SUMMARY
Transposon reactivation is an inherent danger in cells that lose epigenetic silencing during developmental reprogramming. In the mouse, LTR-retrotransposons, or endogenous retroviruses (ERV), account for most novel insertions and are expressed in the absence of histone H3 Lysine 9 trimethylation in preimplantation stem cells. We found abundant, 18 nt tRNA-derived small RNA (tRF) in these cells, and ubiquitously expressed 22 nt tRFs, that include the 3′ terminal CCA of mature tRNAs, and target the tRNA primer binding site (PBS) essential for ERV reverse transcription. We show that the two most active ERV families, IAP and MusD/ETn, are major targets and are strongly inhibited by tRFs in retrotransposition assays. 22 nt tRFs post-transcriptionally silence coding-competent ERVs, while 18 nt tRFs specifically interfere with reverse transcription and retrotransposon mobility. The PBS offers a unique target to specifically inhibit LTR-retrotransposons and tRF-targeting is a potentially highly conserved mechanism of small RNA-mediated transposon control.
Keywords: Epigenetic Reprogramming, Small RNA, tRNA-fragments, Transposon Control, Mouse Endogenous Retroviruses
eTOC BLURB
3′ tRNA fragments limit the mobility of transposable elements in mouse ES cells
INTRODUCTION
Transposable elements (TE) and their repetitive sequences drive transcription, organize chromosomes and induce heterochromatin, but TE mobility driven by intact, active transposons is mutagenic and has to be tightly controlled (Slotkin and Martienssen, 2007). In mice, the most active transposons are LTR-retrotransposons, also known as endogenous retroviruses (ERV). They cause an estimated 10% of germline mutations in today’s inbred laboratory strains, mostly by the ERV-K families IAP (Intracisternal A Particle) and ETn (Early Transposon) (Nellaker et al., 2012). Transcription of TEs is normally repressed by epigenetic marks such as DNA methylation and histone modification. While DNA methylation by DNMT1 represses non-LTR LINE elements and a few LTR-retrotransposons, SETDB1-mediated histone H3K9 trimethylation silences the majority of LTR-retrotransposons (Karimi et al., 2011; Matsui et al., 2010). SETDB1 depleted embryonic stem (ES) cells lose H3K9me3, express ERVs, become trophoblast-like, and contribute to the placenta when transplanted into mouse embryos (Yuan et al., 2009). Trophoblast stem cells (TS) are permissive for ERV expression and ERVs have been instrumental in placental evolution (Chuong et al., 2013; Hemberger, 2010).
Stem cells undergo genome-wide epigenetic reprogramming to gain pluripotency and consequentially release transcriptional control of TEs. In many tissues, small RNAs (sRNA) interfere with transposon expression when epigenetic control is compromised by reprogramming: piRNAs act in the germline and in cancer stem cells (Juliano et al., 2011), while the oocyte is loaded with endogenous siRNAs that prevent transposon damage right after fertilization (Ohnishi et al., 2010; Tam et al., 2008; Watanabe et al., 2008). It is however less clear how transposition is avoided in the early embryo when epigenetic marks are reset to enable pluripotency. We set out to determine sRNA expression in TS and ES cells, including embryonic cell lines that are defective in epigenetic control to better understand which classes of sRNA potentially control TEs in preimplantation embryos. We found very abundant 18 and 22 nucleotide sRNA targeting ERVs that are derived from the 3′ end of mature tRNAs.
tRNA-derived fragments (tRF) are abundant, non-coding small RNAs that are very widespread in most organisms. They have been implicated in stress responses and cancer, cell-cell signaling via exosomes, response to viral infection, and neurological disorders (Keam and Hutvagner, 2015; Raina and Ibba, 2014). At least four cleavage products of mature tRNAs have been reported: 5′ and 3′ halves (30–33 nt) as well as short 18 and 22 nt 3′ fragments. 3′ fragments derived from mature tRNAs end in a CCA trinucleotide. The CCA motif is added during post-transcriptional maturation of tRNAs after cleavage of a 2 nt 3′ trailer sequence and before aminoacylation. A number of biological functions and mechanisms have been ascribed to tRNA halves such as translational inhibition, inhibition of apoptosis, and suppression of breast cancer progression (Kumar et al., 2016).
Recently, a potential role for tRFs in gene silencing has emerged as the RNA interference (RNAi) silencing machinery has been implicated in the biogenesis of many tRF species. Some tRNAs in human are cleaved into 5′ tRFs by DICER (Cole et al., 2009) while 15–30 nt tRFs in mouse ES cells still accumulate in Dicer- and Drosha-knockouts (Li et al., 2012). In Tetrahymena, tRFs are bound by the PIWI protein (Couvillion et al., 2010). Human PIWIL4 binds piRNA-like tRF halves when overexpressed and a number of tRFs match known piRNA sequences (Keam et al., 2014). Human tRFs have been found bound to all four Argonaute (AGO) proteins when overexpressed, with preferential binding to AGO3 and AGO4 over AGO1 and AGO2 (Haussecker et al., 2010; Kumar et al., 2014). Most recently, individual paternal 5′ tRFs have been implicated in apparent downregulation of a small group of LTR-associated genes in mouse by an unknown mechanism (Sharma et al., 2016), and in targeting of some Gypsy elements in Arabidopsis by partial complementarity to various regions (Martinez et al., 2017). Finally, 5′ tRNA processing in Drosophila affects the expression of neighboring piRNA clusters and their DNA- and RNA-transposon targets (Molla-Herman et al., 2015). Little is known about the biological functions of 3′ tRFs. Unlike 5′ tRF, many 3′ CCA tRFs are perfectly complementary to ERVs because ERVs use tRNAs as primers for reverse transcription. Together with the finding that many tRFs are bound by AGO and PIWI proteins, we and others have speculated that 3′ tRFs may restrict TE activity (Kawaji et al., 2008; Li et al., 2012; Schorn and Martienssen, 2011).
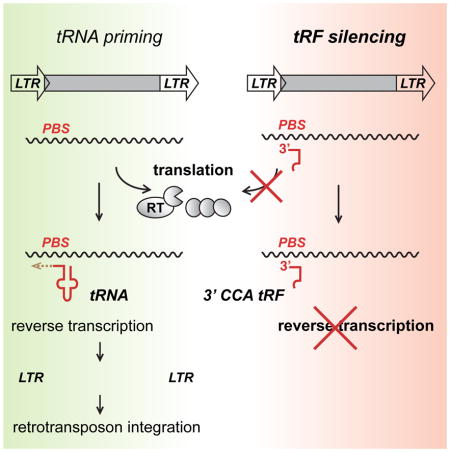
Many small RNA sequencing studies omit RNA fragments shorter than 19 nt or discard sequencing reads that map to multiple loci in the genome (Kumar et al., 2014), thus often discarding reads matching young, potentially active transposons, including tRFs that map to tRNAs and TE at the same time. By modifying small RNA library and analysis procedures to include such fragments, we have found very abundant, 17–19 nt short 3′ CCA tRFs in a variety of mouse stem cell lines. These “18 nt tRFs” matched active ERVs and are especially abundant in cells depleted for H3K9me3 and impaired in transcriptional silencing of ERVs. Using transposition reporter assays, we have found that 3′ CCA tRFs strongly inhibit ERV retrotransposition. 18 nt 3′ CCA tRFs that target the two most active mouse transposon families, IAP and ETn, are enriched in preimplantation stem cells and inhibit these TEs in retrotransposition assays. 18 nt 3′ CCA tRFs specifically interfere with reverse transcription, while transposon expression is unaffected. Therefore, they are able to act on any replication-competent LTR-retrotransposon, including non-coding, non-autonomous elements, which comprise the bulk of TEs in the genome. We also find that transposon expression is affected by 22 nt 3′ tRFs, which decrease RNA and protein level of coding-competent, autonomous elements by post-transcriptional silencing. Silencing by tRFs represents a novel and potentially highly conserved sRNA-based mechanism of transposon control that could serve as an additional line of defense in cells that undergo reprogramming and are incapable of TE silencing by epigenetic transcriptional suppression.
RESULTS
Abundant tRNA-derived small RNA targeting ERVs in mouse stem cells
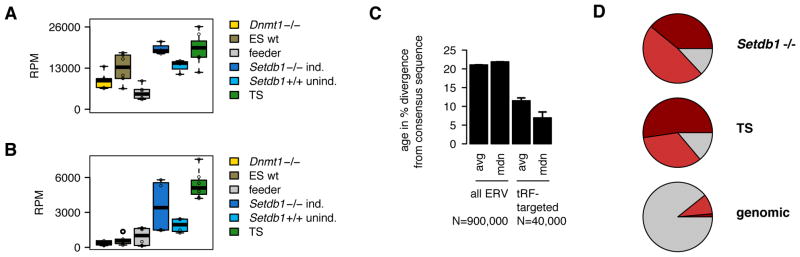
Transposons are released from silencing and transcribed during epigenetic reprogramming in pre-implantation embryos (Fadloun et al., 2013; Peaston et al., 2004). To find out whether sRNA could protect the pre-implantation embryo from transposon insertions, we profiled sRNA in TS cells and in ES cell mutants that mimic epigenetic reprogramming. We found abundant expression of sRNA complementary to LTR-retrotransposons or ERVs (Figure 1A). To our surprise, a substantial proportion of these ERV-targeting sRNA were derived from mature tRNAs and had a 3′ terminal CCA trinucleotide (Figures 1B and 2). The same cell types that had high expression of ERV sRNA generally had high levels of CCA 3′ tRFs (Figure S1A). ERVs are normally silenced by H3K9me3 methylation, and TS cells and Setdb1 inducible knockout ES cells showed the highest levels of tRFs targeting ERVs. In contrast, tRFs targeting ERVs were not elevated in Dnmt1 knockout cells, which primarily reactivate non-LTR elements. Curiously, ERVs targeted by CCA 3′ tRFs were younger than average, indicating this sRNA class could regulate young, potentially active transposons (Figure 1D). We next asked which of the reportedly active ERV-K elements are targeted by CCA 3′ tRFs and found that IAP and ETn transposons were heavily overrepresented when compared to their relatively low genomic abundance (Figure 1E). This supported a functional link between CCA 3′ tRFs and their ERV targets because IAP and ETn were enriched among tRF targets and are the most active transposon families in mouse.
Figure 1. Stem cells with relaxed epigenetic control of LTR-retrotransposons express sRNA targeting ERVs, including 3′ tRNA-derived fragments (tRFs).
