Abstract
Mutational inactivation of BRCA1 confers a cumulative lifetime risk of breast and ovarian cancers. However, the underlying basis for the tissue-restricted tumor-suppressive properties of BRCA1 remains poorly defined. Here we show that BRCA1 mediates ligand-independent transcriptional repression of the estrogen receptor α (ERα), a principal determinant of the growth, differentiation, and normal functional status of breasts and ovaries. In Brca1-null mouse embryo fibroblasts and BRCA1-deficient human ovarian cancer cells, ERα exhibited ligand-independent transcriptional activity that was not observed in Brca1-proficient cells. Ectopic expression in Brca1-deficient cells of wild-type BRCA1, but not clinically validated BRCA1 missense mutants, restored ligand-independent repression of ERα in a manner dependent upon apparent histone deacetylase activity. In estrogen-dependent human breast cancer cells, chromatin immunoprecipitation analysis revealed the association of BRCA1 with ERα at endogenous estrogen-response elements before, but not after estrogen stimulation. Collectively, these results reveal BRCA1 to be a ligand-reversible barrier to transcriptional activation by unliganded promoter-bound ERα and suggest a possible mechanism by which functional inactivation of BRCA1 could promote tumorigenesis through inappropriate hormonal regulation of mammary and ovarian epithelial cell proliferation.
Germline inactivation of the gene that encodes BRCA1 represents a predisposing genetic factor in ≈15–45% of hereditary breast cancers, and minimally 80% of combined hereditary breast and ovarian cancer cases (1). Functionally, BRCA1 has been implicated in the maintenance of global genome stability (2–4), and the underlying basis for this activity likely derives from its central role in the cellular response to DNA damage, wherein it controls both DNA damage repair and the transcription of DNA damage-inducible genes (5–14).
Because the DNA damage-induced signaling pathways that converge on BRCA1 are likely to be conserved in most cell types, BRCA1 is likely to occupy a fundamental and universally conserved role in the mammalian DNA damage response. Nonetheless, germ-line inactivation of BRCA1 leads predominantly to cancer of the breast and ovary, and the underlying basis for its tissue-restricted tumor-suppressive properties thus remains undefined.
At least two hypotheses have been proposed to explain the tissue-specific nature of BRCA1-mediated tumor suppression, both of which invoke a role for estrogen in either the initiation or promotion of tumor formation (15). According to one model, the tissue-specific tumor-suppressive properties of BRCA1 derive, at least in part, from its response to tissue-specific DNA damage. In this regard, certain oxidative metabolites of estrogen itself have been documented to be genotoxic in nature (16), and BRCA1 may therefore play a role in protecting breast and ovarian tissue from estrogen-induced DNA damage.
A second model, not mutually exclusive with the one described above, to account for the this tissue-specific tumor-suppressive function invokes a role for BRCA1 in the modulation of estrogen signaling pathways and, hence, the expression of hormone-responsive genes. In this regard, BRCA1 has been reported to inhibit estrogen-dependent transactivation by the estrogen receptor α (ERα) through its direct interaction with ERα (17, 18). BRCA1 has also been reported to enhance androgen-dependent transactivation by the androgen receptor, allelic variants of which modify cancer penetrance in BRCA1 mutation carriers (19–21). Based on its postulated role in the control of nuclear hormone signaling pathways, BRCA1 could therefore influence epithelial cell proliferation and, by implication, cancer risk in tissues such as breast and ovary.
Herein, we describe a role for BRCA1 in mediating ligand-independent transcriptional repression of the ERα. Initial efforts to elucidate the mechanistic basis for this repression reveal that BRCA1 represents a ligand-reversible barrier to transcriptional activation by unliganded promoter-bound ERα. These findings suggest a potential role for BRCA1 in the proliferative control of normal estrogen-regulated tissues and a potential basis by which its mutational inactivation could promote tumorigenesis through inappropriate hormonal responses.
Materials and Methods
Cell Culture.
p53−/− (Brca1+/+) and p53−/−; Brca1−/− (Brca1−/−) mouse embryonic fibroblasts (MEFs) were cultured as described (14). Human MCF7 cells were maintained in DMEM supplemented with 10% FCS. Human BG-1-derived NEO1 and AS4 cell lines were maintained as described (22). Depletion of hormone ligands for nuclear/steroid receptor activation studies was achieved by cell culture in medium containing either 10% charcoal/dextran-treated serum (HyClone) or defined serum replacement 2 (Sigma).
Plasmids and Transfections.
Transfection assays were performed by using the following conditions.
Reporter plasmids.
Used at 0.5 μg each, including pTRE(F2)-TK-Luc, pGRE-TK-CAT, pERE-TK-Luc, or pPRE-TK-CAT (23); 0.5 μg of pGAL4-SV40-Luc containing five GAL4 DNA-binding sites upstream of the minimal simian virus 40 (SV40) promoter, driving expression of the luciferase reporter gene in the pGL2 vector (Promega); and 0.5 μg of pGAL4-E1B-Luc (24).
Receptor expression plasmids.
Used at 1.0 μg each, including RSV-hTRβ, RSV-hGR, RSV-hERα, and RSV-hPRβ (23).
BRCA1 expression plasmids.
Used at 1.0 μg each, including pcDNA3.1-BRCA1, pcDNA3.1-BRCA1-A1708E, pcDNA3.1-BRCA1-Q356R, and pcDNA3.1-BRCA1-A1708E/Q356R expressing either human wild-type BRCA1 or familial breast cancer-derived BRCA1 mutants (14).
