The mechanisms that regulate self-renewal in hematopoietic stem cells (HSCs) and leukemia stem cells (LSCs) are poorly understood. Herein, Khalaj et al. identify microRNA-99 (miR-99) as a novel noncoding RNA critical for the maintenance of HSCs and LSCs and demonstrate that miR-99 mediates its role by suppressing multiple target genes, including HOXA1.
Abstract
The microRNA-99 (miR-99) family comprises a group of broadly conserved microRNAs that are highly expressed in hematopoietic stem cells (HSCs) and acute myeloid leukemia stem cells (LSCs) compared with their differentiated progeny. Herein, we show that miR-99 regulates self-renewal in both HSCs and LSCs. miR-99 maintains HSC long-term reconstitution activity by inhibiting differentiation and cell cycle entry. Moreover, miR-99 inhibition induced LSC differentiation and depletion in an MLL-AF9–driven mouse model of AML, leading to reduction in leukemia-initiating activity and improved survival in secondary transplants. Confirming miR-99’s role in established AML, miR-99 inhibition induced primary AML patient blasts to undergo differentiation. A forward genetic shRNA library screen revealed Hoxa1 as a critical mediator of miR-99 function in HSC maintenance, and this observation was independently confirmed in both HSCs and LSCs. Together, these studies demonstrate the importance of noncoding RNAs in the regulation of HSC and LSC function and identify miR-99 as a critical regulator of stem cell self-renewal.
Introduction
Acute myeloid leukemia (AML) is composed of functionally heterogeneous cells including leukemic stem cells (LSCs), which exhibit the ability to self-renew and propagate disease (Kreso and Dick, 2014). Because LSCs and normal hematopoietic stem cells (HSCs) display shared functional properties, it is not surprising that they are regulated by similar molecular pathways (Yilmaz and Morrison, 2008). The clinical importance of these observations is highlighted by the finding that AML transcriptomes enriched for HSC and LSC signatures are associated with worse prognoses (Gentles et al., 2010; Eppert et al., 2011; Metzeler et al., 2013). Thus, better understanding the mechanisms that regulate HSC function is likely to improve our understanding of not only HSCs, but also LSC function. Although several studies have identified numerous protein-coding genes that regulate HSCs and LSCs (Yilmaz and Morrison, 2008), it has become increasingly clear that noncoding RNAs also play prominent functional roles in these stem cell populations (Marcucci et al., 2011; Ciccone and Calin, 2015).
MicroRNAs (miRNAs) are small, non–protein-coding RNAs that regulate gene expression predominantly by binding to the 3′ UTR of mRNAs and promoting degradation of transcripts or inhibiting translation (Ha and Kim, 2014). These noncoding elements coordinate expression of targets from multiple signaling pathways, making them potential HSC and LSC regulators. miRNAs demonstrated to support HSC function have typically been studied because of their selective expression in HSCs. For example, miRNAs expressed at the highest levels in HSCs compared with committed progenitors, such as miR-125, miR-146a, and miR-29a/b1, promote HSC self-renewal (Guo et al., 2010; Ooi et al., 2010; Gerrits et al., 2012; Zhao et al., 2013; Hu et al., 2015), whereas others, such as miR-126, the miR-212/132 complex, and miR-193b, suppress HSC function (Lechman et al., 2012; Haetscher et al., 2015; Mehta et al., 2015). Moreover, the role of some of these miRNAs in myeloid leukemogenesis has been demonstrated, as high expression of miR-125 and miR-29a can induce myeloid leukemia (Bousquet et al., 2008, 2012; Han et al., 2010; Klusmann et al., 2010; O’Connell et al., 2010). Furthermore, individual miRNAs, such as miR-126, miR-196, and miR-21, as well as the polycistronic miR-17-92 cluster, promote LSC self-renewal (Wong et al., 2010; Velu et al., 2014; Lechman et al., 2016). Together, these studies indicate that miRNAs are important regulators of normal and malignant stem cells.
Among of the most highly expressed miRNAs in HSCs are members of the miR-99 family, a broadly conserved family that exhibits decreased expression upon differentiation (Ooi et al., 2010; Gerrits et al., 2012). One member, miR-99b, was recently identified as a potential LSC regulator, as it was enriched in AML patients whose transcriptomes exhibited a high “stem cell core enrichment” score (Metzeler et al., 2013). Another member, miR-99a, was the most significantly enriched miRNA in experimentally defined patient LSC populations (Lechman et al., 2016). Although these data suggest a role for miR-99 family members in both HSCs and LSCs, to date, a functional role for miR-99 has not been established. In fact, one study reported that miR-99b overexpression did not cause a significant change in HSC long-term repopulating capacity (Guo et al., 2010). Despite the lack of evidence of miR-99 regulation of HSCs, another group showed that enforced expression of miR-100, a miR-99 family member, inhibited differentiation of AML cells in vitro, suggesting a potential role for the miR-99 family in AML (Zheng et al., 2012); however, studies have yet to be performed to confirm this function in primary AML blasts or in a leukemia model in vivo. Because all miR-99 family members are expressed at high levels in HSCs and LSCs, we sought to determine the role of miR-99 in their maintenance. We used a loss-of-function approach to assess miR-99 function, because it is less prone to experimental artifacts (Concepcion et al., 2012). Using this strategy, we demonstrate that miR-99 is a critical regulator of both HSC and LSC self-renewal, primarily by inhibiting differentiation.
Results
miR-99 supports hematopoietic stem cell clonogenic capacity
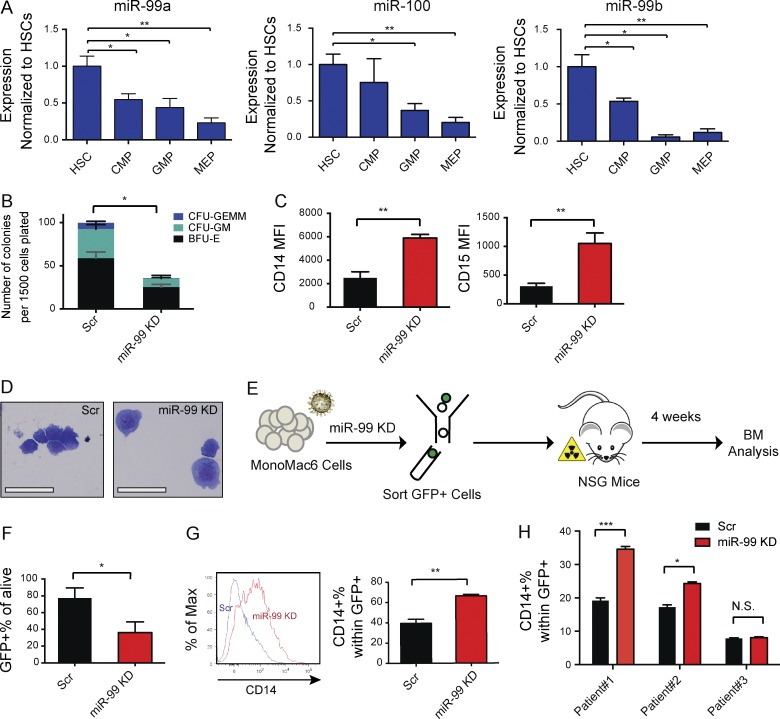
To identify miRNAs that regulate HSC function, we compared miRNA gene expression levels in mouse hematopoietic stem and progenitor cell (HSPC) populations (Chao et al., 2008). Remarkably, we found that all three members of the highly conserved miR-99 family are expressed at significantly higher levels in mouse HSCs compared with more differentiated populations (Fig. 1, A–C), suggesting they might play a role in maintaining HSC function.
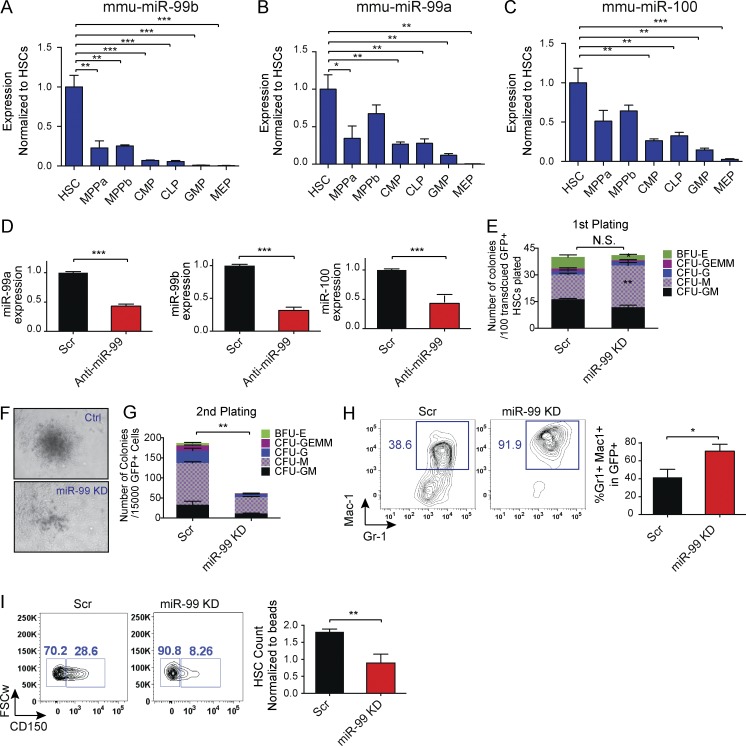
Figure 1.
