Abstract
Despite the widespread use of magnetic resonance imaging (MRI) of the brain, the relative contribution of different biological components (e.g. lipids and proteins) to structural MRI contrasts (e.g., T1, T2, T2*, proton density, diffusion) remains incompletely understood. This limitation can undermine the interpretation of clinical MRI and hinder the development of new contrast mechanisms. Here, we determine the respective contribution of lipids and proteins to MRI contrast by removing lipids and preserving proteins in mouse brains using CLARITY. We monitor the temporal dynamics of tissue clearance via NMR spectroscopy, protein assays and optical emission spectroscopy. MRI of cleared brain tissue showed: 1) minimal contrast on standard MRI sequences; 2) increased relaxation times; and 3) diffusion rates close to free water. We conclude that lipids, present in myelin and membranes, are a dominant source of MRI contrast in brain tissue.
Introduction
Brain structural MRI is a cornerstone of modern medicine and neuroscience. It is, therefore, surprising that the biological source of MRI tissue contrast remains incompletely understood.
While structural MRI contrast mechanisms such as T1-weighted, T2-weighted, proton density and diffusion-weighted MRI can measure different properties of brain tissue, all are based on the interaction of water with surrounding tissue components. What is less well understood is the relative contribution of different tissue components to each structural MRI contrast. The vague term ‘macromolecular composition’ is sometimes used to account for the details of cellular architecture. But what type of cellular and molecular components make up these ‘macromolecules’ driving many types of MRI contrast in the brain? A more complete understanding of the biological basis for MRI contrast has the potential to generate new insights into the pathophysiological mechanisms underlying human diseases and enable technological advancements that improve the diagnostic sensitivity and specificity of MRI.
In consideration of potential biological substrates for structural MRI contrast, the human brain consists of ∼77% water, ∼11% lipids, ∼8% proteins, ∼2% soluble organic substances, ∼1% carbohydrates, and ∼1% inorganic salts (McIlwain and Bachelard, 1985). There are considerable differences between different brain regions with white matter having a ∼60% higher lipid and ∼10–15% lower protein concentration than gray matter (Knaap and J. Valk, 2005; O’Brien and Sampson, 1965; Randall, 1938). Based solely on these concentration levels, lipids and proteins are prime candidates as sources of MRI contrast. In aqueous solutions, both lipids and proteins have separately been shown to affect T1 and T2 relaxation times of water protons (Kucharczyk et al., 1994, 1990; Mäkelä et al., 2001). Among the different types of lipids specifically cholesterol has been shown to have a strong effect on the different relaxation rates of white and gray matter (Koenig, 1991; Koenig et al., 1990). In tissue, both lipids and proteins are found within cell membranes and myelin but their separate effects are more difficult to characterize. Cell membranes are composed of phospholipid bilayers and have been shown to hinder water diffusion and therefore have a strong influence on diffusion contrast (Cooper et al., 1974) and, in the form of nerve fibers, on diffusion anisotropy (Basser et al., 2000, 1994; Conturo et al., 1999; Jones et al., 1999; Mori et al., 1999; Moseley et al., 1990). Myelin consists of ∼70% lipids and ∼30% proteins (Kirschner and A.E., 1992; Norton and Autilio, 1966) and has been shown to have a strong influence on T1, T2 and diffusion contrast (Clark et al., 1992; Eickhoff, 2005; Fatterpekar et al., 2002; Stüber et al., 2014; Walters et al., 2007). Stüber et al. (Stüber et al., 2014) demonstrated in cadaver brain that quantitative maps of T1 and T2* accurately predict subsequent measurements of myelin and iron distribution observed by proton beam microscopy. The degree of myelination has also been shown to lead to an increase of water diffusion anisotropy (Beaulieu and Allen, 1994). In the cuprizone mouse model where mice are fed a cuprizone diet that leads to demyelination of the tissue, a decrease in tissue contrast has been observed for various MRI measures and tissue anisotropy has been shown to go down (Thiessen et al., 2013)
In addition to the protein content within membranes and myelin, proteins such as collagens and elastins are also aggregated in the cell cytoskeleton as well as the extracellular matrix. The effect of the protein-rich cell cytoskeleton on diffusion has been studied in giant squid axons and been shown to be small (Beaulieu and Allen, 1994) but this result has never been verified in brain tissue. Lastly, while inorganic salts make up less than 1% of the brain tissue, they can have a pronounced effect on certain types of MRI contrast (Fukunaga et al., 2010). For example, the paramagnetic properties of metals, such as iron, change the local magnetic field distribution and decreases the transverse relaxation times of the water. T2 and T2* contrast are strongly influenced by iron and T2*-weighted images are often used to detect iron deposition in the brain in relation to diseases such as Parkinson’s (Morawski et al., 2005) and dementia (Zeineh et al., 2015).
In addition to tissue content, the molecular dynamics that govern how macromolecules, such as proteins or lipid chains, interact with water also affect tissue contrast. The mobility of water molecules near macromolecules can be restricted, leading to different relaxation rates for water pools within e.g. extra- or intracellular space and inside the myelin sheaths. Proton exchange between different water pools and magnetization transfer (MT) between macromolecules and water (Helms and Hagberg, 2009; van Gelderen et al., 2016) can lead to cross-relaxation between different pools. Functional groups within macromolecules such as the — OH group found in lipids or the –NH group found in proteins can lead to spin-exchange between macromolecules and water. The relaxivity can thus change with the number of open functional groups and be influenced by tissue fixation or pH of the tissue (Gochberg et al., 1998; Kucharczyk et al., 1994).
Many prior studies focus on myelin when examining macromolecule-water interactions. However, traditionally, it has been difficult to split myelin into its lipids and protein components and study their separate effects on MRI contrast. One way to separate lipids and proteins in brain tissue is to use a tissue clearing technique known as CLARITY (Chung et al., 2013; Epp et al., 2015; Sylwestrak et al., 2016; Tomer et al., 2014). CLARITY is a method that washes out lipids but first uses hydrogel-embedding to preserve the structural integrity of brain tissue and the spatial organization of proteins, nucleic acids and other small molecules (Fig. 1). The clearing process utilizes detergent to remove lipids that render the tissue optically opaque. Refractive index matching of the cleared sample then allows for staining of proteins and optical imaging of non-sectioned tissue samples. In this study we aim to use CLARITY to test the separate effects of lipids and proteins on tissue contrast. Specifically, we acquire brain structural MRI on mouse brain specimens before, during and after the CLARITY process and quantitatively evaluate the timeline of lipid washout by NMR spectroscopy, protein assays and optical emission spectroscopy. At the same time, we assess the effects of tissue clearing on tissue content, tissue deformation and MRI properties, which helps to inform on approaches to MRI-CLARITY comparisons.
Figure 1.
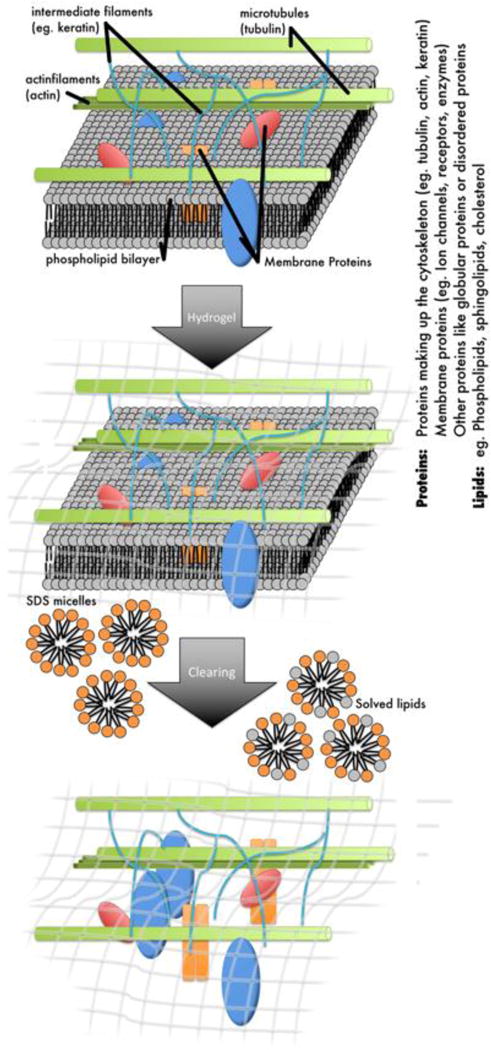
Schematic diagram of the CLARITY protocol. The tissue sample is immersed in a hydrogel that binds to biomolecules with an active NH2 group such as proteins or amino acids. After polymerization of the acrylamide hydrogel within the tissue the sample is cleared using SDS detergent to wash out all tissue components that do not bind to the hydrogel such as the lipids but also other elements like iron and calcium.