Chimeric activators.
Used at 1.0 μg of GAL4-ERα, generated by an amino-terminal fusion of ERα with the GAL4 DNA-binding domain in pM3 (25); 0.1 μg of pVP16-GAL4 or pVP16-GAL4-ERα containing ERα amino acids 251–595, as described (26).
MEFs (6 × 104) or BG-1 cells (2 × 105) cultured in ligand-free medium were transfected by Lipofectin-based methods under serum-free conditions. Culture medium was replaced with fresh ligand-free medium 24 h after transfection, and 10−7 M 17β-estradiol (E2) or 330 nM trichostatin A was added as indicated. Cells were harvested 48 h after transfection for luciferase assay as described (14) or chloramphenicol acetyltransferase (CAT) assay by liquid scintillation counting (Promega).
Reverse Transcription (RT)-PCR Analysis.
BG-1-derived cells were cultured in ligand-free medium for at least 5 days, and treated with 10−7 M E2 for 1 h as indicated. Approximately 15 μg of total cellular RNA was subjected to semiquantitative RT-PCR analysis following a procedure previously described for estrogen-responsive genes (27, 28).
Chromatin Immunoprecipitation (ChIP).
MCF7 cells were cultured in ligand-free medium for at least 5 days and treated with 10−7 E2 for 1 h as indicated. ChIP assays were performed as described (29).
Antibodies.
Antibodies used for soluble and chromatin immunoprecipitations and immunoblot analyses were as follows: BRCA1 (mAb 6B4); ERα (rabbit polyclonal antibody HC-20 or mouse mAb D-12, Santa Cruz Biotechnology); CtIP (mAb 19E8); TFIIH p89 (rabbit polyclonal antibody S-19, Santa Cruz Biotechnology); glutathione S-transferase (MAb 8G11); RNA polymerase II large subunit (mAb 8WG16); cathepsin D (rabbit polyclonal antibody 06-467, Upstate Biotechnology, Lake Placid, NY); pS2 (mouse mAb V3030, Biomeda, Hayward, CA); human progesterone receptor β (mouse mAb PriB-30, Santa Cruz Biotechnology); p84 (mAb 5E10).
Results
BRCA1 has been shown to modulate the ligand-dependent transcriptional activity of specific members of the nuclear hormone receptor family (17–20). However, endogenous BRCA1 present in the transfected cell lines used in previous studies precluded analysis of the effect of BRCA1 on the ligand-independent function of these receptors. Therefore, to more directly assess the role of BRCA1 in nuclear receptor transactivation without competition from endogenous BRCA1, we analyzed a panel of nuclear receptors for their respective ligand-independent transcriptional activities in Brca1-nullizygous MEFs.
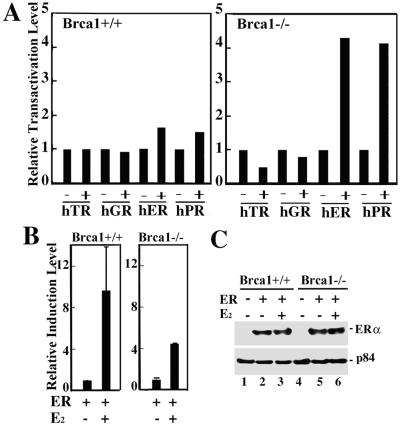
A set of minimal thymidine kinase (TK) promoters, each under control of distinct hormone-response elements specific for either the human thyroid receptor β (TRβ), the glucocorticoid receptor (GR), the ERα, or the progesterone receptor β (PRβ) were individually tested for their respective abilities to direct expression of a reporter gene in the absence or presence of each corresponding receptor (absent ligand) after transfection into Brca1-proficient (Brca1+/+) or Brca1-deficient (Brca1−/−) MEFs (14). Unexpectedly, we observed significant ligand-independent activation of reporter gene expression directed by both the progesterone receptor β and the ERα in Brca1-deficient MEFs compared with Brca1-proficient MEFs (Fig. 1A). By contrast, no ligand-independent stimulation of reporter activity directed by either the thyroid receptor β or the glucocorticoid receptor could be observed in Brca1-deficient MEFs (Fig. 1A). Interestingly, although E2 activated the ERα in both Brca1-proficient and Brca1-deficient MEFs, the relative level of induction observed in Brca1-deficient MEFs was diminished 2-fold relative to Brca1-proficient MEFs (Fig. 1B). We confirmed by immunoblot analysis that the transfected ERα was expressed equivalently in BRCA1-proficient and BRCA1-deficient MEFs, thus excluding the possibility that differences in receptor activity derive from differences in receptor protein expression (Fig. 1C).
Figure 1.
