Synopsis
Human genetic diversity is the result of population genetic forces. This genetic variation influences disease risk and contributes to health disparities. Natural selection is an important influence on human genetic variation. Since immune and inflammatory function genes are enriched for signals of positive selection, the prevalence of rheumatic disease risk alleles seen in different populations is partially the result of differing selective pressures (e.g., due to pathogens). This review summarizes the genetic regions associated with susceptibility to different rheumatic diseases and concomitant evidence for natural selection, including known agents of selection exerting selective pressure in these regions. Integrating rheumatic disease susceptibility studies with population genetics to investigate how natural selection has contributed to genetic variation that influences disease risk will help identify functional variants and elucidate biological mechanisms.
Keywords: rheumatic diseases, population genetics, natural selection, genetic variation, genetic disease association, genetic diversity, adaptation, genetic disease risk
Introduction
Rheumatic diseases are a family of more than 100 chronic, and often disabling, illnesses characterized by inflammation and loss of function, especially in the joints, tendons, ligaments, bones, and muscles. They collectively affect over 20% of US adults, with osteoarthritis, rheumatoid arthritis, spondylarthritides, gout and fibromyalgia being the most prevalent.1,2 Patients often endure lifelong debilitating symptoms, reduced productivity at work, and high medical expenses. Arthritis and related illnesses, as well as back or spine problems are major causes of disability.3 Importantly, since many rheumatic diseases present before or during a woman's reproductive years, they can have effects on fetal and maternal outcomes,4 such as pregnancy loss in women with systemic lupus erythematosus4,5 and vasculitis,4 and infertility in women with rheumatoid arthritis.5
Most rheumatic diseases exhibit marked gender and ethnic disparities. Most predominately afflict women (e.g. rheumatoid arthritis, systemic lupus erythematosus, systemic sclerosis, fibromyalgia), but spondyloarthropathies and gout are more common in men.6 African Americans are at higher risk than European Americans for systemic lupus erythematosus and systemic sclerosis, which they tend to develop earlier in life and experience more severe disease.7 Despite the variation in prevalence, incidence and disease severity that are known to vary among ethnic groups, little is known about the genetic etiology of these diseases in the different populations and the reasons for the ethnic disparities remain elusive.
Left untreated, most rheumatic diseases can affect the ability to raise offspring that successfully reproduce and result in reduced reproductive fitness. Thus, alternative forces must exist that permit the relative high frequency of risk alleles. Since immune and inflammatory responses can be highly sensitive to environmental change,8 evolutionary adaptation to specific environments might have driven selection on immune-related genetic variants, impacting variant frequencies and leaving signatures of selection in the genome. Given that infectious organisms are strong agents of natural selection,9,10 it is plausible that alleles selected for protection against infection confer increased risk of autoimmune and inflammatory diseases, as the “hygiene hypothesis”11 postulates. It is thought that the adaptation to pathogen pressure through functional variation in immune-related genes conferred a specific selective advantage for host survival, including protection from pathogens and tolerance to microbiota.12 However, the emergence of such variation conferring resistance to pathogens is also influencing immune and inflammatory disease risk in specific populations.
In the past decade, multiple genome scans for signatures of selection on common variation have identified many immune-related loci.13-17 Similarly, 90 genome-wide association studies (GWAS) (Table 1) have established rheumatic disease-associated alleles. There is also growing evidence that autoimmune and inflammatory disease-associated variants are under selection.17-21 This review expands on our previous work22 and summarizes the evidence for rheumatic disease-associated loci under selection and the candidate selective pressures. Given that genomic variation can have clinically important consequences,23 elucidating the patterns of variation and the functional role of the selective pressure might contribute to a better understanding of disease etiology and the development of new therapies for improved disease management.
Table 1.
Rheumatic diseases with published GWAS and respective number of associated loci.