Methods
Table 1 summarizes the different methods for measurement of MRI tissue contrast and tissue content before, during and after clearing.
Table 1.
Summary of experiments before, during and after tissue clearing.
| Longitudinal MRI | Longitudinal NMR | Quantitative MRI | ICP-OES | protein analysis (BCA) | pH | Staining | |
|---|---|---|---|---|---|---|---|
| Before clearing | T1w, T2w, MD, FA of 2 mouse brain samples embedded in hydrogel | 1H and 31P measurements of 2 uncleared mouse brains | T1-map, T2-map, T2*w, PDw, MD and FA of mouse brain fixed with PFA and mouse brain fixed in hydrogel | Fe, Ca and P measurement of the olfactory bulb of 4 uncleared mouse brains | Five regions (cortex, corpus callosum, cerebellum, hypocap us, brain stem) were dissected from three hydrogel-embedded but uncleared samples and three cleared brain samples and analyzed for their protein content | pH of uncleared mouse brain | |
| During clearing | Each week T1w, T2w, MD and FA for 4 week clearing period | Each week 1H and 31P spectrum for 4 week clearing period | |||||
| After clearing | T1w, T2w, MD, FA of 2 cleared mouse brain samples | 1H and 31P spectrum of 2 cleared mouse brain samples | T1-map, T2-map, T2*w, PDw, MD and FA of cleared mouse brain and a pure hydrogel and PBS sample | Fe, Ca and P measure ment of the olfactory bulb of 4 cleared mouse brains | The same regions as above of three cleared brains were analyzed for their protein content | pH of cleared mouse brain | Staining of cleared mouse brain for cells using PI and neurofilaments using NF |
ICPOES - Element analysis using Inductively coupled plasma optical emission spectrometry, BCA - Protein analysis using bicinchoninic acid assay, T1w - T1-weighted, T2w - T2-weighted, MD - Mean diffusivity, FA - fractional anisotropy, PDw - Proton density weighted, PFA - 4 % Paraformaldehyde, PI - Propidium Iodide, NF -Neurofilament
CLARITY
The brains of 14 adult C57bl6 mice were perfusion fixed with a hydrogel solution containing 4% acrylamide, 4% paraformaldehyde (PFA), 2% bis-acrylamide, phosphate buffered saline (PBS) and thermal initiator VA-044 (Tomer et al., 2014). Briefly, mice were anaesthetized and transcardially perfused with PBS followed by hydrogel solution. The hydrogel brains were kept in excess of hydrogel solution at 4 °C for 1 day. After fixation, the samples were de-oxygenated, polymerized for 3 hours at 37°C and excess hydrogel was removed. During clearing the samples were incubated with SDS and boric acid clearing buffer (SBC) at 37°C on a shaker. The SBC was replaced each week.
Longitudinal MRI
The brains of two mice to be scanned during different stages of the clearing process were excised and fixed using the CLARITY hydrogel solution described above. The tissue-hydrogel hybrids were cut in 4 coronal slabs of 3 mm thickness each, to speed up the clearing process. The largest coronal brain slab was placed in a container filled with ChristoLube, a proton-free fluid which produces no MR signal. All MRI measurements were performed on a preclinical 7T actively shielded MRI scanner Discovery MR901 (Agilent Technologies-General Electric Healthcare) with 300 mT/m gradients using a 2-channel ‘millipede’ coil. The following types of MR scans were acquired: T1-weighted inversion recovery, TE/TR/TI 12.3/2000/400 ms, 0.12×0.12×0.5 mm resolution, 1 average; T2-weighted inversion recovery, TE/TR/TI 80/5000/2250 ms, 0.24×0.24×0.5 mm resolution, 1 average; Diffusion-weighted MRI sequence with 16-shot EPI readout, TE/TR 13/2000 ms, 0.24×0.24×0.5 mm resolution, 1 average, 30 diffusion directions and 3 b0 images. The diffusion weighting was kept constant throughout the longitudinal study and set at b = 500 s/mm2 in order to get enough signal during all stages of clearing. Evaluation of the MD and FA maps was performed using the FMRIB Software Library (FSL, www.fmrib.ox.ac.uk/fsl). SBC was replaced once a week after the MR measurements. To evaluate white matter (WM) — gray matter (GM) contrast, regions of interest (ROIs) covering the white matter in the corpus callosum and gray matter in the outer cortex of the mouse brain were selected on the T1-weighted image. This was performed for the uncleared sample and week 1. The ROIs were transformed to the MD and FA maps. Mean MD and FA values of these ROIs were measured and the normalized contrast between WM and GM regions was calculated. For week 2 the contrast value was set to zero as no segmentation between WM and GM was possible in the reference T1-weighted dataset.
Longitudinal NMR Spectroscopy
In parallel with the longitudinal MRI measurements, NMR spectroscopy measurements were obtained using the same samples at the same time intervals. The samples were immersed in SBC and inserted within NMR tubes. 1H and 31P spectra of the samples were acquired on a 600 MHz Varian Inova NMR spectrometer with a 10 mm broadband probe. A capillary containing 20% TSP was inserted together with the sample as a reference for the 1H measurements and a capillary containing 0.2 mM medronic acid was inserted as a reference for the 31P measurements. The 1H spectrum was measured with TR = 5 s and 64 repetitions leading to a total scan time of 5.5 min. The 31P spectrum was acquired with TR = 5 s and 11520 repetitions leading to a total scan time of 16 hours. The 31P spectrum for uncleared sample and weeks 1 and 2 was correlated to MD and FA WM-GM contrast by calculating the Pearson coefficient between both datasets.
Quantitative MRI
Cross-sectional MRI scans were performed on six different samples: 1) a slab of an uncleared mouse brain perfusion-fixed in 4% paraformaldehyde (PFA) and kept in PFA for 1 day before being transferred to PBS, 2) an uncleared mouse brain slab embedded in hydrogel and stored in PBS, 3) a cleared mouse brain slab immersed in PBST (PBS + 0.1% TritonX), 4) a cleared mouse brain slab immersed in PBS, 5) a sample containing only polymerized hydrogel and a 6) sample containing only PBS. All the mouse brain samples were immersed in ChristoLube during MRI measurement in order to suppress the background signal. The cleared mouse brain slabs were squeezed between two plastic cover slides in order to better preserve their original shape which otherwise was completely lost once immersed in ChristoLube. The following MRI sequences were applied to all six samples using the same 7T GE scanner as described in the longitudinal MRI protocol:
A T1 map was acquired using a Fast Spin Echo Inversion Recovery (FSE-IR) sequence with TE = 7.17ms, TR = 5000ms, TI = 50, 200, 400, 800, 1600 and 3200ms, 0.23×0.23×0.5mm resolution and 2 averages. A T2 map of the same geometry was acquired using a Spin Echo (SE) sequence with TE = 12.5, 25, 37.5, 50ms, and TR = 3500ms. A gradient echo sequence (GRE) with the same geometry was acquired with TE = 20ms, TR = 3500ms for a T2*-weighted image. The same GRE sequence but with TE = 2.6ms was acquired to get an image with proton density weighted (PD) contrast. A dMRI scan was acquired using a 2D Echo planar imaging (EPI) sequence with 16 shot acquisition, TE = 12.5ms, TR = 1200ms, 0.46×0.46×0.5mm resolution, 3 images with b = 0s/mm2 and 20 linearly independent diffusion directions with b = 500s/mm2.