BRCA1 mediates ligand-independent repression of the receptors for estrogen and progesterone. (A) Brca1+/+ and Brca1−/− MEFs in hormone-free media were transfected with reporter plasmids (pTK-Luc or pTK-CAT) carrying response elements specific for individual hormone receptors without (−) or with (+) plasmids expressing the human thyroid receptor β (hTR), glucocorticoid receptor (hGR), estrogen receptor α (hER), or progesterone receptor β (hPR). Transfections performed without (−) receptor expression plasmids were performed instead with a molar equivalent of the backbone expression plasmid pRSV. The relative transactivation level represents the fold-increase in transfected reporter gene activity measured in cells cotransfected with a specific receptor expression plasmid relative to the level of transfected reporter gene activity measured in cells cotransfected with the backbone pRSV expression plasmid. Reporter gene activity was first normalized to β-galactosidase activity obtained by cotransfection of an internal control pSV40-β-gal expression plasmid as described (14). Expression of the pSV40-β-gal plasmid was not affected by the absence of presence of BRCA1 or any of the nuclear hormone receptors analyzed (data not shown). (B) Brca1+/+ and Brca1−/− MEFs in estrogen-free media were transfected with pERE-TK-Luc carrying three copies of the consensus estrogen response element (ERE) with (+) pRSV-ERα in the absence (−) or presence (+) of E2 (10−7 M) before assay for luciferase activity. The relative induction level represents the relative transactivation level measured in the presence of E2 divided by the relative transactivation level measured in the absence of E2. (C) Brca1+/+ (lanes 1–3) and Brca1−/− (lanes 4–6) MEFs either untransfected (lanes 1 and 4) or transfected (lanes 2, 3, 5, and 6) with an ERα-expressing vector were lysed, and immunoprecipitated ERα was immunoblotted with ERα-specific antibodies (Upper). Immunoblot analysis of the nuclear matrix protein p84 (Lower) indicates that nearly equivalent amounts of each cell lysate were used in the immunoprecipitations.
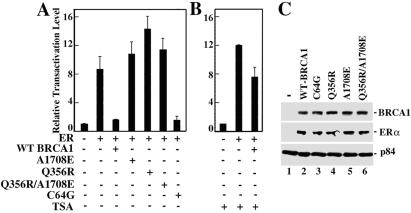
Ectopic expression of wild-type BRCA1 in Brca1-deficient MEFs repressed ligand-independent activation directed by ERα (Fig. 2A). Likewise, a BRCA1 derivative carrying a familial breast cancer-derived missense mutation in the ring finger (C64G) also repressed ligand-independent activation by ERα (Fig. 2A). By contrast, BRCA1 derivatives carrying familial breast cancer-derived missense mutations in either an exon 11-encoded region that binds Rad50 and the transcriptional repressor ZBRK1 (Q356R) or the C-terminal BRCT domain (A1708E) abolished the ability of BRCA1 to repress ligand-independent transactivation directed by ERα (Fig. 2A). Differences in the transcriptional repression activities of the various BRCA1 mutant derivatives could not be attributed to differences in their respective levels of expression because each of the BRCA1 mutant derivatives was expressed at a level comparable to wild-type BRCA1 (Fig. 2C). BRCA1-mediated, ligand-independent repression of ERα was largely reversed by trichostatin A, implicating histone deacetylase (HDAC) activity in this process (Fig. 2B). Collectively, these results reveal a function for BRCA1 as a repressor of ligand-independent, ERα-mediated transactivation.
Figure 2.
Ectopic expression of wild-type BRCA1 in Brca1-deficient MEFs restores ligand-independent repression of ERα transactivation in a histone deacetylase (HDAC)-dependent manner. (A and B) Brca1−/− MEFs in estrogen-free media were transfected with pERE-TK-Luc without (−) or with (+) pRSV-ERα, pCDNA3.1-BRCA1 expressing wild-type human BRCA1 (WT), or pCDNA3.1-BRCA1 derivatives bearing missense mutants A1708E, Q356R, A1708E/Q356R, or C64G before assay for luciferase activity. Where indicated, trichostatin A (TSA; 330 nM) was also included. (C) Brca1−/− MEFs in estrogen-free media were untransfected (lane 1) or cotransfected with expression vectors for ERα and either wild-type BRCA1 (lane 2) or various BRCA1 mutant derivatives (lanes 3–6) as indicated. Cells were lysed, and immunoprecipitated BRCA1 and ERα were subjected to immunoblot analysis using antibodies specific for BRCA1 (Top) or ERα (Middle). Immunoblot analysis of the nuclear matrix protein p84 (Bottom) indicates that nearly equivalent amounts of each cell lysate were used in the immunoprecipitations.
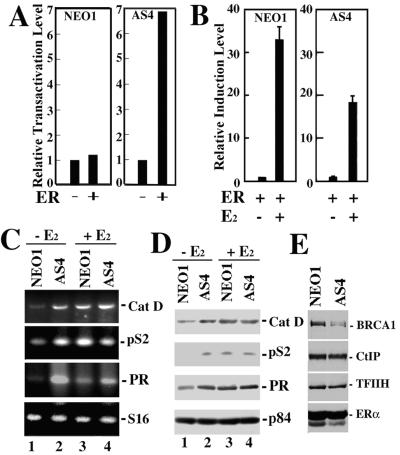
To confirm these results in a biologically relevant cell type, we analyzed the ligand-independent activity of ERα in human ovarian adenocarcinoma BG-1 cells, which are ERα-positive and estrogen-dependent for growth (30). Previously, Annab et al. (22) described the generation of independent BG-1 clonal cell lines that support stably reduced BRCA1 mRNA and protein levels by retroviral-mediated BRCA1 antisense delivery. We tested the ability of ERα to direct ligand-independent transcription of the ERE-TK-Luc reporter gene after transfection into either a control retroviral vector-infected BG-1 clonal cell line (NEO1) or, alternatively, a BRCA1 antisense-infected BG-1 clonal cell line (AS4) exhibiting severely reduced BRCA1 expression levels (Fig. 3E; ref. 22). Consistent with the results obtained in MEF cells, ERα exhibited significantly increased ligand-independent activity in BRCA1-deficient AS4 cells compared with BRCA1-proficient NEO1 cells (Fig. 3A). We also observed a 2-fold reduction in the relative level of E2-mediated induction of reporter gene activity in AS4 cells compared with NEO1 cells, once again consistent with the results obtained in MEF cells (Fig. 3B). These results confirm that in a biologically relevant epithelial cell type, BRCA1 can mediate repression of ligand-independent ERα transactivation activity.
