Abstract
Drug-like molecules targeting allosteric sites in proteins are of great therapeutic interest; however, identification of potential sites is not trivial. A straightforward approach to identify hidden allosteric sites is demonstrated in protein tyrosine phosphatases (PTP) by creation of single alanine mutations in the catalytic acid loop of PTP1B and VHR. This approach relies on the reciprocal interactions between an allosteric site and its coupled orthosteric site. The resulting NMR chemical shift perturbations (CSPs) of each mutant reveal clusters of distal residues affected by acid loop mutation. In PTP1B and VHR, two new allosteric clusters were identified in each enzyme. Mutations in these allosteric clusters detrimentally altered phosphatase activity with reductions in kcat/KM ranging from 30% to nearly 100-fold. This work outlines a simple method for identification of new allosteric sites in PTP, and given the basis of this method in thermodynamics, it is expected to be generally useful in other systems.
Introduction
Protein allostery represents a major mechanism for the regulation of metabolic activity in vivo, whereby enzymatic activity or ligand binding affinity is finely tuned through structural or dynamical perturbations at locations that are spatially distinct from the site of activity. Allosteric perturbations can occur as a result of small molecule or macromolecule binding or post-translational modification. It is widely recognized that designed allosteric ligands can have great therapeutic benefit, and thus, extensive research has been focused in this area [1–3]. The potential benefits are far-reaching because while many proteins are not naturally regulated in vivo by an allosteric mechanism, it has been speculated that all proteins have potential or hidden allosteric sites and that these locations can be exploited for drug design purposes [4]. Experimental and computational methods have been proposed to identify these unknown sites in allosteric and non-allosteric proteins [5–7], and while some approaches have been successful [6,8], the need for additional methods is clear [6].
It has long been known that amino acid mutations alter the NMR chemical shifts of residues both nearby and distant from the site of mutation [9], which often confounds the interpretation of the mutational study. Rather than a nuisance, these distally perturbed chemical shifts can be exploited for the discovery of new allosteric sites in proteins due to the reciprocity that exists in an allosteric system [10,11]. Reciprocity is based on the thermodynamic premise that for two coupled allosteric sites, perturbation at one site should impact the other site. As such, the effects of an amino acid mutation should propagate to distant sites that are allosterically coupled to the site of mutation. Solution NMR spectroscopy is an exquisitely sensitive technique to detect such changes with atomic resolution. Experimental NMR-based methods such CHESCA and RASSMM have relied on this sensitivity to interrogate and elucidate protein networks [12–14]. However, these methods often require prior knowledge of known conformational states or are limited to the detection of altered motions in the millisecond timescale. Here, we describe an approach to identify allosteric networks simply by the examination of protein-wide 1H, 15N chemical shift perturbations (CSPs) for a series of active site mutations. Specifically, analysis of CSP for a series of acid loop alanine mutations in the active sites of protein tyrosine phosphatase (PTP) 1B (PTP1B) and Vaccinia H1-related phosphatase (VHR) identified clusters of distally located residues that are coupled to the active sites of these enzymes. Structural and biochemical studies further verify these newly discovered allosteric sites, confirming the validity of this method in addition to providing mechanistic detail of allostery in these enzymes.
PTP1B plays a major role in regulating glucose homeostasis and leptin signaling and has been an active target for the rational design of inhibitors for the treatment of type II diabetes and obesity [15,16]. VHR is involved in cell cycle regulation [17] and is an enzyme of interest for the treatment of cervical [18], breast [19], and prostate cancers [20]. Competitive inhibition of these enzymes has proven to be a poor strategy toward achieving these goals. There are 107 human PTPs with highly conserved catalytic sites, which present a major hurdle for ligand selectivity [21]. Moreover, ligand binding to the active site is driven by electrostatic complementarity, and anionic inhibitors that mimic substrate binding generally have reduced membrane permeability [21]. Thus, recent drug design efforts have focused on allosteric inhibition of PTPs, because this mechanism is not limited by the aforementioned constraints, and human PTPs have diverse surface features outside of the active site, despite a similar three-dimensional architecture [22]. While research is ongoing, the most promising allosteric inhibitor for PTP1B is a benzofuran-based [23] small molecule (BB3) that binds near α-helix 7, approximately 20 Å from the active site [24]. Additionally, another small molecule inhibitor MSI-1436 has been demonstrated to bind near α-helix 7, validating this secondary structure as an important allosteric site [25]. Similar efforts have not yet been successful for VHR. Given the limited success to date, it is clear that future drug discovery efforts focused on PTP1B and VHR would benefit from identification of novel allosteric sites and an improved understanding of potential allosteric networks present within these enzymes.
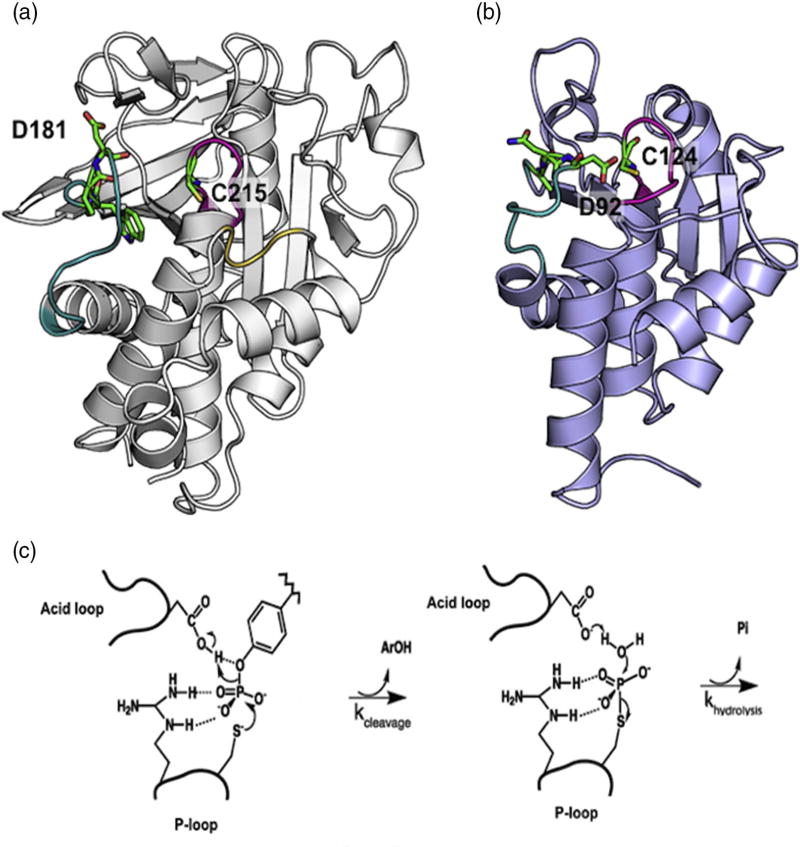
PTP1B and VHR have two conserved catalytic features, the P-loop and acid loop (Fig. 1). The P-loop is critical for binding of the phosphoryl moiety of the polypeptide substrate and contains the cysteine nucleophile (C215 and C124, respectively), which is essential for covalent catalysis [24,26]. The acid loop (also called the WPD loop based on the one-letter amino acid nomenclature for the three N-terminal residues of this loop in PTP1B) contains a conserved aspartic acid (D181 and D92, respectively) that is important for general acid/base catalysis (Fig. 1). Mutation of this aspartic acid in PTP1B and VHR results in a 100,000-fold and 100-fold reduction in catalytic activity, respectively [27,28]. Research has shown that although the P-loop is immobile on the timescale of the chemical reaction, the acid loop in PTP1B moves 9 Å from an open position to a closed conformation to orient D181 for protonation of the tyrosine leaving group [24,29–31]. There is no published evidence for acid loop motion in VHR, but solution NMR data in our lab indicate that this loop also experiences motions on the millisecond timescale, similar to PTP1B (SI Fig. 1). It was recently shown that the timescale of acid loop closure in PTP1B is closely linked to the timescale of phosphoryl cleavage, corresponding to the first step in the mechanism for class I phosphatases (Fig. 1) [32]. These NMR studies also revealed that the acid loop of PTP1B alternates between open and closed conformations in the absence of substrate. This natural motion of the PTP1B acid loop results in alternating interactions with the main body of PTP1B, specifically with residues in the P-loop and α-helix 4 (R221 and T224), the Q-loop and α-helix 6 (T263, D265 and F269), and the E-loop (M109-E115). We reasoned that perturbation of these interactions would be propagated to distal allosteric sites, as any mutation in the acid loop that affects its conformational exchange motion should lead to changes in the ensemble averaged chemical shifts for any distantly coupled amino acid residues. While the importance of conformational motion exchange in the catalytic mechanism of VHR is less established, we believe that mutation of the acid loop in this enzyme would also aid the discovery of unknown allosteric sites in this enzyme based on the millisecond timescale motions we have observed for its acid loop (SI Fig. 1).