| Rheumatic diseases | Number of | |
|---|---|---|
| GWAS | loci | |
| ANCA-associated vasculitis (AAV) | 1 | 18 |
| Ankylosing spondylitis (AS) | 3 | 21 |
| Behçet's disease (BD) | 5 | 9 |
| Dermatomyositis (DM) | 1 | 1 |
| Gout | 4 | 14 |
| Granulomatosis with polyangiitis (GPA) | 1 | 6 |
| Juvenile idiopathic arthritis (JIA) | 3 | 6 |
| Kawasaki disease (KD) | 6 | 16 |
| Osteoarthritis (OA) | 9 | 16 |
| Osteoporosis (OP) | 3 | 3 |
| Paget's disease (PD) | 2 | 9 |
| Psoriasis (PS) | 11 | 60 |
| Psoriatic arthritis (PsA) | 2 | 4 |
| Rheumatoid arthritis (RA) | 19 | 129 |
| Sjögren's syndrome (SS) | 1 | 4 |
| Systemic lupus erythematosus (SLE) | 16 | 124 |
| Systemic sclerosis (SScl) | 3 | 10 |
Numbers compiled from the NHGRI-EBI Catalog of Published Genome-Wide Association Studies (https://www.ebi.ac.uk/gwas) accessed on October 24th, 2016.46
Shared genetic etiology in rheumatic diseases
The family of rheumatic diseases is remarkable for its heterogeneity and similar underlying mechanisms. The genetic heritability of rheumatic diseases is extremely variable, ranging from very high in ankylosing spondylitis to almost negligible in systemic sclerosis.24 GWAS have proved particularly powerful for autoimmune diseases,25 including many autoimmune rheumatic diseases, which might be due to their immune and inflammatory genetic etiology. Table 1 summarizes the rheumatic diseases with published GWAS and the number of disease-associated loci uncovered from these GWAS. The common genetic etiology is exemplified by the sharing of associated loci among rheumatic diseases, such as the Human Leukocyte Antigen (HLA), STAT4, TNIP1, TNFAIP3, and BLK.26 This sharing of risk loci is greater among the groups of diseases characterized by the presence of particular serum autoantibodies (seropositive; such as rheumatoid arthritis, systemic lupus erythematosus, etc.) than it is between the seropositive and seronegative diseases (those typically characterized as not having associated serum autoantibodies.26 This supports the consensus that there is a common genetic background predisposing to autoimmunity and inflammation, and that further combinations of more serologically defined and disease specific variation at HLA and non-HLA genes, in interaction with epigenetic and environmental factors, contribute to disease and its clinical manifestations. It has been suggested that different population genetic factors (e.g., natural selection with coevolution with pathogens, random mutation, isolations, migrations and interbreeding) in similar or distinct environments led to the establishment of the current plethora of loci that predispose to autoimmunity.27 It is thus plausible that population-level phenomena are a reason behind the complexity of gene effects in different autoimmune and rheumatic diseases.
Population genetics, natural selection and adaptation
The genetic basis of disease is influenced by individual and population variation. Population- level phenomena such as mutation, migration, genetic drift and natural selection, have left an imprint on genetic variation that is likely to influence phenotypic expression in specific populations.23 Given its role in driving genetic variation, population genetics can help elucidate human genetic diversity and, consequently, disease etiology.
Natural selection is the process by which a trait becomes either more or less common in a population depending on the differential reproductive success of those with the trait. Natural selection drives adaptation, the evolutionary process whereby over generations the members of a population become better suited to survive and reproduce in that environment. Negative (or purifying) selection is the most common mechanism of selection, usually associated with rare Mendelian disorders. Positive selection increases the prevalence of adaptive traits by increasing the frequency of favorable alleles and is often associated with common complex traits.28 The enrichment for signals of positive selection among genes associated with complex traits is well documented.14,29-31 Balancing selection favors genetic diversity by retaining variation in the population as a result of heterozygote advantage and frequency-dependent advantage. Despite rarer, a pertinent example is the HLA (also known as major histocompatibility complex (MHC)) region,32,33 where highly polymorphic loci play a central role in the recognition and presentation of antigens to the immune system. The high levels of polymorphism are the results of pathogen-driven balancing selection.34 The heterozygote advantage against multiple pathogens contributes to the evolution of HLA diversity, which in turn confers resistance against multiple pathogens and explains the persistence of alleles conferring susceptibility to disease.35 Nevertheless, there is also recent evidence that positive selection might be acting on specific HLA alleles in a local population due to unique environmental pressures.36