Figure 3.
Reduced BRCA1 expression in BG-1 human ovarian adenocarcinoma cells is accompanied by increases in estrogen-independent expression of estrogen-responsive genes. (A) Retroviral vector-infected (NEO1) and BRCA1 antisense-infected (AS4) BG-1 cell clones in estrogen-free media were transfected with pERE-TK-Luc without (−) or with (+) pRSV-ERα before assay for luciferase activity. (B) NEO1 and AS4 cells in estrogen-free media were transfected with pERE-TK-Luc with (+) pRSV-ERα in the absence (−) or presence (+) of E2 (10−7 M) before assay for luciferase activity. (C) NEO1 (lanes 1 and 3) or AS4 (lanes 2 and 4) cells in estrogen-free media were either untreated (lanes 1 and 2) or treated (lanes 3 and 4) with E2 (10−7 M) for 1 h. Cells were harvested and processed for semiquantitative RT-PCR analysis using primers specific for the estrogen-responsive cathepsin D (Cat D), pS2, and progesterone receptor genes, as well as the estrogen-nonresponsive ribosomal S16 gene. (D) NEO1 (lanes 1 and 3) or AS4 (lanes 2 and 4) cells (5 × 106) in estrogen-free media were either untreated (lanes 1 and 2) or treated (lanes 3 and 4) with E2 (10−7 M) for 24 h. Culture medium was concentrated 10-fold by using a Centriprep YM-3 device, and 1/10th of the concentrate was resolved by SDS/15%PAGE and processed for immunoblot analysis using antibodies specific for pS2. Cells were also lysed in RIPA buffer, and 1/10th of the lysate was subjected to immunoblot analysis using antibodies specific for progesterone receptor β (PR), cathepsin D (Cat D), or nuclear matrix protein p84, which served as an internal loading control. (E) Whole cell lysates derived from NEO1 and AS4 cells were resolved by SDS/10%PAGE and processed for immunoblot analysis using antibodies specific for BRCA1, CtIP, and the p89 subunit of the transcription factor IIH (TFIIH), the latter two of which served as internal loading controls. The ERα-positive status of these cells was verified by using an ERα-specific rabbit polyclonal antibody. Densitometric quantitation of the immunoblot and normalization to the CtIP and TFIIH signals revealed BRCA1 expression to be reduced by 70% in AS4 cells compared with NEO1 cells.
To determine whether the reduced BRCA1 expression levels in AS4 cells could be correlated with an increase in the ligand-independent expression of endogenous estrogen-responsive genes, we performed a direct comparative analysis of NEO1 and AS4 cells with respect to their ligand-independent expression of several estrogen-responsive genes. Individual monolayer cultures of NEO1 and AS4 cells were grown in the absence of estrogen for 5 days followed by the addition of either no hormone or, alternatively, E2 (10−7 M) for 1 h. Subsequently, cells were harvested and analyzed by semiquantitative RT-PCR for the expression levels of the endogenous estrogen-responsive pS2, cathepsin D, and progesterone receptor genes.
Relative to the expression level of an internal control ribosomal S16 gene, we observed increases in the ligand-independent expression levels of the pS2, cathepsin D, and progesterone receptor genes of 3-, 5-, and 9-fold, respectively, in BRCA1-deficient AS4 cells compared with BRCA1-proficient NEO1 cells (Fig. 3C). Interestingly, although the addition of E2 stimulated transcription of the pS2, cathepsin D, and the progesterone receptor genes in NEO1 cells, no such E2-dependent increase in the transcription of these genes could be observed in AS4 cells (Fig. 3C). Qualitatively similar results were observed at the protein level by immunoblot analysis. Relative to the level of an internal control protein (nuclear matrix protein p84), E2-independent increases in the steady-state levels of the pS2, cathepsin D, and progesterone receptor proteins could be observed in AS4 cells compared with NEO1 cells (Fig. 3D). Furthermore, although the addition of E2 elevated the steady-state level of each of these proteins in NEO1 cells, no such E2-dependent increase could be observed in AS4 cells (Fig. 3D). Quantitative differences between RT-PCR and immunoblot analyses could reflect the influence of posttranscriptional regulatory processes. Nonetheless, RT-PCR and immunoblot analyses both reveal that the ligand-independent expression of endogenous ERα-target genes is increased in BRCA1-deficient cells. Collectively, these results implicate BRCA1 in the ligand-independent repression of endogenous estrogen-responsive genes.
To explore the mechanism by which BRCA1 mediates ligand-independent repression of ERα, we first determined whether BRCA1 could interact with unliganded ERα in vivo by coimmunoprecipitation of the two proteins in human breast cancer MCF7 cells cultured in the absence of estrogen. Consistent with previous results (18), BRCA1 could be specifically coimmunoprecipitated with unliganded ERα, thus demonstrating that the two proteins can interact in vivo in a ligand-independent manner (data not shown).