(A) Setdb1 knockout mouse ES cells and TS cells have elevated levels of LTR-retrotransposon (ERV) small RNA and (B) 3′ CCA tRFs which target ERVs. Boxplots represent reads per million total mapped reads (RPM) of biological replicates; for a list of all 33 sRNA libraries refer to Figure S3. Setdb1 knockout induced −/−, uninduced +/+; for details see STAR Methods section. (C) tRF-targeted ERVs (in TS cells) are potentially active and younger than the average genomic ERV copy (avg: average; mdn: median; ± SD). (D) The majority of ERV-K targeted by 3′ tRFs are from the ETn and IAP families which are the most active LTR-retrotransposons in mouse (average RPM values of 4 replicate Setdb1 −/− and 7 replicate TS sRNA libraries). For comparison, relative abundance of ERV-K sequences in the mm9 mouse genome: 9% belong to the IAP family, 1% to the ETn family. See also Figure S1.
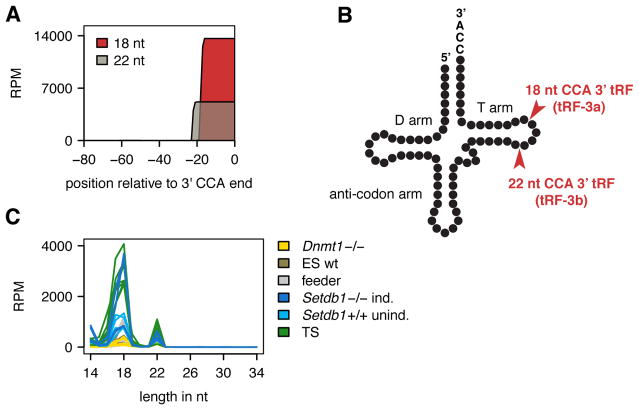
Figure 2. tRFs targeting LTR-retrotransposons (ERV) in mouse are derived from the 3′ end of mature tRNAs.
(A) Alignment of all expressed CCA-tRFs (one representative TS cell sample) along tRNA coordinates (position 0 = CCA end) reveals cleavage of mature tRNAs precisely 22 nt and 17–19 nt from the 3′ CCA end. For tRNA alignments of all samples as well as analysis of other tRNA-derived fragments see Figures S2 and S3. (B) tRNA cleavage sites and nomenclature according to Kumar et al. (2014) are indicated. (C) Size distribution of CCA-tRFs targeting ERVs shows the dominant fragment length in mouse stem cells is 18 nt.
Alignment of tRFs ending in CCA along tRNA coordinates reveals cleavage of mature tRNAs 17–19 nt and precisely 22 nt from the 3′ CCA end, in more than 30 sRNA libraries sequenced (Figures 2A, S2, and S3). Both tRF species have been observed previously and have been named tRF-3a and tRF-3b in human (Kumar et al., 2014). For simplicity, we refer to 17–19 nt tRF-3a as “18 nt tRFs” and to tRF-3b as “22 nt tRFs”. 22 nt tRFs retain two RNA modifications from their parent tRNA: 1-methyladenosine at position 19 counting from the 3′ CCA end and pseudo-uridine at position 22, their 5′ end. Only some of the 22 nt tRFs are complementary to ERVs, the majority of tRFs matching ERVs with 0–1 mismatches are 18 nt long (Figure 2C). Interestingly, we have not found any 3′ tRNA halves in any of the samples sequenced. All tRNA-derived sequences not ending in CCA were 30–33 nt long 5′ halves and had no obvious targets in LTR-retrotransposons (Figure S2). We set out to examine whether and how 3′ CCA tRFs could target and repress LTR-retrotransposons.
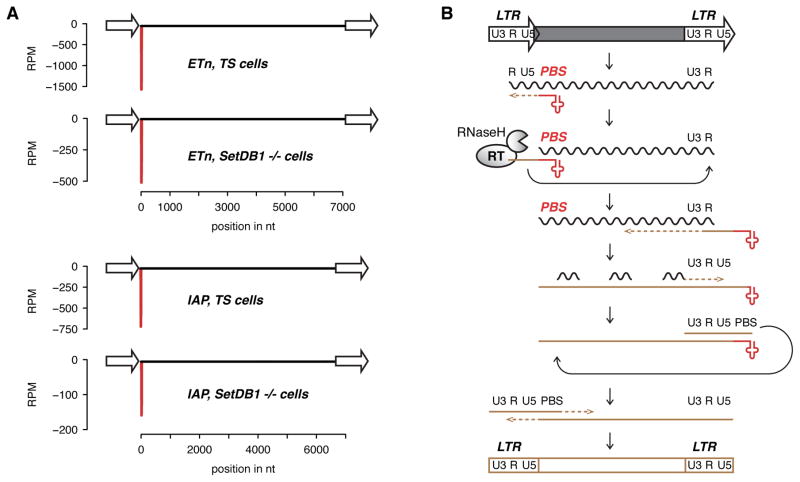
Small RNA reads matching genomic ERVs have a sharp peak in the 5′ UTR precisely at the 18 nt primer binding site (PBS) following the LTR (Figure 3A) where the PBS binds tRNA during LTR-retrotransposon replication (Figure 3B). In general, ERVs have been classified according to the amino acid of the tRNA that primes them, e.g. most ERV-K are primed by Lysine (K) tRNA. However, tRNAs truncated or mutated in key residues are poor primers (Keeney et al., 1995) and the T-loop is important for processive binding of the reverse transcriptase (RT) of most viruses (Le Grice, 2003). Except for the very few LTR-retrotransposons that do not use tRNA priming for proliferation, the PBS is the most highly conserved sequence in any functional ERV making it an excellent target site for regulation by sRNA. Accumulation of tRFs is pronounced for the ERV-K families ETn and IAPEz (Figure 3A). IAPEz is an envelope (E)-containing, autonomous IAP retrovirus whose expression is strongly upregulated upon loss of H3K9me3 in ES cells (Karimi et al., 2011). Importantly, accumulation of tRFs antisense to the PBS was not found in proportion to the abundance of the transposon in the genome but instead was highly enriched for the two most active mouse transposon families IAP and ETn (Figures 1E and S1C).
Figure 3. 3′ tRFs match the primer binding site (PBS) of LTR-retrotransposons.
(A) CCA tRF sequencing reads are complementary to the highly conserved PBS site of LTR-retrotransposons. Major targets are ETn and IAPEz (all genomic loci convoluted, one representative TS and Setdb1 −/− sample out of all replicates). (B) Life cycle of LTR-retrotransposons and -viruses. The long terminal repeats (LTR) encode promoter elements and termination signals. The RNA transcript contains a region repeated at either end (R), a 5′ unique segment (U5), and a segment only included at the 3′ end of the RNA (U3). The 3′ end of cellular tRNAs (red cloverleaf) primes reverse transcription by hybridizing to the primer binding site (PBS). After this segment has been copied into first-strand cDNA (brown line), the RNaseH activity of reverse transcriptase (RT) degrades the complementary RNA and the elongating cDNA is transferred to the 3′ end of the retrotransposon transcript hybridizing to the R region. The remaining RNA is partially degraded by RNaseH leaving behind primers for second-strand cDNA synthesis. After a second transfer event, first- and second-strand synthesis can be completed to result in a full-length, double-stranded retroviral DNA that will be integrated into the host genome.
3′ CCA tRFs inhibit retrotransposition
Often transposon expression is taken as a proxy for transposon activity because mobility of endogenous TEs is difficult to measure. We wanted to test the effect of 3′ CCA tRFs on LTR-retrotransposition. IAP is the most active transposon in mice and responsible for in vivo polymorphisms as well as intergenerational, epiallelic variation (Heidmann and Heidmann, 1991; Morgan et al., 1999). ETn is the second most active transposon family accounting for novel mutations in mice, but ETn are non-autonomous i.e. non-coding and rely on their coding-competent MusD family members for mobility (Ribet et al., 2004). Active copies of both IAP and ETn/MusD have been isolated and used in plasmid-based retrotransposition assays which allow quantification of retrotransposition and study of its mechanism by transfection into human cells (Dewannieux et al., 2004; Ribet et al., 2004) (Fig. 4A, B). This is because human cells lack a confounding background of thousands of endogenous copies found in mouse cells.
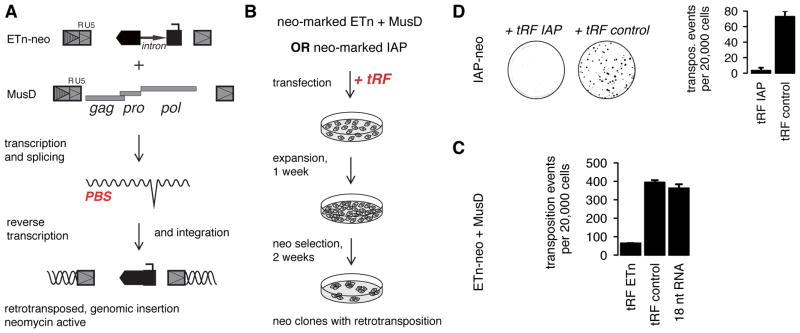
Figure 4. 3′ CCA tRFs inhibit retrotransposition.
(A) The plasmid based retrotransposition assay. Transcription of MusD and ETn is driven by a CMV promoter upstream of the R and U5 portion of the LTR needed for retrovirus replication (see Figure 3B). After transcription and splicing, the retroviral gene products reverse transcribe the retroviral RNA and integrate the cDNA into the host genome. The neomycin (neo) gene becomes active only after splicing and reverse transcription, so that each neo-resistant cell must have had a retrotransposition event. (B) To test for regulation by tRFs, 18 nt tRFs were cotransfected together with MusD and the non-autonomous, neo-marked ETn or together with an autonomous, neo-marked IAP reporter plasmid. IAP transcription is driven by its endogenous promoter. The assays were done in human HeLa cells, as previously described (Dewannieux et al., 2004; Ribet et al., 2004). Neo-resistant clones were fixed, stained, and counted to measure retrotransposon activity. (C) MusD/ETn retrotransposition is inhibited by tRFs against ETn while unaffected by an unrelated 3′ CCA tRF sequence or non-targeting RNA. (D) IAP retrotransposition is strongly inhibited by tRFs targeting IAP but not by an unrelated 3′ CCA tRF sequence. Colony counts are the mean of two replicates ± SD.