The samples were bias-field corrected using the method described in (Li et al., 2015). Profiles were taken along the left-right direction of each image and normalized to compare changes in image contrast between samples. The measured values for the quantitative analysis were taken from a slice in the center of the sample. For quantitative analysis, the FA maps of the cleared, hydrogel and water samples were eroded using a disk with a three-voxel radius to avoid a bright outer contour caused by edge effects.
Inductively coupled plasma optical emission spectrometry (ICP-OES)
Four hydrogel-embedded but uncleared samples and four cleared brain samples were measured with ICP-OES to determine iron, calcium and phosphorus content before and after clearing. The samples were cut into small pieces, dissolved in 50μl concentrated HNO3 and heated to 130°C for 12 hours. When all the liquid had evaporated, the samples were re-suspended in 5 ml 2% HNO3. Standard solutions for iron with 5ppm, 1ppm, 0.5ppm, 0.1ppm, 0.01ppm concentrations, for calcium with 5ppm, 1ppm, 0.5ppm, 0.1ppm, 0.01ppm and for phosphorus with 20ppm, 5ppm, 1ppm, 0.5ppm, 0.01ppm were prepared. ICP-OES was performed using a Thermo Scientific ICAP 6300 Duo View Spectrometer.
Protein analysis
Five regions each (cortex, corpus callosum, cerebellum, hippocampus, brain stem) were dissected from three hydrogel-embedded but uncleared samples and three cleared brain samples and analyzed using a bicinchoninic acid assay (BCA) (Bio-Rad Protein Assay, Bio-Rad) in order to estimate the protein concentration in the samples. Samples were homogenized by mechanical forces (Tissue Ruptor, Qiagen, Hilden, Germany) and incubation with cell lysis buffer (1× Cell lysis buffer, 1× Protease Inhibitor, 1× PMSF all from Cell Signaling, Danvers, USA).
pH
Three hydrogel-fixed but uncleared mouse brains and three cleared mouse brains were mechanically disrupted (Tissue Ruptor, Qiagen) in 1 ml PBS (pH 7.4, ThermoFisher, Grand Island, USA). The pH was measured on each of these brains using pH testing strips. Additionally, a sample containing only SBC and one sample containing only PBS were measured for their pH.
Molecular labeling of cleared tissue
One mouse brain prepared with the same protocol (Tomer et al., 2014) was stained with propidium iodide for cells, anti-neurofilament antibodies for neurofilaments and imaged using a 10× objective on an Olympus FV1200 confocal microscope.
Results
Longitudinal MRI
To show the change of various types of MRI contrast with decreasing lipid content we performed T1-weighted, T2-weighted, and dMRI scans of the same mouse brain sample during different stages of tissue clearing (Fig. 2). The transparency of the brain sample increases during the clearing progress, starting at the cortex and extending over time to the deep brain structures (Fig. 2, 1st column). As the tissue clearing progressed, two main changes were observed in the T1-weighted, T2- and diffusion-weighted MR images: the MRI contrast decreases and the tissue deformation increases. The tissue deforms because it becomes softer and more pliable as it clears. Based on MRI measurements, the tissue volume also expands by 315% between uncleared and week 1. There was no measurable volume increase after week 1. Strong changes in tissue contrast can be observed already after one week of clearing for all imaging modalities with the contrast being lowest in mean diffusivity (MD) and fractional anisotropy (FA) image (Fig. 2, columns 2–4). After two weeks of clearing the only visible contrast in any of the MR images is a signal difference between the central and peripheral regions of the sample. Visual inspection shows that this is due to incomplete clearing around the striatum. The FA and MD values in gray and white matter during the different stages of tissue clearing are reported in Table 2. After three weeks of clearing the contrast in the MR images is entirely gone, and the photographs of the tissue specimen show a uniformly cleared sample. At this time point the sample is very soft, and is easily deformed when the sample is placed in the MRI measurement container with the MR-invisible fluid ChristoLube.
Figure 2.
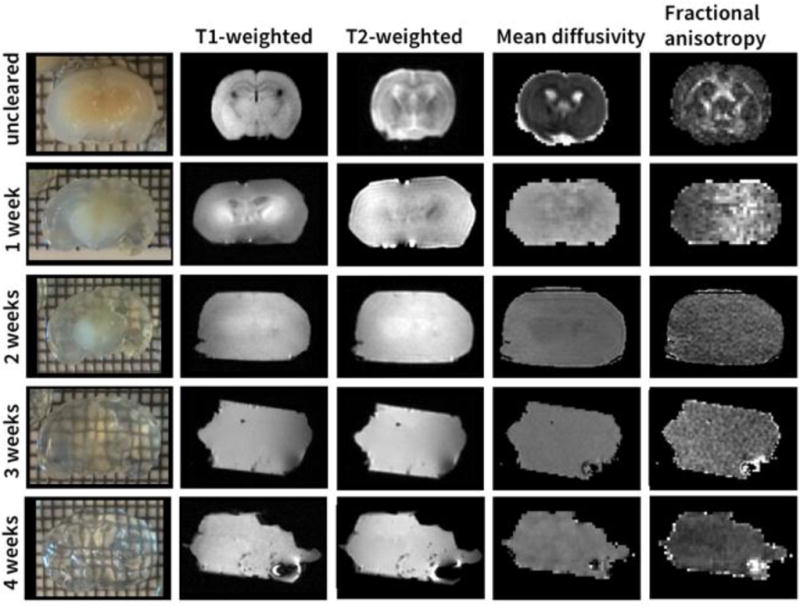
Mouse brain during the different stages of tissue clearing. MRI contrast decreases with increasing transparency of the brain (top to bottom).
Table 2.
Fractional anisotropy (FA) and mean diffusivity (MD) values in white (WM) and gray matter (GM) during different stages of tissue clearing.
| uncleared | week 1 | week 2* | |
|---|---|---|---|
| FA in WM | 0.42 ± 0.20 | 0.13 ± 0.05 | 0.17 ± 0.03 |
| FA in GM | 0.17 ± 0.06 | 0.10 ± 0.05 | |
| MD in WM [10−3 mm2/s] | 0.25 ± 0.11 | 1.44 ± 0.10 | 1.70 ± 0.13 |
| MD in GM [10−3 mm2/s] | 0.42 ± 0.27 | 1.68 ± 0.15 |
contrast too low to differentiate WM and GM regions
Longitudinal NMR Spectroscopy
In parallel with the longitudinal MRI measurements, NMR spectroscopy measurements were made with the same samples and at the same time intervals. Figure 3a (top left) shows the 1H NMR spectra of the mouse brain at one week intervals before during and after clearing. In addition to the endogenous components of the sample, the clearing buffer SBC (detergent SDS and borate acid) within and around the sample also generates large peaks in the 1H NMR spectrum. After one week of clearing, several peaks in the 1H NMR spectrum between 5 and 8 ppm, thought to represent small molecules such as metabolites like adenine complexes (Watanabe et al., 2012), have disappeared. Plotting the integral of of a peak at 2.18 ppm (marked by the red bar in Fig. 3 a) normalized by the integral of the reference peak at 0 ppm shows a decrease of the 2.18 ppm peak after the first week of tissue clearing to less than 40% of the uncleared value (Fig. 3a, top right). Known lipid peaks in the 1H spectrum occur around 1 ppm: however, this region of the spectrum was obscured by the spectrum of the detergent SDS solution.
Figure 3.