miR-99 is highly expressed in hematopoietic stem and progenitors and suppresses myeloid differentiation in vitro. (A–C) Normalized expression levels of miR-99b, miR-99a, and miR-100 as determined by quantitative RT-PCR using miRNA TaqMan probes in mouse hematopoietic cell populations: hematopoietic stem cell (HSC), multipotent progenitor (MPP) Flk−, MPP Flk+, common lymphoid progenitor (CLP), common myeloid progenitor (CMP), granulocyte-macrophage progenitor (GMP), and megakaryocyte-erythroid progenitor (MEP) cells. Expression was normalized against mmu-mir-16. Error bars denote SEM. Representative data from five independent experiments are shown. (D) miR-99 is down-regulated 48 h post-transduction of HSCs with the lentiviral anti–miR-99 vector as shown by quantitative RT-PCR. Expression was normalized against U6 (Student’s t test; n = 3). Representative data from two independent experiments are shown. (E) Comparable number of colonies form after miR-99 KD in first plating, with an increase in the number of CFU macrophage (CFU-M) colonies. 100 GFP+ HSC cells were cultured in methylcellulose. The colonies were scored after 7 d. Data represent mean percentage ± SEM (Student’s t test; n = 3) and are representative of three independent experiments. (F) Smaller colonies were observed after second plating of GFP+ cells derived from miR-99 KD HSCs. Representative data of three independent experiments are shown. (G) Colony-forming capacity of HSCs is reduced after miR-99 KD in a second plating. 15,000 GFP+ cells were replated 7 d after the first plating. Colony types were scored after 7 to 10 d. Data represent mean count ± SEM (Student’s t test; n = 3) and are representative of three independent experiments. (H) miR-99 KD in HSCs induces granulocytic differentiation in methylcellulose colony assays. 7 d after plating, colonies were analyzed for expression of myeloid differentiation markers by flow cytometry. Mean percentage ± SEM (Student’s t test; n = 2). Representative data of three independent experiments are shown. (I) Flow cytometry analysis of LSK cells transduced with anti–miR-99 or Scr vectors and maintained in liquid culture for 8 d reveals a decrease in the absolute number of GFP+Lin−Sca-1+c-Kit+CD150+ HSCs. FSCw denotes forward scatter-width. The data shown are gated on LSK cells. Data represent mean count ± SEM (Student’s t test; n = 3) and are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine if miR-99 regulates HSCs, we first performed overexpression studies. Consistent with previous findings (Guo et al., 2010; Emmrich et al., 2014), enforced expression of miR-99 in HSCs did not significantly alter colony formation (Fig. S1 A). We hypothesized that the absence of a phenotype in the context of overexpression may indicate miR-99 is present in vast excess relative to its target genes and that loss-of-function studies might better reveal its biological function. Thus, we used a lentiviral vector expressing an antisense miR-99 oligonucleotide (anti–miR-99) to knock down miR-99 in Lin−c-Kit+Sca-1+CD34−CD150+ HSCs (Fig. S1 B). This approach allows for simultaneous knockdown (KD) of multiple miR-99 family members, helping address the potential redundant roles of miR-99 family members (Fig. 1 D). Using luciferase reporters containing 3′ UTRs of known miR-99 targets, SMARCA5 and HS2ST3 (Sun et al., 2011; Yang et al., 2015), we confirmed that anti–miR-99 relieves inhibition of miR-99 targets in a dose-dependent manner (Fig. S1, C and D). miR-99 KD in HSCs did not significantly alter the number of colonies in the first plating in methylcellulose. However, more CFU-M colonies were formed at the expense of CFU-GM and BFU-E colonies (Fig. 1 E). Upon secondary plating, miR-99 KD also led to reduced colony size and decreased serial replating capacity (Fig. 1, F and G). miR-99 KD also resulted in increased relative numbers of CFU-M colonies in secondary platings (Fig. S1, E and G), supporting a monocytic differentiation bias in miR-99 KD HSPCs. Comparable reductions in colony number were observed using a second anti–miR-99 construct (vector 2; Fig. S1 F). The decrease in HSC colony formation was accompanied by accelerated myelopoiesis, as evidenced by an increased proportion of Gr1+Mac1+ cells upon miR-99 KD (Fig. 1 H). In addition, total GFP+ miR-99 KD cells exhibited increased apoptosis at the end of the first plating (Fig. S1 G). Consistent with these observations, liquid culture of miR-99 KD HSCs showed a reduced absolute number of HSCs 8 d after transduction (Fig. 1 I). Collectively, these observations demonstrate that miR-99 helps maintain HSPCs in vitro by suppressing their differentiation.
miR-99 regulates HSC long-term reconstitution capacity
Because miR-99 is required to preserve HSC clonogenic capacity in vitro, we sought to test whether miR-99 maintains HSCs in vivo. Transplant of anti–miR-99–transduced HSCs demonstrated a significant reduction in long-term reconstitution capacity compared with scramble (Scr) controls (Fig. 2 A). The difference in engraftment was not caused by defects in homing, as HSCs expressing anti–miR-99 or the Scr control had similar numbers of GFP+ HSCs in the BM 24 h post-transplant (Fig. S2A). In agreement with the in vitro experiments, the peripheral blood (PB) of mice harboring miR-99 KD HSCs showed a significant increase in the proportion of donor-derived Mac1+ myeloid cells (Fig. 2 B). Further analysis of the PB showed that all lineages, including B cells (B220+), T cells (CD3+), granulocytes (Gr1+ Mac1+), and monocytes (Gr1neg Mac1+), exhibited reduced GFP+ chimerism, with the reduction being more prominent in the lymphoid lineage (Fig. S2, B–E). Analysis of the BM in miR-99 KD stable grafts revealed reduced GFP chimerism, consistent with the reduced donor chimerism observed in the PB (Fig. S2 F). Mice transplanted with miR-99 KD HSCs also displayed decreased absolute numbers of GFP+ HSCs (Fig. 2 C) and multipotent progenitors (Fig. 2, D and E). Together, these data demonstrate that miR-99 maintains HSC reconstitution potential.
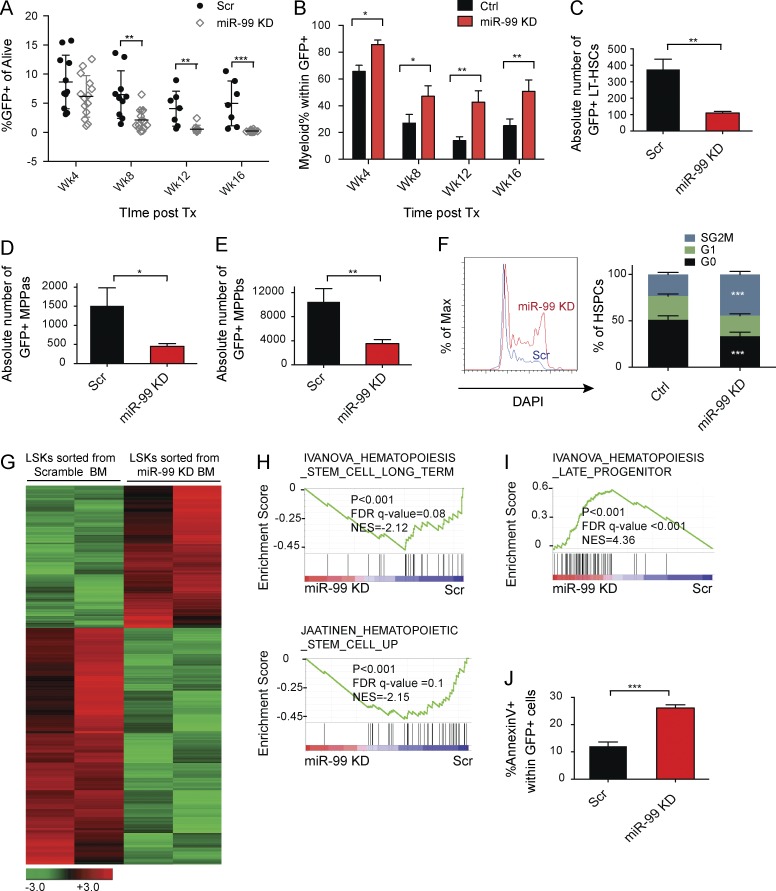
Figure 2.
miR-99 inhibition impairs HSC reconstitution capacity in vivo by inducing differentiation and increased cell cycling. (A) GFP+ chimerism of mice transplanted with miR-99 KD HSCs. HSCs were transduced with anti–miR-99 or scramble (Scr) control vectors, and 48 h later, 5,000 GFP+ cells were transplanted into lethally irradiated recipients along with 300,000 cells from Sca-1–depleted helper BM. Peripheral blood GFP chimerism was analyzed every 4 wk. Data represent mean percentages ± SEM (Student’s t test; n = 11 for Scr and n = 13 for miR-99 KD mice) and are representative of two independent experiments. Post tx, post-transplantation. (B) Flow cytometry analysis of the peripheral blood every 4 wk after transplantation of HSCs transduced with anti–miR-99 or Scr vectors. Data represent mean percentages ± SEM (Student’s t test; n = 11 for Scr and n = 13 for miR-99 KD mice) and are representative of two independent experiments. (C–E) Absolute number of GFP+Lin−c-Kit+Sca-1+CD48−CD150+ HSCs (C), CD48+CD150+ MPPa’s (multipotent progenitors a; (D), and CD48+ CD150neg MPPb's (multipotent progenitors b; E) in bilateral long bones and hips 16 wk post-transplant of HSCs. Data represent mean count ± SEM (Student’s t test; n = 4) and are representative of two independent experiments. (F) Ki-67/DAPI staining of donor-derived GFP+Lin−c-Kit+ HSPCs 6 mo post-transplant of miR-99 KD or Scr HSCs. Data represent mean percentage ± SEM (Student’s t test; n = 7 for Scr and n = 8 for miR-99 KD) and are representative of two independent experiments. (G) RNA-seq analysis of LSK cells FACS sorted from BM of mice transplanted with miR-99 KD or scramble control HSCs 3 mo after transplant (n = 2). (H and I) Gene set enrichment analysis for differentially expressed genes in stably engrafted miR-99 KD versus Scr LSK cells. FDR, false discovery rate; NES, normalized enrichment score. (J) Annexin V staining of total GFP+ cells in the BM 6 mo post-transplant. Data represent mean percentage ± SEM (Student’s t test; n = 7 for Scr and n = 8 for miR-99 KD) and are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
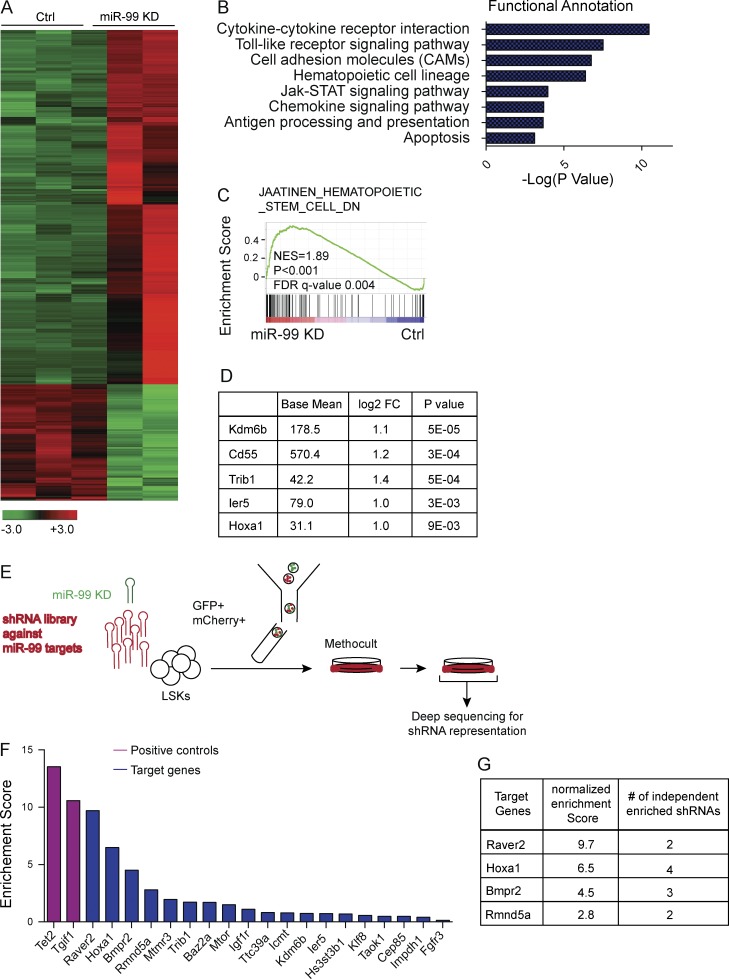
We next sought to determine the mechanistic basis for the alterations in miR-99 KD HSC function. Analysis of the BM revealed that miR-99 KD induced a significant increase in the percentage of cycling lineage-negative Sca1-positive c-Kit–negative (LSK) cells and progenitor cells (Fig. 2 F). To investigate the molecular alterations induced with miR-99 inhibition, we performed RNA sequencing (RNA-seq) on GFP+ miR-99 KD or Scr LSK cells 3 mo after stable engraftment into lethally irradiated recipients (Fig. 2 G). Gene set enrichment analysis (GSEA) revealed depletion of genes constituting previously defined HSC signatures, including c-Kit, Mpl, Hoxa9, Meis1, Tie1, and Angpt1 (Ivanova et al., 2002; Subramanian et al., 2005; Jaatinen et al., 2006). In addition, miR-99 inhibition resulted in enrichment for genes associated with more differentiated progenitors, further confirming miR-99 maintains HSCs in an undifferentiated state (Fig. 2, H and I; Ivanova et al., 2002). Although total GFP+ miR-99 KD BM cells displayed increased apoptosis (Fig. 2 J), there were no differences in apoptosis in Scr control versus miR-99 KD LSK cells or progenitors (Fig. S2, G and H). These findings suggest that at least at the level of LSK and committed progenitors, the major effect of miR-99 KD is to induce differentiation, leading to their depletion, rather than apoptosis. Overall, these results demonstrate that miR-99 maintains mouse HSCs in a quiescent and undifferentiated state.
