Abstract
In this study we showed that constitutive heterochromatin, GC-rich DNA and rDNA are implicated in chromosomal rearrangements during the basic chromosome number changing (dysploidy) in Reichardia genus. This small Mediterranean genus comprises 8–10 species and presents three basic chromosome numbers (x = 9, 8 and 7). To assess genome evolution and differentiation processes, studies were conducted in a dysploid series of six species: R. dichotoma, R. macrophylla and R. albanica (2n = 18), R. tingitana and R. gaditana (2n = 16), and R. picroides (2n = 14). The molecular phylogeny reconstruction comprised three additional species (R. crystallina and R. ligulata, 2n = 16 and R. intermedia, 2n = 14). Our results indicate that the way of dysploidy is descending. During this process, a positive correlation was observed between chromosome number and genome size, rDNA loci number and pollen size, although only the correlation between chromosome number and genome size is still recovered significant once considering the phylogenetic effect. Fluorescent in situ hybridisation also evidenced changes in number, position and organisation of two rDNA families (35S and 5S), including the reduction of loci number and, consequently, reduction in the number of secondary constrictions and nuclear organising regions from three to one per diploid genome. The potential mechanisms of chromosomal and genome evolution, strongly implicating heterochromatin, are proposed and discussed, with particular consideration for Reichardia genus.
Introduction
The Mediterranean genus Reichardia Roth (Asteraceae), made up of annual, biennial or perennial herbs, is a good model for the investigation of genome organisation and evolution since it includes a small number of closely related species (from 8 to 10, depending on the authors) and three basic chromosome numbers: x = 9, 8, and 7. Two tertiary relict species, one from Dinaric Alps and another from Middle East (R. macrophylla Vis. & Pančić and R. dichotoma (DC.) Freyn (R. glauca V.A.Matthews), respectively), have x = 9, in common with the recently described Albanian endemic R. albanica F.Conti & D.Lakušić, which is closely related to the latter taxa [1]. During the quaternary glaciations, certain regions of the Dinaric Alps became refugia for Tertiary flora; some habitats on dolomite substrate are exceptionally rich in endemic or relict species, among which R. macrophylla is included [2, 3]. The second relict species is R. dichotoma, whose geographical distribution is limited to the Eastern Mediterranean (Anatolia, Armenia, Georgia, North-East Iran, Syria and North Lebanon) [1]. Reichardia picroides (L.) Roth and R. intermedia (Sch.Bip.) Cout., with circum-Mediterranean distribution [4], have the lowest basic chromosome number in the genus, x = 7. In this dysploid series other species such as R. tingitana (L.) Roth, with a repartition from the Azores to NW India [5] (which coincides with paleogeographical limits of the Mediterranean basin [6]), the Iberian neoendemic species R. gaditana (Willk.) Cout. [7], and three endemic species from Canary Islands (R. crystallina (Sch.Bip.) Bramwell, R. famarae Bramwell & G.Kunkel ex M.J.Gallego & Talavera and R. ligulata (Vent.) G.Kunkel & Sunding), have an intermediate basic number of x = 8 [5, 8, 9, 10].
According to Flora Europaea, the endemic species from Balkan Peninsula R. macrophylla has been considered to be R. picroides [11]. However, these two taxa have different basic chromosome numbers of x = 9 and x = 7, respectively [12], and different geographical ranges. Reichardia picroides has a large circum-Mediterranean repartition, while R. macrophylla grows in regions considered refugia of Tertiary flora, such as canyons and narrow dry valleys on limestone or dolomite substrata, frequently in Pinus nigra J.F.Arnold communities [2, 3].
Genome size, usually assessed as the 2C value (the amount of DNA in a somatic unreplicated nucleus) [13, 14], is one of the most relevant biological characters, with relationships with many other plant life characters, from morphological to ecological through cytogenetic, phylogenetic and even taxonomical ones [15] (and references therein). Relationships between nuclear DNA amount and chromosomal characters are numerous and clear in the Asteraceae family [16] (and references therein). Of the consequences of genome size in morphological traits, pollen size has been largely understudied, except in the case of the species with different ploidy levels [17].
Among plant species, there is great variability in chromosome number, with variation of basic chromosome number ("x") across a wide range [18] (and references therein). Amongst the rare intra-specific variations, the most frequent are the modifications of ploidy level (very frequent in plants) or Robertsonian mutations. Increases in ploidy level seem to be produced by naturally occurring mutations causing extensive genome rearrangements, resulting in modifications of life cycle, such as flowering time [19], which might lead in some cases to the rise of new species. Although polyploidy is a well-known evolutionary mechanism in plants, in some cases the main evolutionary trend is not a genome multiplication, but a progressive reduction of the basic number, known as dysploidy, namely decreasing, descending or downward dysploidy (from x to x-1, x-2, x-3 etc.) [10, 20, 21].
We previously studied karyotype and constitutive heterochromatin patterns in five of the above-mentioned species of this genus by Giemsa C-banding [10, 22, 23]. There is almost no heterochromatin in R. dichotoma and only a tiny heterochromatic band in R. macrophylla. In contrast, the increase in presence of heterochromatin was observed in two species with x = 8, R. gaditana and R. tingitana, which possess even entire heterochromatic short arms of some chromosome pairs. In R. picroides, whose basic chromosome number is reduced to x = 7, the decrease in heterochromatin and its restriction to centromeres and secondary constrictions (SC) were detected. A possible way to test the role of heterochromatin in governing chromosomal rearrangements during reduction of chromosome number and its impact on genome size changes requires considering closely related species with differentiated karyotypes such as is the case in the genus Reichardia.
Heterochromatin is frequently associated with chromosomal rearrangements [24, 25, 26, 27]. However, it remains to be examined whether it can act upon these rearrangements by making them more likely or less deleterious, considering its well-known properties and its distribution along chromosomes in the Reichardia species.
Heterochromatin is an important constituent of the genome and could be highly informative for untangling the evolutionary histories of closely related species. Constitutive heterochromatin is made up of tandemly repeated sequences which can be AT or GC rich [28, 29, 30] and can be revealed by several stain procedures, such as Giemsa C- [31] and fluorochrome banding [32, 33]. Several authors, especially Schweizer et al. [34], suggest that heterochromatin modification is a rapid process compared to other biological processes ("recent icing on the cake"). At the karyotypic level, it can modify meiotic recombination [26, 35, 36, 37, 38, 39], form interchromosomal connections [10], and affect variation in genome size. In addition, it is frequently associated with translocations, inversions or chromosomal breaks and involved in chromosome segregation [26, 40].
Molecular cytogenetic techniques provide the opportunity to study the fine mechanisms that have acted during the evolution of the chromosome, e.g. dysploidy. In eukaryotes, the rRNA genes can serve as excellent markers in phylogenetic studies. These genes are organised into two distinct families (i.e., 35S and 5S rDNA) that occur as tandem arrays at one or more specific chromosomal regions. Due to their high copy number, detection of the rRNA genes is highly reproducible and provides valuable information concerning chromosomal evolution. Copy number and chromosomal distribution of rDNAs can change rapidly and rDNA transposition or dispersion in plant genomes is observed frequently [41, 42, 43, 44, 45, 46, 47]. These rearrangements generally correlate with species differentiation and speciation. The numbers and locations of rDNA arrays may vary even between infra-specific taxa and can therefore provide chromosomal landmarks for species differentiation [48].
Based on our previous karyological studies [10, 12, 23] we postulated the hypothesis about descending dysploidy in the genus Reichardia. Thus, the main objective of the present work was to understand the mechanism of karyotype evolution by dysploidy in a small cluster of closely related species by checking possible heterochromatin involvement. For this purpose, karyotypes of Reichardia species were characterised using molecular cytogenetic techniques: (1) flow cytometry for DNA quantity and GC% assessment; (2) fluorochrome banding for distribution of GC-rich DNA and neutral heterochromatin; (3) FISH for establishing a physical map of 35S (18S-5.8S-26S) and 5S rRNA genes. In addition, pollen grain size variation relative to basic chromosome number and genome size was measured. The data obtained were analysed in the frame of new molecular phylogenetic evidence.
Material and methods
Origin of material
The origins of the studied populations are shown in Table 1. Six out of the nine species studied are endemics: R. gaditana (Iberian Peninsula), R. macrophylla (Dinaric Alps), R. albanica (from Albania) and R. dichotoma (from the Middle East), plus two endemic species from the Canary Islands, R. crystallina and R. ligulata. The remaining species are more widespread: R. tingitana from the Azores to NW India, and R. picroides and R. intermedia across the whole Mediterranean basin.
Table 1. Origin of studied species and populations.