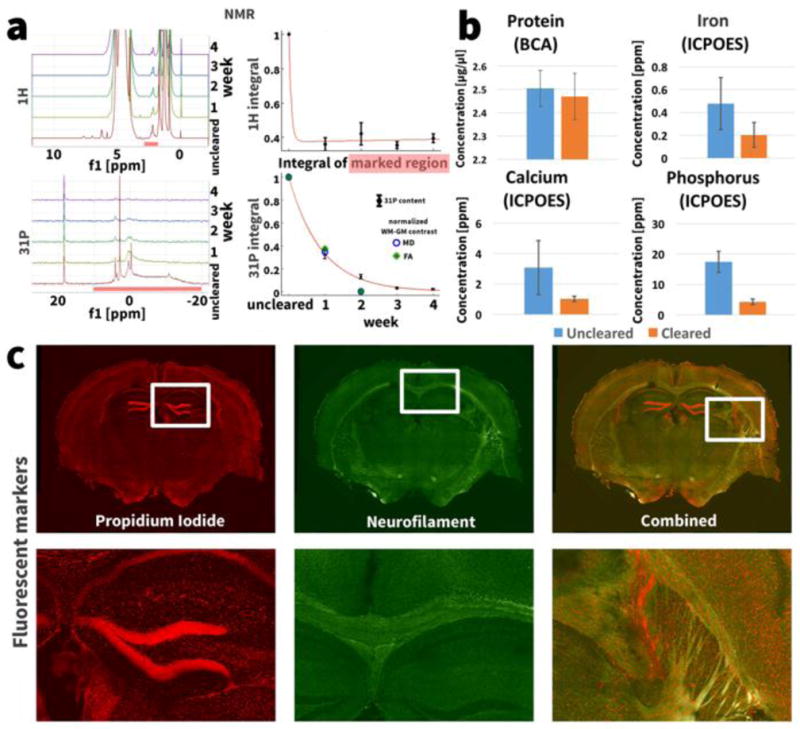
(a) NMR spectra of a mouse brain during different stages of the clearing process. The integral of the marked peak at 2.18 ppm in the 1H spectrum only decreases after the first week of clearing. The integral over all 31P NMR peaks from 10 ppm to −20 ppm decreases exponentially at a rate of [31P]∝eˆ(−0.158t)(R2>0.99) with clearing progress. The same graph shows the normalized WM-GM contrast in the MD and FA images during the longitudinal study. The correlation between WM-GM contrast and 31P content showed a Pearson coefficient of ρ = 0.991 for MD and ρ = 0.986 for FA. (b) The ICP-OES measurements show a trend towards lower iron and calcium concentration in the cleared samples (paired t-test, p(Fe) = 0.2, p(Ca) = 0.17) and a significantly lower phosphorus concentration in the cleared sample (p = 0.01). The bars show mean ± standard deviation over the multiple measured samples. (c) Staining the cleared tissue with propidium iodide for cells (red) and neurofilaments (green) shows that the nucleic acids and proteins are preserved at their original locations in the tissue.
The 31P NMR spectra representing phosphorus content show no peaks from the SBC solution, facilitating an easier interpretation compared to 1H spectra (Fig. 3a, bottom left). Several large peaks are observed in the 31P NMR spectrum of the uncleared sample. These peaks almost completely disappear over the course of tissue clearing. The integral of the peaks in the marked region between 10 and 20 ppm, normalized by the integral of the reference peak at 18.4 ppm, shows a decline in phosphorus content to less than 5% of the uncleared value (Fig. 3a, bottom right). The phosphorus content [31P] is washed out exponentially at a rate of [31P] ∝ e−0.158t (R2>0.99). A small peak at 4 ppm in the 31P NMR spectrum can still be observed after 4 weeks of clearing. The Pearson correlation coefficients between 31P washout and MD and FA contrast measured in the longitudinal MRI experiment were ρ = 0.991 and ρ = 0.986 respectively.
Quantitative MRI
The results of the quantitative MRI in six samples reflecting different stages and components of the clearing process are shown in Figure 4 (pure hydrogel and PBS values shown only in the bar graph) with numerical values in Table 3. The white and grey matter contrast visible in the PFA-fixed (Fig. 4, first row) and the hydrogel-embedded samples (Fig. 4, second row) is no longer present in the cleared samples (Fig. 4, rows four and five) with the exception of some minimal contrast in the T1 and MD maps of the sample immersed in PBS. Similar to the cleared sample in the longitudinal MRI experiment, the cleared sample expanded to 3× the uncleared volume when immersed in PBST. In PBS the expansion was reduced to 1.06× the uncleared volume. The signal intensity profiles highlight the tissue contrast between areas such as cortex (high T1 at the sides of the uncleared profiles), caudoputamen (low T1 left and right of the profile center) and deep gray matter nuclei (high T1 in center of profile) that is present in the uncleared samples but not in the cleared samples. Minimal contrast in the T1 and MD maps is only present for the cleared sample immersed in PBS and not the cleared sampled immersed in PBST. The diffusivity values MD and FA in the cleared samples were similar to free water and independent of tissue expansion (i.e. the same for cleared samples in PBS and PBST). While FA contrast is gone in the cleared samples, the baseline FA for the cleared samples, hydrogel sample and the pure water sample is above 0 potentially due to minor imperfections in gradient calibration of the MRI scanner.
Figure 4.
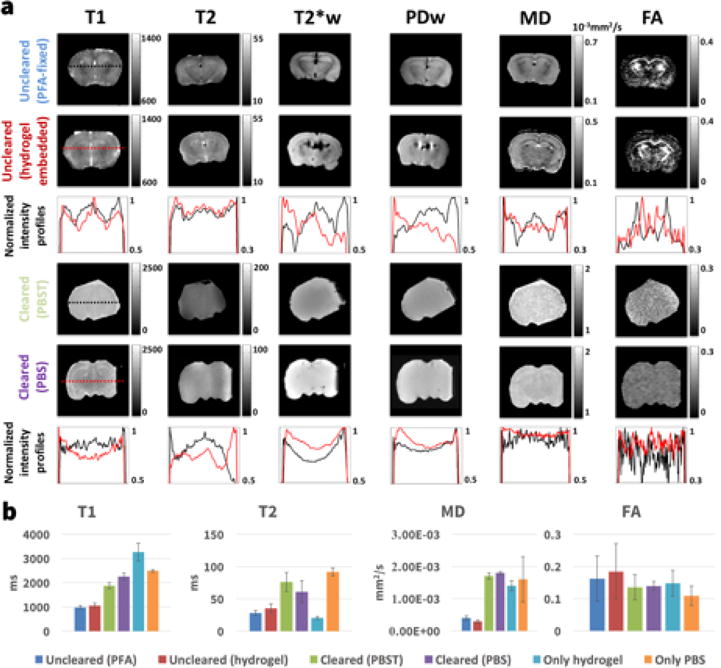
(a) T1-map (T1), T2-map (T2), T2*-weighted image (T2*w), proton density weighted image (PD), mean diffusivity map (MD) and fractional anisotropy map (FA) for an uncleared sample fixed only in PFA (top row), an uncleared sample embedded in acrylamide hydrogel (2nd row), a cleared sample immersed in PBST (4th row) and a cleared sample immersed in PBS (5th row). The second row shows intensity profiles of the images above with black being the intensity profile for the PFA sample and red for the hydrogel sample. The 6th row shows intensity profiles of the cleared brains with black being the profile of the sample immersed in PBST and red the sample in PBS. (b) Comparison of the quantitative MRI measurements for all samples. The bars show the mean value over a whole slice in the center of the sample and the error bars denote one standard deviation. Additionally the T1, T2, MD and FA values for a sample containing only hydrogel and a sample containing only phosphate buffered saline (PBS) are shown.
Table 3.
Overview of the quantitative MRI measurement values shown in Figure 4b. The values shown are mean ± one standard deviation of all voxels in the sample.
| Uncleared (PFA) |
Uncleared (hydrogel) |
Cleared (PBST) |
Cleared (PBS) |
Only hydrogel | Only PBS | ||
|---|---|---|---|---|---|---|---|
| T1 | [ms] | 976 ± 70 | 1050 ± 98 | 1877 ± 127 | 2259 ± 145 | 3264 ± 370 | 2490 ± 55 |
| T2 | [ms] | 28 ± 4 | 35 ± 7 | 76 ± 15 | 61 ± 17 | 21 ± 2 | 92 ± 6 |
| MD | [10− 3mm2/s] | 0.40 ± 0.07 | 0.29 ± 0.04 | 1.70 ± 0.10 | 1.80 ± 0.04 | 1.41 ± 0.16 | 1.61 ± 0.71 |
| FA | 0.16 ± 0.07 | 0.18 ± 0.09 | 0.14 ± 0.04 | 0.14 ± 0.01 | 0.15 ± 0.04 | 0.11 ± 0.03 |
MD - mean diffusivity, FA - fractional anisotropy, PFA - paraformaldehyde, PBS - phosphate buffered saline
Inductively coupled plasma optical emission spectrometry (ICP-OES)
We determined the iron, calcium and phosphorus concentration in 4 uncleared and 4 cleared samples using ICP-OES. For uncleared and cleared samples respectively, the concentrations were as follows: iron 0.48 ± 0.22 ppm (mean ± standard deviation) and 0.20 ± 0.11 ppm (p = 0.2); calcium 3.07 ± 1.77 ppm and 1.02 ± 0.17 ppm (p = 0.17); phosphorus 17.44 ± 3.45 ppm and 4.30 ± 0.92 ppm (p = 0.01) (Fig. 3b).
