Abstract
Type 1 diabetes is a chronic autoimmune disease characterized by pancreatic islet inflammation and β-cell destruction by proinflammatory cytokines and other mediators. Based on RNA sequencing and protein–protein interaction analyses of human islets exposed to proinflammatory cytokines, we identified complement C3 as a hub for some of the effects of cytokines. The proinflammatory cytokines interleukin-1β plus interferon-γ increase C3 expression in rodent and human pancreatic β-cells, and C3 is detected by histology in and around the islets of diabetic patients. Surprisingly, C3 silencing exacerbates apoptosis under both basal condition and following exposure to cytokines, and it increases chemokine expression upon cytokine treatment. C3 exerts its prosurvival effects via AKT activation and c-Jun N-terminal kinase inhibition. Exogenously added C3 also protects against cytokine-induced β-cell death and partially rescues the deleterious effects of inhibition of endogenous C3. These data suggest that locally produced C3 is an important prosurvival mechanism in pancreatic β-cells under a proinflammatory assault.
Complement C3 is at the core of a protein network in human islets. C3 plays an important role as an anti-inflammatory and prosurvival protein during inflammation-induced stress in pancreatic β-cells.
Type 1 diabetes (T1D) is a chronic autoimmune disease characterized by loss of insulin-producing β-cells by apoptosis and pancreatic islet inflammation (insulitis). Insulitis is established when an inadequate “dialog” between the immune system and pancreatic β-cells develops (1, 2) in the context of environmental signals (such as viral infections) acting upon a risk-enhancing genetic background (3). Invading immune cells produce and secrete several cytokines and chemokines in the β-cell surroundings, modulating the expression of key transcription factors, such as nuclear factor κB, signal transducer and activator of transcription, and interferon regulatory factor and their downstream gene networks. The balance between the prosurvival and proapoptotic components of these networks will eventually determine β-cell fate (1). Of interest, β-cells exposed to an inflammation-induced stress express several components of an early innate immune response, including many antiviral proteins (3, 4).
The complement system is a major component of the immune system, acting both in innate and adaptive immune responses. Three distinct pathways can initiate the complement cascade, namely classical, lectin, and alternative pathways; these three pathways of activation culminate with the generation of the C3 convertase enzyme complex, which cleaves complement C3 into C3a and C3b (5). Besides their well-known function in the extracellular system of host defense, recent evidence indicates that complement C3 is also intracellularly activated in immune and nonimmune cell types (6, 7), acting in metabolic organs such as adipose tissue, liver, and pancreas (8).
Elevated serum levels of complement components have been associated with prediabetes (9) and diabetes (10–12). Additionally, genetic and immunohistochemical data indicated a positive correlation between increased expression of complement proteins and higher risk of T1D (13–16). Sera from newly diagnosed T1D patients inhibit insulin secretion in a rat pancreatic β-cell line, BRIN-BD11, an effect dependent on the presence of C1q and C3 (17, 18). Moreover, C3-deficient mice are resistant to multiple low-dose streptozotocin-induced diabetes, and immune cell–derived C3 is required for the development of the disease in this model (19). Alternatively, products of C3 cleavage, such as C3a and C3adesArg (also known as acylation stimulating protein), stimulate insulin secretion both in INS-1 cells and mouse islets (20), and C3a improves β-cell function by increasing mitochondrial oxygen consumption, ATP levels, and cytoplasmic free Ca2+ (21).
In the present study, we performed protein–protein interaction analysis of RNA sequencing data of human islets exposed to the proinflammatory cytokines interleukin (IL)-1β plus interferon (IFN)-γ (22) (unpublished data) and identified C3 as an important hub of a cytokine-modified complement network in human islets. Additional experiments indicated that C3 expression and secretion is increased by proinflammatory cytokines in rodent and human β-cells. Surprisingly, C3 inhibition increased inflammation and apoptosis both under basal conditions and upon cytokine exposure. These effects were mainly due to activation of c-Jun N-terminal kinase (JNK) and inhibition of protein kinase B (AKT) signaling. Treatment with exogenous C3 protected against cytokine-induced β-cell death and partially rescued the effects of C3 inhibition. These data suggest a role of C3, namely as an important prosurvival protein in both rat and human β-cells exposed to inflammation-induced stress.
Materials and Methods
Protein–protein interaction analysis
Protein–protein interaction data were obtained from the InWeb_IM database of physically interacting human proteins (23). For the analysis we used release 2016_02_05 (downloadable via https://www.intomics.com/inbiomap/) and applied a threshold of 0.1 to the confidence score. This yielded a dataset containing the high-confidence subsets of interactions from InWeb_IM, corresponding to the highest scoring third of the total set of interactions, based on the benchmark curve in Figure 2A in Li et al. (23). Additional details are provided in the Supplemental Experimental Procedures (1.7MB, pdf) .
Culture of INS-1E cells, primary rat α- and β-cells, EndoC-βH1 human insulin-producing cells, and human islets
Rat INS-1E cells [research resource identifier (RRID): CVCL_0351, provided by Dr. C. Wollheim, Department of Cell Physiology and Metabolism, University of Geneva, Geneva, Switzerland] were cultured in RPMI 1640 GlutaMAX-I, 10 mM HEPES, 1 mM sodium pyruvate, 5% fetal bovine serum, 50 μM 2-mercaptoethanol, 50 U/mL penicillin, and 50 mg/mL streptomycin.
Adult male Wistar rats (Charles River Laboratories, L’Arbresle, France) were housed and used in agreement with the guidelines of the Belgian Regulations for Animal Care with approval of the Université Libre de Bruxelles Ethical Committee. Islets were isolated, and α- and β-cells were purified as described (24). Preparations containing 96% ± 1% α-cells (n = 6) and 93% ± 1% β-cells (n = 6) were used in this study.
The human β-cell line, EndoC-βH1 (RRID: CVCL_L909, provided by Dr. R. Scharfmann, INSERM U1016, Université Paris-Descartes, Institut Cochin, Paris, France), was cultured in Matrigel/fibronectin-coated plates as described before (25, 26).
Human islets from nondiabetic organ donors (age, 70 ± 3 years; body mass index, 24.4 ± 0.7 kg/m2; Supplemental Table 1 (1.7MB, pdf) ) were isolated in accordance with the local Ethical Committee in the University of Pisa (Pisa, Italy). Human islet isolation was performed by collagenase digestion and density-gradient purification. Isolated islets were cultured in M199 medium containing 5.5 mM glucose (27) and then sent to Brussels within 1 to 5 days of isolation. After overnight recovery, isolated human islets were dispersed and cultured as previously described (28, 29). The percentage of β-cells in the human islet preparations used was 50% ± 4% (n = 16) as determined by insulin immunocytochemistry (22).
RNA interference and adenoviral infection
All small interfering RNAs (siRNAs) used in this study are provided in Supplemental Table 2 (1.7MB, pdf) . Conditions for siRNA transfection and optimal siRNA concentration (30 nM) were established as previously described (30). Lipofectamine RNAiMAX lipid reagent (Invitrogen, Carlsbad, CA) was used to transfect cells and the AllStars negative control siRNA (Qiagen, Venlo, the Netherlands), which does not affect β-cell gene expression, function, or viability, was used as a negative control (29, 30).
For adenovirus infection, INS-1E cells were infected with control adenovirus encoding luciferase (adLuc; Sirion Biotech, Munich, Germany) or with an adenovirus encoding myr-Akt1 (adAkt; Vector Biolabs, Philadelphia, PA) as previously described (31). Cells were transfected with siCtrl or siC3 in the same day and allowed to recover for 24 hours before treatment with cytokines.
Cell treatments
On the basis of previous dose-response experiments done by our group in rodent and human cells, the following cytokine concentrations were used: recombinant human IL-1β (R&D Systems, Abingdon, UK) at 10 or 50 U/mL as indicated; recombinant rat IFN-γ (R&D Systems) at 100 and 500 U/mL for INS-1E cells and primary rat α- and β-cells, respectively; and human IFN-γ (PeproTech, Rocky Hill, NJ) at 1000 U/mL for EndoC-βH1 cells and dispersed human islets (32–34). Where indicated, cells were transfected with 1 μg/mL of the synthetic double-stranded analog polyinosinic-polycytidylic acid (PIC; InvivoGen, San Diego, CA) (28). Forskolin (FK), diazoxide, the phosphoinositide 3-kinase (PI3K) inhibitor wortmannin (Sigma-Aldrich, Bornem, Belgium) and the JNK inhibitor V (Calbiochem, San Diego, CA) were diluted in dimethyl sulfoxide and used at concentrations of 20 μM, 100 μM, 100 nM, and 5 μM, respectively. At the concentration used, diazoxide inhibited insulin secretion by >60% (data not shown).
INS-1E cells and dispersed human islets were pretreated with complement C3 protein (Calbiochem) for 2 hours followed by 24-hour (INS-1E cells) or 48-hour (dispersed human islets) cytokine treatment in the presence of C3. Where indicated, INS-1E cells were treated with the C3aR1 antagonist SB 290157 (Calbiochem) at 10 μM.
Cell viability assessment
Percentage of living, apoptotic, and necrotic cells was determined after staining with DNA-binding dyes Hoechst 33342 (HO) and propidium iodide (PI) as described (29, 35). At least 500 cells were counted per experimental condition. Cell viability was evaluated by two different observers, with one of them being unaware of sample identity, with an agreement between results of >90%.
Messenger RNA extraction and real-time polymerase chain reaction
Poly(A)+ messenger RNA (mRNA) extraction was carried out using a Dynabeads mRNA DIRECT kit (Invitrogen) following the manufacturer’s instructions; reverse transcription was performed as described (36), and quantitative real-time polymerase chain reaction was performed using SYBR Green and compared with a standard curve (37). Expression values were corrected by the housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin. Primers used in this study are provided in Supplemental Table 3 (1.7MB, pdf) .
C3 and chemokine secretion by enzyme-linked immunosorbent assay
Culture supernatants from INS-1E, primary rat α- and β-cells, and dispersed human islets were collected after cytokine treatment of determination of C3 and CXCL10 using commercially available enzyme-linked immunosorbent assay (ELISA) kits for rat and human C3 (Abcam, Cambridge, UK), rat CXCL10 (Abnova, Taoyuan, Taiwan), and human CXCL10 (R&D Systems).
Western blot analysis and immunofluorescence
Cells were washed with cold phosphate-buffered saline, lysed in RIPA buffer containing 1 mM NaF, 0.1 mM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride, and total proteins were run in sodium dodecyl sulfate–polyacrylamide gels. Immunoblot analysis was carried out as described (28) using the antibodies listed in Supplemental Table 4 (1.7MB, pdf) . Phosphorylated (P-) proteins (P-JNK, P–c-Jun, P-AKT, and P-BAD) were corrected for the respective total proteins or α-tubulin and similar results were observed using both approaches (data not shown). Immunofluorescence was performed as described before (28). Images were acquired at ×40 magnification and analyzed using AxiVision software. For antibodies used in this study, see Table 1.
Table 1.