Vouchers are deposited in the following herbaria. BCN: Centre de Documentació de Biodiversitat Vegetal, Universitat de Barcelona. BC: Institut Botànic de Barcelona. BEOU: University of Belgrade. SY: Sonja Siljak-Yakovlev (personal collection), Orsay.
| Species | Locality | Collectors and herbarium where voucher is deposited |
|---|---|---|
| R. dichotoma (DC.) Freyn (R. glauca A.Matthews) | 1. Mountain pass Tigranashen and Sovetashen, Armenia | G. Fajvush, E. Gabrielian, N. Garcia-Jacas, M. Hovanyssian, A. Susanna, J. Vallès (BCN) |
| 2. Marand, Iran | N. Garcia-Jacas, A. Susanna, V. Mozaffarian, J. Vallès (BCN) | |
| 3. Mt Ehden, Lebanon | S. Siljak-Yakovlev, M. Bou Dagher-Kharrat, (SY) | |
| R. macrophylla Vis. & Pančić | 4. Near Konjic, Bosnia & Herzegovina | S. Siljak-Yakovlev (SY) |
| 5. Diva Grabovica, Bosnia & Herzegovina | ||
| 6. Mt Orjen, Montenegro | ||
| 7. Lastva, Bosnia & Herzegovina | ||
| 8. Sutjeska canyon, Bosnia & Herzegovina | ||
| R. albanica F. Conti & D. Lakušić | 9. Mali i Cikes, Llogara, Albania | D. Lakušić, N. Kuzmanović, M. Lazarevic, A. Alegro, F. Conti (BEOU) |
| R. tingitana (L.) Roth | 10. Canary Islands, Spain | From Puerto de la Cruz botanic garden (SY) |
| 11. Oriola, Spain | J. Vallès (BCN) | |
| R. gaditana (Willk.) Cout. | 12. Portugal | M. Queirós (from Coimbra botanical garden) (SY) |
| R. crystallina (Sch.Bip.) Bramwell | 13. Porís de Abona,Tenerife, Canary Islands, Spain | A. Santos-Guerra, J. Vallès (BCN) |
| R. ligulata (Vent.) G.Kunkel & Sunding | 14. Punta de Teno,Tenerife, Canary Islands, Spain | A. Santos-Guerra, J. Vallès (BCN) |
| 15. Roque de las Bodegas, Tenerife, Canary Islands, Spain | A. Santos-Guerra, J. Vallès (BCN) | |
| 16. Andén Verde, Gran Canaria, Canary Islands, Spain | A. Santos-Guerra, J. Vallès (BCN) | |
| R. intermedia (Sch.Bip.) Cout. | 17. Oran, Algeria | K. Abdeddaim (SY) |
| 18. Spain | T. Garnatje (from Barcelona botanical garden) (BC) | |
| R. picroides (L.) Roth | 19. Gornji Okrug, Dalmatia, Croatia | S. Siljak-Yakovlev (SY) |
| 20. Dubrovnik, Dalmatia, Croatia | ||
| 21. Lavandou, Côte d’Azur, France |
From cytogenetic and palynological points of view we have studied five well representative species, comprising all three basic chromosome numbers, among the nine species of genus. Out of the four not studied from this viewpoint, three are endemic to the Canary Islands and are close to R. tingitana (a nearly circum-Mediterranean species with 2n = 16), and one (R. intermedia) is very close to R. picroides (2n = 14). For molecular phylogenetic study and genome size estimation all species of the genus were considered except Reichardia famarae Bramwell & G.Kunkel ex Gallego & Talavera, endemic from Canary Islands, for which we missed the material.
Estimation of nuclear DNA content and base composition by flow cytometry
Total DNA amounts were assessed by flow cytometry according to Marie and Brown [49]. Petunia hybrida Vilm. ‘PxPc6’ (2C = 2.85 pg, 41.0% GC) and Lycopersicon esculentum Mill. ‘Roma’ (2C = 1.99 pg, 40.0% GC) were used as internal standards. Leaves of both the studied species and the internal standard were chopped up using a razor blade in a plastic Petri dish with 600 μl of Galbraith nucleus-isolation buffer [50] containing 0.1% (w/v) Triton X-100, 10 mM sodium metabisulphite and 1% polyvinylpyrrolidone 10,000. The suspension was passed through a 48 μm mesh nylon filter. The nuclei were stained with 50 μg/ml propidium iodide, after 15 min RNase treatment (2.5 U/ml). Base composition was assessed using AT-specific fluorochrome bisbenzimide Hoechst 33342 (5 μg/ml; Aldrich) and GC-specific fluorochrome mithramycin (50 μg/ml). DNA content of 5,000–10,000 stained nuclei was determined for each sample using an Elite ESP flow cytometer (Beckman-Coulter, Roissy, France) with a water-cooled argon laser. Total 2C DNA value was calculated using the linear relationship between the fluorescent signals from the stained nuclei of the Reichardia specimen and the internal standard. Base composition (GC percentage) was calculated using the nonlinear model established by Godelle et al. [51]. Each studied population comprised at least five individuals, measured separately and with two replicates.
Chromosome preparation
Root tips obtained from germinated seedlings or from living plants (growing in experimental garden, Orsay, France), were pre-treated with 2 mM 8-hydroxyquinoline during 2 h (R. picroides and R. gaditana), 2 h 15 min (R. tingitana) or 3 h (R. macrophylla, R. dichotoma and R. albanica) at approximately 16°C, and then fixed in 3:1 absolute ethanol:glacial acetic acid at 4°C for at least one day. Chromosome plates for fluorochrome banding and FISH experiments were prepared using the air-drying technique of Geber and Schweizer [52], with slight modifications. Root tips were washed in citrate buffer (pH 4.6) for 10 min and then transferred into the enzyme mixture [4% cellulase “Onozuka” R-10 (Yakult Honsha Co. Tokyo, Japan), 1% pectolyase Y-23 (Seishin Co. Tokyo, Japan), 4% hemicellulase (Sigma)] at 37°C for 15 min. The resulting protoplast suspension was washed three times in citrate buffer and fixed in 3:1 absolute ethanol:glacial acetic acid. Centrifugation was performed at 4000 rpm (1500 g) for 5 min. The final pellet was resuspended in 50 μl of fixative and protoplasts were transferred onto a clean slide, air-dried, and kept at room temperature until use.
Fluorochrome banding and rDNA mapping by fluorescent in situ hybridisation (FISH)
GC-rich DNA region staining with chromomycin A3 (CMA3, Sigma Aldrich Co., Steinheim, Germany) was performed according to Schweizer [32] with minor modifications as described by Siljak-Yakovlev et al. [33].
A double FISH experiment was carried out following the method of Heslop-Harrison et al. [53]. Slides were counterstained and mounted in Vectashield medium containing DAPI (4′, 6-diamidino-2-phenylindole, Vector Laboratories) which also revealed neutral or nonspecific heterochromatin (rich neither in AT nor in GC bases) as DAPI+ bands. This type of heterochromatin mainly corresponded to constitutive heterochromatin stained by Giemsa C-banding.
For rDNA analyses and CMA fluorochrome banding, a minimum of 10 well-spread metaphases were analysed for each species.
Microscopy and chromosome analysis
Chromosome observations were performed using an epifluorescence Zeiss Axiophot microscope with different combinations of excitation and emission filter sets (01, 07, 15 and triple filter set 25). The signals were analysed using the highly sensitive CCD camera (RETIGA 2000R; Princeton Instruments, Evry, France) and an image analyser (Metavue, Evry, France).
The construction of idiograms and the Giemsa C-banding for detection of constitutive heterochromatin in five representatives have been published in two our previous works [10, 23].
Pollen grain measurement
Pollen grains were acetolysed according to Erdtman [54]. The measurements of pollen grains’ polar axis (P) and equatorial (E) diameter were performed on 100 acetolysed grains mounted for at least three weeks in glycerine jelly. All measurements were made on well-formed pollen grains under a 40× objective lens on Zeiss Axiophot microscope.
DNA extraction, amplification and sequencing
Total genomic DNA was extracted following the CTAB method of Doyle and Doyle [55] as modified by Soltis et al. [56], from silica gel-dried leaves collected in the field or fresh leaves of plants cultivated in the Botanical Institute of Barcelona. In some cases, herbarium material was used. Double-stranded DNA was amplified from ITS regions with the 1406F [57] and ITS4 [58] primers. In some cases, we used the ITS1 [58] as forward primer. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Valencia, California, U.S.A.). Both strands were sequenced with 1406F or ITS1 as forward primers and ITS4 as the reverse primer. Direct sequencing of the amplified DNA segments was performed using Big Dye Terminator Cycle sequencing v2.0 (PE Biosystems, Foster City, California, U.S.A.). Nucleotide sequencing was carried out at the Centres Científics i Tecnològics, University of Barcelona on an ABI PRISM 3700 DNA analyser (PE Biosystems, Foster City, California, U.S.A.).