miR-99 maintains LSCs in MLL-AF9+ acute myeloid leukemia
To investigate whether miR-99 may play a role in malignant hematopoiesis, we analyzed the miRNA-sequencing data from 153 AML patients from The Cancer Genome Atlas (TCGA) database. Our analysis revealed that expression of miR-99 family members inversely correlates with bulk blast differentiation status as reflected by the French–American–British classification scheme (Fig. 3 A), compatible with a role for miR-99 as an inhibitor of differentiation in AML (Ley et al., 2013).
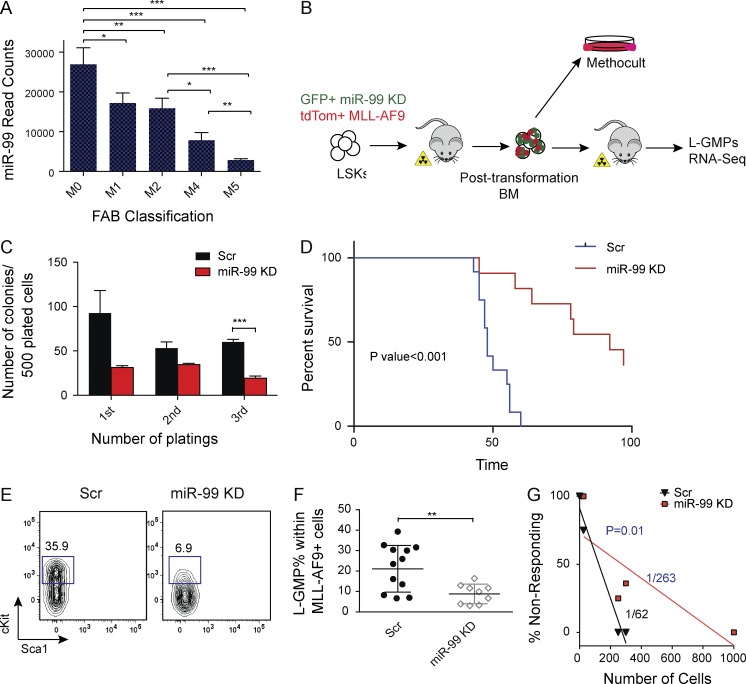
Figure 3.
miR-99 KD improves survival in an MLL-AF9 model of leukemia by depleting leukemia stem cells. (A) miRNA-sequencing data from 153 AML patients in the TCGA database. Sum of read counts of all miR-99 family members (miR-99a, miR-99b, and miR-100) is graphed as a function of the French–American–British (FAB) classification. (B) Schematic for miR-99 KD experiments in the MLL-AF9 mouse model of AML. LSK cells were cotransduced with GFP+ miR-99 KD and tdTom+ MLL-AF9 overexpressing vectors, and GFP+ tdTom+ cells were transplanted into sublethally irradiated mice. BM from transplanted animals was used for secondary transplants and methylcellulose replating assays. (C) Methylcellulose colony plating assay of post-transformed BM from primary AML recipients with miR-99 KD. 500 GFP+ tdTom+ cells were plated into methylcellulose media and replated every 7 d. Data represent mean count ± SEM (Student’s t test; n = 3) and are representative of two independent experiments. (D) Kaplan–Meier curves of recipients of secondary leukemia transplants. 300 GFP+ tdTom+ leukemic BM cells from primary recipients were transplanted into sublethally irradiated C57BL/6 recipients (Mantel–Cox test; n = 12 for Scr and n = 11 for miR-99 KD). Representative data from four independent experiments are shown. (E and F) Flow cytometry analysis of L-GMPs from the BM of secondary recipients. Gates were drawn on the GFP+ tdTom+ CD16/32+ population. Mean percentage ± SEM (Student’s t test; n = 12 for Scr and n = 9 for miR-99 KD). Representative data from four independent experiments are shown. (G) GFP+ tdTom+ cells were FACS sorted from the BM of primary recipients and transplanted in limiting dilutions into secondary recipients. Limiting dilution analysis was performed using ELDA software. See also Fig. S3 D. Representative data two independent experiments are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
AML samples with MLL-AF9 rearrangements have been shown to express higher levels of miR-99b (Garzon et al., 2008). Thus, we used the retroviral MLL-AF9 model of AML to test the role of miR-99 in leukemogenesis (Krivtsov et al., 2006). We used anti–miR-99 vector 2 to perform these studies because it knocked down miR-99a to levels comparable to the anti–miR-99 vector 1 but was a more efficient inhibitor of miR-99b expression in human AML cells (Fig. S2 I and not depicted). To generate leukemia, LSK cells were transduced concomitantly with an MLL-AF9–expressing vector (tandem dimer Tomato [tdTom]–positive MIT retrovirus) and the retroviral anti–miR-99 vector (GFP+ LMN vector; Dickins et al., 2005; Chen et al., 2013; Fig. S2 J). Efficient KD of both miR-99a and miR-99b by anti–miR-99 was confirmed by quantitative RT-PCR (Fig. S2, K and L). Transduced cells were transplanted into sublethally irradiated animals. Although miR-99 KD mice displayed reduced splenic involvement, evidenced by decreased c-Kit+ cells and fewer LSCs (Fig. S3, A and B), these primary recipients did not exhibit differences in overall survival (Fig. S3 C). Colony-formation assays using leukemic blasts from the BM of primary recipients revealed significant reduction in colony-forming ability upon miR-99 KD, demonstrating that miR-99 regulates MLL-AF9+ AML maintenance (Fig. 3, B and C).
To assess the role of miR-99 in the maintenance of established post-transformed AML, we secondarily transplanted leukemic blasts from the BM of primary recipients. Mice transplanted with miR-99 KD blasts exhibited a significant improvement in survival (median survival 48 d in Scr vs. 92 d in miR-99 KD recipients; Fig. 3 D). miR-99 family members have been shown to be highly expressed in LSC-enriched populations from patient AML samples (Metzeler et al., 2013; Lechman et al., 2016). Thus, we asked whether the improved survival is mediated through depletion of the LSC compartment. As leukemic granulocyte-macrophage progenitors (L-GMPs) are the LSCs in the MLL-AF9 model of AML (Krivtsov et al., 2006; Somervaille and Cleary, 2006; Stubbs et al., 2008), we analyzed the BM of secondary recipients at the time of sacrifice for alterations in L-GMP frequency. This analysis revealed a reduction in the percentage of L-GMPs in mice transplanted with miR-99 KD blasts (Fig. 3, E and F). To further evaluate the effect of miR-99 KD on LSC function, we performed limiting dilution assays using MLL-AF9 blasts from primary recipients. Leukemia-initiating cell frequency was significantly reduced after transduction with anti–miR-99 (Fig. 3 G). Collectively, these data demonstrate that miR-99 is a critical positive regulator of LSC self-renewal.
miR-99 maintains LSCs self-renewal by inhibiting differentiation
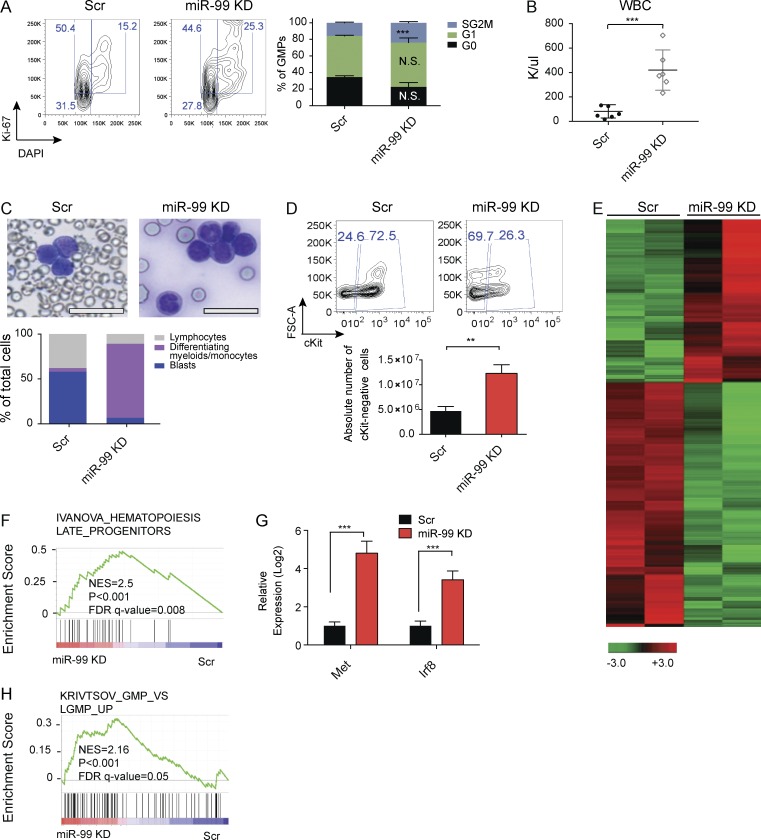
We next asked whether the decrease in L-GMP induced by miR-99 KD was caused by alterations in differentiation and/or cell cycling of these cells, similar to the phenotype observed in LSK cells. Consistent with this hypothesis, L-GMPs from animals transplanted with miR-99 KD MLL-AF9 AML displayed an increased proportion of cycling cells (Fig. 4 A). This phenotype was associated with a dramatic increase in the number of peripheral blood white blood cells (WBCs; Fig. 4 B). Because secondary transplantation of miR-99 KD blasts resulted in improved survival, we hypothesized that the WBCs were composed of more differentiated cells as opposed to blasts, a phenomenon that has been described with AML differentiation-inducing agents used in the clinic, such as ATRA and IDH1 inhibitors (Nowak et al., 2009). As expected, Wright–Giemsa staining of PB smears revealed that WBCs from miR-99 KD mice displayed greater differentiation (Fig. 4 C). Moreover, miR-99 KD mice displayed a significant increase in the absolute number of c-Kit− cells in the BM, compatible with induction of differentiation (Fig. 4 D).