Antibodies Used in This Study
| Peptide/Protein Target | Antigen Sequence | Name of Antibody | Manufacturer and Catalog No. | Species Raised in, Monoclonal or Polyclonal | Dilution Used | RRID |
|---|---|---|---|---|---|---|
| C3 | NA | Anti-C3 antibody | Abcam, Cambridge, UK, catalog no. ab97462 | Rabbit, polyclonal | 1:1000 (WB) or 1:100 (ICC) | AB_10679468 |
| Cleaved caspase-3 | NA | Cleaved caspase-3 (Asp175) | Cell Signaling Technology, Danvers, MA, catalog no. 9661 | Rabbit, polyclonal | 1:1000 | AB_2341188 |
| P-SAPK/JNK | NA | P-SAPK/JNK (Thr183/Tyr185) antibody | Cell Signaling Technology, Danvers, MA, catalog no. 9251 | Rabbit, polyclonal | 1:1000 | AB_331659 |
| JNK1 | NA | JNK1 (2C6) mouse mAb | Cell Signaling Technology, Danvers, MA, catalog no. 3708S | Mouse, monoclonal | 1:1000 | AB_1904132 |
| P-AKT | NA | P-Akt (Ser473) (D9E) XP® Rabbit mAb | Cell Signaling Technology, Danvers, MA, catalog no. 4060 | Rabbit, monoclonal | 1:10000 | AB_2315049 |
| AKT | NA | Akt antibody | Cell Signaling Technology, Danvers, MA, catalog no. 9272 | Rabbit, polyclonal | 1:5000 | AB_329827 |
| P-BAD | NA | P-Bad (Ser136) (D25H8) rabbit mAb | Cell Signaling Technology, Danvers, MA, catalog no. 4366 | Rabbit, monoclonal | 1:500 | AB_10547878 |
| BAD | NA | Bad antibody | Cell Signaling Technology, Danvers, MA, catalog no. 9292 | Unknown | 1:500 | AB_331419 |
| Glucagon | NA | Monoclonal anti-Glucagon antibody produced in mouse | Sigma, Bornem, Belgium, catalog no. G2654 | Mouse, monoclonal | 1:1000 | AB_259852 |
| Insulin | NA | Monoclonal anti-insulin antibody produced in mouse | Sigma-Aldrich, Bornem, Belgium, catalog no. I2018 | Mouse, monoclonal | 1:1000 | AB_260137 |
| Insulin (for histology) | NA | Polyclonal guinea pig anti-insulin antibody | DAKO, Glostrup, Denmark, catalog no. A056401-2 | Guinea pig, polyclonal | 1:250 | AB_2617169 |
| Insulin (for histology) | NA | Guinea pig anti-insulin polyclonal Antibody | Abcam, Cambridge, UK, catalog no. ab7842 | Guinea pig, polyclonal | 1:100 | AB_306130 |
| α-Tubulin | NA | Monoclonal anti–α-tubulin antibody | Sigma-Aldrich, Bornem, Belgium, catalog no. T9026 | Mouse, monoclonal | 1:5000 | AB_477593 |
| Anti-mouse IgG | NA | Peroxidase AffiniPure F(ab′)2 fragment donkey anti-mouse IgG (H+L) | Jackson ImmunoResearch Laboratories, West Grove, PA, catalog no. 715-036-150 | Polyclonal | 1:5000 | AB_2340773 |
| Anti-rabbit IgG | NA | Peroxidase AffiniPure F(ab′)2 fragment donkey anti-rabbit IgG (H+L) | Jackson ImmunoResearch Laboratories, West Grove, PA, catalog no. 711-036-152 | Polyclonal | 1:5000 | AB_2340590 |
| Goat anti-mouse IgG | NA | Goat anti-mouse IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 488 | Life Technologies, Carlsbad, CA, catalog no. A11029 | Goat, polyclonal | 1:1000 | AB_2534088 |
| Goat anti–guinea pig IgG | NA | Goat anti–guinea pig IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 488 | Life Technologies, Carlsbad, CA, catalog no. A11073 | Goat, polyclonal | 1:1000 | AB_2534117 |
| Donkey anti-rabbit IgG | NA | Alexa Fluor 488–AffiniPure donkey anti-rabbit IgG (H+L) antibody | Jackson ImmunoResearch Laboratories, West Grove, PA, catalog no. 711-545-152 | Donkey, polyclonal | 1:200 | AB_2313584 |
| Donkey anti–guinea pig | NA | DyLight 594–conjugated AffiniPure donkey anti–guinea pig | Jackson ImmunoResearch Laboratories, West Grove, PA, catalog no. 706-515-148 | Donkey, polyclonal | 1:200 | The product has been discontinued |
| Goat anti-rabbit IgG | NA | Goat anti-rabbit IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 568 | Life Technologies, Carlsbad, CA, catalog no. A11036 | Goat, polyclonal | 1:1000 | AB_2534094 |
Abbreviations: ICC, immunocytochemistry; IgG, immunoglobulin G; mAb, monoclonal antibody; NA, not applicable; WB, western blot.
Histology
Pancreas sections from nondiabetic donors and type 1 diabetic patients were obtained from two cohorts: the University of Pisa donors and previously characterized samples from the nPOD collection (38) (Supplemental Table 5 (1.7MB, pdf) ). Following antigen retrieval with 10 mM citrate buffer (pH 6.0), immunofluorescence was performed using primary antibodies against C3 and insulin and different secondary antibodies. Images were acquired at ×10, ×20, and ×40 magnification and analyzed using AxiVision or Leica MetaMorph software. See Supplemental Experimental Procedures (1.7MB, pdf) for further details.
Glucose-stimulated insulin secretion and insulin accumulation in the medium
Glucose-stimulated insulin secretion was performed in INS-1E cells as previously described (29). Briefly, INS-1E cells were preincubated for 1 hour in glucose-free RPMI 1640 GlutaMAX-I medium (Life Technologies, Carlsbad, CA) and then incubated with Krebs–Ringer solution for 30 minutes. Afterward, cells were exposed to 1.7, 17, or 17 mM glucose plus FK (20 μM) for 30 minutes. Insulin release and insulin content were measured using a rat insulin ELISA kit (Mercodia, Uppsala, Sweden) in cell-free supernatants and acid ethanol–extracted cell lysates, respectively. Results were normalized by total protein concentration, which was measured in cell lysates using protein assay dye reagent (Bio-Rad Laboratories, Hercules, CA). Insulin accumulation in the medium was measured 16 hours after addition of cytokines and/or siRNAs targeting C3.
Statistics
Data are shown as mean ± standard error of the mean (SEM) or as plotted box plot, indicating lower quartile, median, and higher quartile, with whiskers representing the range of the remaining data points. Comparisons were performed using a two-tailed paired Student t test or by analysis of variance (ANOVA) followed by paired t test with Bonferroni correction, as indicated. Results were considered as statistically significant when a P value was ≤0.05.
Data availability
The RNA sequencing data sets used in the present study are available online at http://lmedex.ulb.ac.be/data.php.
Results
Overview of the study
Using RNA sequencing data of human islets exposed to the proinflammatory cytokines IL-1β plus IFN-γ, we carried out a protein–protein interaction analysis, which indicated networks containing complement C3 to be highly significantly regulated in human islets. Subsequently, we performed mechanistic studies to assess C3 role in β-cell survival (see Supplemental Fig. 1 (1.7MB, pdf) for overview of our experimental approach).
Protein–protein interaction analysis
Based on an integrative analysis of the RNA sequencing data (case vs control) of 10 preparations of human pancreatic islets treated or not with proinflammatory cytokines (22) (unpublished data), we performed an initial unbiased scan of the entire human interactome (network collection no. 1; see Supplemental Data (1.7MB, pdf) ) using the InWeb_IM resource, recently benchmarked and found to have a higher coverage and better functional biological relevance than do comparable resources (23). For this we used an algorithmic approach based on P value integration of the gene expression data and Monte Carlo simulation rather than overrepresentation analysis, which allows for identification of more subtle patterns in the data. From the initial analysis, multiple highly significant networks (P < 10−6, which is as significant as it is possible to get with 1 million iterations in the Monte Carlo simulation–based approach used) containing C3 were identified. Investigating this observation further, we performed an exhaustive search of the near neighborhood of C3 (network collections nos. 2 and 3; see Supplemental Data (1.7MB, pdf) ) and found most of these (including the full first-order network around C3) to be significant. For completeness, note that additional networks not containing C3 were also identified as significant, but we have deliberately chosen to focus on the C3 networks in this study owing to the novelty and potential biological relevance of C3-related networks. The full set of findings of the global network analysis is being addressed in the course of a follow-up study.
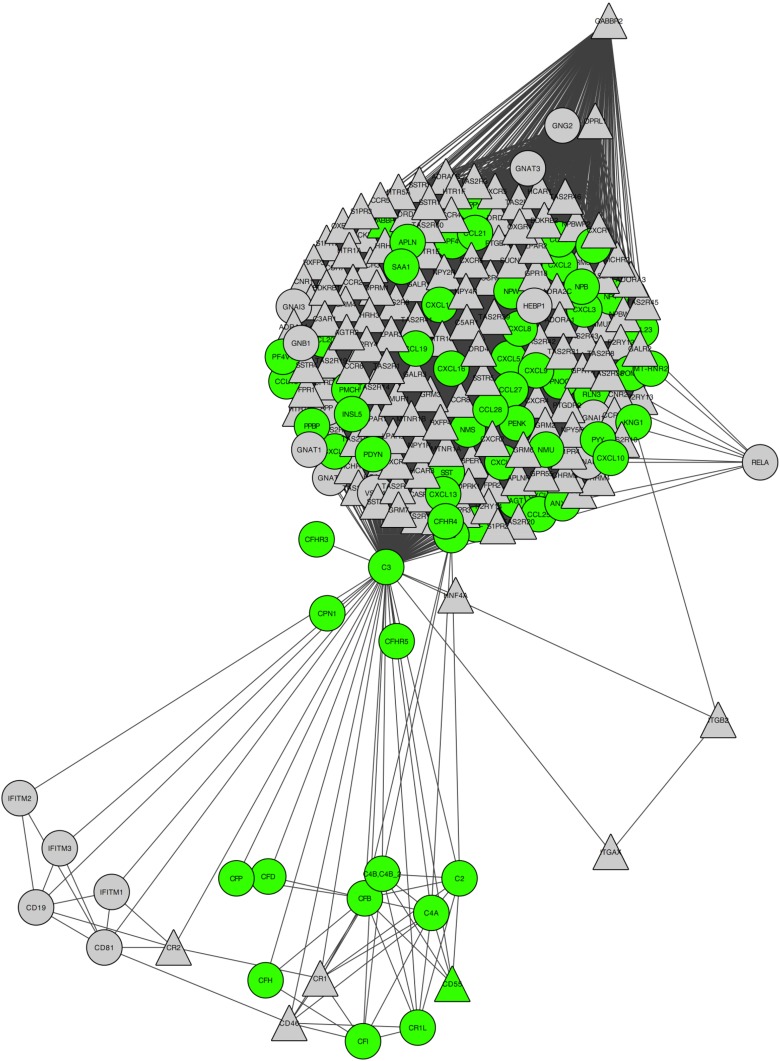
Figure 1 shows the first-order network around C3 (significance, P < 10−6) with visual indication of intracellular/extracellular parts as well as receptor vs nonreceptor, whereas Supplemental Tables 6 (1.7MB, pdf) and 7 (1.7MB, pdf) show the results of the remaining 216 networks investigated (209 out of 217 networks containing C3 had the lowest possible P value, and we chose the network with C3 as the central protein, as the main illustration, because this network highlights the entire near neighborhood around C3). For easy in-depth inspection of details in the network, we also included the network and its visualization as a data file for the Open Source network visualization tool Cytoscape (http://www.cytoscape.org) in the Supplemental Experimental Procedures (1.7MB, pdf) .
Figure 1.
Network of proteins capable of physically interacting with C3. The network in its entirety is significantly enriched (P < 10−6) for signals in gene expression (case vs control) of 10 independent human islet preparations left untreated or treated with IL-1β plus IFN-γ (50 and 1000 U/mL, respectively) for 48 hours. Protein–protein interactions are based on a high-confidence subset of InWeb_IM (confidence score > 0.1). Green symbols indicate extracellular; triangle symbols indicate receptor.
Ingenuity pathway analysis of C3 interacting partners
To better understand how C3 protein partners affect human pancreatic β-cells, we analyzed the 216 proteins identified as C3 partners using Ingenuity Pathway Analysis (Ingenuity Systems, http://www.ingenuity.com) (Supplemental Fig. 2 (1.7MB, pdf) ). The analysis of C3 protein partners identified several proinflammatory signaling pathways among the top 40 canonical pathways (Supplemental Fig. 2 (1.7MB, pdf) ; Supplemental Table 8 (1.7MB, pdf) ). “Complement system” was also observed in the analysis, showing the presence of other complement components, such as factor B, factor D, and C3aR1, in human islets.
