Abstract
KLF11 (TIEG2) is a pancreas-enriched transcription factor that has elicited significant attention because of its role as negative regulator of exocrine cell growth in vitro and in vivo. However, its functional role in the endocrine pancreas remains to be established. Here, we report, for the first time, to our knowledge, the characterization of KLF11 as a glucose-inducible regulator of the insulin gene. A combination of random oligonucleotide binding, EMSA, luciferase reporter, and chromatin immunoprecipitation assays shows that KLF11 binds to the insulin promoter and regulates its activity in beta cells. Genetic analysis of the KLF11 gene revealed two rare variants (Ala347Ser and Thr220Met) that segregate with diabetes in families with early-onset type 2 diabetes, and significantly impair its transcriptional activity. In addition, analysis of 1,696 type 2 diabetes mellitus and 1,776 normoglycemic subjects show a frequent polymorphic Gln62Arg variant that significantly associates with type 2 diabetes mellitus in North European populations (OR = 1.29, P = 0.00033). Moreover, this variant alters the corepressor mSin3A-binding activity of KLF11, impairs the activation of the insulin promoter and shows lower levels of insulin expression in pancreatic beta cells. In addition, subjects carrying the Gln62Arg allele show decreased plasma insulin after an oral glucose challenge. Interestingly, all three nonsynonymous KLF11 variants show increased repression of the catalase 1 promoter, suggesting a role in free radical clearance that may render beta cells more sensitive to oxidative stress. Thus, both functional and genetic analyses reveal that KLF11 plays a role in the regulation of pancreatic beta cell physiology, and its variants may contribute to the development of diabetes.
Keywords: insulin, polymorphisms, TGF-β, type 2 diabetes
Components of both the exocrine and endocrine pancreas are affected by diseases, e.g., pancreatic cancer and type 2 diabetes mellitus (T2DM), which severely compromise both the quality and span of human life. Both glandular compartments share the same cellular origin and early morphogenetic pathways, suggesting a close functional and pathophysiological relationship. For instance, the exocrine-specific transcription factor p48 and the endocrine-specific pancreatic duodenal homeobox gene 1 (PDX-1) are both expressed in the common cell precursor (1); and, under pathological conditions their compartmentalization may be lost, as exemplified by the detection of PDX-1 in pancreatic cancer (2). In fact, T2DM is both a common feature and a risk factor for the subsequent development of pancreatic cancer (3, 4). The TGF-β-inducible transcription factor KLF11 regulates exocrine cell growth and behaves as a tumor suppressor in pancreatic cancer (ref. 5 and M.E.F.-Z. and R.U., unpublished observation). Because the TGF-β signaling pathway is also a major regulator of endocrine cell fate (1, 6), the current study has been designed to define the role of the Sp1-like transcription factor KLF11 in the biology of the endocrine beta cell. Interestingly, several beta cell-specific genes display a potential binding site for Sp1 proteins in their promoter, suggesting that KLF11 is a potential endocrine regulator. Because the TGF-β signaling pathway is crucial for pancreatic development and the impaired beta cell function observed in maturity-onset diabetes of the young (MODY) is caused by mutations in transcription factor genes that are also important in pancreas development [hepatocyte nuclear factors and PDX-1 (7), KLF11 may also be a candidate gene for predisposition to diabetes. Indeed, here we show that KLF11 is a glucose-induced regulator of the insulin gene and functional KLF11 gene variants are significantly associated with diabetes. These data, in combination with previous studies on KLF11, outline a molecular pathway important in both pancreatic cancer and diabetes.
Methods
Promoter Sequence Analysis. For EMSAs, purified GST-fusion proteins of KLF11 (8) were incubated in a buffer containing 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl, 10 mM Tris·HCl (pH 7.5), and 50 μg/ml poly dI-dC (Promega) for 10 min at room temperature, and incubated for an additional 20 min with 0.035 pmol of end-labeled probes. The reactions were analyzed by electrophoresis on 4% nondenaturing polyacrylamide gel followed by autoradiography. Site-directed mutagenesis of the GC box was generated by replacing each base with A, T, or C and densitometric units of each protein/probe complex were determined by using nih image 1.6.1 software. The relative binding specificity for an individual nucleotide at each position was calculated by using the following equation: (dNTP units)n/(SdNTPs units)n, where n = nucleotide position within the 9 bp GC box and is graphically represented by the height of letters, using the program weblogo (9).
Chromatin Immunoprecipitation (ChIP). At 24 h after transfection with KLF11 FLAG-tagged plasmids (8), βTC3 cells were cross-linked with formaldehyde for 20 min at 25°C, harvested in SDS-lysis buffer (Upstate Biotechnology, Lake Placid, NY), and sheared to fragment DNA to ≈500 bp. Samples were then immunoprecipitated by using an agarose-conjugated anti-FLAG antibody (Sigma-Aldrich, St. Louis) or agarose beads alone at 4°C overnight, washed, and eluted by using the ChIP kit (Upstate Biotechnology). Crosslinks were removed at 65°C for 4 h, and immunoprecipitated DNA was purified by using phenol/chloroform extraction and ethanol precipitation. Then, a 200-bp region of the insulin promoter was amplified by PCR, using the following primers 5′-GGCCATCTGCCTACCCACCC and 5′-AGGCCCAAAGAGGAGAGTACATAC and visualized by 2.5% agarose gel.
