Abstract
The R1 origin region contains many symmetrical DNA sequence elements which allow the formation of complex secondary structures. A 218-bp in vivo deletion in a cloned R1 origin fragment removes most of them. As this deletion was never observed in plasmids containing all R1 replication functions, it was introduced by BglI in vitro recombination into the `basic replicon' of R1 cloned into pBR322. The recombinant plasmid with the 218-bp deletion and its derivatives unambiguously show that the deleted symmetrical elements are not absolutely essential for R1 replication as was previously assumed though they seem to determine a more efficient origin function. Likewise, a hypothetical protein of a mol. wt. of 14 000 daltons, the major part of which would be encoded by the deleted sequences, does not seem to be of particular importance for R1-specific replication. This is the first report of an alteration in the origin region of an IncFII plasmid which affects plasmid replication without abolishing it completely.
Keywords: antibiotic resistance factor R1, BglI recombination, DNA secondary structure, origin of replication, plasmid replication
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Brawner M. E., Jaskunas S. R. Identification of polypeptides encoded by the replication of resistance factor R100. J Mol Biol. 1982 Jul 25;159(1):35–55. doi: 10.1016/0022-2836(82)90030-4. [DOI] [PubMed] [Google Scholar]
- Burger K. J., Steinbauer J., Röllich G., Kollek R., Goebel W. Copy number control and incompatibility of plasmid R1: identification of a protein that seems to be involved in both processes. Mol Gen Genet. 1981;182(1):44–52. doi: 10.1007/BF00422765. [DOI] [PubMed] [Google Scholar]
- Diaz R., Staudenbauer W. L. Origin and direction of mini-R1 plasmid DNA replication in cell extracts of Escherichia coli. J Bacteriol. 1982 Jun;150(3):1077–1084. doi: 10.1128/jb.150.3.1077-1084.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobom G., Grosschedl R., Lusky M., Scherer G., Schwarz E., Kössel H. Functional analysis of the replicator structure of lambdoid bacteriophage DNAs. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):165–178. doi: 10.1101/sqb.1979.043.01.023. [DOI] [PubMed] [Google Scholar]
- Kollek R., Oertel W., Goebel W. Isolation and characterization of the minimal fragment required for autonomous replication ("basic replicon") of a copy mutant (pKN102) of the antibiotic resistance factor R1. Mol Gen Genet. 1978 Jun 1;162(1):51–57. doi: 10.1007/BF00333850. [DOI] [PubMed] [Google Scholar]
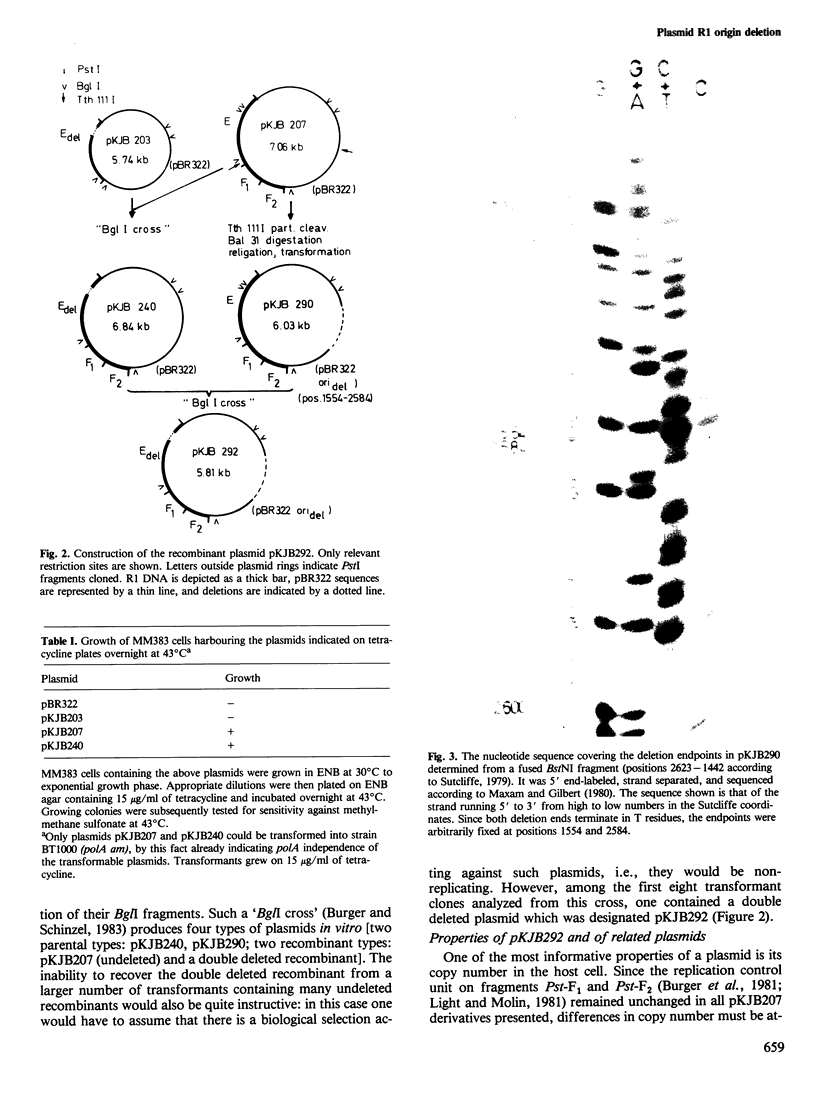
- Kollek R., Oertel W., Goebel W. Site-specific deletion at the replication origin of the antibiotic resistance factor R1. Mol Gen Genet. 1980 Feb;177(3):413–419. doi: 10.1007/BF00271479. [DOI] [PubMed] [Google Scholar]
- Light J., Molin S. Replication control functions of plasmid R1 act as inhibitors of expression of a gene required for replication. Mol Gen Genet. 1981;184(1):56–61. doi: 10.1007/BF00271195. [DOI] [PubMed] [Google Scholar]