DNA sequences were edited with Chromas 1.56 (Technelysium PTy, Tewantin, Queensland, Australia) and aligned visually. The sequences were deposited in GenBank (see the Appendix for the accession numbers). The sequence alignment is available from the corresponding author.
Phylogenetic analysis
To determine model under the Akaike Information Criterion (AIC) [59] the data set was analysed using MrModeltest 2.2 [60]. This model was used to perform a Bayesian analysis using MrBayes 3.2.1 [61]. Four Markov chains were run simultaneously for two million generations, and these were sampled every 100 generations. Data from the first 1000 generations were discarded as the burn-in period, after confirming that likelihood values had stabilised prior to the 1000th generation. Posterior probabilities were estimated through the construction of a 50% majority rule consensus. The outgroup (Sonchus kirkii Hamlin) has been chosen on the basis of the work of Kim et al. [62].
Ancestral character states reconstructions
The Phytools package of R [63] was used to perform ancestral state reconstructions, using the consensus tree resulting from Bayesian analysis reduced to the set of ingroup taxa. The ancestral 2C-values were reconstructed under maximum likelihood with the fastAnc and contMap commands, and ancestral chromosome numbers were inferred with the re-rooting method. Alternatively, ancestral GS were also reconstructed using maximum parsimony for continuous traits in Mesquite v.3.04 software [64].
Correlation analyses
Phylogenetic generalised least squares analyses (PGLS) were conducted under the Brownian motion model and Pagel model of evolution using the ape and nlme packages of R [65, 66, 67] on the log-transformed dataset of Bayesian tree, ultrametricised and pruned to the five species with available data for all traits.
Results
Genome size
The genome size of the studied populations ranged from 2.86 (R. picroides) to 5.54 pg (R. dichotoma) for holoploid (2C) DNA (or from 1399 to 2709 Mbp for monoploid genome size (1Cx). Coefficients of variation were under 5, accounting for a reliable quality of the measurements (Table 2). These data were in agreement with the basic chromosome number: species with x = 9 showed the highest DNA content (mean value of 5.21 pg/2C) and those with x = 8 and x = 7 showed lower DNA content (mean values of 3.52 and 2.92 pg/2C, respectively). The percent of G-C bases (GC %) ranges from 39.4 in R. picroides to 41.2 in R. gaditana (Table 2).
Table 2. DNA content and GC percentage of Reichardia species from different populations.
| Species (2n) | Population number (see Table 1) or name | 2C DNA in pga (SD; CV) | 1Cx in Mbp [68] | GC% |
|---|---|---|---|---|
| R. dichotoma (18) | 1 | 5.20 (0.04; 1.23) | 2543 | 40.7 |
| 2 | 5.08 (0.03; 0.79) | 2484 | 40.4 | |
| 3 | 5.54 (0.07; 0.83) | 2709 | ||
| Mean for species | 5.27 | 40.6 | ||
| R. macrophylla (18) | 4 | 5.25 (0.04; 3.66) [69] | 2567 | |
| 5 | 5.13 (0.10; 0.77) | 2509 | ||
| 6 | 5.20 (0.09; 0.75) | 2543 | ||
| 7 | 5.10 (0.10; 0.66) [70] | 2494 | 40.9 | |
| 8 | 5.05 (0.07; 0.79) | 2469 | ||
| Mean for species | 5.15 | |||
| R. albanica (18) | 9 | 5.22 (0.02; 0.43) [1] | 2553 | |
| R. tingitana (16) | 10 | 3.50 (0.07; 0.83) [70] | 1712 | 40.5 |
| 11 | 3.23 (0.02; 0.58) | 1579 | ||
| Mean for species | 3.37 | |||
| R. gaditana (16) | 12 | 3.40 (0.08; 0.67) [70] | 1663 | 41.2 |
| R. crystallina (16) | 13 | 3.43 (0.08; 2.35) [71] | 1677 | - |
| R. ligulata (16) | 14 | 3.84 (0.32; 2.42) [71] | 1878 | |
| 15 | 3.62 (0.14; 2.29) [71] | 1770 | ||
| 16 | 3.95 (0.50; 2.73) [71] | 1932 | ||
| Barranco de Roque | 3.78 (0.01; 2.21) [72] | 1852 | ||
| Bermejo [72] | ||||
| Mean for species | 3.80 | |||
| R. intermedia (14) | 17 | 2.90 (0.02; 1.11) [70] | 1418 | |
| 18 | 2.93 (0.04; 1.24) | 1433 | ||
| Mean for species | 2.92 | |||
| R. picroides (14) | 19 | 2.90 (0.05; 0.45) | 1418 | 39.4 |
| 20 | 3.00 (0.03; 0.78) [70] | 1467 | 39.5 | |
| 21 | 2.86 (0.02; 0.38) | 1399 | 39.9 | |
| Mean for species | 2.92 | 39.6 |
aMean value for population; SD = standard deviation; CV = coefficient of variation
Mapping of heterochromatin and rRNA genes
Neutral (unspecific) heterochromatin pattern (DAPI+ bands)
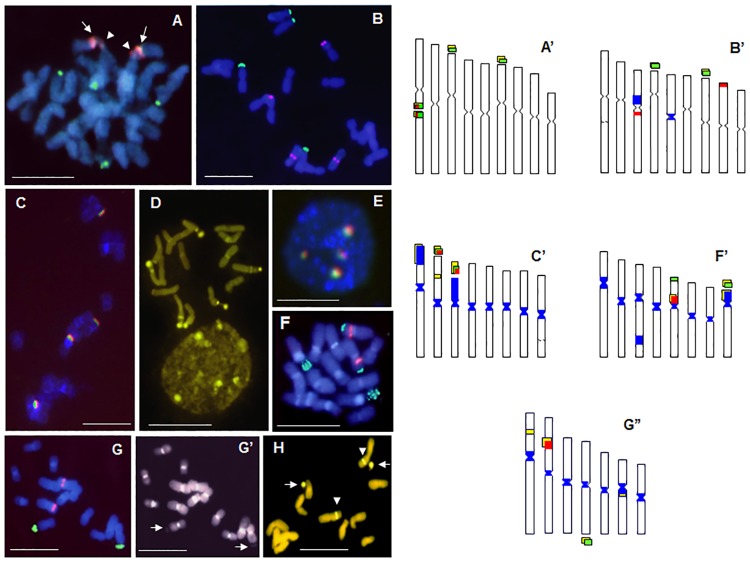
The band positions of neutral (or unspecific) heterochromatin, revealed by DAPI after FISH, are shown in Table 3 and Fig 1. In R. dichotoma this type of heterochromatin was not observed (Fig 1A’). In R. macrophylla, the presence of neutral heterochromatin was restricted to two chromosome pairs; a large paracentromeric band on short arm of pair 3 and a centromeric band on pair 5 (Fig 1B’). In R. tingitana all the centromeres are heterochromatic, and also a part (pair 1) or the totality (pair 3) of the short arms (Fig 1C’). In R. gaditana, not only the centromeres, but also one intercalary region (pair 3) and the whole short arm of pair 8 are stained (Fig 1F and 1F’). The unspecific heterochromatin only remains in centromeres of R. picroides (Fig 1G’ and 1G”).
Table 3. Distribution of DAPI bands after FISH experiment.
| Species (2n) | Centromeric region | Pericentromeric region | Paracentromeric region | Intercalary band | Terminal band |
|---|---|---|---|---|---|
| R. dichotoma (18) | 0 | 0 | 0 | 0 | 0 |
| R. macrophylla (18) | pairs 3, 5 | 0 | pair 3 | 0 | 0 |
| R. tingitana (16) | all pairs | pair 3a | 0 | 0 | pair 1 |
| R. gaditana (16) | all pairs | pair 1 | 0 | pairs 3, 8b | 0 |
| R. picroides (14) | all pairs | 0 | pairs 1, 6 | 0 | 0 |
| R. intermedia (14) | all pairs | 0 | pairs 1, 6 | 0 | 0 |
pair = chromosome pair,
aentire DAPI positive chromosome arm comprising secondary constriction,
bentire DAPI positive chromosome arm except satellite
Fig 1.