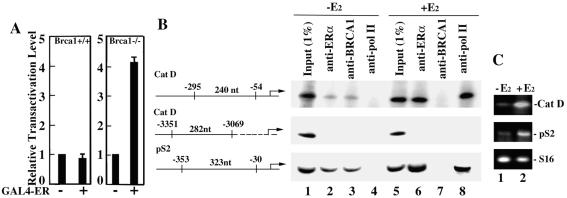
To explore the possibility that BRCA1 represses the transactivation function of promoter-bound, unliganded ERα, we first tested the effect of BRCA1 on the ligand-independent transcriptional activity of ERα tethered to the yeast GAL4 DNA-binding domain by using a reporter template bearing GAL4 DNA-binding sites. This approach permitted us to assess the effect of BRCA1 on the transactivation function of unliganded ERα independent of any effects that BRCA1 might have on the DNA-binding activity of unliganded ERα. GAL4-ERα was cotransfected along with a GAL4-SV40-luciferase reporter template into Brca1-proficient and Brca1-deficient MEFs. We observed significant ligand-independent stimulation of reporter activity in Brca1-deficient, but not in Brca1-proficient, MEFs (Fig. 4A), suggesting one mechanism by which BRCA1 mediates ligand-independent repression of ERα is through direct repression of the DNA-bound receptor.
Figure 4.
BRCA1 represses unliganded promoter-bound ERα-mediated transactivation. (A) Brca1+/+ and Brca1−/− MEFs were transfected with a pGAL4-SV40-Luc reporter plasmid either without (−) or with (+) a pGAL4-ERα expression plasmid before assay for luciferase activity. (B) Schematic diagram of the cathepsin D (Cat D) and pS2 gene regions targeted for ChIP analysis. Negative numbers refer to sequence coordinates that delimit PCR amplicons defined by gene-specific primer pairs relative to the transcription initiation site (right-angled arrow). Numbered nucleotides (nt) refer to the expected sizes of PCR-amplified products. MCF-7 cells, cultured the absence of estrogen, were treated without (−E2) or with (+E2) E2 (10−7 M) for 1 h. Soluble chromatin was prepared and subjected to immunoprecipitation by using monoclonal antibodies specific for ERα (anti-ERα), BRCA1 (anti-BRCA1), or the RNA polymerase II large subunit (anti-pol II). Immunoprecipitated DNA was PCR-amplified by using primers that span the indicated regions of the cathepsin D and pS2 gene promoters. Input (1%) of the soluble chromatin subjected to immunoprecipitation was PCR-amplified directly by using each primer pair as indicated. (C) MCF-7 cells, cultured in the absence of estrogen, were treated without (−E2) or with (+E2) E2 (10−7 M) for 1 h before harvest and processing for semiquantitative RT-PCR analysis using primers specific for the estrogen-responsive cathepsin D (Cat D) and pS2 genes, as well as the estrogen-nonresponsive ribosomal S16 gene.
To confirm this observation under biologically relevant conditions in vivo, we used ChIP analyses to determine whether BRCA1 can be recruited directly to estrogen-responsive promoters in the absence of ligand. MCF-7 cells were grown in the absence of estrogen for 5 days followed by the addition of either no hormone or, alternatively, E2 (10−7 M) for 1 h. Promoter occupancy before and after E2 treatment at the estrogen response elements within the endogenous pS2 and cathepsin D gene promoters by ERα, BRCA1, and RNA polymerase II was then monitored by ChIP using antibodies specific for each of the three proteins and semiquantitative PCR with primers flanking the estrogen response elements of the pS2 and cathepsin D promoters. In the absence of E2, ERα could be detected in association with both the pS2 and cathepsin D promoters, and this level was increased dramatically by the addition of E2 (Fig. 4B, lanes 2 and 6). Strikingly, we also observed pS2 and cathepsin D promoter occupancy by BRCA1 in the absence of E2, and a reduction in such occupancy after E2 treatment (Fig. 4B, lanes 3 and 7). By contrast, RNA polymerase II could be detected only following, but not before, E2 treatment, consistent with its ligand-dependent recruitment concomitant with transcriptional activation of the pS2 and cathepsin D genes (Fig. 4B, lanes 4 and 8 and C, lanes 1 and 2). The specificity of factor association within the estrogen-responsive region of the pS2 and cathepsin D promoters was confirmed by ChIP analysis using antibodies specific for ZBRK1, a sequence-specific DNA-binding transcriptional repressor that does not bind to pS2 or cathepsin D promoter sequences (14). ZBRK1-specific antibodies failed to immunoprecipitate pS2 and cathepsin D promoter sequences (data not shown). Further specificity of the ChIP assay was demonstrated by the inability to detect occupancy by ERα, BRCA1, or RNA polymerase II of a region ≈3 kb upstream of the cathepsin D promoter (Fig. 4B). These results thus reveal the association of BRCA1 with unliganded ERα at endogenous estrogen-responsive promoters under physiologically relevant conditions in vivo.