Natural selection leaves a distinctive molecular signature in the targeted genomic region, and different statistical methods have been developed to detect signatures of selection.12 It has been hypothesized by Klironomos and colleagues that, in addition to genetic (sequence) variation, heritable epigenetic modifications can affect rates of fitness increase, as well as patterns of genotypic and phenotypic change during adaptation.37 However, the role of epigenetic variation in the response to natural selection has not been formally assessed, as the methodology to test signatures of natural selection on epigenetic variation is just emerging.38
Natural selection in rheumatic disease
Given that, if untreated, rheumatic diseases can diminish reproductive potential and impair the ability to raise offspring that successfully reproduce, some evolutionary process must sustain the relative high frequency of risk alleles seen in current populations around the world. Since the human genome is shaped by adaptation to environmental pressures at the population level, one plausible reason for the higher frequency of disease-risk alleles may be the direct effect of population-specific natural selection. This hypothesis is supported by the experimental evidence for MHC heterozygote superiority against multiple pathogens, a mechanism that would contribute to the evolution of HLA diversity and explain the persistence of alleles conferring susceptibility to disease.35
There is compelling evidence that natural selection is acting on a significant fraction of the human genome.15,39-43 Immune function genes and pathways are consistently reported in tests for natural selection. As a result of several genome-wide scans, over 300 immune-related genes have been suggested as putative targets of positive selection.13-17 Although the challenge in validating the true signals remains,44 several genes involved in immune-related functions have been shown to be under selection.20,45
A total of 61 regions with evidence for selection and association with at least one rheumatic disease are shown in Table 2. This table includes 35 regions previously reported as being under selection in the literature,22 plus rheumatic disease-associated loci from current GWAS (in Table 1) and evidence of recent positive selection from Hapmap phase II data.15 Specifically, a region published in the GWAS Catalog46 as associated with a rheumatic disease was considered as exhibiting evidence for natural selection if it contained at least two SNPs within 200 kb with an absolute integrated Haplotype Score (iHS) value in the top 0.1% of the genome-wide distribution in one population (Asian, European or African). A total of 39 regions that met these criteria are included in Table 2, 14 of which were previously reported. These 39 regions with evidence for selection represent about 10% of all regions associated with a rheumatic disease in a GWAS: 13% for systemic lupus erythematosus (SLE), 9% for rheumatoid arthritis (RA), 7% for psoriasis (PS), and 5% for ankylosing spondylitis (AS). This fraction of disease-associated loci with concomitant evidence for selection is higher than previous reports focusing on SNPs instead of regions. Notably, when using the top 1% of iHS variants, Raj and colleagues21 reported that inflammatory diseases (which included AS, RA and SLE) have 5% of SNPs targeted by positive selection. Limiting comparisons to SNPs instead of regions might miss regions with both evidence for disease association and selection at different SNPs. The numbers of GWAS-associated loci, including those with and without concomitant evidence for recent positive selection, are illustrated in Figure 1. Among all regions in Table 2, a higher number of signals of selection were found in European (36%), followed by Asian (32%) and African (32%) populations. This is consistent with previous reports of enrichment of inflammatory-disease SNPs targeted by positive selection in subjects of European ancestry.21,47