Fig. 1.
PTP structure and mechanism. (a) Apo WT PTP1B (PDB ID: 2HNP) is represented in gray ribbon [50]. The acid loop is shown in teal, P-loop in magenta, and the Q-loop in yellow. W179, P180, D181, and C215 are shown in stick configuration. (b) Apo WT VHR (PDB ID: 1VHR) is represented in blue ribbon [51]. The acid loop is shown in teal and the P-loop is in magenta. A90, N91, D92, and C124 are shown in stick configuration. (c) The catalytic reaction occurs in two steps. First, a cysteine from the P-loop acts as a nucleophile to attack the phosphorus atom of the phosphorylated tyrosine of the substrate. The P–O bond is cleaved and the acid loop closes to facilitate the protonation of the leaving aryl group by aspartic acid. In the second step, the aspartate residue aids in the hydrolysis of the covalent intermediate with concomitant release of inorganic phosphate.
Results and Discussion
Acid loop mutations in PTP1B reveal a hidden allosteric network
To map the hidden allosterically coupled network to the active site of PTP1B, we made 13 single-site alanine mutations at each position of the acid loop (176-YTTWPDFGVPESP-188). By investigating the frequency of CSPs, we aimed to identify regions that are distally linked to the active site. Phosphatase kinetics were measured for each mutant using the p-nitrophenylphosphate (pNPP) cleavage assay, and the catalytic efficiencies of the mutants were found to range from a 105-fold reduction to a 2.4-fold increase, in agreement with many prior studies that highlighted the importance of specific acid loop interactions in regulating the catalytic efficiency of PTP1B (SI Table 1) [27,33,34]. Each mutant and the wild-type enzyme were subsequently analyzed by collecting two-dimensional transverse relaxation optimized spectroscopy 1H, 15N heteronuclear single quantum coherence (HSQC) spectra and comparing NMR chemical shifts for amide resonances (SI Fig. 2). As expected, analysis of the NMR spectra revealed that residues near the site of mutation had amide resonances that shifted significantly, relative to the wild-type spectrum, consistent with local perturbations due to the alanine mutations. These sites included amino acids in the acid loop, P-loop (C215-R221), and Q-loop (R254-T263; SI Fig. 3A). It was also observed that a majority of amino acids distant from the active site were unaffected by acid loop mutation (SI Fig. 3B). However, significant changes in chemical shifts were also identified for amino acids located up to 30 Å from the active site (Fig. 2 and SI Fig. 3C), suggesting the potential network of residues linked to the acid loop.
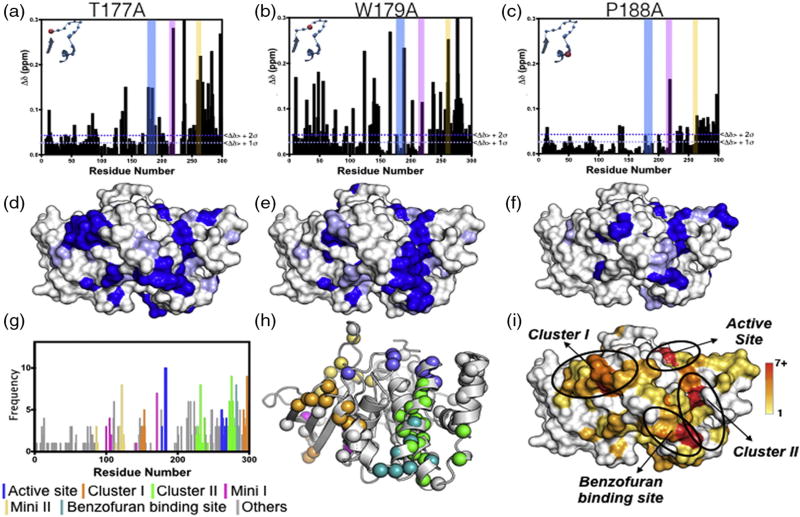
Fig. 2.
CSPs from alanine mutations in the WPD acid loop. (a) T177A, (b) W179A, and (c) P188A are shown. The vertical colored bars represent residues in the acid loop (blue), P-loop (pink), and the Q-loop (yellow), and the inset shows the location of the mutation in the acid loop. The corresponding surface renderings of significant CSPs are shown in (d–f). Blue represents residues with CSP values greater than <Δδ> + 2σ, and light blue represents <Δδ> + 1σ. (g) Cumulative frequency of 2σ CSPs for residues from all 13 loop mutations plotted as a histogram and color-coded by protein location. (h) PTP1B structural representation (PDB ID: 1PTT) of CSP colored by protein location as determined in (g). (i) Surface rendering of clusters highlighted in (g and h). Colors correspond to the frequency of a residue significantly perturbed by 1 of the 13 mutations as indicated by the color bar. Protein figures were generated with PyMOL [52]. Data for the remaining 10 acid loop mutations are provided in SI Fig. 4.
