Abstract
Exome and whole-genome sequencing are becoming increasingly routine approaches in Mendelian disease diagnosis. Despite their success, the current diagnostic rate for genomic analyses across a variety of rare diseases is approximately 25 to 50%. We explore the utility of transcriptome sequencing [RNA sequencing (RNA-seq)] as a complementary diagnostic tool in a cohort of 50 patients with genetically undiagnosed rare muscle disorders. We describe an integrated approach to analyze patient muscle RNA-seq, leveraging an analysis framework focused on the detection of transcript-level changes that are unique to the patient compared to more than 180 control skeletal muscle samples. We demonstrate the power of RNA-seq to validate candidate splice-disrupting mutations and to identify splice-altering variants in both exonic and deep intronic regions, yielding an overall diagnosis rate of 35%. We also report the discovery of a highly recurrent de novo intronic mutation in COL6A1 that results in a dominantly acting splice-gain event, disrupting the critical glycine repeat motif of the triple helical domain. We identify this pathogenic variant in a total of 27 genetically unsolved patients in an external collagen VI–like dystrophy cohort, thus explaining approximately 25% of patients clinically suggestive of having collagen VI dystrophy in whom prior genetic analysis is negative. Overall, this study represents a large systematic application of transcriptome sequencing to rare disease diagnosis and highlights its utility for the detection and interpretation of variants missed by current standard diagnostic approaches.
INTRODUCTION
The advent of whole-exome sequencing (WES) and whole-genome sequencing (WGS) has greatly accelerated our capacity to identify variants that explain many Mendelian diseases in both known and new disease genes. Although these technologies are mainstays in Mendelian disease diagnosis, their success rate for detecting causal variants is far from complete, ranging from 25 to 50% (1–4). The primary challenge of these genome-based diagnostics is that the capacity of WES and WGS to discover genetic variants substantially exceeds our ability to interpret their functional and clinical impact (5–7).
One approach to improve the interpretation of genetic variation is to integrate functional genomic information such as RNA sequencing (RNA-seq), which provides direct insight into transcriptional perturbations caused by genetic changes (8, 9). Analysis of the complementary DNA (cDNA) of single genes has proven useful on a case-by-case basis to provide diagnoses to patients with Mendelian disorders (10–13), and RNA-seq has previously been used to observe the effect of pathogenic variants, which were identified through DNA sequencing (14, 15). However, the use of transcriptome sequencing has not yet been assessed for the discovery of pathogenic variants in a cohort of Mendelian disease patients. Such approaches have already proven useful for elucidating mechanisms of cancer and common disease (16, 17) but are not currently systematically applied to rare disease diagnosis.
Here, we describe the application of this technology to the diagnosis of patients with a range of primary muscle disorders, including myopathies and muscular dystrophies, using RNA obtained from affected muscle tissue (table S1). To investigate the value of RNA-seq for diagnosis, we obtained primary muscle RNA from 63 patients with putatively monogenic muscle disorders. Thirteen of these cases had been previously diagnosed with variants expected to have an effect on transcription, such as loss-of-function or essential splice site variants, allowing us to validate the capability of RNA-seq to identify transcriptional aberrations (table S2). The remaining cohort of 50 genetically undiagnosed patients included cases for whom DNA sequencing had prioritized variants predicted to alter RNA splicing or strong candidate genes, as well as cases with no strong candidates from genetic analysis (see Fig. 1A and Materials and Methods for inclusion criteria).
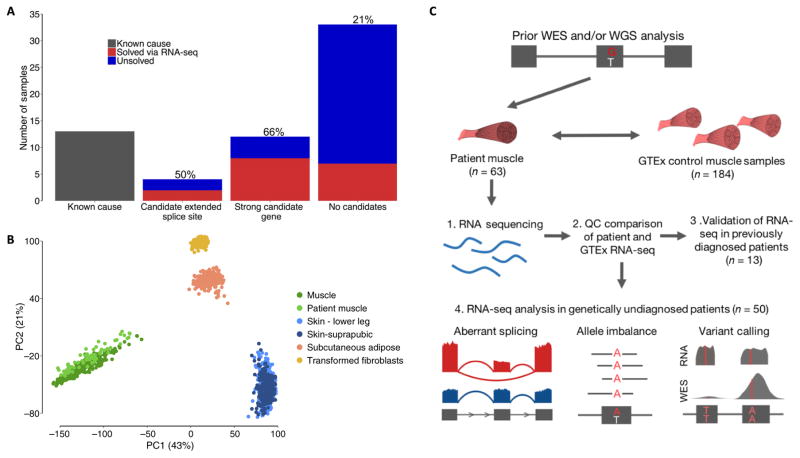
Fig. 1. Experimental design and quality control.
(A) Overview of the number of samples that underwent RNA-seq. We performed RNA-seq on 13 previously genetically diagnosed patients, 4 patients in whom previous genetic analysis had identified an extended splice site variant of unknown significance (VUS), 12 patients in whom genetic analysis had identified a strong candidate gene, and 34 patients with no strong candidates from previous analysis. RNA-seq enabled the diagnosis of 35% of patients overall, with the rate, shown above the bar plots, varying depending on previous evidence from genetic analysis. (B) PCA based on gene expression profiles of patient muscle samples passing quality control (n = 61) and GTEx samples of tissues that potentially contaminate muscle biopsies shows that patient samples cluster closely with GTEx skeletal muscle. (C) Overview of experimental setup and RNA-seq analyses performed. Our framework is based on identifying transcriptional aberrations that are present in patients and missing in GTEx controls. Upon ensuring that GTEx and patient RNA-seq data were comparable, we validated the capacity of RNA-seq to resolve transcriptional aberrations in previously diagnosed patients and performed analyses of aberrant splicing, allele imbalance, and variant calling in our remaining cohort of genetically undiagnosed muscle disease patients.
RESULTS
Importance of sequencing the disease-relevant tissue
Recent large-scale studies have shown that gene expression and mRNA isoforms vary widely across tissues, indicating that for many diseases, sequencing the disease-relevant tissue will be valuable for the correct interpretation of genetic variation (18, 19). This is illustrated by the relative expression of known muscle disease genes in skeletal muscle, whole-blood, and fibroblast samples from the Genotype-Tissue Expression (GTEx) Consortium project (fig. S1) (20). A majority of the most commonly disrupted genes in muscle disease are poorly expressed in blood and fibroblasts, suggesting that RNA-seq from these easily accessible tissues may be underpowered to detect relevant transcriptional aberrations in certain genes. For these reasons, we chose to pursue RNA-seq from primary muscletissue biopsies, which are routinely performed as part of the diagnostic evaluation of undiagnosed muscle disease patients (21, 22).
Comparison of patient RNA-seq to a muscle RNA-seq reference panel
Patient muscle samples were sequenced using the same protocol as in the GTEx project (20) and analyzed using identical pipelines to minimize technical differences, with patients sequenced at or above the same coverage as GTEx controls. From 430 skeletal muscle RNA-seq samples available through GTEx, we selected a subset of 184 samples based on RNA-seq quality metrics including RNA integrity score and ischemic time, as well as phenotypic features such as age, body mass index (BMI), and cause of death to more closely match our patient samples.
Comparison between our GTEx reference panel and patient muscle RNA-seq samples showed analogous quality metrics (table S3). Principal component analysis (PCA) of expression and splicing profiles demonstrated that patient muscle RNA-seq closely resembled control muscle when compared to tissues that potentially contaminate muscle biopsies, such as skin or fat, despite variation in the site of muscle biopsy across patients (Fig. 1B, fig. S2A, and table S1). On the basis of this clustering, we removed two samples from analysis because their expression patterns clustered more closely with GTEx adipose tissue than muscle, consistent with tissue contamination or late-stage degenerative muscle pathology (fig. S2B). We also performed fingerprinting on patient WES, WGS, and RNA-seq data to ensure that the source of DNA sequencing and muscle RNA-seq data was the same individual.
We explored the utility of analyzing patient RNA-seq data to detect aberrant splice events and allele-specific expression and performed variant calling from RNA-seq data to identify pathogenic events or to prioritize genes for closer analysis (Fig. 1C). We also identified outlier gene expression status in patients; however, this analysis was under-powered to prioritize candidate genes in our study (fig. S3). The resulting diagnoses were made primarily through the detection of aberrant splice events in patients, with information on gene-level allele imbalance playing a complementary role.
In previously diagnosed cases, manual evaluation of pathogenic essential splice site variants revealed a splice aberration, such as exon skipping or extension, demonstrating that RNA-seq can help resolve the effect of variants on transcription (fig. S4, A to F). To detect aberrant transcriptional events genome-wide, we developed an approach based on identifying high-quality exon-exon splice junctions present in patients or groups of patients and missing in GTEx controls (code available at https://github.com/berylc/MendelianRNA-seq). We performed splice junction discovery from split-mapped reads, considering only those that were uniquely aligned and nonduplicate. To account for library size and stochastic gene expression differences between samples, we performed local normalization of read counts based on read support for overlapping annotated junctions (fig. S5, A and B). We then performed filtering of splice junctions based on the number of samples in which a splice junction is observed and the number of reads and normalized value supporting that junction in each sample. Our approach successfully reidentified all known pathogenic events in patients in whom manual evaluation had revealed aberrant splicing around splice variants previously identified through genomic testing. We defined filtering parameters that selectively identified these previously known aberrant splice events and applied them to our remaining cohort of undiagnosed patients. This method resulted in the identification of a median of 5, 26, and 190 potentially pathogenic splice events per sample in ~190 neuromuscular disease associated genes, Online Mendelian Inheritance in Man (OMIM) genes, and all genes, respectively (fig. S6), which required manual curation to interpret pathogenicity and led to the diagnoses made in this study.