Table 2.
Rheumatic disease regions with evidence for selection and implicated agents of selection.
| Gene region | Position | Rheumatic disease association | References for evidence of natural selection | Population | Selective pressure | References for pathogen-driven selection |
|---|---|---|---|---|---|---|
| TNFRSF14, MMEL1* | 1p36.32 | RA | YRI | |||
| IL23R | 1p31.3 | AS | 18 | protozoa | 53 | |
| MAGI3, PTPN22* | 1p13.2 | RA, SLE | 19,20 | YRI | protozoa | 53 |
| FCGR2B | 1q23.3 | SLE | 57 | Plasmodium falciparum | 57 | |
| TNFSF4 | 1q25.1 | RA, SS, SLE | 19 | |||
| NCF2, RGL1* | 1q25.3 | SLE | ASI | |||
| CR1 | 1q32 | SLE | 65 | Plasmodium falciparum | 65 | |
| TLR5 | 1q41-q42 | SLE | 20 | YRI | Salmonella enterica ser. Typhimurium and other exposures | 20 |
| PELI1* | 2p14 | KD | ASI | |||
| ALMS1P, DGUOK* | 2p13.1 | SLE | 19 | CEU | ||
| PARD3B* | 2q33.3 | OA | 20 | CEU | ||
| CNTN6* | 3p26.3 | SLE | ASI | |||
| XCR1, CCR3* | 3p21.31 | BD | YRI | |||
| CCDC66, ARHGEF3 | 3p14.3 | RA | 20 | YRI | ||
| BTLA | 3q13.2 | RA | ASI | |||
| ARHGAP31, CD80 | 3q13.33 | JIA, SLE | 21 | YRI | ||
| MRPS22* | 3q23 | KD | 20 | ASI | ||
| SLC2A9* | 4p16.1 | Gout | YRI | |||
| KCNIP4* | 4p15.2 | RA | 20 | CEU, YRI | ||
| TECRL* | 4q13.1 | KD | CEU | |||
| ANTRX2 | 4q21 | AS | 21 | |||
| IL2, IL21* | 4q27 | RA | 21,47 | YRI | ||
| Intergenic* | 4q28.3 | SLE | ASI | |||
| PTGER4 | 5p13.1 | AS | 21,53 | protozoa | 53 | |
| COMMD10, SEMA6A* | 5q23.1 | GPA | ASI, CEU | |||
| ALDH7A1* | 5q23.2 | OP | CEU | |||
| TNIP1 | 5q33.1 | SLE, SScl, PsA | 19 | |||
| PTTG1 | 5q33.3 | SLE | 19 | |||
| IRF4* | 6p25.3 | RA | CEU | |||
| ITPR3 | 6p21.31 | SLE | 20 | YRI | ||
| HLA* | 6p22.1-6p21.31 | AAV, AS, BD, GPA, JIA, KD, OA, PS, PsA, RA, SS, SLE, SScl | 20,21,66-69 | ASI, CEU, YRI | bacterial infection | 34,54,55 |
| SNRPC, UHRF1BP1* | 6p21.31 | SLE | 19-21 | CEU | Mycobacterium tuberculosis | 70 |
| VARS, LSM2 | 6p21 | SLE | 21 | |||
| CCDC167, MIR4462* | 6p21.2 | SLE | YRI | |||
| PRDM1, ATG5* | 6q21 | RA, SLE | YRI | |||
| TRAF3IP2* | 6q21 | PS, PsA | YRI | |||
| IKZF1 | 7p12.2 | SLE | 19 | |||
| GTF2I* | 7q11.23 | SS | 20 | ASI | ||
| HIP1* | 7q11.23 | SLE | 21 | YRI | ||
| LSMEM1, NPM1P14* | 7q31.1 | OA | ASI | |||
| XKR6, BLK* | 8p23.1 | KD, RA, SS, SLE, SSc | 19,20 | ASI | ||
| GRHL2* | 8q22.3 | RA | CEU | |||
| KDM4C* | 9p24.1 | SLE | ASI | |||
| NTNG2, SETX* | 9q34.13 | SLE | 20 | CEU | ||
| FAM171A1* | 10p13 | SLE | ASI | |||
| CTNNA3* | 10q21.3 | PsA | CEU | |||
| CD5 | 11q12.2 | RA | 71 | |||
| GRM5* | 11q14.3 | RA | CEU | |||
| OSBPL8* | 12q21.2 | SLE | CEU | |||
| SH2B3, NAA25 | 12q24.12-q24.13 | RA | 21,56 | bacterial infection | 56 | |
| KIAA0391* | 14q23.1 | PS | ASI | |||
| PRKCH, HIF1A* | 14q23.1 | RA | 20 | CEU, YRI | ||
| CLEC16A, CIITA | 16p13.13 | RA, SLE | 19,21,72 | |||
| ITGAM, ITGAX | 16p11.2 | SLE | 19,20 | |||
| PRSS54* | 16q21 | SLE | YRI | |||
| WWOX | 16q23.2 | OA | 20 | CEU | ||
| IRF8 | 16q24.1 | RA, SScl | 73 | |||
| RABEP1, NUP88* | 17p13.2 | RA | CEU | |||
| BCAS3, NACA2* | 17q23.2 | Gout, OA | 20 | ASI | ||
| TYK2 | 19p13.2 | RA, SLE | 53 | protozoa | 53 | |
| PAK7* | 20p12.2 | PS | YRI |
Rheumatic disease associations were reported in the literature (column “references for evidence of natural selection”), and/or in the NHGRI-EBI GWAS Catalog accessed on October 24th, 2016. In addition to the disease-associated regions with evidence for selection reported in the literature, rheumatic disease-associated loci from the GWAS catalog with evidence of recent positive selection from HapMap phase II data (assessed by the presence of at least two SNPs within approximately 200 kb with an absolute iHS value in the top 0.1% of the genome-wide distribution in one population) are also included and denoted by the asterisk. See Table 1 for disease abbreviations. ASI: Asian, CEU: European, YRI: African populations.
Figure 1.
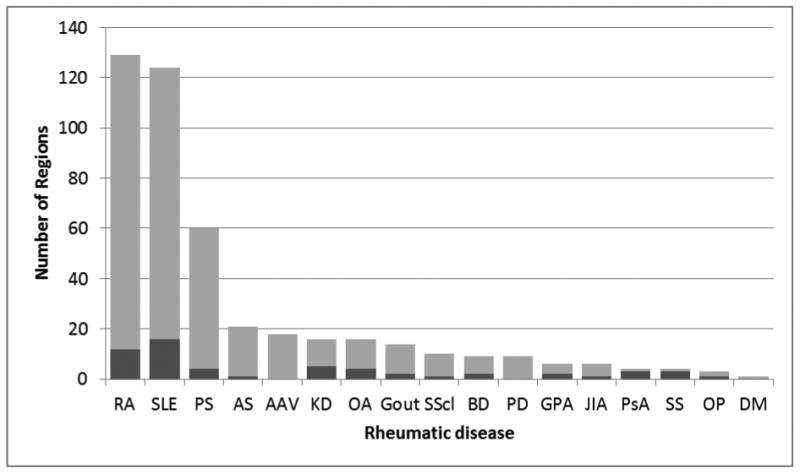
Number of rheumatic disease GWAS-associated loci, including those with concomitant evidence for recent positive selection (dark shaded area). See Table 1 for disease abbreviations.