To quantify residues affected by mutation, we calculated CSPs relative to the WT enzyme for each amide resonance for all 13 acid loop mutants (see Materials and Methods). Residues with CSP greater than 2 SD (2σ) from the 10% trimmed mean were plotted onto the structure of PTP1B. Representative CSP plots and their location on the PTP1B structure are shown in Fig. 2a–f for acid loop mutants, T177A, W179A, and P188A. The remaining 10 CSP plots are shown in SI Fig. 4. To better visualize regions cumulatively affected by all 13 of the acid loop mutations, we combined residues with CSP greater than 2σ (□Δδ ≥ 0.047 ppm) from the 10% trimmed mean value for all mutants in a frequency plot (Fig. 2g and SI Table 2). This plot illustrates the number of times the chemical shift of a particular resonance is significantly perturbed by 1 of the 13 mutations. Residues with CSP above the 2σ cutoff are mapped onto the PTP1B structure (Fig. 2h). As expected, the active site was most affected by acid loop mutations; however, five distal clusters of residues were also identified. These included three prominent regions that we designated Cluster I, Cluster II, and the BB3 binding site [23], and two minor regions designated Mini I and Mini II. As mentioned above, BB3 is a well-characterized allosteric inhibitor of PTP1B, and reassuringly, its binding location between α-helices 6 and 7 was predicted by the cumulative CSP analysis [23]. Residues Y152 and Y153 and the C-terminal α-helix 7 were previously identified to participate in an allosteric network, which has been suggested to affect the product release step [35]. Here, we have identified 11 residues residing along α-helix 7 and β-sheets 9 and 10 including Y152 and Y153 in Cluster I that have varying sensitivities to perturbations in the acid loop (SI Table 2). Interestingly, Cluster II is a previously unidentified allosteric site, which consists of 19 residues residing along α-helices 4 and 6 (SI Table 3). In addition, Clusters I and II have significant solvent-accessible surface areas (SASAs) of 613 Å2 and 310 Å2, respectively, and Cluster II forms a depression on the protein surface (Fig. 2i), supporting the notion that these clusters are viable targets for the rational design of small allosteric inhibitors. The two minor regions, Mini I and II, have 5 and 8 residues in each cluster, which both consist of residues found in the E-loop and are buried within the protein body, resulting in nominal SASAs of 29 Å2 and 122 Å2, respectively. While regions with such small SASA are unlikely binding sites for allosteric ligands [36], they are likely informative for understanding the allosteric pathways in PTP1B, but they are not considered further in this work. Ultimately, Clusters I and II represent allosteric binding sites that could be exploited for the inhibition of PTP1B, which has been recalcitrant to such efforts to date.
By altering the finely tuned balance between flexibility and rigidity in the acid loop, the alanine scanning method provides atomistic detail of the network that governs the conformational transitions between the open and closed state of PTP1B. Importantly, it provides information on the sensitivity of the distal residues to acid loop perturbations, which enables further targeting of hotspots for allosteric drug and mechanistic studies.
Reciprocal distal allosteric alanine mutations affect PTP1B catalytic activity
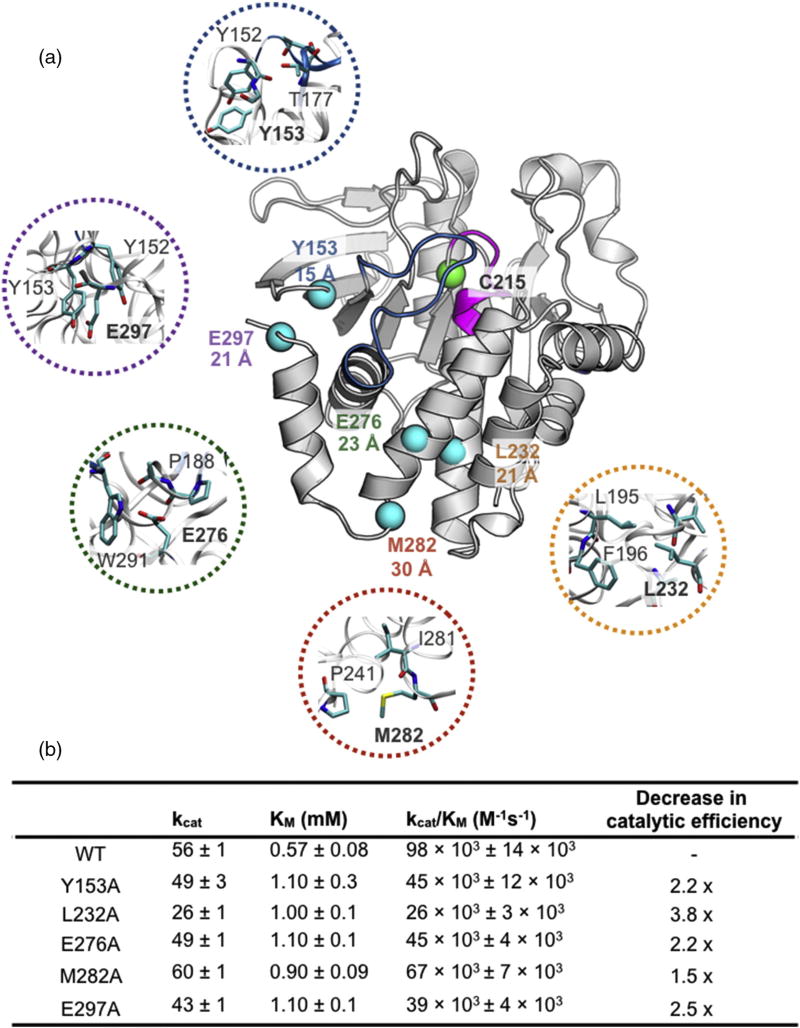
To test for reciprocal interactions of these newly identified allosteric sites and the connectivity to the PTP1B active site and thus the validity of this approach, we individually mutated residues within Cluster I (Y153, E297), Cluster II (L232, E276), and the BB3 site (M282) to alanine and, their effect on catalytic activity and active site chemical shifts was examined. These allosteric residues range from 15 to 30 Å from the catalytic nucleophile, C215 (Fig. 3a). Clusters I and II both contain a residue localized near the N-terminal and C-terminal hinges of the acid loop. The amide of Cluster I residue Y153 forms a hydrogen bond (3.6 Å) with the hydroxyl side chain of T177 at the N-terminal hinge of the catalytic acid loop. The carboxyl oxygen atom of Cluster II residue E276 forms a weak electrostatic interaction (4.9 Å) with the amide nitrogen atom of A189 at the C-terminal acid loop hinge (Fig. 3a). The structural context of other allosteric residues chosen for mutation is shown in the expanded regions of Fig. 3a.
Fig. 3.
Allosteric sites selected for mutagenesis in PTP1B. (a) Locations of allosteric residues selected for mutations to alanine are shown as spheres. The catalytic nucleophile C215 is shown as a green sphere. The distance between the Cα atom of C215 and the Cα of the indicated residue is displayed underneath the residue label. Closeup view of the allosteric residues selected for mutation is color-coded with respect to each mutant. (PDB ID: 1PTT) [26]. Table of Michaelis–Menten kinetic constants of allosteric alanine mutants is shown below. Thermal stability of each mutation is shown in SI Fig. 5.
The effects of these distal mutations on catalytic activity were measured using the pNPP assay (see Materials and Methods), and the results are summarized in Fig. 3b. Both Y153A (Cluster I) and E276A (Cluster II) mutations exhibit a 2.2-fold decrease in catalytic efficiency, indicating that the catalytic activity is sensitive to the overall packing surrounding the acid loop hinge and direct interaction with the hinge. However, direct hinge contacts are not the only interactions important for catalytic activity as seen in mutations further away from the acid loop. E297A (Cluster I) and L232A (Cluster II) are over 20 Å away from C215 and exhibit a decrease in kcat/KM of 2.5 and 3.8-fold, respectively. It is notable that the kinetic perturbations seen in the E297A mutation are similar in scale to the altered kinetics measured for an α-helix 7 truncated construct [35]. This suggests that the allosteric network propagates through specific residue-based interactions rather than consolidated domain interactions. Lastly, M282 (BB3) is solvent exposed and located on the end of α-helix 6, 30 Å from C215. The M282A mutant resulted in a decrease in phosphatase catalytic efficiency by a factor of 1.5. To ensure that the observed kinetic perturbations were not due to an artifact of decreased enzyme stability, we measured the stability of each mutant by thermal melting detected by CD spectroscopy. The observed midpoint of the unfolding (Tm) of each distal mutant was equivalent to or higher than WT PTP1B (SI Fig. 5). These kinetic data support the notion that there is a protein network that extends from the active site to distant residues on the protein surface, which can influence the integrity of the catalytic activity of PTP1B, suggesting that targeting these sites with small molecules could alter PTP1B function.