Metaphase chromosome plates and interphase nuclei of Reichardia species after double target FISH with 5S (red signals) and 35S (green signals) rDNA loci, CMA (yellow signals) and DAPI staining (blue): R. dichotoma showing 5S and 35S colocalised loci at both sites of intercalary secondary constriction (arrows and arrowheads) and four terminal 35S signals (A); R. macrophylla with 4 terminal 35S signals and two intercalary and two terminal 5S signals (B); R. tingitana showing four colocalised 5S and 35S signals (C), numerous CMA+ bands in metaphase chromosomes and interphase nucleus (D), nucleus after FISH with four 35S/5S spots (E); R. gaditana with four terminal 35S (two being very intense) and two intercalary 5S signals (F); R. picroides showing two terminal sat 35S on long chromosome arms and two intercalary 5S signals on short arms (G), DAPI+ centromeric bands and DAPI negative satellites (arrows) in the same metaphase plate as FISH, better visible on black and white photograph (G’) and CMA+ satellites (arrows) corresponding to 35S and intercalary bands (arrowheads) that corresponds to 5S signals (H). Scale bar 10 μm. Idiograms of R. dichotoma (A’), R. macrophylla (B’), R. tingitana (C’), R. gaditana (F’), R. picroides (G”) showing distribution of CMA+ (yellow) and DAPI+ (blue) bands, 35S (green) and 5S (red) rDNA signals.
G-C rich heterochromatin distribution (CMA+ bands)
The number and the distribution of CMA+ bands are presented in Table 4. Chromomycin positive bands were visible in all satellites and secondary constrictions (Fig 1). Additional terminal or intercalary bands were observed in R. tingitana (Fig 1D and 1C’), R. gaditana (Fig 1F) and R. picroides (Figs 1H and 1G”).
Table 4. Number and distribution of CMA+ bands in diploid chromosome set.
| Species (2n) | Total CMA+ bands number | Intercalary bands in SC | Terminal bands | Satellite SC | Intercalary bands | PC bands |
|---|---|---|---|---|---|---|
| R. dichotoma (18) | 6 | pair 1 | 0 | pairs 3, 6 | 0 | 0 |
| R. macrophylla (18) | 4 | 0 | 0 | pairs 4, 7 | 0 | 0 |
| R. tingitana (16) | 8 | 0 | pairs 1, 3 | pair 2 | pair 2 | 0 |
| R. gaditana (16) | 6 | 0 | pair 8* | pair 8 | 0 | pair 5 |
| R. picroides (14) | 8 | 0 | 0 | pair 4 | pairs 1, 2 | pair 6 |
| R. intermedia (14) | 8 | 0 | 0 | pair 4 | pairs 1, 2 | pair 6 |
pair = chromosome pair,
*entire short chromosome arm,
PC = paracentromeric bands
Physical mapping of 35 and 5S rRNA genes
The number and position of 35S and 5S rDNA loci are shown in Table 5. In R. dichotoma there were three 35S rDNA loci (two in satellite and one in intercalary SC) and one 5S rDNA locus (Fig 1A and 1A’). The only 5S rRNA site was colocalised with the 35S in intercalary SC on the long arm of chromosome pair 1. Both 35S and 5S signals were present on both sides of this SC (Fig 1A and 1A’). In the second species with x = 9, R. macrophylla, two 35S rDNA loci, both in satellite SCs, and also two 5S loci, one on intercalary position near the centromere on the long arm of the pair 3, and another terminal on the short arm of pair 8, were observed (Fig 1B and 1B’). Reichardia tingitana displayed four colocalised 5S and 35S signals in metaphase chromosomes (Fig 1C and 1C’) and four 35S/5S signals in nucleus (Fig 1E).
Table 5. Number and position of 35S and 5S rDNA loci.
| Species (2n) | 35S rDNA loci | 5S rDNA loci | ||
|---|---|---|---|---|
| number | position | number | position | |
| R. dichotoma (18) | 3 | pair 1 –intercalary on both sides of SC;pairs 3 and 6 –sat SCs | 1 | pair 1 –intercalary on both sides of SC |
| R. macrophylla (18) | 2 | pairs 4 and 7 –sat SCs | 2 | pair 3 –intercalary; pair 8 –terminal |
| R. tingitana (16) | 2 | pairs 2 and 3 –sat SCs | 2 | pairs 2 and 3 –sat SCs |
| R. gaditana (16) | 2 | pair 5 –terminal; pair 8 –sat SC | 1 | pair 5 –intercalary |
| R. picroides (14) | 1 | pair 4 –sat SC long arm | 1 | pair 2 –intercalary |
| R. intermedia (14) | 1 | pair 4 –sat SC long arm | 1 | pair 2 –intercalary |
Reichardia gaditana possessed one terminal and one satellite 35S locus (pair 5 and 8 respectively) and one 5S locus near the centromere on the short arm of pair 5 (Fig 1F and 1F’). The 5S and one of the 35S sites are positioned on the same chromosome arm. Only one 35S (at the satellite SC of long arm of chromosome pair 4) and one 5S (intercalary on the pair 2) loci were found in R. picroides (Fig 1G and 1G”). Size dimorphism of the satellites and their detachment from the chromosomes (Fig 1H, arrows), chromomycin bands that correspond to 5S signals (Fig 1H, arrow-heads) and DAPI negative satellites corresponding to 35S signals (Fig 1G’) were also observed.
According to obtained results one hypothetical schema of karyotype evolution in the genus Reichardia is presented in Fig 2.
Fig 2. Hypothetical schema of overarching karyotype evolution in the genus Reichardia involving heterochromatin, rDNA and genome size changes during descending dysploidy.
Pollen grain dimensions
The mean values with maximal and minimal measures of polar axis (P) and equatorial diameter E are presented in Table 6. Pollen size decreases with the reduction of the basic chromosome number from E = 40.52 and 38.28 μm for two species with x = 9 to 31.72 μm for species with x = 7.
Table 6. Comparison among data concerning genome size, total length of diploid chromosome set, pollen grain dimensions and Giemsa C-bands.
| Species | 2n | 2C DNA (pg) | TKL1 in μm [23] | Pollen size (μm) [10] | Number of Giemsa C-bands [23] | |
|---|---|---|---|---|---|---|
| E2 | P3 | |||||
| R. dichotoma | 18 | 5.27 | 67.92 (0.35)4 | 40.52 (0.55) 44–395 | 35.60 (0.50) 38–34 | 6 |
| R. macrophylla | 18 | 5.15 | 83.32 (0.55) | 38.28 (0.53) 40–36 | 32.12 (0.63)37-30 | 18 |
| R. albanica | 18 | 5.22 | 56.60 (0.27) | - | - | - |
| R. tingitana | 16 | 3.37 | 55.32 (0.40) | 33.16 (0.42) 35–32 | 28.16 (0.43) 30–27 | 24 |
| R. gaditana | 16 | 3.40 | 51.54 (0.35) | 34.84 (0.39) 36–33 | 30.04 (0.37) 31–28 | 16 |
| R. picroides | 14 | 2.92 | 51.70 (0.30) | 31.72 (0.45) 34–30 | 26.72 (0.44) 30–25 | 16 |
1TKL = total karyotype length (2n);
2E = pollen equatorial diameter;
3P = pollen polar axis;
4standard deviation;
5Max—Min of E and P.
Molecular phylogeny, ancestral character states reconstruction and trait correlation
The phylogenetic tree resulting from the Bayesian analysis of the ITS dataset is presented in Fig 3. It shows the taxa sharing a common chromosome number clustered in well supported clades, with the clade containing the species with x = 9 recovered as sister to the two other clades of x = 8 and x = 7. This topology is compatible with decreasing dysploidy, which was confirmed by the reconstruction of ancestral chromosome numbers (Fig 3). Since ancestral genome size values inferred using Bayesian and parsimony reconstruction methods were similar, we presented here only the results obtained with Bayesian method (Fig 3). From the ancestral state reconstructions, it is noticeable that chromosome number and genome size apparently evolved in parallel. In this sense, a positive and significant relationship between the somatic chromosome number (2n) and the holoploid (2C) genome size was supported by the phylogenetic generalised least squares (PGLS) regression analyses (Table 7), regardless of whether the phylogenetic signal was taken into account (pBM = 0.0164) or not (pPagel = 0.0145). The positive correlation detected between 2n and 35S (pPagel = 0.0464), P (pPagel = 0.0284) and E (pPagel = 0.0194) loses its significance when considering the phylogenetic signal (Table 7).
Fig 3. Majority-rule consensus phylogeny of post-burn trees of Reichardia obtained through Bayesian analysis of the ITS dataset, plotted on geographic map and showing reconstruction of ancestral genome size and chromosome number.
Posterior probabilities are indicated on branches. Values in boxes represent the ancestral genome sizes and their corresponding variances. Dots on the map depict the origin of the sequenced samples. Data on the presence of Reichardia species across Mediterranean countries were retrieved from Euro+Med [7] for R. gaditana, from Blamey and Grey-Wilson [4] for R. intermedia and for R. macrophylla and R. albanica from Conti et al. [1].
