Abstract
Voltage-gated potassium channels of the KCNQ (Kv7) subfamily are essential for control of cellular excitability and repolarization in a wide range of cell types. Recently, we and others found that some KCNQ channels functionally and physically interact with sodium-dependent solute transporters, including myo-inositol transporters SMIT1 and SMIT2, potentially facilitating various modes of channel-transporter signal integration. In contrast to indirect effects such as channel regulation by SMIT-transported, myo-inositol-derived phosphatidylinositol 4,5-bisphosphate (PIP2), the mechanisms and functional consequences of the physical interaction of channels with transporters have been little studied. Here, using co-immunoprecipitation with different channel domains, we found that SMIT1 binds to the KCNQ2 pore module. We next tested the effects of SMIT1 co-expression, in the absence of extracellular myo-inositol or other SMIT1 substrates, on fundamental functional attributes of KCNQ2, KCNQ2/3, KCNQ1, and KCNQ1-KCNE1 channels. Without exception, SMIT1 altered KCNQ ion selectivity, sensitivity to extracellular K+, and pharmacology, consistent with an impact on conformation of the KCNQ pore. SMIT1 also altered the gating kinetics and/or voltage dependence of KCNQ2, KCNQ2/3, and KCNQ1-KCNE1. In contrast, SMIT1 had no effect on Kv1.1 (KCNA1) gating, ion selectivity, or pharmacology. We conclude that, independent of its transport activity and indirect regulatory mechanisms involving inositol-derived increases in PIP2, SMIT1, and likely other related sodium-dependent solute transporters, regulates KCNQ channel ion selectivity, gating, and pharmacology by direct physical interaction with the pore module.
Introduction
Voltage-gated potassium (Kv) channel α-subunits of the KCNQ (Kv7) gene family serve a variety of functions in excitable and non-excitable cells. In mammalian neurons, heteromeric KCNQ2/3 (Kv7.2/Kv7.3) complexes generate a non-inactivating, muscarinic-inhibited Kv current (M-current) that raises the threshold for neuronal activation (1). Kv channel α-subunits, including those in the KCNQ family, also form complexes with single transmembrane domain ancillary (β) subunits encoded by the KCNE gene family, leading to various channel subunit compositions with diverse functional characteristics. One such example is found in the human heart, where KCNQ1 (Kv7.1) forms complexes with KCNE1 to generate the slow-activating, non-inactivating IKs current important for ventricular repolarization (2, 3). KCNQ1-KCNE2, in contrast, forms a constitutively active K+ current in parietal cells, thyroid cells, and the choroid plexus epithelium (4, 5, 6).
Inherited sequence variants in human KCNQ2 or KCNQ3 genes encoding loss-of-function mutations in KCNQ2 and KCNQ3 channel proteins cause epilepsy syndromes such as benign familial neonatal convulsions, illustrating the importance of the M-current in maintaining normal neuronal excitability (7). In addition, gain-of-function mutations in human KCNQ2 can cause nonepileptic myoclonus, hypoventilation at birth (associated with central hypoventilation rather than seizure activity), and hypomyelination (8). In mice, Kcnq2 deletion also results in hypoventilation, which is severe enough to cause the pups to die within minutes after birth. This is strikingly reminiscent of the phenotype of mice with germline deletion of Slc5a3, which encodes the sodium-coupled myo-inositol transporter, SMIT1; mice lacking SMIT1 exhibit depleted brain myo-inositol and central apnea, and die shortly after birth from hypoventilation (9). These findings suggest the possibility of a molecular mechanistic link between hypoventilation caused by disruption of KCNQ2 and that caused by disruption of SMIT1.
Accordingly, we previously found that KCNQ1-KCNE2 and KCNQ2/3 channels can form complexes with SMIT1 and SMIT2 to form “chansporter” complexes involving reciprocal regulation of both partners. In the case of KCNQ1-KCNE2-SMIT1, deletion of Kcne2 in mice resulted in dysregulation of cerebrospinal fluid myo-inositol and increased seizure susceptibility (10). KCNQ2/3-SMIT1/2 chansporter complexes protected SMIT1 activity from the inhibitory effects of cellular depolarization, and KCNQ2 was required for potentiation of SMIT activity in the presence of myo-inositol in vitro (11).
Phosphatidylinositol 4,5-bisphosphate (PIP2), a phosphoinositide critical to the regulation of ion channels including KCNQ2/3 (12) and KCNQ1 (13, 14) is generated from myo-inositol, the primary substrate of SMIT1. Myo-inositol is an important osmolyte that can be transported into cells by SMIT1 to facilitate osmoregulation—an activity that in turn raises intracellular PIP2 levels. Because the resulting PIP2 alters gating of ion channels, including KCNQ2/3, negative-shifting their activation such that they open at more negative potentials, SMIT1 can influence both osmoregulation and electrical excitability (15).
Because of binding to the actin cytoskeleton, membrane-proximal PIP2 diffuses relatively slowly, with a reported diffusion coefficient of 0.0004 μm2/s. Disruption of the actin cytoskeleton using cytochalasin D permits much faster PIP2 diffusion away from the site of its production (16). Accordingly, we previously found that cytoskeletal disruption to allow free diffusion of PIP2 attenuated KCNQ2/3 currents in cells co-transfected with KCNQ2/3 and SMIT1 (but not those transfected with KCNQ2/3 alone), presumably by negating the effects of co-expressed SMIT1-mediated myo-inositol uptake, which could otherwise ensure locally high PIP2 levels (11). This suggested close KCNQ-SMIT juxtaposition, an hypothesis supported by co-localization and/or co-immunoprecipitation of KCNQ1-SMIT1 and KCNQ2/3-SMIT1/2 complexes (10, 11). However, the channel domains required for transporter interaction, and the functional consequences of physical interaction on fundamental channel properties have not been previously elucidated.
Here, we address this gap in knowledge, identifying the KCNQ2 domain required for SMIT1 interaction and uncovering unexpected effects of SMIT1 on fundamental functional attributes of KCNQ1-3 potassium channels. Importantly, to ensure that we were studying the consequences upon channel function of channel-transporter physical interaction, and not the effects of PIP2 generated from transported myo-inositol, we omitted myo-inositol and other SMIT1 substrates from the extracellular solution. In addition, key findings were recapitulated in the presence of a SMIT1 inhibitor, phlorizin, in the bath solution. Thus, in this study, we demonstrate that physical interaction with SMIT1, even in the absence of SMIT1 transport activity, exerts marked effects on KCNQ channel ion selectivity, gating, and pharmacology.
Materials and Methods
KCNQ2 fragment generation
KCNQ2-YFP and KCNQ2-GFP, kind gifts from Dr. Naoto Hoshi (University of California, Irvine), were modified by restriction digest/polymerase chain reaction (PCR) amplification to create (1–97)-green fluorescent protein (GFP), GFP-(321–852), and (1–549)-yellow fluorescent protein (YFP). KCNQ2(1–224)-YFP was constructed from KCNQ2(1–549)-YFP by excising the undesired fragments using a single AccI digest and re-ligating with T4 DNA ligase. KCNQ2(222–323)-YFP was constructed using KCNQ2(1–549)-YFP as a template and later as a backbone as well. The target fragment was amplified by PCR and added restriction sites for EcoRI and KpnI using the primer pair 5′-GAATTCCTGGGATACGTGGTCTACGCTCACA-3′ and 5′-GGGTACCCTGCTCTTGGACTTTCA-3′. PCR products were re-amplified with the primer pairs 5′-GGATCCACCGGTCCCACCATGGTGAG-3′ and 5′-CTCACCATGGTGGGACCGGTGGATCC-3′ to improve the 5′ Kozak sequence. PCR products and backbone (KCNQ2(1–549)-YFP) were then digested with EcoRI and KpnI, and then ligated using T4 DNA ligase before transformation into DH5α cells and subsequent purification by Maxiprep (Qiagen, Hilden, Germany). KCNQ2(G279S) was made from full-length KCNQ2 in pCDNA3 using the QuikChange mutagenesis kit (Agilent, Santa Clara, CA) with primers 5′-AGATCTCGAGCTCAAGCTTCGCCRCCATGGGATCGGTGGTCTACGCTC-3′ and 5′-GAGCGTAGACCACCGATCCCATGGYGGCGAAGCTTGAGCTCGAGATCT-3′. The mutated plasmids were transformed and purified as per QuikChange kit instructions.
Protein biochemistry
CHO cells were cultured, transfected, and lysed as reported in our previous article (11). In brief, cells were transfected using Mirus LT-1 transfection reagent with a total of 15 μg cDNA per 10 cm plate and allowed 36–48 h expression at 37°C before lysis. Lysis buffer was composed of 1% IGEPAL, 0.1% sodium dodecyl sulfate, 50 mM Tris (pH 8.0), 150 mM NaCl, and a protease-inhibitor cocktail tablet (Thermo Fisher Scientific, Waltham, MA). Total protein was quantified by bicinchoninic acid assay (Thermo Fisher Scientific). Proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidine fluoride membranes for immunoblotting with the following antibodies, as noted: KCNQ2 (Santa Cruz Biotechnology, Santa Cruz, CA), DDK (Origene, Rockville, MD), FLAG (Sigma-Aldrich, St. Louis, MO), SMIT1 (Santa Cruz, CA), YFP (Santa Cruz), GFP (Santa Cruz; Rockland Immunochemicals, Pottstown, PA). For secondary detection, horseradish-peroxidase -conjugated antibodies (BioRad, Hercules, CA) were used in conjugation with Luminata Forte horseradish peroxidase substrate (Millipore, Billerica, MA). Imaging was performed using Syngene (Frederick, MA) Gbox hardware and software. For co-immunoprecipitations (co-IPs), all samples were first pre-cleared of non-specific interaction by incubating the total lysate with protein A/G PLUS-coated agarose beads (Santa Cruz) for 1 h. Beads were then pelleted and discarded. Total protein was quantified by bicinchoninic acid assay, as above. Immunoprecipitating antibodies were then added at a dilution of 1:100 for overnight pulldown at 4°C. The next day, antibody-antigen complexes were pulled down with fresh protein A/G PLUS agarose beads.