Rodent and human pancreatic cells express and release complement C3
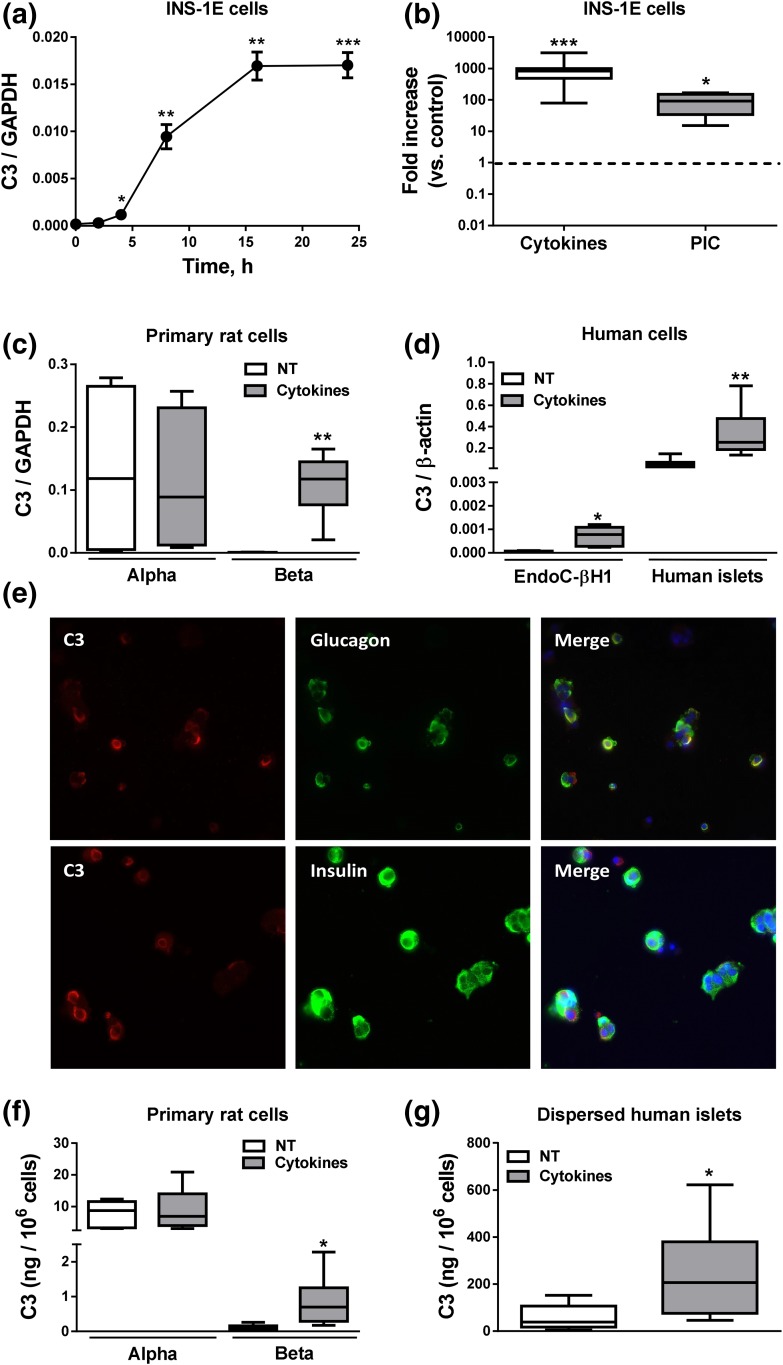
The proinflammatory cytokines IL-1β plus IFN-γ induced a progressive increase in C3 mRNA expression in insulin-producing INS-1E cells, with the maximum effect observed at 16 hours of treatment [Fig. 2(a)]. We compared cytokine-induced C3 expression with PIC, a mimic of double-stranded RNA, produced during viral replication. After 24 hours of treatment, cytokines induced a 10-fold higher increase in C3 expression (930-fold) as compared with PIC (90-fold) [Fig. 2(b)]. FACS-purified rat α- and β-cells showed distinct profiles of C3 expression. Thus, α-cells have higher basal expression of C3 mRNA (250-fold) than do β-cells [Fig. 2(c)], but after exposure to cytokines there were no changes in C3 expression in α-cells, whereas C3 expression strongly increased in β-cells [255-fold, Fig. 2(c)], reaching expression levels similar to those observed in α-cells.
Figure 2.
Complement C3 expression in pancreatic cells. (a) INS-1E cells were left untreated or treated with IL-1β plus IFN-γ (10 and 100 U/mL, respectively) for 2, 4, 8, 16, and 24 hours. (b) INS-1E cells were left nontreated or treated with cytokines (10 and 100 U/mL, respectively) or PIC (1 μg/mL) for 24 hours. (c) Primary rat α- and β-cells were left untreated or treated with IL-1β plus IFN-γ (50 and 500 U/mL, respectively) for 48 hours. (d) EndoC-βH1 cells and dispersed human islets were left untreated or treated with IL-1β plus IFN-γ (50 and 1000 U/mL, respectively) for 48 hours. C3 mRNA expression was analyzed by real-time polymerase chain reaction (RT-PCR) and normalized by the housekeeping genes GAPDH or β-actin. (e) Immunocytochemistry of C3 (red), insulin or glucagon (green), and HO (blue) was performed to confirm C3 expression in three dispersed human islet preparations (images are representative of three independent experiments; original magnification, ×40). (f and g) C3 secretion in (f) primary rat α- and β-cells and (g) dispersed human islets was measured by ELISA. C3 secretion was normalized by number of cells. Results are means ± SEM of 4 to 23 independent experiments. *P ≤ 0.05, **P < 0.01, and ***P < 0.001 vs treated with cytokines or PIC by Student t test. NT, nontreated.
C3 expression was also enhanced by cytokines in human insulin-producing EndoC-βH1 cells and in dispersed human islets. Dispersed human islets expressed much higher basal C3 expression than did EndoC-βH1 cells, but C3 expression increased more in EndoC-βH1 cells exposed to IL-1β plus IFN-γ than in dispersed human islets (14-fold vs sixfold) [Fig. 2(d)]. C3 is expressed both in glucagon- and insulin-positive cells in human islets as well as in the neighbor ductal cells, as evaluated by immunofluorescence [Fig. 2(e) and data not shown].
As shown for mRNA expression, FACS-purified rat α-cells released more C3 to the medium than did β-cells (7.69 ng/106 cells vs 0.13 ng/106 cells). Cytokine treatment led to a sevenfold increase in C3 release by β-cells, but it did not affect C3 secretion in α-cells [Fig. 2(f)]. C3 secretion followed the same pattern observed for mRNA expression in dispersed human islets, with IL-1β plus IFN-γ increasing the amount of C3 secreted into the culture medium [Fig. 2(g)].
C3 expression in pancreas sections from nondiabetic and type 1 diabetic donors
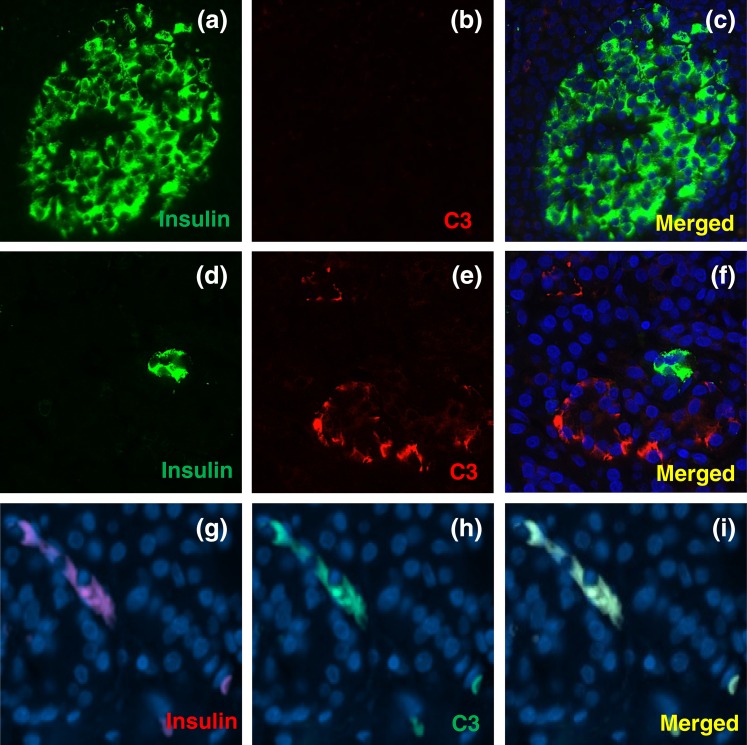
We next evaluated C3 expression in pancreas sections obtained from four nondiabetic and three type 1 diabetic donors from two different cohorts (University of Pisa and nPOD collection) (Fig. 3; Supplemental Fig. 3 (1.7MB, pdf) ). Pancreas sections from type 1 diabetic donors seemed to present a higher number of C3+ cells [Fig. 3(a–f)]. C3 expression was observed in acinar and ductal cells as well as in other cells located close to islets, but clusters positive for both insulin and C3, as well as isolated C3-positive insulin-positive cells, were observed only in the pancreas from one of the type 1 diabetic donors [Fig. 3(g–i); Supplemental Fig. 3 (1.7MB, pdf) ]. Owing to the scarcity of the available human material it was not possible to perform quantification of the number/types of C3+ cells. These observations need now to be confirmed by evaluation of a larger number of samples, specially taking into account the heterogeneity of pancreas histology observed in T1D patients.
Figure 3.
C3 expression in pancreas sections from nondiabetic and type 1 diabetic donors. (a–f) Pancreas sections from the nPOD cohort were analyzed by immunofluorescence. (a–c) Nondiabetic control and (d–f) type 1 diabetic donors were stained for (a and d; green) insulin, (b and e; red) C3, and (c and f; blue) HO. (g–i) Pancreas sections from a type 1 diabetic patient from the University of Pisa cohort were stained for (g; red) insulin, (h; green) C3, and (i; yellow) merged. DAPI staining is shown in blue. Images are representative of four nondiabetic subjects and three individuals with T1D. Original magnification, ×40.