Diagnoses made via RNA-seq
RNA-seq allowed the diagnosis of 17 previously unsolved families, yielding an overall diagnosis rate of 35% in this challenging subset of rare disease patients for whom extensive prior analysis of DNA sequencing data had failed to return a genetic diagnosis. We also identified splice disruption in other known and putatively novel disease genes in several patients; however, due to unavailability of additional information, such as parental DNA, we could not pursue these cases further (fig. S7). Detection of aberrant splicing led to the identification of a broad class of both coding and noncoding pathogenic variants, resulting in a range of splice defects such as exon skipping, exon extension, and exonic and intronic splice gain, which were validated by reverse transcription polymerase chain reaction (RT-PCR) analysis (see Fig. 2, Table 1, and the Supplementary Materials and Methods). RNA-seq patterns also helped pinpoint three structural variants in DMD that were subsequently confirmed by WGS (fig. S8).
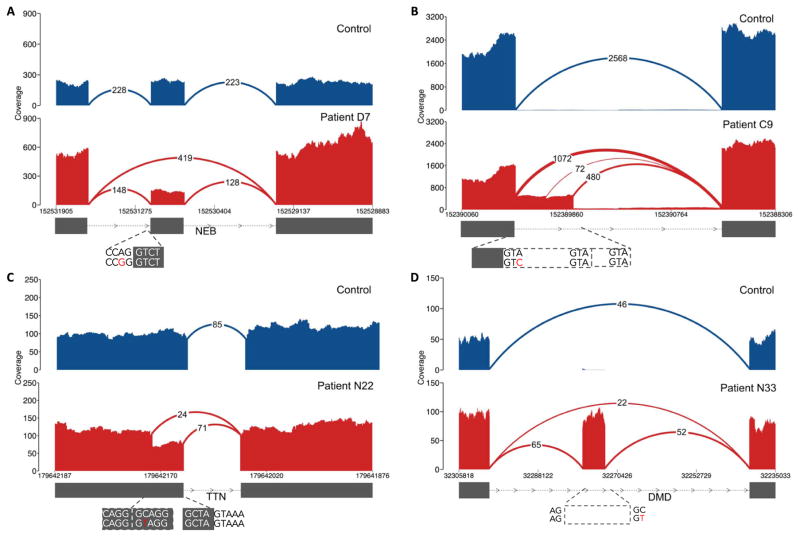
Fig. 2. Types of pathogenic splice aberrations discovered in patients.
RNA-seq identified a range of aberrations caused by both coding and noncoding variants, such as (A) exon skipping caused by an essential splice site variant in patient D7, (B) exon extension caused by a donor +3 A>C extended splice site variant in nemaline myopathy patient C9 (where disruption of splicing at the canonical splice site results in splicing from intact GTA motifs from the intron), (C) exonic splice gain caused by a C>T donor splice site–creating variant in patient N22 with a donor +5-G sequence context, resulting in a stronger splice motif than the existing canonical splice site, and (D) intronic splice gain in patient N33 caused by a C>T donor splice site–creating deep intronic variant. Evidence for wild-type splicing in addition to the inclusion of the pseudoexon in the patient is in line with the milder Becker’s muscular dystrophy phenotype. Splice aberrations shown in (B) to (D) result in the introduction of a premature stop codon to the transcript.
Table 1.
Diagnoses made in the study via patient muscle RNA-seq.
| Patient | Phenotype | Gene | Variants | Variant class | Effect |
|---|---|---|---|---|---|
| E2 | Nemaline myopathy | NEB | chr2: 152,544,805 C>T chr2: 152,520,057 C>T |
Essential splice, extended splice | Exon skipping + exon extension, exon extension |
| C9 | Nemaline myopathy | NEB | chr2: 152,581,432 TG>T chr2: 152,389,953 A>C |
Frameshift, extended splice | Exon extension |
| E4 | Fetal akinesia | TTN | chr2: 179,586,600 CAT>C chr2: 179,446,219 ATACT>A |
Frameshift, extended splice | Exon skipping |
| C6 | Duchenne muscular dystrophy | DMD | chrX: 32,366,860 A>C | Intronic variant | Intronic splice gain |
| N33 | Myalgia, myoglobinuria | DMD | chrX: 32,274,692 G>A | Intronic variant | Intronic splice gain |
| C7 | Becker muscular dystrophy | DMD | chrX: 31,613,687 G>T | Intronic variant | Intronic splice gain |
| N29 | Collagen VI–related dystrophy | COL6A1 | chr21: 47,409,881 C>T | Intronic variant | Intronic splice gain |
| N30 | Collagen VI–related dystrophy | COL6A1 | chr21: 47,409,881 C>T | Intronic variant | Intronic splice gain |
| N31 | Collagen VI–related dystrophy | COL6A1 | chr21: 47,409,881 C>T | Intronic variant | Intronic splice gain |
| N32 | Collagen VI–related dystrophy | COL6A1 | chr21: 47,409,881 C>T | Intronic variant | Intronic splice gain |
| N25 | Nemaline myopathy | NEB | chr2: 152,355,017 G>T chr2: 152,449,646G>A |
Intronic variant, nonsense | Intronic splice gain |
| C11 | Congenital fiber-type disproportion | RYR1 | chr19: 38,958,362 C>T chr19: 38,958,372 G>A |
Synonymous, missense | Exonic splice gain |
| N22 | Multi/minicore congenital myopathy | TTN | chr2: 179,642,185 G>A chr2: 179,523,240 CTTCT>C |
Missense, frameshift | Exonic splice gain |
| C1 | α-Dystroglycanopathy | POMGNT1 | chr1: 46,655,129 C>A chr1: 46,660,532 G>A |
Essential splice, synonymous | Exonic splice gain, exon skipping |
| C3 | Duchenne muscular dystrophy | DMD | chrX: 31,790,694–31,798,498 | Inversion-deletion | Exon skipping |
| C2 | Duchenne muscular dystrophy | DMD | chrX: 31,378,946–151,194,962 | Inversion | Splice disruption |
| C4 | Duchenne muscular dystrophy | DMD | chrX: 32,521,820–35,180,380 | Inversion | Splice disruption |
Cases diagnosed in this study highlight several key advantages of RNA-seq in rare disease diagnosis to confirm the pathogenicity of variants and to detect previously unidentified variation. In four patients with previously detected extended splice site VUS, RNA-seq confirmed splice disruption in two patients (Fig. 1A and fig. S9, A and B). The variants had no observable effect on local splicing patterns in the remaining two patients, emphasizing the value of RNA-seq in ruling out non-pathogenic VUS (fig. S9, C and D).
RNA-seq also led to the identification of an additional disruptive extended splice site variant missed by exome sequencing. In a nemaline myopathy patient with one previously detected recessive frameshift variant in the NEB gene, RNA-seq identified an exon extension event caused by an underlying variant at the +3 position of the donor site, which led to the introduction of a premature stop codon to the transcript as the second recessive allele (Fig. 2B). The exon harboring this variant was not captured in the exome kit used to screen the patient (fig. S10), underlining the utility of RNA-seq at complementing WES to identify previously undetected variants.
Synonymous and missense variants in large, variation-rich genes, such as TTN, are exceptionally challenging to interpret and are often filtered out in DNA sequencing pipelines (23, 24). With RNA-seq, we were able to assign pathogenicity to a missense variant in TTN and two synonymous variants in RYR1 and POMGNT1 (fig. S11). In patient N22, the identified missense variant created a GT donor splice site for which the consensus motif included a G nucleotide in the +5 position, known to contribute to the strength of the splice site (25, 26). The well-conserved donor +5-G motif was missing in the competing canonical splice site, thus resulting in a stronger novel splice site and gain of splicing from the exon body (Fig. 2C). A similar mechanism was observed in RYR1, caused by a synonymous variant in a patient carrying a second pathogenic allele in the gene (fig. S11A). In an additional patient carrying an essential splice site variant in POMGNT1, we identified a synonymous variant disrupting an exonic splice motif and resulting in exon skipping (fig. S11, B to D).
In eight cases, RNA-seq aided in the identification of noncoding pathogenic variants. We identified splice site–creating hemizygous deep intronic variants in DMD that resulted in the creation of a pseudoexon and led to a premature stop codon in the coding sequence in three patients (Fig. 2D and fig. S12). Although RNA-seq from a patient with severe Duchenne muscular dystrophy showed only splicing to the pseudoexon (fig. S12), wild-type splicing between annotated exons was observed in two patients with a milder Becker muscular dystrophy phenotype, indicating the presence of residual functional DMD transcripts that explain the milder disease course. Such intronic variants are unobservable with WES and too abundant to be interpretable with WGS alone, emphasizing the utility of RNA-seq at resolving pathogenicity of these noncoding variants.