Channel subunit cRNA preparation and Xenopus laevis oocyte injection
Complementary RNA (cRNA) transcripts encoding human KCNQ1, KCNQ2, KCNQ3, KCNA1, KCNE1, KCNE3, or SMIT1 were generated by in vitro transcription using the T7 polymerase mMessage mMachine kit (Thermo Fisher Scientific), after vector linearization, from cDNA sub-cloned into plasmids incorporating Xenopus laevis β-globin 5′ and 3′ untranslated regions flanking the coding region to enhance translation and cRNA stability. cRNA was quantified by spectrophotometry. Defolliculated stage V and VI Xenopus laevis oocytes (Ecocyte Bioscience, Austin, TX) were injected with Kv channel α-subunit cRNAs (1–10 ng), alone or with KCNEx cRNA (2 ng) and/or SMIT1 cRNA (10 ng). Oocytes were incubated at 16°C in Bath’s saline solution (Ecocyte) containing penicillin and streptomycin, with daily washing, for 3–5 days before two-electrode voltage-clamp (TEVC) recording.
TEVC
TEVC recording was performed at room temperature with an OC-725C amplifier (Warner Instruments, Hamden, CT) and pClamp8 software (Molecular Devices, Sunnyvale, CA) 43–5 days after cRNA injection, as described in the previous section. Oocytes were placed in a small-volume oocyte bath (Warner Instruments) and viewed with a dissection microscope. The bath solution was (in mM) 96 NaCl, 4 KCl, 1 MgCl2, 1 CaCl2, and 10 HEPES (pH 7.6). Pipettes were of 1–2 MΩ resistance when filled with 3 M KCl. Currents were recorded in response to pulses between −80 and +40 mV at 20 mV intervals, or to a single pulse to +40 mV from a holding potential of −80 mV, to yield current-voltage relationships and current magnitude and to quantify the activation rate. TEVC data analysis was performed with Clampfit (Molecular Devices) and Origin 6.1 (OriginLab; Northampton, MA) software; values are stated as the mean ± SE. Normalized tail currents were plotted versus pre-pulse voltage and fitted with a single Boltzmann function:
| (1) |
where g is the normalized tail conductance, A1 is the initial value at −∞, A2 is the final value at +∞, V1/2 is the half-maximal voltage of activation, and Vs is the slope factor. Where informative, currents were compared with one another using one-way analysis of variance (ANOVA) to assess statistical significance (p < 0.05). If multiple comparisons were performed, a post hoc Tukey’s honest significant difference test was performed after ANOVA.
Relative permeability calculations
According to the Goldman-Hodgkin-Katz (GHK) voltage equation,
| (2) |
where Erev is the absolute reversal potential and P is permeability. This permits calculation of the relative permeability of each ion if concentrations on either side of the membrane are known. A modified version of this equation was used here to determine the relative permeability of two ions in a system in which only the extracellular ion concentration was known. Thus, relative permeability of Rb+, Cs+, and Na+ compared to K+ ions was calculated for all channels by plotting the I/V relationships for each channel with each extracellular ion (100 mM) and comparing them to that with 100 mM extracellular K+ ion to yield a change in reversal potential (ΔErev) for each ion compared to that of K+. Permeability ratios for each ion (X) compared to K+ were then calculated as
| (3) |
Values were compared between channel types and statistical significance was assessed using ANOVA.
Calculating the effect of extracellular potassium on current ratios
The GHK flux equation calculates the theoretical reduction in current as a function of extracellular potassium concentration:
| (4) |
where is the ionic flux of potassium, I is the current, A is the membrane area, is the permeability of potassium, is the membrane potential, F is Faraday’s number, R is the gas constant, T is the absolute temperature, and [K+]i and [K+]o are the intra- and extra-cellular concentrations of potassium, respectively. Using the values for [K+]i in Xenopus laevis oocytes from Weber (17), and assuming that the channel in question is an ideal extracellular potassium-independent Kv channel, the theoretical ratio of the currents measured at two different concentrations of [K+]o can be calculated using the “independence relation equation”:
| (5) |
Data presentation and statistics
Data were compared using Bonferroni’s correction or a post hoc Tukey’s honest significant difference test (GraphPad Prism and Microsoft Excel). Error bars on figures indicate the mean ± SE.
Results
SMIT1 binds to the KCNQ2 pore module
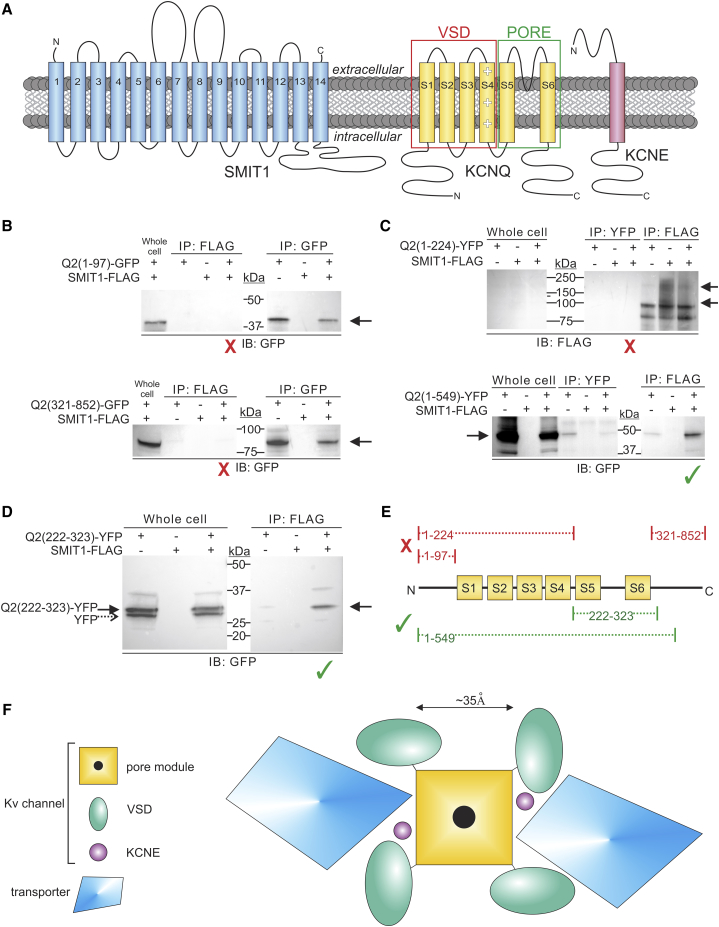
We previously discovered KCNQ2-SMIT1 complex formation (11). The SMIT1 sequence suggests as many as 14 transmembrane domains, whereas Kv α-subunits such as KCNQ2 each have six, and KCNE subunits one (Fig. 1 A).
Figure 1.
The KCNQ2 pore module is required for binding to SMIT1. (A) Topologies of SMIT1 (putative), KCNQ α-subunits, and KCNE (experimentally confirmed). (B–E). CHO cells were transfected with KCNQ2 fragments and/or SMIT1-FLAG, as indicated. All blots are each representative of n = 2 experiments. The immunoprecipitating (IP) antibodies are labeled FLAG, YFP, or GFP, and the immunoblot (IB) antibodies are labeled GFP or FLAG. The red “X” denotes fragments that did not bind to SMIT1, and the green check mark signifies SMIT1-binding fragments. (B) Cytosolic Q2 fragments. (Top) The N-terminal fragment KCNQ2(1–97) does not bind to SMIT1-FLAG. (Bottom) The C-terminal fragment KCNQ2 (321–852) does not bind to SMIT1-FLAG. (C) Transmembrane Q2 fragments. (Top) The N-terminus through the S4-S5 linker fragment (KCNQ2(1–224)) does not bind to SMIT1. (Bottom) Extension of the previous fragment to include the pore-forming S5-S6 region (KCNQ2(1–549)) enables binding to SMIT1-FLAG. (D) The S5–S6 pore-forming region fragment (KCNQ2(222–323)) binds to SMIT1-FLAG. (E) Summary of binding results. (F) Cartoon view from the extracellular side comparing the Kv channel and solute transporter approximate dimensions and hypothetical docking configurations.