C3 inhibition increases basal- and cytokine-induced apoptosis in β-cells but does not affect β-cell function
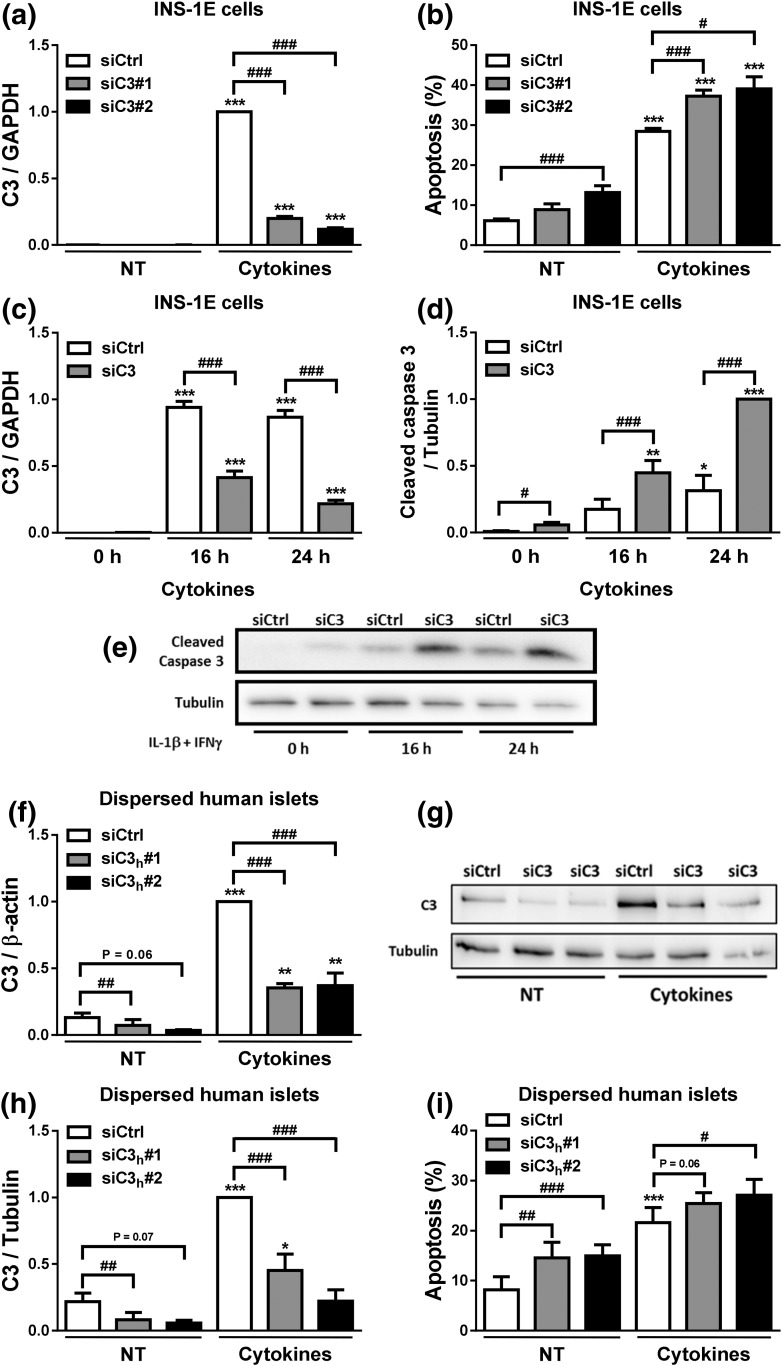
To investigate whether C3 plays a role in β-cell death, we used two independent siRNAs for each species to inhibit C3 expression in INS-1E cells, primary rat α- and β-cells, and dispersed human islets (Fig. 4; Supplemental Fig. 4 (1.7MB, pdf) ). C3 knockdown (KD) [Fig. 4(a)] increased both basal- and cytokine-induced apoptosis in INS-1E cells [Fig. 4(b)]. These results were confirmed by the increased expression of cleaved caspase-3 in INS-1E cells KD for C3 [Fig. 4(c)] and exposed or not for 16 or 24 hours to cytokines [Fig. 4(d) and 4(e)]. C3 inhibition (Supplemental Fig. 4a and 4c (1.7MB, pdf) ) did not affect cell death in primary rat α-cells (Supplemental Fig. 4b (1.7MB, pdf) ), but it slightly increased cytokine-induced cell death in primary rat β-cells (Supplemental Fig. 4d (1.7MB, pdf) ). Importantly, and in line with the findings in INS-1E cells and rat β-cells, C3 silencing (induced by two independent siRNAs and confirmed at both the mRNA [Fig. 4(f)] and protein levels [Fig. 4(g) and 4(h)] induced higher apoptosis in dispersed human islets, the “gold standard” in the field, under both basal conditions and following cytokine treatment [Fig. 4(i)]. These results suggest that C3 plays an important role in preserving β-cell survival. To evaluate the β-cell function in C3 KD cells, we measured INS2 and PDX1 mRNA expression and medium insulin accumulation (Supplemental Fig. 3e–g (1.7MB, pdf) ). No significant differences were observed between control and C3 KD cells under both basal condition and upon cytokine treatment (Supplemental Fig. 4e–g (1.7MB, pdf) ). Next, we assessed glucose-stimulated insulin secretion in INS-1E cells left untreated or treated with proinflammatory cytokines in the absence or presence of exogenous C3 (Supplemental Fig. 4h and 4i (1.7MB, pdf) ; see below additional information on exogenous C3). As previously shown (39–41), cytokine treatment impaired glucose-induced insulin secretion (Supplemental Fig. 4h and 4i (1.7MB, pdf) ). Exogenous C3 addition, however, did not alter insulin secretion or content (Supplemental Fig. 4h and 4i (1.7MB, pdf) ). As a whole, these findings suggest that modulation of the C3 pathway affects β-cell survival through a mechanism that is probably independent of β-cell function.
Figure 4.
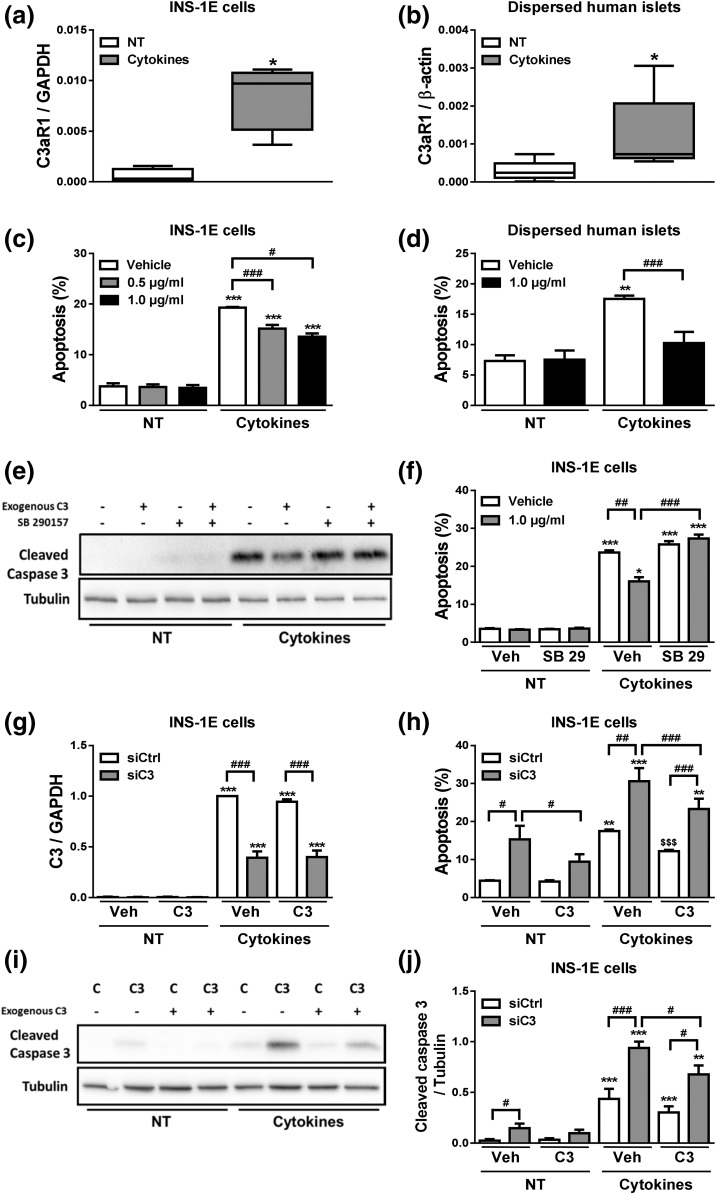
C3 inhibition increases β-cell apoptosis. (a–e) INS-1E cells were transfected with siCtrl or siRNAs targeting rat C3 (siC3). Cells were then left untreated (NT) or treated with IL-1β plus IFN-γ (10 and 100 U/mL, respectively) for 24 hours. (a) C3 mRNA expression was analyzed by real-time polymerase chain reaction (RT-PCR) and normalized by the housekeeping gene GAPDH. (b) Apoptosis was evaluated using HO and PI staining in INS-1E cells. (c–e) INS-1E cells were transfected with siCtrl or siRNA targeting rat C3 (siC3 no. 1). (c) C3 mRNA expression was analyzed by RT-PCR and normalized by the housekeeping gene GAPDH. (d and e) Cleaved caspase-3 and α-tubulin were measured by western blot (images are representative of seven independent experiments). (f–i) Dispersed human islets were transfected with siCtrl or siRNA targeting human C3 (siC3h). Then, cells were left untreated or treated with IL-1β plus IFN-γ (50 and 1000 U/mL, respectively) for 48 hours. (f) C3 mRNA expression was analyzed by RT-PCR and normalized by the housekeeping gene β-actin. (g and h) C3 and α-tubulin were measured by western blot. (i) Apoptosis was evaluated using HO and PI staining in dispersed human islets. Results are means ± SEM of four to seven independent experiments. *P ≤ 0.05, **P < 0.01, and ***P < 0.001 vs nontreated with cytokines (NT) and transfected with the same siRNA; #P ≤ 0.05, ##P < 0.01, ###P < 0.001 as indicated by bars; ANOVA followed by Student t test with Bonferroni correction.
Exogenously added C3 prevents cytokine-induced β-cell apoptosis
To assess whether C3 may have an autocrine-like effect, we evaluated the expression of the C3a receptor, namely C3aR1. C3aR1 mRNA was expressed in INS-1E cells [Fig. 5(a)] and dispersed human islets [Fig. 5(b)], and its expression was upregulated after cytokine exposure [Fig. 5(a) and 5(b)]. We next evaluated whether exogenous C3 protects β-cells against cytokine-induced cell death. INS-1E cells and dispersed human islets were exposed to IL-1β plus IFN-γ in the absence or presence of C3 [Fig. 5(c) and 5(d)]. In both cases, addition of C3 to the medium partially prevented cytokine-induced apoptosis [Fig. 5(c) and 5(d)]. Addition of the C3aR1 antagonist SB 290157 prevented the protective effects of exogenously added C3 against cytokine-induced apoptosis, suggesting that C3aR1 receptor activation by C3 is indeed required for C3 effects on β-cells [Fig. 5(e) and 5(f)]. These results led us to investigate whether exogenously added C3 could rescue C3-deficient cells from apoptosis. Cells exposed to siC3, and with a >60% inhibition of C3 expression [Fig. 5(g)], had an increase in both basal- and cytokine-induced apoptosis as evaluated by nuclear dyes and cleavage of caspase-3 [Fig. 5(h–j)]; interestingly, this was partially but not completely prevented by addition of exogenous C3 [Fig. 5(h–j)]. These findings suggest that C3 may promote β-cell survival via both intracellular production and signaling and also via an autocrine–paracrine way, that is, released by the β-cells themselves and/or by the neighbor cells (e.g., α-cells).
Figure 5.
Exogenous C3 prevents cytokine- and C3 deficiency–induced apoptosis. (a and b) C3aR1 mRNA expression was analyzed by real-time polymerase chain reaction (RT-PCR) and normalized by the housekeeping gene GAPDH or β-actin in, respectively, (a) INS-1E cells and (b) dispersed human islets. (c) INS-1E cells and (d) dispersed human islets were pretreated with the indicated C3 concentrations for 2 hours. Cells were then left untreated (NT) or treated with IL-1β plus IFN-γ in the absence or presence of exogenously added C3 (0.5 or 1.0 µg/mL) for (c) 24 hours or (d) 48 hours; apoptosis was evaluated using HO and PI staining. (e and f) INS-1E were left untreated (NT) or treated with IL-1β plus IFN-γ in the absence or presence of exogenously added C3 (1.0 µg/mL) and/or C3aR1 antagonist SB 290157 (10 μM) for 24 hours. (e) Cleaved caspase-3 and α-tubulin were measured by western blot. (f) Apoptosis was evaluated using HO and PI staining in INS-1E cells. (g–j) INS-1E cells were transfected with siCtrl (C) or siRNA targeting rat C3 (siC3 no. 1, C3). Cells were then left untreated (NT) or treated with IL-1β plus IFN-γ (10 and 100 U/mL, respectively) in the absence or presence of exogenously added C3 (1.0 µg/mL) for 16 hours. (g) C3 mRNA expression was analyzed by RT-PCR and normalized by the housekeeping gene GAPDH. (h) Apoptosis was evaluated using HO and PI staining in INS-1E cells. (i and j) Cleaved caspase-3 and α-tubulin were measured by western blot. Data represent the means ± SEM of four to seven independent experiments. *P ≤ 0.05, **P < 0.01, and ***P < 0.001 vs nontreated with cytokines (NT) and transfected with the same siRNA for (g–j); #P ≤ 0.05 and ###P < 0.001 as indicated by bars; ANOVA followed by Student t test with Bonferroni correction.