In two patients with no strong candidates from WES and WGS (N22 and N25), we identified heterozygous splice disruption in two commonly disrupted recessive muscle disease genes, NEB and TTN. These genes harbor regions with highly similar sequences, the so-called triplicate repeat regions (27, 28). Because of high sequence similarity, the region has poor mapping quality, resulting in low-quality variant calls that are filtered by the most current diagnostic pipelines. To identify possible pathogenic variants in the triplicated regions of NEB and TTN in these two patients, we developed a method based on remapping the triplicate regions to a detriplicated pseudoreference and performing hexaploid variant calling (fig. S13, A to C). This method was applied to available WES/WGS and RNA-seq data for all patients and identified one novel nonsense and one novel frameshift variant in NEB and TTN in these two patients, which finalized their diagnoses (fig. S13D, N25, and fig. S13E, N22).
Identification of a recurrent splice site–creating variant in collagen VI–related dystrophy
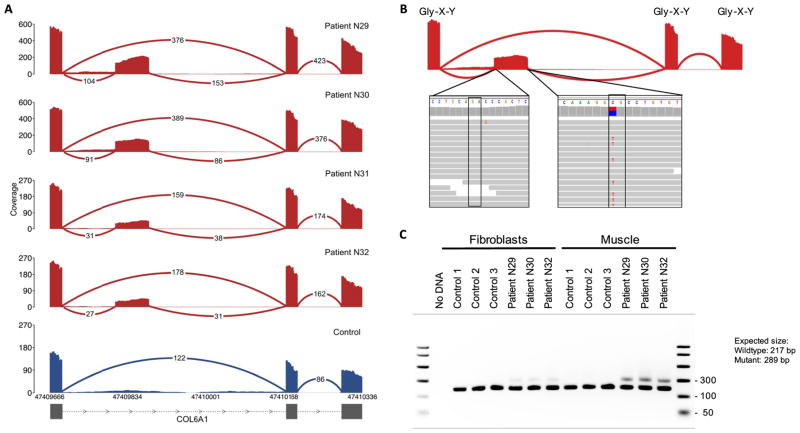
A notable example of the power of transcriptome sequencing is our discovery of a genetic subtype of severe collagen VI–related dystrophy, which is caused by mutations in one of the three collagen VI genes (COL6A1, COL6A2, and COL6A3) (21). In four patients who had previously tested negative with deletion/duplication testing and fibroblast cDNA sequencing of the collagen VI genes as well as clinical WES and WGS, we identified an intron inclusion event in COL6A1 using RNA-seq (Fig. 3A). The splicing-in of this intronic segment, which is missing in GTEx controls and all other patients in our cohort, is caused by a donor splice site–creating GC>GT variant that pairs with a cryptic acceptor splice site 72 base pairs (bp) upstream, creating an in-frame pseudoexon (Fig. 3B). This variant is missing in the 1000 Genomes Project data set (29) as well as an in-house data set of 5500 control WGS samples. The resulting inclusion of 24 amino acids occurs within the N-terminal triple-helical collagenous G-X-Y repeat region of the COL6A1 gene, the disruption of which has been well established to cause dominant-negative pathogenicity in a variety of collagen disorders (30). Notably, cDNA analysis shows that the aberrant transcript is observable in muscle but in much smaller amounts in cultured dermal fibroblasts, making the event identifiable by muscle transcriptome analysis despite being previously missed by fibroblast cDNA sequencing (Fig. 3C). Using this information, we genotyped the variant in a larger, genetically undiagnosed collagen VI–like dystrophy cohort and identified 27 additional patients carrying the intronic variant. We confirmed that the variant had occurred as an independent de novo mutation in all 16 families for whom trio DNA was available. On the basis of this screening, we estimate that up to a quarter of all cases clinically suggestive of collagen VI–related dystrophy but negative by exon-based sequencing are due to this recurrent de novo mutation (see the Supplementary Materials and Methods).
Fig. 3. Identification of a recurrent splice site–creating variant in four collagen VI–related dystrophy patients.
(A) Splicing-in of the pseudoexon was observed in four patients in our cohort (red) and missing in all other patients and GTEx samples (blue). (B) Inclusion of the 24–amino acid segment is caused by a C>T donor splice site–creating variant, which pairs with an AG splice acceptor site 72 bp upstream. The variant is found in a CpG nucleotide context, which likely explains its recurrent de novo status, and disrupts the Gly-X-Y repeat motifs of COL6A1. (C) The inclusion event is observable in RT-PCR amplicons from patient muscle but is found at comparatively lower levels in cultured dermal fibroblasts derived from the patients, explaining why the pathogenic event was missed in all four patients through previous fibroblast cDNA sequencing.
Evaluation of splice prediction algorithms and RNA-seq in alternative tissues
Exons harboring the pathogenic variants identified in this study show low coverage in GTEx whole-blood and fibroblast samples, indicating that a majority of these diagnoses likely could not have been made using RNA-seq from these tissues (fig. S14). Furthermore, many of the diagnoses made in this study could not have been made on genotype information alone, because splice prediction algorithms alone are currently insufficient to classify variants as causal (31, 32). Although existing in silico algorithms correctly predicted disruption for the two extended splice site VUS in our study, they also generated false-positive predictions for the remaining two extended splice site variants with no effect on splicing (see fig. S15A and the Supplementary Materials and Methods). In addition, existing algorithms showed poor specificity in identifying splice site–creating coding variants, identifying on average more than 100 putative splice site–creating rare variants [<1% population frequency in Exome Aggregation Consortium (ExAC)] exome-wide (fig. S15B).
DISCUSSION
Our results show that RNA-seq is valuable for the interpretation of coding as well as noncoding variants and can provide a substantial increase in diagnosis rate in patients for whom exome or whole-genome analysis has not yielded a molecular diagnosis. In our cohort, RNA-seq led to the diagnosis of 66% of patients where clinical phenotyping and DNA sequencing prioritized a strong candidate gene. In comparison, through identifying aberrant splice events found in patients and missing in GTEx controls, we were able to diagnose 21% of patients with no strong candidates from WGS or WES.
Our work illustrates the value of large multitissue transcriptome data sets such as GTEx to serve as a reference to facilitate the identification of extreme splicing or allele balance outlier events in patients. In the case of muscle disorders, our diagnoses were made primarily through direct identification of aberrations in splicing using the GTEx skeletal muscle RNA-seq data set as a reference panel. Our present work focused on identifying such aberrations in known muscle disease genes, and the considerably lower number of putatively pathogenic events identified in neuromuscular disease genes versus all genes underlines the advantage of a candidate gene list for this analysis. Further improvements in filtering identified splice junctions to obtain a smaller list of candidate events will be useful to expand this work for new disease gene discovery. In addition, with increasing sample sizes and improvements in methods, RNA-seq can also be used to identify somatic variants and to detect regulatory variants upstream, through analysis of expression status and allelic imbalance.
Access to the disease-relevant tissue for many Mendelian disorders remains a major barrier for the use of transcriptome sequencing in genetic diagnosis. The RNA-seq framework developed in this study can be adapted for rare diseases where biopsies are available, such as Mendelian disorders affecting the heart, kidney, liver, skin, and other tissues. For example, during the preparation of this paper, the application of RNA-seq to fibroblast samples for the genetic diagnosis of mitochondrial disease was reported in an unpublished preprint (33). For disorders where biopsy of the disease-relevant tissue is unattainable, analyses are possible through identification of proxy tissues using databases such as GTEx and careful consideration of the expression status of the relevant genes in the proxy tissue. Alternatively, the framework developed in this study can also enable diagnoses through reprogramming patient cells into induced pluripotent stem cells and differentiation into disease-relevant tissues of interest.
Evaluation of existing splice prediction algorithms for the splice-disrupting variants identified in the study highlights that information on DNA sequence alone does not currently match the ability of RNA-seq to identify the transcriptional consequences of variants on a genome-wide scale. The diagnoses made in our study with RNA-seq, particularly the discovery of the highly recurrent mutation in COL6A1, demonstrate that other such cryptic splice-affecting variants may contribute substantially to undiagnosed diseases that have evaded prior detection with exome or whole-genome analysis. Overall, this work suggests that RNA-seq is a valuable component of the diagnostic toolkit for rare diseases and can aid in the identification of new pathogenic variants in known genes as well as new mechanisms for Mendelian disease.
MATERIALS AND METHODS
Study design
We sought to explore the utility of transcriptome sequencing as a complementary diagnostic tool to exome and whole-genome analysis. We reasoned that RNA-seq would allow us to interpret variants previously identified through genetic analysis and may pinpoint genetic lesions that may have eluded DNA sequencing. To interpret transcriptional aberrations seen in patients, we obtained a reference panel of 184 sets of skeletal muscle RNA-seq data from the GTEx project. Our framework was based on identifying transcriptional aberrations present in patients but missing in GTEx controls. We first validated the capacity of RNA-seq to resolve transcriptional aberrations in 13 patients with prior genetic diagnosis and then analyzed the remaining 50 genetically undiagnosed patients to detect aberrant splice events and allele-specific expression and performed variant calling from RNA-seq data to identify pathogenic events or to prioritize genes for closer analysis.