Here, to identify specific KCNQ2 domains involved in interaction, we generated and expressed KCNQ2 fragments tagged with either YFP or GFP in Chinese hamster ovary (CHO) cells and tested for interaction with co-expressed SMIT1-FLAG co-IPs and western blotting. Cytosolic N- and C-terminal fragments of KCNQ2 contain known protein-binding sites (18), yet neither the N-terminal (1–97) nor the C-terminal (321–852) fragments demonstrated interaction with SMIT1 (Fig. 1 B). Similarly, the KCNQ2(1–224) construct, which includes the entire KCNQ2 N-terminus and voltage-sensor domain through the S4-S5 linker, also failed to co-IP with SMIT1 (Fig. 1 C, upper). In contrast, KCNQ2(1–549), starting at the N-terminus and ending midway in the cytosolic C-terminus, interacted with SMIT1 (Fig. 1 C, lower). By process of elimination, addition of the pore module (S5-S6) appeared to restore KCNQ2 interaction with SMIT1. To verify this, we generated the KCNQ2(222–323) fragment, which encodes only the pore module, from the S4-S5 linker through the first few amino acids of the C-terminus. As predicted, the 222–323 construct interacted with SMIT1 (Fig. 1 D), confirming that the KCNQ2 pore module was necessary and sufficient for SMIT1 binding. An additional, lower-molecular-weight protein (dashed arrow) was observed in the lysate Western blot (Fig. 1 D). As this protein was detectable with anti-GFP antibody, was at the correct migration distance for YFP, and did not co-IP with the anti-FLAG antibody, we suggest that it is YFP expressed without the Q2 fragment, owing to the presence of a comparatively strong Kozak consensus sequence in front of the YFP tag, 3′ of the Q2 fragment. It is also feasible that the lower migration distance arose from altered post-translational processing; in any case, unlike the full-length YFP-tagged KCNQ2(222–323) protein (Fig. 1 D, solid arrow), the lower-molecular-weight form did not co-IP along with the transporter. The results are summarized in Fig. 1 E.
Examining approximate dimensions from previous high-resolution structures of Kv channels (19, 20) and vSGLT (21), a prokaryotic ortholog of Na+-coupled solute transporters such as SMIT1, it is evident that a sufficiently large docking site for SMIT1 to access the KCNQ2 pore could exist between neighboring KCNQ2 voltage-sensor domains. In this configuration, channel-transporter interaction might influence, or be influenced by, associated KCNE subunits, which also sit in this putative docking site, in a pocket between the pore module and the voltage-sensor domain ((22); Fig. 1 F). One would predict that transporter docking in this position to access the pore module would affect fundamental channel properties such as gating and also, potentially, pore conformation and pharmacology, which we therefore tested extensively, as described below.
SMIT1 modifies KCNQ2/3 voltage dependence and ion selectivity
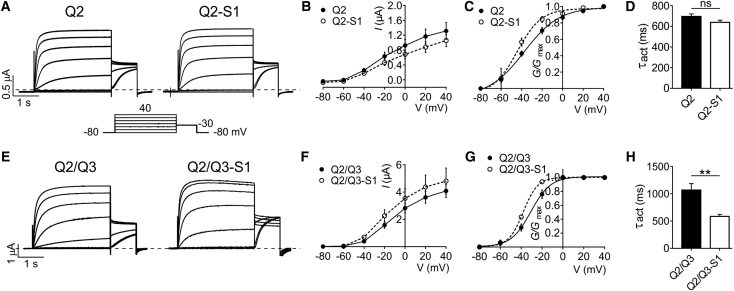
We and others previously found that after several hours of pre-incubation in myo-inositol (to provide a substrate for PIP2 production), SMIT1 co-expression left-shifted the voltage dependence of KCNQ2/3 activation (e.g., by −21.5 mV) (11, 15). In this study, we conducted all electrophysiological analyses in the absence of known extracellular SMIT1 substrates such as myo-inositol to isolate the effects of physical interaction of SMIT1 with KCNQ channels in the absence of SMIT1 transport activity. Under these conditions, SMIT1 left-shifted the voltage dependence of KCNQ2 by −6.7 mV and increased the slope by 6.2 mV (Fig. 2, A–C), with negligible effects on activation rate (Fig. 2 D).
Figure 2.
SMIT1 negative-shifts and speeds KCNQ2/3 activation. (A) Representative traces from TEVC recordings of Xenopus laevis oocytes injected with cRNA encoding KCNQ2, alone or co-injected with cRNA encoding SMIT1; n = 29–32. (B) Mean current-voltage relationship for subunit combinations as in (A); n = 29–32. Error bars indicate the mean ± SE. (C) Mean normalized tail current (at −30 mV) versus pre-pulse potential for oocytes as in (A); n = 29–32. Error bars indicate the mean ± SE. (D) Mean activation rate reported as τ of a single-exponential fit at −40 mV for the traces in (A); n = 29–32. Error bars indicate the mean ± SE; ns = p > 0.05. (E) Representative traces from TEVC recordings of Xenopus laevis oocytes injected with cRNA encoding KCNQ2/KCNQ3, alone or co-injected with cRNA encoding SMIT1; n = 10–13. (F) Mean current-voltage relationship for subunit combinations as in (E); n = 10–13. Error bars indicate the mean ± SE. (G) Mean normalized tail current (at −30 mV) versus pre-pulse potential for oocytes as in (D); n = 10–13. Error bars indicate the mean ± SE. (H) Mean activation rate reported as τ of a single exponential fit at −40 mV for traces in (E); n = 10–13. Error bars indicate the mean ± SE; ∗∗p < 0.01.
SMIT1 also left-shifted KCNQ2/3 activation voltage dependence (by −7.2 mV) and increased the slope by 2.5 mV (Fig. 2, E–G). In addition, SMIT1 speeded KCNQ2/3 activation rate (twofold at −40 mV) (Fig. 2 H; see Table S1 for summary).
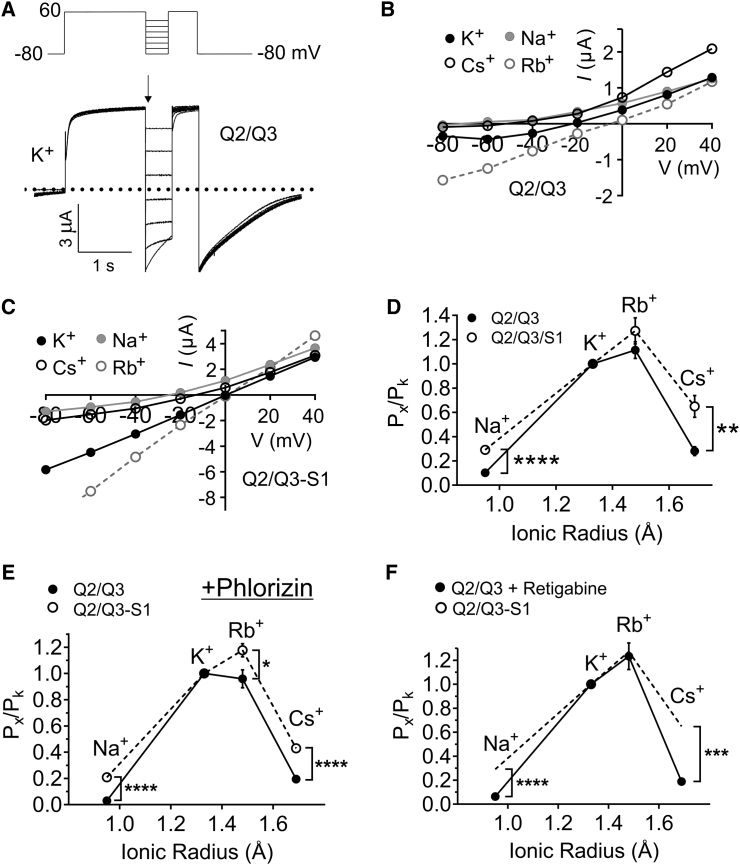
Because we found that SMIT1 co-assembles with the KCNQ2 pore module (Fig. 1), we also determined SMIT1 effects on KCNQ2/3 ion selectivity, changes in which are a potential consequence of conformational changes in the pore, near or at the selectivity filter. We exploited shifts in reversal potential via pseudo-bi-ionic substitution with monovalent cations to estimate permeability ratios for KCNQ2/3 and KCNQ2/3-SMIT1 using a modification of the GHK voltage equation (Eq. 3) and a voltage protocol in which, after depolarization to +60 mV for 1 s to achieve a uniformly high channel-open probability (Po), currents were measured during the early phase of a subsequent voltage “family” step to provide an I/V relationship for each ion essentially isolated from voltage-dependent differences in Po (Fig. 3 A).
Figure 3.
SMIT1 alters KCNQ2/Q3 ion selectivity. (A) Representative traces from TEVC recordings of Xenopus laevis oocytes injected with cRNA encoding KCNQ2/3 in 100 mM K+ (inset shows the voltage protocol: 1 s pulse to 60 mV followed by a 500 ms inactivation recovery step to voltages between −80 and 40 mV, then a 500 ms −80 mV tail pulse). (B) Mean current-voltage relationship for KCNQ2/3 alone in 100 mM K+ (black circles), Cs+ (open circles), and Na+ (gray circles); n = 12. (C) Mean current-voltage relationship for KCNQ2/3 in combination with SMIT1 100 mM K+ (black circles), Cs+ (open circles), and Na+ (gray circles); n = 14. (D) Estimated mean permeability relative to that of K+ versus ionic radius (Pauling) for Na+, Rb+, and Cs+ through KCNQ2/Q3 (solid circles) and KCNQ2/Q3 + SMIT1 (open circles, dotted lines) channels; n = 12–14. Error bars indicate the mean ± SE. The statistical significance for comparison of the relative permeability of Na+ was ∗∗∗∗p < 0.0001, and that for Cs+ was ∗∗p < 0.05. Values for relative permeability ratios, in the order Na+, Rb+, and Cs+, were 0.10, 1.1, and 0.28 for KCNQ2/Q3 and 0.29, 1.27, and 0.65 for KCNQ2/Q3 + SMIT1. (E) Estimated mean permeability relative to that of K+ versus ionic radius (Pauling) for Na+, Rb+, and Cs+ through KCNQ2/Q3 (solid circles) and KCNQ2/Q3 + SMIT1 (open circles, dotted lines) channels in phlorizin (500 μM); n = 13–14. Error bars indicate the mean ± SE. Statistical significance for comparison of the relative permeability of Na+ was ∗∗∗∗p < 0.0001 and that for Cs+ was ∗∗∗∗p < 0.0001. Values for relative permeability ratios, in the order Na+, Rb+, and Cs+, were 0.03, 0.96, and 0.19 for KCNQ2/Q3 and 0.21, 1.2, and 0.43 for KCNQ2/Q3 + SMIT1. (F) Estimated mean permeability relative to that of K+ versus ionic radius (Pauling) for Na+, Rb+, and Cs+ through KCNQ2/Q3 (solid circles) in retigabine (10 μM) and KCNQ2/Q3 + SMIT1 without retigabine (dotted lines; data taken from (D); n = 8–14. Error bars indicate the mean ± SE. Statistical significance for comparison of the relative permeability of Na+ was ∗∗∗∗p < 0.0001 and that for Cs+ was ∗∗∗p < 0.0002. Values for relative permeability ratios, in the order Na+, Rb+, and Cs+, were 0.06, 1.23, and 0.12 for KCNQ2/Q and 0.21, 1.2, and 0.43 for KCNQ2/Q3 + SMIT1,.
