2 domain of CHIRs resembles the membrane proximal (D2) domains of KIRs and NCR1 (Fig. 4 and data not shown). The analysis suggests that the single Ig-like domain of the 1DLA receptors probably corresponds to the h-type, whereas almost all of the remaining domains probably correspond to the s-type. Major differences of the predicted models, in comparison with the KIR and LILR domains, were found mainly in connecting loops, some of which contain residues that have been implicated in ligand binding. Analysis of ESTs suggests differences in tissue expression patterns between the different CHIR groups (groups B and C in Fig. 2).
Fig. 4.
Predicted folding of CHIR Ig-like domains. (A) Structural model of the Ig-like domain predicted from the CHIR1DLA10 sequence (in blue) and superimposed on the D1 Ig-like domain of the human KIR2DL1 sequence (in yellow; PDB ID code 1IM9). (B) Structural model of the Ig-like domain predicted from the CHIR2DL15 sequence (in blue) and superimposed on the D1 Ig-like domain of the human LILRB1 sequence (in yellow; PDB ID code 1P7Q). Amino acid residues implicated in ligand binding are shown in red.
Discussion
The CHIR genes show two interesting characteristics. First, in contrast to the human LRC genes (27), many CHIR genes encode both positive residue in the TM region and a long CYT tail with ITIMs. In this regard, they resemble some of the Ly49 genes in rats, which belong to the lectin-type natural-killer cell receptors, the functional homologues of KIRs in rodents (23). Second, in contrast to the mammalian LRC genes, phylogenetic analyses of the extracellular (Ig-like) domains or the intracellular (TM and CYT) portions of the CHIR proteins show that, in most cases, genes with similar structure and potentially similar function in terms of activation or inhibition are closely related.
The CHIR genes encode one or two Ig-like domains. Previously, we showed that the membrane distal domains of all of the CHIR2D genes (D1) and the single domain of the CHIR1D genes form a monophyletic group named CI, whereas the membrane proximal domains (D2) form a second monophyletic group named CII (see figure 3 of ref. 7). The mammalian LRC genes encode up to six Ig-like domains (2), and phylogenetic analyses suggest that all of these domains can be divided into two major monophyletic groups named MI and MII (2, 7). Our results indicate that the first group of CHIR domains contains both the s- and h-type of Ig-like domains, whereas the second group contains only the s-type (Fig. 4). The functional significance of the inferred differences (Figs. 4 and 8) between the CHIR groups is not known, but an obvious possibility is that the differences influence ligand-binding specificities of the CHIR domains. By contrast, in the mammalian LRC, the MI group contains the s-type of domains, whereas the MII group contains both s- and h-types (7, 28, 29). These observations suggest that Ig-like domains with different structures (s or h) shared the most recent common ancestor and that the h-type of Ig-like domain has evolved from the s-type independently in both the mammalian and avian lineages.
Taking into account available information on the mammalian LRC genes, the following interpretation of the CHIR data can be put forward. At least two kinds of information indicate that the CHIR region is the chicken homologue of the human LRC. First, from the phylogenetic analysis of sequences encoding Ig-like domains in the chicken genome, the CHIR genes emerge as the closest relatives of the mammalian LRC genes (Fig. 8, ref. 6, and figure 1 in ref. 7). Second, the CHIR genes are syntenic to G protein-coupled receptor genes homologous to genes found in the human extended LRC region. Because the human LRC and the CHIR genes form distinct monophyletic groups on phylogenetic trees, they are apparently the result of separate expansions in the human and chicken lineage, respectively. Furthermore, because the human genes are divided into two monophyletic subgroups (ignoring singleton genes), there must have been at least two separate expansions in the human lineage, one producing the KIR genes and the other giving rise to the LILR genes. The inclusion of mouse LRC genes in the analysis also reveals the existence of two subgroups, one related to the human KIRs and the other related to the human LILR genes. Hence, presumably the divergence of the KIR and LILR groups preceded the human–mouse divergence.