Agents of selection
The wide variety of environments inhabited by human populations is likely exerting different selective pressures that lead to adaptation through natural selection. Climatic factors such as altitude, latitude, ultra-violet radiation levels, temperature, as well as diet and pathogens have been reported as agents of selection driving adaptations to these environments and lifestyles. As recently reviewed,22 some relevant examples include signals of natural selection driven by annual photoperiod variation reported for restless leg syndrome risk variants,48 correlation between climate variables and SNPs involved in immune response, as well as pathways related to UV radiation, infection and immunity, and cancer,49 and correlations between worldwide migration trajectories and variants associated with, among other, systemic lupus erythematosus and systemic sclerosis.50 Interestingly, expression QTLs (eQTLs) (see article by Laufer et al elsewhere in this issue for definition) from immune function and metabolism genes are enriched in signals of environmental adaptation,51 which highlights the importance of regulatory variations in local adaptation.
Nevertheless, the strongest effect of climate is in shaping the spatial pattern and species diversity of human pathogens,9 which is directly relevant to immune and inflammatory disease predisposition. As recently reviewed,52 in the constant co-evolutionary battle between host and pathogen, pathogens that diminish reproductive potential, either through death or poor health, drive selection on genetic variants that affect pathogen resistance. As Hancock suggested,49 it is likely that selection signals in immune-related loci may implicate variants evolving under a model of antagonistic pleiotropy, where the selective pressure was pathogen resistance, and the inflammatory disorder is a pleiotropic consequence of the resistance allele. This could hence be a mechanism explaining the prevalence of immune risk alleles that are common in the population.
Indeed, pathogens have been the main selective pressure through human evolution.10 In an analysis that included climate, diet regimes, and pathogen loads, Fumagalli and colleagues10 showed that the diversity of the local pathogenic environment is the predominant driver of local adaptation, and that climate conditions only played a relatively minor role. In addition, they reported an enrichment of genes associated to SLE, RA and AS, which supports the hypothesis that some susceptibility alleles for rheumatic diseases may be maintained in human population due to past selective processes.10 The enrichment for signals of positive selection in inflammatory-disease susceptibility loci has been recently corroborated.21 Reviews of selection signatures left by pathogen-exerted pressure, including immune-related genes, can be found elsewhere.52,53
Genetic regions associated with susceptibility to different rheumatic diseases and evidence of selection that has been attributed to host-pathogen coevolution are shown in Table 2. In a fraction of the regions with evidence for selection and disease-association, known pathogens have been implicated as the selective pressure. Variation in the HLA and SH2B3 has been reported as a protective factor against bacterial infection.34,54-56 Resistance to protozoa and tuberculosis infection have been implicated as the selective pressures for PTPN22 and UHRF1BP1, respectively. Interestingly, the SLE susceptibility allele in UHRF1BP1 is associated with decreased UHRF1BP1 RNA expression in different cell subsets, suggesting that the disease risk allele under selection has a regulatory effect.21 In the context of SLE predisposing loci, Clatworthy et al.57 has shown that FCGR2B is important in controlling the immune response to Plasmodium falciparum, the parasite responsible for the most severe form of malaria, and suggests that the higher frequency of human FCGR2B polymorphisms predisposing to SLE in Asians and Africans may be maintained because these variants reduce susceptibility to malaria. Grossman et al.20 implicated Salmonella typhimurium and other exposures that directionally drive selection of the toll-like receptor 5 (TLR5) gene,58 which is involved in recognition of flagellated bacteria. Unlike endosomal TLRs, such as TLR8 and TLR8, that have been subject to purifying selection, cell-surface TLRs involved in pathogen recognition experienced more relaxed constraints.59 The non-synonymous variant in PTPN22 shows complex signatures of selection, increasing the risk of SLE, RA, and other autoimmune diseases, but being protective against Crohn's disease.60 Karlsson et al.61 have recently reported that cholera has exerted strong selective pressure on pro-inflammatory pathways. Despite the modest number of examples that offer clear functional hypotheses (e.g. SH2B3, TRL5), collectively this list supports the hypothesis that the increased prevalence of rheumatic disease may result, at least partially, from past events of selection that increased host resistance to infection.62
Discussion
This review summarizes the genetic regions associated with susceptibility to different rheumatic diseases and concomitant evidence for selection, including known agents of selection exerting selective pressure in these regions. Uncovering these rheumatic disease-associated loci under selection underscores the importance of population genetics and how the understanding of human genetic diversity is crucial to understanding disease etiology or treatment response at both the population and individual levels.