We performed ETn/MusD retrotransposition assays (Fig. 4A, B) in the presence of 18 nt 3′ CCA tRFs targeting ETn, an unrelated 3′ CCA tRF sequence, or a non-targeting 18 nt RNA control (Figure 4C). 18 nt ETn tRFs derived from the 3′ end of tRNALys3-AAA that match the specific ETnIIbeta element used in the assay are abundantly expressed in many cell types. The MusD6 element used in the assay does not have perfect PBS complementarity to its cellular tRNALys primer and has two mismatches to ETn tRFs. We found that ETn retrotransposition occurred at high frequencies in HeLa cells as expected. ETn was strongly inhibited when plasmids were co-transfected with tRFs targeting ETn, but was not inhibited when co-transfected with unrelated tRF nor non-targeting control (Fig. 4C). We also tested IAP retrotransposition to see whether inhibition by tRFs is conserved among ERV retrotransposons. We found that IAP transposition was strongly downregulated in the presence of tRFs that target the specific IAP element of the reporter assay (Figure 4D). The IAP copy used in the plasmid assay belongs to the IAPEz family, which is the IAP family most highly targeted by endogenous tRFs. However, we decided to study ETn/MusD inhibition in more detail because the combination of a coding-competent (MusD) and a noncoding, non-autonomous (ETn) transposon would give further insight into the mechanism of tRF inhibition.
Mechanism of ERV inhibition by tRFs
We considered three possible ways in which tRFs could suppress retrotransposition: (i) transcriptional silencing by tRFs guiding H3K9me3 deposition, (ii) post-transcriptional silencing by RNAi, and (iii) inhibition of retroviral intermediates by blocking reverse transcription. Transcriptional silencing of ERVs is mediated by SETDB1/KAP1-mediated deposition of H3K9me3, and the same young ERV elements that are targeted by tRFs also accumulate K9me3 in ES cells (Matsui et al., 2010; Rowe et al., 2010). H3K9me3-mediated heterochromatin is nucleated at the PBS and spreads from there (Rebollo et al., 2011; Wolf et al., 2008). KRAB zinc finger proteins (ZFP) confer target specificity for many different classes of ERVs (Ecco et al., 2016; Tan et al., 2013), but it was tempting to speculate that tRFs targeting the PBS could also recruit H3K9me3 resembling sRNA-mediated transcriptional silencing at repeats in yeast, flies and mice (Aravin et al., 2008; Sienski et al., 2012; Volpe et al., 2002; Watanabe et al., 2011). We found however only modestly elevated H3K9me3 levels at tRF-targeted ETn loci in TS cells, and very low levels at ERVs altogether despite high expression levels of both 3′ tRFs and SETDB1 (Figure S4). Indeed, relaxed transcriptional repression of ERVs in TS cells allows ERVs to function as species-specific enhancer elements and may contribute to rapid placental evolution (Chuong et al., 2013).
To examine whether tRFs could act post-transcriptionally via RNAi we first looked at overall RNA levels. Neither MusD nor ETn total RNA level changed during tRF-mediated inhibition of retrotransposition (Figure 5A). MusD encodes the proviral gag, pro, and pol genes that are expressed as fusion precursor proteins and become processed into mature viral components by the encoded protease. We probed lysates of transfected HeLa cells with antiserum that recognizes the 26 kD Gag subunit associated with active protease and virus. MusD Gag levels did not change in the presence of 18 nt 3′ tRFs when compared to control tRFs that have no sequence complementarity to ETn or MusD (Figure 5B). This indicated that 18 nt 3′ tRFs do not induce miRNA-like mRNA degradation or translational inhibition. Knock-down of each of the four AGO proteins with siRNAs had no effect on the inhibition of ERV retrotransposition by 18 nt 3′ tRFs (data not shown), and neither did knock-down of all four AGOs compared with control siRNAs (Figure S4). This led us to conclude that albeit processing and binding of tRFs by AGOs may be important during biogenesis, AGO proteins did not seem to be essential for the observed repression of ERV retrotransposition by 18 nt tRFs.
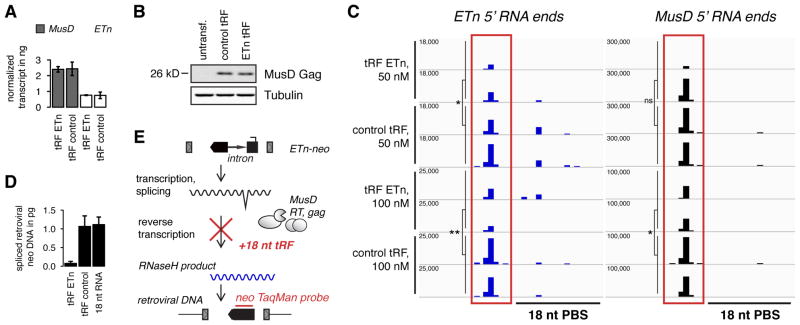
Figure 5. 18 nt 3′ tRFs do not interfere with primary transcript or protein levels but inhibit reverse transcription.
(A) MusD and ETn RNA transcript levels are not affected in cells which showed decreased MusD/ETn activity after transfection of ETn tRFs. Mean transcript levels ± SD were determined by quantitative Taqman RT-PCR. (B) MusD Gag protein levels are not affected by co-transfection of 18 nt, targeting tRFs. (C) Uncapped 5′-P RNA ends were sequenced by high-throughput, modified RACE. Each track represents one replicate. The position of all 5′ RNA ends including RT-RNaseH products (boxed red) and potential RNA cleavage products within ~40 bp surrounding the PBS and 5′ UTR are shown. The decrease of RNaseH products indicates that 18 nt targeting tRFs specifically inhibit accumulation of retroviral RNA intermediates. Note that for visibility, y-axis RPM maxima differ between MusD, ETn, and tRF concentrations. Welch two sample t-test, p-value ** < 0.05, * < 0.1, ns = not significant (D) Downstream retroviral DNA intermediates are decreased by ETn tRFs. TaqMan primers and probe detect extrachromosomal, retroviral DNA only (position indicated in E). Data represented as mean ± SD, normalized to total transfected plasmid DNA. (E) Outline of ETn-neo retroviral intermediates and model of retrotransposon silencing by 18 nt tRFs. See also Figure S4.
Inhibition of reverse transcription by tRFs
Next, we tested whether tRFs have an effect on downstream intermediates of retrotransposition for example by inhibiting reverse transcription. We reasoned that mapping of uncapped retroviral RNA 5′ ends spanning the PBS could be used to answer the following questions: (i) What are the natural 5′ ends of retroviral ETn and MusD RNaseH intermediates (Figure 3B)? (ii) Do we see changes in any retroviral RNA intermediates, i.e. do tRFs interfere with reverse transcription? (iii) Is there any signature of RNAi-like cleavage by tRFs at or near the PBS? We used a modified RACE-protocol to map uncapped, 5′ monophosphate RNA ends as they occur in retroviral intermediates and RNAi cleavage products from samples with tRF silencing of ETn/MusD. Briefly, we reverse transcribed 5′ adapter-ligated RNA with an ETn/MusD-specific primer and amplified the resulting cDNA with Illumina primers to sequence these rare retrotransposition intermediates to high depth. We were able to unambiguously map the RNaseH cleavage site of MusD and ETn to 6 nt upstream of the PBS (Figure 5C, all lanes). Inhibition of retrotransposition by tRFs was in striking correlation with the abundance of RNaseH intermediates: only in the presence of ETn tRFs were there less retroviral RNaseH intermediates that result from first-strand cDNA synthesis by RT. Thus, tRFs interfere with reverse transcription before RNaseH products are formed. The PBS target sequences of MusD and ETn are identical except for two mismatches. The inhibition of reverse transcription is most pronounced for perfectly complementary ETn intermediates (Figure 5C, left panel) but ETn tRFs affect MusD at high concentrations (right panel). If tRFs prevent RT and RNaseH activity, we expected a decrease of downstream retroviral DNA intermediates as well. We isolated extrachromosomal DNA from cells that underwent tRF inhibition, capturing transfected transposon plasmids and retroviral cDNA. Retroviral cDNA levels were detected by quantitative TaqMan PCR amplification of the spliced neomycin reporter gene and precisely reflected the outcome of retrotransposition efficiency (Figure 5D). This confirmed that 18 nt 3′ tRF inhibition acts upstream of first-strand retroviral cDNA synthesis by RT (Figure 5E). Therefore, tRFs are able to specifically restrict mobility of otherwise harmless transposon transcripts.
Endogenous tRFs and 22 nt tRFs
We set out to see whether we can test the effect of endogenous rather than synthetic 3′ tRFs. Inhibition of endogenous 3′ tRFs by antisense oligo “sponges” as often used to interrogate miRNA function, were difficult to interpret because antisense RNAs mimic 5′ tRFs which are able to inhibit translation (Kumar et al., 2016) and bind to the unwound 3′ portion of the tRNA primer. Instead, we mutated the tRF target site, the PBS of the LTR-retrotransposon. As expected, mutation of the ETn PBS resulted in reduced retrotransposition due to reduced tRNA priming of the element carrying the retrotransposition indicator (Figure S5A). However, replacement of the MusD PBS with an unrelated sequence resulted in a drastic increase of ETn retrotransposition (Figure 6A) suggesting wildtype MusD might be silenced through the PBS target sequence, preventing ETn mobilization in trans. MusD Gag protein levels (Figure 6B) and RNA transcript levels were strikingly elevated in the MusD-PBS* mutant (Figure 6C), suggesting sRNA-mediated silencing of transposons with intact PBS by complementary, endogenous tRFs. Repression of a luciferase reporter gene in place of the MusD coding sequence (Figure 6D) confirmed that the silencing effect was not due to an indirect effect of tRF or tRNA binding on allocation of viral RNA for translation versus reverse transcription in viral particles. The observed silencing had all the hallmarks of sRNA-mediated silencing, including a miRNA-like tolerance for mismatches given that the MusD PBS has two mismatches with endogenous “ETn” Lys-AAA 3′ tRFs.