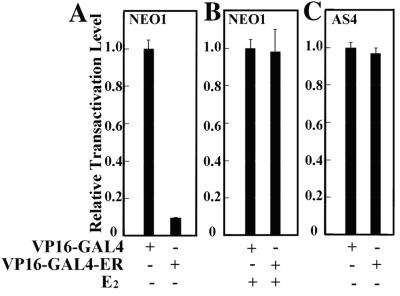
Like other steroid receptors, ERα contains two transactivation domains, an N-terminal ligand-independent activation function (AF-1) that is targeted by a variety of steroid-independent cell-signaling pathways, and a C-terminal ligand-inducible activation function (AF-2) that resides within the receptor ligand-binding domain (31, 32). Previous analyses of ERα suggest a model whereby repressive factors binding to sequences within its C-terminal ligand-binding domain repress constitutively active AF-1 in the absence of an agonist or in the presence of an antagonist (26, 33). To determine whether ligand-independent repression of ERα by BRCA1 is mediated through the ERα ligand-binding domain, we tested the ligand-independent activity of a VP16-GAL4-ERα receptor chimera after its expression in both BRCA1-proficient and BRCA1-deficient BG-1 clonal cell lines. This chimera encodes ERα amino acids 251–595, including the hinge region and the ligand-binding domain, fused C-terminally to the hybrid transactivator VP16-GAL4 (26).
Previously, deletion analysis of this receptor chimera revealed that constitutive VP16-GAL4-ERα activity could be recovered by the removal of sequences within the ligand-binding domain of the ERα moiety, thereby implicating the ERα ligand-binding domain in ligand-independent transcriptional repression of a neighboring constitutive activation domain (26). To determine whether this ligand-independent repression is mediated by BRCA1, we transfected the VP16-GAL4-ERα chimera along with a reporter template bearing GAL4 DNA binding sites into both BRCA1-proficient NEO1 cells and BRCA1-deficient AS4 cells. In NEO1 cells, the VP16-GAL4-ERα chimera exhibited minimal constitutive transactivation activity in the absence of E2; in response to E2, this level was dramatically increased to one approaching that of the potent VP16-GAL4 activator alone (Fig. 5 A and B). By contrast, in AS4 cells the VP16-GAL4-ERα chimera exhibited constitutive transactivation activity comparable to that exhibited by the VP16-GAL4 activator alone (Fig. 5C). The addition of E2 had a minimal effect on the elevated constitutive transactivation activity of the ERα chimera in AS4 cells (data not shown), suggesting that the principle effect of E2 is to override a ligand-independent barrier to the transactivation activity of the chimeric receptor. This barrier is present in NEO1 cells, but deficient in AS4 cells. Similar results were also observed by using isogenic Brca1-proficient and Brca1-deficient MEFs, eliminating the possibility that cell type-specific peculiarities contribute to the differential transactivation properties of the VP16-GAL4-ERα chimera in the presence and absence of BRCA1 (data not shown). Collectively, these results reveal the ERα ligand-binding domain to be a platform for the recruitment of BRCA1 from which the latter may confer ligand-independent repression on a linked activation domain. Hence, we conclude that BRCA1-mediated ligand-independent repression of ERα is likely to be mediated through the ERα ligand-binding domain.
Figure 5.
VP16-GAL4-ERα exhibits hormone-dependent activity in BRCA1-proficient cells and constitutive activity in BRCA1-deficient cells. NEO1 (A and B) and AS4 (C) cells in estrogen-free media were transfected with a GAL4-E1B-Luc reporter plasmid along with (+) plasmids expressing either VP16-GAL4 or VP16-GAL4-ERα. Subsequently, transfected cells were either untreated (−) or treated (+) with E2 (10−7 M) before assay for luciferase activity.
Discussion
Recently, BRCA1 has been proposed to inhibit the ligand-dependent transcriptional activity of ERα through a direct interaction between the two proteins (18). Our current analysis of ERα transcriptional activity in Brca1-nullizygous MEFs revealed BRCA1 to be a ligand-reversible barrier to transcriptional activation by unliganded ERα. The biological relevance of this finding is further strengthened by the observation that BRCA1 also mediates ligand-independent repression of the ERα in human ovarian adenocarcinoma cells.
The underlying mechanism by which BRCA1 mediates ligand-independent repression of ERα transcriptional activity appears to involve targeted recruitment by unliganded, promoter-bound ERα of a BRCA1-associated HDAC activity. This conclusion is based first on the observation that the HDAC inhibitor trichostatin A can effectively reverse ligand-independent repression mediated by BRCA1 and, second, on the results of ChIP analyses, which revealed the association of unliganded ERα with BRCA1 on endogenous estrogen-response elements in vivo. A likely target of BRCA1-mediated ligand-independent ERα repression is the constitutive AF-1 activation domain within ERα. Previous studies have indicated that antagonist-bound AF-2 can repress AF-1 activity through the recruitment of the nuclear corepressor N-CoR (33), whereas the ligand-binding domain of unliganded ERα can repress a linked heterologous activation domain in a ligand-reversible manner, presumably by the recruitment of a soluble corepressor (26). Our observation that an estrogen-dependent VP16-GAL4 chimeric transactivator carrying the ERα ligand-binding domain exhibits constitutive activity in BRCA1-deficient, but not in BRCA1-proficient BG-1 cells, reveals the ERα ligand-binding domain to be a potential site of BRCA1 recruitment for ligand-independent repression of a linked activation domain. Hence, BRCA1 could be recruited to the ERα ligand-binding domain as part of a larger repression complex to silence AF-1 function in the absence of ligand. The recent report of a direct interaction between BRCA1 and the ERα ligand-binding domain (18) lends additional support to this model.