A combination of population-level phenomena, including possibly bottlenecks, migration, admixture, natural selection, and random genetic drift, are likely contributors to this complexity of gene effects in different rheumatic diseases. Given the complex history of selective pressures acting on humans, unequal selective pressures and a diverse spectrum of plausible evolutionary models are expected to be exerted on susceptibility loci for rheumatic diseases.28 It is likely that several pathogens have exerted pressure on the same loci and that selection can vary in form, intensity, time and space, which is consistent with the observation that both risk and protective alleles for rheumatic diseases increased in frequency due to selection.17 For most regions, the exact selective pressure leaving the signature of selection is unclear. Clearly, these signatures are not necessarily the result of adaptation, but might be a consequence of random genetic drift. In any case, regardless of the population phenomenon shaping current human genetic diversity, this genetic variation is the basis clinically relevant traits at both the individual and population levels.23
An important next step to delineate the selective advantage conferred by these rheumatic disease risk variants are functional studies using in vitro experiments and model organisms to identify the underlying functional variants and quantify the phenotypic consequences of the candidate adaptive alleles. Human- pathogen coevolution is ongoing and, despite the emergence of new pathogens (e.g. HIV), potential pathogens driving these host-specific adaptations are expected to have long-standing relationships with humans, including those that cause malaria, smallpox, cholera, tuberculosis and leprosy,63 as well as the human microbiome.64 Regardless of the agent of selection and the reasons for the emergence of both common and rare rheumatic disease-causing alleles, incorporating population genetics to understand human genetic diversity will lead to a better understanding of the causes of health disparities, identification of functional variants and discovery of cellular mechanisms, and contribute to the development of new therapies.
Key Points.
If untreated, rheumatic diseases can diminish reproductive potential and impair the ability to raise offspring that successfully reproduce. Thus, it is likely that the frequency of disease-risk alleles seen in populations around the world is influenced by population-specific natural selection.
Both autoimmune and non-autoimmune rheumatic disorders show genetic associations in regions with signatures of selection.
The prevalence of rheumatic disease may result, at least partially, from past events of selection that increased host resistance to infection.
Many of the complexities of gene effects in different rheumatic diseases can be explained by population genetics phenomena.
Acknowledgments
This study was supported by the US National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (NIH) under Award Numbers K01 AR067280, R03 AR065801, and P60 AR062755. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Disclosure statement: The author has nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Helmick CG, Felson DT, Lawrence RC, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum. 2008 Jan;58(1):15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence RC, Felson DT, Helmick CG, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum. 2008 Jan;58(1):26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. Prevalence and Most Common Causes of Disability Among Adults - United States, 2005. Morbidity and Mortality Weekly Report. 2009;58:421–426. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5816a2.htm. [PubMed] [Google Scholar]
- 4.Ostensen M, Andreoli L, Brucato A, et al. State of the art: Reproduction and pregnancy in rheumatic diseases. Autoimmun Rev. 2015 May;14(5):376–386. doi: 10.1016/j.autrev.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Clowse ME, Chakravarty E, Costenbader KH, Chambers C, Michaud K. Effects of infertility, pregnancy loss, and patient concerns on family size of women with rheumatoid arthritis and systemic lupus erythematosus. Arthritis care & research. 2012 May;64(5):668–674. doi: 10.1002/acr.21593. [DOI] [PubMed] [Google Scholar]
- 6.NIH. Arthritis and Rheumatic Diseases. National Institute of Health Publication No 14-4999. 2014 http://www.niams.nih.gov/Health_Info/Arthritis/arthritis_rheumatic.pdf.
- 7.NIH. Progress in Autoimmune Diseases Research. National Institute of Health Publication No 05-514. 2005 www.niaid.nih.gov/topics/autoimmune/documents/adccfinal.pdf.
- 8.Okin D, Medzhitov R. Evolution of inflammatory diseases. Current biology : CB. 2012 Sep 11;22(17):R733–740. doi: 10.1016/j.cub.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guernier V, Hochberg ME, Guegan JF. Ecology drives the worldwide distribution of human diseases. PLoS biology. 2004 Jun;2(6):e141. doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admetlla A, Pattini L, Nielsen R. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011 Nov;7(11):e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989 Nov 18;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quintana-Murci L, Clark AG. Population genetic tools for dissecting innate immunity in humans. Nature reviews Immunology. 2013 Apr;13(4):280–293. doi: 10.1038/nri3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickrell JK, Coop G, Novembre J, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome research. 2009 May;19(5):826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007 Oct 18;449(7164):913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS biology. 2006 Mar;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008 Mar;40(3):340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 17.Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010 Jan;11(1):17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- 18.