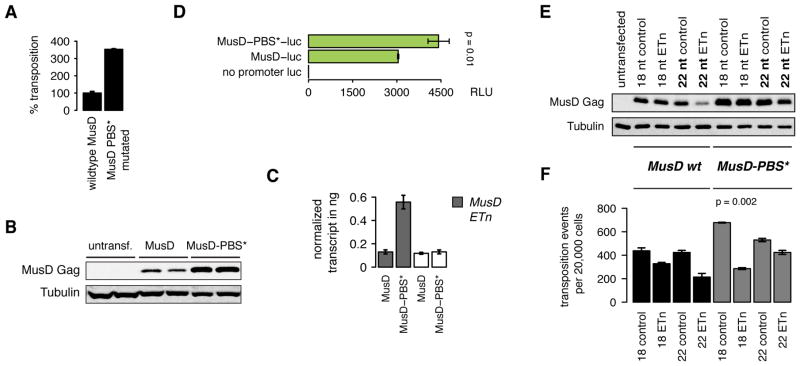
Figure 6. MusD lacking a tRNA primer binding site is released from silencing by endogenous tRFs.
(A) The MusD PBS was replaced by an unrelated sequence to destroy the tRF target site (MusD-PBS*). Relative increase of transposition was higher for low amounts of transfected transposon plasmids (here 25 ng) in agreement with endogenous level of tRFs being the effector. (B) MusD Gag protein and (C) MusD RNA level are higher in the MusD-PBS* mutant. (D) Likewise, luciferase reporter gene expression is released from silencing by endogenous tRFs when the PBS is mutated. The luciferase ORF was cloned exactly in place of the first MusD ORF. RLU = relative light units. (E) MusD protein expression is decreased by 22 nt ETn tRFs but not by control tRF oligos or 18 nt tRFs. (F) Retrotransposition efficiencies are affected according to silencing of coding competent MusD by 22 nt tRFs and inhibition of reverse transcription of ETn-neo by 18 nt tRFs. Colony counts are the mean of two replicates ± SD. p-values Welch two sample t-test, all data represented as mean ± SD. See also Figure S5 and S6.
Deletion of the PBS abrogates tRNA binding. Consequently, the MusD-PBS* mutant RNA cannot be reverse transcribed and no RNaseH products are detectable (data not shown). Instead, only ETn-neo is reverse transcribed and integrated by the MusD enzymatic machinery resulting in the observed neomycin resistant colony counts. Consequently, the MusD-PBS* mutant cannot be used to test for tRF inhibition during reverse transcription but was able to reveal upstream effects of endogenous tRFs. We found that MusD lacking a PBS can act entirely in trans on non-autonomous ETn copies (without producing MusD tRNA-primed retroviral intermediates) and that endogenous tRFs can repress autonomous, coding competent transposons by post-transcriptional sRNA-mediated silencing.
Since 18 nt 3′ tRFs had no effect on MusD transcript or protein levels (Figures 5A and 5B) they were unlikely to mediate post-transcriptional silencing. Instead, 22 nt 3′ tRFs (Figures 2 and S2) were more likely to be responsible, as HeLa cells express the same highly conserved 18 and 22 nt tRFs found in murine pre-implantation stem cells (Figure S6A). Indeed, co-transfection of 22 nt tRFs induced post-transcriptional silencing of MusD Gag protein (Figure 6E) and decreased transposition (Figure 6F) while control tRFs and 18 nt tRFs had no effect on RNA or protein levels (Figures S5B and 6E). 22 nt tRFs reduced protein levels specifically of wildtype MusD, but not MusD-PBS*, confirming silencing through the PBS target site. In contrast, 18 nt tRFs still inhibited retrotransposition mediated by the MusD-PBS* mutant since 18 nt tRFs interfere with successful reverse transcription of ETn-neo (Figure 6F). Thus, 18 nt tRFs are able to target very prolific non-autonomous, non-coding elements like ETn that have escaped all other means of transposon defense, while 22 nt tRFs prevent translation of coding competent LTR-retrotransposons as long as they carry the tRNA primer binding site (Figure 7).
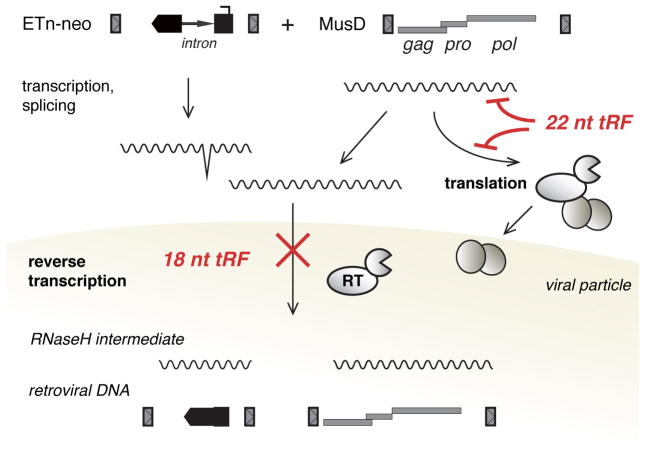
Figure 7. 22 nt tRFs mediate post-transcriptional gene silencing while 18 nt tRFs interfere with reverse transcription including non-coding, mobile elements.
Model of retrotransposon silencing by 3′ tRFs: 22 nt tRFs target coding-competent LTR-retrotransposons (here MusD) at the level of retroviral protein production. 18 nt tRFs inhibit reverse transcription of any element with perfect complementarity at the PBS, including non-autonomous elements (here ETn). tRFs specifically promote silencing of retrotransposition-competent elements which maintain a functional PBS.
DISCUSSION
18 nt 3′ tRFs as a novel sRNA-based mechanism of transposon control
Transcriptional activation of LTR-retrotransposons is a hallmark of early embryonic reprogramming in the mouse (Fadloun et al., 2013; Macfarlan et al., 2012; Peaston et al., 2004) which suggests there should be some alternative means of transposon control. We profiled small RNAs that could potentially restrict transposon mobility in cells that lack epigenetic suppression of TEs. We found abundant ERV-targeting 18 nt 3′ CCA tRFs in Setdb1 knockout ES cells as well as extraembryonic TS cells (Figure 1B) both of which have low H3K9me3 at ERVs and are reportedly permissive for ERV transcription. By focusing on unique mapping reads, tRFs mapping to the repeat portion of the genome have often been excluded from analysis. ERV-K is the most active TE superfamily in mouse, and the top two most active mouse TE families, IAP and ETn, match ~75% of ERV-K targeting tRF reads (Figure 1D). This is in contrast to the relatively small genomic space these two families occupy in the genome. Together with the young age of tRF-targeted ERVs (Figure 1C), our data strongly suggests a function for tRF in targeting active, endogenous elements.
The tRF targets we found reflect exactly those transposons that need to be silenced in the developing embryo and are repressed in ES cells by ZFP-guided, SETDB1-mediated deposition of H3K9me3. Setdb1 knockout ES cells release IAP, MusD, and ETn from transcriptional silencing (Karimi et al., 2011). Indeed, expression of IAP tRFs and IAP transcripts are well correlated in the cell lines we studied (Figure S1C). Interestingly, ETn and MusD expression was much lower in the presence of SETDB1 in TS cells (Figure S1C), and did not correlate with matching tRFs, although we cannot exclude the possibility that some ETn elements went undetected due to their high sequence divergence. The main ERV target of ZFP809 is the ERV1 family member RLTR6-int (Wolf et al., 2015) which is also a major tRF target (Figure S1D). However, this ERV1 element has not been reported to be mobile and lacks a gag ORF making it a less obvious threat to genome integrity. One idea is that tRFs are produced in response to expression of LTR-retroelements. Indeed, the same 18 nt Lys3-AAA 3′ tRF that targets ETnIIbeta (Table S1) has been found as a non-coding sRNA that was upregulated upon human HIV infection, and suppressed HIV RT activity primed by tRNALys3-AAA (Yeung et al., 2009). Similarly, an 18 nt Proline tRF targeting HTLV-1 has been found upregulated in infected T-cells (Ruggero et al., 2014). However, the strictly intracellular murine ERVs MusD and ETn did not induce additional tRF production in human HeLa cells. We sequenced sRNA from HeLa cells after transfection with MusD, ETn, and a control plasmid, respectively, and did not find elevated levels of 3′ tRFs or ETn tRFs specifically (Figure S6B). Thus ERV expression did not promote tRNA cleavage into tRFs. Rather, it is possible that genome-wide H3K9me3 reprogramming that releases ERVs affects tRF production indirectly. The fact that we find overall 3′ CCA tRF level elevated in cells with low H3K9me3 argues for this scenario (Figure S1A). The top tRF-targeted ERV families (IAP, ETn, ERV1-RLTR6) and tRNA genes are all enriched for the histone variant H3.3 which has recently been shown to function upstream of KAP1/SETDB1 in repressing ERVs in mouse ES cells (Elsasser et al., 2015; Kraushaar et al., 2013). Thus loss of H3.3 could change tRNA regulation while releasing ERVs. While transposon mobility is hazardous to the host, ERV sequences have provided important regulatory elements for stem cells and development (Faulkner et al., 2009; Fort et al., 2014; Kunarso et al., 2010) and function as species-specific enhancers in murine trophoblasts (Chuong et al., 2013). Many imprinted genes are derived from ERVs and are epigenetically regulated during development (Hemberger, 2010), in some cases by small RNAs (Ito et al., 2015; Watanabe et al., 2011). ERV-derived viral envelope proteins drive the fusion of trophoblast cells to the mother tissue (Dupressoir et al., 2009). Without a doubt ERV sequences have been important building blocks for evolution but their mobility must be tightly controlled and we identify tRFs as important non-coding small RNAs that are able to restrict retrotransposition during reprogramming.
Reverse transcription as a target for retrotransposon control
LTR-retrotransposons depend on tRNA priming of reverse transcription for proliferation. We found that 18 nt 3′ CCA tRFs suppress replication of IAP and ETn LTR-retrotransposons (Figures 4 and 5). High resolution mapping and quantification of RNaseH retroviral intermediates, revealed that 18 nt 3′ CCA tRFs prevent the very first step of retroviral cDNA synthesis, the formation of RNaseH products that accompany minus-strand strong-stop DNA synthesis (Figures 3C and 5C). This conclusion is supported by the fact that downstream retroviral DNA intermediates were strongly depleted (Figure 5D) while transcription of MusD and ETn as well as MusD translation were not affected (Figures 5A and 5B). Furthermore, retroviral enzymes and RT priming by tRNALys were still functional and produced RNaseH intermediates (Figure 5C). Therefore, it seems unlikely that tRFs interfere with recruitment of tRNALys or ERV RNA to viral particles for reverse transcription. Instead, we conclude 18 nt 3′ tRFs most likely interfere with tRNA priming of RT by competing for binding of the PBS (Figure 5E). This is consistent with the findings that successful retroviral priming and elongation requires full-length tRNA.