Should BRCA1 function to inhibit the ligand-dependent transcriptional activity of ERα (17, 18), it seems unlikely to do so through a mechanism that involves promoter-bound ERα. Our ChIP analysis revealed the association of BRCA1 with ERα at endogenous estrogen-response elements before, but not after, estrogen stimulation. Thus, we favor a model in which BRCA1, along with an associated corepressor(s) that minimally includes an HDAC activity, is recruited by unliganded, promoter-bound ERα to effectively silence the constitutive AF-1 activation domain and thereby repress estrogen-responsive target gene transcription. After estrogen stimulation, a ligand-induced conformational change within ERα could lead to enhanced affinity of the ERα for its cognate binding site and release of a BRCA1-containing repression complex, thereby liberating AF-1 and AF-2 to synergistically recruit coactivators and the RNA polymerase II holoenzyme to promote transcription (29). It is also possible that BRCA1 could function additionally as a barrier to the productive association of either unliganded and/or liganded ERα with promoter DNA, and this could underlie the previous observation that BRCA1 can inhibit ligand-dependent ERα transactivation (17, 18).
Interestingly, we observed that a deficiency of BRCA1 also leads to a reduction in the relative level of E2-mediated ERα activation. In both Brca1-nullizygous MEFs and BRCA1-deficient BG-1 (AS4) cells, the relative level of E2-mediated activation of a transfected ERα-responsive reporter gene was diminished when compared with Brca1-proficient cells. Furthermore, in AS4 cells, the endogenous estrogen-response genes that we monitored exhibited increased estrogen-independent expression and little or no estrogen-dependent stimulation when compared with BRCA1-proficient BG-1 (NEO1) cells. It is possible that the expression of these genes is largely derepressed in a BRCA1-deficient background and cannot therefore be increased substantially in response to estrogen.
Previously, Annab et al. (22) demonstrated that relative to parental or retroviral vector-infected BG-1 cell clones, BRCA1 antisense-infected BG-1 cell clones exhibit enhanced estrogen-independent growth in culture (22). Furthermore, BG-1 clone AS4, which exhibits severely reduced BRCA1 expression levels, exhibited increased tumorigenicity in ovariectomized nude mice compared with the retroviral vector-infected NEO1 cell clone (22). These observations suggest that forced reduction of BRCA1 in BG-1 ovarian adenocarcinoma cells may influence estrogen-independent growth both in vitro and in vivo. Our observation that AS4 cells support significant increases in the estrogen-independent expression levels of different ERα-target genes compared with BRCA1-proficient NEO1 cells may provide a mechanistic basis for the estrogen-independent growth advantages that AS4 cells exhibit.
The finding that BRCA1 can function as a ligand-reversible barrier to transcriptional activation by unliganded ERα suggests the potential involvement of BRCA1 in the proliferative control of normal estrogen-regulated tissues. Thus, mutational inactivation of BRCA1 could result in persistent expression of estrogen-responsive genes in the absence of threshold levels of estrogenic stimulation. In this way, inappropriate hormonal responses brought about by BRCA1 mutation might possibly promote the proliferation of transformation-initiated cells.
Previous analyses have revealed that a significant proportion of BRCA1-associated breast tumors are negative for ERα expression (34). However, the loss of ERα expression in BRCA1-associated tumors is likely to represent a relatively late event in breast tumor progression, one that may have occurred after any proliferative advantages conferred upon transformation-initiated cells by homozygous BRCA1 mutation have ensued. Possibly, the down-regulation of ERα expression in BRCA1-mutated tumors could derive in part from negative feedback control enlisted by BRCA1-mutated breast epithelial cells to restrict the promiscuous expression of estrogen-responsive genes. Future studies should illuminate the mechanistic basis for BRCA1-mediated transcriptional repression of ERα and clarify its functional role in the larger network of hormone signaling pathways that control the growth, differentiation, and homeostasis of breast and ovary.
Acknowledgments
We thank D. Jones and P. Garza for technical assistance, Drs. M. J. Tsai and B. W. O'Malley for the receptor expression and reporter plasmids, Dr. J. H. White for the GAL4-VP16-ERα expression plasmid, and Dr. P.-L. Chen, Dr. P. R. Yew, and W. Tan for advice and comments. This work was supported by National Institutes of Health Grants P01CA30195 and P01CA81020, the McDermott Endowment Fund, and a San Antonio Cancer Institute Pilot Project Grant.