Distal network perturbations shift the acid loop into an inactive state
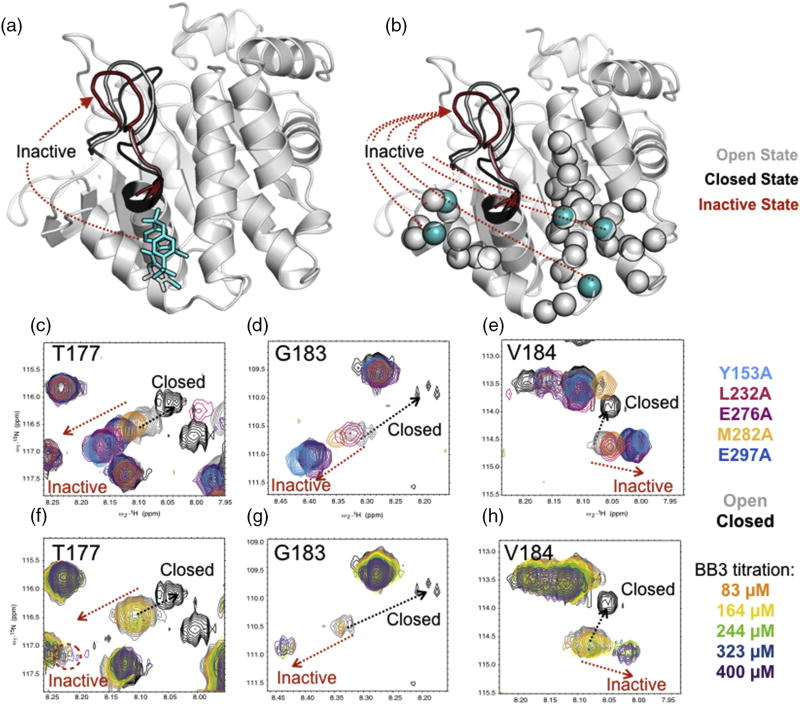
In addition to altered catalytic activity, the reciprocal mutations also perturb the NMR resonances of the acid loop. A closer analysis of the perturbations observed in the acid loop (residues T177, G183, and V184) indicates that the distal mutations shift the acid loop into an inactive conformational state (Fig. 4). For residues involved in a two-site (open/closed) conformational exchange motions, their NMR chemical shift represents the population-weighted average of the two conformations. Thus, if the chemical shifts of the two conformations are known, the observed chemical shift lies in a linear path between the endpoint chemical shifts and is an indicator of the equilibrium poise between the two conformations. An overlay of L232A, E297A M282A, Y153A, and E276A mutants along with WT apo- (open loop) and tungstate-bound (closed loop) HSQC spectra showed a linear trend of chemical shifts moving away from the trajectory of the closed tungstate-bound state (Fig. 4c–e). Interestingly, the titration of the allosteric inhibitor BB3 into WT PTP1B indicated that the acid loop was in slow exchange and that the ligand-saturated peaks appeared along the same path as the distal mutant spectra (Fig. 4f–h). This suggests that the mechanism of inhibition of BB3 is through the perturbation of the allosteric network consisting of Cluster I and II, consequently redirecting the loop into an inactive state and altering the loop conformational dynamics. Assuming that the acid loops of the distal mutants are sampling open and closed conformations [32], the open/close equilibrium is altered by the distal perturbations. The magnitude of the perturbation in the loop residues T177, G183, and V184 between each distal mutation varies, suggesting that the two clusters may have different functional mechanisms in the modulation of the acid loop.
Fig. 4.
Allosteric mutants alter acid loop conformation. BB3 titration and distal mutants of PTP1b both shift the acid loop into an inactive state. Cartoon rendering of PTP1B (PDB ID: 1PTT, 2HNP) is shown where the acid loop is affected by (a) BB3 ligand shown in stick and (b) distal mutations shown in cyan spheres. Gray spheres represent the network detected through alanine loop mutant analysis. (c–e) Acid loop resonances of distal mutations overlaid with WT apo (open) and closed conformations. (f–h) Acid loop resonances of a BB3 titration overlaid with resonances in the closed conformation from a tungstate titration. The protein backbone was rendered in Pymol [52].
PTP1B acid loop communicates allosteric signals through interior hydrophobic residues
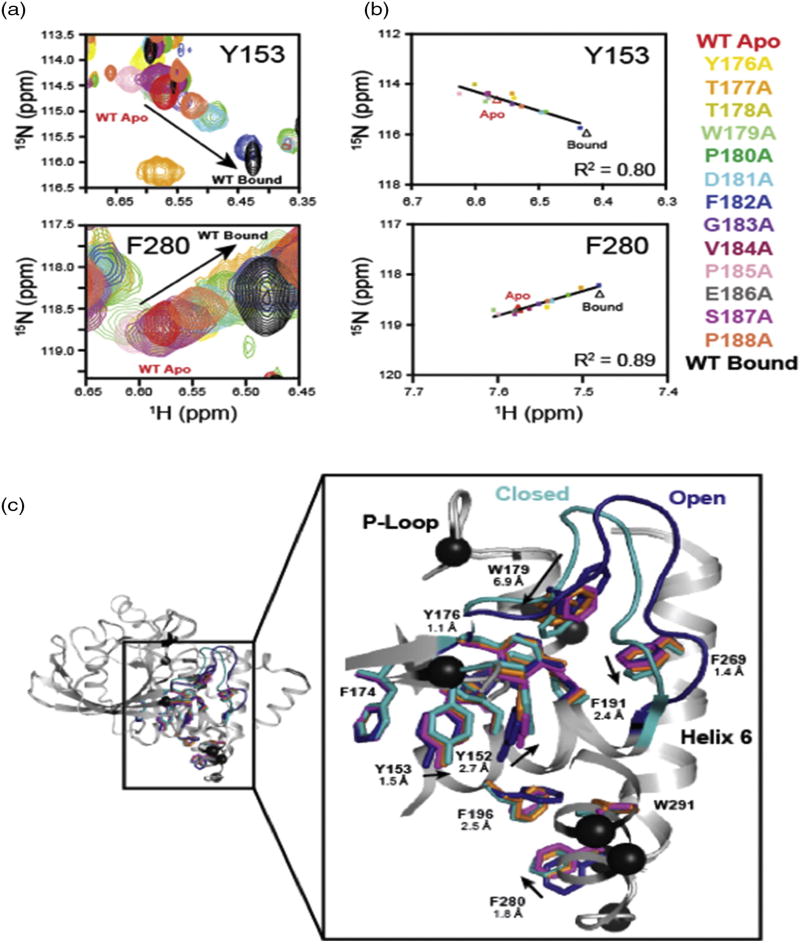
Many of the amino acids identified in our CSP cluster analysis reside in or near an internal aromatic hydrophobic network that extends 20 Å from the active site to the C terminus of α-helix 6. We examined these clusters in detail through analysis of the pattern of mutation-induced chemical shifts. Intriguingly, the chemical shifts exhibited by apo WT, ligand-bound WT, and the 13 acid loop mutants showed 11 residues residing in the active or distal cluster sites with CSP that follow a linear trend (R2 = 0.66–0.94), suggesting that acid loop mutations alter the equilibrium conformation of the acid loop and that its conformation is sensed by other residues in PTP1B. Overlays of NMR spectra for two of the residues in the allosteric network, Y153 (Cluster I) and F280 (BB3 Cluster), are shown in Fig. 5a and b and SI Fig. 6. There is an emerging pattern where the chemical shifts for distal residues in the T177A, P180A, D181A, F182A, and P188A mutants are on average, distributed closer to the bound WT shifts (Fig. 5b and SI Fig. 6). However, the linear trend of these shifts differs for different residues, indicating that the hydrophobic cluster does not respond uniformly to acid loop mutations, suggesting non-concerted motions and a complex energy landscape.