Transposon mobility is often strongly inhibited in their adapted host species, and even in permissive tissues, endogenous retrotransposition is rare and not clonal. Further, given the thousands of ERV copies in the mouse genome we would not be able to determine which IAP or ETn copies were affected by tRFs in the mouse. By studying mouse ERV activity in human cells we were able to assay mobility of a discrete element and dissect the mechanism of tRF inhibition. tRNAs and tRFs are highly conserved between species including 22 nt 3′ tRFs with (imperfect) complementarity to the PBS of LTR-retrotransposons. Deleting the MusD PBS revealed their ability to interfere with RNA and protein levels of coding-competent LTR-retrotransposons (Figure 6). 18 nt 3′ tRFs do not change RNA or protein levels, but poison reverse transcription of ETn instead. In contrast, 22 nt 3′ tRFs induce post-transcriptional silencing of MusD (Figures 6 and 7). Resembling miRNAs, 22 nt tRFs were highly effective silencing agents despite mismatches to their target, and co-immunoprecipitate with endogenous AGO2 (Li et al., 2012). 18 nt tRFs have also been found bound by AGO proteins (see Introduction), but knock down of all four AGOs did not reduce inhibition by 18 nt tRFs (Figure S4B). While 18 nt tRFs may not need RNAi enzymes for RT inhibition, cleavage of tRNAs into 18 and 22 nt small RNAs and the observed post-transcriptional silencing by 22 nt tRFs (Figures 7A and 7B) strongly suggest they are products or components of the RNAi machinery, consistent with their incorporation into RISC and PIWI complexes. While we cannot exclude the possibility that, along with 22 nt tRFs, factors such as KRAB-ZFP also mediate silencing at the PBS, proteins that recognize MusD PBS from mouse have not been found in humans.
tRNA primer binding distinguishes self- from non-self
Binding to endogenous, human AGO2 has been confirmed for 22 nt 3′ tRFs (Li et al., 2012), and can downregulate reporter constructs with a complementary target site in the 3′ UTR (Haussecker et al., 2010; Maute et al., 2013). A DICER-dependent 22 nt 3′ tRF (tRF-3b) has been shown to downregulate RPA1 expression in a miRNA-like fashion (Maute et al., 2013), but no biological function had been demonstrated for 18 nt CCA 3′ tRFs (tRF-3a). We found 18 nt CCA 3′ tRFs abundantly expressed in mouse stem cells in which ERVs are released from epigenetic silencing. They target a large number of ERVs, particularly young and active ones that cause ongoing mutagenesis, and inhibit retrotransposition by interfering with reverse transcription. By deleting the PBS, we found that MusD can mobilize non-autonomous ETn elements entirely in trans, even though retrotransposons such as IAP and L1 have a strong cis preference for their own transcript assuring propagation of the active, coding master copy (Dewannieux et al., 2004; Esnault et al., 2000; Wei et al., 2001). LTR-retrotransposons are thought to reverse transcribe from RNA multimer templates undergoing inter- and intramolecular strand transfers (Heidmann and Heidmann, 1991; Panganiban and Fiore, 1988). The MusD mutant lacking the PBS cannot be reverse transcribed, but ETn is efficiently retrotransposed with no MusD retroviral intermediates (Figures 6A and 7B). Thus any MusD copy that escapes epigenetic silencing and RNAi inhibition can still successfully replicate abundant non-coding ETn transcripts which by-pass splicing and translation and may “go incognito” for transcriptional- and post-transcriptional silencing. Indeed, ETn elements are much less of a target of epigenetic silencing in the mouse genome than the coding competent MusD (Maksakova et al., 2009), and ETn is much more successful in terms of genome penetrance and novel insertions (Nellaker et al., 2012). Notably, tRF are able to safeguard genome integrity against LTR-retrotransposons which escape other means of detection by specifically recognizing RNA that has the ability to move, i.e. that has a highly complementary PBS to prime reverse transcription (Figure 7).
Transposon defense is complicated for the host by the challenge to discriminate self from non-self: many genes have functional domains derived from transposons (Feschotte, 2008) or harbor TE sequences in their non-coding, regulatory regions (Faulkner et al., 2009). This means transposon silencing mediated by siRNAs and piRNAs derived from the entire TE comes with potential “off-target” effects on gene regulation. The PBS target site of 3′ CCA tRFs is downstream of the LTR promoter but upstream of their protein coding sequences. It is highly conserved across LTR-retrotransposons but not in genes and is thereby a unique target sequence to recognize and inhibit endogenous as well as infectious LTR-retrotransposons with the cell’s readily available repertoire of 3′ CCA tRFs. Potential target sites of 3′ CCA tRFs are also found at the PBS of human ERVs (Kawaji et al., 2008; Li et al., 2012), and abundant 3′ CCA tRFs with perfect complementarity to HERVs are found in cancer cell lines (A.J.S. and R.M., unpublished data). Highly conserved Lys3-AAA 3′ tRFs in human cells restrict horizontally introduced murine MusD in our study but also abate exogenous HIV virus infection (Yeung et al., 2009). Our results suggest that viral defense could have evolved from a much more widespread role for tRFs in inhibiting LTR retroelements.
The PBS is the Achilles heel of retrotransposition: if this 18 nt sequence is mutated the TE evades tRF silencing but also loses its ability to replicate using host tRNAs. Indeed, tRNA metabolism and TE defense could be linked: TE mobility has to be especially restricted in tissues with high proliferation that offer plenty of tRNA retrotransposition primers but also templates for tRFs. Since all organisms have tRNAs, tRFs provide an innate immunity during horizontal entry of LTR-retrotransposons because they are present no matter if the individual has been exposed to that transposon before. Thus the ability of 3′ CCA tRFs to inhibit LTR-retrotransposons could very well be a highly conserved mechanism to control mobility of transposons that have escaped epigenetic repression.
STAR-METHODS
KEY RESOURCES TABLE
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| goat anti-rabbit, HRP-coupled | Jackson Immuno Research | Cat #111-035-144 |
| rabbit MusD1 gag antiserum | Thierry Heidmann lab | #58 |
| rabbit polyclonal anti-beta tubulin | Abcam | Cat #ab6046 |
| rabbit polyclonal H3K9me3 antibody | Active Motif | Cat #39161 |
| Bacterial and Virus Strains | ||
| One Shot Stbl3 Competent E. coli | Invitrogen | Cat #C737303 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1000 U/ml LIF | ESGRO/Millipore | Cat #ESG1106 |
| 2-βmercaptoethanol | Sigma | Cat #M7522 |
| DMEM | GIBCO/Thermo Fisher Scientific | Cat #10566-016 |
| FBS | Stem Cell Technologies | Cat #06952 |
| FGF-4 | R&D Systems | Cat #235-F4-025 |
| G418, geneticin | Thermo Fisher Scientific | Cat #10131027 |
| Glo-lysis buffer | Promega | Cat #E266A |
| Knockout-DMEM | GIBCO/Thermo Fisher Scientific | Cat #10829018 |
| L-glutamine | Thermo Fisher Scientific | Cat #25030081 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat #11668019 |
| MEM non-essential amino acids | Thermo Fisher Scientific | Cat #11140050 |
| Restore Stripping Buffer | Thermo Fisher Scientific, Pierce | Cat #21059 |
| Trizol | Thermo Fisher Scientific | Cat #15596018 |
| Critical Commercial Assays | ||
| Bradford Protein Assay | Biorad | Cat #5000006 |
| Dual-Glo Luciferase Assay System | Promega | Cat #E2920 |
| ECL Western Blotting Substrate | Thermo Fisher Scientific, Pierce | Cat #32106 |
| First Choice RLM Race kit | Thermo Fisher Scientific, Ambion | Cat #AM1700 |
| Illumina Small RNA kit v1.5 | Illumina | discontinued |
| Illumina TruSeq Small RNA kit | Illumina | Cat #RS-200-0012 |
| KOD polymerase | Millipore | Cat #71975-3 |
| Power SYBR Green PCR Master Mix | Thermo Fisher Scientific | Cat #4367659 |
| QIAGEN DNA miniprep kit | QIAGEN | Cat #27104 |
| SuperScriptII, SuperScriptIII | Thermo Fisher Scientific | Cat #18064014, Cat #18080044 |
| TaqMan Universal Master Mix II, no UNG | Thermo Fisher Scientific | Cat #4440040 |
| TurboDNase | Thermo Fisher Scientific, Ambion | Cat #AM2239 |
| Deposited Data | ||
| raw and analyzed data | this paper | GSE82199 |
| Experimental Models: Cell Lines | ||
| mouse embryonic fibroblast | MTI Global Stem | Cat #GSC-6001G |
| J1 mouse embryonic stem cells (mESC) | ATCC | Cat #SCRC-1010 |
| J1 mouse Dnmt1 knockout mESC | Wolf Reik (Babraham Institute, Cambridge/UK) | Dnmt1tm1Enl, MGI:1857601 |
| mouse Setdb1 inducible knockout mESC | Yoichi Shinkai (Kyoto University, Japan | clone 33.6 |
| mouse trophoblast stem cells | Janet Rossant (Sick Children Hospital, Toronto/Canada | #TS6.5 |
| human HeLa cells | ATCC | CCL-2 |
| Oligonucleotides | ||
| all oligonucleotide sequences see Table S1 | this paper | N/A |
| Recombinant DNA | ||
| pCMV-beta | Clontech | U02451.1 |
| pCMV-ETnIIbeta-TNFneo | (Ribet et al., 2004) | AC126548 |
| pCMV-MusD6 | (Ribet et al., 2004) | AC124426 |
| pIAP92L23-TNFneo | (Dewannieux et al., 2004) | AC012382.14 |
| pCMV-MusD6-PBS* | this paper | N/A |
| pCMV-ETn-PBSMusD | this paper | N/A |
| pGL4.21[luc2P/Puro] | Promega | E6761 |
| pGL4.74[hRluc/TK] | Promega | E6921 |
| pGL4.21-MusD | this paper | N/A |
| pGL4.21-MusD-PBS* | this paper | N/A |
| Software and Algorithms | ||
| Bamtools | (Barnett et al., 2011) | https://sourceforge.net/projects/bamtools/ |
| Bedtools | (Quinlan and Hall, 2010) | http://bedtools.readthedocs.io/en/latest/index.html |
| Bowtie | (Langmead et al., 2009) | http://bowtie-bio.sourceforge.net/index.shtml |
| Bowtie2 | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Broad Institute IGV browser | (Robinson et al., 2011) | http://software.broadinstitute.org/software/igv/download |
| Cutadapt | (Martin, 2011) | https://pypi.python.org/pypi/cutadapt |
| fastx-toolkit | Assaf Gordon | https://github.com/agordon/fastx_toolkit |
| Samtools | (Li et al., 2009) | http://samtools.sourceforge.net/ |
| Other | ||
| 15% Novex TBE urea gels | Thermo Fisher Scientific | Cat #EC6885BOX |
| Amersham Protran nitrocellulose membrane, 45 uM | GE Healthcare | Cat #10600072 |
CONTACT FOR REAGENTS AND RESOURCE SHARING
Requests for reagents will be fulfilled by the corresponding, lead author Rob Martienssen (martiens@cshl.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines and Tissue Culture
Mouse embryonic fibroblast (MEF) feeder cells to support stem cell growth were obtained from MTI Global Stem (GSC-6001G). Mouse trophoblast stem cells (TS6.5 derived from B5/EGFP transgenic mice) were grown as previously reported with FGF4 (235-F4-025, R&D systems) and in MEF-conditioned medium (Tanaka et al., 1998). Setdb1 inducible knockout mouse embryonic stem cells were treated with 4-hydroxytamoxifen as previously described (clone 33.6; Matsui et al., 2010). Downregulation of SETDB1 was confirmed by quantitative PCR (Figure S1B). Setdb1 inducible mESC, as well as wildtype J1 (ATCC SCRC-1010) and J1 Dnmt1 knockout mESC (Dnmt1tm1Enl, MGI:1857601) were grown in Knockout-DMEM (GIBCO/Thermo Fisher Scientific), 1000 U/ml LIF (ESGRO/Millipore), 0.1 mM 2-βmercaptoethanol, 15% FBS (Stem Cell Technologies), 1 mM MEM non-essential amino acids, and 20 mM L-glutamine, and weaned off MEF cells 2–24 h before lysis and RNA harvest.