Reporter Assays. The KLF11 variants were generated following the recommendations of the Stratagene QuikChange kit. Then, βTC3 cells were transfected by using TransFast transfection (Promega, Charbonnieres, France). The KLF11 constructs were cotransfected with the Gal4 reporter construct (10), a luciferase reporter vector containing six tandem repeats of KLF11-binding sites (10), the human insulin promoter, or catalase 1 promoter (5), and the luciferase activity was measured as described (5). Association of endogenous mSin3A with KLF11 was analyzed by immunoprecipitation assays (ref. 8 and Supporting Methods, which is published as supporting information on the PNAS web site).
Genetic Analysis and Study Population. The identification and genotyping of genetic variants and calculation of logarithm of odds scores are described in Supporting Methods and Table 4, which is published as supporting information on the PNAS web site. The 96 normoglycemic control and 66 T2DM subjects of the pilot study were included in the familial study with 313 French cases from T2DM diabetes multiplex families that had at least one affected first-degree relative, and 313 control subjects (spouses) that were selected to match the cases for T2DM risk factors (Table 5, which is published as supporting information on the PNAS web site). The second study consisted of 1,383 consecutive T2DM patients and 1,463 control subjects older than 45 years of age from general populations (Table 5). Association was assessed by χ2 analysis (11). Logistic regression and ANOVA were performed by using SPSS software (SPSS, Chicago). Tests for deviation from Hardy-Weinberg equilibrium (12) and for association were adapted from Sasieni (13).
Results
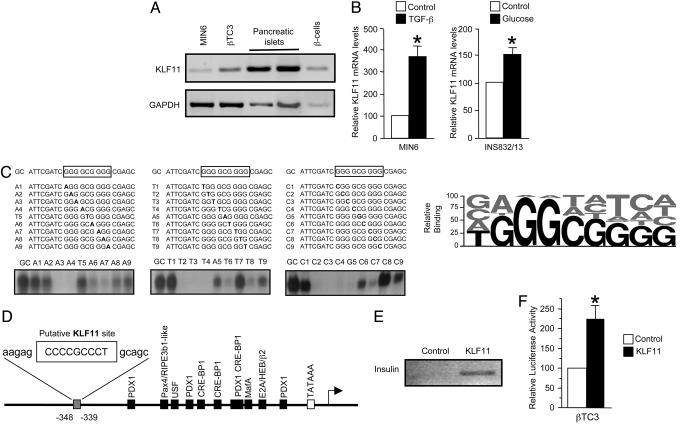
KLF11 Plays a Role in Glucose Signaling in Pancreatic Beta Cells. KLF11 is expressed in human pancreatic islets and in the pancreatic beta cell lines HIT-T15, INS832/13, βTC3, and Min6 (Fig. 1A and data not shown). Similar to its inducible expression in exocrine cells, KLF11 mRNA is up-regulated by TGF-β in beta cells (Fig. 1B). Under these conditions, mRNA insulin levels are significantly higher as compared with those at basal conditions. Moreover, high glucose levels induced KLF11 mRNA expression in beta cells (Fig. 1B). KLF11, as a member of the Sp1/KLF family, has been predicted to bind to either GC-rich or CACC sequences. Data from random oligonucleotide binding and EMSA define the most probable GC-box sequence that binds KLF11 as T/G-GGGCGGGG/A (Fig. 1C). Interestingly, such a sequence is present in the promoter region of the insulin gene (Fig. 1D). Both ChIP and luciferase reporter assays showed that KLF11 binds and activates the human insulin promoter in beta cells under high-glucose levels (Fig. 1 E and F). Thus, these data indicate that in pancreatic beta cells KLF11 is inducible by glucose and up-regulates levels of insulin expression. Therefore, KLF11 may be involved in a positive regulation loop that is important in glucose homeostasis, raising the question whether aberrant KLF11 function may predispose to diabetes. To address this issue, we analyzed the association of KLF11 gene variants with diabetes.
Fig. 1.
Role of KLF11 in pancreatic beta cells. (A) KLF11 mRNA expression in the beta cell lines Min6 and βTC3 in two independent isolations of human pancreatic islets, and in human beta cells isolated by zinc selection (14). (B) Quantative PCR measurement of KLF11 mRNA expression in both Min6 and INS832/13 cells. (C) EMSA with the KLF11-GST fusion protein showing binding to the wild-type and mutated GC boxes. (Right) A representation of the relative percentage of DNA binding (Lower). (D) Position of the putative KLF11-binding sequence of the insulin promoter. (E) Binding of KLF11 to insulin promoter analyzed by ChIP assays in βTC3 cells. (F) Luciferase assays of Flag-tagged KLF11 transiently cotransfected with the human insulin promoter reporter construct in βTC3 cells cultured in high-glucose medium (n = 6). *, significantly different (P < 0.05).