Fig. 5.
Analysis of the hydrophobic core network in PTP1B. (a) Overlays of NMR spectra for acid loop mutants highlighting the range of chemical shifts for Y153 and F280 in the aromatic network. (b) Linear correlation of acid loop mutational effects on Y153 and F280 in the aromatic network. The color of the data points indicates the chemical shift for the acid loop mutant listed to the right of panel (b). (c) Superimposed structures for WT andW179F PTP1B in the apo- and substrate-bound forms. The acid loop and aromatic side chains for WT apo PTP1B (PDB ID: 2HNP) are shown in blue [24]. The acid loop and aromatic side chains for WT vanadate-bound PTP1B (PDB ID: 3I80) are shown in cyan [53]. Aromatic side chains for W179F are shown in magenta (apo, PDB ID: 3QKP) and orange (vanadate bound, PDB ID: 3QKQ) [33]. Residues exhibiting linear chemical shifts for acid loop mutants with R2 = 0.65–0.98 are shown as black spheres. WT apo and bound PTP1B are shown in light gray ribbons.
A comparison of the apo- (open acid loop) to ligand-bound (closed acid loop) crystal structures for WT PTP1B and W179F mutant construct demonstrates that the hydrophobic network responds to substrate binding and subsequent acid loop closure, as aromatic side chains shift by as much as 2.5 Å upon ligand binding to the active site. The tryptophan in the acid loop undergoes a rotation in which the C5 atom of the indole ring moves 6.9 Å into the hydrophobic pocket upon loop closure. This appears to drive the rearrangement of side-chain packing in the hydrophobic network. The importance of this packing interaction is consistent with experiments where W179 was mutated to a phenyalanine in which the open:close equilibrium of the acid loop is significantly altered [33]. The loss of packing interactions caused by this mutation is propagated to the residues throughout the aromatic network, as side chains for apo- and vanadate-bound W179F exhibit smaller magnitude conformational changes than does WT PTP1B under identical conditions (Fig. 5c). These observations suggest that the volume occupied by W179 is important for stabilizing the closed form of the acid loop and results in conformational changes that are conveyed throughout the aromatic network, which links the allosteric clusters identified in this work. Peti and coworkers also demonstrated the importance of hydrophobic interactions in the activity and allostery in PTP1B [35].
The hydrophobic core of enzymes has been previously demonstrated to be of importance in the allosteric signal propagation. In 1991, Vliet et al. demonstrated that a single tyrosine-to-phenylalanine mutation in the hydrophobic interface of aspartate transcarbamoylase reverses the ATP binding effect [37]. In this Y-to-F mutant, ATP acts as an allosteric inhibitor rather than activator. Interestingly, the sensitivity of this mutant to CTP, as a feedback inhibitor, is unchanged. These studies showed the importance of the hydrophobic core in distinct allosteric responses. A more recent study of protein kinase A also demonstrated the importance of the hydrophobic protein core in allostery [38]. The hydrophobic core of PTP1B exhibits species conservation among PTP1B enzymes; however, many of the hydrophobic residues identified in our work as part of the allosteric network are not conserved among human tyrosine-specific phosphatases [22].
The approach we described here exploits the dynamic plasticity that exists within an enzyme and the reciprocity of perturbations in allosterically linked sites. At a fundamental level, altered residue interactions and packing upon mutation lead to conformational changes that are propagated from the active site to the distal residues through an intramolecular network. These changes can be monitored as perturbations in the population-averaged chemical shift to reveal previously unidentified sites of allosteric regulation. Using this method, we have identified a hydrophobic network in PTP1B that extends through the core of the protein to three primary surface clusters more than 20 Å away. We have demonstrated that these regions communicate with the active site and appear to modulate the conformational equilibrium of the acid loop between open and closed states with subsequent effects on catalytic activity. Together, these findings provide support for the idea that novel effector binding sites can be identified in the manner described herein to aid drug design efforts. With this goal in mind, Cluster II is of particular interest for future pharmacological inhibitor design for PTP1B due to a motif in this cluster near the end of α-helix 4 (P241, S242, and S243) that is not found in the homologous T-cell PTP, a PTP that has been implicated in regulating immune response pathways and has been shown to be an unintended target of PTP1B inhibitors [39].
Allosteric sites are also identified in VHR
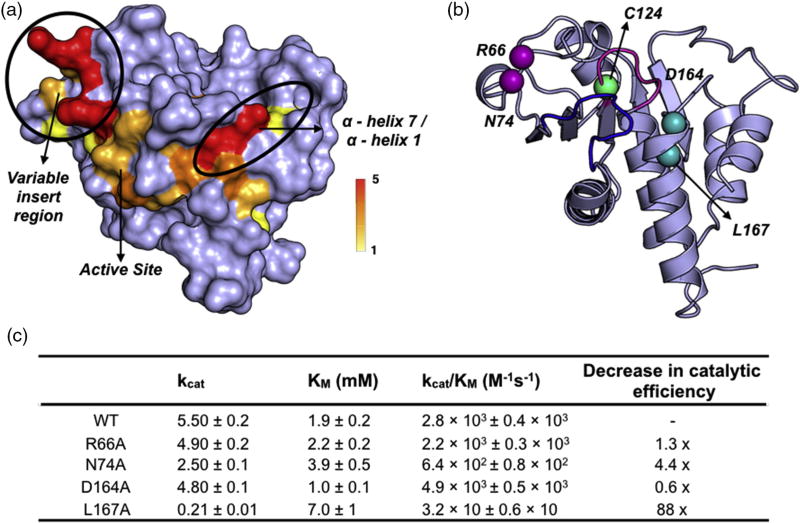
To test the generality of our approach, we performed similar experiments on the dual specificity phosphatase, VHR. Currently, there are no known allosteric networks in this enzyme, which presents a perfect candidate for the validation of this method. As with PTP1B, acid loop residues (91-NDTQE-95) were mutated to alanine, the CSP were calculated relative to the WT enzyme, and residues with CSP greater than 2σ from the overall trimmed mean (Δδ ≥ 0.065 ppm) were mapped onto the enzyme structure (Fig. 6). Like in PTP1B, the residues with the most significantly affected CSP lie near the sites of mutation including the P-loop (C124, R125) and the acid loop (I88, K89, A90, L98). The data also illuminate two clusters of perturbed residues emanating from the acid loop. There are significant CSPs that extend past both acid loop hinges to the variable insert region (A62-G65, N72-A75) and to α-helix 4 (A100–A105) and α-helix 7 (N163-Q169, C171; Fig. 6). The variable insert region and α-helix 7 are regions of interest for allosteric modulation, as they are solvent exposed and ~13 Å and ~17 Å away from the active site cysteine, respectively (Fig. 6b).