HeLa cells (ATCC CCL-2) were grown in DMEM, 10% FCS. All cell lines used in this study were grown at 37°C and 5% CO2.
METHOD DETAILS
RNA Preparation and Small RNA Sequencing
RNA was extracted using Trizol (Thermo Fisher Scientific) and 80% EtOH washes during precipitation. For small RNA cloning, total RNA was size selected for 14–38 nt on 15% Novex TBE urea gels (Thermo Fisher Scientific). Small RNA libraries were prepared using the Illumina Small RNA kit v1.5 and Illumina TruSeq Small RNA kit and sequenced on Illumina platforms GAII, HiSeq 2000, and NextSeq 500 as outlined in the GEO submission accompanying this study. To test for tRF induction during transposition (Figure S6), HeLa cells were transfected with Lipofectamine 2000 (Thermo Fisher Scientific) and 0.6 ug MusD6, or ETnIIbeta-neo, or mock plasmid (pCMVbeta) respectively, and 14–38 nt small RNA were sequenced using the Illumina TruSeq Small RNA kit and the Illumina Miseq platform.
Retrotransposition Assays
2.5x 105 HeLa cells were seeded in 6-well plates the day before transfection and transfected in triplicates with plasmid DNA and 200 pmol tRFs (unless otherwise mentioned) using Lipofectamine 2000 (Thermo Fisher Scientific) resulting in a final concentration of 100 nM tRFs. For MusD/ETn assays, CMV-driven MusD6 and ETnIIbeta-TNFneo plasmids (0.6 ug each, unless otherwise mentioned) were cotransfected (Ribet et al., 2004). The IAP assay was done using 1.2 ug IAP-92L23-TNFneo plasmid driven by its endogenous promoter (Dewannieux et al., 2004). Transposon sequence accession numbers are AC124426, AC126548, AC012382.14. ETnIIbeta is most closely related to the MMETn consensus sequence of the RepeatMasker annotation, MusD6 is an ETnERV2 element and IAP-92L23 belongs to the IAPEz family. Sequences of all transfected tRF RNA oligonucleotides are listed in Table S1. The ETnIIbeta oligo reflects an abundant, endogenously expressed ETn tRF sequence derived from tRNALys. The neo-tagged IAP-92L23 element and hence the tRF oligo used for transfection have two mismatches compared to the highly abundant, endogenously expressed IAPEz tRF derived from tRNAPhe. After transfection, cells were expanded for two passages and 1/10 of the cells were selected for 10–14 days with 500 ug/ml geneticin (Thermo Fisher Scientific). Geneticin resistant colonies were fixed with 10% v/v formaldehyde in PBS, stained with 0.1% w/v bromophenol blue in PBS, and counted for retrotransposition events. Retrotransposition inhibition by tRFs was stronger for higher concentrations of tRFs (data not shown) thus silencing was dose-dependent. Oligonucleotide sequences are listed in Table S1.
H3K9me3 ChIPseq
H3K9me3 native ChIP on TS cells was done as described in Karimi et al., 2011 using an Active Motif antibody (#39161). Specificity of the H3K9me3 pull-down was verified by SYBRgreen qPCR for Prtg, Pik3r3, Cdx2, and Tcfap2a previously shown to be H3K9 trimethylated in TS cells (Rugg-Gunn et al., 2010). Primers are listed in Table S1. Mono- and dinucleosomal fractions were cloned using Ethan Ford’s protocol (https://ethanomics.wordpress.com/chip-seq-library-construction-using-the-illumina-TruSeq-adapters/) and Illumina TruSeq DNA adapters. Libraries were sequenced on the Illumina NextSeq 500 platform.
Modified 5′ RACE
An initial few hundred sequences of 5′ RNA ends of MusD and ETn were determined by Sanger sequencing and using the Ambion First Choice RLM Race kit, omitting the TAP/CIP treatment to clone only uncapped 5′ RNA from cleavage events and RNaseH intermediates. Ligated RNA was reverse transcribed using SuperScriptII (Thermo Fisher Scientific) and random hexamers to include non-polyadenylated sequences. To obtain greater sequencing depth, cDNA was amplified in nested PCR (i) with MusD/ETn-specific primers, (ii) to include Illumina TruSeq adapters (for primers see Table S1) and subsequently sequenced on the Illumina MiSeq platform.
Quantitative RT-PCRs
RNA was treated with TurboDNase (Thermo Fisher Scientific) and reverse transcribed using random hexamers and SuperscriptIII (Thermo Fisher Scientific). SYBR Green PCR (Thermo Fisher Scientific) was used to measure SETDB1, IAP, ETn, and MusD expression in preimplantation stem cells (Figures S1B and S1C). To measure MusD and ETn expression in transfected cells (Figures 5A, 6C, and S5B) mean transcript levels ± SD were determined by quantitative TaqMan RT-PCR specific for MusD or ETn, respectively, and normalized to endogenous actin levels. RNA was harvested two days (Figure 5A) and four days (Figures 6C, S5B) post transfection when MusD and ETn are expressed and retroviral intermediates detectable. All primer and probe sequences are listed in Table S1.
Retroviral DNA Quantification
Extrachromosomal DNA was isolated from HeLa cells three days post transfection using the QIAGEN DNA miniprep kit. Mean values ± SD using custom-designed TaqMan primers/probe against the neo splice junction were normalized to total transfected plasmid DNA using primers/probe against the CMV-promoter driving ETn and MusD. Primer and probe sequences used for quantitative TaqMan PCR are listed in Table S1.
Western Blot Analysis
HeLa cells were lysed two days after transfection in 1x Laemmli buffer with 4 M urea and 50 mM DTT. Proteins were separated by 12% SDS-PAGE and transferred onto Amersham Protran nitrocellulose membrane (0.45 uM, GE Healthcare). Rabbit antiserum raised against MusD1 Gag and recognizing MusD6 Gag proteins was a kind gift from the Heidmann lab (Ribet et al., 2004). Anti-rabbit HRP-coupled IgG was used as secondary antibody (Jackson Immuno Research) and ECL Western Blotting Substrate (Pierce) for detection. To control for equal loading, membranes were stripped with Restore Stripping Buffer (Pierce) and incubated with rabbit anti-beta Tubulin antibody (Abcam). All antibody incubations were performed in 5% milk in TBS buffer with 0.05% v/v Tween20.
Cloning
For pCMV-MusD6-PBS*, the MusD6 PBS sequence was replaced by a non-targeting siRNA sequence (Figure S5C). MusD6 contained a potential alternative tRNA binding site 9 bp downstream of the canonical PBS, which we mutated at the same time. The replacement was introduced by cloning a gBlock Gene Fragment (Integrated DNA Technologies) with the desired sequence SnaBI/AvrII into pCMV-MusD6. pCMV-ETnIIbeta was PCR mutagenized to contain the MusD6-PBS using KOD polymerase (Millipore) (primers see Table S1) to result in pCMV-ETn-PBSMusD-neo after digestion of parental plasmid with DpnI. All sequences that contained LTRs were propagated in Stbl3 E. coli (Thermo Fisher Scientific, Invitrogen).
To generate luciferase reporter constructs, the CMV promoter and the entire 5′ UTR, including the LTR and PBS, were PCR amplified from pCMV-MusD6 and pCMV-MusD6-PBS*, respectively, (primers see Table S1) and KpnI/ApaI inserted into pGL4.21 (Promega). This resulted in pGL4.21-MusD6 and pGL4.21-MusD6-PBS*, in which expression of firefly luciferase is driven by the promoter and the transposon UTR, conserving the first six amino acids of the gag ORF followed directly by the luciferase reporter gene (see Figure 6D “MusD6-luc”, “MusD6-PBS*-luc”, “no promoter-luc” = empty pGL4.21). All sequences that underwent PCR amplification or DNA synthesis were verified by Sanger sequencing.
Luciferase Assays
HeLa cells were transfected with 100 ng of each pGL4.21 firefly luciferase reporter construct and 100 ng renilla luciferase pGL4.74 per 6-well and lysed in 500 ul Glo-lysis buffer (Promega, #E266A) one day post transfection. 70 ul lysate was measured with the Dual-Glo Luciferase Assay System (Promega) using a GloMax-Multi+ Microplate Multimode Reader (Promega). Firefly luciferase luminescence was normalized with renilla luciferase luminescence, and relative light units (RLU) were calculated by normalizing luminescence values for total protein amount determined by Bradford Protein Assay (Biorad) at 595 nm.