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012 Nov 1;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos PS, Shaftman SR, Ward RC, Langefeld CD. Genes associated with SLE are targets of recent positive selection. Autoimmune diseases. 2014;2014 doi: 10.1155/2014/203435. 2014. (ArticleID:203435):Article ID 203435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossman SR, Andersen KG, Shlyakhter I, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013 Feb 14;152(4):703–713. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj T, Kuchroo M, Replogle JM, Raychaudhuri S, Stranger BE, De Jager PL. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am J Hum Genet. 2013 Apr 4;92(4):517–529. doi: 10.1016/j.ajhg.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos PS, Shedlock AM, Langefeld CD. Genetics of autoimmune diseases: insights from population genetics. Journal of human genetics. 2015 Nov;60(11):657–664. doi: 10.1038/jhg.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torkamani A, Pham P, Libiger O, et al. Clinical implications of human population differences in genome-wide rates of functional genotypes. Frontiers in genetics. 2012;3:211. doi: 10.3389/fgene.2012.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selmi C, Lu Q, Humble MC. Heritability versus the role of the environment in autoimmunity. J Autoimmun. 2012 Dec;39(4):249–252. doi: 10.1016/j.jaut.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 25.Hu X, Daly M. What have we learned from six years of GWAS in autoimmune diseases, and what is next? Current opinion in immunology. 2012 Oct;24(5):571–575. doi: 10.1016/j.coi.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Kirino Y, Remmers EF. Genetic architectures of seropositive and seronegative rheumatic diseases. Nature reviews Rheumatology. 2015 Jul;11(7):401–414. doi: 10.1038/nrrheum.2015.41. [DOI] [PubMed] [Google Scholar]
- 27.Ramos PS, Criswell LA, Moser KL, et al. A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. PLoS Genet. 2011 Dec;7(12):e1002406. doi: 10.1371/journal.pgen.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Rienzo A. Population genetics models of common diseases. Curr Opin Genet Dev. 2006 Dec;16(6):630–636. doi: 10.1016/j.gde.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Bustamante CD, Fledel-Alon A, Williamson S, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005 Oct 20;437(7062):1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 30.Blekhman R, Man O, Herrmann L, et al. Natural selection on genes that underlie human disease susceptibility. Current biology : CB. 2008 Jun 24;18(12):883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torgerson DG, Boyko AR, Hernandez RD, et al. Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence. PLoS Genet. 2009 Aug;5(8):e1000592. doi: 10.1371/journal.pgen.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andres AM, Hubisz MJ, Indap A, et al. Targets of balancing selection in the human genome. Molecular biology and evolution. 2009 Dec;26(12):2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gineau L, Luisi P, Castelli EC, et al. Balancing immunity and tolerance: genetic footprint of natural selection in the transcriptional regulatory region of HLA-G. Genes Immun. 2015 Jan-Feb;16(1):57–70. doi: 10.1038/gene.2014.63. [DOI] [PubMed] [Google Scholar]
- 34.Prugnolle F, Manica A, Charpentier M, Guegan JF, Guernier V, Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Current biology : CB. 2005 Jun 7;15(11):1022–1027. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 35.McClelland EE, Penn DJ, Potts WK. Major histocompatibility complex heterozygote superiority during coinfection. Infection and immunity. 2003 Apr;71(4):2079–2086. doi: 10.1128/IAI.71.4.2079-2086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawashima M, Ohashi J, Nishida N, Tokunaga K. Evolutionary analysis of classical HLA class I and II genes suggests that recent positive selection acted on DPB1*04:01 in Japanese population. PLoS One. 2012;7(10):e46806. doi: 10.1371/journal.pone.0046806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klironomos FD, Berg J, Collins S. How epigenetic mutations can affect genetic evolution: model and mechanism. BioEssays : news and reviews in molecular, cellular and developmental biology. 2013 Jun;35(6):571–578. doi: 10.1002/bies.201200169. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Fan C. A neutrality test for detecting selection on DNA methylation using single methylation polymorphism frequency spectrum. Genome biology and evolution. 2015 Jan;7(1):154–171. doi: 10.1093/gbe/evu271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberle MA, Rieder MJ, Kruglyak L, Nickerson DA. Allele frequency matching between SNPs reveals an excess of linkage disequilibrium in genic regions of the human genome. PLoS Genet. 2006 Sep 8;2(9):e142. doi: 10.1371/journal.pgen.0020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002 Oct 24;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 41.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genetical research. 1974 Feb;23(1):23–35. [PubMed] [Google Scholar]
- 42.Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007 Jun;3(6):e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gulko B, Hubisz MJ, Gronau I, Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat Genet. 2015 Mar;47(3):276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome research. 2009 May;19(5):711–722. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fumagalli M, Cagliani R, Pozzoli U, et al. Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome research. 2009 Feb;19(2):199–212. doi: 10.1101/gr.082768.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hindorff LA, MacArthur J, Morales J, et al. A Catalog of Published Genome-Wide Association Studies. [Accessed july 22nd, 2016]; Available at: www.genome.gov/gwastudies.