Table 7. Phylogenetic generalised least squares (PGLS) regression statistics between somatic chromosome number (2n) and other cytogenetic and pollen traits.
| Pagel (λ = 0) | Brownian motion (λ = 1) | |||
|---|---|---|---|---|
| Slope | p | Slope | p | |
| CMA | -0.1122 | 0.4345 | -0.2765 | 0.1402 |
| 35S | 0.2326 | 0.0464* | 0.1966 | 0.0683 |
| 5S | 0.0925 | 0.5793 | 0.022 | 0.7728 |
| TKL | 0.4116 | 0.0864 | 0.3334 | 0.2017 |
| 2C | 0.3649 | 0.0145* | 0.3987 | 0.0164* |
| P | 0.8672 | 0.0284* | 0.6651 | 0.1171 |
| E | 0.9659 | 0.0194* | 0.7851 | 0.0946 |
| GC | 4.7031 | 0.2132 | 2.2225 | 0.3742 |
| AS | 1.2811 | 0.2472 | 0.2703 | 0.6165 |
| R | -0.128 | 0.5787 | -0.3145 | 0.4849 |
CMA: chromomycin-positive regions; 35S: 35S rDNA loci; 5S: 5S rDNA loci; TKL: total karyotype length; 2C: holoploid genome size; P: pollen grain polar axis; E: pollen grain equatorial diameter; GC: percentage of GC bases; AS: chromosome asymmetry index; R: chromosome arm ratio. Significance: *P < 0.05.
All our results are summarized on a phylogenetic framework and presented in Fig 4.
Fig 4. Karyological, cytogenetic and pollen traits plotted on the Reichardia phylogeny.
Posterior probabilities are indicated on branches.
Discussion
Reduction of basic chromosome number—Descending dysploidy
According to Watanabe’s Index to Plant Chromosome Numbers in Asteraceae (http://www.lib.kobe-u.ac.jp/infolib/meta_pub/G0000003asteraceaeresult-en, accessed August 22nd, 2016) [73], the chromosome number (always indicating a diploid level) has already been reported for R. dichotoma (2n = 18), R. macrophylla (2n = 18), R. albanica (2n = 18), R. tingitana (2n = 16), R. gaditana (2n = 16) and R. picroides and R. intermedia (2n = 14). A few discordant counts, most probably due to plant misidentifications, are also reported.
The basic numbers reported in the five species studied here support the hypothesis of decreasing dysploidy (i.e. progressive reduction of the basic chromosome number) as the most likely mechanism driving the karyotypic evolution in Reichardia. Several findings presented here provide further evidence of a reduction of chromosome number, such as:
The basic number x = 9 is generally considered as ancestral in the Asteraceae family [74, 75, 76, 77] and in the Cichorieae tribe [78]. According to Semple and Watanabe [77], x = 10 was the ancestral basic number out of the two dominant numbers in the subfamily Cichorioideae, x = 10 and x = 9; as for the tribe Cichorieae, which Reichardia belongs to, only x = 9 is indicated as the main basic number by these authors.
In the genus Reichardia, the three species with x = 9 are tertiary relicts, their distribution areas being restricted to habitats known as refugia for Tertiary flora [2, 3]. Consequently, the ancestral features are expected in these species.
Reichardia picroides, which presents the lowest basic number (x = 7), has a modern distribution area (the whole Mediterranean basin). The low value of its asymmetry index (As) may therefore reflect acquired symmetry or secondary symmetry [23] of the karyotype, resulting from rearrangements during chromosome number reduction.
The two species with the biggest base chromosome number and genome size (R. dichotoma and R. macrophylla) are exclusively perennial, whereas the others are perennial, biennial or annual. In the sister groups of Reichardia (Launaea, Sonchus), annual, biennial and perennial taxa exist as well.
The inference of ancestral chromosome numbers confirms the descending direction of dysploidy (Fig 3).
Changes in heterochromatin pattern and ribosomal genes mapping
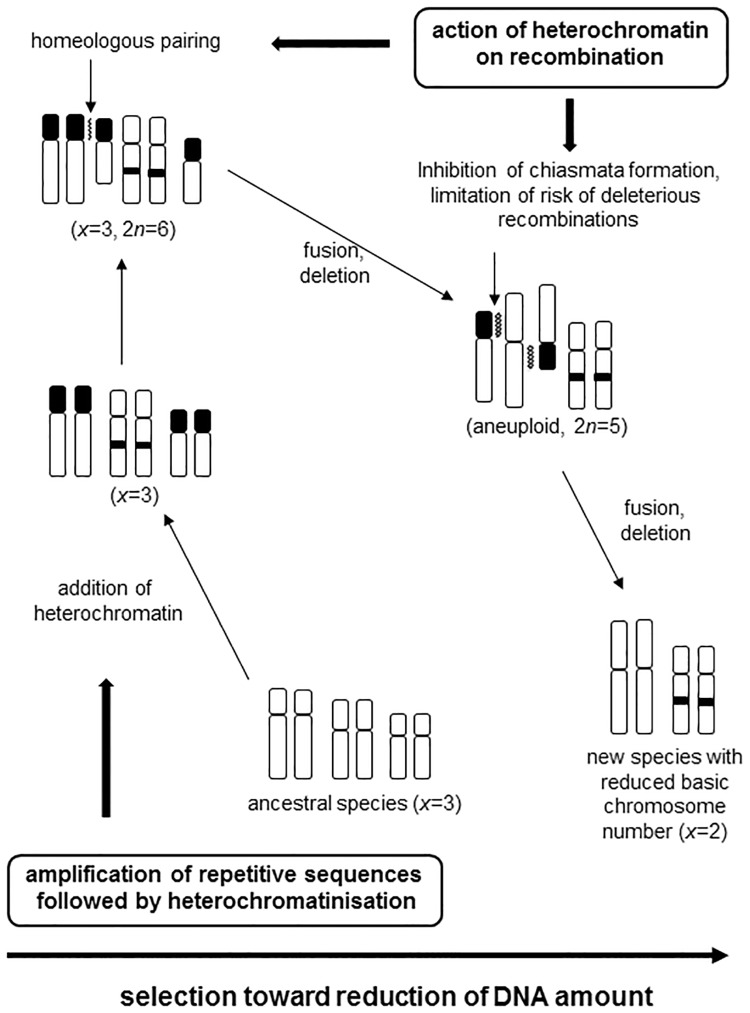
The role of heterochromatin chromosomal restructuring during reduction of the chromosome number and decreases in DNA content was revealed for Reichardia in the present study. One general hypothetical schema of this evolutionary process is proposed (Fig 5).
Fig 5. Hypothetical schema of the implication of heterochromatin in chromosomal restructuring during reduction of the basic chromosome number and decrease of DNA content.
Evolution by decreasing dysploidy requires a transitory homeologous state (Fig 5). The probability of a chromosome rearrangement is relatively low. Thus, it is unlikely that both chromosomes in a pair are subject to the same change at the same time. This homeologous state is generally considered as a deleterious state (heterozygous disadvantage). The genetic models which describe this kind of transition frequently involve the role of population structuring [79]. The karyotype polymorphism and the high frequency of homeologous karyotypes in R. macrophylla, an endemic species with a fragmented distribution area [10, 12], may be considered as arguments supporting this hypothesis.
Another evolutionary pattern in the karyotype of the genus Reichardia are the position (terminal or intercalary, or both) and the number of secondary constrictions (SC)—the diploid set of chromosomes in R. dichotoma presents six SC, but only four have been observed in R. macrophylla, R. tingitana and R. gaditana, with two in R. picroides and R. intermedia (Fig 2).
Neutral (unspecific), GC and AT-rich heterochromatin distribution
In our previous studies [10, 23], Giemsa C-banding revealed different distribution patterns of constitutive heterochromatin for five Reichardia species (Table 6). An increase in heterochromatin was observed in R. tingitana and R. gaditana and a decrease was observed in R. picroides. In the latter, heterochromatin is present only in centromeric regions and satellites, while other species also possess intercalary and terminal heterochromatic bands. DAPI counterstaining after FISH, in which there is a denaturation of the DNA as during C-banding technique, demonstrates the constitutive heterochromatin. Conventional DAPI staining without denaturation reveals regions of DNA rich in AT bases. Thus, the DAPI staining after FISH essentially confirmed C-banding data, revealing a different distribution of constitutive heterochromatin for each Reichardia species. Most of the C-bands were DAPI positive except those associated with NORs and some intercalary bands which were GC-rich and CMA+ (Table 5; Fig 1G, 1G’ and 1H). C-bands in SC (NORs) were always CMA positive and DAPI negative. Presence of unspecific or GC-rich heterochromatin and SCs fragility facilitates chromosome breakages at these sites and favors restructuring or rearrangement of the chromosomes. During this process the loss of heterochromatin blocks (entire chromosome arms in R. gaditana and R. tingitana) and SCs contribute to the reduction of chromosome number and genome size (Fig 5).