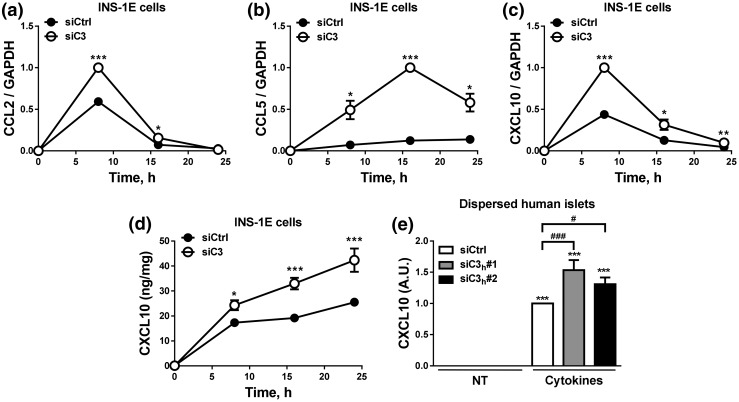
C3 silencing increases cytokine-induced chemokine expression
Our protein–protein analysis showed that C3 binds to several chemokines, and “chemokine signaling” was one of the top 20 canonical pathways identified (Fig. 1; Supplemental Fig. 1 (1.7MB, pdf) ; Supplemental Tables 6–8 (1.7MB, pdf) ). In line with this, C3 KD exacerbated cytokine-induced mRNA expression of CCL2, CCL5, and CXCL10 in INS-1E cells [Fig. 6(a–c)]. This was confirmed at protein level in INS-1E cells and in dispersed human islets, where C3-inhibited cells secreted higher amounts of CXCL10 as compared with control cells [Fig. 6(d) and (e)].
Figure 6.
C3 inhibition exacerbates chemokine expression and release. (a–d) INS-1E cells were transfected with siCtrl or siRNA targeting rat C3 (siC3 no. 1). Cells were then left untreated or treated with IL-1β plus IFN-γ (10 and 100 U/mL, respectively) as indicated under the figure. CCL2 (a), CCL5 (b), and CXCL10 (c) mRNA expression was analyzed by real-time polymerase chain reaction (RT-PCR) and normalized by the housekeeping gene GAPDH. (d and e) CXCL10 release was measured by ELISA in (d) INS-1E cells and (e) dispersed human islets. (e) Dispersed human islets were transfected with siCtrl or siRNA targeting human C3 (siC3h). Cells were then left untreated or treated with IL-1β plus IFN-γ (50 and 1000 U/mL, respectively) for 48 hours. In (d), CXCL10 release was normalized by protein content, whereas CXCL10 release in (e) was normalized by number of cells. Results are means ± SEM of three to six independent experiments. *P ≤ 0.05, **P < 0.01, and ***P < 0.001 vs transfected with (a–d) siCtrl vs (e) nontreated with cytokines (NT) and transfected with the same siRNA; #P ≤ 0.05, ###P < 0.001 as indicated by bars; ANOVA followed by Student t test with Bonferroni correction.
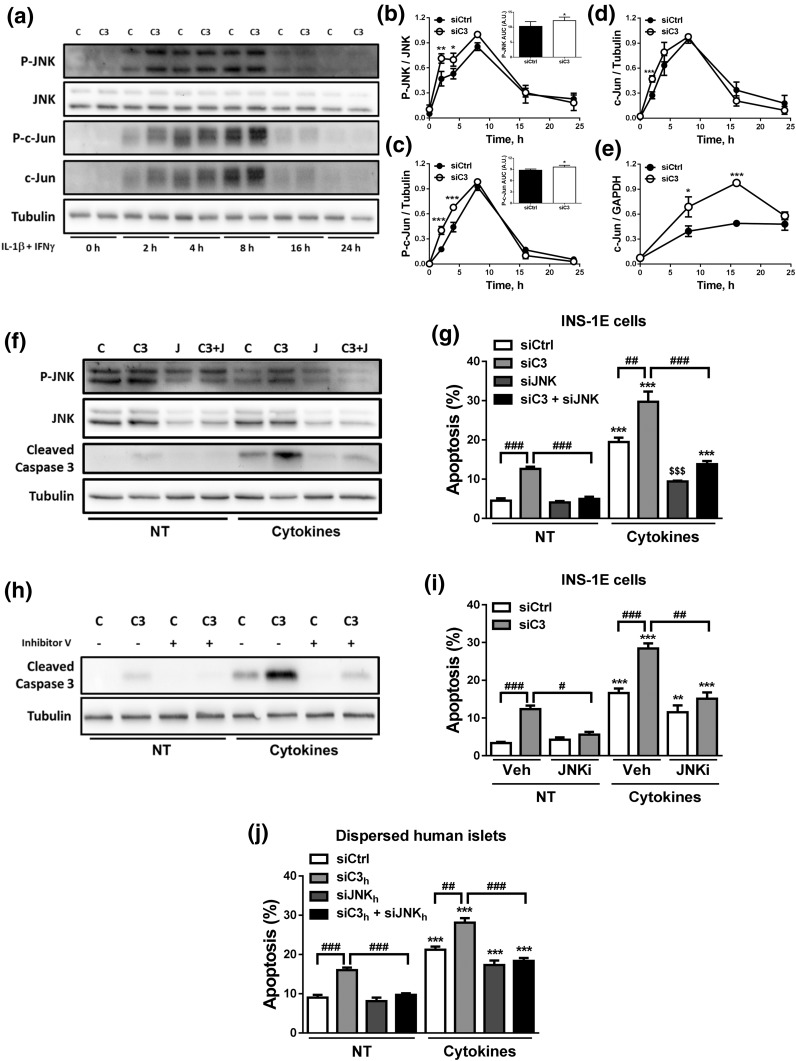
JNK pathway is activated in C3-silenced cells
We next examined the mechanisms by which C3 inhibition leads to increased β-cell death. JNK activation plays a key role in cytokine-induced β-cell apoptosis (31, 42, 43), and we observed that C3 KD in INS-1E cells augmented cytokine-induced JNK and c-Jun phosphorylation at early time points (2 to 8 hours) [Fig. 7(a–c)]. This augmented JNK activation in the context of C3 KD, and cytokine treatment was confirmed by analyzing the area under the curve of the P-JNK [Fig. 7(b), inset] and P–c-Jun [Fig. 7(c), inset] time course and by the increased expression of c-Jun, a P-JNK target gene [Fig. 7(d) and 7(e)]. To determine whether JNK activation contributes to β-cell death observed in C3-inhibited cells, we silenced C3 and JNK1 in parallel in INS-1E cells [Fig. 7(f) and 7(g); Supplemental Fig. 5 (1.7MB, pdf) ]. As shown in Fig. 4(b), C3 KD increased both basal- and cytokine-induced apoptosis. JNK1 KD prevented both basal- and cytokine-induced cell death in C3 KD cells, as evaluated by both nuclear dyes and cleavage of caspase-3 [Fig. 7(f) and 7(g); Supplemental Fig. 5d (1.7MB, pdf) ]. JNK inhibition using the chemical JNK inhibitor V also prevented C3 KD-induced apoptosis [Fig. 7(h) and 7(i)]. Similar results were obtained in dispersed human islets, in which JNK inhibition in parallel to C3 KD (Supplemental Fig. 5e and 5f (1.7MB, pdf) ) prevented C3 KD-induced apoptosis under both basal condition and following cytokine exposure [Fig. 7(j)]. These data suggest that activation of JNK1 phosphorylation is a necessary component of the cell death process induced by C3 inhibition and/or cytokine exposure.
Figure 7.
C3 inhibition activates the JNK pathway. (a–e) INS-1E cells were transfected with siCtrl (C) or siRNA targeting rat C3 (siC3 no. 1, C3). Cells were then left untreated or treated with IL-1β plus IFN-γ (10 and 100 U/mL, respectively) as indicated under the figure. (a–d) P-JNK, JNK, P–c-Jun, c-Jun, and α-tubulin were measured by western blot. (a) Images are representative of four independent experiments. (b–d) Densitometry analysis of the western blots for (b) P-JNK, (c) P–c-Jun, and (d) c-Jun. Quantification of the area under curve (AUC) of P-JNK and P–c-Jun are shown in the inset graphs in (b) and (c), respectively. (e) c-Jun mRNA expression was analyzed by real-time polymerase chain reaction (RT-PCR) and normalized by the housekeeping gene GAPDH. (f and g) INS-1E cells transfected with siCtrl (C) or siRNAs targeting rat C3 (siC3 no. 1, C3), JNK (J), or both (C3 + J). Cells were then left untreated (NT) or treated with IL-1β plus IFN-γ (10 and 100 U/mL, respectively) for 16 hours. (f) P-JNK, JNK, cleaved caspase-3, and α-tubulin were measured by western blot. Images are representative of four independent experiments. (g) Apoptosis was evaluated using HO and PI staining in INS-1E cells. (h and i) INS-1E cells were transfected with siCtrl (C) or siRNA targeting rat C3 (siC3 no. 1, C3). Cells were then left untreated (NT) or treated with IL-1β plus IFN-γ in the absence or presence of JNK inhibitor V (JNKi). (h) Cleaved caspase-3 and α-tubulin were measured by western blot. Images are representative of four independent experiments. (i) Apoptosis was evaluated using HO and PI staining in INS-1E. (j) Apoptosis was evaluated using HO and PI staining in dispersed human islets transfected with siCtrl, siC3 no. 2h, siJNKh, or siC3 no. 2h + siJNKh and exposed for 48 hours to cytokines. Results are means ± SEM of three to five independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ***P < 0.001 vs transfected with (b–e) siCtrl vs (g, i, and j) nontreated with cytokines (NT) and transfected with the same siRNA; #P < 0.05, ##P < 0.01, and ###P < 0.001 as indicated by bars; ANOVA followed by Student t test with Bonferroni correction.
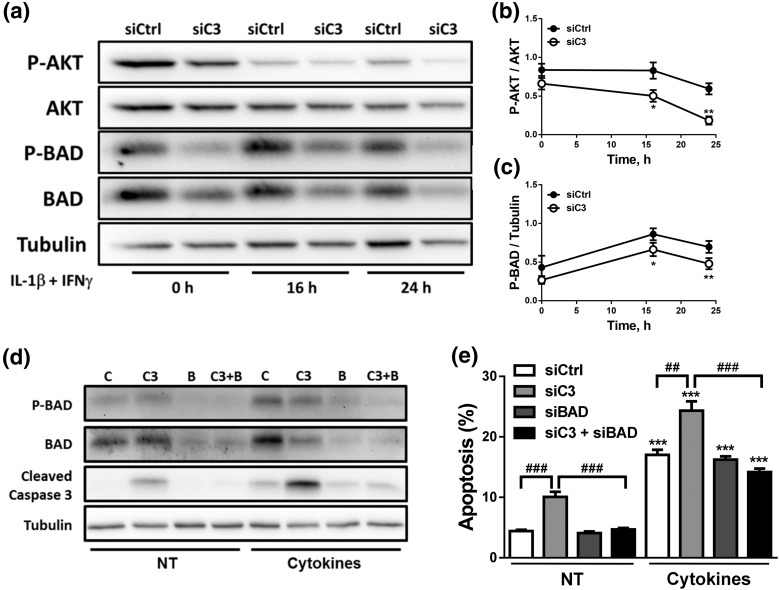
The AKT pathway is downregulated in C3-silenced cells, contributing to β-cell death
AKT signaling protects pancreatic β-cells against different stress conditions (44, 45). As C3aR1 signals via AKT (46), we investigated whether AKT signaling is disturbed in C3-silenced cells. The proapoptotic protein BAD, whose phosphorylation by AKT leads to its inhibition, was also evaluated (Fig. 8). AKT phosphorylation was decreased in C3-inhibited cells, and, consequently, P-BAD levels were also lower under these conditions [Fig. 8(a–c)]. Importantly, C3/BAD double KD [Fig. 8(d); Supplemental Fig. 6a–c (1.7MB, pdf) ) diminished apoptosis secondary to C3 KD under both basal condition and after cytokine exposure as confirmed by both nuclear dyes and cleavage of caspase-3 [Fig. 8(d) and 8(e); Supplemental Fig. 6d (1.7MB, pdf) ].
Figure 8.