The permeability series for both KCNQ2/3 and KCNQ2/3-SMIT1 favored the order Rb+, K+, Cs+, Na+. However, the mean permeability of KCNQ2/3-SMIT1 for Cs+ relative to K+ was 0.65 ± 0.09 (n = 14), compared to 0.28 ± 0.04 (n = 12) for KCNQ2/3, whereas the mean permeability of KCNQ2/3/SMIT1 for Na+ relative to K+ was 0.30 ± 0.03 (n = 14), compared to 0.12 ± 0.02 (n = 12) for KCNQ2/3—a 132% (p < 0.05) and 141% (p < 0.0001) increase in permeability, respectively (Fig. 3, B–D). The data suggest that although the pore remains relatively selective for Rb+ and K+ over other monovalent cations, SMIT1 binding is exerting enough of a perturbation in the KCNQ2/3 selectivity filter or other pore elements to produce a relative increase in the permeability of Na+ and Cs+.
The absence of myo-inositol (or any other substrates of SMIT1) in the bath solution in all experiments in this study ensured that effects of SMIT1 on KCNQ channel properties were not arising from myo-inositol uptake increasing intracellular PIP2. However, as an additional control, in a different batch of oocytes, we compared the effects of SMIT1 on KCNQ2/3 channel ion selectivity in the absence of extracellular myo-inositol but in the presence of phlorizin, a competitive inhibitor of SMIT1, at a concentration (500 μM) chosen to inhibit SMIT1 transport activity by >90% (10). In the presence of phlorizin (Figs. 3 E and S1), SMIT1 altered the ion selectivity of KCNQ2/3 in the same manner as observed in the absence of phlorizin (Fig. 3 D), further reinforcing our hypothesis that physical interaction with SMIT1, without the necessity for SMIT1 transport activity, alters fundamental pore properties of KCNQ2/3.
SMIT1 effects on KCNQ2/3 gating and ion selectivity are independent of one another
Because SMIT1 co-expression produced a negative shift in the voltage dependence of KCNQ2/3 activation (Fig. 2 G), we examined whether this effect alone could explain the altered ion selectivity described above. Retigabine, a KCNQ2/3 channel opener and anti-seizure medication, negatively shifts KCNQ2/3 voltage dependence to an even greater degree (−20 mV at 10 μM) than that achieved by SMIT1 co-expression (23, 24). We therefore compared the relative ion permeability of KCNQ2/3 channels (measured using 100 mM bath concentration of each ion tested) in the presence of 10 μM bath retigabine (Fig. S2), and plotted the permeability series against non-retigabine-treated KCNQ2/3-SMIT1 complexes (Fig. 3 F). Although retigabine increased the KCNQ2/3 Rb+ permeability relative to K+ conductance, similar to the effects of SMIT1 co-expression, retigabine did not increase relative Na+ or Cs+ permeability, suggesting that the most dramatic effects of SMIT1 co-expression on KCNQ2/3 ion selectivity did not arise simply from a negative shift in open probability.
KCNQ1 channels are notable for their higher permeability of Rb+ compared to K+ ((25); Fig. S10), but KCNQ2/3 channel Rb+ permeability was previously reported as lower than that of K+ when using 15 mM concentrations of the permeating ions (26). As that does not tally with our data for KCNQ2/3 alone using 100 mM bath Rb+ or K+, we repeated our experiments using 15 mM bath concentrations of permeating ions. These studies recapitulated both the previous report of the KCNQ2/3 selectivity series using 15 mM instead of 100 mM permeant ions (K+ > Rb+ > Cs+ > Na+) (26) and our findings herein that SMIT1 co-expression, in the absence of its substrate, substantially increases Na+ and Cs+ permeation, relative to K+ permeation, of KCNQ2/3 channels (Fig. S3).
These data indicate that SMIT1 alters KCNQ2/3 ion selectivity and pore occupancy, effects that can in themselves alter ion channel gating, for instance, by occupying a “gating site” to interfere with ion channel deactivation (27). We therefore next tested the influence of altering pore occupancy on SMIT1 effects on KCNQ2/3 gating by quantifying G/V relationships in the presence of 100 mM bath K+ instead of the 4 mM K+ we used above (Fig. 2 G). The increased extracellular K+ shifted KCNQ2/3 V1/2 activation by ∼−20 mV independent of SMIT1 co-expression, thus preserving the ∼7 mV negative shift in KCNQ2/3 V1/2 activation caused by SMIT1 co-expression (Fig. S4; Table S1). This suggests that SMIT1 alters KCNQ2/3 gating independent of SMIT1 effects on KCNQ2/3 ion occupancy.
SMIT1 modifies KCNQ2/3 pharmacology
The pharmacology of Kv channel α-subunits is in some cases sensitive to the various types of non-pore (β) subunits with which Kv channel α-subunits co-assemble (28, 29). Here, we found that SMIT1 alters the sensitivity of KCNQ2/3 channels to the canonical KCNQ family channel antagonist, XE991. The altered pharmacology was most apparent at +40 mV, at which membrane potential SMIT1 co-expression increased by 2.5-fold the degree of KCNQ2/3 inhibition by 50 μM XE991 (Fig. S5, A and B).
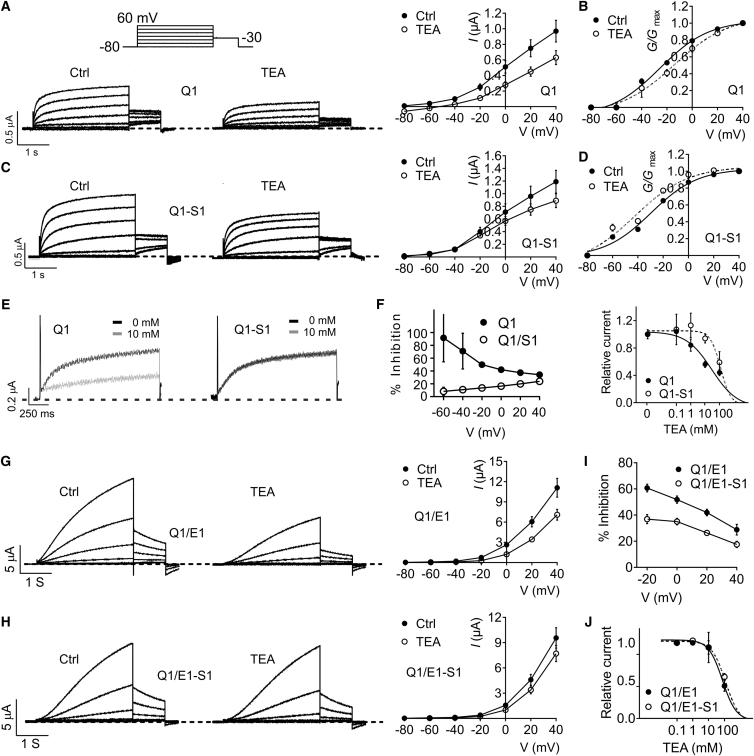
Different KCNQ channel family members exhibit contrasting sensitivity to the classic open-channel Kv channel blocker, tetraethylammonium (TEA) (30). Here, we found that SMIT1 exerted a protective effect against TEA block of KCNQ2/3 (Fig. S5, C and D). The differential effects of SMIT1 on voltage dependence of KCNQ2/3 block by XE991 versus TEA are illustrated in Fig. S5, E and F. When measured at 0 mV, SMIT1 decreased KCNQ2/3 TEA sensitivity fourfold, shifting the IC50 from 6.9 to 28.6 mM (Fig. S5 G).
SMIT1 does not alter Kv1.1 gating, ion selectivity, or TEA sensitivity
As a control for possible non-specific effects of SMIT1 co-expression on Kv channel function in general, we next quantified effects of SMIT1 co-expression on functional properties of Kv1.1 (KCNA1), a Kv channel we previously found not to regulate SMIT1 activity when the two are co-expressed (11). Here, we found that SMIT1 had no effects on Kv1.1 I/V or G/V relationships (Fig. S6), ion selectivity (Fig. S7), or TEA sensitivity (Fig. S8) (see also Table S1). Thus, SMIT1 effects on KCNQ2/3 function and pharmacology are not overexpression artifacts or other artifacts applicable to all Kv channels in general.