Abbreviations
- ERα
estrogen receptor α
- MEF
mouse embryonic fibroblast
- E2
17β-estradiol
- RT-PCR
reverse transcription–PCR
- HDAC
histone deacetylase
- ChIP
chromatin immunoprecipitation
- AF-1
N-terminal ligand-independent activation function
- AF-2
C-terminal ligand-inducible activation function
References
- 1.Martin A M, Weber B L. J Natl Cancer Inst. 2000;92:1126–1135. doi: 10.1093/jnci/92.14.1126. [DOI] [PubMed] [Google Scholar]
- 2.Tirkkonen M, Johannsson O, Agnarsson B A, Olsson H, Ingvarsson S, Karhu R, Tanner M, Isola J, Barkardottir R B, Borg A, Kallioniemi O. Cancer Res. 1997;57:1222–1227. [PubMed] [Google Scholar]
- 3.Tomlinson G E, Chen T T, Stastny V A, Virmani A K, Spillman M A, Tonk V, Blum J L, Schneider N R, Wistuba I I, Shay J W, et al. Cancer Res. 1998;58:3237–3243. [PubMed] [Google Scholar]
- 4.Xu X, Weaver Z, Linke S P, Li C, Gotay J, Wang X W, Harris C C, Ried T, Deng C X. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 5.Zheng L, Li S, Boyer T G, Lee W-H. Oncogene. 2000;19:6159–6175. doi: 10.1038/sj.onc.1203968. [DOI] [PubMed] [Google Scholar]
- 6.Welsch P L, Owens K N, King M C. Trends Genet. 2000;16:69–74. doi: 10.1016/s0168-9525(99)01930-7. [DOI] [PubMed] [Google Scholar]
- 7.Gowen L C, Avrutskaya A V, Latour A M, Koller B H, Leadon S A. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 8.Moynahan M E, Chiu J W, Koller B H, Jasin M. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 9.Zhong Q, Chen C F, Li S, Chen Y, Wang C-C, Xiao J, Chen P-L, Sharp Z D, Lee W-H. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 10.Harkin D P, Bean J M, Miklos D, Song Y-H, Truong VB, Englert C, Christians F C, Ellisen L W, Maheswaran S, Oliner J D, Haber D A. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 11.Hollander M C, Sheikh M S, Bulavin D V, Lundgren K, Augeri-Henmueller L, Shehee R, Molinaro T A, Kim K E, Tolosa E, Ashwell J D, et al. Nat Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 12.Somasundaram K, Zhang H, Zeng Y X, Houvras H, Peng Y, Zhang H, Wu G S, Licht J D, Weber B L, El-Deiry W S. Nature (London) 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 13.Wang X W, Zhan Q, Coursen J D, Khan M A, Kontny U, Yu L, Hollander M C, O'Conner P M, Fornace A J, Jr, Harris C. Proc Natl Acad Sci USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng L, Pan H, Li S, Fleskin-Nikitin A, Chen P-L, Boyer T G, Lee W-H. Mol Cell. 2000;6:757–768. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]
- 15.Hilakivi-Clarke L. Cancer Res. 2000;60:4993–5001. [PubMed] [Google Scholar]
- 16.Liehr J G. Endocr Rev. 2000;21:40–54. doi: 10.1210/edrv.21.1.0386. [DOI] [PubMed] [Google Scholar]
- 17.Fan S, Wang J-A, Yuan R, Meng Q, Yuan R-Q, Ma Y X, Erdos M R, Pestell R G, Yuan F, Auborn K J, et al. Science. 1999;284:1354–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 18.Fan S, Yong X, Wang C, Yuan R-Q, Meng Q, Wang J-A, Erdos M, Goldberg I D, Webb P, Kushner P J, et al. Oncogene. 2001;20:77–87. doi: 10.1038/sj.onc.1204073. [DOI] [PubMed] [Google Scholar]
- 19.Yeh S, Hu Y-C, Rahman M, Lin H-K, Hsu C-L, Ting H-J, Kang H-Y, Chang C. Proc Natl Acad Sci USA. 2000;97:11256–11261. doi: 10.1073/pnas.190353897. . (First Published October 3, 2000; 10.1073/pnas.19053897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J J, Irvine R A, Buchanan G, Koh S S, Park J M, Tilley W D, Stallcup M R, Press M F, Coetzee G A. Cancer Res. 2000;60:5946–5949. [PubMed] [Google Scholar]
- 21.Rebbeck T R, Kantoff P W, Krithivas K, Neuhausen S, Blackwood M A, Godwin A K, Daly M B, Narod S A, Garber J E, Lynch H T, et al. Am J Hum Genet. 1999;64:1371–1377. doi: 10.1086/302366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Annab L A, Hawkins R E, Solomon G, Barrett J C, Afshari C A. Breast Cancer Res. 2000;2:139–148. doi: 10.1186/bcr45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leng X, Blanco J, Tsai S Y, Ozato K, O'Malley B W, Tsai M J. J Biol Chem. 1994;269:31436–31442. [PubMed] [Google Scholar]
- 24.Hsu H-L, Wadman I, Baer R. Proc Natl Acad Sci USA. 1994;91:3181–3185. doi: 10.1073/pnas.91.8.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadowski I, Bell B, Broad P, Hollis M. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 26.Lee H S, Aumais J, White J H. J Biol Chem. 1996;271:25727–25730. [PubMed] [Google Scholar]
- 27.Tong D, Schneeberger C, Leodolter S, Zeilinger R. Anal Biochem. 1997;251:173–177. doi: 10.1006/abio.1997.2280. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Brattain M G, Appert H. Biochem Biophys Res Commun. 1997;231:283–289. doi: 10.1006/bbrc.1997.6083. [DOI] [PubMed] [Google Scholar]
- 29.Shang Y, Hu X, DiRenzo J, Lazar M A, Brown M. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 30.Geisinger K R, Kute T E, Pettenati M J, Welander C E, Dennard Y, Collins L A, Berens M E. Cancer. 1989;63:280–288. doi: 10.1002/1097-0142(19890115)63:2<280::aid-cncr2820630213>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 31.Weigel N, Zhang Y. J Mol Med. 1998;76:469–479. doi: 10.1007/s001090050241. [DOI] [PubMed] [Google Scholar]
- 32.Klinge C M. Steroids. 2000;65:227–251. doi: 10.1016/s0039-128x(99)00107-5. [DOI] [PubMed] [Google Scholar]
- 33.Lavinsky R M, Jepsen K, Heinzel T, Torchia J, Mullen T M, Schiff R, Del-Rio A L, Ricote M, Ngo S, Gemsch J, et al. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loman N, Johannsson O, Bendahl P O, Borg A, Ferno M, Olsson H. Cancer. 1998;83:310–319. [PubMed] [Google Scholar]