Clinical sample selection
Patient cases with available muscle biopsies were referred by clinicians from March 2013 through June 2016. Samples fell into four broad categories:
Patients for whom previous genetic analysis had resulted in a diagnosis with at least one loss-of-function or essential splice site variant, serving as positive controls to assess the capability of RNA-seq to identify the transcriptional effect of the variants (n = 13; patient IDs starting with “D”).
Patients with candidate extended splice site variants that had been categorized as VUS, for which assignment of pathogenicity would result in a complete diagnosis for the patient (n = 4; patient IDs starting with “E”).
Patients for whom a strong candidate gene was implicated because of either a well-defined monogenic disease phenotype, such as patients with clear Duchenne muscular dystrophy evidenced by clinical diagnosis and loss of dystrophin expression (n = 6), or the presence of one pathogenic heterozygous variant identified in a gene matching the patient’s phenotype, without a second pathogenic variant in that gene (n = 6; patient IDs starting with “C”).
Patients with no strong candidates based on previous genetic analysis such as WES or WGS (n = 34; patient IDs starting with “N”).
Patients who fit categories 2 to 4 are referred to as undiagnosed before RNA-seq and constitute the denominator for the 35% diagnosis rate. All patients had prior analysis of WES and/or WGS data, except two cases (patients E4 and D11) for whom targeted sequencing had identified candidate extended and essential splice site variants, respectively. We favored cases with previous trio WES or WGS: 29 of 63 patients had complete trios, with 3 additional patients having one parent sequenced. Although age of onset was not considered as an exclusion criterion, most of the patients in the cohort had a congenital or early childhood–onset primary muscle disorder.
Muscle biopsies or RNA were shipped frozen from clinical centers via a liquid nitrogen dry shipper and stored in liquid nitrogen cryogenic storage. Before submission to the sequencing platform, all muscle samples were visually inspected, photographed, cut into 50-μm sections on a Leica CM1950 model cryostat, and transferred to prechilled cryotubes in preparation for RNA extraction. When muscle arrived embedded in optimum cutting temperature compound, 8-μm transverse cryosections were mounted on positively charged Superfrost Plus slides (VWR, 48311–703) and stained with hematoxylin and eosin (H&E) to assess the relative proportion of muscle versus fibrosis and adipose infiltration as well as the presence of overt freeze-thaw artifact. All samples analyzed with H&E showed muscle quality sufficient to proceed to RNA-seq.
RNA sequencing
RNA was extracted from muscle biopsies via the miRNeasy Mini Kit from QIAGEN according to the kit’s instructions. All RNA samples were measured for quantity and quality. Samples had to meet the minimum cutoff of 250 ng of RNA and RNA quality score (RQS) of 6 to proceed with RNA-seq library preparation. A fraction of samples falling below an RQS of 6 were also submitted for sequencing. All samples submitted had a range of RQSs between 3.5 and 8.
Sequencing was performed at the Broad Institute Genomics Platform using the same non–strand-specific protocol with poly-A selection of mRNA (Illumina TruSeq) used in the GTEx sequencing project (20) to ensure consistency of our samples with GTEx control data. Paired-end 76-bp sequencing was performed on Illumina HiSeq 2000 instruments, with sequence coverage of 50 million or 100 million reads. One sample (patient N33) was sequenced to a higher depth at 500 million reads to permit downsampling analysis of the effects of increasing RNA-seq depth.
Selection of GTEx controls
GTEx data were downloaded from the Database of Genotypes and Phenotypes (dbGaP) (www.ncbi.nlm.nih.gov/gap) under accession phs000424.v6.p1. From 430 available GTEx skeletal muscle RNA-seq samples, we selected 184 samples on the basis of RNA integrity score (between 6 and 9), number of nonduplicate uniquely mapped read pairs (between 35 million and 75 million reads), and ischemic time (<12 hours) to remove any samples that were outliers for these quality metrics. GTEx samples were further filtered to remove those with known clinical conditions such as Klinefelter’s syndrome or those for whom death followed after long- or intermediate-term illness or medical intervention (Hardy scale 0, 3, or 4). Overall, approximately 80% of GTEx samples with available muscle RNA-seq are older than 40 (median age, 54) and have a BMI over 25 (median BMI, 27). Thus, we selected samples to enrich for younger GTEx donors to more closely match our patient cohort. All samples younger than 50 were selected, resulting in 76 samples with high-quality RNA-seq data. We then added older samples back on the criterion that their BMI was below 30. This resulted in a total of 184 GTEx control samples for our reference panel, with comparable male and female sample count (105 males and 79 females). This filtering method also enriched the RNA-seq data from organ donors and surgical donors as opposed to postmortem samples (72% of selected GTEx controls are derived from surgical or organ donors versus 45% in the unfiltered data set). A full list of GTEx sample IDs used as the reference panel can be found in table S4.
RNA-seq alignment and quality control
GTEx BAM files downloaded from dbGaP were realigned after conversion to FASTQ files with Picard SamToFastq. Both patient and GTEx reads were aligned via STAR 2-Pass version v.2.4.2a using hg19 as the genome reference and GENCODE V19 annotations. Briefly, first-pass alignment was performed for novel junction discovery, and the identified junctions were filtered to exclude unannotated junctions with less than five uniquely mapped read supports, as well as junctions found on the mitochondrial genome. These junctions were then used to create a new annotation file, and second-pass alignment was performed as recommended by the STAR manual to enable sensitive junction discovery. Duplicate reads were marked with Picard MarkDuplicates (v.1.1099).
Quality metrics for patient and GTEx RNA-seq data were obtained by running RNA-SeQC (v1.1.8) on STAR-aligned BAM files (34). PCA on gene expression was performed on the basis of RPKM (reads per kilobase of transcript per million mapped reads) values calculated by RNA-SeQC. Two samples (D6 and N3) were removed because of outlier status in PCA, consistent with a high proportion of nonmuscle tissue in the samples (fig. S2B). For GTEx samples, the expression and exon-level read count data were downloaded from dbGaP under accession phs000424.v6. For PCA of exon inclusion metrics, we obtained PSI (percentage spliced in) values for GTEx samples as described in (35).
To ensure that patient DNA and RNA data were identity-matched, we compared variants identified in WES, WGS, and RNA-seq data. WES, WGS, and RNA-seq data were joint-genotyped for a set of ~5800 common single nucleotide polymorphisms (SNPs) collated by Purcell et al. (36) using the Genome Analysis Toolkit (GATK) HaplotypeCaller package version 3.4. We then calculated pairwise inheritance by descent estimates between DNA sequencing and RNA-seq data using PLINK (v1.08p). Relatedness coefficients for WES, WGS, and RNA-seq data from the same individual ranged from 0.67 to 1.00 across our samples (mean, 0.9), compared to a range of 0 to 0.18 (mean, 0.001) for non-matching individuals, confirming that the sources for DNA sequencing and RNA-seq were the same for each patient in our data set.
Exome sequencing and WGS
WES on DNA samples (>250 ng of DNA, at >2 ng/μl) was performed using Illumina or Agilent SureSelect v2 exome capture. The exome sequencing pipeline included sample plating, library preparation (2-plexing of samples per hybridization), hybrid capture, sequencing (76-bp paired reads), and sample identification quality control check. Hybrid selection libraries covered >80% of targets at 20× with a mean target coverage of >80×. The exome sequencing data were demultiplexed, and each sample’s sequence data were aggregated into a single Picard BAM file. WGS was performed on 500 ng to 1.5 μg of genomic DNA using a PCR-free protocol. These libraries were sequenced on the Illumina HiSeq X10 with 151-bp paired-end reads and a target mean coverage of >300×.
Exome and genome sequencing data were processed through a Picard-based pipeline using base quality score recalibration and local realignment at known insertions/deletions (indels). The Burrows-Wheeler Aligner was used for mapping reads to the human genome build 37 (hg19). SNPs and indels were jointly called across all samples using GATK HaplotypeCaller. Default filters were applied to SNP and indel calls using the GATK variant quality score recalibration, and variants were annotated using Variant Effect Predictor (v78); additional information on this pipeline is provided in the first supplementary section of (37). The variant call set was uploaded to the seqr analysis platform (seqr.broadinstitute.org) to perform variant filtering using inheritance patterns, functional annotation, and variant frequency in reference databases including ExAC (37) and 1000 Genomes (29).
Identification of pathogenic splice events
Splice junctions were identified from split-mapped reads, considering only uniquely aligned, nonduplicate reads that passed platform/vendor quality controls. For each splice junction, we noted the following:
the genomic coordinates
the gene in which the junction was observed based on GENCODE v.19
the number of samples in which the splice junction was observed
the number of total reads supporting the junction in 245 samples (184 GTEx and 61 patient samples)
the per-sample read support for the junction.
We then performed local normalization of per-sample read support on the basis of the support for the highest shared annotated junction (fig. S5A). For example, an exon-skipping event harbors two annotated exon-intron junctions, and we normalized this by the maximum of read count support for canonical splicing at these two wild-type junctions. This local normalization allows for filtering low-level mapping noise and accounts for stochastic gene expression and library size differences between samples (fig. S5B).