The phylogenetic tree of the CHIR genes divides them into three monophyletic groups (Figs. 2 and 5 and data not shown), presumably resulting from three separate expansions in the chicken lineage. The genes in the three groups are distinguished by their structure: one group encodes receptors with a single Ig-like domain, and the other two groups specify receptors with two Ig-like domains. The latter two groups differ in that one of them encodes proteins with short CYT tails, and the other encodes proteins with long CYT tails. Because the 2DL-2DLA group diverged before the divergence of the 2DS and 1DLA groups (Fig. 2), we assume that the ancestral gene of the three groups was of the 2DLA type, the 2DS group evolved from this ancestor by shortening of the CYT tail, and the 1D group probably arose by the loss of one domain. The three groups of chicken genes occupy single genomic segments (although it is unclear whether the individual segments are all in one region), but, within this segment, the genes of the different groups intermingle. Both the differences in the gene structure and the intermingling of genes that form different groups suggest a high degree of intraregional and intragenic rearrangements during and after the expansion of the three groups. The randomness of the transcriptional orientation of the genes throughout the region is consistent with this supposition. The relatively short branches leading to the CHIR genes (in comparison with the LRC genes) suggest that the expansion of the entire cluster probably took place relatively recently.
In contrast to the CHIR genes, no grouping according to gene structure is recognizable in the human LRC region. Here, the KIR genes encode two or three Ig-like domains (2D or 3D), and the LILR genes encode two (2D) or four (4D) domains (2). This situation could have arisen by evolution from an ancestral gene that, like the ancestor of the CHIR genes, encoded two Ig-like domains. Subsequently, after the separation of the KIR from the LILR genes, either two duplications of a one-domain-encoding gene produced first a three- and then a four-domain-encoding gene, or a two-domain duplication produced a four-domain-encoding gene, which then, by loss of one domain-encoding segment, gave rise to a three-domain-encoding gene. The 2D, 3D, and 4D genes can encode either a long (L) or a short (S) CYT domain (2). In this case, too, the loss or gain of the segment encoding the CYT extension seems to have occurred repeatedly. The variation in the gene structure presupposes high frequency of shuffling of gene segments. It is therefore surprising that the region (in contrast to the CHIR regions) contains three segments in which multiple genes have the same transcriptional orientation. In both the LRC and CHIR regions, the inhibitory and activating forms (L and S, respectively) of the genes appear to be intermingled within and between different genomic clusters. Apparently, the change from one form to another occurs with relative ease, perhaps by nucleotide substitutions and/or recombination.
The clustering of sequences described thus far was based on whole-gene sequences. When the individual Ig-like domains are subjected to phylogenetic analysis separately, a somewhat different clustering pattern emerges (2, 7). Both the mammalian and the chicken domain sequences fall into two groups (MI and MII, and CI and CII) (figure 3 of ref. 7). All of the mammalian KIR Ig-like domains and the LILR D2–D4 domains are of MII type, and only the D1 domain, which is shared by all of the LILR proteins, is of the MI type (ref. 2 and figure 3 of ref. 7). This distribution suggests that the ancestor of the KIR and LILR genes had both the MI and MII domains and that in the KIR lineage, the MI domain type was lost before the expansion of the genes. In the CHIR proteins in the 1D group (Group B in Fig. 2), the single domain is of CI type, whereas in the two 2D groups, the D1 domain is of CI type and the D2 domain is of the CII type, regardless of whether the genes are of the L or S type. Thus, the ancestor of the three groups probably had both the CI and CII encoding segments, and in the ancestor of the 1D group, the one of the two domains that was lost was of the CII type. The relationship between the mammalian and the chicken domains remains somewhat ambiguous (figures 1 and 3 of ref. 7). Although the MI and CI domains appear to be monophyletic, the relationship between the MII and CII domains remains unresolved; they are either monophyletic or polyphyletic. In any case, however, the divergence of at least the CI-MI domains apparently predated the mammalian–avian divergence (7). This conclusion does not contradict the observed monophyly of the LRC and CHIR genes. Each of these monophyletic groups could have originated from a single ancestral gene, which encoded multiple Ig-like domains, some of which were of type CI-MI and others of type CII and/or MII. The scenario described above required repeated expansions of genes by duplications from one ancestral gene (birth of new gene clusters) and the contraction (or death) of old clusters (30, 31) combined with abundant gene exchanges and shuffling of exons. The study of the LRC genes and their avian homologues thus offers a view into the workshop of a very busy evolutionary tinker.
Supplementary Material
Acknowledgments
We thank John Trowsdale, Takeshi Shiina, and Yoko Satta for their valuable comments. This work was supported by National Institutes of Health Grant GM20293 (to M.N.).
Author contributions: N.N., J.K., and M.N. designed research; N.N., I.M., and D.C. performed research; N.N., I.M., D.C., and W.M. analyzed data; and N.N., D.C., J.K., and M.N. wrote the paper.
Abbreviations: CHIR, chicken Ig-like receptor; CYT, cytoplasmic; ITIM, immunoreceptor tyrosine-based inhibitory motif; LRC, leukocyte receptor complex; KIR, killer cell Ig-like receptors; LILR, leukocyte Ig-like receptor; TM, transmembrane.