Fig. 6.
Allosteric sites in VHR. (a) Surface rendering of CSP values greater than +2σ of the 10% trimmed mean (0.065 ppm) with the frequency of occurrence (out of the 5 loop mutants) indicated by the color bar. (b) Cartoon rendering of VHR (PDB ID: 1VHR) with selected distal mutations shown in spheres. Purple spheres represent residues in the variable insert region, while cyan spheres represent residues in α-helix 7. The acid loop is shown in blue, while the P-loop is represented in magenta.
To confirm the importance of these affected regions as sites of allosteric regulation, we subjected amino acids in these locations to alanine mutagenesis, and their catalytic efficiencies were assessed, as was previously done with PTP1B. In support of their allosteric coupling with the VHR active site, mutation of surface residues in the variable insert loop had a measurable effect on activity, as both the N74A and R66A mutations exhibit 4.4- and 1.3-fold decreases in catalytic activity relative to WT VHR, respectively. Surprisingly, mutation of residues in α-helix 7 had opposing effects on VHR activity in which the D164A mutation caused a 2-fold increase in catalytic efficiency and L167A resulted in a decrease of 2 orders of magnitude (Fig. 6c). These contrasting effects may result from differences in how these residues interact with the adjacent N-terminal α-helix 1. L167 forms interhelical hydrophobic interactions with V2, L12, and L16 in α-helix 1 just before the recognition region (SI Fig. 7A). Loss of efficient hydrophobic packing for this residue upon mutation to a smaller alanine side chain might be sufficient to displace the N-terminal α-helix 1 and alter the recognition region, resulting in the observed decrease in activity. The importance of α-helix 1 can be seen in the structure (PDB ID: 1J4X) of VHR complexed with the biphosphorylated MAP kinase peptide, where residue T13 in the peptide is shown to form electrostatic interactions with N13 in α-helix 1 of VHR, indicating that this helix may be important for selectivity of peptide substrates (SI Fig. 7B) [40]. It is less clear why the D164A mutation leads to a twofold increase in catalytic efficiency; however, D164 is at the N terminus of α-helix 7 and is adjacent to N163 in the Q-loop, which hydrogen bonds with the side chain of D92 in the acid loop (SI Fig. 7C). Given the high propensity for alanine amino acids to form an alpha helix, it is possible that the D164A mutation may stabilize the N terminus of α-helix 7 and, in turn, promote hydrogen bond formation with D92 to aid substrate binding and catalysis.
In VHR, acid loop mutations demonstrated that distal residues that interact with the acid loop hinges propagate outward from both N- and C-terminal ends of the loop. Mutation of these newly identified surface residues at the periphery of this network alters VHR catalytic activity and confirms the broad applicability of this method for identifying novel allosteric effector binding sites. The allosteric clusters identified for the variable insert region and α-helix 7 have SASAs of 194 Å2 and 97 Å2. The challenge for designing potential ligands in these regions is the small SASA; therefore, in the case of VHR, designed allosteric modulators might make use of fragment linking to engage both of these allosteric sites [21].
Conclusion
The challenge for the development of a potent and selective drug candidate remains for PTPs due to low bioavailability of charged ligands and limited selectivity between structural homologs. Elucidating potential allosteric sites in PTPs is important for the ongoing efforts in developing effective therapeutic strategies for Type II diabetes, obesity, and cancer. Since the discovery of the BB3 allosteric binding site in PTP1B, many computational studies have alluded to its effect on acid loop flexibility when the benzofuran ligand occupies the BB3 site [41,42]. However, there has been little success in identifying other potential allosteric sites or in the elucidation of the allosteric network in PTP1B. An approach was used here to identify allosteric sites in PTP1B and in a dual-specificity phosphatase, VHR, by searching for amino acid residues whose chemical shift is affected by the introduction of active site loop mutations. This study demonstrates a simple method by which unknown allosteric networks in two PTPs have been identified. The CSP highlighted in PTP1B and VHR resembles a blueprint of the dynamic network present within each phosphatase. For both PTP1B and VHR, we have demonstrated that alanine mutations of surface residues in distal regions can alter the catalytic activity even though the sites are separated by tens of Angstroms. The effects on catalysis are somewhat modest in some cases; however, one would expect larger synergistic effects from small molecules that can interact with multiple residues in the newly identified allosteric sites. Given the differences between PTP1B and VHR and the universality of the principles exploited, we expect that this method can be applied to numerous proteins. The wealth of information gained from this simple experiment can be used to develop new allosteric inhibitors and dissect the mechanistic features of allostery.
Materials and Methods
Materials
Deuterium oxide for labeled protein was purchased from Cambridge Isotope Laboratories (Tewksbury, MA), and 15N-ammonium chloride was purchased from Sigma-Aldrich (St. Louis, MO). pNPP disodium salt was purchased from Santa Cruz Biotechnology. PTP1B inhibitor [3-(3,5-dibromo-4-hydroxybenzoyl)-2-ethyl-N-[4-[(2-thiazolylamino)sulfonyl]phenyl]-6-benzofuransulfonamide] was purchased from Cayman Chemical (Ann Arbor, MI). Sodium tungstate dehydrate was purchased from MP biomedical.
Site-directed mutagenesis
The oligonucleotide primers utilized for site-directed mutagenesis of PTP1B and VHR are shown in Supplementary Tables 7 and 8. All oligonucleotide sequences were purchased from the Keck Biotechnology Resource Laboratory (Yale University). Confirmation of the desired DNA sequences was confirmed by DNA sequencing (Keck Biotechnology Resource Laboratory, Yale University).
Protein expression and purification
The PTP1B (1–301) and VHR were expressed in Escherichia coli BL21(DE3) cells supplemented with 100 mg/mL ampicillin (LB expression) or carbenicillin (M9 expression). For isotopic enrichment, cells were grown in M9 minimal media supplemented with 15N-ammonium chloride (1.0 g/L). PTP1B was expressed and purified as previously described [25,32]. The VHR gene sequence contains a polyHis-tag and a TEV cleavage site at the N terminus; thus, VHR was purified using a Ni-NTA column. The cell pellet was resuspended in lysis buffer [20 mM Tris Base, 500 mM NaCl, 20 mM imidazole, 5 mM 2-mercaptoethanol,5%glycerol (pH 7.4)], loaded onto the nickel column, and subsequently eluted with elution buffer [20 mM Tris Base, 500 mM NaCl, 500 mM imidazole, 5 mM 2-mercaptoethanol, 5% glycerol (pH 7.4)]. For NMR studies, pure VHR fractions were dialyzed into 20 mM Bis-Tris propane, 100 mM NaCl, 1 mM TCEP, 7% D2O, and 0.02% NaN3 (pH 6.5) for VHR, into 50 mM Hepes (pH 6.8) with 150 mM NaCl, 0.5 mM TCEP, 7% D2O, and 0.02% NaN3 (NMR buffer) for PTP1B, and concentrated to 0.2–0.4 mM.