To identify pathogenic splice events, splice junctions in protein coding genes were filtered in terms of the number of samples a splice junction is present in and the number of reads and the normalized value supporting that junction. Specifically, we defined a sensitive cutoff at which an aberrant splice event is seen with at least 5% of the read support as compared to the shared annotated junction, with at least two reads supporting the event. We also required a splice junction to contain at least one annotated exon-exon junction, indicating that the event was spliced into an existing transcript (fig. S5A). We performed analysis on a per-sample basis, each time requiring the normalized value of a given splice junction to be maximum in that sample and twice that of the next highest sample, allowing us to search for unique events in the patient.
All candidate pathogenic splice events were manually evaluated using the Integrative Genomics Viewer. This resulted in the identification of aberrant splicing at eight of nine pathogenic essential splice site variants and resulted in the diagnosis of 10 of 17 patients in the study. A splice aberration was not observed around an essential splice site variant found in TTN in patient D5 because of insufficient number of reads mapping to the local region (fig. S4E). We extended filtering parameters to identify splice junctions present in fewer than 10 samples, but with high read support in each sample, allowing us to identify the intronic splice-gain event present in four patients in COL6A1 (Fig. 3A). We note that this approach would also identify putatively pathogenic splice aberrations, for which there are GTEx carriers. The remaining three Duchenne muscular dystrophy patients were diagnosed through manual analysis of splicing patterns in DMD and resulted in the identification of splice disruption. Overlapping structural variants at these regions were confirmed by subsequent WGS (fig. S8).
Statistical analysis and code availability
Our approach to evaluating outlier status for allele imbalance in patients involved defining the 95% confidence interval (means ± 2 SD) of mean allele balance in GTEx individuals for each gene and identifying patients for whom the gene-level allele balance fell outside of the range. Comparison between GTEx and patient RNA-seq data quality metrics relied on a t test for significance. Data processing, analysis, and figure generation were performed using scripts written in Python 2.7 and R 3.2; code for identifying and filtering splice junctions and for variant calling in the triplicate regions of NEB and TTN is available at https://github.com/berylc/MendelianRNA-seq.
Supplementary Material
Acknowledgments
Sequencing and analysis were provided by the Broad Institute of Massachusetts Institute of Technology (MIT) and Harvard Center for Mendelian Genomics (Broad CMG). We thank H. Brooks, D. Sookiasian, M. E. Leach, D. Ezzo, J. Dastgir, A. Rutkowski, C. Grosmann, C. Konermans, S. Ceulemans, M.-L. Chu, E. Moran, and K. Matthews for sample collection and quality control. We also thank C. Miceli, S. Nelson, V. Rusu, and D. Altshuler for sharing control cell lines and plasmids.
Funding: This project was supported by funding from the Broad Institute’s BroadIgnite and Broadnext10 programs. B.B.C. is supported by the NIH GM096911 training grant. T.T. is supported by the Academy of Finland, the Finnish Cultural Foundation, the Orion-Farmos Research Foundation, and the Emil Aaltonen Foundation. M.L. is supported by the Australian NHMRC (National Health and Medical Research Council) CJ Martin Fellowship, the Australian American Association Sir Keith Murdoch Fellowship, and a Muscular Dystrophy Association/American Association of Neuromuscular and Electrodiagnostic Medicine (MDA/AANEM) development grant. L.B.W., S.A.S., N.G.L., N.F.C., K.N.N., and E.C.O. are supported by the NHMRC of Australia (1080587, 1075451, 1002147, 1113531, 1022707, 1031893, and 1090428). K.J.K. is supported by a National Institute of General Medical Sciences (NIGMS) fellowship grant (F32GM115208). A.H.O.-L. is supported by an NIGMS fellowship grant (4T32GM007748). A.H.B. is supported by the NIH R01 HD075802 and R01 AR044345 and by MDA383249 from the Muscular Dystrophy Association. P.B.K., E.E., and H.K.M. are supported by NIH R01NS080929. J.J.D. is supported in part by funding from Genome Canada (a Disruptive Innovations in Genomics grant). Funding relevant to this research includes fellowship support of S.T.C. and a project grant supporting an Australian-wide program about gene discovery in inherited neuromuscular disorders performed in collaboration with D.G.M. [NHMRC APP1048816 (2013–2017) and NHMRC APP1080587 (2015–2019)]. The Broad CMG was funded by the National Human Genome Research Institute (NHGRI), the National Eye Institute, and the National Heart, Lung, and Blood Institute (NHLBI) grant UM1 HG008900 to D.G.M. and H. Rehm. The GTEx project was supported by the Common Fund of the Office of the Director of the NIH (http://commonfund.nih.gov/GTEx). Additional funds were provided by the National Cancer Institute (NCI), NHGRI, NHLBI, National Institute on Drug Abuse (NIDA), National Institute of Mental Health (NIMH), and National Institute of Neurological Disorders and Stroke (NINDS). Donors were enrolled at Biospecimen Source Sites that were funded by NCI/Science Applications International Corporation (SAIC)–Frederick Inc. (SAIC-F) subcontracts to the National Disease Research Interchange (10XS170) and the Roswell Park Cancer Institute (10XS171). The Laboratory, Data Analysis, and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to the Broad Institute Inc. Biorepository operations were funded through an SAIC-F subcontract to the Van Andel Institute (10ST1035). Additional data repository and project management were provided by SAIC-F (HHSN261200800001E). The Brain Bank was supported by a supplement to the University of Miami grant DA006227.
Members of the GTEx Consortium
LDACC–Analysis Working Group (AWG): Kristin G. Ardlie,1 Gad Getz,1,2 Ellen T. Gelfand,1 Ayellet V. Segrè,1 François Aguet,1 Timothy J. Sullivan,1 Xiao Li,1 Jared L. Nedzel,1 Casandra A. Trowbridge,1 Daniel G. MacArthur,1,3 Monkol Lek,1,3 Taru Tukiainen,3,4 Kane Hadley,4 Katherine H. Huang,4 Michael S. Noble,4 Duyen T. Nguyen,4 Beryl B. Cummings;3,4 Funded Statistical Methods groups–AWG: Andrew B. Nobel,5 Fred A. Wright,6 Andrey A. Shabalin,7 John J. Palowitch,8 Yi-Hui Zhou,9 Emmanouil T. Dermitzakis,10,11,12 Mark I. McCarthy,13,14,15 Anthony J. Payne,13 Tuuli Lappalainen,16,17 Stephane Castel,16,17 Sarah Kim-Hellmuth,16,17 Pejman Mohammadi,16,17 Alexis Battle,18 Princy Parsana,18 Sara Mostafavi,19 Andrew Brown,10,11,12 Halit Ongen,10,11,12 Olivier Delaneau,10,11,12 Nikolaos Panousis,10,11,12 Cedric Howald,10,11,12 Martijn van de Bunt,13,14 Roderic Guigo,20,21,22 Jean Monlong,20,21,23 Ferran Reverter,20,24 Diego Garrido,20,21 Manuel Munoz,20,21 Gireesh Bogu,20,21 Reza Sodaei,20,21 Panagiotis Papasaikas,20,21 Anne W. Ndungu,13 Stephen B. Montgomery,25 Xin Li,25 Laure Fresard,25 Joe R. Davis,25 Emily K. Tsang,25,26 Zachary Zappala,25 Nathan S. Abell,25 Michael J. Gloudemans,25,26 Boxiang Liu,25,27 Farhan N. Damani,28 Ashis Saha,28 Yungil Kim,18 Benjamin J. Strober,29 Yuan He,29 Matthew Stephens,30,31 Jonathan K. Pritchard,30,32,33 Xiaoquan Wen,34 Sarah Urbut,30 Nancy J. Cox,35,36 Dan L. Nicolae,37 Eric R. Gamazon,35,36 Hae Kyung Im,38 Christopher D. Brown,39 Barbara E. Engelhardt,40 YoSon Park,39 Brian Jo,41 Ian C. McDowell,42 Ariel Gewirtz,41 Genna Gliner,43 Don Conrad,44,45 Ira Hall,46,47,48 Colby Chiang,46 Alexandra Scott,46 Chiara Sabatti,49 Eleazar Eskin,50 Christine Peterson,51 Farhad Hormozdiari,52 Eun Yong Kang,52 Serghei Mangul,52 Buhm Han,53 Jae Hoon Sul;54 Enhancing GTEx funded group: Andrew P. Feinberg,55 Lindsay F. Rizzardi,56 Kasper D. Hansen,57 Peter Hickey,58 Joshua Akey,59 Manolis Kellis,4,60 Jin Billy Li,61 Michael Snyder,61 Hua Tang,61 Lihua Jiang,61 Shin Lin,61,62 Barbara E. Stranger,63 Marian Fernando,64 Meritxell Oliva,64 John Stamatoyannopoulos,65 Rajinder Kaul,65 Jessica Halow,65 Richard Sandstrom,65 Eric Haugen,65 Audra Johnson,65 Kristen Lee,65 Daniel Bates,65 Morgan Diegel,65 Brandon L. Pierce,66 Lin Chen,66 Muhammad G. Kibriya,66 Farzana Jasmine,66 Jennifer Doherty,67 Kathryn Demanelis,66 Stephen B. Montgomery,25 Emily K. Tsang,25 