QUANTIFICATION AND STATISTICAL ANALYSIS
Small RNA Analysis
Small RNA reads were quality filtered using Gordon Assaf’s fastx-toolkit. Cutadapt was used to clip Illumina adapters, size select for 14–38 nt read length and remove any Truseq Illumina stop oligo sequences. Reads from the mouse stem cell samples were aligned to the mouse mm9 UCSC genome annotation using Bowtie2 which assigns multi-mapping reads in an unbiased way. Since young, potentially active transposable elements include highly conserved sequences, multimappers were included and randomly assigned to a single locus. Aligned reads were filtered for 0–1 mismatches using Samtools and Bamtools. For repeat analysis, aligned reads were intersected with the RepeatMasker annotation (Repeat Library 20090604, http://www.repeatmasker.org) using Bedtools. Reads mapping within annotated ERVs are shown in Figure 1A. For tRF analysis, reads were sorted into non-CCA and CCA-ending sequences (CCA clipped), and aligned against the RepeatMasker tRNA annotation (remaining mm9 masked). CCA-tRNA reads (Figures S1A and S2B) aligned exactly to the 3′ end of tRNAs minus the tRNA trailer sequence in the genomic annotation but comprising a terminal CCA like mature tRNAs do (Figures 2A, S2A and S3). Non-CCA tRNA reads (Figure S2E) were plotted along tRNA coordinates defining the 5′ end as zero position (Figure S2D). Subsequently, tRFs (including terminal CCA nucleotides for CCA reads) were aligned against mm9 (tRNA sequences masked) and intersected with the RepeatMasker ERV annotation to determine read counts and size distributions of ERV tRFs (Figures 1B-D, S1D, 2C, S2C/F). All read counts were normalized to total aligned reads per library including 3′ CCA tRNA reads that aligned to mature tRNAs but not the reference genome. Read counts per million (RPM) and position of CCA tRFs within ERVs were collected and plotted along ERV coordinates of selected mm9 subfamilies (Figure 3A).
Small RNA reads from sequencing of transfected HeLa cells underwent the same filtering and quality control of raw reads as described above. Reads with a 3′ terminal CCA were clipped for CCA (as above) and aligned against the human hg38 RepeatMasker tRNA annotation (Repeat Library 20140131) with the remaining human genome masked. Positions within tRNA coordinates were plotted (not shown) to verify reads align to the very 3′ end of the annotation minus the genomic tRNA trailer sequence that is missing in mature tRNAs and 3′ CCA tRFs. Subsequently, 3′ CCA tRF read counts were normalized as described above for the mm9 analysis and plotted by size for Figures S6A and S6B. Read counts of all sequences containing the last 16 nt of the “ETn” Lys-AAA tRF were plotted by size for Figure S6C.
R scripts were used for data visualization. All analysis was done using custom Perl and Bash scripts including Awk/Sed/Unix commands and the above described software (references see STAR Key Resources Table).
Statistical Analysis
Where indicated, p values were calculated using the Welch Two Sample t-test and 95% confidence intervals in R (version 3.2.2). Boxplots were plotted with R, the edges being the 25th and 75th percentiles and the middle line representing the median. Whiskers represent minimum and maximum values within 1.5 times the interquartile range and all outliers are plotted. For small RNA data, outliers and all data points are plotted.
ChIPseq Analysis
Paired-end reads were aligned with Bowtie2 and processed using Bamtools, Samtools, Bedtools, and custom scripts. For the analysis of tRF loci tested in H3K9me3 enrichment, no mismatches were allowed for small RNA alignment. tRF-targeted PBS (+/− 100 bp) that had more than 2 RPM (~20 raw reads) and occurred in at least two of eight replicates of sRNA TS data sets were defined as “tRF-targeted”, all other PBS of that ERV family were defined as control. Average read counts of four TS cell ChIPseq H3K9me3 replicates were calculated for targeted versus control tRF loci and plotted.
5′ RACE Analysis
105 bp single-end reads were quality-filtered using the fastx toolkit and mapped against the MusD6 and ETnIIbeta sequences with Bowtie allowing 0 mismatches. The PBS region contains several SNPs between MusD6 and ETnIIbeta allowing unambiguous mapping of reads. Read counts were normalized to total mapped reads per million (RPM) and coverage for all reads >100 nt was calculated using Bedtools to display 5′ RNA ends in the Broad Institute IGV browser with MusD6 and ETnIIbeta as “genome”. Welch Two Sample t-test was performed for each pair of replicates after adding up RPM values for each RNaseH product peak (three consecutive nucleotide positions).
DATA AVAILABILITY
The high-throughput sequencing data reported in this paper was deposited at NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE82199, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=wlsnsguczliddyt&acc=GSE82199
Supplementary Material
(A) Setdb1 knockout mouse ES cells and TS cells show elevated overall levels of 3′ CCA tRFs. Amounts and sizes of 3′ CCA tRFs for all samples and replicates are shown in Figure S3. (B) Induction of Setdb1 knockout was efficient and abolished SETDB1 expression (primers span exon 14–15). (B) IAP expression is only modestly increased in Dnmt1 −/− ES cells but strongly upregulated in Setdb1 −/− and TS cells. MusD and its non-coding ETn partner are released in Setdb1 −/− as previously reported. All values are mean transcript levels normalized to actin expression +/− SD as determined by quantitative SYBRgreen RT-PCR. All primer sequences are listed in Table S1. (C) All ERV families targeted by 3′ CCA tRFs in Setdb1 knockout and TS cells. Pie charts represent average RPM values of 4 replicate Setdb1 −/−, and 7 replicate TS sRNA libraries. For comparison, relative abundance of ERV sequences in the mm9 mouse genome: 3% of all ERVs belong to the IAP family, 0.4% to the ETn family. IAP and ETn both belong to the ERV-K superfamily.
(A) CCA-tRFs align to the very 3′ end of mature tRNAs (one TS cell sample shown, all samples see Figure S3) and (B) are 18 and 22 nt in length. (C) CCA tRFs that match ERVs are primarily 18 nt in length. (D) Non-CCA tRFs are all 5′ end derived halves (one representative TS cell sample shown) and (E) 30–33 nt long but also include degraded halves, so are less stable than CCA-tRFs. (F) ERVs are no obvious targets of 5′ tRF halves in mouse based on sequence alignment.
Alignment of CCA reads along tRNA coordinates for all samples of this study show they map to the very 3′ end of mature tRNAs and are ~18 and 22 nt in length. 18 and 22 nt 3′ tRFs appear in different ratios for different tissue types but ratios also slightly depend on which Illumina platform and adapters were used for sRNA sequencing. Technical replicates, designated a/b, are separate library preparations from the same RNA sample; biological replicates are designated by integer count.
(A) H3K9me3 levels are extremely low at ERVs in TS cells, at targeted loci (yellow) as well as all other genomic ERV loci (control, khaki). MusD6 is an ETnERV2 element and ETnIIbeta belongs to the MMETn family. Boxplots include 4 biological replicates of H3K9me3 ChIPseq reads in RPM within +/− 100 bp of the ERV PBS. For details see STAR methods section ChIPseq. (B) Knock-down of all four AGOs had no significant effects on tRF retrotransposition inhibition. Note that transposition inhibition was more modest in the presence of high levels of artificial siRNA, which lower the final absolute amount of co-transfected transposon plasmids and tRFs. 0.2x 106 cells were transfected with 0.6 ug of MusD and ETn-neo plasmids, 50 nM tRFs and 4x25 nM siGenome pool AGO1/2/3/4 (Thermo Scientific Dharmacon, see Supplemental Table S1). Knockdown of any single AGO had no consistent effect on tRF inhibition of MusD/ETn retrotransposition (data not shown). Colony counts are the mean of three replicates +/− SD.
(A) Retrotransposition of mutated ETn, ETn-PBSMusD-neo, is decreased. To test the impact of endogenous tRFs on RT priming, we replaced the PBS with the MusD PBS which is known to sufficiently prime autonomous MusD but has two mismatches with the tRNALys primer. While this mutation may have relieved ETn from RT inhibition by Lys-tRFs, it decreases tRNALys priming at the same time and resulted in a net reduction of transposition compared to wildtype. As expected, ETn tRF transfection did not significantly suppress retrotransposition of the ETn-PBSMusD-neo mutant, since reverse transcription is more strongly inhibited by perfectly complementary tRFs. Colony counts are the mean of two biological replicates +/− SD, wildtype MusD with control tRFs was set to 100%. One biological replicate had standard amounts of ETn/MusD and tRFs transfected, the other one 0.3 ug ETn/MusD. p-value Welch two sample t-test; * p-value = 0.1–0.05, ns: not significant. (B) ETn tRFs do not change total RNA level of MusD or ETn in transposition assays with either wildtype MusD or MusD-PBS*. Mean transcript levels +/− SD were determined by quantitative Taqman RT-PCR. (C) Sequences of ETn, MusD and MusD-PBS* are shown 5′ to 3′ within a 40 nt window around the PBS. Nucleotides different from the MusD sequence are bold and underlined.
(A) HeLa cells have similar expression level of 17–19 nt and 22 nt tRFs as mouse preimplantation stem cells (Figures S2 and S3). (B) Overall 3′ tRFs did not change after transfection of HeLa cells with MusD, ETn-neo or control plasmid (pCMVbeta), respectively. pCMVbeta was used as control because MusD and ETn-neo are in a pCMVbeta plasmid backbone. (C) Transfection with ERVs did also not trigger ETn tRFs (Lysine3-AAA) specifically. Boxplots represent RPM of three biological replicates per condition.
Table S1, related to STAR Methods section. Oligonucleotide sequences
HIGHLIGHTS.
highly abundant tRNA fragments (tRF) in mouse stem cells
3′ CCA tRFs target and inhibit endogenous retroviruses (ERV) active in mouse
tRFs target the highly conserved primer binding site (PBS) of LTR-retrotransposons
18 nt tRFs block reverse transcription, 22 nt tRFs induce RNAi
Acknowledgments
We thank Janet Rossant (Sick Children Hospital, Toronto/Canada) for the mouse trophoblast cell line. The inducible Setdb1 knockout ES cell line was a kind gift of Yoichi Shinkai (Kyoto University, Japan). We are very grateful to Marie Dewannieux and Thierry Heidmann (CNRS-IGR, Villejuif/France) for providing us with the ETn/MusD and IAP retrotransposition reporter plasmids and the Gag MusD antiserum. We thank Wolf Reik, Wendy Dean, and Myriam Hemberger (Babraham Institute, Cambridge/UK) for discussion and for providing us with the Dnmt1 knockout line as well as additional TS cell clones.