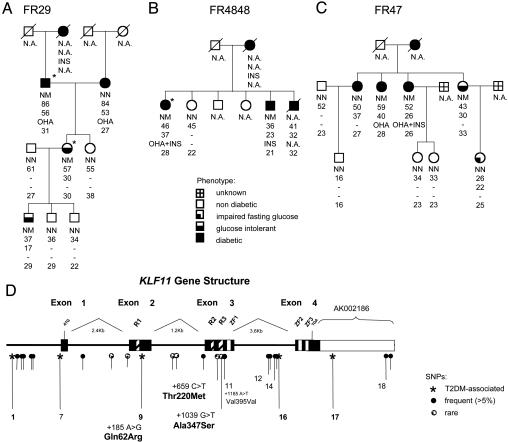
KLF11 Gene Variants Are Associated with Diabetes. Sequencing of the KLF11 gene in 190 probands of early-onset T2DM families identified two rare KLF11 variants present in three distinct families, but absent in 313 late-onset T2DM patients and 313 normoglycemic subjects: A MODY-like family with four diabetic generations (FR29) and a bilinear transmission of T2DM in the second generation presented the [+1,039 G>T (Ala347Ser)] variant that was transmitted with diabetes/glucose intolerance in all three generations examined (Fig. 2A). Linkage analysis under a dominant inheritance MODY-like (12) and late-onset T2DM model (15) revealed the maximal, theoretically possible logarithm of odds scores (0.6 and 0.68, respectively). The evidence for linkage is not likely to be an at random observation, because 10,000 simulations under the no-linkage hypothesis resulted in an empirical P value of 0.030 and 0.035, respectively. The second rare variant [+659 C>T (Thr220Met)] was identified in two diabetic sibling of family FR4848, and in the three sisters of family FR47 with either glucose intolerance or frank diabetes (Fig. 2 B and C, respectively). However, in the latter pedigree, this variant was absent in one diabetic family member with an onset of diabetes at a later age. Interestingly, the Ala347Ser variant is located within the third repressor domain of KLF11, and Thr220Met between two repressor domains. Bioinformatics analysis [using PIX, a peptide identification system, which can be accessed at www.hgmp.mrc.ac.uk/Registered/Webapp/pix/, and SOPMA (16)] predicted both variants alter the secondary protein structure of these domains, which may have functional implications on KLF11 transcriptional activity.
Fig. 2.
Identification of KLF11 variants. (A) Pedigree of the MODY-like family with SNP [+1,039 G>T Ala347Ser]. (B and C) Pedigree of early-onset diabetes families with SNP [+659 C>T Thr220Met]. Below the symbols are genotype, age at medical examination, age of onset, diabetic treatment, and BMI are indicated. OHA, oral hypoglycemic agent; INS, insulin. Asterisks indicate presence of diabetic complications (e.g., neuropathy). (D) KLF11 gene structure with the position of SNPs. R, repressor domain; Zn, zinc finger domain.
Analysis of the KLF11 gene-related sequences revealed 19 frequent KLF11 SNPs (minor allele frequency of >5%, Fig. 2D). Differences between allele frequencies in normoglycemic and T2DM subjects were first assessed in a pilot study with a cutoff P value of 0.2 to prevent exclusion of potentially T2DM associated SNPs elusive in this pilot (Table 6, which is published as supporting information on the PNAS web site). SNP 1, 9, 16, and 17 showed a highly significant association with T2DM in the subsequently analyzed cohort (P < 0.0002, Table 1). The T2DM-associated SNPs are in strong linkage disequilibrium (LD) with each other (D′ = 0.96-0.98), resulting in an association of combined haplotypes similar to that of individual SNPs (data not shown). To exclude that the causal, T2DM-associated variants reside in a neighboring gene, we genotyped 47 additional SNPs (Fig 4, which is published as supporting information on the PNAS web site). Both the variants [KLF11 AS + 2541 A>G] and [AK091299 UTR + 2422 A>T] in cilia-associated protein-1 (CYS1) that are in strong LD with the KLF11 variants, show a significant association with T2DM (Table 1). The T2DM-associated LD block of SNPs does not include other SNPs of CYS1, suggesting that the functional variant(s) is within KLF11 sequences. Thus, the underlying cause of association may result from combined effects of several KLF11 variants, or could depend on functionality of one SNP. A likely functional SNP is the nonsynonymous coding variant SNP9, which is responsible for a Gln to Arg conversion at amino acid 62, and which was predicted (PIX program) to affect the α-helix structure in proximity of the first repressor domain that is responsible for the interaction with the corepressor mSin3A. Furthermore, this SNP displays a highly significant T2DM association that is independent of age, sex, and body mass index (BMI) differences between subjects (binominal logistic regression, P = 0.0003). Further analysis of 1,463 normoglycemic and 1,383 T2DM subjects from North European populations shows that the allele frequency of [KLF11+185 A>G (Gln62Arg)] in T2DM subjects was significantly higher than in controls (Table 2, P = 0.033). Overall, the initial and second case-control study results in a combined OR of 1.29 (Table 3, 95% CI of OR = 1.12-1.49, P = 0.00033). The fraction of attributed risk for T2DM susceptibility of Gln62Arg in the followup study is 2% and 7% under an additive and a dominant model, respectively; and is similar to values recently reported for the CAP10 at-risk haplotype (17). Thus, genetic analyses using family-based and case-control studies reveal that KLF11 variants have an association with early-onset diabetes and T2DM.
Table 1.