C3 silencing inhibits the AKT pathway. (a–c) INS-1E cells were transfected with siCtrl or siRNA targeting rat C3 (siC3 no. 1). Cells were then left untreated or treated with IL-1β plus IFN-γ (10 and 100 U/mL, respectively) for 16 or 24 hours. (a–c) P-BAD, BAD, P-AKT, AKT, and α-tubulin were measured by western blot. (a) Images are representative of four to seven independent experiments. (b and c) Densitometry analysis of the western blots for (b) P-AKT and (c) P-BAD. (d and e) INS-1E cells were transfected with siCtrl (C) or siRNAs targeting rat C3 (siC3 no. 1, C3), BAD (B), or both (C3 + B). Cells were then left untreated (NT) or treated with IL-1β plus IFN-γ (10 and 100 U/mL, respectively) for 16 hours. (d) P-BAD, BAD, cleaved caspase-3, and α-tubulin were measured by western blot. Images are representative of four independent experiments. (e) Apoptosis was evaluated using HO and PI staining in INS-1E cells. Results are means ± SEM of four to seven independent experiments. *P ≤ 0.05, **P < 0.01, and ***P < 0.001 vs transfected with (b and c) siCtrl vs (e) nontreated with cytokines (NT) and transfected with the same siRNA; ##P < 0.01 and ###P < 0.001 as indicated by bars; ANOVA followed by Student t test with Bonferroni correction.
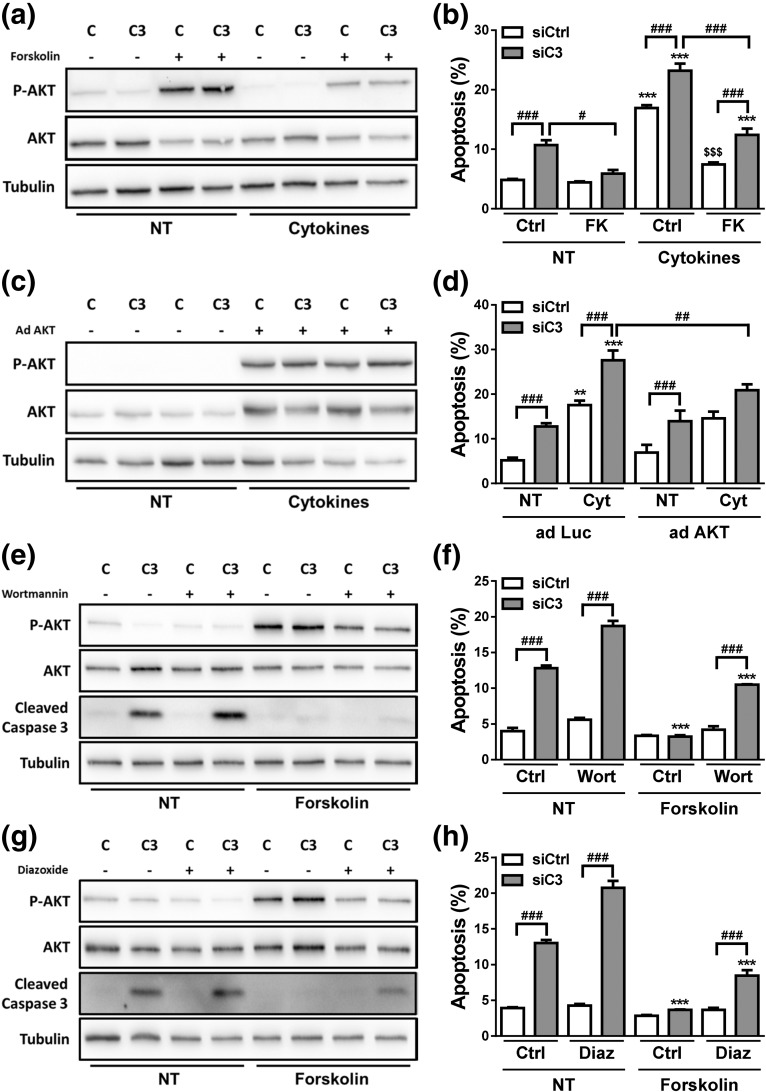
In mirror experiments, we investigated whether increasing AKT function protects β-cells against death secondary to C3 deficiency. For this purpose, cells exposed to siCtrl or siC3 (Supplemental Fig. 7a and 7b (1.7MB, pdf) ) were treated with the adenylate cyclase stimulator FK [Fig. 9(a) and 9(b)] or infected with an adenovirus encoding a constitutively active mutant form of AKT (AdAKT) [Fig. 9(c) and 9(d)]. FK increased AKT phosphorylation under both basal condition and following cytokine exposure and it prevented C3 KD-induced apoptosis under both basal condition and following exposure to cytokines [Fig. 9(a) and 9(b)]. Infection with AdAKT increased P-AKT and AKT under all conditions tested [Fig. 9(c)], and it protected C3 KD cells against cytokine-induced apoptosis [Fig. 9(d)]. The AdAKT, however, neither prevented the increase in apoptosis under basal condition in C3 KD cells nor the apoptosis induced by cytokines in cells exposed to cytokines and siCtrl [Fig. 9(d)]. This discrepancy between some of the effects of FK and AdAKT suggests that FK has additional beneficial effects besides upregulation of AKT pathway (47, 48). As a whole, the results shown above suggest that C3 inhibition impairs AKT signaling and consequently leads to upregulation of BAD, thus contributing to β-cell apoptosis. To further characterize the involvement of the AKT pathway in C3 KD cells, control and C3-silenced cells were treated with the PI3K inhibitor wortmannin or the KATP channel agonist diazoxide in the absence or presence of FK [Fig. 9(e–h)]. These two compounds have been shown to decrease FK-induced AKT phosphorylation by different mechanisms (49, 50). We confirmed that wortmannin and diazoxide decreased P-AKT expression in INS-1E cells as expected (Supplemental Fig. 7c (1.7MB, pdf) ). Under these conditions, diazoxide inhibited insulin release into the culture medium by at least 60%, whereas wortmannin had no effects on insulin accumulation (Supplemental Fig. 7d (1.7MB, pdf) ). FK failed to protect β-cells from cytokine-induced apoptosis when used in combination with wortmannin, suggesting that FK-induced AKT phosphorylation is a relevant mechanism for β-cell protection upon cytokine treatment (Supplemental Fig. 7e and 7f (1.7MB, pdf) ). A more marked increase in cell death was observed when C3-inhibited cells were treated with either wortmannin [Fig. 9(f)] or diazoxide [Fig. 9(h)], which may be explained by the more severe inhibition of AKT phosphorylation in C3 KD cells [Fig. 9(e) and 9(g)]. As shown in Fig. 9(b), FK fully prevented C3 KD-induced apoptosis in vehicle-treated cells, but FK could only partially protect C3-silenced cells from wortmannin and diazoxide effects on cell death.
Figure 9.
AKT stimulation protects against C3 deficiency–induced apoptosis. (a and b) INS-1E cells were transfected with siCtrl (C) or siRNA targeting rat C3 (siC3 no. 1, C3). Cells were then treated with vehicle only (DMSO, Ctrl) or FK (20 µM) in the absence (NT) or presence of IL-1β plus IFN-γ (10 and 100 U/mL, respectively) for 16 hours. (a) P-AKT, AKT, and α-tubulin were measured by western blot. Images are representative of four independent experiments. (b) Apoptosis was evaluated using HO and PI staining in INS-1E cells. (c and d) siCtrl (C) and siC3 no. 1 (C3) INS-1E cells were infected with a control adenovirus encoding luciferase (adLuc) or with an adenovirus encoding myr-Akt1 (adAKT). Cells were then left untreated (NT) or treated with IL-1β plus IFN-γ (10 and 100 U/mL, respectively) for 16 hours. (c) P-AKT, AKT, and α-tubulin were measured by western blot in INS-1E cells transfected with siCtrl (C) or a siRNA targeting rat C3 (siC3 no. 1, C3). Images are representative of 4 independent experiments. (d) Apoptosis was evaluated using HO and PI staining in INS-1E cells. (e and f) INS-1E cells were transfected with siCtrl (C) or siRNA targeting rat C3 (siC3 no. 1, C3). Cells were then treated with vehicle only (DMSO, Ctrl) or wortmannin (Wort, 100 nM) in the absence (NT) or presence of FK (20 µM) for 16 hours. (e) P-AKT, AKT, cleaved caspase-3, and α-tubulin were measured by western blot. Images are representative of four independent experiments. (f) Apoptosis was evaluated using HO and PI staining in INS-1E cells. (g and h) INS-1E cells were transfected with siCtrl (C) or siRNA targeting rat C3 (siC3 no. 1, C3). Cells were then treated with vehicle only (DMSO, Ctrl) or diazoxide (Diaz, 200 µM) in the absence (NT) or presence of FK (20 µM) for 16 hours. (g) P-AKT, AKT, cleaved caspase-3, and α-tubulin were measured by western blot. Images are representative of four independent experiments. (f) Apoptosis was evaluated using HO and PI staining in INS-1E cells. Results are means ± SEM of four independent experiments. **P < 0.01 and ***P < 0.001 vs nontreated with cytokines or FK (NT) and transfected with the same siRNA; $$$P < 0.001 vs siCtrl plus cytokines; #P ≤ 0.05, ##P < 0.01, and ###P < 0.001 as indicated by bars; ANOVA followed by Student t test with Bonferroni correction.
Discussion
Proinflammatory cytokines released by the immune system modulate β-cell gene networks and transcription factors (e.g., nuclear factor κB, interferon regulatory factors, and signal transducer and activator of transcription), leading to β-cell dysfunction and activation of both proapoptotic and antiapoptotic pathways (1). Gene expression signatures induced by some of these cytokines, that is, IL-1β plus IFNγ (22), are remarkably similar to the gene signatures observed in islets isolated from patients in the early stages of T1D (51), validating the use of the present in vitro model.
Several of these cytokine-induced genes belong to immune-related pathways whose functions in β-cells remain to be elucidated. Using a novel computational framework (23) to identify cytokine-induced human protein–protein interaction networks in human islets, we identified complement C3 as a central component of a network involving 216 proteins that are modified by cytokine exposure. Pathway analysis of this network identified several proteins related to G protein–coupled receptor signaling (e.g., C3aR1 and CXCR3), complement system (e.g., factor B and factor D), and proinflammatory pathways (e.g., TF65 and CCL3) as canonical pathways. In the present study, we provide evidence that complement C3 plays an important role in β-cell survival, and that C3 deficiency activates the mitochondrial pathway of apoptosis via different mechanisms, besides upregulating production of chemokines that will further attract immune cells.
Complement proteins can be synthesized by several human cells, including the exocrine pancreas (52) (present data) and islet α- and β-cells (present data). Importantly, we detected an augmented C3 expression in the pancreas and islet region in histological material from T1D patients. Besides its supportive role in defense against invading pathogens (5), the complement system participates in several nonimmune processes, such as lipogenesis (53), food intake (54), and neurogenesis (55).
Interestingly, complement components can be intracellularly activated (6, 7), and they play important roles in cell and tissue development, homeostasis, and survival (7). In liver, for instance, complement C3a and C5a are essential for cell survival during hepatocyte regeneration (56). In this study, we observed that C3 promotes β-cell survival via both intracellular production and signaling and also via an autocrine–paracrine way, that is, via release by the β-cells themselves and/or by the neighbor cells, such as α-cells and exocrine cells, and consequent signaling via surface receptors, such as C3aR1.
C3 deficiency leads to β-cell death under basal conditions and upon cytokine exposure via activation of caspase-3 and consequent apoptosis. The JNK pathway is augmented in C3-deficient cells, and JNK1 inhibition completely prevents basal and cytokine-induced apoptosis, indicating that JNK1 contributes to the deleterious effects of C3 inhibition. In line with these observations, administration of C3adesArg, a product of C3 activation, modulates JNK expression in adipose tissue and skeletal muscle from C3 knockout mice fed on a high-fat diet (57).
AKT acts as a crucial prosurvival pathway in pancreatic β-cells exposed to different stressors via BAD phosphorylation and consequent inhibition of its proapoptotic activity (45, 58, 59). We observed that AKT is inhibited in C3-silenced cells, leading to higher levels of unphosphorylated BAD and subsequent activation of apoptosis. Experiments using FK, an adenovirus encoding a constitutively active mutant form of AKT, and pharmacological AKT inhibition (wortmannin and diazoxide) confirmed that AKT signaling counteracts the induction of apoptosis in C3-deficient cells, particularly following exposure to proinflammatory cytokines. It is interesting that assembly of another member of the complement family, namely the terminal complement complex C5b-9 (composed of C5b, C6, C7, C8, and C9 proteins), protects oligodendrocytes from apoptosis via activation of PI3K/AKT signaling and consequent BAD inhibition (60).