Kinetic assay
Steady-state kinetics were measured for PTP1B and VHR using pNPP (0.4, 1.0, 2.0, 4.0, 8.0, 12.0, and 20.0 mM for PTP1B and VHR) in a three-component buffer system [100 mM NaAc, 50 mM Bis-Tris, 50 mM Tris (pH 5.5)]. The reaction was quenched at various time points (10 to 40 s) by adding 1 M NaOH. The rate of reaction was calculated from the slope of the absorbance measured from different time points monitored at 405 nm using a molar extinction coefficient for pNPP of 18,000 M−1 cm−1. Kinetic parameters were determined by fitting the initial reaction rates using the Michaelis–Menten equation in GraphPad Prism version 7.0a for Mac OS X. Kinetic experiments were performed twice for each pNPP concentration.
Visualization and characterization of PTP1B and VHR crystal structures
Figures were rendered in PyMOL [43] or VMD [44] as indicated. The Multiple Alignment Plugin [45] was used to align crystal structures, and Tachyon [46] ray tracing was used to render figures in VMD. Bond angle, length, and SASA (probe radius = 1.4 Å) were measured using VMD.
NMR assignment
Amide resonances in two-dimensional 1H, 15N HSQC spectra were assigned using data deposited in the BMRB (BMRB: 5474, 19,224, and 25,375) [25,47,48]. Assignments were confirmed for select mutants using HNCACB triple resonance data collected at 19 °C on a Varian 600 MHz spectrometer equipped with pulsed field gradients and a triple resonance probe.
NMR titrations from alanine mutation
To identify regions with significant CSPs, we calculated composite chemical shifts as follows:
| (1) |
where δHN and δN are the changes in amide proton and nitrogen chemical shifts from initial positions, respectively [49]. The number of resonances used for each CSP analysis are 214, 210, 204, 202, 209, 203, 207, 204, 205, 210, 204, 206, and 203 for PTP1B (Y176A, T177A, T178A, P180A, D181A, F182A, G183A, V184A, P185A, E186A, S187A, and P188A, respectively). For VHR 151, resonances were used to calculate the CSP for each of the five mutants. CSPs were deemed significant if the value was greater than 2 SD from the 10% trimmed mean of all the data sets.
Tungstate and Benzobromarone NMR titration
Structural mapping of benzofuran analog binding was completed by monitoring changes in 1H, 15N transverse relaxation optimized spectroscopy HSQC spectra for 15N2H-labeled PTP1B upon the addition of the inhibitor dissolved in d6-DMSO. Spectra were collected at various points up to a concentration of 0.400 mM inhibitor using a 9.20-mM inhibitor stock into 0.29 mM 15N2H-PTP1B (550 µL starting volume).
Tungstate binding was completed by the addition of tungstate from stock solution of 1.36 M tungstate dissolved in NMR buffer into 0.21 mM WT (492 µL), 0.26 mM Y153A (526 µL), and 0.32 mM E276A (500 µL).
Data availability
All experimental data will be made available upon request. DNA sequences are provided in the Supplementary Data.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant GM112781 and NSF MCB 1615415 to J.P.L. and T32 GM008283 to V.B.
Abbreviations used
- CSP
chemical shift perturbation
- PTP
protein tyrosine phosphatase
- PTP1B
protein tyrosine phosphatase 1B
- VHR
Vaccinia H1-related phosphatase
- BB30
benzofuran-based small molecule
- pNPP
p-nitrophenylphosphate
- SASA
solvent-accessible surface areas
Footnotes
Appendix A. Supplementary Data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2017.06.009.
References
- 1.Skovpen YV, Conly CJT, Sanders DAR, Palmer DRJ. Biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni. J. Am. Chem. Soc. 2016;138:2014–2020. doi: 10.1021/jacs.5b12695. [DOI] [PubMed] [Google Scholar]
- 2.Kasbekar M, Fischer G, Mott BT, Yasgar A, Hyvonen M, Boshoff HIM, et al. Selective small moleculae inhibitor of the Mycobacterium tuberculosis fumarate hydratase reveals an allosteric regulatory site. Proc. Natl. Acad. Sci. U. S. A. 2016;113:7503–7508. doi: 10.1073/pnas.1600630113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dokholyan NV. Controlling allosteric networks in proteins. Chem. Rev. 2016;116:6463–6487. doi: 10.1021/acs.chemrev.5b00544. [DOI] [PubMed] [Google Scholar]
- 4.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 5.Guarnera E, Berezovsky IN. Allosteric sites: remote control in regulation of protein activity. Curr. Opin. Struct. Biol. 2016;37:1–8. doi: 10.1016/j.sbi.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Curr. Opin. Struct. Biol. 2004;14:706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Lu S, Huang W, Zhang J. Recent computational advances in the identification of allosteric sites in proteins. Drug Discov. Today. 2014;19:1595–1600. doi: 10.1016/j.drudis.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Reynolds KA, McLaughlin RN, Ranganathan R. Hot spots for allosteric regulation on protein surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurle MR, Eads CD, Pearlman DA, Seibel GL, Thomason J, Kosen PA, et al. Comparison of solution structures of mutant bovine pancreatic trypsin inhibitor proteins using two-dimensional nuclear magnetic resonance. Protein Sci. 1992;1:91–106. doi: 10.1002/pro.5560010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimmel JL, Reinhart GD. Reevaluation of the accepted allosteric mechanism of phosphofructokinase from Bacillus stearothermophilus. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3844–3849. doi: 10.1073/pnas.050588097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockless S, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 12.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holliday MJ, Camilloni C, Armstrong GS, Vendruscolo M, Eisenmesser EZ. Networks of dynamic allostery regulate enzyme function. Structure. 2017;25:276–286. doi: 10.1016/j.str.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Axe JM, Yezdimer EM, O'Rourke KF, Kerstetter NE, You WL, Chang CEA, et al. Amino acid networks in a (beta/alpha)(8) barrel enzyme change during catalytic turnover. J. Am. Chem. Soc. 2014;136:6818–6821. doi: 10.1021/ja501602t. [DOI] [PubMed] [Google Scholar]
- 15.Feldhammer M, Uetani N, Miranda-Saavedra D, Tremblay ML. PTP1B: a simple enzyme for a complex world. Crit. Rev. Biochem. Mol. Biol. 2013;48:430–445. doi: 10.3109/10409238.2013.819830. [DOI] [PubMed] [Google Scholar]
- 16.Tonks NK, Diltz CD, Fischer EH. Characterization of the major protein-tyrosine-phosphatases of human placenta. J. Biol. Chem. 1988;263:6731–6737. [PubMed] [Google Scholar]
- 17.Rahmouni S, Cerignoli F, Alonso A, Tsutji T, Henkens R, Zhu C, et al. Loss of the VHR dual-specific phosphatase causes cell-cycle arrest and senescence. Nat. Cell Biol. 2006;8:524–531. doi: 10.1038/ncb1398. [DOI] [PubMed] [Google Scholar]