Figure 4.
miR-99 KD depletes LSCs in the MLL-AF9 mouse model of AML by inducing differentiation. (A) Representative flow cytometry graph and the corresponding bar graph for the cell cycle staining performed on L-GMPs FACS sorted from the BM of secondary MLL-AF9 transplantation recipients. Data represent mean percentage ± SEM (Student’s t test; n = 3) and are representative of two independent experiments. (B) White blood cell (WBC) count of the secondary transplantation recipients. Data represent mean count ± SEM (Student’s t test; n = 6) and are representative of three independent experiments. (C) Wright–Giemsa staining of the peripheral blood from secondary transplant recipients, and summary of cytological features based on a 200-cell manual count per condition (bars, 25 µm). (D) miR-99 KD increases the absolute number of normal c-Kit− GFP+ tdTom+ cells in the BM. Data represent mean count ± SEM (Student’s t test; n = 5 for Scr and n = 3 for miR-99 KD) and are representative of three independent experiments. (E) Heat map of RNA-seq data generated from L-GMPs derived from the BM of secondary transplantation recipients with or without miR-99 KD at the time of death. (n = 2). (F) GSEA of differentially expressed genes in miR-99 KD and scramble control L-GMPs shows induction of a more differentiated signature. (G) TaqMan quantitative RT-PCR analysis of Met and Irf8 in miR-99 KD and Scr L-GMPs sorted from secondary recipients. Expression data are normalized to Actb (Student’s t test; n = 3) and are representative of two independent experiments. (H) GSEA of miR-99 KD and scramble control L-GMPs reveals induction of genes present in a signature up-regulated in normal GMPs compared with L-GMP (Table S1). **, P < 0.01; ***, P < 0.001.
To investigate the molecular mechanisms underlying miR-99 KD–induced AML differentiation, we performed RNA-seq on L-GMPs sorted from secondary transplant recipients (Fig. 4 E). GSEA revealed that miR-99 KD induces genes up-regulated in normal hematopoietic progenitors compared with HSCs, including Irf8 and Met (Ivanova et al., 2002), and this finding was validated by quantitative RT-PCR (Fig. 4, F and G). To identify an expression signature that enriches for leukemia differentiation genes, we used publicly available RNA-seq data to generate a gene set that is up-regulated in normal GMPs compared with MLL-AF9 L-GMPs (Padj < 0.05; Krivtsov et al., 2006; Table S1). Remarkably, L-GMPs from miR-99 KD mice exhibited enrichment for this gene set, confirming that miR-99 suppresses differentiation in LSCs (Fig. 4 H). Together, these data indicate that miR-99 maintains LSCs in an MLL-AF9 mouse model of AML in the undifferentiated and quiescent state.
miR-99 suppresses differentiation in human AML
Given that the mature sequences of all miR-99 family members are identical in mouse and human, we hypothesized miR-99 might exert similar biological effects in both mouse and human. To directly assess the roles of miR-99 in human hematopoiesis, we first evaluated miR-99 expression in different HSPC populations from normal human BM, and we found that miR-99 family members display a trend in expression similar to that seen in mice, with the highest expression levels in HSPCs (Fig. 5 A). To determine whether miR-99 maintains human HSPCs, we knocked down miR-99 in CD34+ umbilical cord blood cells and performed methylcellulose colony assays. miR-99 KD resulted in decreased colony formation, similar to studies using mouse HSPCs (Fig. 5 B).
Figure 5.
miR-99 functionally suppresses human AML differentiation. (A) Normalized expression levels of miR-99a, miR-99b and miR-100 by quantitative PCR using miRNA TaqMan probes in human HSPCs, including hematopoietic stem cells (HSCs), multipotent progenitors (MPPs), common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs). Expression was normalized to sno-R2. Data represent mean ± SEM and are representative of five independent experiments. (B) The colony-forming capacity of CD34+ human cord blood cells is reduced after miR-99 KD. CD34+ cells were transduced with lentiviral anti–miR-99 or scramble control. GFP+ cells were isolated and cultured in complete methylcellulose, and the colonies were scored after 14 d. Data represent mean ± SEM (Student’s t test; n = 3) and are representative of two independent experiments. (C) Flow cytometric evaluation of myeloid differentiation marker expression on MonoMac6 AML cells 5 d after transduction with anti–miR-99 or scramble control. Data represent mean ± SEM (Student’s t test; n = 3) and are representative of three independent experiments. (D) Wright–Giemsa stains of cytospin preparations of MonoMac6 cells 8 d post-transduction with lentiviral anti–miR-99 or Scr reveals induction of differentiation upon miR-99 KD (bars, 25 µm). (E) Overview of the xenotransplantation experiment performed on MonoMac6 AMLs. Cells were transduced with anti–miR-99 or Scr. After 48 h, GFP+ cells were flow sorted, and 800,000 cells were transplanted into sublethally irradiated NSGs. BM was analyzed 4 wk after the transplant. (F) miR-99 KD reduces the of GFP+ engraftment of MonoMac6 cells in the BM of recipients 4 wk post-transplantation. Data represent mean percentage ± SEM (Student’s t test; n = 4) and are representative of two independent experiments. (G) Representative histogram and aggregated data from flow cytometric evaluation of CD14 expression on GFP+ xenografted cells in the BM of the recipient animals 4 wk post-transplantation. Data represent mean percentage ± SEM (Student’s t test; n = 4) and are representative of two independent experiments. (H) Flow cytometry analysis of three AML patient samples after miR-99 KD. Patient samples were transduced with anti–miR-99 or scramble control and analyzed for the expression of differentiation markers 5–8 d later. Data represent mean percentage ± SEM (Student’s t test; n = 2) and are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine whether miR-99 also regulates human AML blast function, we knocked down miR-99 in the MonoMac6 AML cell line, which harbors an MLL-AF9 rearrangement (Super et al., 1995). miR-99 KD increased CD14+ and CD15+ myelomonocytic differentiation (Fig. 5 C). Evaluation of Wright–Giemsa–stained cytospins confirmed maturation of blasts with reduced nuclear to cytoplasmic ratios and increased cytoplasmic granules (Fig. 5 D). These observations demonstrate miR-99 suppresses differentiation in MonoMac6 cells. Similar to the effects observed in normal mouse HSPCs, miR-99 KD also induced apoptosis (Fig. S3 E).
To test whether miR-99 is required for the maintenance of human AML in vivo, we xenotransplanted miR-99 KD and Scr MonoMac6 cells (Fig. 5 E). After 4 wk, miR-99 KD leukemic cells showed a significant reduction in engraftment levels as well as increased CD14+ monocytic differentiation (Fig. 5, F and G). To investigate the effect of miR-99 KD on patient primary AML blasts, we performed in vitro differentiation assays. miR-99 KD induced CD14+ monocytic differentiation in two of three samples (Fig. 5 H). Collectively, these results replicate our findings in the MLL-AF9+ mouse model of AML and demonstrate that miR-99 similarly suppresses differentiation in human AML blasts.
An shRNA screen identifies functional miR-99 targets
We next sought to identify miR-99 targets that mediate its effects on Lin−Sca-1+c-Kit+ (LSKs) cells by performing an shRNA library screen against candidate miR-99 targets to test their ability rescue miR-99 KD-induced HSPC clonogenic defects; such shRNAs would be predicted to be enriched upon serial replating of miR-99 KD LSK cells. To design a list of genes for inclusion in the shRNA library, we acutely knocked down miR-99 in LSK cells followed by RNA-seq after 48 h to determine which of the predicted target genes are likely direct targets of miR-99 (Fig. 6 A). Consistent with induction of differentiation, genes up-regulated after miR-99 KD included cytokine and Toll-like receptor signaling pathways (Miranda and Johnson, 2007; Fig. 6 B). Furthermore, GSEA revealed significant induction of the NF-κB pathway and chemokine signaling (Fig. S3 F). We also observed genes induced with differentiation of HSCs into committed progenitors including Cd14, Cd86, Il7R, and Ccl5 (Fig. 6 C; Jaatinen et al., 2006). To include all potential genes mediating the phenotype, we compiled miR-99 predicted target genes from multiple algorithms and generated a list of 344 miR-99 targets (Table S2 A). 4,179 genes showed increased expression in the miR-99 KD experimental group (log2 fold change >0.2; Table S2 B), including 81 predicted target genes (Fig. S3 G and Table S2 C). We generated shRNAs against 45 candidate genes based on their miRNA-binding scores and also included shRNAs against two genes (Tet2 and Tgif1) previously shown to increase colony formation when knocked down as positive controls (Moran-Crusio et al., 2011; Yan et al., 2013). LSK cells were cotransduced with the shRNA library and anti–miR-99 vectors and then FACS sorted into methylcellulose and replated after 10 d. Genomic DNA (gDNA) was extracted from the resulting colonies to assess retroviral integrants (Fig. 6 E). The relative representation (enrichment score) of each shRNA was calculated immediately after transduction (T0) and at the end of the secondary plating for each replicate. Enrichment scores after secondary plating were normalized to T0 enrichment scores (i.e., the normalized enrichment score). Average normalized enrichment scores from three biological replicates were used to rank the genes, and genes predicted to be targeted by miR-99 in both mouse and human were prioritized for functional validation. As expected, the positive controls were among the most highly enriched shRNAs, validating the robustness of the screen (Fig. 6 F). Among the top enriched target genes shared between mouse and human were a number of genes including the ribonucleoprotein Raver2, bone morphogenetic protein receptor type II (Bmpr2), and the transcription factor Hoxa1 (Fig. 6 F and Table S2 G).
Figure 6.