Changes in number, position and organisation of 35 and 5S rRNA genes
The particular organisation of overlapping 5S and 35S rRNA genes in R. dichotoma and R. tingitana has already been observed in numerous plants [80, 81], and for certain genera this colocalisation is the predominant pattern of rRNA genes, as is the case of Artemisia. In this Asteraceae genus, the colocalisation was first observed at cytological level [82, 83, 84] and then validated by molecular techniques [85]. Our cytological observations for two Reichardia species (R. dichotoma and R. tingitana) should be also verified by the DNA fibre mapping technique and by molecular methods, which we plan to use in future investigation of these taxa.
In R. dichotoma, 35S and 5S rRNA genes are colocalised in intercalary SC (Fig 1A’), while those in R. macrophylla are separated on different chromosome pairs (Fig 1B’). This disposition could be explained by a break in the intercalary SC of R. dichotoma followed by a translocation and inversion on two other chromosomes in the terminal position in R. macrophylla (Fig 2). In R. tingitana, 35S and 5S colocalised (Fig 1C’) while in R. gaditana these two rDNA families were located on the same chromosome arm of pair V (another 35S locus is located in chromosome pair VIII) (Fig 1F’) and separated on different chromosomes pairs in R. picroides (Fig 1G”). Changes in organisation and position of rRNA genes in Reichardia species were also followed by the reduction of the number of 35S and 5S loci from 3 to 1 per diploid genome. All these changes indicate substantial restructuring during dysploidy, suggesting that the process occurred over a long period of time.
Genome downsizing and reduction of total chromosome length and pollen size with decreasing dysploidy
Genome size and pollen size (E and P) were closely correlated in this dysploid series (Table 3). The most important genome downsizing was observed between the species with 2n = 18 and 2n = 16. Pollen size decreases perceptibly with the reduction of the basic chromosome number. Whereas the relationship between polyploidy and pollen size has been abundantly reported [86, 87, 88], it is, to our knowledge, the first time that the correlation between pollen size and dysploidy has been established. However, the positive correlation detected between 2n and pollen size loses its significance when considering the phylogenetic signal, suggesting that it could rather reflect a shared evolutionary history. Further analyses on an extended sampling of dysploid lineages are necessary to could shed light on the relation between dysploidy and pollen size.
Genome downsizing and the cell cycle: Evolutionary forces at genomic level
Fundamental properties of the cell cycle are modified by variations in the DNA amount [89, 90, 91]. For example, rapid cell division is needed to facilitate a short life cycle, for which a small nuclear DNA amount is favoured [92, 93]. By this means, natural selection is acting on the DNA amount (at the genomic level); this process can be identified as a main evolutionary force determining the pattern of heterochromatin content. This trend is verified in Reichardia, where R. dichotoma, R. albanica and R. macrophylla, the three species with the highest DNA amount are perennial, while the others species show a tendency toward reduced genome size and shorter life cycle.
The increase in the heterochromatic content of the two intermediate species (R. tingitana and R. gaditana) in an overall context of reduction in genome size, rDNA loci number and shorter life cycle could appear paradoxical. However, the expansion of heterochromatin area, while resulting from purely molecular processes involving the amplification of certain types of tandemly repeated sequences [94], also multiplies chromosomic regions particularly sensitive to chromosome breakages and as such favours genome restructuring that can led to genome size decrease.
Concluding remarks
Reichardia constitutes a model genus for studies on genome evolution, since it presents three basic chromosome numbers for only ca. 10 taxa. This study has shown that descending dysploidy was coupled with a high genomic dynamism involving decrease in genome size, changes in heterochromatin pattern, and modifications of the location and organisation of ribosomal genes. By facilitating translocations, and especially the centric fusions frequently observed during descending dysploidy, chromosome breakage in heterochromatin area was highlighted as an important contributor to genome restructuring. In Reichardia, dysploidy is accompanied with pollen size reduction, a trend that should be further addressed in an extended taxonomic sampling.
Acknowledgments
We thank all our colleagues quoted in Table 1 for helping us in material collection. Odile Robin, Mike Bourge, Spencer Brown, Paula Bonaventura and Daniel Vitales are thanked for technical assistance in cytogenetic, flow cytometric and molecular phylogenetic experiments, and two anonymous reviewers for helpful comments on a previous version of the manuscript. Alastair Plant is thanked for the revision of the English text. The supports of CNRS (France) and of projects 2014SGR514 (Catalan government) and CGL2013-49097-C2-2-P (Spanish government) were also thanked.
Appendix
GenBank accession numbers: Reichardia dichotoma: MF114121; Reichardia macrophylla: MF114123; Reichardia albanica: MF114126; Reichardia gaditana: MF114122; Reichardia tingitana: MF114119; Reichardia crystallina: MF114125; Reichardia ligulata: MF114118; Reichardia intermedia: MF114124; Reichardia picroides: MF114120.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by CNRS France; Catalan government project n° 2014SGR514 and the Spanish government project n° CGL2013-49097-C2-2-P. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Conti F, Niketić M, Vukojičić S, Siljak-Yakovlev S, Barina Z, Lakušić D. A new species of Reichardia (Asteraceae, Cichorieae) from Albania and reevaluation of R. macrophylla. Phytotaxa. 2015; 236: 121–134. http://dx.doi.org/10.11646/phytotaxa.236.2.2 [Google Scholar]
- 2.Ritter-Studnicka H. Flora i vegetacija na dolomitima Bosne i Hercegovine. God Biol Inst Sarajevo. 1957; 10: 129–161. [Google Scholar]
- 3.Ritter-Studnicka H. Reliktgesellschaften auf Dolomitböden in Bosnien und der Hercegovina. Vegetatio. 1967; 15: 190–212. doi: 10.1007/BF01963748 [Google Scholar]
- 4.Blamey M, Grey-Wilson C. Flores silvestres del Mediterráneo. Barcelona: Omega; ISBN 978-84-282-1450-6; 2008. [Google Scholar]
- 5.Gallego MJ. Estudio cariológico de las especies españolas del género Reichardia Roth (Compositae). Lagascalia. 1980; 9: 149–158. [Google Scholar]
- 6.Ovchinnikov PN. Flora i rastitel’nost ushchel’ya reki Varzob. K problem osvoeniya biologicheskikh resursov Pamiro-Altaya. Instit. Botan. Izdatel Nauk. Leningrad, 1971.
- 7.Euro+Med PlantBase—the information resource for Euro-Mediterranean plant diversity. Published on the Internet http://ww2.bgbm.org/EuroPlusMed/ [22/08/2016]; 2006-.
- 8.Nazarova AE. K kariologii podsem. Cichorioideae Kitam. sem. Asteraceae. Biol Z Arm. 1968; 21: 93–98. [Google Scholar]
- 9.Siljak S. I. O. P. B. chromosome numbers reports LVII. Taxon. 1977; 26: 447–448. [Google Scholar]
- 10.Siljak-Yakovlev S. Etude cytogénétique et palynologique de Compositae endémiques ou reliques de la flore yougoslave. Thèse d'Etat, Université Paris XI, Orsay; 1986.