Several chemokines were identified as C3 partners in our protein–protein interaction analysis, and “chemokine signaling” was one of the canonical pathways identified in the Ingenuity Pathway Analysis. C3 silencing exacerbated CCL2, CCL5, and CXCL10 mRNA expression and CXCL10 release after cytokine exposure by islet cells. CXCL10 is upregulated in islets from NOD mice (61) and type 1 diabetic patients (62) and it may contribute to the exacerbation of insulitis and β-cell destruction via attraction of additional immune-competent cells (63).
Although our data suggest that both extracellular and intracellular complement C3 activation are important for β-cell survival, it remains to be clarified how C3 is activated in the islet milieu. The alternative pathway, one of the ways by which the complement system is activated, is permanently activated via the “tick-over mechanism” (i.e., spontaneous hydrolysis of C3) in healthy individuals (64). It has been recently proposed that adipose tissue–produced adipsin (or factor D) can reach islets via the circulation and participate in C3 activation in the β-cell vicinity (21). Moreover, factor B is expressed and secreted by ductal cells, which may contribute to this process (65). Therefore, it is conceivable that components of the complement system secreted by pancreatic cells (e.g., C3 by α- and β-cells in the present study) culminate in C3 activation that protects β-cells against proinflammatory assaults. This protection may be sufficient to prevent β-cell death during mild activation of the innate immune response, but it will probably not be sufficient to prevent β-cell death in the context of a protracted autoimmune assault.
In conclusion, we provide evidence that complement C3 is a key regulator of a cytokine-modulated complement network in human islets, with anti-inflammatory and prosurvival roles in rodent and human pancreatic β-cells. Local production and secretion of C3 into the β-cell vicinity, as well as intracellular C3 activation, may play an important protective role in β-cells exposed to the autoimmune assault leading to T1D.
Acknowledgments
The authors thank M. Pangerl, A. M. Musuaya, N. Pachera, and I. Millard from the Université Libre de Bruxelles Center for Diabetes Research, Université Libre de Bruxelles, for excellent technical support; Dr. J.-V. Turantzine, Université Libre de Bruxelles Center for Diabetes Research, Université Libre de Bruxelles, for help in the analysis of the RNA sequencing of human islets; and the Flow Cytometry Facility of the Erasmus campus of the Université Libre de Bruxelles and Christine Dubois for the cell sorting.
Acknowledgments
This work was supported by grants from the Fonds National de la Recherche Scientifique, Belgium, and by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases/Human Islet Research Network Consortium Grant 1UC4DK104166-01 (to D.L.E.). D.L.E., S.R.H., and R.W. have received funding from the Horizon 2020 Program [T2Dsystems (GA667191)]. D.L.E. and P.M. have received funding from the Innovative Medicines Initiative 2 Joint Undertaking under Grant 115797 [Innovative Approach Towards Understanding and Arresting Type 1 Diabetes (INNODIA)]. This Joint Undertaking receives support from the Union’s Horizon 2020 research and innovation program and European Federation of Pharmaceutical Industries and Association (EFPIA), Juvenile Diabetes Research Foundation (JDRF), and the Leona M. and Harry B. Helmsley Charitable Trust. L.M. is supported by a Fonds National de la Recherche Scientifique postdoctoral fellowship. This research was performed with the support of the Network for Pancreatic Organ Donors with Diabetes (nPOD), a collaborative T1D research project sponsored by JDRF. Organ procurement organizations partnering with nPOD to provide research resources are listed at http://www.jdrfnpod.org/for-partners/npod-partners/.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ANOVA
- analysis of variance
- ELISA
- enzyme-linked immunosorbent assay
- FK
- forskolin
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HO
- Hoechst 33342
- IFN
- interferon
- IL
- interleukin
- JNK
- c-Jun N-terminal kinase
- KD
- knockdown
- mRNA
- messenger RNA
- P-
- phosphorylated
- PI
- propidium iodide
- PIC
- polyinosinic-polycytidylic acid
- PI3K
- phosphatidylinositol 3-kinase
- RRID
- research resource identifier
- SEM
- standard error of the mean
- siRNA
- small interfering RNA
- T1D
- type 1 diabetes.
References
- 1.Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and β-cell loss in type 1 diabetes. Nat Rev Endocrinol. 2009;5(4):219–226. [DOI] [PubMed] [Google Scholar]
- 2.Morgan NG, Leete P, Foulis AK, Richardson SJ. Islet inflammation in human type 1 diabetes mellitus. IUBMB Life. 2014;66(11):723–734. [DOI] [PubMed] [Google Scholar]
- 3.Op de Beeck A, Eizirik DL. Viral infections in type 1 diabetes mellitus—why the β cells? Nat Rev Endocrinol. 2016;12(5):263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marroqui L, Lopes M, dos Santos RS, Grieco FA, Roivainen M, Richardson SJ, Morgan NG, Op de Beeck A, Eizirik DL. Differential cell autonomous responses determine the outcome of coxsackievirus infections in murine pancreatic α- and β-cell cells. eLife. 2015;4:e06990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liszewski MK, Kolev M, Le Friec G, Leung M, Bertram PG, Fara AF, Subias M, Pickering MC, Drouet C, Meri S, Arstila TP, Pekkarinen PT, Ma M, Cope A, Reinheckel T, Rodriguez de Cordoba S, Afzali B, Atkinson JP, Kemper C. Intracellular complement activation sustains T cell homeostasis and mediates effector differentiation. Immunity. 2013;39(6):1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolev M, Le Friec G, Kemper C. Complement—tapping into new sites and effector systems. Nat Rev Immunol. 2014;14(12):811–820. [DOI] [PubMed] [Google Scholar]
- 8.Phieler J, Garcia-Martin R, Lambris JD, Chavakis T. The role of the complement system in metabolic organs and metabolic diseases. Semin Immunol. 2013;25(1):47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao X, Xia Y, Zhang Q, Wu HM, Du HM, Liu L, Wang CJ, Shi HB, Guo XY, Liu X, Li CL, Su Q, Meng G, Yu B, Sun SM, Wang X, Zhou M, Jia QY, Song K, Niu KJ. Elevated serum complement C3 levels are related to the development of prediabetes in an adult population: the Tianjin Chronic Low-Grade Systematic Inflammation and Health Cohort Study. Diabet Med. 2016;33(4):446–453. [DOI] [PubMed] [Google Scholar]
- 10.Bottazzo GF, Dean BM, Gorsuch AN, Cudworth AG, Doniach D. Complement-fixing islet-cell antibodies in type-I diabetes: possible monitors of active β-cell damage. Lancet. 1980;1(8170):668–672. [PubMed] [Google Scholar]
- 11.Engström G, Hedblad B, Eriksson KF, Janzon L, Lindgärde F. Complement C3 is a risk factor for the development of diabetes: a population-based cohort study. Diabetes. 2005;54(2):570–575. [DOI] [PubMed] [Google Scholar]
- 12.Wlazlo N, van Greevenbroek MM, Ferreira I, Feskens EJ, van der Kallen CJ, Schalkwijk CG, Bravenboer B, Stehouwer CD. Complement factor 3 is associated with insulin resistance and with incident type 2 diabetes over a 7-year follow-up period: the CODAM Study. Diabetes Care. 2014;37(7):1900–1909. [DOI] [PubMed] [Google Scholar]
- 13.Planas R, Carrillo J, Sanchez A, de Villa MC, Nuñez F, Verdaguer J, James RF, Pujol-Borrell R, Vives-Pi M. Gene expression profiles for the human pancreas and purified islets in type 1 diabetes: new findings at clinical onset and in long-standing diabetes. Clin Exp Immunol. 2010;159(1):23–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowe P, Wasserfall C, Croker B, Campbell-Thompson M, Pugliese A, Atkinson M, Schatz D. Increased complement activation in human type 1 diabetes pancreata. Diabetes Care. 2013;36(11):3815–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallionpää H, Elo LL, Laajala E, Mykkänen J, Ricaño-Ponce I, Vaarma M, Laajala TD, Hyöty H, Ilonen J, Veijola R, Simell T, Wijmenga C, Knip M, Lähdesmäki H, Simell O, Lahesmaa R. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes. 2014;63(7):2402–2414. [DOI] [PubMed] [Google Scholar]
- 16.Törn C, Liu X, Hagopian W, Lernmark Å, Simell O, Rewers M, Ziegler AG, Schatz D, Akolkar B, Onengut-Gumuscu S, Chen WM, Toppari J, Mykkänen J, Ilonen J, Rich SS, She JX, Sharma A, Steck A, Krischer J; TEDDY Study Group . Complement gene variants in relation to autoantibodies to β cell specific antigens and type 1 diabetes in the TEDDY Study. Sci Rep. 2016;6:27887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conroy SJ, Abdel-Wahab YH, Caraher EM, Byrne PM, Murphy E, Nolan J, Flatt PR, Newsholme P. Evidence for complement-dependent and -independent inhibition of insulin secretion from clonal β-cells incubated in the presence of sera of newly diagnosed IDDM patients. J Endocrinol. 2000;164(2):139–147. [DOI] [PubMed] [Google Scholar]
- 18.Conroy SJ, Green I, Dixon G, Byrne PM, Nolan J, Abdel-Wahab YH, McClenaghan N, Flatt PR, Newsholme P. Evidence for a sustained increase in clonal β-cell basal intracellular Ca2+ levels after incubation in the presence of newly diagnosed type-1 diabetic patient sera. Possible role in serum-induced inhibition of insulin secretion. J Endocrinol. 2002;173(1):53–62. [DOI] [PubMed] [Google Scholar]
- 19.Lin M, Yin N, Murphy B, Medof ME, Segerer S, Heeger PS, Schröppel B. Immune cell-derived C3 is required for autoimmune diabetes induced by multiple low doses of streptozotocin. Diabetes. 2010;59(9):2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahrén B, Havel PJ, Pacini G, Cianflone K. Acylation stimulating protein stimulates insulin secretion. Int J Obes Relat Metab Disord. 2003;27(9):1037–1043. [DOI] [PubMed] [Google Scholar]
- 21.Lo JC, Ljubicic S, Leibiger B, Kern M, Leibiger IB, Moede T, Kelly ME, Chatterjee Bhowmick D, Murano I, Cohen P, Banks AS, Khandekar MJ, Dietrich A, Flier JS, Cinti S, Blüher M, Danial NN, Berggren PO, Spiegelman BM. Adipsin is an adipokine that improves β cell function in diabetes. Cell. 2014;158(1):41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eizirik DL, Sammeth M, Bouckenooghe T, Bottu G, Sisino G, Igoillo-Esteve M, Ortis F, Santin I, Colli ML, Barthson J, Bouwens L, Hughes L, Gregory L, Lunter G, Marselli L, Marchetti P, McCarthy MI, Cnop M. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8(3):e1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T, Wernersson R, Hansen RB, Horn H, Mercer J, Slodkowicz G, Workman CT, Rigina O, Rapacki K, Stærfeldt HH, Brunak S, Jensen TS, Lage K. A scored human protein–protein interaction network to catalyze genomic interpretation. Nat Methods. 2017;14(1):61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marroqui L, Masini M, Merino B, Grieco FA, Millard I, Dubois C, Quesada I, Marchetti P, Cnop M, Eizirik DL. Pancreatic α cells are resistant to metabolic stress-induced apoptosis in type 2 diabetes. EBioMedicine. 2015;2(5):378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravassard P, Hazhouz Y, Pechberty S, Bricout-Neveu E, Armanet M, Czernichow P, Scharfmann R. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J Clin Invest. 2011;121(9):3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brozzi F, Nardelli TR, Lopes M, Millard I, Barthson J, Igoillo-Esteve M, Grieco FA, Villate O, Oliveira JM, Casimir M, Bugliani M, Engin F, Hotamisligil GS, Marchetti P, Eizirik DL. Cytokines induce endoplasmic reticulum stress in human, rat and mouse β cells via different mechanisms. Diabetologia. 2015;58(10):2307–2316. [DOI] [PubMed] [Google Scholar]