Kevin S. Smith,25 Qin Li,61 Rui Zhang;61 National Institutes of Health (NIH) Common Fund: Concepcion R. Nierras;68 NIH/NCI: Helen M. Moore,69 Abhi Rao,69 Ping Guan,69 Jimmie B. Vaught,69 Philip A. Branton,69 Latarsha J. Carithers;70 NIH/NHGRI: Simona Volpi,71 Jeffery P. Struewing,71 Casey G. Martin,71 Lockhart C. Nicole;71 NIH/NIMH: Susan E. Koester,72 Anjene M. Addington;72 NIH/NIDA: A. Roger. Little;73 Biospecimen Collection Source Site–National Disease Research Interchange: William F. Leinweber,74 Jeffrey A. Thomas,74 Gene Kopen,74 Alisa McDonald,74 Bernadette Mestichelli,74 Saboor Shad,74 John T. Lonsdale,74 Michael Salvatore,74 Richard Hasz,75 Gary Walters,76 Mark Johnson,76 Michael Washington,76 Lori E. Brigham,77 Christopher Johns,78 Joseph Wheeler,78 Brian Roe,79 Marcus Hunter,79 Kevin Myer;79 Biospecimen Collection Source Site–Roswell Park Cancer Institute: Barbara A. Foster,80 Michael T. Moser,80 Ellen Karasik,80 Bryan M. Gillard,80 Rachna Kumar,80 Jason Bridge,81 Mark Miklos;81 Biospecimen Core Resource–Van Andel Research Institute: Scott D. Jewell,82 Daniel C. Rohrer,82 Dana Valley,82 Robert G. Montroy;82 Brain Bank Repository–University of Miami: Deborah C. Mash,83 David A. Davis;84 Leidos Biomedical Project Management: Anita H. Undale,85 Anna M. Smith,86 David E. Tabor,86 Nancy V. Roche,86 Jeffrey A. McLean,86 Negin Vatanian,86 Karna L. Robinson,86 Leslie Sobin,86 Mary E. Barcus,87 Kimberly M. Valentino,86 Liqun Qi,86 Stephen Hunter,86 Pushpa Hariharan,86 Shilpi Singh,86 Ki Sung Um,86 Takunda Matose,86 Maria M. Tomadzewski;86 Ethical, Legal, and Social Implications Study: Laura A. Siminoff,88 Heather M. Traino,89 Maghboeba Mosavel,90 Laura K. Barker;91 Genome Browser Data Integration, and Visualization–European Bioinformatics Institute: Daniel R. Zerbino,92 Thomas Juettmann,92 Kieron Taylor,92 Magali Ruffier,92 Dan Sheppard,92 Steven Trevanion,92 Paul Flicek;92 Genome Browser Data Integration and Visualization–Genomics Institute, University of California, Santa Cruz: W. James Kent,93 Kate R. Rosenbloom,93 Maximilian Haeussler,93 Christopher M. Lee,93 Benedict Paten,93 John Vivan,93 Jingchun Zhu,93 Mary Goldman,93 Brian Craft;93 Other members of the AWG: Gen Li,94 Pedro G. Ferreira,95,96 Esti Yeger-Lotem,97,98 Matthew T. Maurano,99 Ruth Barshir,97 Omer Basha,97 Hualin S. Xi,100 Jie Quan,100 Michael Sammeth,101 Judith B. Zaugg102
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/9/386/eaal5209/DC1
Materials and Methods
Fig. S1. Expression of commonly disrupted muscle disease genes in muscle, blood, and fibroblasts.
Fig. S2. PCA based on PSI metrics and gene expression of GTEx and patient samples.
Fig. S3. Overview of results from expression outlier analysis.
Fig. S4. Evaluation of RNA-seq around pathogenic essential splice site variants previously identified by genetic analysis.
Fig. S5. Overview of splice junction filtering approach.
Fig. S6. Number of potentially pathogenic splice events identified per patient.
Fig. S7. Examples of splice disruption in patients with no diagnosis at the completion of the study.
Fig. S8. Identification of aberrant splicing overlapping structural variants with RNA-seq.
Fig. S9. Resolving the effect of extended splice site variants with RNA-seq.
Fig. S10. Coverage of exon harboring splice-disrupting variant identified in patient C9 in RNA-seq and WES.
Fig. S11. Assignment of pathogenicity to missense and synonymous variants with RNA-seq.
Fig. S12. Identification of pathogenic noncoding varants with RNA-seq.
Fig. S13. Overview of triplicate region remapping.
Fig. S14. Comparison of the number of reads aligning to exons harboring pathogenic variants identified in the study in GTEx muscle, whole blood, and fibroblast tissues.
Fig. S15. Evaluation of splice prediction algorithms.
Fig. S16. Identification of allele imbalance with RNA-seq.
Table S1. Overview of clinical cases that underwent RNA-seq.
Table S2. Summary of patients previously diagnosed by genetic analysis with variants expected to result in transcriptional aberrations and the corresponding effect seen in the RNA-seq data.
Table S3. Comparison of quality metrics between patient and GTEx RNA-seq samples showing correspondence between patients and controls.
Table S4. List of 184 GTEx control skeletal muscle RNA-seq samples.
Table S5. PCR conditions and primers used for RT-PCR validation of splice aberrations identified via RNA-seq and Sanger sequencing of cDNA.
Table S6. PCR conditions and primers used for genomic Sanger sequence validation of variants identified in patients.
Competing interests
C.G.B., V.B., D.G.M., M.L., B.B.C., and S. Wilton are inventors on U.S. Provisional Patent Application no. 62/358,482, which covers “Diagnosing COL6-related disorders and methods for treating same,” and was filed on 5 July 2016 by NINDS. D.G.M. is a founder and owns stock in Goldfinch Biopharma, but this work is unrelated to the company. All other authors declare that they have no competing interests.
Data and materials availability
Patient sequencing data generated as part of this study were deposited in dbGaP under accession ID phs000655.v3.p1. GTEx transcriptome sequencing data can be obtained from dbGaP under accession ID phs000424.v6.p1. Code for splice junction discovery, normalization, and filtering is available on https://github.com/berylc/MendelianRNA-seq. List of OMIM and neuromuscular disease genes used for splice detection and allele-specific expression analysis can be found at https://github.com/macarthur-lab/omim and https://github.com/berylc/MendelianRNA-seq, respectively.
Author contributions
B.B.C., T.T., and D.G.M. conceived and designed the experiments. B.B.C. and T.T. analyzed the RNA-seq data. J.L.M., Y.H., A.B., and M.R.D. designed and performed validation experiments. B.B.C., M.L., S.D., A.R.F., L.B.W., S.A.S., G.L.O., H.M.R., E.C.O., R.G., S.T.C., and C.G.B. analyzed the exome and whole-genome data. S.D., A.R.F., V.B., L.B.W., S.A.S., G.L.O., E.E., H.M.R., A.S., H.G., K.C., E.C.O., R.G., N.G.L., A.T., A.H.B., P.B.K., K.N.N., V.S., J.J.D., F.M., N.F.C., S.T.C., and C.G.B. provided patient samples and clinical information. The Broad CMG and GTEx provided sequencing support for patient and control DNA sequencing and RNA-seq. F.Z., B.W., K.J.K., A.H.O.-L., D.B., and H.J. contributed reagents, materials, and analysis tools. J.L.M., T.T., M.L., S.D., A.R.F., V.B., L.B.W., S.A.S., K.J.K., A.H.O.-L., E.C.O., N.G.L., A.T., J.J.D., C.G.B., and S.T.C. critically evaluated the manuscript. B.B.C. and D.G.M. wrote the manuscript.
Footnotes
Broad Institute of MIT and Harvard University, Cambridge, MA 02142, USA.
Massachusetts General Hospital Cancer Center and Department of Pathology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA 02114, USA.
Broad Institute of MIT and Harvard University, Cambridge, MA 02142, USA.
Department of Statistics and Operations Research and Department of Biostatistics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599–3260, USA.
Bioinformatics Research Center and Departments of Statistics and Biological Sciences, North Carolina State University, Raleigh, NC 27695, USA.
Center for Biomarker Research and Personalized Medicine, Virginia Commonwealth University, Richmond, VA 23298–0581, USA.
Department of Statistics and Operations Research, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599–3260, USA.
Bioinformatics Research Center and Department of Biological Sciences, North Carolina State University, Raleigh, NC 27695, USA.
Department of Genetic Medicine and Development, University of Geneva Medical School, 1211 Geneva, Switzerland.
Institute for Genetics and Genomics in Geneva (iGE3), University of Geneva, 1211 Geneva, Switzerland.
Swiss Institute of Bioinformatics, 1211 Geneva, Switzerland.
Wellcome Trust Centre for Human Genetics Research, Nuffield Department of Clinical Medicine, University of Oxford, Oxford OX3 7BN, U.K.
Oxford Centre for Diabetes, Endocrinology and Metabolism, Churchill Hospital, University of Oxford, Oxford OX3 7LE, U.K.
Oxford National Institute for Health Research Biomedical Research Centre, Churchill Hospital, Oxford OX3 7LJ, U.K.
New York Genome Center, 101 Avenue of the Americas, New York, NY 10013, USA.
Department of Systems Biology, Columbia University Medical Center, New York, NY 10032, USA.
Department of Computer Science, Johns Hopkins University, Baltimore, MD 21218, USA.