SMIT1 modifies the voltage dependence and ion selectivity of KCNQ1-based channels
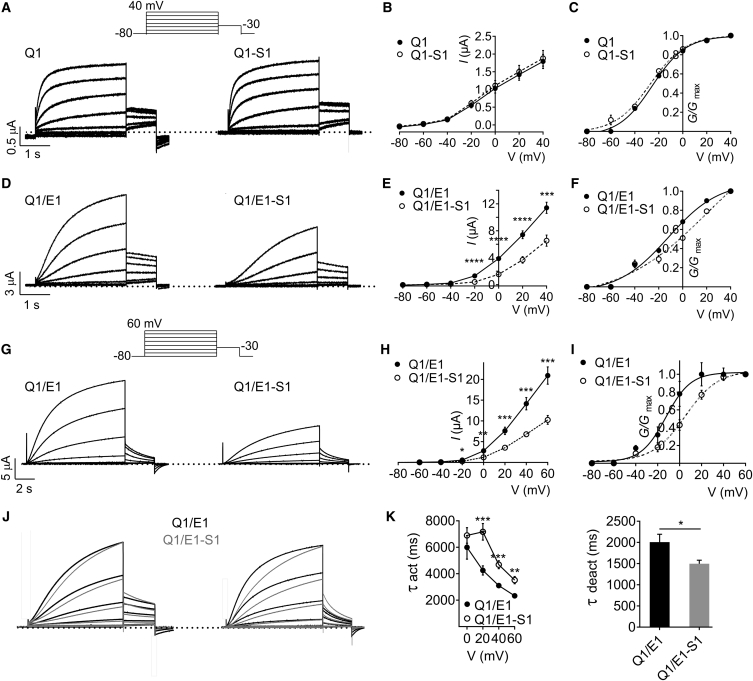
KCNQ1 is a close relative of KCNQ2 and KCNQ3, yet it exhibits markedly different tissue expression, pharmacology, and greater functional flexibility when co-assembled with KCNE β-subunits (31). We previously showed that KCNQ1 forms complexes with SMIT1 in vivo and in vitro, but did not pursue a systematic examination of the effects of SMIT1 on KCNQ1 pore properties (10). Here, SMIT1 had little to no effect on KCNQ1 voltage dependence or macroscopic current amplitude (Fig. 4, A–C), although the resting membrane potential (RMP) of oocytes expressing KCNQ1-SMIT1 was on average 4 mV more depolarized than KCNQ1 alone (−48.5 ± 0.70 mV versus −52.7 ± 0.53 mV, respectively; n = 97–102, p < 0.0001).
Figure 4.
SMIT1 positive-shifts and slows KCNQ1-KCNE1 activation. (A) Representative traces from TEVC recordings of Xenopus laevis oocytes injected with cRNA encoding KCNQ1 (Q1), alone or co-injected with cRNA encoding SMIT1 (S1); n = 97–102. The inset shows the voltage protocol. (B) Mean peak current-voltage relationship for subunit combinations as in (A); n = 97–102. Error bars indicate the mean ± SE. (C) Mean normalized tail current (at −30 mV) versus pre-pulse potential for oocytes as in (A); n = 97–102. Error bars indicate the mean ± SE. (D) Representative traces from TEVC recordings of Xenopus laevis oocytes injected with cRNA encoding KCNQ1/KCNE1 (Q1/E1), alone or co-injected with cRNA encoding SMIT1 (S1); n = 90–100. The inset shows the voltage protocol. (E) Mean peak current-voltage relationship for subunit combinations as in (D); n = 90–100. Error bars indicate the mean ± SE; ∗∗∗p < 0.0001; ∗∗∗∗p < 0.00001. (F) Mean normalized tail current (at −30 mV) versus pre-pulse potential for oocytes as in (D); n = 90–100. Error bars indicate the mean ± SE. (G) Representative traces from TEVC recordings of Xenopus laevis oocytes injected with cRNA encoding KCNQ1/KCNE1 (Q1/E1), alone or co-injected with cRNA encoding SMIT1 (S1); n = 12, using longer (10 s) prepulses than in (D) and pulsing to +60 mV (voltage protocol in lower inset). (H) Mean peak current-voltage relationship for subunit combinations as in (G); n = 12. Error bars indicate the mean ± SE; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.0001. (I) Mean normalized tail current (at −30 mV) versus pre-pulse potential for oocytes as in (G); n = 12. Error bars indicate the mean ± SE. (J) Normalized, averaged traces from recordings as in (A) (left) and (G) (right), to illustrate slowed KCNQ1/KCNE1 (Q1/E1) activation due to SMIT1 (S1) co-expression; n = 12. (K) Mean activation (left) and deactivation (right) rates for traces as in (G); n = 12. Error bars indicate the mean ± SE; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
In vivo, KCNQ1 is thought to always be co-assembled with one or more types of KCNE β-subunit (10). KCNQ1 forms complexes with the KCNE1 β-subunit in human heart and inner ear, generating an unusually slow activating Kv current (2, 3). Here, using the same voltage protocol we used for homomeric KCNQ1 (with a 3 s pre-pulse), we found that SMIT1 attenuated KCNQ1-KCNE1 channel activity (by 42% at 40 mV) (Fig. 4, D and E; n = 90–100, p < 0.0001) and right-shifted the voltage dependence of KCNQ1-KCNE1 activation (Fig. 4 F). To better quantify the latter effect, we increased the pre-pulse duration to 10 s and also depolarized to +60 mV. With this protocol, we observed a twofold attenuation of KCNQ1-KCNE1 current by SMIT1 (Fig. 4, G and H), and observed a large (17 mV) positive shift in the voltage dependence of KCNQ1-KCNE1 activation arising from SMIT1 co-assembly (Fig. 4 I; n = 12).
In addition, SMIT1 co-expression caused a 9 mV depolarizing shift in the RMP of oocytes expressing KCNQ1-KCNE1 (from −57.4 ± 0.65 to −48.3 ± 1.4 mV; n = 90–100, p < 0.0001). SMIT1 also altered KCNQ1-KCNE1 gating kinetics, slowing activation and speeding deactivation, easily apparent when overlaying normalized average traces for KCNQ1/KCNE1 ± SMIT1 using the voltage protocols shown in Fig. 4, A, G, and J). SMIT1 slowed KCNQ1-KCNE1 activation by ∼75% at +20 mV and speeded deactivation by 25% at −30 mV (Fig. 4 K). See Table S1 for a summary of parameters.
In the intestine, KCNQ1 is regulated by KCNE3, with contrasting effects to those imposed by KCNE1. Thus, KCNQ1-KCNE3 complexes are constitutively active and deactivate very slowly at strongly hyperpolarized potentials, giving an almost linear I/V relationship between −120 and +40 mV (32, 33). Here, we found that SMIT1 co-expression did not alter KCNQ1-KCNE3 peak current, but negative-shifted its G/V relationship by 7 mV without altering the slope (Fig. S9). Thus, for both KCNE1 and KCNE3, SMIT1 co-expression exaggerated their opposite effects on the voltage dependence of KCNQ1 activation (Table S1).
Modulation of KCNQ1 by KCNE1 was previously investigated by comparing the ratio of Rb+ to K+ conductance (GRb/GK), a straightforward yet sensitive way to probe the conformation of the K+-channel selectivity filter (25, 34). We hypothesized that if SMIT1 was binding close to the pore of KCNQ1, then the KCNQ1 GRb/GK ratio should be affected. GRb/GK ratios for KCNQ1 and KCNQ1-KCNE1 were previously reported as >2 and <1, respectively (25). Here, KCNQ1 had a GRb/GK ratio of 4.2 (n = 12), and KCNQ1-KCNE1 a ratio of 0.96 (n = 20) (Fig. S10, A–C). In contrast, KCNQ1-SMIT1 had a GRb/GK ratio of 3.0, reduced compared to homomeric KCNQ1 (Fig. S10, B and C, n = 18, p < 0.05), whereas KCNQ1-KCNE1-SMIT1 had a GRb/GK ratio of 1.31, larger than that of KCNQ1-KCNE1 (Fig. S10, B and C, n = 16, p < 0.05).
We next further investigated effects of SMIT1 on estimated permeability ratios for KCNQ1 and KCNQ1-KCNE1 using the approach described earlier for KCNQ2/3 (see Fig. 3). The permeability series for all subunit configurations favored the order Rb+, K+, Cs+, Na+ (Fig. S11). However, the mean permeability of KCNQ1-SMIT1 for Cs+ relative to K+ was 0.42 ± 0.01 (n = 10), compared to 0.26 ± 0.01 (n = 7) for KCNQ1, a 61% increase in relative Cs+ permeability (Fig. S11, A–D). Likewise, the mean permeability of KCNQ1-KCNE1-SMIT1 for Cs+ relative to K+ was 0.25 ± 0.04 (n = 9), compared to 0.13 ± 0.01 (n = 6) for KCNQ1-KCNE1, a 92% increase in relative Cs+ permeability (Fig. S11, E–H). Together, the data suggest that SMIT1 alters the selectivity filter conformations of KCNQ1 and KCNQ1-KCNE1.