- 11.Sell PD. Reichardia In: Flora Europaea (Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA, eds.). Cambridge, United Kingdom, Cambridge University Press, v. 4, 325; 1976. [Google Scholar]
- 12.Siljak-Yakovlev S. Analyse comparative des caryotypes de deux espèces du genre Reichardia Roth. (R. macrophylla Vis et Pancic et R. picroides (L.) Roth) et leur relation taxonomique. Caryologia. 1981; 34: 267–274. doi: 10.1080/00087114.1981.10796891 [Google Scholar]
- 13.Swift H. The constancy of deoxyribose nucleic acid in plant nuclei. P Natl Acad Sci-Biol. 1950; 36: 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greilhuber J, Doležel J, Lysák MA, Bennett MD. The origin, evolution and propos estabilization of the terms ‘Genome size’ and ‘C-value’ to describe nuclear DNA contents. Ann Bot-London. 2005; 95: 255–260. doi: 10.1093/aob/mci019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: progress, problems, prospects. Ann Bot-London. 2005; 95: 45–90. doi: 10.1093/aob/mci003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallès J, Canela MÁ, Garcia S, Hidalgo O, Pellicer J, Sánchez-Jiménez I. et al. Genome size variation and evolution in the family Asteraceae. Caryologia. 2013; 66: 221–235. doi: 10.1080/00087114.2013.829690 [Google Scholar]
- 17.Knight CA, Clancy RA, Götzenberger L, Dann L, Beaulieu JM. On the Relationship between Pollen Size and Genome Size. J Bot. 2010; Article ID 612017, 7 pages. doi: 10.1155/2010/612017 [Google Scholar]
- 18.Peruzzi L. “x” is not a bias, but a number with real biological significance. Plant Biosyst. 2013; 147: 1238–1241. [Google Scholar]
- 19.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005; 8: 135–141. doi: 10.1016/j.pbi.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 20.Moraes AP, Souza-Chies T, Stiehl-Alves EM, Piccolli P, Eggers L, Siljak-Yakovlev S. et al. Evolutionary trends in Iridaceae: new cytogenetic findings from the New World. Bot J Linn Soc. 2015; 177: 27–49. doi: 10.1111/boj.12232 [Google Scholar]
- 21.Mas de Xaxars G, Garnatje T, Pellicer J, Siljak-Yakovlev S, Vallès J, Garcia S. Impact of dysploidy and polyploidy on the diversification of high mountain Artemisia (Asteraceae) and allies. Alpine Bot. 2016; 126:35–48. doi: 10.1007/s00035-015-0159-x [Google Scholar]
- 22.Siljak-Yakovlev S. Chromosome number reports. Taxon. 1982; 31: 768. [Google Scholar]
- 23.Siljak-Yakovlev S. La dysploïdie et l’évolution du caryotype. Bocconea. 1996; 5: 211–220. [Google Scholar]
- 24.Wichman HA, Payne CT, Ryder OA, Hamilton MJ, Maltbie M, Baker RJ. Genomic distribution of heterochromatic sequences in equids: implications to rapid chromosomal evolution. J Hered. 1991; 82: 369–377. [DOI] [PubMed] [Google Scholar]
- 25.Dimitri P, Junakovic N. Revising the selfish DNA hypothesis: new evidence on accumulation of transposable elements in heterochromatin. Trends Genet. 1999; 15:123–4. [DOI] [PubMed] [Google Scholar]
- 26.Grewal SIS, Songtao J. Heterochromatin revisited. Nat Rev Gen. 2007; 8: 35–46. doi: 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- 27.Faulkner GJ, Carninci P. Altruistic functions for selfish DNA. Cell Cycle. 2009; 8: 2895–2900. doi: 10.4161/cc.8.18.9536 [DOI] [PubMed] [Google Scholar]
- 28.Brutlag DL. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1981; 14: 121–144. doi: 10.1146/annurev.ge.14.120180.001005 [DOI] [PubMed] [Google Scholar]
- 29.Stupar RM, Song J, Tek AL, Cheng Z, Dong F, Jiang J. Highly Condensed Potato Pericentromeric Heterochromatin Contains rDNA-Related Tandem Repeats. Genetics. 2002; 162: 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanrobay E, Thomas M, Tatout Ch. Heterochromatin positioning and nuclear architecture. In: Evans DE, Graumann K, Bryant JA (eds.) Plant nuclear structure, genome architecture and gene regulation. Wiley-Backwell. Ann Plant Rev. 2013; 46: 157–190. [Google Scholar]
- 31.Marks GE. The Giemsa staining of centromeres of Nigella damascena. J Cell Sci. 1975; 18: 19–25. [DOI] [PubMed] [Google Scholar]
- 32.Schweizer D. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma (Berl.). 1976; 58: 307–324. [DOI] [PubMed] [Google Scholar]
- 33.Siljak-Yakovlev S, Cerbah M, Coulaud J, Stoian V, Brown SC, Jelenic S. et al. Nuclear DNA content, base composition, heterochromatin and rDNA in Picea omorika and Picea abies. Theor Appl Genet. 2002; 104: 505–512. doi: 10.1007/s001220100755 [DOI] [PubMed] [Google Scholar]
- 34.Schweizer D, Strehl S, Hagemann S. Plant repetitive DNA elements and chromosome structure. Chromosomes Today. 1989; 10: 33–43. [Google Scholar]
- 35.Miklos GLG, Nankivell RN. Telomeric satellite DNA function in regulating recombination. Chromosoma. 1976; 56: 143–167. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto M, Miklos GLG. Genetic studies on heterochromatin in Drosophila melanogaster and their implications for the function DNA. Chromosoma. 1978; 66: 71–98. [DOI] [PubMed] [Google Scholar]
- 37.Navas-Castillo J, Cabrero J.. Camacho JPM. Chiasma redistribution in presence of supernumerary chromosome segments in grasshoppers: dependence on the size of the extra segment. Heredity. 1987; 58: 409–412. doi: 10.1038/hdy.1987.69 [Google Scholar]
- 38.de la Torre J, Torroja E, Gosálvez J, López-Fernández C. A model for quantifying genetic recombination in chromosome polymorphisms due to supernumerary heterochromatic segments. Heredity. 1987; 58: 345–349. doi: 10.1038/hdy.1987.61 [DOI] [PubMed] [Google Scholar]
- 39.Ellermeier Ch, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G. et al. RNAi and heterochromatin repress centromeric meiotic recombination. P Natl Acad Sci-Biol. 2010; 107: 8701–8705. doi: 10.1073/pnas.0914160107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.John B. The biology of heterochromatin In: Heterochromatin (Verma RS, ed.). Cambridge, United Kingdon: Cambridge University Press; 1988. pp. 1–147. [Google Scholar]
- 41.Schubert I, Wobus U. In-situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma. 1985; 92: 143–148. doi: 10.1007/BF00328466 [Google Scholar]
- 42.Raina SN, Mukai Y. Genomic in situ hybridization in Arachis (Fabaceae) identifies the diploid wild progenitors of cultivated (A. hypogaea) and related wild (A. monticola) peanut species. Pl Syst Evol. 1999; 214: 251–262. doi: 10.1007/BF00985743. [Google Scholar]
- 43.Raskina O, Belyayev A, Nevo E. Quantum speciation in Aegilops: molecular cytogenetic evidence from rDNA cluster variability in natural populations. Proc Natl Acad Sci U S A. 2004; 101: 14818–14823. doi: 10.1073/pnas.0405817101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Datson PM, Murray BG. Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosome Res. 2006; 14: 845–857. doi: 10.1007/s10577-006-1092-z [DOI] [PubMed] [Google Scholar]
- 45.Hidalgo O, Garcia-Jacas N, Garnatje N, Romaschenko K, Susanna A, Siljak-Yakovlev S. Extreme environmental conditions and phylogenetic inheritance: systematics of Myopordon and Oligochaeta (Asteraceae, Cardueae-Centaureinae). Taxon. 2008; 57: 769–778. [Google Scholar]
- 46.Muratovic E, Robin O, Bogunic F, Soljan D, Siljak-Yakovlev S. Karyotype Evolution and Speciation of European Lilies from Lilium sect. Liriotypus. Taxon. 2010; 59: 165–175. [Google Scholar]
- 47.Garnatje T, Hidalgo O, Vitales D, Pellicer J, Vallès J, Robin O. et al. Swarm of terminal 35S in Cheirolophus (Asteraceae, Centaureinae). Genome. 2012; 55, 529–535. doi: 10.1139/g2012-041 [DOI] [PubMed] [Google Scholar]
- 48.Siljak-Yakovlev S, Pustahija F, Vicic V, Robin O. Molecular cytogenetics (FISH and fluorochrome banding): resolving species relationships and genome organization In: Methods in Molecular Biology (Besse P, ed.). Clifton, NJ, USA: Springer-Verlag, v. 1115, 389–323; 2014. [DOI] [PubMed] [Google Scholar]
- 49.Marie D, Brown SC. A cytometric exercise in plant DNA histograms, with 2C values of 70 species. Biol Cell. 1993; 78: 41–51. doi: 10.1016/0248-4900(93)90113-S [DOI] [PubMed] [Google Scholar]
- 50.Galbraith D, Harkins K, Maddox J, Ayres N, Sharma D, Firoozabady E. Rapid flow cytometry analysis of the cell cycle in intact plant tissue. Science. 1983; 220: 1049–1051. doi: 10.1126/science.220.4601.1049 [DOI] [PubMed] [Google Scholar]
- 51.Godelle B, Cartier D, Marie D, Brown SC, Siljak-Yakovlev S. Heterochromatin study demonstrating the non-linearity of fluorometry useful for calculating genomic base composition. Cytometry. 1993; 14: 618–626. doi: 10.1002/cyto.990140606 [DOI] [PubMed] [Google Scholar]
- 52.Geber G, Schweizer D. Cytochemical heterochromatin differentiation in Sinapis alba (Cruciferae) using a simple air-drying technique for producing chromosome spreads. Plant Syst Evol. 1988; 158: 97–106. doi: 10.1007/BF00936336 [Google Scholar]
- 53.Heslop-Harrison JS, Schwarzacher T, Anamthawat-Jonsson K, Leitch IJ. In situ hybridization with automated chromosome denaturation techniques. Methods Cell. Mol. Biol. 1991; 3: 109–116. [Google Scholar]
- 54.Erdtman G. Pollen Morphology and Plant Taxonomy Angiosperms (An Introduction to Palynology). Stockholm, Sweden: Almqvist & Wiksel; 1952. [Google Scholar]
- 55.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987; 19:11–15. [Google Scholar]
- 56.Soltis DE, Soltis PS, Collier TG, Edgerton ML. Chloroplast DNA variation within and among the Heuchera group (Saxifragaceae); evidence for chloroplast transfer and paraphyly. Amer J Bot. 1991; 78:1091–1112. [Google Scholar]
- 57.Nickrent DL, Schuette KP, Starr EM. A molecular phylogeny of Arceuthobium (Viscaceae) based on nuclear ribosomal DNA internal transcribed spacer sequences. Am J Bot. 1994; 81:1149–1160. [Google Scholar]
- 58.White T J, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: PCR Protocols: A Guide to Methods and Applications (Innis MA, Gelfand DH, Sninsky JJ, White TH, eds.). New York: Academic Press, Inc; 1990. Pp. 315–322. [Google Scholar]
- 59.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike Information Criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004; 53: 793–808. doi: 10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- 60.Nylander JAA. MrModeltest, version 2. Program distributed by the author Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- 61.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S. et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012. May; 61(3): 539–542. doi: 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim SC, Crawford DJ, Jansen RK. Phylogenetic relationships among the genera of the subtribe Sonchinae (Asteraceae): evidence from ITS sequences. Syst Bot. 1996; 21: 417–432. doi: 10.2307/2419668 [Google Scholar]
- 63.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things). Method Ecol Evol. 2012; 3: 217–223.23467194 [Google Scholar]
- 64.Maddison WP, Maddison DR. Mesquite: A modular system for evolutionary analysis, version 3.04 2015; http://mesquiteproject.org [Google Scholar]
- 65.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004; 20: 289–290. doi: 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 66.Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1–122. http://CRAN.R-project.org/package=nlme; 2015.