A.J.S. was supported by NIH grant R01GM076396 to R.A.M. M.J.G. was supported by a Bristol-Myers Squibb fellowship from the Watson School of Biological Sciences. This work was supported by the Howard Hughes Medical Institute-Gordon and Betty Moore Foundation (GMBF3033). The authors acknowledge assistance from the Cold Spring Harbor Laboratory Shared Resources, which are funded in part by the Cancer Center Support Grant (5PP30CA045508).
Footnotes
Supplemental information includes six Figures and Table S1 containing all oligonucleotide sequences used in the study.
AUTHORS CONTRIBUTIONS
Conceptualization, A.J.S. and R.A.M.; Formal Analysis, A.J.S.; Methodology, A.J.S. and M.J.G.; Investigation, A.J.S., M.J.G, and C.LB.; Writing – Original Draft, A.J.S.; Writing – Review & Editing, R.A.M., A.J.S., M.J.G.; Funding Acquisition, R.A.M.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24:2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat Genet. 2004;36:534–539. doi: 10.1038/ng1353. [DOI] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev Cell. 2016;36:611–623. doi: 10.1016/j.devcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann O, Heidmann T. Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell. 1991;64:159–170. doi: 10.1016/0092-8674(91)90217-m. [DOI] [PubMed] [Google Scholar]
- Hemberger M. Genetic-epigenetic intersection in trophoblast differentiation: implications for extraembryonic tissue function. Epigenetics. 2010;5:24–29. doi: 10.4161/epi.5.1.10589. [DOI] [PubMed] [Google Scholar]
- Ito M, Sferruzzi-Perri AN, Edwards CA, Adalsteinsson BT, Allen SE, Loo TH, Kitazawa M, Kaneko-Ishino T, Ishino F, Stewart CL, et al. A trans-homologue interaction between reciprocally imprinted miR-127 and Rtl1 regulates placenta development. Development. 2015;142:2425–2430. doi: 10.1242/dev.121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam SP, Hutvagner G. tRNA-Derived Fragments (tRFs): Emerging New Roles for an Ancient RNA in the Regulation of Gene Expression. Life (Basel) 2015;5:1638–1651. doi: 10.3390/life5041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam SP, Young PE, McCorkindale AL, Dang TH, Clancy JL, Humphreys DT, Preiss T, Hutvagner G, Martin DI, Cropley JE, et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014;42:8984–8995. doi: 10.1093/nar/gku620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JB, Chapman KB, Lauermann V, Voytas DF, Astrom SU, von Pawel-Rammingen U, Bystrom A, Boeke JD. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol Cell Biol. 1995;15:217–226. doi: 10.1128/mcb.15.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushaar DC, Jin W, Maunakea A, Abraham B, Ha M, Zhao K. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 2013;14:R121. doi: 10.1186/gb-2013-14-10-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs) Trends Biochem Sci. 2016;41:679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Le Grice SF. “In the beginning”: initiation of minus strand DNA synthesis in retroviruses and LTR-containing retrotransposons. Biochemistry. 2003;42:14349–14355. doi: 10.1021/bi030201q. [DOI] [PubMed] [Google Scholar]
- Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40:6787–6799. doi: 10.1093/nar/gks307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Zhang Y, Mager DL. Preferential epigenetic suppression of the autonomous MusD over the nonautonomous ETn mouse retrotransposons. Mol Cell Biol. 2009;29:2456–2468. doi: 10.1128/MCB.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Choudury SG, Slotkin RK. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla-Herman A, Valles AM, Ganem-Elbaz C, Antoniewski C, Huynh JR. tRNA processing defects induce replication stress and Chk2-dependent disruption of piRNA transcription. EMBO J. 2015;34:3009–3027. doi: 10.15252/embj.201591006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Nellaker C, Keane TM, Yalcin B, Wong K, Agam A, Belgard TG, Flint J, Adams DJ, Frankel WN, Ponting CP. The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol. 2012;13:R45. doi: 10.1186/gb-2012-13-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, Sakaki Y, Sasaki H, Hohjoh H. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res. 2010;38:5141–5151. doi: 10.1093/nar/gkq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban AT, Fiore D. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science. 1988;241:1064–1069. doi: 10.1126/science.2457948. [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5:171. doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, Goyal P, Keane TM, Jones S, Hirst M, et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011;7:e1002301. doi: 10.1371/journal.pgen.1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Dewannieux M, Heidmann T. An active murine transposon family pair: retrotransposition of “master” MusD copies and ETn trans-mobilization. Genome Res. 2004;14:2261–2267. doi: 10.1101/gr.2924904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci U S A. 2010;107:10783–10790. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero K, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G, Grassi A, Zanovello P, Bronte V, Ciminale V, et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol. 2014;88:3612–3622. doi: 10.1128/JVI.02823-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn A, Martienssen RA. F1000Prime Recommendation of [Couvillion MT et al. Genes Dev 2010. 2011;24(24):2742–7. doi: 10.3410/f.8668956.9177054. ]. (In F1000Prime, 03 Mar 2011 F1000Prime.com/8668956#eval9177054 : F1000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Xu X, Elkenani M, Smorag L, Zechner U, Nolte J, Engel W, Pantakani DV. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res. 2013;11:1045–1059. doi: 10.1016/j.scr.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Hug K, Goff SP. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc Natl Acad Sci U S A. 2008;105:12521–12526. doi: 10.1073/pnas.0805540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer EM, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015;29:538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37:6575–6586. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Setdb1 knockout mouse ES cells and TS cells show elevated overall levels of 3′ CCA tRFs. Amounts and sizes of 3′ CCA tRFs for all samples and replicates are shown in Figure S3. (B) Induction of Setdb1 knockout was efficient and abolished SETDB1 expression (primers span exon 14–15). (B) IAP expression is only modestly increased in Dnmt1 −/− ES cells but strongly upregulated in Setdb1 −/− and TS cells. MusD and its non-coding ETn partner are released in Setdb1 −/− as previously reported. All values are mean transcript levels normalized to actin expression +/− SD as determined by quantitative SYBRgreen RT-PCR. All primer sequences are listed in Table S1. (C) All ERV families targeted by 3′ CCA tRFs in Setdb1 knockout and TS cells. Pie charts represent average RPM values of 4 replicate Setdb1 −/−, and 7 replicate TS sRNA libraries. For comparison, relative abundance of ERV sequences in the mm9 mouse genome: 3% of all ERVs belong to the IAP family, 0.4% to the ETn family. IAP and ETn both belong to the ERV-K superfamily.
(A) CCA-tRFs align to the very 3′ end of mature tRNAs (one TS cell sample shown, all samples see Figure S3) and (B) are 18 and 22 nt in length. (C) CCA tRFs that match ERVs are primarily 18 nt in length. (D) Non-CCA tRFs are all 5′ end derived halves (one representative TS cell sample shown) and (E) 30–33 nt long but also include degraded halves, so are less stable than CCA-tRFs. (F) ERVs are no obvious targets of 5′ tRF halves in mouse based on sequence alignment.
Alignment of CCA reads along tRNA coordinates for all samples of this study show they map to the very 3′ end of mature tRNAs and are ~18 and 22 nt in length. 18 and 22 nt 3′ tRFs appear in different ratios for different tissue types but ratios also slightly depend on which Illumina platform and adapters were used for sRNA sequencing. Technical replicates, designated a/b, are separate library preparations from the same RNA sample; biological replicates are designated by integer count.
(A) H3K9me3 levels are extremely low at ERVs in TS cells, at targeted loci (yellow) as well as all other genomic ERV loci (control, khaki). MusD6 is an ETnERV2 element and ETnIIbeta belongs to the MMETn family. Boxplots include 4 biological replicates of H3K9me3 ChIPseq reads in RPM within +/− 100 bp of the ERV PBS. For details see STAR methods section ChIPseq. (B) Knock-down of all four AGOs had no significant effects on tRF retrotransposition inhibition. Note that transposition inhibition was more modest in the presence of high levels of artificial siRNA, which lower the final absolute amount of co-transfected transposon plasmids and tRFs. 0.2x 106 cells were transfected with 0.6 ug of MusD and ETn-neo plasmids, 50 nM tRFs and 4x25 nM siGenome pool AGO1/2/3/4 (Thermo Scientific Dharmacon, see Supplemental Table S1). Knockdown of any single AGO had no consistent effect on tRF inhibition of MusD/ETn retrotransposition (data not shown). Colony counts are the mean of three replicates +/− SD.
(A) Retrotransposition of mutated ETn, ETn-PBSMusD-neo, is decreased. To test the impact of endogenous tRFs on RT priming, we replaced the PBS with the MusD PBS which is known to sufficiently prime autonomous MusD but has two mismatches with the tRNALys primer. While this mutation may have relieved ETn from RT inhibition by Lys-tRFs, it decreases tRNALys priming at the same time and resulted in a net reduction of transposition compared to wildtype. As expected, ETn tRF transfection did not significantly suppress retrotransposition of the ETn-PBSMusD-neo mutant, since reverse transcription is more strongly inhibited by perfectly complementary tRFs. Colony counts are the mean of two biological replicates +/− SD, wildtype MusD with control tRFs was set to 100%. One biological replicate had standard amounts of ETn/MusD and tRFs transfected, the other one 0.3 ug ETn/MusD. p-value Welch two sample t-test; * p-value = 0.1–0.05, ns: not significant. (B) ETn tRFs do not change total RNA level of MusD or ETn in transposition assays with either wildtype MusD or MusD-PBS*. Mean transcript levels +/− SD were determined by quantitative Taqman RT-PCR. (C) Sequences of ETn, MusD and MusD-PBS* are shown 5′ to 3′ within a 40 nt window around the PBS. Nucleotides different from the MusD sequence are bold and underlined.
(A) HeLa cells have similar expression level of 17–19 nt and 22 nt tRFs as mouse preimplantation stem cells (Figures S2 and S3). (B) Overall 3′ tRFs did not change after transfection of HeLa cells with MusD, ETn-neo or control plasmid (pCMVbeta), respectively. pCMVbeta was used as control because MusD and ETn-neo are in a pCMVbeta plasmid backbone. (C) Transfection with ERVs did also not trigger ETn tRFs (Lysine3-AAA) specifically. Boxplots represent RPM of three biological replicates per condition.
Table S1, related to STAR Methods section. Oligonucleotide sequences
Data Availability Statement
The high-throughput sequencing data reported in this paper was deposited at NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE82199, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=wlsnsguczliddyt&acc=GSE82199