SMIT1 modifies the sensitivity of KCNQ1-based channels to extracellular potassium ions
Increasing extracellular potassium ([K+]o) results in inhibition of KCNQ1 channels via stabilization of the inactivated state, suggesting that [K+]o regulates KCNQ1 activity in addition to the more obvious effects of driving force (35). Here, we investigated whether SMIT1 alters this phenomenon. Using the independence relation equation (Eq. 5), we determined that the K+-mediated inhibition of steady-state current at +60 mV for KCNQ1 alone, and in complex with SMIT1, at concentrations from 4 to 50 mM [K+]o—plotted here alongside the theoretical ratios calculated from the GHK flux equation for comparison (Fig. S12, A and B). Although currents generated by KCNQ1 or KCNQ1-SMIT1 were each attenuated compared to the value predicted from the GHK flux equation, SMIT1 presence was partially protective across the [K+]o range evaluated (10–50 mM) (Fig. S12 B, n = 8, p < 0.0001 at 50 mM [K+]o). These results further support the hypothesis that SMIT1 influences the KCNQ1 pore conformation.
SMIT1 modifies KCNQ1 and KCNQ1-KCNE1 pharmacology
Application of 100 mM TEA to KCNQ1 resulted in inhibition across all voltage steps tested, with relatively less inhibition at higher positive voltages, arising from knockoff of TEA from its extracellularly located binding site by K+ moving outward through the KCNQ1 pore (Fig. 5, A and B). Strikingly, although 100 mM TEA block of KCNQ1 was little altered by SMIT1 at +40 mV, at more negative voltages, SMIT1 co-assembly desensitized KCNQ1 to TEA block. This reversed the polarity of the G/Gmax shifts exerted by TEA, from 11.3 mV more positive (KCNQ1 + TEA) to 13.2 mV more negative (KCNQ1-SMIT1 + TEA) (Fig. 5, A–D). The protective effect of SMIT1 progressively increased the more negative the voltage to the extent that the voltage dependence of block was reversed by SMIT1 (Fig. 5, E and F). At 0 mV, SMIT1 increased the KCNQ1 TEA IC50 fourfold, from 29.3 ± 0.1 to 121 ± 0.1 mM (Fig. 5 F).
Figure 5.
SMIT1 protects KCNQ1 from TEA block. (A) Left: Representative traces. Right: Mean current-voltage relationship, from TEVC recordings of Xenopus laevis oocytes injected with cRNA encoding KCNQ1 (Q1) in ND96 or 100 mM TEA; n = 16. The inset shows the voltage protocol: 3 s pulse to voltages between −80 and 40 mV followed by a 1 s −30 mV tail pulse. Error bars indicate the mean ± SE. (B) Mean normalized tail current (at −30 mV) versus pre-pulse potential for oocytes as in (A); n = 16. Error bars indicate the mean ± SE. (C) Left: Representative traces. Right: Mean current-voltage relationship, from TEVC recordings of Xenopus laevis oocytes expressing KCNQ1-SMIT1 (Q1–S1) in ND96 or 100 mM TEA; n = 13, using voltage protocol as in (A). Error bars indicate the mean ± SE. (D) Mean normalized tail current (at −30 mV) versus pre-pulse potential for oocytes as in (C); n = 13. Error bars indicate the mean ± SE. (E) Representative traces recorded at 0 mV for Q1 and Q1-S1 in 0 mM (black) vs. 10 mM (gray) TEA; n = 5. (F). Left: Mean % inhibition (100 mM TEA) versus voltage for KCNQ1 (Q1) with or without SMIT1 (S1); n = 5. Error bars indicate the mean ± SE. Right: TEA dose response for KCNQ1 (Q1) with or without SMIT1 (S1); n = 5. Error bars indicate the mean ± SE. (G) Left: Representative traces. right, mean current-voltage relationship, from two-electrode voltage-clamp recordings of Xenopus laevis oocytes expressing KCNQ1/KCNE1 (Q1/E1) in ND96 or 100 mM TEA; n = 6. The voltage protocol was as in (A). (H) Left: Representative traces. Right: Mean current-voltage relationship, from TEVC recordings of Xenopus laevis oocytes expressing KCNQ1/KCNE1-SMIT1 (Q1/E1-S1) in ND96 or 100 mM TEA; n = 6. Voltage protocol was as in (A). (I) Mean % inhibition (100 mM TEA) versus voltage for KCNQ1 (Q1) with or without SMIT1 (S1); n = 6. Error bars indicate the mean ± SE. (J) TEA dose response for KCNQ1-KCNE1 (Q1-E1) with or without SMIT1 (S1); n = 4–5. Error bars indicate the mean ± SE.
SMIT1 had a weaker protective effect against block of KCNQ1-KCNE1 by 100 mM TEA, and the directionality of the voltage dependence of block for KCNQ1-KCNE1 was preserved, with less TEA block at more positive voltages (Fig. 5, G–I). SMIT1 altered the KCNQ1-KCNE1 TEA IC50 at 0 mV by only 8%, from 95 ± 0.3 to 103 ± 0.1 mM (Fig. 5 J).
The adamantane compound AC-1 (2-(4-chlorophenoxy)-2-methyl-N-[5-[(methylsulfonyl)amino]tricyclo[3.3.1.13,7]dec-2-yl]-propanamide30, also known as JNJ303, was recently shown to have relatively high potency and specificity in inhibiting KCNQ1-KCNE1 complexes, yet does not inhibit homomeric KCNQ1 because JNJ303 binds within fenestrations made available when KCNE1 co-assembles with KCNQ1 (29). Here, we also found homomeric KCNQ1 to be insensitive to 1 μM JNJ303 (Fig. S13, A and B; n = 8). KCNQ1-SMIT1 currents were likewise not inhibited by 1 μM JNJ303, but, interestingly, instead exhibited a −8 mV shift in their voltage dependence of activation when exposed to 1 μM JNJ303, compared to currents recorded in normal bath solution (Fig. S13, C and D; n = 8). This resulted in a 70% increase in KCNQ1-SMIT1 currents provoked by 1 μM JNJ303, at −20 mV (Fig. S13 E). KCNE1 endowed KCNQ1 with sensitivity to JNJ303 block, as previously reported (29). Thus, KCNQ1-KCNE1 channels were inhibited by 1 μM JNJ303 (e.g., ∼50% at 0 mV) (Fig. S13 F). SMIT1 co-expression slightly and voltage-dependently altered this inhibition (Fig. S13 G), being mildly protective at positive voltages against peak current inhibition (Fig. S13 H) and slowing activation (Fig. S13, I and J) by 1 μM JNJ303 (although KCNQ1-KCNE1 currents were smaller and slower-activating at baseline in the presence of SMIT1).
Discussion
Channel-transporter complexes
KCNQ-SMIT1 complexes are part of an emerging class of channel-transporter co-assemblies (11, 36, 37, 38, 39, 40). We previously found that co-assembly of SMIT1 with KCNQ1/KCNE2 channels was important for the regulation of myo-inositol transport between the blood and cerebrospinal fluid, and that KCNQ1 upregulated SMIT1 activity in oocytes, whereas KCNQ1-KCNE2 downregulated it (10). More recently, we found that reciprocal regulation between SMIT1 or SMIT2 occurs with neuronal KCNQ2/3 channels (11). Together, these data suggested that there is direct, consequential physical contact between these unlikely partners. Here, we tested this hypothesis, after first discovering that SMIT1 physically interacts with the pore module of KCNQ2 channels, and found that this interaction results in unique biophysical and pharmacological characteristics for the KCNQ channels analyzed. Our finding that SMIT1 shifts the voltage dependence of activation of KCNQ2, KCNQ2/3, and KCNQ1/KCNE1 in the absence of externally applied myo-inositol or other SMIT1 substrates underscores the notion that channel-transporter co-assembly in and of itself can have physiologically relevant consequences for channel function, without the necessity for solute transport by the transporter. This is further supported by the changes in KCNQ2/3 and KCNQ1/KCNE1 activation rate caused by SMIT1 co-assembly (Figs. 2 and 4). These data suggest that SMIT1 interacts with the S4-S5 linker and S6 (which are both included in our KCNQ2 pore-module construct in Fig. 1), and/or with the voltage-sensor module (S1-S4) of some KCNQ channels. Although the latter interaction was not detected in our co-IPs, it could involve electrostatic interactions easily disrupted by our co-IP conditions, which are by necessity stringent to avoid false positives.
Our finding that co-assembly of SMIT1 with KCNQ1, KCNQ1/KCNE1, or KCNQ2/3 channels alters their ion selectivity strongly supports the idea that SMIT1 alters KCNQ pore conformation via close physical juxtaposition of the transporter with the channel pore. Previous examples of altered selectivity include mutation of the S4-S5 linker of the Shaker K+ channel, increasing the permeability of Rb+ relative to K+ (41). In addition, a point mutation in Orai, the plasma membrane component of the store-operated Ca2+ influx and Ca2+-release-activated Ca2+ channel, was shown to transform the Ca2+-release-activated Ca2+ channel from a Ca2+-selective inward rectifier to a monovalent cation-selective outward rectifier (42). Importantly, the fact that mutations in KCNE1 altered ion selectivity of the current it generates when expressed in Xenopus laevis oocytes (by modulating endogenous Xenopus KCNQ1) presented early evidence that KCNE1 contributes to a K+-selective pore (43).