Forward genetic screen identifies miR-99 target genes. (A) Heat map shows genes differentially expressed between scramble control and miR-99 KD LSK cells. RNA-Seq was performed on LSK cells 48 h after transduction with miR-99 KD or scramble control lentivirus. Infected cells were FACS sorted based on GFP expression (n = 3 for control and n = 2 for miR-99 KD). (B) Functional annotation of genes up-regulated in LSK cells after miR-99 KD using the database for annotation, visualization, and integrated discovery (DAVID). The top functional groups are depicted as a function of −log (p-value). (C) GSEA of the differentially expressed genes in LSK cells after miR-99 KD reveals induction of a differentiation gene signature. The gene set denotes genes down-regulated in CD133+ cells (hematopoietic stem cells [HSCs]) compared with CD133− cells. (D) Top up-regulated miR-99 target genes identified from RNA-seq experiments in LSK cells transduced with miR-99 KD or scramble control vectors. FC, fold change. (E) Schematic representation of the shRNA library screen experiment designed to identify miR-99 target genes that rescue the hematopoietic phenotype induced by miR-99 KD. LSK cells were cotransduced with lentiviral anti–miR-99 and the retroviral shRNA library. 48 h later, the resulting GFP+mCherry+ cells were FACS sorted into complete methylcellulose and allowed to form colonies. 10 d later, colonies were resuspended, and cells were replated a second time and cultured for an additional 7 d. gDNA from the resulting GFP+ mCherry+ cells was deep sequenced to identify integrated shRNAs. Representative data from three independent experiments are shown. (F) Pooled shRNA library screen identifies genes that are positively selected in LSKs transduced with anti–miR-99 KD. The y axis depicts normalized enrichment scores, which is defined as the enrichment score for each shRNA divided by its enrichment score at T0, calculated as the mean of three independent experiments, and shown in descending order. The x axis denotes genes from the screen that are predicted to be targeted by miR-99 both in mouse and human. Positive controls included shRNAs targeting Tet2 and Tgif1. Average data from three independent experiments are shown. (G) Normalized enrichments scores and the number of independent enriched shRNAs for each gene, depicted for the top genes identified in the shRNA screen. Average data from three independent experiments are shown.
miR-99 inhibits differentiation by targeting HOXA1
Among the top enriched genes identified in our shRNA screen, we prioritized Hoxa1 for functional validation, because all four shRNAs designed against this gene exhibited enrichment (Fig. 6 G). In addition, Hoxa1 has been shown to exhibit an inverse expression pattern to miR-99 in several contexts: (1) HOXA1 is induced upon ATRA-induced differentiation of the NB4 AML cell line (Park et al., 2003), (2) it is up-regulated as BM precursors differentiate into macrophages (De Santa et al., 2007), and (3) its expression is highest in the more differentiated M5 AMLs (Fig. S3I ). Finally, previous work in the nonhematopoietic context has shown that Hoxa1 is a bona fide target of the miR-99 family using luciferase assays (Chen et al., 2014; Wang et al., 2015). Moreover, Hoxa1 is shown to be a conserved target of the miR-99 family in mouse and human (Fig. S3 J).
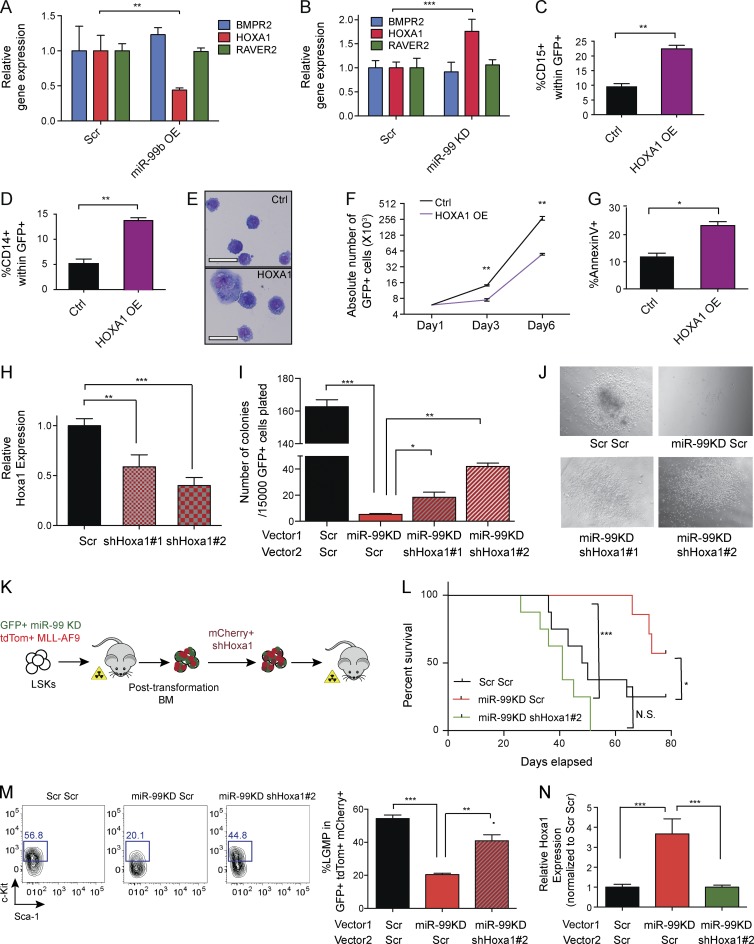
To test whether HOXA1 mediates the effects of miR-99 on differentiation, we overexpressed HOXA1 in MonoMac6 cells, where we previously demonstrated miR-99 normally inhibits differentiation. Confirming miR-99 targeting of HOXA1 in the hematopoietic context, miR-99 overexpression reduced HOXA1 mRNA expression (Fig. 7 A), whereas miR-99 KD increased HOXA1 mRNA expression (Fig. 7 B). Although Hox family members are generally thought to promote self-renewal (Argiropoulos and Humphries, 2007), enforced expression of HOXA1 (Fig. S3 K) induced myeloid differentiation in MonoMac6 cells as evidenced by increased expression of CD14 and CD15 by flow cytometry (Fig. 7 C, D). Wright–Giemsa staining confirmed increased differentiation of leukemic blasts demonstrated by acquisition of cytological features consistent with myelomonocytic maturation (Fig. 7 E). As expected, enforced expression of HOXA1 fully mimicked the miR-99 KD phenotype, as the increased differentiation was also associated with reduced cell growth and induction of apoptosis (Figs. 7 F, G). Together, these data demonstrate that miR-99 suppresses differentiation in MonoMac6 cells by targeting HOXA1.
Figure 7.
HOXA1 mediates miR-99 function in normal and malignant hematopoiesis. (A) TaqMan quantitative RT-PCR for HOXA1, BMPR2, and RAVER2 expression upon miR-99 overexpression in MonoMac6 AML cells 48 h post-transduction. Expression was normalized to ACTB (Student’s t test; n = 3). Representative data from two independent experiments are shown. (B) TaqMan quantitative RT-PCR for HOXA1, BMPR2, and RAVER2 expression upon miR-99 KD in MonoMac6 AML cells 48 h post-transduction. Expression was normalized to ACTB (Student’s t test; n = 3). Representative data from two independent experiments are shown. (C and D) Flow cytometry analysis of MonoMac6 cells 4 d after transduction with HOXA1-overexpressing virus. miR-99 KD induces the myeloid differentiation markers CD15 (C) and CD14 (D). Data represent mean percentage ± SEM (Student’s t test; n = 3) and are representative of two independent experiments. (E) Wright–Giemsa staining of MonoMac6 cells 4 d post-transduction with miR-99 KD (bars, 25 µm). (F) Growth curve for MonoMac6 cells as a function of time after transduction. Data represent mean count ± SEM (Student’s t test; n = 3) and are representative of two independent experiments. (G) Representative flow cytometry graph and the corresponding diagram depicting Annexin V apoptosis assay with HOXA1 overexpression in MonoMac6 cells. Data represent mean percentage ± SEM (Student’s t test; n = 3) and are representative of two independent experiments. (H) TaqMan quantitative RT-PCR for Hoxa1 expression 48 h after shHoxa1 transduction of LSK cells. Expression was normalized to Actb (Student’s t test; n = 3). Representative data from two independent experiments are shown. (I) Hoxa1 KD partially rescues colonies reduced upon miR-99 KD. LSK cells were infected with anti–miR-99 (GFP+) and shHoxa1 (mCherry+) viruses. 2 d post-transduction, the resulting GFP+ mCherry+ cells were sorted into Methocult M3434 and replated after 7 d. Shown are the results from secondary colonies, which were scored 10 d after plating. Data represent mean count ± SEM (Student’s t test; n = 3) and are representative data of two independent experiments. (J) Simultaneous KD of Hoxa1 in miR-99 KD LSKs increases the size of miR-99 KD colonies. Shown are representative colonies from secondary platings. (K) Schematic for Hoxa1 KD LSC rescue experiments. miR-99 KD MLL-AF9 (GFP+ tdTom+) BM blasts from primary recipients were transduced with shHoxa1 (mCherry+), and the resulting GFP+ tdtom+ mCherry+ cells were transplanted into sublethally irradiated mice. (L) Kaplan–Meier curves of mice transplanted with MLL-AF9 blasts. 100,000 BM blasts from primary recipients of the indicated genotypes were transplanted into sublethally irradiated secondary recipients (Mantel–Cox test; n = 8 per condition). Representative data from two independent experiments are shown. (M) Flow cytometry analysis of L-GMPs from the BM of secondary recipients. Cells were pregated on GFP+ tdTom+ mCherry+ CD16/32+ cells. Data represent mean percentage ± SEM (Student’s t test; n = 3 for Scr and shHoxa1#1 and n = 4 for miR-99 KD and shHoxa1#2) and are representative of two independent experiments. (N) TaqMan quantitative RT-PCR for Hoxa1 expression in L-GMPs sorted from secondary recipients at the time of death. Expression was normalized to Actb. Data represent mean ± SEM (Student’s t test; n = 3) and are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To investigate whether Hoxa1 mediates miR-99 function in the stem cell compartment, colony-forming assays were performed on Lin−c-Kit+Sca-1+ cells in which miR-99 and Hoxa1 were simultaneously suppressed. Hoxa1 KD efficiency was confirmed by quantitative RT-PCR (Fig. 7 H). Consistent with our shRNA library screen results, Hoxa1 KD in LSK cells rescued the reduction in colonies formed upon secondary plating of miR-99 KD cells (Fig. 7 I). Furthermore, simultaneous KD of Hoxa1 increased the size of colonies that were smaller with miR-99 KD alone (Fig. 7 J). Together, these data confirm Hoxa1’s central role in mediating miR-99 KD phenotypes in normal hematopoiesis.
To investigate the role of Hoxa1 in MLL-AF9 AML, we transduced blasts from primary MLL-AF9 miR-99 KD AMLs with the more potent Hoxa1 shRNA virus (shHoxa1-#2; Fig. 7 H). The infected cells (GFP+ tdTom+ mCherry+) were transplanted into sublethally irradiated mice (Fig. 7 K). miR-99 KD resulted in improved survival as expected, whereas simultaneous KD of Hoxa1 rescued this phenotype and shortened survival (Fig. 7 L). To test whether Hoxa1 KD affects LSC function, we analyzed the BM of the leukemia-engrafted animals at the time of death. miR-99 KD resulted in decreased L-GMPs, whereas simultaneous KD of Hoxa1 reversed this effect (Fig. 7 M). Analysis of L-GMPs from secondary recipients confirmed the expected changes in Hoxa1 mRNA expression, with miR-99 KD resulting in higher levels of Hoxa1 and Hoxa1 KD reversing this effect (Fig. 7 N). Collectively, these data demonstrate that miR-99 mediates its role in MLL-AF9 AML LSCs by suppressing Hoxa1.