SNP frequencies in French Caucasian case-control study
|
X2P values
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variants | Subject | NN | NM | MM | Multiple allele frequency | HWE* | Additive† | Codominant‡ | Recessive§ | Dominant¶ | |
| KLF11 | |||||||||||
| 1 | Adjacent sequence (AS) | Control | 244 | 62 | 4 | 0.11 | 0.98 | ||||
| −1,659 G>C | T2DM | 202 | 88 | 8 | 0.17 | 0.67 | 0.0022 | 0.0084 | 0.22 | 0.0023 | |
| 7 | Promoter −86 del/ins | 5-5 | 5-4/5-6 | 4-4/4-6/6-6 | 4/6 | ||||||
| (GCC)∥ | Control | 145 | 127/16 | 20/2/0 | 0.27/0.03 | 0.24 | 0.08‡‡ | 0.15‡‡ | |||
| T2DM | 130 | 129/6 | 31/1/0 | 0.32/0.01 | 0.76 | 0.03** | 0.17†† | 0.31§§ | 0.07§§ | ||
| 9 | Exon 2 + 185 A>G, | Control | 252 | 58 | 3 | 0.10 | 0.87 | ||||
| Gln62Arg | T2DM | 211 | 95 | 7 | 0.17 | 0.33 | 0.00023 | 0.00083 | 0.20 | 0.00019 | |
| 11 | Exon 3 + 1185 A>T, | Control | 202 | 96 | 13 | 0.20 | 0.71 | ||||
| Val395Val | T2DM | 192 | 100 | 7 | 0.19 | 0.15 | 0.81 | 0.39 | 0.20 | 0.85 | |
| 12 | IVS3 + 976 T>C | Control | 198 | 96 | 14 | 0.20 | 0.59 | ||||
| T2DM | 191 | 102 | 8 | 0.20 | 0.19 | 0.82 | 0.39 | 0.21 | 0.83 | ||
| 14 | IVS3 + 1,331 T>C | Control | 282 | 30 | 1 | 0.05 | 0.83 | ||||
| T2DM | 262 | 22 | 0 | 0.04 | 0.50 | 0.30 | 0.46 | 0.34 | 0.35 | ||
| 16 | IVS3 + 1,398 del(A) | Control | 239 | 57 | 2 | 0.10 | 0.48 | ||||
| T2DM | 192 | 93 | 7 | 0.17 | 0.27 | 0.00055 | 0.0021 | 0.09 | 0.000077 | ||
| 17 | AS + 343 G>T | Control | 240 | 64 | 2 | 0.11 | 0.56 | ||||
| T2DM | 207 | 89 | 7 | 0.17 | 0.68 | 0.0031 | 0.0096 | 0.09 | 0.0047 | ||
| 18 | AS + 1,357 C>T | Control | 194 | 91 | 13 | 0.20 | 0.58 | ||||
| T2DM | 195 | 101 | 9 | 0.20 | 0.34 | 0.96 | 0.56 | 0.36 | 0.76 | ||
| Non-KLF11 | |||||||||||
| 2 | TAF1B-SL1 + 16 T>G, | Control | 88 | 138 | 64 | 0.46 | 0.48 | ||||
| Ser6Ala | T2DM | 97 | 139 | 61 | 0.44 | 0.41 | 0.51 | 0.81 | 0.65 | 0.55 | |
| 6 | LBP32 + 1189 C>T, | Control | 180 | 111 | 14 | 0.23 | 0.63 | ||||
| Ile419Val | T2DM | 183 | 108 | 12 | 0.22 | 0.50 | 0.67 | 0.90 | 0.70 | 0.73 | |
| 17 | KLF11 AS + 2,541 A>G | Control | 240 | 61 | 3 | 0.11 | 1.00 | ||||
| T2DM | 214 | 88 | 6 | 0.16 | 0.53 | 0.0069 | 0.022 | 0.32 | 0.0064 | ||
| 21 | AK091299 UTR | Control | 245 | 59 | 4 | 0.11 | 0.77 | ||||
| +2,422 A>T | T2DM | 207 | 87 | 7 | 0.17 | 0.68 | 0.0028 | 0.095 | 0.34 | 0.0024 | |
| 25 | AK091299 UTR | Control | 174 | 123 | 10 | 0.23 | 0.04§§ | ||||
| +3,107 G>A | T2DM | 154 | 128 | 22 | 0.28 | 0.57 | 0.05 | 0.06 | 0.03 | 0.14 | |
| 44 | rs7587317 (genomic) | Control | 263 | 145 | 19 | 0.21 | 1.00 | ||||
| T2DM | 180 | 108 | 16 | 0.17 | 1.00 | 0.08 | 0.22 | 0.10 | 0.33 | ||
| 47 | RRM2 IVS8-177 A/G | Control | 163 | 142 | 26 | 0.29 | 0.60 | ||||
| T2DM | 171 | 118 | 17 | 0.25 | 0.65 | 0.07 | 0.19 | 0.10 | 0.07 | ||
Frequency analysis of variants showing a suggestive T2DM association in a pilot study (P < 0.2).
X2 test for deviations from the Hardy-Weinberg equilibrium.
X2 test for differences of allele frequencies between control and T2DM subjects.