Physiological and mechanistic implications of altered K+ channel selectivity
Altered selectivity of K+ channels is observed in pathophysiology. Hypokalemia is an electrolyte disorder caused by low-blood-serum K+ concentrations, with moderate hypokalemia (2.5–3 mM K+) leading to cardiac arrhythmias, and severe hypokalemia (<2.5 mM K+) resulting in cardiac arrest and sudden death (44). In response to hypokalaemic conditions, the RMP of cardiac cells becomes more depolarized and bi-stable and is restored to its natural hyperpolarity once physiological K+ concentrations are restored (45, 46, 47, 48, 49). This aberrant behavior has been shown to involve the two-pore-domain K+ (K2P) channel, TWIK1 (50). During hypokalemia, TWIK1 reversal potentials are more depolarized, suggesting that TWIK1 becomes more permeable to Na+ when K+ is sparse (51). Altered selectivity of TWIK1 in response to hypokalemia has been hypothesized as a way of regulating cardiac excitability in response to pathophysiological conditions (50). Likewise, the increased relative permeability of KCNQ2/3 channels induced by SMIT1 co-assembly (Fig. 3) might impact the physiology of cells in which they (or KCNQ2/3 with SMIT2) interact. We very recently found KCNQ2/3-SMIT1/2 co-localization in sciatic nerve nodes of Ranvier and in axon initial segments (11). Especially in these relatively tight confines that act as gatekeepers for nervous signaling, increased Na+ permeability of KCNQ2/3 would be expected to favor excitability, although it is also important to note that SMIT1 left-shifts the voltage dependence of KCNQ2/3 activation (Fig. 2), which would have the opposite effect, as the prevailing selectivity still favors K+.
In the case of Rb+, our oocyte-based quantification of KCNQ2/3 ion selectivity using 15 mM of permeating ions matches quite well (with or without SMIT1 co-expression) previous reports for human embryonic kidney (HEK) cells expressing KCNQ2/3 or native neuronal M-current (26, 52, 53). However, we observe higher Na+ and Cs+ permeability upon SMIT1 co-expression, regardless of ion concentration, and also in the presence of the SMIT1 inhibitor phlorizin (ensuring lack of transport activity for SMIT1, in addition to the lack of extracellular SMIT1 substrate, which also guarantees that the effects cannot arise from, e.g., transported myo-inositol raising intracellular PIP2 concentration). Interestingly, previous comparisons indicated that native rat sympathetic neuronal M-current has >2-fold higher relative Cs+ permeability than cloned KCNQ2/3 channels in HEK cells (0.2 vs. 0.086), prompting prior speculation that another regulatory subunit might be involved (26, 52, 53). Although the permeabilities we calculated from oocyte recordings do not match exactly previous reports for KCNQ2/3 channels in HEK or native neurons, it is of interest that co-expression of SMIT1 likewise doubles Cs+ permeability, recapitulating this specific mismatch between cloned KCNQ2/3 and native M-current.
The results from GRb/GK ratio experiments also suggested that SMIT1 influences pore conformation, reducing the GRb/GK ratio of KCNQ1 while increasing it for KCNQ1/KCNE1. It appears that SMIT1 may both allosterically modulate the pore of KCNQ1 and impede the ability of KCNE1 to do the same, suggesting that the interaction of SMIT1 is at a position near to that of a bound KCNE1 β-subunit. KCNE1 is thought to interact with the S4-S5 linker and S6 of KCNQ1, hindering conformational changes around these positions, thus uncoupling the movement of S4 from S6 gate opening (25). Interestingly, our protein biochemistry data suggest this same general vicinity as the binding site for SMIT1 (Fig. 1). Because of the limited data available regarding the topology and structure of SMIT1, we are not yet in a position to speculate in a meaningful manner regarding which elements of SMIT1 interact with KCNQs; neither do we yet understand the stoichiometry of SMIT1 in complexes with KCNQs. Both of these important questions, together with narrowing down specific interaction sites within the pore, are topics of ongoing and future analyses.
Mechanistic implications of altered K+ channel pharmacology
Co-assembly of SMIT1 with KCNQ1 also reduced the efficacy of external TEA and, strikingly, dampened or even reversed the voltage dependence of TEA block. The results of previous experiments and simulations agree that the major binding sites for TEA are located at the external and internal entrances to the selectivity filter (54, 55, 56) (in our case, we did not access the internal site, because TEA is relatively membrane impermeable and we only used extracellular application). Moreover, the binding efficacy of TEA is dictated by the loading state of K+ ions inside the channel at any given point (55). If, as our data strongly support, SMIT1 is allosterically modulating the pore, then the binding affinity of K+ within the selectivity filter would conceivably be altered, thus effecting how TEA binds at the external and internal interface of the KCNQ1 pore domain; SMIT1 co-assembly may also affect the TEA binding site(s) by direct physical interaction.
SMIT1 had more subtle effects on KCNQ2/3 TEA sensitivity but was nevertheless partially protective. Less is known about the mechanisms by which XE991 inhibits KCNQ2/3, but we can surmise that the dramatic effects of SMIT1 on this phenomenon are consistent with alteration of pore architecture and/or direct perturbation of the XE991 binding site (which remains to be identified) by SMIT1. JNJ303 is considered a highly selective inhibitor, only binding to fenestrations revealed in S6 of KCNQ1 brought about via interaction with KCNE1 (29). Unexpectedly, JNJ303 was an agonist of KCNQ1-SMIT1, suggesting that SMIT1 may also bind to or affect the conformation of a site in S6 similar to that occupied by KCNE1. KCNE1 protected KCNQ1 from the SMIT1-induced agonist effect of JNJ303, possibly indicating that KCNE1 shields this portion of S6 from SMIT1 influence (Fig. S13). Reciprocally, JNJ303 normalized KCNQ1-KCNE1-SMIT1 current levels and activation rate so that they were more similar to those of KCNQ1-KCNE1, suggesting that JNJ303 might interfere with the functional interaction between, or might intercalate between, KCNQ1-KCNE1 and SMIT1.
In conclusion, our findings provide extensive evidence that SMIT1 alters KCNQ function and pore conformation via direct physical interaction, without the requirement for SMIT1 transport activity. The unique biophysical and pharmacological characteristics that result from this interaction raise new questions as to the impact that chansporter complexes have upon physiology, disease, and channel-targeted therapy. Future studies will be aimed at refining our understanding of the molecular mechanisms, and sub-domains, that mediate chansporter complex formation.
Author Contributions
R.W.M., D.L.N., and G.W.A. performed experiments, analyzed data, prepared figures, and contributed to writing the manuscript.
Acknowledgments
We gratefully acknowledge the generous gift of GFP-tagged wild-type KCNQ2 cDNA from Dr. Naoto Hoshi (University of California, Irvine). This work was funded by the National Institutes of Health (GM115189 to G.W.A.) and an American Heart Association Predoctoral Fellowship (14PRE20040013 to D.L.N.).
Editor: William Kobertz.
Footnotes
Thirteen figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30739-7.
Supporting Material
References
- 1.Wang H.S., Pan Z., McKinnon D. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science. 1998;282:1890–1893. doi: 10.1126/science.282.5395.1890. [DOI] [PubMed] [Google Scholar]
- 2.Barhanin J., Lesage F., Romey G. KVLQT1 and IsK (minK) proteins associate to form the IKS cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 3.Sanguinetti M.C., Curran M.E., Keating M.T. Coassembly of KVLQT1 and minK (IsK) proteins to form cardiac IKS potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 4.Roepke T.K., Anantharam A., Abbott G.W. The KCNE2 potassium channel ancillary subunit is essential for gastric acid secretion. J. Biol. Chem. 2006;281:23740–23747. doi: 10.1074/jbc.M604155200. [DOI] [PubMed] [Google Scholar]
- 5.Roepke T.K., Kanda V.A., Abbott G.W. KCNE2 forms potassium channels with KCNA3 and KCNQ1 in the choroid plexus epithelium. FASEB J. 2011;25:4264–4273. doi: 10.1096/fj.11-187609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roepke T.K., King E.C., Abbott G.W. Kcne2 deletion uncovers its crucial role in thyroid hormone biosynthesis. Nat. Med. 2009;15:1186–1194. doi: 10.1038/nm.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biervert C., Schroeder B.C., Steinlein O.K. A potassium channel mutation in neonatal human epilepsy. Science. 1998;279:403–406. doi: 10.1126/science.279.5349.403. [DOI] [PubMed] [Google Scholar]
- 8.Mulkey S.B., Ben-Zeev B., Cilio M.R. Neonatal nonepileptic myoclonus is a prominent clinical feature of KCNQ2 gain-of-function variants R201C and R201H. Epilepsia. 2017;58:436–445. doi: 10.1111/epi.13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry G.T., Wu S., Greer J.J. Loss of murine Na+/myo-inositol cotransporter leads to brain myo-inositol depletion and central apnea. J. Biol. Chem. 2003;278:18297–18302. doi: 10.1074/jbc.M213176200. [DOI] [PubMed] [Google Scholar]