Discussion
Using a combination of in vitro and in vivo approaches, our studies demonstrate that miR-99 is a critical regulator of self-renewal in both HSC and LSCs. miR-99 maintains stem cell function by suppressing differentiation, and this role is conserved in both mouse and human hematopoiesis. Although gene expression studies including our own had suggested that miR-99 might serve a functional role in HSCs and LSCs, prior studies using overexpression approaches failed to demonstrate a function for miR-99 in HSC regulation. However, our loss-of-function approach reveals a significant effect of miR-99 on HSC and LSC self-renewal. These findings suggest targeting miR-99 is a promising therapeutic strategy in contexts where depletion of one or both populations is desirable, such as eradication of preleukemic HSCs and LSCs in AML as well as HSC differentiation therapies for diseases that arise in HSCs, such as CML and MDS.
miRNAs exert their biological effects by inhibiting multiple downstream target genes (Lewis et al., 2005; Bartel, 2009), making it difficult to identify the genes that mediate their phenotypes. In this study, we used an shRNA library–based forward genetic screen to identify miR-99 targets that mediate the HSPC replating defect induced upon miR-99 KD. Among the top hits was the transcription factor HOXA1, which we confirmed mediates the differentiation phenotype induced after miR-99 KD in AML cell lines, hematopoietic stem and progenitor cells as well as leukemic stem cells; this is an unexpected finding considering that Hox family members are generally known for their role in promoting self-renewal and leukemogenesis (Argiropoulos and Humphries, 2007). This finding emphasizes the need to perform unbiased screens to identify miRNA mechanisms of action rather than a candidate gene approach based on previously defined roles. Although this screen sheds light on the mechanism of miR-99 function, our data do not exclude the possibility that miR-99 may mediate HSC and LSC function by targeting more than one gene. Although two other top hits from the shRNA screen, Raver2 and Bmpr2, were up-regulated upon miR-99 KD in LSKs, they did not display an increase after miR-99 inhibition in MonoMac6 cells, suggesting they may not contribute significantly to the differentiation phenotype in this cell line. Nonetheless, it will be important to further test the functional interactions between Hoxa1 and these genes and additional targets identified in our screen, in both the normal and leukemic context.
Approximately 60% of mammalian miRNAs are members of polycistrons (Chiang et al., 2010). As such, miRNAs with different seed sequences are transcribed simultaneously, and this coordinate expression is presumed to confer important biologic advantages (Han et al., 2015). miR-99 family members are located on polycistrons that also include members of the miR-125 and let7 families. As miR-125 also promotes HSC self-renewal (Guo et al., 2010; Ooi et al., 2010; Gerrits et al., 2012), our findings suggest that miR-125 and miR-99 have coevolved to support HSC function, which would potentially provide the basis for their coevolution and gene duplication as members of the same miRNA polycistron. Given that ectopic overexpression of the polycistron did not increase HSC function more than overexpression of miR-125 only, the action of miR-99 may depend on the expression of miR-125 or may be further modulated by let7. Intriguingly, prior studies suggest that let-7 may balance the effects of miR-125 and miR-99 by suppressing HSC function (Copley et al., 2011, 2013; Zipeto et al., 2016), a possibility also suggested by its ability to promote differentiation of AML cells (Pelosi et al., 2013). It will be important to dissect these interactions in more depth in the future, as well as post-transcriptional mechanisms that control expression of each of the miRNAs that constitute this cluster. Future experiments may also address the transcriptional regulation of this polycistron. Although HOXA10 and CDX2 have been shown to activate expression of this cluster, these studies were not performed in primary normal and malignant hematopoietic populations (Lin et al., 2011; Emmrich et al., 2014).
Although our data reveal for the first time that miR-99 promotes HSC and LSC self-renewal by inhibiting differentiation, a prior study using overexpression approaches failed to demonstrate a role for miR-99 in HSC function (Guo et al., 2010), a finding we confirmed in our own studies (Fig. S1 A). There are several potential reasons for these differences. First, whereas miR-99 is highly expressed in HSCs, it is predicted to target a relatively small number of genes (only 47 conserved targets, in contrast to miR-125 and let-7, which are predicted by the TargetScan algorithm to target 1,032 and 740 targets, respectively; Lewis et al., 2003, 2005; Grimson et al., 2007). Thus, miR-99 expression may be sufficiently high in HSCs for maximal target inhibition, rendering ectopic expression of miR-99 incapable of inducing further phenotypic change. In this scenario, knocking down miR-99 would still be able to release inhibition of miR-99 targets and induce phenotypes. Supporting this possibility, studies have established the importance of miRNA/target mRNA stoichiometry in determining the physiological consequences of miRNA expression, as the presence of fewer predicted target transcripts allows miRNAs to down-regulate their target genes more efficiently (Arvey et al., 2010). Therefore, bioinformatic approaches identifying relative miRNA/target gene expression levels in HSCs might help resolve these paradoxical results. Finally, whereas miR-99 overexpression studies evaluate the effect of overexpressing individual miRNAs, antagomir targeting strategies that inhibit multiple family members may induce more dramatic phenotypes (Fig. 1 D). This phenomenon has been observed in studies of other miRNAs in the hematopoietic system, as miR-146a KD in mouse HSPCs increased megakaryopoiesis in vivo (Starczynowski et al., 2010), whereas overexpression failed to alter megakaryocyte development or platelet production (Opalinska et al., 2010; Starczynowski et al., 2011). Collectively, our studies highlight the importance of using complementary approaches to evaluate miRNA function in the hematopoietic system.
Materials and methods
Cell sorting
Antibody staining and enrichment procedures for HSCs and multipotent progenitor, common myeloid progenitor, common lymphoid progenitor, granulocyte-macrophage progenitor, and megakaryocyte-erythroid progenitor cell sorting and analyses were performed as previously described (Akashi et al., 2000; Christensen and Weissman, 2001; Kiel et al., 2005).
Constructs
Lentiviral vectors (Fig. S1 B) were constructed in the miR-Zip backbone (Systems Biosciences) using the following sequences for anti–miR-99 vector 1: 5′-GAACCCGTGGATCCGATCCTGCGCTTCCTGTCAGCACAAGATCGGATCTACGGGTTTTTTT-3′ and anti–miR-99 vector 2: 5′-GCACCCGTGGAACCGACCGTGAGCTTCCTGTCAGCGCAAGGTCGGTTCTACGGGTGTTTTT-3′.
Retroviral vectors (Fig. S3 B) were constructed using previously described miR-30 shRNA expression vectors (Dickins et al., 2005). The classical shRNA sequences used in lentiviral backbones were modified into sequences compatible with miR-30 constructs. The sequence used for vector 2 is 5′-AAGGTATATTGCTGTTGACAGTGAGCGAACCCGTGGAACCGACCGTGAGTAGTGAAGCCACAGATGTACGCAAGGTCGGTTCTACGGGTGTGCCTACTGCCTCG-3′.
shRNA sequences used to knock down Hoxa1 included the following 97-mer oligo sequences: shHoxa1#1, 5′-TGCTGTTGACAGTGAGCGCTCCACTTCAACAAGTACCTTATAGTGAAGCCACAGATGTATAAGGTACTTGTTGAAGTGGAATGCCTACTGCCTCGGA-3′; shHoxa1#2, 5′-TGCTGTTGACAGTGAGCGACCAGTACATTCACCACTCATATAGTGAAGCCACAGATGTATATGAGTGGTGAATGTACTGGGTGCCTACTGCCTCGGA-3′.
Methylcellulose assays
For colony replating assays, mouse LT-HSCs (c-Kit+Sca-1+Lin−CD150+CD34−) cells were transduced with lentiviral anti–miR-99. Cells were maintained in DMEM-F12 with 10% FCS, 1× penicillin/streptomycin, 1× Glutamax, 10 ng/ml mSCF, 10 ng/ml mFlt3 ligand, 10 ng/ml thrombopoietin (mTPO), 10 ng/ml mIL-3, and 10 ng/µL mIL-6.
48 h after transduction, 100 GFP+ cells were plated into complete methylcellulose media containing mIL-3, mIL-6, mSCF, and mEPO (Methocult M3434; STEMCELL Technologies) for 7 d. After counting the colonies, all cells were resuspended in FACS media and analyzed for expression of cell surface markers. 20,000 GFP+ cells were used for replating assays, and the colonies were scored 7 to 10 d afterwards.
For human colony-forming assays, CD34+ cells were enriched using CD34 magnetic beads (catalog no. 130–046-702; Miltenyi Biotec). Prior to adding the CD34+ cells, lentiviral miR-Zip virus was spinofected for 2 h at 2,300 g in a 24-well plate precoated with 100 µg/ml Retronectin according to the manufacturer’s guidelines (catalog no. T100A; Clontech). The cells were then added to the plate and spun at 400 g for 10 min. Cells were maintained in StemSpan (STEMCELL Technologies) supplemented with 10% FCS, 20 ng/ml TPO, 20 ng/ml IL-3, 20 ng/ml granulocyte colony-stimulating factor, and 1× penicillin/streptomycin overnight. 1,500 GFP+ cells were plated into H4435 media (STEMCELL Technologies), which contains SCF, IL-3, IL-6, EPO, granulocyte colony-stimulating factor, and granulocyte-macrophage colony-stimulating factor. The colonies were scored after 14 d in culture.
For MLL-AF9 colony-formation assays, 1,000 GFP+ tdTom+ blasts from primary recipients were plated into M3434 media (STEMCELL Technologies). 1,000 GFP+ tdTom+ cells were replated every 7 d.
AML cell line differentiation in vitro
MonoMac6 cells were obtained from ATCC and maintained according to their guidelines. MonoMac6 cells were supplemented with RPMI containing 10% FCS, 1× penicillin/streptomycin, and 1× Glutamax (Thermo Fisher Scientific). 105 cells were plated in 12-well plates and transduced with lentiviral anti–miR-99 or Hoxa1 overexpression viruses by spinofection at 1,200 g at 37°C for 1 h. Cells were washed the next day and then cultured in RPMI as described above. Expression of myeloid differentiation makers was analyzed by flow cytometry 5–8 d after infection.