Department of Computer Science, Stanford University, Stanford, CA 94305, USA.
Center for Genomic Regulation, Barcelona, Catalonia, Spain.
Universitat Pompeu Fabra, 08003 Barcelona, Catalonia, Spain.
Institut Hospital del Mar d’Investigacions Mèdiques, 08003 Barcelona, Spain.
Department of Human Genetics, McGill University, Montréal, Québec, Canada.
Universitat de Barcelona, 08028 Barcelona, Catalonia, Spain.
Departments of Genetics and Pathology, Stanford University, Stanford, CA 94305, USA.
Biomedical Informatics Program, Stanford University, Stanford, CA 94305, USA.
Department of Biology, Stanford University, Stanford, CA 94305, USA.
Department of Computer Science, Johns Hopkins University, Baltimore, MD 21218, USA.
Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21218, USA.
Department of Human Genetics, University of Chicago, Chicago, IL 60637, USA.
Department of Statistics, University of Chicago, 5734 South University Avenue, Chicago, IL 60637, USA.
Departments of Genetics and Biology, Stanford University, Stanford, CA 94305, USA.
Howard Hughes Medical Institute, Chevy Chase, MD 10032, USA.
Department of Biostatistics, University of Michigan, 1415 Washington Heights, Ann Arbor, MI 48109, USA.
Division of Genetic Medicine, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN 37232, USA.
Department of Clinical Epidemiology, Biostatistics and Bioinformatics and Department of Psychiatry, Academic Medical Center, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, Netherlands.
Section of Genetic Medicine, Department of Medicine, Department of Statistics, and Department of Human Genetics, University of Chicago, 900 East 57th Street KCBD 3220, Chicago, IL 60637, USA.
Section of Genetic Medicine, Department of Medicine, University of Chicago, 900 East 57th Street KCBD 3220, Chicago, IL 60637, USA.
Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Department of Computer Science, Center for Statistics and Machine Learning, Princeton University, 35 Olden Street, Princeton, NJ 08540, USA.
Lewis-Sigler Institute, Princeton University, Princeton, NJ 08540, USA.
Computational Biology and Bioinformatics Graduate Program, Duke University, Durham, NC 27708, USA.
Department of Operations Research and Financial Engineering, Princeton University, Princeton, NJ 08540, USA.
Department of Genetics, Washington University School of Medicine, St. Louis, MO 63108, USA.
Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO 63108, USA.
McDonnell Genome Institute, Washington University School of Medicine, Saint Louis, MO 63108, USA.
Department of Medicine, Washington University School of Medicine, Saint Louis, MO 63108, USA.
Department of Genetics, Washington University School of Medicine, Saint Louis, MO 63108, USA.
Departments of Biomedical Data Science and Statistics, Stanford University, Health Research and Policy Redwood building, Stanford, CA 94305–5404, USA.
Departments of Computer Science and Human Genetics, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Department of Biostatistics, University of Texas MD Anderson Cancer Center, 1400 Pressler Street, Houston, TX 77030, USA.
Department of Computer Science, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Department of Convergence Medicine, University of Ulsan College of Medicine, Asan Medical Center, Mugeo-dong, Nam-gu, Ulsan, Korea.
Department of Psychiatry and Biobehavioral Sciences, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Center for Epigenetics, Johns Hopkins University School of Medicine, and Departments of Medicine, Biomedical Engineering, and Mental Health, Johns Hopkins University Schools of Medicine, Engineering, and Public Health, Baltimore, MD 21205, USA.
Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
McKusick-Nathans Institute of Genetic Medicine, Center for Epigenetics, Johns Hopkins School of Medicine, and Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD 21205, USA.
Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD 21205, USA.
Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA.
Computer Science and Artificial Intelligence Laboratory, MIT, Cambridge, MA 02139, USA.
Department of Genetics, Stanford University, Stanford, CA 94305, USA.
Division of Cardiology, University of Washington, Seattle, WA 98195, USA.
Section of Genetic Medicine, Department of Medicine, Institute for Genomics and Systems Biology, Center for Data Intensive Science, University of Chicago, Chicago, IL 60637, USA.
Section of Genetic Medicine, Department of Medicine, Institute for Genomics and Systems Biology, University of Chicago, Chicago, IL 60637, USA.
Altius Institute for Biomedical Sciences, Seattle, WA 98121, USA.
Department of Public Health Sciences, University of Chicago, Chicago, IL 60637, USA.
Department of Epidemiology, Geisel School of Medicine at Dartmouth, Lebanon, NH 03756, USA.
Office of Strategic Coordination, Division of Program Coordination, Planning, and Strategic Initiatives, Rockville, MD 20852–9305, USA.
Biorepositories and Biospecimen Research Branch, Division of Cancer Treatment and Diagnosis, NCI, Bethesda, MD 20892, USA.
National Institute of Dental and Craniofacial Research, 6701 Democracy Boulevard, Bethesda, MD 20892, USA.
Division of Genomic Medicine, NHGRI, Rockville, MD 20892, USA.
Division of Neuroscience and Basic Behavioral Science, NIMH, NIH, Bethesda, MD 20892, USA.
NIDA, NIH, U.S. Department of Health and Human Services, Bethesda, MD 20892, USA.
National Disease Research Interchange, Philadelphia, PA 19103, USA.
Gift of Life Donor Program, Philadelphia, PA 19103, USA.
LifeNet Health, Virginia Beach, VA 23453, USA.
Washington Regional Transplant Community, Annandale, VA 22003, USA.
Center for Organ Recovery and Education, Pittsburgh, PA 15238, USA.
LifeGift, Houston, TX 77054, USA.
Roswell Park Cancer Institute Pharmacology and Therapeutics, Buffalo, NY 14263, USA.
Unyts, 110 Broadway, Buffalo, NY 14203, USA.
Van Andel Research Institute, Grand Rapids, MI 49503, USA.
Department of Neurology, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Brain Endowment Bank, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
National Institute of Allergy and Infectious Diseases, NIH, 5601 Fishers Lane, Rockville, MD 20852, USA.
Biospecimen Research Group, Clinical Research Directorate, Leidos Biomedical Research Inc., Rockville, MD 20852, USA.
Frederick National Laboratory for Cancer Research, 8560 Progress Drive, Room C3021, Frederick, MD 21701, USA.
Temple University, Philadelphia, PA 19122, USA.
Temple University, Ritter Annex 9th Floor, 1301 Cecil B. Moore Avenue, Philadelphia, PA 19122, USA.
Virginia Commonwealth University, Richmond, VA 23219, USA.
Temple University, Philadelphia, PA 19122, USA.
European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, U.K.
Genomics Institute, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.
Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, NY 10032, USA.
i3S Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Rua Alfredo Allen, 208, 4200–135 Porto, Portugal.
IPATIMUP–Institute of Molecular Pathology and Immunology, University of Porto, Rua Dr. Roberto Frias sin número, 4200–625 Porto, Portugal.
Ben-Gurion University of the Negev, Beer-Sheva, 84105 Israel.
National Institute for Biotechnology in the Negev, Beer-Sheva 84105, Israel.
Institute for Systems Genetics, New York University Langone Medical Center, New York, NY 10016, USA.
Computational Sciences, Pfizer Inc., 610 Main Street, Cambridge, MA 02140, USA.
Institute of Biophysics Carlos Chagas Filho, Federal University of Rio de Janeiro (UFRJ), 21941902 Rio de Janeiro, Brazil.
European Molecular Biology Laboratory, Meyerhofstrasse 1, 69117 Heidelberg, Germany.