- 18.Henkens R, Delvenne P, Arafa M, Moutschen M, Zeddou M, Tautz L, et al. Cervix carcinoma is associated with an up-regulation and nuclear localization of the dual-specificity protein phosphatase VHR. BMC Cancer. 2008;8:147. doi: 10.1186/1471-2407-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao L, ElShamy WM. BRCA1-IRIS activates cyclin D1 expression in breast cancer cells by downregulating the JNK phosphatase DUSP3/VHR. Int. J. Cancer. 2007;121:39–46. doi: 10.1002/ijc.22597. [DOI] [PubMed] [Google Scholar]
- 20.Arnoldussen YJ, Lorenzo PI, Pretorius ME, Waehre H, Risberg B, Maelandsmo GM, et al. The mitogen-activated protein kinase phosphatase Vaccinia H1-related protein inhibits apoptosis in prostate cancer cells and is over-expressed in prostate cancer. Cancer Res. 2008;68:9255–9264. doi: 10.1158/0008-5472.CAN-08-1224. [DOI] [PubMed] [Google Scholar]
- 21.Zhang ZY. Drugging the undruggable: therapeutic potential of targeting protein tyrosine phosphatases. Acc. Chem. Res. 2016 doi: 10.1021/acs.accounts.6b00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, et al. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W, et al. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat. Struct. Mol. Biol. 2004;11:730–737. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 24.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- 25.Krishnan N, Koveal D, Miller DH, Xue B, Akshinthala SD, Kragelj J, et al. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat. Chem. Biol. 2014;10:558–566. doi: 10.1038/nchembio.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 27.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denu JM, Zhou GC, Guo YP, Dixon JE. The catalytic role of aspartic acid-92 in a human dual-specific protein-tyrosine-phosphatase. Biochemistry. 1995;34:3396–3403. doi: 10.1021/bi00010a031. [DOI] [PubMed] [Google Scholar]
- 29.Jia ZC, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein-tyrosine-phosphatase 1b. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 30.Peters GH, Frimurer TM, Andersen JN, Olsen OH. Molecular dynamics simulations of protein-tyrosine phosphatase 1B. I. Ligand-induced changes in the protein motions. Biophys. J. 1999;77:505–515. doi: 10.1016/S0006-3495(99)76907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters GH, Frimurer TM, Andersen JN, Olsen OH. Molecular dynamics simulations of protein-tyrosine phosphatase 1B. II. Substrate-enzyme interactions and dynamics. Biophys. J. 2000;78:2191–2200. doi: 10.1016/S0006-3495(00)76768-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittier SK, Hengge AC, Loria JP. Conformational motions regulate phosphoryl transfer in related protein tyrosine phosphatases. Science. 2013;341:899–903. doi: 10.1126/science.1241735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandao TAS, Johnson SJ, Hengge AC. The molecular details of WPD-loop movement differ in the protein-tyrosine phosphatases YopH and PTP1B. Arch. Biochem. Biophys. 2012;525:53–59. doi: 10.1016/j.abb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang ZY, Dixon JE. Protein tyrosine phosphatases: mechanism of catalysis and substrate specificity. Adv. Enzymol. Relat. Areas Mol. Biol. 1994;68:1–36. doi: 10.1002/9780470123140.ch1. [DOI] [PubMed] [Google Scholar]
- 35.Choy MS, Li Y, Machado LESF, Kunze MBA, Connors CR, Wei XY, et al. Conformational rigidity and protein dynamics at distinct timescales regulate PTP1B activity and allostery. Mol. Cell. 2017;65:644–658. doi: 10.1016/j.molcel.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng AC, Coleman RG, Smyth KT, Cao Q, Soulard P, Caffrey DR, et al. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007;25:71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 37.Van Vliet F, Xi XG, De Staercke C, de Wannemaeker B, Jacobs A, Cherfils J, et al. Heterotropic interactions in aspartate transcarbamoylase: turning allosteric ATP activation into inhibition as a consequence of a single tyrosine to phenylalanine mutation. Proc. Natl. Acad. Sci. U. S. A. 1991;88:9180–9183. doi: 10.1073/pnas.88.20.9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Ahuja LG, Chao FA, Xia Y, McClendon CL, Kornev AP, et al. A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci. Adv. 2017;3:e1600663. doi: 10.1126/sciadv.1600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ibarra-Sanchez MD, Simoncic PD, Nestel FR, Duplay P, Lapp WS, Tremblay ML. The T-cell protein tyrosine phosphatase. Semin. Immunol. 2000;12:379–386. doi: 10.1006/smim.2000.0220. [DOI] [PubMed] [Google Scholar]
- 40.Schumacher MA, Todd JL, Rice AE, Tanner KG, Denu JM. Structural basis for the recognition of a bisphosphorylated MAP kinase peptide by human VHR protein phosphatase. Biochemistry. 2002;41:3009–3017. doi: 10.1021/bi015799l. [DOI] [PubMed] [Google Scholar]
- 41.Cui W, Cheng YH, Geng LL, Liang DS, Hou TJ, Ji MJ. Unraveling the allosteric inhibition mechanism of PTP1b by free energy calculation based umbrella sampling. J. Chem. Inf. Model. 2013;53:1157–1167. doi: 10.1021/ci300526u. [DOI] [PubMed] [Google Scholar]
- 42.Kamerlin LS, Rucker R, Boresch S. A molecular dynamics study of WPD-loop flexibility in PTP1b. Biochem. Biophys. Res. Commun. 2007;356:1011–1016. doi: 10.1016/j.bbrc.2007.03.093. [DOI] [PubMed] [Google Scholar]
- 43.The PyMOL Molecular Graphics System, Version 1.7.6.0. Schrödinger: LLC; [Google Scholar]
- 44.Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 45.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinf. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone J. An Efficient Library for Parallel ray Tracing and Animation. University of Missouri-Rolla; 1998. [Google Scholar]
- 47.Meier S, Li YC, Koehn J, Vlattas I, Wareing J, Jahnke WG, et al. Letter to the editor: backbone resonance assignment of the 298 amino acid catalytic domain of protein tyrosine phosphatase 1B (PTP1B) J. Biomol. NMR. 2002;24:165–166. doi: 10.1023/a:1020949923448. [DOI] [PubMed] [Google Scholar]
- 48.Krishnan N, Krishnan K, Connors CR, Choy MS, Page R, Peti W, et al. PTP1B inhibition suggests a therapeutic strategy for Rett syndrome. J. Clin. Invest. 2015;125:3163–3177. doi: 10.1172/JCI80323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10,256–10,261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 50.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein-tyrosine-phosphatase 1b. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- 51.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 52.Hakim F, Wang Y, Carreras A, Hirotsu C, Zhang J, Peris E, et al. Chronic sleep fragmentation during the sleep period induces hypothalamic endoplasmic reticulum stress and PTP1b-mediated leptin resistance in male mice. Sleep. 2015;38:31–U367. doi: 10.5665/sleep.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brandao TAS, Hengge AC, Johnson SJ. Insights into the reaction of protein-tyrosine phosphatase 1B crystal structures for transition state analogs of both catalytic steps. J. Biol. Chem. 2010;285:15,874–15,883. doi: 10.1074/jbc.M109.066951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental data will be made available upon request. DNA sequences are provided in the Supplementary Data.