Mice and transplantations
LT-HSCs (c-Kit+Sca-1+Lin−CD150−CD34−) were double-FACS sorted and transduced with lentiviral anti–miR-99. The cells were kept in DMEM-F12 with 10% FCS, 10 ng/ml SCF, 10 ng/ml Flt3 ligand, 10 ng/ml TPO, 10 ng/ml IL-3, and 10 ng/ml IL-6. 48 h after transduction, GFP+ cells were sorted and used for transplants along with 300,000 Sca-1–depleted BM helper cells. Transplantations were performed by retro-orbital injection into lethally irradiated (2 × 475 rad) C57BL/6J mice (5,000 GFP+ cells per mouse). Helper cells were Sca-1 depleted using Sca-1–Biotin (catalog no. 108104; BioLegend) antibody and streptavidin beads (catalog no. 130–048-102; Miltenyi Biotec) according to the manufacturer’s instructions. All mice were maintained under pathogen-free conditions according to a Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Animal Care and Use Committee–approved protocol.
shRNA library screen
The shRNA library against candidate miR-99 target genes was generated by designing four shRNA sequences per gene (Table S2, D and E) and cloning them into an miR-E–based shRNA expressing retrovirus (Fig. S3 H; Fellmann et al., 2013), including 192 shRNAs against 47 target and control genes. 4–7 × 106 LSK cells were sorted from 10 to 15 C57BL/6J mice per replicate and transduced simultaneously with a lentiviral GFP+ anti–miR-99 and retroviral mCherry+ shRNA library. Cells were maintained in DMEM-F12 with 10% FCS, 10 ng/ml mSCF, 10 ng/ml mFlt3 ligand, 10 ng/ml mTPO, 10 ng/ml mIL-3, and 10 ng/ml mIL-6. 48 h post-transduction, mCherry+ GFP+ cells were sorted into M3434 Methocult media (STEMCELL Technologies). More than 2,000 colonies were obtained at the end of the first plating (shRNA coverage >10-fold) after 10 d. The colonies were resuspended in FACS buffer, and the GFP+ mCherry+ cells were used for secondary plating. After 7 d, the cells were harvested for gDNA extraction and sequenced. The experiment was repeated in three independent biological replicates.
Lentiviral transduction of patient AML samples
AML patient samples were thawed and plated in StemSpan (STEMCELL Technologies) supplemented with 10% FCS, 20 ng/ml hTPO, 20 ng/ml hIL-3, 20 ng/ml granulocyte colony-stimulating factor (hG-CSF), and 1× penicillin/streptomycin overnight. Lentiviral anti–miR-99 viral particles were spun for 2 h at 1,200 g in 24-well plates precoated with 100 μg/ml Retronectin (Takara). Cells were resuspended in media, added to the plate, and the spun at 400 g for 10 min.
Luciferase assays
HS3ST2 and SMARCA5 3′ UTRs were amplified by PCR using human genomic DNA and the following primers: HS3ST2 forward, 5′-GCTGCTCGAGCCGTGAGATTTGCTCCCAGA-3′; HS3ST2 reverse, 5′-ATAGCGGCCGCTTAAGGACACGAGAGAGCCA-3′; SMARCA5 forward, 5′-GCTGCTCGAGCAGTAGTTCTTTAATTTACGGGTCT-3′; SMARCA5 reverse, 5′-ATAGCGGCCGCGAAGCCTATTTTATTACTTCAAGTGTT-3′.
The resulting PCR products were then cloned in the PsiCHECK-2 vector for measurement of luciferase activity (catalog no. C8021; Promega).
Cell cycle analysis
BM cells were sorted for the indicated cell populations and subsequently fixed and permeabilized using 80% ethanol. The cells were incubated with anti–Ki67-PE antibody (catalog no. 556027; BD PharMingen) and resuspended in 500 µl PBS and 0.2 µg/ml DAPI before flow cytometry analysis.
Reverse transcription quantitative PCR
Total RNA was extracted from cells using TRIzol (Sigma) or an RNeasy kit (QIAGEN). cDNA was generated using a First-strand cDNA Synthesis Kit (Thermo Fisher Scientific), a TaqMan MicroRNA Reverse Transcript Kit (Applied Biosystems), or Quantimir (Systems Biosciences). TaqMan probes for individual miRNAs were purchased from Applied Biosystems. β-Actin and miR-16 were used as internal housekeeping controls.
Online supplemental material
Fig. S1 shows results from luciferase reporter assays to identify miR-99 targets as well as in vitro experiments after miR-99 KD and overexpression in HSPCs. Fig. S2 includes data for in vivo transplant studies of miR-99 KD HSCs as well as studies characterizing the retroviral anti–miR-99 vector in MLL-AF9 AML. Fig. S3 shows data related to characterization of MLL-AF9 AML primary recipients, limiting dilution transplants, snf shRNA library screen results. Tables S1 and S2 are provided as Excel files. Table S1 lists genes up-regulated comparing LGMPs with normal GMPs. Table S2 shows RNA-Seq and DNA deep sequencing data regarding the shRNA library experiment on primary hematopoietic stem and progenitors.
Supplementary Material
Acknowledgments
We wish to thank Dr. Scott Lowe (MSKCC) and Dr. Andrea Ventura (MSKCC) for scientific advice and all members of the Park laboratory for constructive discussions. We thank Dr. Ralph Garippa, Dr. Qing Xiang, and Sanjoy Mehta from the MSKCC RNAi core for assistance with shRNA library deep sequencing and analysis and Dr. Nicholas Socci for RNA-seq analysis.
This work was supported by the National Institutes of Health National Cancer Institute (R01 grant CA164120-01A1, to C.Y. Park). B.H. Durham was supported by the American Society of Hematology Senior Research Training Award for Fellows and a New York State Council on Graduate Medical Education Empire Clinical Research Investigator Program Fellowship.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- gDNA
- genomic DNA
- HSC
- hematopoietic stem cell
- HSPC
- hematopoietic stem and progenitor cell
- KD
- knockdown
- LSC
- leukemic stem cell
- LSK
- lineage negative Sca-1 positive c-Kit negative
- miRNA
- microRNA
- PB
- peripheral blood
- RNA-seq
- RNA sequencing
- Scr
- scramble
- tdTom
- tandem dimer Tomato
- TPO
- thrombopoietin
- WBC
- white blood cell
References
- Akashi K., Traver D., Miyamoto T., and Weissman I.L.. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Argiropoulos B., and Humphries R.K.. 2007. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 26:6766–6776. 10.1038/sj.onc.1210760 [DOI] [PubMed] [Google Scholar]
- Arvey A., Larsson E., Sander C., Leslie C.S., and Marks D.S.. 2010. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 6:363 10.1038/msb.2010.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet M., Quelen C., Rosati R., Mansat-De Mas V., La Starza R., Bastard C., Lippert E., Talmant P., Lafage-Pochitaloff M., Leroux D., et al. . 2008. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J. Exp. Med. 205:2499–2506. 10.1084/jem.20080285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet M., Nguyen D., Chen C., Shields L., and Lodish H.F.. 2012. MicroRNA-125b transforms myeloid cell lines by repressing multiple mRNA. Haematologica. 97:1713–1721. 10.3324/haematol.2011.061515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M.P., Seita J., and Weissman I.L.. 2008. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb. Symp. Quant. Biol. 73:439–449. 10.1101/sqb.2008.73.031 [DOI] [PubMed] [Google Scholar]
- Chen D., Sun Y., Yuan Y., Han Z., Zhang P., Zhang J., You M.J., Teruya-Feldstein J., Wang M., Gupta S., et al. . 2014. miR-100 induces epithelial-mesenchymal transition but suppresses tumorigenesis, migration and invasion. PLoS Genet. 10:e1004177 10.1371/journal.pgen.1004177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Deshpande A.J., Banka D., Bernt K.M., Dias S., Buske C., Olhava E.J., Daigle S.R., Richon V.M., Pollock R.M., and Armstrong S.A.. 2013. Abrogation of MLL-AF10 and CALM-AF10-mediated transformation through genetic inactivation or pharmacological inhibition of the H3K79 methyltransferase Dot1l. Leukemia. 27:813–822. 10.1038/leu.2012.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H.R., Schoenfeld L.W., Ruby J.G., Auyeung V.C., Spies N., Baek D., Johnston W.K., Russ C., Luo S., Babiarz J.E., et al. . 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 24:992–1009. 10.1101/gad.1884710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J.L., and Weissman I.L.. 2001. Flk-2 is a marker in hematopoietic stem cell differentiation: A simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA. 98:14541–14546. 10.1073/pnas.261562798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone M., and Calin G.A.. 2015. MicroRNAs in myeloid hematological malignancies. Curr. Genomics. 16:336–348. 10.2174/138920291605150710122815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion C.P., Han Y.-C., Mu P., Bonetti C., Yao E., D’Andrea A., Vidigal J.A., Maughan W.P., Ogrodowski P., and Ventura A.. 2012. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 8:e1002797 10.1371/journal.pgen.1002797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley M.R., Kent D.G., Benz C., Wohrer S., Rowe K.M., Day C.W., and Eaves C.J.. 2011. Inhibition of Let-7 processing in adult murine hematopoietic stem cells induces a fetal-like high self-renewal pattern in their progeny. Blood. 118:45–45. [Google Scholar]
- Copley M.R., Babovic S., Benz C., Knapp D.J.H.F., Beer P.A., Kent D.G., Wohrer S., Treloar D.Q., Day C., Rowe K., et al. . 2013. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 15:916–925. 10.1038/ncb2783 [DOI] [PubMed] [Google Scholar]
- De Santa F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., and Natoli G.. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 130:1083–1094. 10.1016/j.cell.2007.08.019 [DOI] [PubMed] [Google Scholar]
- Dickins R.A., Hemann M.T., Zilfou J.T., Simpson D.R., Ibarra I., Hannon G.J., and Lowe S.W.. 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37:1289–1295. 10.1038/ng1651 [DOI] [PubMed] [Google Scholar]
- Emmrich S., Rasche M., Schöning J., Reimer C., Keihani S., Maroz A., Xie Y., Li Z., Schambach A., Reinhardt D., and Klusmann J.H.. 2014. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes Dev. 28:858–874. 10.1101/gad.233791.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J., et al. . 2011. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17:1086–1093. 10.1038/nm.2415 [DOI] [PubMed] [Google Scholar]
- Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A.M., Kwon J.S., Guan Y., et al. . 2013. An optimized microRNA backbone for effective single-copy RNAi. Cell Reports. 5:1704–1713. 10.1016/j.celrep.2013.11.020 [DOI] [PubMed] [Google Scholar]