Protein analysis
Protein analysis of the uncleared and cleared samples using a bicinchoninic acid assay (BCA) showed protein concentrations of 2.50 ± 0.08 μg/μl and 2.47μg ± 0.10 μg/μl respectively (Fig. 3b). A paired t-test showed no significant difference between these concentrations (p = 0.41).
pH
pH measurements of the uncleared and cleared tissue samples showed a neutral pH between 7–8 for all six measured tissue samples. Measurement of a pure SBC sample showed a pH slightly above 8 while the pH of PBS was 7.
Molecular labeling of cleared tissue
Figure 3c shows a cleared mouse brain slab stained for DNA using propidium iodide and for neurofilaments using anti-neurofilament antibodies. Both DNA and neurofilament proteins are preserved at their original locations in the cleared specimen.
Discussion
In tissue, the separate effect of lipids has traditionally been difficult to measure and only the combined influence of lipids and proteins in the form of cell membranes, organelles or myelin on MRI contrast has previously been shown (Clark et al., 1992; Eickhoff, 2005; Fatterpekar et al., 2002; Stüber et al., 2014; Walters et al., 2007). In this study, we combined MRI and CLARITY measurements and NMR, BCA and ICP-OES to demonstrate the effect of tissue clearing on tissue content and the accompanying changes in relaxivity, diffusivity and MRI tissue contrast. Specifically, we found that lipid washout led to a change in quantitative relaxation times (T1, T2) and diffusion values (MD, FA) towards those of free water, and a dramatic decrease in MR tissue contrast in the cleared tissue samples (T1, T2, T2*, PD, MD, FA). We also confirm the successful washout of lipids and retention of proteins using NMR, BCA and ICP-OES.
Lipids
The strong decrease in MRI tissue contrast with clearing shows that lipids not only have a strong influence on tissue relaxivity but that they are responsible for the relaxivity differences between white matter and grey matter. This agrees with prior findings that glycolipids have a strong effect on water proton relaxivity in aqueous solutions and that there are significantly different concentrations in white matter versus grey matter (Kucharczyk et al., 1994). The resultant increase in the relaxation times of the cleared sample are also in good agreement with Kucharczyk et al. who showed that phospholipids alone, and more so in combination with other lipids like cholesterol, decreased T1 and T2 by more than 50% within an ionic buffer solution (Kucharczyk et al., 1994). The property of cholesterol to form so called “potholes” in the phospholipid bilayer membrane can lead to long-lived hydrogen bonds that could lead to the strong effect on relaxation times and tissue contrast (Koenig et al., 1990). A decrease in phosphorus concentration has also been shown to decrease the proton exchange rate (Liepinsh, E., Otting, 1996), which could contribute to the observed increased relaxation times with lipid washout.
Lipid washout also led to diminished diffusion contrast and anisotropic water diffusion. There was also a strong correlation between lipid washout and MD and FA WM-GM contrast within the first two weeks of tissue clearing. Post-clearing MD increased to a value comparable to free water. Further, no diffusion anisotropy was detected after lipid removal and FA maps displayed no tissue contrast in cleared samples. Other groups have previously shown that cell membranes are a dominant source of hindered water diffusion in tissue (Cooper et al., 1974; Novikov et al., 2011) and experiments in a giant squid axon demonstrated that the anisotropic water diffusion within the brain is primarily the result of axonal membranes aligned in parallel (Beaulieu and Allen, 1994). Multiple layers of cell membranes such as myelin wrapped around the axons can increase this effect by ∼20% (Beaulieu, 2002). Our results suggest that it is specifically the lipid components of the membranes and myelin that are the major contributor to the differential rates and patterns of diffusion observed by MRI.
Lipid washout was confirmed with 31P NMR, which showed that more than 90% of the phospholipids were washed out during the clearing process (Fig. 3a). It is expected that other types of lipids, not bound to the CLARITY hydrogel, are washed out to a similar degree (Chung et al., 2013). The small amount of phosphorus (<3%) detected in the NMR spectra after clearing could be attributed to phosphomonoesters and phosphodiesters that are part of the DNA and expected to attach to the hydrogel. The ICP-OES measurements showed about 25% of phosphorus remaining in the cleared tissue (Fig. 3b). Since the uncleared and cleared samples were stored in phosphate buffered saline (PBS) prior to the ICP-OES preparations, remaining PBS in the samples could lead to this larger amount of residual phosphorus compared to NMR.
Proteins
The protein analysis using a BCA array showed that more than 83% of proteins that make up the cytoskeleton were retained in a cleared specimen. This is in agreement with previous measurements of protein retention in cleared samples (Chung et al., 2013). Staining and imaging for cells and neurofilaments showed nucleic acids and proteins fixed at their original position. The significant increase in relaxation times suggests, however, that proteins in brain tissue do not shorten T1 and T2 values as much as lipids. Furthermore, the flat MR images observed postclearing show that the retained proteins and nucleic acids are not responsible for the usual signal differences that distinguish white and gray matter and other tissue structures. The fact that proteins contribute minimally to tissue contrast is a new finding that has previously not been possible to measure due to the difficulty in separating lipids and proteins in tissue. In aqueous solutions, proteins have been shown to affect relaxivity by proton exchange through –NH, –OH, – SH or amide groups (Mäkelä et al., 2001) and without proton exchange, protein macromolecules in close proximity to water molecules can affect the tumbling rates of water and thereby affect water relaxivity (Koenig and Brown, 1990).
There are several possible reasons why proteins would not contribute significantly to T1 and T2 contrast. For example, the effect of proteins on tissue contrast would be relatively weak if the affected T2 component is extremely short or if the net non-specific distribution of proteins within brain tissue is relatively homogeneous and different types of proteins affect the MRI signal in similar ways, leading to homogeneous relaxivity properties across the brain.
Since proteins also make up the cytoskeleton, our diffusion MRI data indicates that the cytoskeleton and components such as microfilaments do not play a significant role in creating the anisotropic diffusion patterns typically observed in brain tissue. Proteins showed very little effect on water diffusivity given that the mean diffusivity increased to a value comparable to free water after the lipids were removed. With CLARITY we have demonstrated this explicitly for the first time in the central nervous system of a mammal, over a large extent of brain tissue covering complete tissue structures. This is in good agreement with prior studies in giant squid axons where the effect of the cytoskeleton has been shown to be small compared to the axonal membranes (Beaulieu and Allen, 1994).
Effects of fixation and tissue clearing
In our experiments, tissue contrast and relaxation times are not only influenced by changing lipid and protein concentrations but also several aspects of the fixation and clearing protocol. Firstly, we must consider the effects of protein-crosslinking induced by both tissue fixation and hydrogel embedding. PFA forms methylene bridges between active –NH groups of the proteins (Birkl et al., 2016) and hydrogel embedding leads to cross-links between proteins and the surrounding acrylamide network in the presence of PFA (Tomer et al., 2014). This reduction of active –NH groups and decreased protein mobility was previously studied within protein solutions and shown to have varied affects on T1 and T2 depending on the buffer solution, pH and whether or not the proteins had been denatured (Mäkelä et al., 2001). Within human tissue, fixation has been shown to decrease in T1 and T2 relaxation times (Birkl et al., 2016). Specifically, at 3T, T1 in fixation was shown to decrease T1 by 67–76% in GM and 67% in WM and T2 by 5–11% in GM and 26% in WM. Longer fixation times have also been shown to further decrease T1 and T2 (Tovi and Ericsson, 1992). The effects of fixation in combination with a decrease in temperature (room temperature, 21°C vs. physiological temperature, 37°C) cause a decrease in diffusion rates compared to in vivo measurements (Sun et al., 2003). Despite this, it is well established that diffusion anisotropy effects are well-preserved in fixed tissue (McNab et al., 2009; Miller et al., 2012, 2011; Sun et al., 2003) and this is evident in our pre-clearing images (Figs. 2 & 4). Since all of our experiments start with fixed tissue, the effects of tissue fixation reflect a difference between in vivo and fixed tissue and possible changes between PFA-fixed and hydrogel embedded tissue but not between cleared and uncleared samples.