- Messer W., Meijer M., Bergmans H. E., Hansen F. G., von Meyenburg K., Beck E., Schaller H. Origin of replication, oriC, of the Escherichia coli K12 chromosome: nucleotide sequence. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):139–145. doi: 10.1101/sqb.1979.043.01.020. [DOI] [PubMed] [Google Scholar]
- Monk M., Kinross J. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol. 1972 Mar;109(3):971–978. doi: 10.1128/jb.109.3.971-978.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel W., Kollek R., Beck E., Goebel W. The nucleotide sequence of a DNA fragment from the replication origin of the antibiotic resistance factor R1drd19. Mol Gen Genet. 1979 Mar 27;171(3):277–285. doi: 10.1007/BF00267582. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Feingold J., Ohtsubo H., Mickel S., Bauer W. Unidirectional replication in Escherichia coli of three small plasmids derived from R factor R12. Plasmid. 1977 Nov;1(1):8–18. doi: 10.1016/0147-619x(77)90004-x. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ohtsubo H., Ohtsubo E. The nucleotide sequence of the region surrounding the replication origin of an R100 resistance factor derivative. Mol Gen Genet. 1979 Mar 27;171(3):287–293. doi: 10.1007/BF00267583. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Inokuchi H., Ohtsubo H., Ohtsubo E. Genes and sites involved in replication and incompatibility of an R100 plasmid derivative based on nucleotide sequence analysis. Mol Gen Genet. 1980;179(3):527–537. doi: 10.1007/BF00271742. [DOI] [PubMed] [Google Scholar]
- Ryder T. B., Davidson D. B., Rosen J. I., Ohtsubo E., Ohtsubo H. Analysis of plasmid genome evolution based on nucleotide-sequence comparison of two related plasmids of Escherichia coli. Gene. 1982 Mar;17(3):299–310. doi: 10.1016/0378-1119(82)90146-9. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe P. M., Ely S., Timmis K. N. Plasmid replication functions. IV. Promoters in the replication region of plasmid R6-5. Nucleic Acids Res. 1979 Nov 24;7(6):1469–1484. doi: 10.1093/nar/7.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Synenki R. M., Nordheim A., Timmis K. N. Plasmid replication functions. III. Origin and direction of replication of a "mini" plasmid derived from R6-5. Mol Gen Genet. 1979 Jan 5;168(1):27–36. doi: 10.1007/BF00267930. [DOI] [PubMed] [Google Scholar]
- Takanami M. RNA polymerase nascent product analysis. Methods Enzymol. 1980;65(1):497–499. doi: 10.1016/s0076-6879(80)65058-7. [DOI] [PubMed] [Google Scholar]
- Timmis K. N., Danbara H., Brady G., Lurz R. Inheritance functions of group IncFII transmissible antibiotic resistance plasmids. Plasmid. 1981 Jan;5(1):53–75. doi: 10.1016/0147-619x(81)90077-9. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Ohmori H., Bird R. E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A. 1977 May;74(5):1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky S. L., Marians K. J. Escherichia coli factor Y sites of plasmid pBR322 can function as origins of DNA replication. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6111–6115. doi: 10.1073/pnas.78.10.6111. [DOI] [PMC free article] [PubMed] [Google Scholar]