- Garzon R., Volinia S., Liu C.-G., Fernandez-Cymering C., Palumbo T., Pichiorri F., Fabbri M., Coombes K., Alder H., Nakamura T., et al. . 2008. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 111:3183–3189. 10.1182/blood-2007-07-098749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Plevritis S.K., Majeti R., and Alizadeh A.A.. 2010. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 304:2706–2715. 10.1001/jama.2010.1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits A., Walasek M.A., Olthof S., Weersing E., Ritsema M., Zwart E., van Os R., Bystrykh L.V., and de Haan G.. 2012. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood. 119:377–387. 10.1182/blood-2011-01-331686 [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K.-H., Johnston W.K., Garrett-Engele P., Lim L.P., and Bartel D.P.. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 27:91–105. 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Lu J., Schlanger R., Zhang H., Wang J.Y., Fox M.C., Purton L.E., Fleming H.H., Cobb B., Merkenschlager M., et al. . 2010. MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. USA. 107:14229–14234. 10.1073/pnas.0913574107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., and Kim V.N.. 2014. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15:509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Haetscher N., Feuermann Y., Wingert S., Rehage M., Thalheimer F.B., Weiser C., Bohnenberger H., Jung K., Schroeder T., Serve H., et al. . 2015. STAT5-regulated microRNA-193b controls haematopoietic stem and progenitor cell expansion by modulating cytokine receptor signalling. Nat. Commun. 6:8928 10.1038/ncomms9928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-C., Park C.Y., Bhagat G., Zhang J., Wang Y., Fan J.-B., Liu M., Zou Y., Weissman I.L., and Gu H.. 2010. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J. Exp. Med. 207:475–489. 10.1084/jem.20090831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-C., Vidigal J.A., Mu P., Yao E., Singh I., González A.J., Concepcion C.P., Bonetti C., Ogrodowski P., Carver B., et al. . 2015. An allelic series of miR-17 ∼ 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nat. Genet. 47:766–775. 10.1038/ng.3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Dooley J., Chung S.S., Chandramohan D., Cimmino L., Mukherjee S., Mason C.E., de Strooper B., Liston A., and Park C.Y.. 2015. miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood. 125:2206–2216. 10.1182/blood-2014-06-585273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N.B., Dimos J.T., Schaniel C., Hackney J.A., Moore K.A., and Lemischka I.R.. 2002. A stem cell molecular signature. Science. 298:601–604. 10.1126/science.1073823 [DOI] [PubMed] [Google Scholar]
- Jaatinen T., Hemmoranta H., Hautaniemi S., Niemi J., Nicorici D., Laine J., Yli-Harja O., and Partanen J.. 2006. Global gene expression profile of human cord blood-derived CD133+ cells. Stem Cells. 24:631–641. 10.1634/stemcells.2005-0185 [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., and Morrison S.J.. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121:1109–1121. 10.1016/j.cell.2005.05.026 [DOI] [PubMed] [Google Scholar]
- Klusmann J.-H., Li Z., Böhmer K., Maroz A., Koch M.L., Emmrich S., Godinho F.J., Orkin S.H., and Reinhardt D.. 2010. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 24:478–490. 10.1101/gad.1856210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A., and Dick J.E.. 2014. Evolution of the cancer stem cell model. Cell Stem Cell. 14:275–291. 10.1016/j.stem.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., et al. . 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442:818–822. 10.1038/nature04980 [DOI] [PubMed] [Google Scholar]
- Lechman E.R., Gentner B., van Galen P., Giustacchini A., Saini M., Boccalatte F.E., Hiramatsu H., Restuccia U., Bachi A., Voisin V., et al. . 2012. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 11:799–811. 10.1016/j.stem.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechman E.R., Gentner B., Ng S.W.K., Schoof E.M., van Galen P., Kennedy J.A., Nucera S., Ciceri F., Kaufmann K.B., Takayama N., et al. . 2016. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 29:214–228. (published erratum appears in Cancer Cell. 2016. http://dx.doi.org/10.1016/j.ccell.2016.03.015) 10.1016/j.ccell.2015.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., and Burge C.B.. 2003. Prediction of mammalian microRNA targets. Cell. 115:787–798. 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., and Bartel D.P.. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J. Jr, Laird P.W.; Cancer Genome Atlas Research Network., et al. . 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K.-Y., Zhang X.-J., Feng D.-D., Zhang H., Zeng C.-W., Han B.-W., Zhou A.-D., Qu L.-H., Xu L., and Chen Y.-Q.. 2011. miR-125b, a target of CDX2, regulates cell differentiation through repression of the core binding factor in hematopoietic malignancies. J. Biol. Chem. 286:38253–38263. 10.1074/jbc.M111.269670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Mrózek K., Radmacher M.D., Garzon R., and Bloomfield C.D.. 2011. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 117:1121–1129. 10.1182/blood-2010-09-191312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A., Zhao J.L., Sinha N., Marinov G.K., Mann M., Kowalczyk M.S., Galimidi R.P., Du X., Erikci E., Regev A., et al. . 2015. The microRNA-132 and MicroRNA-212 cluster regulates hematopoietic stem cell maintenance and survival with age by buffering FOXO3 expression. Immunity. 42:1021–1032. 10.1016/j.immuni.2015.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzeler K.H., Maharry K., Kohlschmidt J., Volinia S., Mrózek K., Becker H., Nicolet D., Whitman S.P., Mendler J.H., Schwind S., et al. . 2013. A stem cell-like gene expression signature associates with inferior outcomes and a distinct microRNA expression profile in adults with primary cytogenetically normal acute myeloid leukemia. Leukemia. 27:2023–2031. 10.1038/leu.2013.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M.B., and Johnson D.E.. 2007. Signal transduction pathways that contribute to myeloid differentiation. Leukemia. 21:1363–1377. 10.1038/sj.leu.2404690 [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K., Reavie L., Shih A., Abdel-Wahab O., Ndiaye-Lobry D., Lobry C., Figueroa M.E., Vasanthakumar A., Patel J., Zhao X., et al. . 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 20:11–24. 10.1016/j.ccr.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak D., Stewart D., and Koeffler H.P.. 2009. Differentiation therapy of leukemia: 3 decades of development. Blood. 113:3655–3665. 10.1182/blood-2009-01-198911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.M., Chaudhuri A.A., Rao D.S., Gibson W.S.J., Balazs A.B., and Baltimore D.. 2010. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. USA. 107:14235–14240. 10.1073/pnas.1009798107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi A.G.L., Sahoo D., Adorno M., Wang Y., Weissman I.L., and Park C.Y.. 2010. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc. Natl. Acad. Sci. USA. 107:21505–21510. 10.1073/pnas.1016218107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalinska J.B., Bersenev A., Zhang Z., Schmaier A.A., Choi J., Yao Y., D’Souza J., Tong W., and Weiss M.J.. 2010. MicroRNA expression in maturing murine megakaryocytes. Blood. 116:e128–e138. 10.1182/blood-2010-06-292920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.J., Vuong P.T., de Vos S., Douer D., and Koeffler H.P.. 2003. Comparative analysis of genes regulated by PML/RAR α and PLZF/RAR α in response to retinoic acid using oligonucleotide arrays. Blood. 102:3727–3736. 10.1182/blood-2003-02-0412 [DOI] [PubMed] [Google Scholar]
- Pelosi A., Careccia S., Lulli V., Romania P., Marziali G., Testa U., Lavorgna S., Lo-Coco F., Petti M.C., Calabretta B., et al. . 2013. miRNA let-7c promotes granulocytic differentiation in acute myeloid leukemia. Oncogene. 32:3648–3654. 10.1038/onc.2012.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille T.C.P., and Cleary M.L.. 2006. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 10:257–268. 10.1016/j.ccr.2006.08.020 [DOI] [PubMed] [Google Scholar]
- Starczynowski D.T., Kuchenbauer F., Argiropoulos B., Sung S., Morin R., Muranyi A., Hirst M., Hogge D., Marra M., Wells R.A., et al. . 2010. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 16:49–58. 10.1038/nm.2054 [DOI] [PubMed] [Google Scholar]
- Starczynowski D.T., Kuchenbauer F., Wegrzyn J., Rouhi A., Petriv O., Hansen C.L., Humphries R.K., and Karsan A.. 2011. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp. Hematol. 39:167–178.e4. 10.1016/j.exphem.2010.09.011 [DOI] [PubMed] [Google Scholar]
- Stubbs M.C., Kim Y.M., Krivtsov A.V., Wright R.D., Feng Z., Agarwal J., Kung A.L., and Armstrong S.A.. 2008. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: Development of a model for rapid therapeutic assessment. Leukemia. 22:66–77. 10.1038/sj.leu.2404951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Lee Y.S., Malhotra A., Kim H.K., Matecic M., Evans C., Jensen R.V., Moskaluk C.A., and Dutta A.. 2011. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 71:1313–1324. 10.1158/0008-5472.CAN-10-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Super H.J., Martinez-Climent J., and Rowley J.D.. 1995. Molecular analysis of the Mono Mac 6 cell line: Detection of an MLL-AF9 fusion transcript. Blood. 85:855–856. [PubMed] [Google Scholar]
- Velu C.S., Chaubey A., Phelan J.D., Horman S.R., Wunderlich M., Guzman M.L., Jegga A.G., Zeleznik-Le N.J., Chen J., Mulloy J.C., et al. . 2014. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J. Clin. Invest. 124:222–236. 10.1172/JCI66005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li Y., Qi W., Zhang N., Sun M., Huo Q., Cai C., Lv S., and Yang Q.. 2015. MicroRNA-99a inhibits tumor aggressive phenotypes through regulating HOXA1 in breast cancer cells. Oncotarget. 6:32737–32747. 10.18632/oncotarget.5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P., Iwasaki M., Somervaille T.C.P., Ficara F., Carico C., Arnold C., Chen C.-Z., and Cleary M.L.. 2010. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 70:3833–3842. 10.1158/0008-5472.CAN-09-3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Womack B., Wotton D., Guo Y., Shyr Y., Davé U., Li C., Hiebert S., Brandt S., and Hamid R.. 2013. Tgif1 regulates quiescence and self-renewal of hematopoietic stem cells. Mol. Cell. Biol. 33:4824–4833. 10.1128/MCB.01076-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Gong Y., Wang Q., Wang Y., and Zhang X.. 2015. The role of miR-100-mediated Notch pathway in apoptosis of gastric tumor cells. Cell. Signal. 27:1087–1101. 10.1016/j.cellsig.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Yilmaz O.H., and Morrison S.J.. 2008. The PI-3kinase pathway in hematopoietic stem cells and leukemia-initiating cells: A mechanistic difference between normal and cancer stem cells. Blood Cells Mol. Dis. 41:73–76. 10.1016/j.bcmd.2008.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.L., Rao D.S., O’Connell R.M., Garcia-Flores Y., and Baltimore D.. 2013. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. eLife. 2:e00537 10.7554/eLife.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.-S., Zhang H., Zhang X.-J., Feng D.-D., Luo X.-Q., Zeng C.-W., Lin K.-Y., Zhou H., Qu L.-H., Zhang P., and Chen Y.Q.. 2012. MiR-100 regulates cell differentiation and survival by targeting RBSP3, a phosphatase-like tumor suppressor in acute myeloid leukemia. Oncogene. 31:80–92. 10.1038/onc.2011.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipeto M.A., Court A.C., Sadarangani A., Delos Santos N.P., Balaian L., Chun H.-J., Pineda G., Morris S.R., Mason C.N., Geron I., et al. . 2016. ADAR1 activation drives leukemia stem cell self-renewal by impairing Let-7 biogenesis. Cell Stem Cell. 19:177–191. 10.1016/j.stem.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.