- 67.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 68.Doležel J, Bartoš J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry. 2003; 51: 127–128. [DOI] [PubMed] [Google Scholar]
- 69.Siljak-Yakovlev S, Pustahija F, Šolić EM, Bogunić F, Muratović E, Bašić N. et al. Towards a database of genome size and chromosome number of Balkan flora: C-values in 343 taxa with novel values for 252. Adv Sci Lett (U.S.A.). 2010; 3:190–213. [Google Scholar]
- 70.Siljak-Yakovlev S, Cerbah M, Zoldoš V, Godelle B. Heterochromatin and rDNA organisation and evolution in the genus Reichardia. Cytogenet Cell Genet. 1998; 81: 114. [Google Scholar]
- 71.Garcia S, Hidalgo O, Jakovljević I, Siljak-Yakovlev S., Vigo J., Garnatje T. et al. New data on genome size in 128 Asteraceae species and subspecies, with first assessments for 40 genera, three tribes and two subfamilies. Plant Biosyst. 2013; 147: 1219–1227. doi: 10.1080/11263504.2013.863811 [Google Scholar]
- 72.Suda J, Kyncl T, Freiová R. Nuclear DNA amounts in Macaronesian angiosperms. Ann Bot-London. 2003; 92: 153–164. doi: 10.1093/aob/mcg104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watanabe K. Index to chromosome numbers in the Asteraceae. http://www-asteraceae.cla.kobe-u.ac.jp/index.html. 2013; Accessed August 2016.
- 74.Solbrig TO, Anderson CL, Kyhos DW, Raven PH, Rudenberger L. Chromosome numbers in Compositae V. Asteraceae II. Am J Bot. 1964; 51: 513–519. [Google Scholar]
- 75.Stucky J, Jackson RC. DNA content of seven species of Asteraceae and its significance to theories of chromosome evolution in the tribe. Am J Bot. 1975; 62: 509–518. [Google Scholar]
- 76.Solbrig TO. Chromosomal cytology and evolution in the family Compositae In: The biology and Chemistry of Compositae. (Heywood VH, Harborne JB, Turner BL, eds.). London, United Kingdom: Academic Press, v. I, 267–281; 1977. [Google Scholar]
- 77.Semple JC, Watanabe K. A review of chromosome numbers in Asteraceae with hypotheses on chromosomal base number evolution In: Systematics, evolution and biogeography of Compositae (Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds.). Vienna, Austria: International Association of Plant Taxonomy; 2009. pp. 61–72. [Google Scholar]
- 78.Stebbins GL. A new classification of the tribe Cichorieae, family Compositae. Madroño. 1953; 12: 65–81. [Google Scholar]
- 79.Lande R. The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity. 1985; 54: 323–332. [DOI] [PubMed] [Google Scholar]
- 80.Zoldos V, Muratovic E, Bogunic F, Birus I, Robin O, Horvat T. et al. Anatomic, cytogenetic and molecular variations between non-serpentine and serpentine populations of endemic Lilium bosniacum. Chromosome Res. 2007; 15: 38. [Google Scholar]
- 81.Siljak-Yakovlev S, Temunovic M, Robin O, Raquin Ch, Frascaria-Lacoste N. Genome size and physical mapping of rDNA and G-C rich heterochromatin in three European Fraxinus species. Tree Genet Genomes. 2013; 10: 231–239. [Google Scholar]
- 82.Torrell M, Cerbah M, Siljak-Yakovlev S, Vallès J. Etude cytogénétique de trois taxons du complexe d’Artemisia campestris L. (Asteraceae, Anthemideae): localisation de l’hétérochromatine et de l’ADN ribosomique. Bocconea. 2001; 13: 623–628. [Google Scholar]
- 83.Torrell M, Cerbah M, Siljak-Yakovlev S, Vallès J. Molecular cytogenetics of the genus Artemisia (Asteraceae, Anthemideae): fluorochrome banding and fluorescence in situ hybridization. I. Subgenus Seriphidium and related taxa. Plant Syst Evol. 2003; 239: 141–153. doi: 10.1007/s00606-002-0259-0 [Google Scholar]
- 84.Garcia S, Garnatje T, Hidalgo O, McArthur ED, Siljak-Yakovlev S, Vallès J. Extensive ribosomal DNA (35S and 5S) colocalization in the North American endemic sagebrushes (subgenus Tridentatae, Artemisia, Asteraceae) revealed by FISH. Plant Syst Evol. 2007; 267: 79–92. doi: 10.1007/s00606-007-0558-6 [Google Scholar]
- 85.Garcia S, Lim KY, Chester M, Garnatje T, Pellicer J, Vallès JA. et al. Linkage of 35 S and 5 S rRNA genes in Artemisia (family Asteraceae): first evidence from angiosperms. Chromosoma. 2009; 118:85–97. doi: 10.1007/s00412-008-0179-z [DOI] [PubMed] [Google Scholar]
- 86.Brochmann C. Pollen and seed morphology of Nordic Draba (Brassicaceae): phylogenetic and ecological implications. Nord J Bot. 1992; 12: 657–673 doi: 10.1111/j.1756-1051.1992.tb01843.x [Google Scholar]
- 87.Oliveira VM, Forni-Martins ER, Magalhães PM, Alves MN. Chromosomal and morphological studies of diploid and polyploid cytotypes of Stevia rebaudiana (Bertoni) Bertoni (Eupatorieae, Asteraceae). Genet Mol Biol. 2004; 27: 215–222. [Google Scholar]
- 88.Marinho RC, Mendes-Rodrigues C, Bonetti AM, Oliveira PE. Pollen and stomata morphometrics and polyploidy in Eriotheca (Malvaceae-Bombacoideae). Plant Biology. 2014; 16:508–511. doi: 10.1111/plb.12135 [DOI] [PubMed] [Google Scholar]
- 89.Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. P Roy Soc Lond B Bio. 1972; 178: 259–275. doi: 10.1098/rspb.1972.0042 [DOI] [PubMed] [Google Scholar]
- 90.Price HJ. Evolution of DNA content in higher plants. Bot Rev. 1976; 42: 27–52. [Google Scholar]
- 91.Rees H, Narayan RKJ. Chromosomal DNA in higher plants. Philos T Roy Soc B. 1981; 292: 569–578. doi: 10.1098/rstb.1981.0051 [Google Scholar]
- 92.Jones RN, Brown LM. Chromosome evolution and DNA variation in Crepis. Heredity. 36: 1976; 91–104. doi: 10.1038/hdy.1976.10 [Google Scholar]
- 93.Labani RM, Elklington TT. Nuclear DNA variation in the genus Allium L. (Liliaceae). Heredity. 1987; 59: 119–128. doi: 10.1038/hdy.1987.103 [Google Scholar]
- 94.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994; 371: 215–220. doi: 10.1038/371215a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.