X2 test for differences of genotype frequencies between control and T2DM subjects under a codominant model.
X2 test for differences of genotype frequencies between control and T2DM subjects under a recessive model.
X2 test for differences of genotype frequencies between control and T2DM subjects under a dominant model.
Triallelic variant with four, five, and six GCC repeats that was analyzed in a 2 × 3 contingency table with genotype frequencies.
Triallelic variant with four, five, and six GCC repeats that was analyzed with allele frequencies and a 2 × 6 contingency table with genotype frequencies.
Analyses in the recessive model of 4-4 alleles vs. all others, and in the dominant model of 4-4, 4-5, and 4-6 alleles vs. 5-5 and 5-6 alleles.
Analyses in the recessive model of 6-6 alleles vs. all others, and in the dominant model of 5-6, 4-6, and 6-6 alleles vs. 5-5, 5-4, and 4-4 alleles.
Deviated from HWE, 3.2% of the genotypes were reanalyzed without observing errors. By using an Armitage's trend test, which is robust to Hardy-Weinberg (13), this SNP is associated with T2DM, P = 0.04.
Table 2. Frequency of Gln62Arg KLF11 in additional population samples.
| OR (95% Cl), X2P value
|
|||||||
|---|---|---|---|---|---|---|---|
| Sample | NN | NM | MM | n | Minor allele frequency | Allele frequency | Dominant model |
| Initial familial study | |||||||
| Normoglycemic | 252 | 58 | 3 | 313 | 0.102 | 1.85(1.33-2.57) | 2.00(1.39-2.87) |
| T2DM subjects | 211 | 95 | 7 | 313 | 0.174 | 0.00023 | 0.00019 |
| Second set | |||||||
| Normoglycemic | 1,138 | 307 | 18 | 1,463 | 0.117 | 1.18(1.01-1.38) | 1.18(1.00-1.40) |
| T2DM subjects | 1,032 | 326 | 25 | 1,383 | 0.136 | 0.034 | 0.057 |
| Overall | |||||||
| Normoglycemic | 1,390 | 365 | 21 | 1,776 | 0.115 | 1.29(1.12-1.49) | 1.32(1.13-1.54) |
| T2DM subjects | 1,243 | 421 | 32 | 1,696 | 0.143 | 0.00033* | 0.00054* |
NN, homozygous wild-type allele carriers; NM, heterozygous allele carriers; MM, homozygous mutant allele carriers; OR, odds ratio.
P value and combined OR for allelic effects calculated by a Mantel-Haenszel test (25).
Table 3.
Analysis of normoglycemic Gln62Arg carriers during OGTT
| AA allele | AG allele | P value† | |
|---|---|---|---|
| No. of subjects | 57 | 13 | |
| Age | 56 ± 1.4 | 51 ± 3.7 | 0.82 |
| Sex (M/W) | 25/32 | 3/10 | |
| BMI | 24.9 ± 0.7 | 21.4 ± 2.0 | 0.057 |
| OGTT time, min | |||
| Glucose, mM/I | |||
| 0 | 5.1 ± 0.1 | 5.0 ± 0.1 | 0.53 |
| 30 | 8.1 ± 0.2 | 7.3 ± 0.5 | 0.14 |
| 60 | 7.1 ± 0.3 | 6.3 ± 0.7 | 0.25 |
| 90 | 5.8 ± 0.2 | 5.4 ± 0.4 | 0.51 |
| 120 | 5.1 ± 0.2 | 4.7 ± 0.3 | 0.36 |
| ΔAUCglu | 170 ± 19 | 116 ± 43 | 0.22 |
| Insulin, mU/I | |||
| 0 | 8.6 ± 1.6 | 5.3 ± 1.1 | 0.31 |
| 30 | 43.9 ± 4.1 | 28.6 ± 5.5 | 0.084 |
| 60 | 50.7 ± 4.1 | 27.0 ± 4.8 | 0.0081 |
| 90 | 38.1 ± 4.1 | 26.5 ± 5.1 | 0.21 |
| 120 | 36.6 ± 4.4 | 18.3 ± 3.6 | 0.049 |
| ΔAUCins | 3,811 ± 331 | 2,065 ± 338 | 0.013 |
| Assesement of beta cell function and insulin sensitivity‡ | |||
| IG of 30 min | 4.4 ± 0.4 | 3.7 ± 0.8 | 0.38 |
| IG of 120 min | 95.8 ± 72.0 | 24.3 ± 22.5 | 0.62 |
| First-phase Stumvoll index | 285.0 ± 29.5 | 337.6 ± 76.4 | 0.46 |
| Second-phase Stumvoll index | 99.4 ± 5.8 | 107.9 ± 14.8 | 0.54 |
| ISI-Belfiore | 0.13 ± 0.03 | 0.08 ± 0.04 | 0.50 |
| ISI-Matsuda | 229.6 ± 41.3 | 220.7 ± 56.1 | 0.93 |
| ISI-Stumvoll | 0.12 ± 0.003 | 0.14 ± 0.007 | 0.045 |
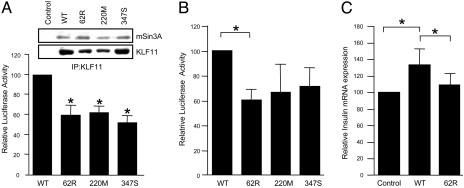
Biochemical and Functional Characterization of KLF11 Variants. To assess the putative physiopathological link between T2DM and KLF11 variants, we analyzed the transcriptional regulatory activity of both the rare Thr220Met and Ala347Ser variants and the nonsynonymous SNP9 (Gln62Arg). Studies using the Gal4 reporter assay have previously shown that the N-terminal domain of KLF11 harbors three repression domains (8). Interestingly, 62Arg-, 220Met- and 347Ser-KLF11 variants demonstrate a significantly increased repression activity compared with the wild-type N-terminal KLF11-GAL4 chimeric construct (Fig. 3A). Similarly, the 62Arg- and 220Met-KLF11 variants show increased repression activity on the reporter plasmid containing repetitive KLF11-binding sites compared with wild-type control (data not shown). Interestingly, 62Arg-, 220Met-, and 347Ser-KLF11 are gain-of-function variants, suggesting they may affect corepressor affinity. Indeed, an increased binding of the corepressor mSin3A to 62-Arg KLF11 variant compared with wild type was observed by coimmunoprecipitation (Fig. 3A). The Thr220Met and Ala347Ser mutations show no alteration in binding affinity, suggesting that other not-yet-identified corepressors may mediate the transcriptional effects of these proteins. Taken together, these results suggest that the association of impaired KLF11 function with diabetes may, under specific conditions, result from altered KLF11 transcriptional activity.