- 10.Abbott G.W., Tai K.K., Berry G.T. KCNQ1, KCNE2, and Na+-coupled solute transporters form reciprocally regulating complexes that affect neuronal excitability. Sci. Signal. 2014;7:ra22. doi: 10.1126/scisignal.2005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neverisky D.L., Abbott G.W. KCNQ-SMIT complex formation facilitates ion channel-solute transporter cross talk. FASEB J. 2017;31:2828–2838. doi: 10.1096/fj.201601334R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K.S., Duignan K.M., Tzingounis A.V. The voltage activation of cortical KCNQ channels depends on global PIP2 levels. Biophys. J. 2016;110:1089–1098. doi: 10.1016/j.bpj.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y., Zaydman M.A., Cui J. KCNE1 enhances phosphatidylinositol 4,5-bisphosphate (PIP2) sensitivity of IKs to modulate channel activity. Proc. Natl. Acad. Sci. USA. 2011;108:9095–9100. doi: 10.1073/pnas.1100872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loussouarn G., Park K.H., Escande D. Phosphatidylinositol-4,5-bisphosphate, PIP2, controls KCNQ1/KCNE1 voltage-gated potassium channels: a functional homology between voltage-gated and inward rectifier K+ channels. EMBO J. 2003;22:5412–5421. doi: 10.1093/emboj/cdg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai G., Yu H., Hille B. Osmoregulatory inositol transporter SMIT1 modulates electrical activity by adjusting PI(4,5)P2 levels. Proc. Natl. Acad. Sci. USA. 2016;113:E3290–E3299. doi: 10.1073/pnas.1606348113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho H., Kim Y.A., Ho W.K. Low mobility of phosphatidylinositol 4,5-bisphosphate underlies receptor specificity of Gq-mediated ion channel regulation in atrial myocytes. Proc. Natl. Acad. Sci. USA. 2005;102:15241–15246. doi: 10.1073/pnas.0408851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber W. Ion currents of Xenopus laevis oocytes: state of the art. Biochim. Biophys. Acta. 1999;1421:213–233. doi: 10.1016/s0005-2736(99)00135-2. [DOI] [PubMed] [Google Scholar]
- 18.Schwake M., Jentsch T.J., Friedrich T. A carboxy-terminal domain determines the subunit specificity of KCNQ K+ channel assembly. EMBO Rep. 2003;4:76–81. doi: 10.1038/sj.embor.embor715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyle D.A., Morais Cabral J., MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 20.Long S.B., Campbell E.B., Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 21.Faham S., Watanabe A., Abramson J. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321:810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroncke B.M., Van Horn W.D., Sanders C.R. Structural basis for KCNE3 modulation of potassium recycling in epithelia. Sci. Adv. 2016;2:e1501228. doi: 10.1126/sciadv.1501228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Main M.J., Cryan J.E., Burbidge S.A. Modulation of KCNQ2/3 potassium channels by the novel anticonvulsant retigabine. Mol. Pharmacol. 2000;58:253–262. doi: 10.1124/mol.58.2.253. [DOI] [PubMed] [Google Scholar]
- 24.Wickenden A.D., Yu W., Wagoner P.K. Retigabine, a novel anti-convulsant, enhances activation of KCNQ2/Q3 potassium channels. Mol. Pharmacol. 2000;58:591–600. doi: 10.1124/mol.58.3.591. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Zhang M., Tseng G.N. Probing the structural basis for differential KCNQ1 modulation by KCNE1 and KCNE2. J. Gen. Physiol. 2012;140:653–669. doi: 10.1085/jgp.201210847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prole D.L., Marrion N.V. Ionic permeation and conduction properties of neuronal KCNQ2/KCNQ3 potassium channels. Biophys. J. 2004;86:1454–1469. doi: 10.1016/S0006-3495(04)74214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demo S.D., Yellen G. Ion effects on gating of the Ca2+-activated K+ channel correlate with occupancy of the pore. Biophys. J. 1992;61:639–648. doi: 10.1016/S0006-3495(92)81869-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panaghie G., Abbott G.W. The impact of ancillary subunits on small-molecule interactions with voltage-gated potassium channels. Curr. Pharm. Des. 2006;12:2285–2302. doi: 10.2174/138161206777585175. [DOI] [PubMed] [Google Scholar]
- 29.Wrobel E., Rothenberg I., Seebohm G. KCNE1 induces fenestration in the Kv7.1/KCNE1 channel complex that allows for highly specific pharmacological targeting. Nat. Commun. 2016;7:12795. doi: 10.1038/ncomms12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadley J.K., Noda M., Brown D.A. Differential tetraethylammonium sensitivity of KCNQ1-4 potassium channels. Br. J. Pharmacol. 2000;129:413–415. doi: 10.1038/sj.bjp.0703086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abbott G.W. Biology of the KCNQ1 potassium channel. New J. Sci. 2014;2014:237431. [Google Scholar]
- 32.Abbott G.W. KCNE1 and KCNE3: the yin and yang of voltage-gated K+ channel regulation. Gene. 2016;576:1–13. doi: 10.1016/j.gene.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroeder B.C., Waldegger S., Jentsch T.J. A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature. 2000;403:196–199. doi: 10.1038/35003200. [DOI] [PubMed] [Google Scholar]
- 34.Liu X.S., Zhang M., Tseng G.N. Probing the interaction between KCNE2 and KCNQ1 in their transmembrane regions. J. Membr. Biol. 2007;216:117–127. doi: 10.1007/s00232-007-9047-7. [DOI] [PubMed] [Google Scholar]
- 35.Larsen A.P., Steffensen A.B., Olesen S.P. Extracellular potassium inhibits Kv7.1 potassium channels by stabilizing an inactivated state. Biophys. J. 2011;101:818–827. doi: 10.1016/j.bpj.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mistry A.C., Wynne B.M., Hoover R.S. The sodium chloride cotransporter (NCC) and epithelial sodium channel (ENaC) associate. Biochem. J. 2016;473:3237–3252. doi: 10.1042/BCJ20160312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neverisky D.L., Abbott G.W. Ion channel-transporter interactions. Crit. Rev. Biochem. Mol. Biol. 2015;51:257–267. doi: 10.3109/10409238.2016.1172553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J.Y., Dus M., Suh G.S. Drosophila SLC5A11 mediates hunger by regulating K+ channel activity. Curr. Biol. 2016;26:1965–1974. doi: 10.1016/j.cub.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shcheynikov N., Ko S.B., Muallem S. Regulatory interaction between CFTR and the SLC26 transporters. Novartis Found. Symp. 2006;273:177–186. discussion 186–192, 186. [PubMed] [Google Scholar]
- 40.Singh H., Li M., Toro L. MaxiK channel interactome reveals its interaction with GABA transporter 3 and heat shock protein 60 in the mammalian brain. Neuroscience. 2016;317:76–107. doi: 10.1016/j.neuroscience.2015.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slesinger P.A., Jan Y.N., Jan L.Y. The S4-S5 loop contributes to the ion-selective pore of potassium channels. Neuron. 1993;11:739–749. doi: 10.1016/0896-6273(93)90083-4. [DOI] [PubMed] [Google Scholar]
- 42.Yeromin A.V., Zhang S.L., Cahalan M.D. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldstein S.A., Miller C. Site-specific mutations in a minimal voltage-dependent K+ channel alter ion selectivity and open-channel block. Neuron. 1991;7:403–408. doi: 10.1016/0896-6273(91)90292-8. [DOI] [PubMed] [Google Scholar]
- 44.Zaza A. Serum potassium and arrhythmias. Europace. 2009;11:421–422. doi: 10.1093/europace/eup005. [DOI] [PubMed] [Google Scholar]
- 45.Christé G. Effects of low [K+]o on the electrical activity of human cardiac ventricular and Purkinje cells. Cardiovasc. Res. 1983;17:243–250. doi: 10.1093/cvr/17.4.243. [DOI] [PubMed] [Google Scholar]
- 46.Gadsby D.C., Cranefield P.F. Two levels of resting potential in cardiac Purkinje fibers. J. Gen. Physiol. 1977;70:725–746. doi: 10.1085/jgp.70.6.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCullough J.R., Baumgarten C.M., Singer D.H. Intra- and extracellular potassium activities and the potassium equilibrium potential in partially depolarized human atrial cells. J. Mol. Cell. Cardiol. 1987;19:477–486. doi: 10.1016/s0022-2828(87)80399-1. [DOI] [PubMed] [Google Scholar]
- 48.McCullough J.R., Chua W.T., Singer D.H. Two stable levels of diastolic potential at physiological K+ concentrations in human ventricular myocardial cells. Circ. Res. 1990;66:191–201. doi: 10.1161/01.res.66.1.191. [DOI] [PubMed] [Google Scholar]
- 49.Sheu S.S., Korth M., Fozzard H.A. Intra- and extracellular K+ and Na+ activities and resting membrane potential in sheep cardiac Purkinje strands. Circ. Res. 1980;47:692–700. doi: 10.1161/01.res.47.5.692. [DOI] [PubMed] [Google Scholar]
- 50.Ma L., Zhang X., Chen H. TWIK-1 two-pore domain potassium channels change ion selectivity and conduct inward leak sodium currents in hypokalemia. Sci. Signal. 2011;4:ra37. doi: 10.1126/scisignal.2001726. [DOI] [PubMed] [Google Scholar]
- 51.Chen H., Chatelain F.C., Lesage F. Altered and dynamic ion selectivity of K+ channels in cell development and excitability. Trends Pharmacol. Sci. 2014;35:461–469. doi: 10.1016/j.tips.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Block B.M., Jones S.W. Ion permeation and block of M-type and delayed rectifier potassium channels. Whole-cell recordings from bullfrog sympathetic neurons. J. Gen. Physiol. 1996;107:473–488. doi: 10.1085/jgp.107.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cloues R., Marrion N.V. Conduction properties of the M-channel in rat sympathetic neurons. Biophys. J. 1996;70:806–812. doi: 10.1016/S0006-3495(96)79620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heginbotham L., MacKinnon R. The aromatic binding site for tetraethylammonium ion on potassium channels. Neuron. 1992;8:483–491. doi: 10.1016/0896-6273(92)90276-j. [DOI] [PubMed] [Google Scholar]
- 55.Luzhkov V.B., Aqvist J. Mechanisms of tetraethylammonium ion block in the KcsA potassium channel. FEBS Lett. 2001;495:191–196. doi: 10.1016/s0014-5793(01)02381-x. [DOI] [PubMed] [Google Scholar]
- 56.Yellen G., Jurman M.E., MacKinnon R. Mutations affecting internal TEA blockade identify the probable pore-forming region of a K+ channel. Science. 1991;251:939–942. doi: 10.1126/science.2000494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.