In addition to fixation and hydrogel embedding, the clearing process can also strongly change the protein environment as it relates to relaxation and diffusion rates. Specifically, lipid washout and tissue expansion during clearing can lead to changes in water diffusion and the number of water binding sites, thereby affecting the relaxivity of the sample. The increase in tissue volume during clearing leads to a decrease in local protein density which can impact relaxation times, tissue contrast and diffusion properties. For the longitudinal measurements, there was a 315% volume increase between the uncleared sample and week 1 but no measurable increase between week 1 and week 3. Since we observe a strong effect on tissue contrast between week 1 and 3, this suggests that a large part of the contrast decrease cannot be attributed to the volume increase. However, we did observe that the residual contrast (T1 and MD maps) in a cleared sample immersed in PBS (+6% expansion) was not present for a cleared sample immersed in PBST (+300% expansion). Quantitative comparisons of the +6% versus 300% expanded samples revealed differences in T1 and T2 relaxation times of −20% and +20% respectively but no difference in FA or MD. The solutions for the +300% increased volume (PBST) and +6% volume (PBS), were kept as similar as possible, but the 0.1% of Triton-X in the PBST sample could also have influenced the protein environment and relaxation times. Since the sample volumes can be affected not only by immersion solution but also by the cover slips fixing sample shape and position in the ChristoLube container, volumes were measured based on the MRI data to ensure that the presented volumes were the effective volumes during the MRI scan. To thoroughly test the effects of volume on tissue contrast, a study using a different clearing protocol such as iDISCO (Renier et al., 2014), which leads to a decrease of tissue volume after clearing, could provide additional information. We also controlled for the potential effects of a pH change during clearing but found pH to be neutral in both uncleared and cleared samples.
Myelin
While the separate effects of lipids and protein have traditionally been difficult to measure, there are many studies that measure myelination through MRI (Laule et al., 2007). Koenig et al. have shown that myelin can relax adjacent water molecules seven times more efficiently than the corresponding protein-water interface (Koenig et al., 1990). In multiple sclerosis strong correlations have been found between histologically-defined myelin content and MRI measurements of T1 (Mottershead et al., 2003), T2 (Whittall et al., 1997) FA (Schmierer et al., 2007), magnetization transfer ratio (MTR) (Schmierer et al., 2004) and myelin water fraction (MWF) (Laule et al., 2006). For T2 measurements, water pools with a very short T2 component are attributed to water trapped between the myelin sheaths (Whittall et al., 1997). Demyelination effects on MRI contrast have also been studied via mouse models such as the cuprizone (Nathoo et al., 2014; Thiessen et al., 2013) and shiverer mouse models (Song et al., 2002). In one study, the demyelination status of a lesion in cuprizone-fed mice was found to be accurately predicted by a combination of T1, T2 and MTR (Merkler et al., 2005). In shiverer mice, diffusion perpendicular to the fiber tracts was found to be elevated relative to controls (Song et al., 2002). Our tissue clearing results are in good agreement with these MRI studies of demyelination but also pinpoint more specifically the lipids within the myelin as the key component leading to the observed changes.
Iron & calcium
Although our study focuses on lipids and proteins, we take into account the effect of other components in brain tissue. Paramagnetic properties of metals, such as iron, change the local magnetic field distribution and decrease the transverse relaxation times T2 and T2* of water (Fukunaga et al., 2010). The lack of T2 and T2* contrast in the cleared samples suggests that iron was also washed out during the experiments. This was confirmed by the ICP-OES measurements that showed a decrease in iron concentration in cleared samples from 0.48 ± 0.23 ppm to 0.20 ± 0.11 ppm. Less than 5% of iron in the brain is free iron (Kakhlon and Cabantchik, 2002). Most iron is stored in proteins such as ferritin or transferrin. Ferritin and transferrin should be preserved by the hydrogel embedding but it is not clear if the iron associated with these proteins is also retained. Our ICP-OES measurements provide evidence that at least some iron associated with these proteins is also washed out during tissue clearing. The decrease in tissue contrast, however, shows that these low concentrations do not lead to any significant tissue contrast in the cleared samples. Accumulation of calcium has also been shown to influence tissue contrast (Wehrli, 2013). The ICP-OES measurements showed a decrease in calcium concentration in the cleared samples, which again confirms that these low calcium concentrations do not influence tissue contrast.
CLARITY
Our experiment relied on an accurate knowledge of which tissue components are washed out or retained and to what degree. We, therefore, measured lipid washout by measuring changes in phosphorus content using NMR spectroscopy and ICP-OES. Lipid washout has not previously been quantified for CLARITY and therefore our results demonstrating an exponential decay of lipid content over time and washout of more than 95% of tissue lipids content by 4 weeks, provide new insights into the temporal characteristics of tissue clearing that may help refine tissue clearing approaches.
Another interesting observation is that the protein-hydrogel network in the cleared tissue had different MRI properties from the pure hydrogel. In tissue, proteins linking acrylamide strands of the gel network create a different configuration of the gel network compared to a pure hydrogel (Fig. 1 in (Tomer et al., 2014)). This difference in the hydrogel configuration could be one of the causes of the different relaxation times and mean diffusivity observed in the cleared tissue versus the pure hydrogel sample. Another effect is that the pure hydrogel sample has not undergone the clearing process. The clearing process prevents the hydrogel within the tissue from making further connections, while the pure hydrogel sample can continue to form new connections over time, leading to a different rigidity of the network compared to the cleared tissue sample.
Several other aspects of the fixation and clearing protocol can also affect the hydrogel structure and clearing outcome. This includes variations in tissue fixation time before polymerization, polymerization time and oxygen content within the sample container during polymerization. In the current study, efforts have been made to control for each of these effects.
The observed MRI contrast decrease and tissue deformation of the cleared samples have important implications for studies comparing MRI and CLARITY data in the same samples (Aswendt et al., 2017). Firstly, the strong decrease of MRI tissue contrast after clearing indicates that MRI measurements must be performed before tissue clearing. Secondly, the significant tissue expansion and distortion observed during the clearing process demonstrates the need for non-linear co-registration algorithms to align the CLARITY images to MRI or a common space such as the Allen Brain Atlas (Hawrylycz et al., 2012). MRI is often compared to histology as a means to understand the biological substrate of the MRI signal on a cellular level. Comparing MRI against conventional 2D histological sections allows examination of cell or myelin density (Geyer et al., 2011) or fiber directionality in 2 dimensions (Leuze et al., 2014). To compare 3D structures, such as fibers running through-plane, a comparison of MRI with a 3D histological technique is preferred. Tissue clearing enables visualization of tissue samples up to 10–100× thicker than histological sections and as such provides new opportunities for validating MRI results.
Conclusion
The recent advent of the CLARITY technique has allowed us for the first time to separate the relative contribution of lipids and proteins to brain MRI contrast. Our results show that although lipids and proteins account for approximately the same percentage of brain matter by weight, the lipids are by far the dominant source of tissue contrast. While proteins may influence relaxivity, we observed minimal T1 and MD contrast and no contrast at all for all the other MRI sequences when the lipids are removed by clearing. Our data also provides evidence that hindered water diffusion in brain tissue is mainly caused by lipids found within bi-lipid cell membranes, while the preserved cytoskeleton has no observable effect on water diffusion rates or anisotropy.