Fig. 3.
Functional characterization of nonsynonymous KLF11 variants. (A) GAL4 reporter assay with N-terminal KLF11-GAL4DBD constructs cotransfected in CHO cells (n = 4). (Upper) A representative Western blot of coimmunoprecipitation experiments (n = 2) showing mSin3a binding of Flag-tagged KLF11 (WT and variants) or empty vector (control). (B) Luciferase assays of Flag-tagged KLF11 variants cotransfected with human insulin promoter reporter constructs in βTC3 cells (n = 3). (C) Real-time PCR quantification of insulin mRNA expression in βTC3 cells cultured in high-glucose medium (25 mM) and transfected with control, WT KLF11, or 62R-KLF11. Values are corrected for GAPDH levels (n = 4). *, significantly different (P < 0.05).
We also anticipated a direct link between the altered KLF11 function and the transcriptional regulation of the insulin gene. In the pancreatic beta cells, wild-type KLF11 induced the insulin promoter activity by 2-fold (Fig. 1F), but its activation was significantly impaired by 62Arg-KLF11 variant under high-glucose levels (Fig. 3B). The 220Met- and 347Ser-KLF11 variants show no effect on the activation of the insulin promoter. Interestingly, under similar conditions, 62Arg-KLF11 variant showed a decreased induction of endogenous insulin mRNA expression compared with the wild-type protein (Fig. 3C). Thus, these results suggest that an altered 62Arg-KLF11 function may impair insulin levels by increasing the repression activity on this promoter. In fact, analysis of the available oral glucose tolerance tests (OGTTs) of 70 normoglycemic subjects from the familial case-control cohort show a significantly decreased plasma insulin level at 60 and 120 min after an oral glucose load for AG-allele carriers compared with AA-allele carriers (Table 3 G and A allele encoding 62Arg- and wild-type KLF11, respectively). In addition, the normoglycemic AG allele carriers are lean and have increased insulin sensitivity that may protect them against impaired glucose tolerance. Analysis in prediabetic subjects also suggests that the AG allele is associated with decreased plasma insulin levels (Data not shown; P < 0.05 at 30 and 90 min for 33 French AA allele vs. 12 AG allele carriers).
In addition, KLF11 may affect pancreatic beta cells function by modulating the expression of free radical scavengers such as superoxide dismutase (SOD)2 and catalase 1 that were recently identified as KLF11 target genes (5). These antioxidant enzymes have a very low expression level in pancreatic islets compared with other metabolic tissues, e.g., liver and skeletal muscle, and overexpression of these enzymes protects beta cells against glucolipotoxicity (18). In pancreatic beta cells, KLF11 represses the promoter activity of the SOD2 (data not shown) and catalase 1 (Fig. 5, which is published as supporting information on the PNAS web site). The KLF11 variants show no changes in transcriptional activity of the SOD2 promoter (data not shown), but all three significantly increase repression of catalase 1 promoter under high-glucose levels (Fig. 5). Taken together, these results suggest that the association of altered functional KLF11 variants with diabetes may result from an altered KLF11 transcriptional activity.
Discussion
In this study, we addressed the questions as to whether KLF11 is involved in the regulation of pancreatic beta cell function, and whether it plays a role as gene modifier in predisposition to diseases of beta cell origin such as diabetes. Our study has yielded evidence that this is indeed the case. Regarding the biology of KLF11, our data support a model where in pancreatic beta cells the product of the KLF11 gene is induced by glucose to regulate in turn insulin transcription. Beside this important observation, further additional data point to an involvement of KLF11 in the biology of pancreatic beta cells. In these cells, KLF11 is also regulated by TGF-β, a pathway playing a critical role in the development and homeostasis of both the exocrine and endocrine pancreas. In addition, KLF11 regulates key genes encoding scavengers of oxidative stress, including SOD2 and catalase 1. This observation is extremely important in the context of this study, because a tight control of oxidative stress is critical for maintaining the homeostasis of pancreatic beta cells. In fact, oxidative stress is believed to be involved in the progression of pancreatic beta cell dysfunction found in T2DM.