- 47.Brinkworth JF, Barreiro LB. The contribution of natural selection to present-day susceptibility to chronic inflammatory and autoimmune disease. Current opinion in immunology. 2014 Dec;31:66–78. doi: 10.1016/j.coi.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forni D, Pozzoli U, Cagliani R, et al. Genetic adaptation of the human circadian clock to day-length latitudinal variations and relevance for affective disorders. Genome biology. 2014 Oct 30;15(10):499. doi: 10.1186/s13059-014-0499-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hancock AM, Witonsky DB, Alkorta-Aranburu G, et al. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 2011 Apr;7(4):e1001375. doi: 10.1371/journal.pgen.1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corona E, Chen R, Sikora M, et al. Analysis of the genetic basis of disease in the context of worldwide human relationships and migration. PLoS Genet. 2013 May;9(5):e1003447. doi: 10.1371/journal.pgen.1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye K, Lu J, Raj SM, Gu Z. Human expression QTLs are enriched in signals of environmental adaptation. Genome biology and evolution. 2013;5(9):1689–1701. doi: 10.1093/gbe/evt124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nat Rev Genet. 2014 Jun;15(6):379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cagliani R, Sironi M. Pathogen-driven selection in the human genome. International journal of evolutionary biology. 2013;2013:204240. doi: 10.1155/2013/204240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988 Sep 8;335(6186):167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 55.Qutob N, Balloux F, Raj T, et al. Signatures of historical demography and pathogen richness on MHC class I genes. Immunogenetics. 2012 Mar;64(3):165–175. doi: 10.1007/s00251-011-0576-y. [DOI] [PubMed] [Google Scholar]
- 56.Zhernakova A, Elbers CC, Ferwerda B, et al. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet. 2010 Jun 11;86(6):970–977. doi: 10.1016/j.ajhg.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clatworthy MR, Willcocks L, Urban B, et al. Systemic lupus erythematosus-associated defects in the inhibitory receptor FcgammaRIIb reduce susceptibility to malaria. Proc Natl Acad Sci U S A. 2007 Apr 24;104(17):7169–7174. doi: 10.1073/pnas.0608889104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hawn TR, Wu H, Grossman JM, Hahn BH, Tsao BP, Aderem A. A stop codon polymorphism of Toll-like receptor 5 is associated with resistance to systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2005 Jul 26;102(30):10593–10597. doi: 10.1073/pnas.0501165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barreiro LB, Ben-Ali M, Quach H, et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009 Jul;5(7):e1000562. doi: 10.1371/journal.pgen.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013 Sep;14(9):661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 61.Karlsson EK, Harris JB, Tabrizi S, et al. Natural selection in a bangladeshi population from the cholera-endemic ganges river delta. Science translational medicine. 2013 Jul 3;5(192):192ra186. doi: 10.1126/scitranslmed.3006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sironi M, Clerici M. The hygiene hypothesis: an evolutionary perspective. Microbes and infection / Institut Pasteur. 2010 Jun;12(6):421–427. doi: 10.1016/j.micinf.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 63.Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982 Oct;85(Pt 2):411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- 64.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cockburn IA, Mackinnon MJ, O'Donnell A, et al. A human complement receptor 1 polymorphism that reduces Plasmodium falciparum rosetting confers protection against severe malaria. Proc Natl Acad Sci U S A. 2004 Jan 6;101(1):272–277. doi: 10.1073/pnas.0305306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cagliani R, Riva S, Pozzoli U, et al. Balancing selection is common in the extended MHC region but most alleles with opposite risk profile for autoimmune diseases are neutrally evolving. BMC evolutionary biology. 2011;11:171. doi: 10.1186/1471-2148-11-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Black FL, Hedrick PW. Strong balancing selection at HLA loci: evidence from segregation in South Amerindian families. Proc Natl Acad Sci U S A. 1997 Nov 11;94(23):12452–12456. doi: 10.1073/pnas.94.23.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Fu Y, Liu Z, et al. An ancient balanced polymorphism in a regulatory region of human major histocompatibility complex is retained in Chinese minorities but lost worldwide. Am J Hum Genet. 2006 Mar;78(3):393–400. doi: 10.1086/500593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan Z, Shon AM, Ober C. Evidence of balancing selection at the HLA-G promoter region. Hum Mol Genet. 2005 Dec 1;14(23):3619–3628. doi: 10.1093/hmg/ddi389. [DOI] [PubMed] [Google Scholar]
- 70.Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci U S A. 2012 Jan 24;109(4):1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carnero-Montoro E, Bonet L, Engelken J, et al. Evolutionary and functional evidence for positive selection at the human CD5 immune receptor gene. Molecular biology and evolution. 2012 Feb;29(2):811–823. doi: 10.1093/molbev/msr251. [DOI] [PubMed] [Google Scholar]
- 72.Swanberg M, Lidman O, Padyukov L, et al. MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat Genet. 2005 May;37(5):486–494. doi: 10.1038/ng1544. [DOI] [PubMed] [Google Scholar]
- 73.Choudhury A, Hazelhurst S, Meintjes A, et al. Population-specific common SNPs reflect demographic histories and highlight regions of genomic plasticity with functional relevance. BMC Genomics. 2014;15:437. doi: 10.1186/1471-2164-15-437. [DOI] [PMC free article] [PubMed] [Google Scholar]