Acknowledgments
We would like to thank Ryan Spitler for help with the ICP-OES measurements and Brian Edlow, Raju Tomer, Dan Spielman, Brian Rutt, Joonas Autio and Michelle Cheng for valuable discussions. This study was funded by the Stanford Neurosciences Institute, the Gatsby Foundation, the Oxford-Stanford Big Data in Biomedicine Seed Grant, Stanford Radiology Angel Funds, GE Healthcare, the Dana Foundation, DARPA (Cooperative Agreement Number W911NF-14-2-0013), the NIMH, NIDA, HHMI, NSF and the NIH (R01-NS095985, R01-MH111444, P41-EB015891 & S10-RR02635).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aswendt M, Schwarz M, Abdelmoula WM, Dijkstra J, Dedeurwaerdere S. Whole-Brain Microscopy Meets In Vivo Neuroimaging: Techniques, Benefits, and Limitations. Mol Imaging Biol. 2017;19:1–9. doi: 10.1007/s11307-016-0988-z. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson Ser B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magn Reson Med. 2000;44:625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Beaulieu C. The basis of anisotropic water diffusion in the nervous system — a technical review. NMR Biomed. 2002;15:435–455. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- Beaulieu C, Allen PS. Water diffusion in the giant axon of the squid: Implications for diffusion-weighted MRI of the nervous system. Magn Reson Med. 1994;32:579–583. doi: 10.1002/mrm.1910320506. [DOI] [PubMed] [Google Scholar]
- Birkl C, Langkammer C, Golob-schwarzl N, Leoni M, Haybaeck J, Goessler W, Fazekas F, Ropele S. Effects of formalin fixation and temperature on MR relaxation times in the human brain. 2016:458–465. doi: 10.1002/nbm.3477. [DOI] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky Ka, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–7. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark VP, Courchesne E, Grafe M. In vivo myeloarchitectonic analysis of human striate and extrastriate cortex using magnetic resonance imaging. Cereb Cortex. 1992;2:417–424. doi: 10.1093/cercor/2.5.417. [DOI] [PubMed] [Google Scholar]
- Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder aZ, Shimony JS, McKinstry RC, Burton H, Raichle ME. Tracking neuronal fiber pathways in the living human brain. Proc Natl Acad Sci U S A. 1999;96:10422–7. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RL, Chang DB, Young AC, Martin CJ, Ancker-Johnson D. Restricted diffusion in biophysical systems: experiment. Biophys J. 1974;14:161–177. doi: 10.1016/S0006-3495(74)85904-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S. High-Resolution MRI Reflects Myeloarchitecture and Cytoarchitecture of Human Cerebral Cortex. Hum Brain Mapp. 2005;24:206–215. doi: 10.1002/hbm.20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp JR, Niibori Y, Liz Hsiang H-L, Mercaldo V, Deisseroth K, Josselyn Sa, Frankland PW. Optimization of CLARITY for clearing whole brain and other intact organs. eNeuro. 2015 doi: 10.1523/ENEURO.0022-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatterpekar GM, Naidich TP, Delman BN, Aguinaldo JG, Gultekin SH, Sherwood CC, Hof PR, Drayer BP, Fayad ZA. Cytoarchitecture of the human cerebral cortex: MR microscopy of excised specimens at 9.4 Tesla. Am J Neuroradiol. 2002;23:1313–1321. [PMC free article] [PubMed] [Google Scholar]
- Fukunaga M, Li TQ, van Gelderen P, de Zwart Ja, Shmueli K, Yao B, Lee J, Maric D, Aronova Ma, Zhang G, Leapman RD, Schenck JF, Merkle H, Duyn JH. Layer-specific variation of iron content in cerebral cortex as a source of MRI contrast. Proc Natl Acad Sci U S A. 2010;107:3834–3839. doi: 10.1073/pnas.0911177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer S, Weiss M, Reimann K, Lohmann G, Turner R. Microstructural Parcellation of the Human Cerebral Cortex — From Brodmann’s Post-Mortem Map to in vivo Mapping with High-Field Magnetic Resonance Imaging. Front Hum Neurosci. 2011;5:19. doi: 10.3389/fnhum.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gochberg DF, Kennan RP, Maryanski MJ, Gore JC. The role of specific side groups and pH in magnetization transfer in polymers. J Magn Reson. 1998;131:191–198. doi: 10.1006/jmre.1998.1371. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, Daly BD, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SGN, Jones AR. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–9. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms G, Hagberg GE. In vivo quantification of the bound pool T1 in human white matter using the binary spin-bath model of progressive magnetization transfer saturation. Phys Med Biol. 2009;54:N529–40. doi: 10.1088/0031-9155/54/23/N01. [DOI] [PubMed] [Google Scholar]
- Jones DK, Simmons A, Williams SC, Horsfield MA. Non-invasive assessment of axonal fiber connectivity in the human brain via diffusion tensor MRI. Magn Reson Med. 1999;42:37–41. doi: 10.1002/(sici)1522-2594(199907)42:1<37::aid-mrm7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Kakhlon O, Cabantchik ZI. The labile iron pool: Characterization, measurement, and participation in cellular processes. Free Radic Biol Med. 2002 doi: 10.1016/S0891-5849(02)01006-7. [DOI] [PubMed] [Google Scholar]
- Kirschner DA, AE B. Myelin Structure and Components. In: Martenson RE, editor. Myelin: Biology and Structure. CRC Press; 1992. pp. 3–80. [Google Scholar]
- Knaap M, van der Valk J. Magnetic Resonance of Myelination and Myelin Disorders 2005 [Google Scholar]
- Koenig SH. Cholesterol of myelin is the determinant of gray???white contrast in MRI of brain. Magn Reson Med. 1991;20:285–291. doi: 10.1002/mrm.1910200210. [DOI] [PubMed] [Google Scholar]
- Koenig SH, Brown RD. Field-cycling relaxometry of protein solutions and tissue: Implications for MRI. Prog Nucl Magn Reson Spectrosc. 1990;22:487–567. doi: 10.1016/0079-6565(90)80008-6. [DOI] [Google Scholar]
- Koenig SH, Brown RD, Spiller M, Lundbom N. Relaxometry of brain: Why white matter appears bright in MRI. Magn Reson Med. 1990;14:482–495. doi: 10.1002/mrm.1910140306. [DOI] [PubMed] [Google Scholar]
- Kucharczyk W, Lenkinski RE, Kucharczyk J, Henkelman MR. The effect of phospholipid vesicles on the NMR relaxation of Water: An explanation for the MR appearance of the neurohypophysis? Am J Neuroradiol. 1990;11:693–700. [PMC free article] [PubMed] [Google Scholar]
- Kucharczyk W, Macdonald M, Stanisz GJ, Henkelman RM. Relaxivity at MR Imaging: of Cerebrosides and pH. Radiology. 1994;192:521–529. doi: 10.1148/radiology.192.2.8029426. [DOI] [PubMed] [Google Scholar]
- Laule C, Leung E, Lis DKB, Traboulsee AL, Paty DW, MacKay AL, Moore GRW. Myelin water imaging in multiple sclerosis: quantitative correlations with histopathology. Mult Scler. 2006;12:747–53. doi: 10.1177/1352458506070928. [DOI] [PubMed] [Google Scholar]
- Laule C, Vavasour IM, Kolind SH, Li DKB, Traboulsee TL, Moore GRW, MacKay AL. Magnetic Resonance Imaging of Myelin. Neurotherapeutics. 2007;4:460–484. doi: 10.1016/j.nurt.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuze C, Anwander A, Bazin PL, Dhital B, Stüber C, Reimann K, Geyer S, Turner R. Layer-specific intracortical connectivity revealed with diffusion MRI. Cereb Cortex. 2014;24:328–339. doi: 10.1093/cercor/bhs311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gore J, Davatzikos C. Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation. Magn Reson Imaging. 2015;32:913–923. doi: 10.1038/nbt.3121.ChIP-nexus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepinsh E, Otting G. Proton Exchange Rates from Amino Acid Side Chains -Implications for Image Control. Magn Reson Med. 1996;35:30–42. doi: 10.1002/mrm.1910350106. [DOI] [PubMed] [Google Scholar]