In light of these data, KLF11 is a truly valid candidate gene for genetic predisposition to T2DM. Indeed, our comprehensive genetic analysis by using family-based and case-control studies provide evidence for a causal association between three nonsynonymous KLF11 variants, early-onset and polygenic T2DM. The overall P value for association of the KLF11 variant Gln62Arg with T2DM in our study, including 1,696 T2DM and 1,776 normoglycemic subjects, is significant (0.00033), and even the modest association observed in the second study alone (0.03) is of similar order, as recently shown in replication studies of validated candidate genes for T2DM such as PPARγ (19) and KIR 6.2 (20). It remains to be established whether the 62Arg mutant is the only functional KLF11 variant among the group of late-onset T2DM-associated variants. For the three nonsynonymous variants, we showed that they affect the function of KLF11, and consequently, they may influence primary beta cell functions, and the balance between proliferation and differentiation of beta cells. Moreover, KLF11 has recently been shown to be up-regulated after refeeding in mouse skeletal muscles (21), suggesting a role for KLF11 in postprandial glucose metabolism of this tissue. In addition, the caveolin-1 gene that is highly expressed in adipose tissue is repressed by KLF11 in a cholesterol-dependent manner (22). The fact that caveolin-1 binding to the insulin receptor stimulates both the kinase activity and recruitment of the insulin receptor to lipid rafts at the plasma membrane, and the fact that insulin receptor mutations impairing the calveolin binding result in T2DM (23), suggest that calveolin 1 is essential for insulin signaling. Therefore, we do not exclude that an altered KLF11 function in skeletal muscle, adipose tissue, or liver may also play a role in susceptibility to diabetes.
In light of the evidence, it is tempting to speculate on the possibilities of targeting KLF11 for the therapy of diabetes. The initial structural and molecular modeling data on mSin3A interaction with KLF11 (24), which shows that this interaction differs from other transcription factors that bind to mSin3A, may present a fundamental step toward a rational drug design effort. Thus, further studies to unravel the mechanisms underlying the association of altered KLF11 function with the pathogenesis and severity of diabetes may also contribute with novel targets to the development of antidiabetic agents.
Supplementary Material
Acknowledgments
We thank all families and patients who participated to this study. We thank K. Borch-Johnsen and T. Drivsholm (Steno Diabetes Center and Research Center for Prevention and Health) for recruiting and characterization of part of the Danish control subjects. We thank F. Pattou and J. Kerr-Conte (Université de Lille) for their kind gift of human pancreatic islet and beta cell RNA. We thank T. Gurevitch, (Hadassah University Hospital), J. Hiddinga (Mayo Clinic), and C. Wachter and P. Boutin (Unité Mixte de Recherche 8090) for assistance with transfection experiments, pancreatic beta cell culture, and advice on the manuscript, respectively. We thank T. J. Woodage for providing TaqMan SNP genotyping assays. This work was supported by Mayo Clinic Cancer Center Specialized Program of Research Excellence Pancreatic Cancer Grant P50 CA10270 and National Institute of Health Grants DK52913 and DK56620 (to R.U.), Juvenile Diabetes Foundation Grant 1-2001-325 (to D.M.), the European Regional Development Fund, Region Nord-Pas de Calais (ARCir) (to B.N. and P.F.), the European Economic Community (Genomic Integrated Force for Type II Diabetes 1998-2002), and Eli Lilly through the Lilly Consortium for Diabetes and Obesity.
Author contributions: B.N., C.D., D.M., and P.F. designed research; B.N., M.E.F.-Z., V.A.-K., Y.H.H., E.J., E.V., Y.B., E.D., N.B., and T.C. performed research; B.N., M.E.F.-Z., V.A.-K., E.J., M.V., T.C., G.M.D.-T., H.J., M.-A.C., K.C., P.G., S.H., N.H., G.C., M.P., T.H., O.P., R.U., D.M., and P.F. contributed new reagents/analytic tools; B.N., M.E.F.-Z., V.A.-K., C.D., Y.H.H., E.V., V.D., M.V., T.C., M.P., and T.H. analyzed data; and B.N., M.E.F.-Z., R.U., and P.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP, chromatin immunoprecipitation; T2DM, type 2 diabetes mellitus; MODY, maturity-onset diabetes of the young; OGTT, oral glucose tolerance test; SOD, superoxide dismutase; LD, linkage disequilibrium; BMI, body mass index; PDX-1, pancreatic duodenal homeobox gene 1.