REFERENCES AND NOTES
- 1.Ankala A, da Silva C, Gualandi F, Ferlini A, Bean LJ, Collins C, Tanner AK, Hegde MR. A comprehensive genomic approach for neuromuscular diseases gives a high diagnostic yield. Ann Neurol. 2015;77:206–214. doi: 10.1002/ana.24303. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, Eng CM. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor JC, Martin HC, Lise S, Broxholme J, Cazier J-B, Rimmer A, Kanapin A, Lunter G, Fiddy S, Allan C, Aricescu AR, Attar M, Babbs C, Becq J, Beeson D, Bento C, Bignell P, Blair E, Buckle VJ, Bull K, Cais O, Cario H, Chapel H, Copley RR, Cornall R, Craft J, Dahan K, Davenport EE, Dendrou C, Devuyst O, Fenwick AL, Flint J, Fugger L, Gilbert RD, Goriely A, Green A, Greger IH, Grocock R, Gruszczyk AV, Hastings R, Hatton E, Higgs D, Hill A, Holmes C, Howard M, Hughes L, Humburg P, Johnson D, Karpe F, Kingsbury Z, Kini U, Knight JC, Krohn J, Lamble S, Langman C, Lonie L, Luck J, McCarthy D, McGowan SJ, McMullin MF, Miller KA, Murray L, Németh AH, Andrew MN, Nutt D, Ormondroyd E, Bang Oturai A, Pagnamenta A, Patel SY, Percy M, Petousi N, Piazza P, Piret SE, Polanco-Echeverry G, Popitsch N, Powrie F, Pugh C, Quek L, Robbins PA, Robson K, Russo A, Sahgal N, van Schouwenburg PA, Schuh A, Silverman E, Simmons A, Sørensen PS, Sweeney E, Taylor J, Thakker RV, Tomlinson I, Trebes A, Twigg SRF, Uhlig HH, Vyas P, Vyse T, Wall SA, Watkins H, Whyte MP, Witty L, Wright B, Yau C, Buck D, Humphray S, Ratcliffe PJ, Bell JI, Wilkie AOM, Bentley D, Donnelly P, McVean G. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet. 2015;47:717–726. doi: 10.1038/ng.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, Harrell TM, McMillin MJ, Wiszniewski W, Gambin T, Coban Akdemir ZH, Doheny K, Scott AF, Avramopoulos D, Chakravarti A, Hoover-Fong J, Mathews D, Witmer PD, Ling H, Hetrick K, Watkins L, Patterson KE, Reinier F, Blue E, Muzny D, Kircher M, Bilguvar K, López-Giráldez F, Sutton VR, Tabor HK, Leal SM, Gunel M, Mane S, Gibbs RA, Boerwinkle E, Hamosh A, Shendure J, Lupski JR, Lifton RP, Valle D, Nickerson DA, Bamshad MJ Centers for Mendelian Genomics. The genetic basis of Mendelian phenotypes: Discoveries, challenges, and opportunities. Am J Hum Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacArthur D, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein DB, Allen A, Keebler J, Margulies EH, Petrou S, Petrovski S, Sunyaev S. Sequencing studies in human genetics: Design and interpretation. Nat Rev Genet. 2013;14:460–470. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lek M, MacArthur D. The challenge of next generation sequencing in the context of neuromuscular diseases. J Neuromuscul Dis. 2014;1:135–149. [PubMed] [Google Scholar]
- 8.Wang Z, Gerstein M, Snyder M. RNA-seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat Rev Genet. 2016;17:257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colapietro P, Colapietro P, Gervasini C, Natacci F, Rossi L, Riva P, Larizza L. NF1 exon 7 skipping and sequence alterations in exonic splice enhancers (ESEs) in a neurofibromatosis 1 patient. Hum Genet. 2003;113:551–554. doi: 10.1007/s00439-003-1009-2. [DOI] [PubMed] [Google Scholar]
- 12.Morel CF, Thomas MA, Cao H, O’Neil CH, Pickering JG, Foulkes WD, Hegele RA. A LMNA splicing mutation in two sisters with severe Dunnigan-type familial partial lipodystrophy type 2. J Clin Endocrinol Metabol. 2006;91:2689–2695. doi: 10.1210/jc.2005-2746. [DOI] [PubMed] [Google Scholar]
- 13.Eriksson M, Ted Brown W, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonorazky H, Liang M, Cummings B, Lek M, Micallef J, Hawkins C, Basran R, Cohn R, Wilson MD, MacArthur D, Marshall CR, Ray PN, Dowling JJ. RNAseq analysis for the diagnosis of muscular dystrophy. Ann Clin Transl Neurol. 2016;3:55–60. doi: 10.1002/acn3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Kim C, Bradfield J, Guo Y, Toskala E, Otieno FG, Hou C, Thomas K, Cardinale C, Lyon GJ, Golhar R, Hakonarson H. Whole-genome DNA/RNA sequencing identifies truncating mutations in RBCK1 in a novel Mendelian disease with neuromuscular and cardiac involvement. Genome Med. 2013;5:67. doi: 10.1186/gm471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung H, Lee D, Lee J, Park D, Jeong Kim Y, Park W-Y, Hong D, Park PJ, Lee E. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 2015;47:1242–1248. doi: 10.1038/ng.3414. [DOI] [PubMed] [Google Scholar]
- 17.Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, Johnson R, Segrè AV, Djebali S, Niarchou A, Wright FA, Lappalainen T, Calvo M, Getz G, Dermitzakis ET, Ardlie KG, Guigó R GTEx Consortium. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bönnemann CG, Wang CH, Quijano-Roy S, Deconinck N, Bertini E, Ferreiro A, Muntoni F, Sewry C, Béroud C, Mathews KD, Moore SA, Bellini J, Rutkowski A, North KN Members of the International Standard of Care Committee for Congenital Muscular Dystrophies. Diagnostic approach to the congenital muscular dystrophies. Neuromuscul Disord. 2014;24:289–311. doi: 10.1016/j.nmd.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald CM. Clinical approach to the diagnostic evaluation of hereditary and acquired neuromuscular diseases. Phys Med Rehabil Clin N Am. 2012;23:495–563. doi: 10.1016/j.pmr.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begay RL, Graw S, Sinagra G, Merlo M, Slavov D, Gowan K, Jones KL, Barbati G, Spezzacatene A, Brun F, Di Lenarda A, Smith JE, Granzier HL, Mestroni L, Taylor M Familial Cardiomyopathy Registry. Role of titin missense variants in dilated cardiomyopathy. J Am Heart Assoc. 2015;4:e002645. doi: 10.1161/JAHA.115.002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roca X, Krainer AR, Eperon IC. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev. 2013;27:129–144. doi: 10.1101/gad.209759.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, Maller JB, Kukurba KR, DeLuca DS, Fromer M, Ferreira PG, Smith KS, Zhang R, Zhao F, Banks E, Poplin R, Ruderfer DM, Purcell SM, Tukiainen T, Minikel EV, Stenson PD, Cooper DN, Huang KH, Sullivan TJ, Nedzel J, Bustamante CD, Billy Li J, Daly MJ, Guigo R, Donnelly P, Ardlie K, Sammeth M, Dermitzakis ET, McCarthy MI, Montgomery SB, Lappalainen T, MacArthur DG GTEx Consortium, Geuvadis Consortium. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiiski K, Lehtokari VL, Löytynoja A, Ahlstén L, Laitila J, Wallgren-Pettersson C, Pelin K. A recurrent copy number variation of the NEB triplicate region: Only revealed by the targeted nemaline myopathy CGH array. Eur J Hum Genet. 2015;24:574–580. doi: 10.1038/ejhg.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bang M-L, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. The complete gene sequence of titin, expression of an unusual ≈700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 29.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butterfield RJ, Foley AR, Dastgir J, Asman S, Dunn DM, Zou Y, Hu Y, Donkervoort S, Flanigan KM, Swoboda KJ, Winder TL, Weiss RB, Bönnemann CG. Position of glycine substitutions in the triple helix of COL6A1, COL6A2, and COL6A3 is correlated with severity and mode of inheritance in collagen VI myopathies. Hum Mutat. 2013;34:1558–1567. doi: 10.1002/humu.22429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spurdle AB, Couch FJ, Hogervorst FB, Radice P, Sinilnikova OM IARC Unclassified Genetic Variants Working Group. Prediction and assessment of splicing alterations: Implications for clinical testing. Hum Mutat. 2008;29:1304–1313. doi: 10.1002/humu.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duzkale H, Shen J, McLaughlin H, Alfares A, Kelly MA, Pugh TJ, Funke BH, Rehm HL, Lebo MS. A systematic approach to assessing the clinical significance of genetic variants. Clin Genet. 2013;84:453–463. doi: 10.1111/cge.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kremer LS, Bader DM, Mertes C, Kopajtich R, Pichler G, Iuso A, Haack TB, Graf E, Schwarzmayr T, Terrile C, Konafikova E, Repp B, Kastenmüller G, Adamski J, Lichtner P, Leonhardt C, Funalot B, Donati A, Tiranti V, Lombes A, Jardel C, Gläser D, Taylor RW, Ghezzi D, Mayr JA, Rötig A, Freisinger P, Distelmaier F, Strom TM, Meitinger T, Gagneur J. Genetic diagnosis of Mendelian disorders via RNA sequencing. 2016;2016 doi: 10.1101/066738. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schafer S, Miao K, Benson CC, Heinig M, Cook SA, Hubner N. Alternative splicing signatures in RNA3seq data: Percent spliced in (PSI) Curr Protoc Hum Genet. 2015;87:11.16.1–11.16.14. doi: 10.1002/0471142905.hg1116s87. [DOI] [PubMed] [Google Scholar]
- 36.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kähler A, Duncan L, Stahl E, Genovese G, Fernández E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PKE, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SGN, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, Sklar P. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Levy Moonshine A, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won H-H, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:1–12. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 40.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 41.Pertea M, Lin X, Salzberg SL. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 2001;29:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mersch B, Gepperth A, Suhai S, Hotz-Wagenblatt A. Automatic detection of exonic splicing enhancers (ESEs) using SVMs. BMC Bioinformatics. 2008;9:369. doi: 10.1186/1471-2105-9-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chasin LA. Searching for splicing motifs. Adv Exp Med Biol. 2008;623:85–106. doi: 10.1007/978-0-387-77374-2_6. [DOI] [PubMed] [Google Scholar]
- 45.Cooper ST, Lo HP, North KN. Single section Western blot Improving the molecular diagnosis of the muscular dystrophies. Neurology. 2003;61:93–97. doi: 10.1212/01.wnl.0000069460.53438.38. [DOI] [PubMed] [Google Scholar]
- 46.Chan Y, Tong H-Q, Beggs AH, Kunkel LM. Human skeletal muscle-specific α-actinin-2 and-3 isoforms form homodimers and heterodimers in vitro and in vivo. Biochem Biophys Res Commun. 1998;248:134–139. doi: 10.1006/bbrc.1998.8920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.