- 27.Lupi R, Del Guerra S, Tellini C, Giannarelli R, Coppelli A, Lorenzetti M, Carmellini M, Mosca F, Navalesi R, Marchetti P. The biguanide compound metformin prevents desensitization of human pancreatic islets induced by high glucose. Eur J Pharmacol. 1999;364(2-3):205–209. [DOI] [PubMed] [Google Scholar]
- 28.Marroqui L, Dos Santos RS, Fløyel T, Grieco FA, Santin I, Op de Beeck A, Marselli L, Marchetti P, Pociot F, Eizirik DL. TYK2, a candidate gene for type 1 diabetes, modulates apoptosis and the innate immune response in human pancreatic β-cells. Diabetes. 2015;64(11):3808–3817. [DOI] [PubMed] [Google Scholar]
- 29.Santin I, Dos Santos RS, Eizirik DL. Pancreatic β-cell survival and signaling pathways: effects of type 1 diabetes-associated genetic variants. Methods Mol Biol. 2016;1433:21–54. [DOI] [PubMed] [Google Scholar]
- 30.Moore F, Naamane N, Colli ML, Bouckenooghe T, Ortis F, Gurzov EN, Igoillo-Esteve M, Mathieu C, Bontempi G, Thykjaer T, Ørntoft TF, Eizirik DL. STAT1 is a master regulator of pancreatic β-cell apoptosis and islet inflammation. J Biol Chem. 2011;286(2):929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marroquí L, Santin I, Dos Santos RS, Marselli L, Marchetti P, Eizirik DL. BACH2, a candidate risk gene for type 1 diabetes, regulates apoptosis in pancreatic β-cells via JNK1 modulation and crosstalk with the candidate gene PTPN2. Diabetes. 2014;63(7):2516–2527. [DOI] [PubMed] [Google Scholar]
- 32.Ortis F, Cardozo AK, Crispim D, Störling J, Mandrup-Poulsen T, Eizirik DL. Cytokine-induced proapoptotic gene expression in insulin-producing cells is related to rapid, sustained, and nonoscillatory nuclear factor-κB activation. Mol Endocrinol. 2006;20(8):1867–1879. [DOI] [PubMed] [Google Scholar]
- 33.Kutlu B, Cardozo AK, Darville MI, Kruhøffer M, Magnusson N, Ørntoft T, Eizirik DL. Discovery of gene networks regulating cytokine-induced dysfunction and apoptosis in insulin-producing INS-1 cells. Diabetes. 2003;52(11):2701–2719. [DOI] [PubMed] [Google Scholar]
- 34.Eizirik DL, Mandrup-Poulsen T. A choice of death—the signal-transduction of immune-mediated β-cell apoptosis. Diabetologia. 2001;44(12):2115–2133. [DOI] [PubMed] [Google Scholar]
- 35.Marroqui L, Dos Santos RS, Op de Beeck A, Coomans de Brachène A, Marselli L, Marchetti P, Eizirik DL. Interferon-α mediates human beta cell HLA class I overexpression, endoplasmic reticulum stress and apoptosis, three hallmarks of early human type 1 diabetes. Diabetologia. 2017;60(4):656–667. [DOI] [PubMed] [Google Scholar]
- 36.Liu D, Darville M, Eizirik DL. Double-stranded ribonucleic acid (RNA) induces β-cell Fas messenger RNA expression and increases cytokine-induced β-cell apoptosis. Endocrinology. 2001;142(6):2593–2599. [DOI] [PubMed] [Google Scholar]
- 37.Overbergh L, Valckx D, Waer M, Mathieu C. Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine. 1999;11(4):305–312. [DOI] [PubMed] [Google Scholar]
- 38.Marhfour I, Lopez XM, Lefkaditis D, Salmon I, Allagnat F, Richardson SJ, Morgan NG, Eizirik DL. Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia. 2012;55(9):2417–2420. [DOI] [PubMed] [Google Scholar]
- 39.Eizirik DL, Strandell E, Bendtzen K, Sandler S. Functional characteristics of rat pancreatic islets maintained in culture after exposure to human interleukin 1. Diabetes. 1988;37(7):916–919. [DOI] [PubMed] [Google Scholar]
- 40.Eizirik DL, Sandler S, Welsh N, Cetkovic-Cvrlje M, Nieman A, Geller DA, Pipeleers DG, Bendtzen K, Hellerström C. Cytokines suppress human islet function irrespective of their effects on nitric oxide generation. J Clin Invest. 1994;93(5):1968–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cetkovic-Cvrlje M, Eizirik DL. TNF-α and IFN-γ potentiate the deleterious effects of IL-1β on mouse pancreatic islets mainly via generation of nitric oxide. Cytokine. 1994;6(4):399–406. [DOI] [PubMed] [Google Scholar]
- 42.Bony C, Roche S, Shuichi U, Sasaki T, Crackower MA, Penninger J, Mano H, Pucéat M. A specific role of phosphatidylinositol 3-kinase γ. A regulation of autonomic Ca2+ oscillations in cardiac cells. J Cell Biol. 2001;152(4):717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurzov EN, Ortis F, Cunha DA, Gosset G, Li M, Cardozo AK, Eizirik DL. Signaling by IL-1β+IFN-γ and ER stress converge on DP5/Hrk activation: a novel mechanism for pancreatic β-cell apoptosis. Cell Death Differ. 2009;16(11):1539–1550. [DOI] [PubMed] [Google Scholar]
- 44.Drucker DJ. The biology of incretin hormones. Cell Metab. 2006;3(3):153–165. [DOI] [PubMed] [Google Scholar]
- 45.Cunha DA, Gurzov EN, Naamane N, Ortis F, Cardozo AK, Bugliani M, Marchetti P, Eizirik DL, Cnop M. JunB protects β-cells from lipotoxicity via the XBP1-AKT pathway. Cell Death Differ. 2014;21(8):1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strainic MG, Liu J, Huang D, An F, Lalli PN, Muqim N, Shapiro VS, Dubyak GR, Heeger PS, Medof ME. Locally produced complement fragments C5a and C3a provide both costimulatory and survival signals to naive CD4+ T cells. Immunity. 2008;28(3):425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunha DA, Ladrière L, Ortis F, Igoillo-Esteve M, Gurzov EN, Lupi R, Marchetti P, Eizirik DL, Cnop M. Glucagon-like peptide-1 agonists protect pancreatic β-cells from lipotoxic endoplasmic reticulum stress through upregulation of BiP and JunB. Diabetes. 2009;58(12):2851–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Igoillo-Esteve M, Gurgul-Convey E, Hu A, Romagueira Bichara Dos Santos L, Abdulkarim B, Chintawar S, Marselli L, Marchetti P, Jonas JC, Eizirik DL, Pandolfo M, Cnop M. Unveiling a common mechanism of apoptosis in β-cells and neurons in Friedreich’s ataxia. Hum Mol Genet. 2015;24(8):2274–2286. [DOI] [PubMed] [Google Scholar]
- 49.Misra UK, Pizzo SV. Coordinate regulation of forskolin-induced cellular proliferation in macrophages by protein kinase A/cAMP-response element-binding protein (CREB) and Epac1-Rap1 signaling: effects of silencing CREB gene expression on Akt activation. J Biol Chem. 2005;280(46):38276–38289. [DOI] [PubMed] [Google Scholar]
- 50.Cornu M, Yang JY, Jaccard E, Poussin C, Widmann C, Thorens B. Glucagon-like peptide-1 protects β-cells against apoptosis by increasing the activity of an IGF-2/IGF-1 receptor autocrine loop. Diabetes. 2009;58(8):1816–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lundberg M, Krogvold L, Kuric E, Dahl-Jørgensen K, Skog O. Expression of interferon-stimulated genes in insulitic pancreatic islets of patients recently diagnosed with type 1 diabetes. Diabetes. 2016;65(10):3104–3110. [DOI] [PubMed] [Google Scholar]
- 52.Andoh A, Fujiyama Y, Sumiyoshi K, Bamba T. Local secretion of complement C3 in the exocrine pancreas: ductal epithelial cells as a possible biosynthetic site. Gastroenterology. 1996;110(6):1919–1925. [DOI] [PubMed] [Google Scholar]
- 53.MacLaren R, Cui W, Cianflone K. Adipokines and the immune system: an adipocentric view. Adv Exp Med Biol. 2008;632:1–21. [DOI] [PubMed] [Google Scholar]
- 54.Ohinata K, Yoshikawa M. Food intake regulation by central complement system. Adv Exp Med Biol. 2008;632:35–46. [PubMed] [Google Scholar]
- 55.Shinjyo N, Ståhlberg A, Dragunow M, Pekny M, Pekna M. Complement-derived anaphylatoxin C3a regulates in vitro differentiation and migration of neural progenitor cells. Stem Cells. 2009;27(11):2824–2832. [DOI] [PubMed] [Google Scholar]
- 56.Markiewski MM, DeAngelis RA, Strey CW, Foukas PG, Gerard C, Gerard N, Wetsel RA, Lambris JD. The regulation of liver cell survival by complement. J Immunol. 2009;182(9):5412–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Munkonda MN, Lapointe M, Miegueu P, Roy C, Gauvreau D, Richard D, Cianflone K. Recombinant acylation stimulating protein administration to C3−/− mice increases insulin resistance via adipocyte inflammatory mechanisms. PLoS One. 2012;7(10):e46883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wrede CE, Dickson LM, Lingohr MK, Briaud I, Rhodes CJ. Fatty acid and phorbol ester-mediated interference of mitogenic signaling via novel protein kinase C isoforms in pancreatic β-cells (INS-1). J Mol Endocrinol. 2003;30(3):271–286. [DOI] [PubMed] [Google Scholar]
- 59.Srinivasan S, Ohsugi M, Liu Z, Fatrai S, Bernal-Mizrachi E, Permutt MA. Endoplasmic reticulum stress-induced apoptosis is partly mediated by reduced insulin signaling through phosphatidylinositol 3-kinase/Akt and increased glycogen synthase kinase-3β in mouse insulinoma cells. Diabetes. 2005;54(4):968–975. [DOI] [PubMed] [Google Scholar]
- 60.Soane L, Cho HJ, Niculescu F, Rus H, Shin ML. C5b-9 terminal complement complex protects oligodendrocytes from death by regulating Bad through phosphatidylinositol 3-kinase/Akt pathway. J Immunol. 2001;167(4):2305–2311. [DOI] [PubMed] [Google Scholar]
- 61.Cardozo AK, Proost P, Gysemans C, Chen MC, Mathieu C, Eizirik DL. IL-1β and IFN-γ induce the expression of diverse chemokines and IL-15 in human and rat pancreatic islet cells, and in islets from pre-diabetic NOD mice. Diabetologia. 2003;46(2):255–266. [DOI] [PubMed] [Google Scholar]
- 62.Roep BO, Kleijwegt FS, van Halteren AG, Bonato V, Boggi U, Vendrame F, Marchetti P, Dotta F. Islet inflammation and CXCL10 in recent-onset type 1 diabetes. Clin Exp Immunol. 2010;159(3):338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lasch S, Müller P, Bayer M, Pfeilschifter JM, Luster AD, Hintermann E, Christen U. Anti-CD3/anti-CXCL10 antibody combination therapy induces a persistent remission of type 1 diabetes in two mouse models. Diabetes. 2015;64(12):4198–4211. [DOI] [PubMed] [Google Scholar]
- 64.Merle NS, Church SE, Fremeaux-Bacchi V, Roumenina LT. Complement system part I—molecular mechanisms of activation and regulation. Front Immunol. 2015;6:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sumiyoshi K, Andoh A, Fujiyama Y, Sakumoto H, Bamba T. Biosynthesis and secretion of MHC class III gene products (complement C4 and factor B) in the exocrine pancreas. J Gastroenterol. 1997;32(3):367–373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA sequencing data sets used in the present study are available online at http://lmedex.ulb.ac.be/data.php.