References
- 1.Kim, S. K. & Hebrok, M. (2001) Genes Dev. 15, 111-127. [DOI] [PubMed] [Google Scholar]
- 2.Song, S. Y., Gannon, M., Washington, M. K., Scoggins, C. R., Meszoely, I. M., Goldenring, J. R., Marino, C. R., Sandgren, E. P., Coffey, R. J. J., Wright, C. V. & Leach, S. D. (1999) Gastroenterology 117, 1416-1426. [DOI] [PubMed] [Google Scholar]
- 3.Fisher, W. E. (2001) World J. Surg. 25, 503-508. [DOI] [PubMed] [Google Scholar]
- 4.Coughlin, S. S., Calle, E. E., Teras, L. R., Petrelli, J. & Thun, M. J. (2004) Am. J. Epidemiol. 159, 1160-1167. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Zapico, M. E., Mladek, A., Ellenrieder, V., Folch-Puy, E., Miller, L. & Urrutia, R. (2003) EMBO J. 22, 4748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanvito, F., Herrera, P. L., Huarte, J., Nichols, A., Montesano, R., Orci, L. & Vassalli, J. D. (1994) Development (Cambridge, U.K.) 120, 3451-3462. [DOI] [PubMed] [Google Scholar]
- 7.McCarthy, M. I. & Froguel, P. (2002) Am. J. Physiol. 283, E217-E225. [DOI] [PubMed] [Google Scholar]
- 8.Zhang, J. S., Moncrieffe, M. C., Kaczynski, J., Ellenrieder, V., Prendergast, F. G. & Urrutia, R. (2001) Mol. Cell. Biol. 21, 5041-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. (2004) Genome Res. 14, 1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaczynski, J., Zhang, J. S., Ellenrieder, V., Conley, A., Duenes, T., Kester, H., van Der Burg, B. & Urrutia, R. (2001) J. Biol. Chem. 276, 36749-36756. [DOI] [PubMed] [Google Scholar]
- 11.Sham, P. C. & Curtis, D. (1995) Ann. Hum. Genet. 59, 97-105. [DOI] [PubMed] [Google Scholar]
- 12.Mendell, N. R. & Simon, G. A. (1984) Ann. Hum. Genet. 48, 283-286. [DOI] [PubMed] [Google Scholar]
- 13.Sasieni, P. D. (1997) Biometrics 53, 1253-1261. [PubMed] [Google Scholar]
- 14.Lukowiak, B., Vandewalle, B., Riachy, R., Kerr-Conte, J., Gmyr, V., Belaich, S., Lefebvre, J. & Pattou, F. (2001) J. Histochem. Cytochem. 49, 519-528. [DOI] [PubMed] [Google Scholar]
- 15.Waeber, G., Delplanque, J., Bonny, C., Mooser, V., Steinmann, M., Widmann, C., Maillard, A., Miklossy, J., Dina, C., Hani, E., et al. (2000) Nat. Genet. 24, 291-295. [DOI] [PubMed] [Google Scholar]
- 16.Geourjon, C. & Deleage, G. (1995) Comput. Appl. Biosci. 11, 681-684. [DOI] [PubMed] [Google Scholar]
- 17.Cox, N. J., Hayes, M. G., Roe, C. A., Tsuchiya, T. & Bell, G. I. (2004) Diabetes 53, S19-S25. [DOI] [PubMed] [Google Scholar]
- 18.Robertson, R. P., Harmon, J., Tran, P. O. & Poitout, V. (2004) Diabetes 53, S119-S124. [DOI] [PubMed] [Google Scholar]
- 19.Altshuler, D., Hirschhorn, J. N., Klannemark, M., Lindgren, C. M., Vohl, M. C., Nemesh, J., Lane, C. R., Schaffner, S. F., Bolk, S., Brewer, C., et al. (2000) Nat. Genet. 26, 76-80. [DOI] [PubMed] [Google Scholar]
- 20.Love-Gregory, L., Wasson, J., Lin, J., Skolnick, G., Suarez, B. & Permutt, M. A. (2003) Diabetologia 46, 136-137. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto, J., Ikeda, Y., Iguchi, H., Fujino, T., Tanaka, T., Asaba, H., Iwasaki, S., Ioka, R. X., Kaneko, I. W., Magoori, K., et al. (2004) J. Biol. Chem. 279, 16954-16962. [DOI] [PubMed] [Google Scholar]
- 22.Cao, S., Fernandez-Zapico, M. E., Jin, D., Puri, V., Cook, T. A., Lerman, L. O., Zhu, X. Y., Urrutia, R. & Shah, V. (November 5, 2004) J. Biol. Chem., 10.1074/jbc.M407941200. [DOI] [PubMed]
- 23.Cohen, A. W., Combs, T. P., Scherer, P. E. & Lisanti, M. P. (2003) Am. J. Physiol. 285, E1151-E1160. [DOI] [PubMed] [Google Scholar]
- 24.Pang, Y. P., Kumar, G. A., Zhang, J. S. & Urrutia, R. (2003) FEBS Lett. 548, 108-112. [DOI] [PubMed] [Google Scholar]
- 25.Mantel, N. & Haenszel, W. (1959) J. Natl. Cancer Inst. 22, 719-748. [PubMed] [Google Scholar]
- 26.Albareda, M., Rodriguez-Espinosa, J., Murugo, M., de Leiva, A. & Corcoy, R. (2000) Diabetologia 43, 1507-1511. [DOI] [PubMed] [Google Scholar]
- 27.Stumvoll, M., Mitrakou, A., Pimenta, W., Jenssen, T., Yki-Jarvinen, H., Van Haeften, T., Renn, W. & Gerich, J. (2000) Diabetes Care 23, 295-301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.