- Mäkelä HI, Gröhn OH, Kettunen MI, Kauppinen Ra. Proton exchange as a relaxation mechanism for T1 in the rotating frame in native and immobilized protein solutions. Biochem Biophys Res Commun. 2001;289:813–818. doi: 10.1006/bbrc.2001.6058. [DOI] [PubMed] [Google Scholar]
- McIlwain H, Bachelard H. Biochemistry and the Central Nervous System. Churchill Livingstone; Edinburgh: 1985. [Google Scholar]
- McNab Ja, Jbabdi S, Deoni SCL, Douaud G, Behrens TEJ, Miller KL. High resolution diffusion-weighted imaging in fixed human brain using diffusion-weighted steady state free precession. Neuroimage. 2009;46:775–85. doi: 10.1016/j.neuroimage.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Merkler D, Boretius S, Stadelmann C, Ernsting T, Michaelis T, Frahm J, Brück W. Multicontrast MRI of remyelination in the central nervous system. NMR Biomed. 2005;18:395–403. doi: 10.1002/nbm.972. [DOI] [PubMed] [Google Scholar]
- Miller KL, McNab Ja, Jbabdi S, Douaud G. Diffusion tractography of post-mortem human brains: Optimization and comparison of spin echo and steady-state free precession techniques. Neuroimage. 2012;59:2284–2297. doi: 10.1016/j.neuroimage.2011.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KL, Stagg CJ, Douaud G, Jbabdi S, Smith SM, Behrens TE, Jenkinson M, Chance SA, Esiri MM, Voets NL, Jenkinson N, Aziz TZ, Turner MR, JohansenBerg H, McNab JA. Diffusion imaging of whole, post-mortem human brains on a clinical MRI scanner. Neuroimage. 2011;57:167–181. doi: 10.1016/j.neuroimage.2011.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawski M, Meinecke C, Reinert T, Dörffel AC, Riederer P, Arendt T, Butz T. Determination of trace elements in the human substantia nigra. Nuclear Instruments and Methods in Physics Research, Section B: Beam Interactions with Materials and Atoms. 2005:224–228. doi: 10.1016/j.nimb.2005.01.061. [DOI] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::AID-ANA21>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Moseley ME, Cohen Y, Kucharczyk J, Mintorovitch J, Asgari HS, Wendland MF, Tsuruda J, Norman D. Diffusion-weighted MR imaging of anisotropic water diffusion in cat central nervous system. Radiology. 1990;176:439–445. doi: 10.1148/radiology.176.2.2367658. [DOI] [PubMed] [Google Scholar]
- Mottershead JP, Schmierer K, Clemence M, Thornton JS, Scaravilli F, Barker GJ, Tofts PS, Newcombe J, Cuzner ML, Ordidge RJ, McDonald WI, Miller DH. High field MRI correlates of myelin content and axonal density in multiple sclerosis— a post-mortem study of the spinal cord. J Neurol. 2003;250:1293–301. doi: 10.1007/s00415-003-0192-3. [DOI] [PubMed] [Google Scholar]
- Nathoo N, Yong VW, Dunn JF. Understanding disease processes in multiple sclerosis through magnetic resonance imaging studies in animal models. NeuroImage Clin. 2014;4:743–756. doi: 10.1016/j.nicl.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton WT, Autilio La. The lipid composition of purified bovine brain myelin. J Neurochem. 1966;13:213–222. doi: 10.1111/j.1471-4159.1966.tb06794.x. [DOI] [PubMed] [Google Scholar]
- Novikov DS, Fieremans E, Jensen JH, Helpern Ja. Random walks with barriers. Nat Phys. 2011;7:508–514. doi: 10.1038/Nphys1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien JS, Sampson EL. Lipid composition of the normal human brain: gray matter, white matter, and myelin. J Lipid Res. 1965;6:537–544. [PubMed] [Google Scholar]
- Randall L. Chemical Topography of Brain. J Biol Chem. 1938;124 [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-lavigne M. Resource iDISCO: A Simple, Rapid Method to Immunolabel Large Tissue Samples for Volume Imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Schmierer K, Scaravilli F, Altmann DR, Barker GJ, Miller DH. Magnetization transfer ratio and myelin in postmortem multiple sclerosis brain. Ann Neurol. 2004;56:407–415. doi: 10.1002/ana.20202. [DOI] [PubMed] [Google Scholar]
- Schmierer K, Wheeler-Kingshott CAM, Boulby PA, Scaravilli F, Altmann DR, Barker GJ, Tofts PS, Miller DH. Diffusion tensor imaging of post mortem multiple sclerosis brain. Neuroimage. 2007;35:467–477. doi: 10.1016/j.neuroimage.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SK, Sun SW, Ramsbottom MJ, Chang C, Russell J, Cross AH. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage. 2002;17:1429–36. doi: 10.1006/nimg.2002.1267. [DOI] [PubMed] [Google Scholar]
- Stüber C, Morawski M, Schäfer A, Labadie C, Wähnert M, Leuze C, Streicher M, Barapatre N, Reimann K, Geyer S, Spemann D, Turner R. Myelin and iron concentration in the human brain: a quantitative study of MRI contrast. Neuroimage. 2014;93(Pt 1):95–106. doi: 10.1016/j.neuroimage.2014.02.026. [DOI] [PubMed] [Google Scholar]
- Sun SW, Neil JJ, Song SK. Relative indices of water diffusion anisotropy are equivalent in live and formalin-fixed mouse brains. Magn Reson Med. 2003;50:743–8. doi: 10.1002/mrm.10605. [DOI] [PubMed] [Google Scholar]
- Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A. Multiplexed Intact-Tissue Transcriptional Analysis at Resource Multiplexed Intact-Tissue Transcriptional Analysis at Cellular Resolution. Cell. 2016;164:792–804. doi: 10.1016/j.cell.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiessen JD, Zhang Y, Zhang H, Wang L, Buist R, Del MR, Kong J, Li X, Martin M. Quantitative MRI and ultrastructural examination of the cuprizone mouse model of demyelination. 2013:1562–1581. doi: 10.1002/nbm.2992. [DOI] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovi M, Ericsson A. Measurements of T1 and T2 over time in formalin-fixed human whole-brain specimens. Acta Radiol. 1992;33:400–4. doi: 10.1080/02841859209172021. [DOI] [PubMed] [Google Scholar]
- van Gelderen P, Jiang X, Duyn JH. Effects of magnetization transfer on T1 contrast in human brain white matter. Neuroimage. 2016;128:85–95. doi: 10.1016/j.neuroimage.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters NB, Eickhoff SB, Schleicher A, Zilles K, Amunts K, Egan GF, Watson JDG. Observer-independent analysis of high-resolution MR images of the human cerebral cortex: in vivo delineation of cortical areas. Hum Brain Mapp. 2007;28:1–8. doi: 10.1002/hbm.20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Sheriff S, Lewis KB, Cho J, Tinch SL, Balasubramaniam A, Michael a. Metabolic Profiling Comparison of Human Pancreatic Ductal Epithelial Cells and Three Pancreatic Cancer Cell Lines using NMR Based Metabonomics. Mol Biomarkers Diagnosis. 2012 doi: 10.4172/2155-9929.S3-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli FW. Magnetic resonance of calcified tissues. J Magn Reson. 2013;229:35–48. doi: 10.1016/j.jmr.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittall KP, MacKay AL, Graeb DA, Nugent RA, Li DK, Paty DW. In vivo measurement of T2 distributions and water contents in normal human brain. Magn Reson Med. 1997;37:34–43. doi: 10.1002/mrm.1910370107. [DOI] [PubMed] [Google Scholar]
- Zeineh MM, Chen Y, Kitzler HH, Hammond R, Vogel H, Rutt BK. Activated iron-containing microglia in the human hippocampus identified by magnetic resonance imaging in Alzheimer disease. Neurobiol Aging. 2015 doi: 10.1016/j.neurobiolaging.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