References
- 1.Lanier, L. L. (2003) Curr. Opin. Immunol. 15, 308–314. [DOI] [PubMed] [Google Scholar]
- 2.Martin, A. M., Kulski, J. K., Witt, C., Pontarotti, P. & Christiansen, F. T. (2002) Trends Immunol. 23, 81–88. [DOI] [PubMed] [Google Scholar]
- 3.Trowsdale, J., Barten, R., Haude, A., Stewart, C. A., Beck, S. & Wilson, M. J. (2001) Immunol. Rev. 181, 20–38. [DOI] [PubMed] [Google Scholar]
- 4.Dennis, G. J., Kubagawa, H. & Cooper, M. D. (2000) Proc. Natl. Acad. Sci. USA 97, 13245–13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volz, A., Wende, H., Laun, K. & Ziegler, A. (2001) Immunol. Rev. 181, 39–51. [DOI] [PubMed] [Google Scholar]
- 6.Viertlboeck, B. C., Crooijmans, R. P. M. A., Groenen, M. A. M. & Gobel, T. W. F. (2004) J. Immunol. 173, 7385–7393. [DOI] [PubMed] [Google Scholar]
- 7.Nikolaidis, N., Klein, J. & Nei, M. (February 9, 2005) Immunogenetics, 10.1007/s00251-004-0764-0. [DOI] [PMC free article] [PubMed]
- 8.Altschul, S. F., Gish, W., Miller, W., Meyers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 9.International Chicken Genome Sequence Consortium (2004) Nature 432, 695–715.15592404 [Google Scholar]
- 10.Yeh, R.-F., Lim, L. P. & Burge, C. B. (2001) Genome Res. 11, 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makalowska, I., Sood, R., Faruque, M. U., Hu, P., Robbins, C. M., Eddings, E. M., Baxevanis, A. D. & Carpten, J. D. (2002) Gene 284, 203–213. [DOI] [PubMed] [Google Scholar]
- 12.Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., Ponting, C. P. & Bork, P. (2004) Nucleic Acids Res. 32, D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E. L. L., et al. (2004) Nucleic Acids Res. 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 16.Nei, M. & Kumar, S. (2000) Molecular Evolution and Phylogenetics (Oxford Univ. Press, New York).
- 17.Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. (2004) Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwede, T., Kopp, J., Guex, N. & Peitsch, M. C. (2003) Nucleic Acids Res. 31, 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley, L. A., MacCallum, R. M. & Strernberg, M. J. E. (2000) J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 20.Shiina, T., Shimizu, S., Hosomichi, K., Kohara, S., Watanabe, S., Hanzawa, K., Beck, S., Kulski, J. K. & Inoko, H. (2004) J. Immunol. 172, 6751–6763. [DOI] [PubMed] [Google Scholar]
- 21.Marsh, S. G. E., Parham, P., Dupont, B., Geraghty, D. E., Trowsdale, J., Middleton, D., Vilches, C., Carrington, M., Witt, C., Guethlein, L. A., et al. (2003) Tissue Antigens 62, 79–86. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J. G., Bashirova, A., Palmer, S., Sims, S., Trowsdale, J., Abi-Rached, L., Parham, P., Carrington, M. & Beck, S. (2005) Genome Res. 15, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao, L. & Nei, M. (2004) Immunogenetics 56, 343–354. [DOI] [PubMed] [Google Scholar]
- 24.Hughes, A. L. (2002) Mol. Phylogenet. Evol. 25, 330–340. [DOI] [PubMed] [Google Scholar]
- 25.Hao, L. & Nei, M. (2005) Gene, in press.
- 26.Klein, J. & Hořejší, V. (1997) Immunology (Blackwell Scientific, Oxford).
- 27.Faure, M. & Long, E. O. (2002) J. Immunol. 168, 6208–6214. [DOI] [PubMed] [Google Scholar]
- 28.Chapman, T. L., Heikema, A. P., West, J., Anthony P. & Bjorkman, P. J. (2000) Immunity 13, 727–736. [DOI] [PubMed] [Google Scholar]
- 29.Boyington, J. C., Brooks, A. G. & Sun, P. D. (2001) Immunol. Rev. 181, 66–78. [DOI] [PubMed] [Google Scholar]
- 30.Ota, T. & Nei, M. (1995) Mol. Biol. Evol. 12, 94–102. [DOI] [PubMed] [Google Scholar]
- 31.Sitnikova, T. & Su, C. (1998) Mol. Biol. Evol. 15, 617–625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.