Wang et al. show that deletion of autophagy gene FIP200 in neural stem cells (NSCs) leads to microglia infiltration into the subventricular zone (SVZ), inhibiting NSC differentiation non–cell autonomously in a p53-independent manner. Microglia infiltration into the SVZ is caused by increased chemokine expression in FIP200-null NSCs.
Abstract
Recent studies have shown important roles for autophagy genes in the regulation of different tissue stem cells, including neural stem/progenitor cells (NSCs). However, little is known about whether autophagy can regulate NSCs through cell-extrinsic mechanisms. Here, we show that deletion of an essential autophagy gene, FIP200, in NSCs increased expression of Ccl5 and Cxcl10 in a p53-independent manner, mediating increased infiltration of microglia into the subventricular zone of both FIP200hGFAP conditional knockout (cKO) and FIP200;p53hGFAP 2cKO mice. The microglia exhibited an activated M1 phenotype consistent with their potential to inhibit differentiation of FIP200-null NSCs. Blocking either microglia infiltration or activation rescued the deficient differentiation of FIP200-null NSCs from FIP200;p53hGFAP 2cKO mice. Lastly, we showed that increased chemokine expression in FIP200-null NSCs was induced by abnormal p62 aggregate formation and activation of NF-κB signaling. Our results suggest that autophagy plays a crucial role in regulating neurogenesis and restricting local immune response in postnatal NSCs through non–cell autonomous mechanisms.
Introduction
Postnatal neural stem cells (NSCs)/progenitor cells reside in the subventricular zone (SVZ) of the lateral ventricle and subgranular zone of dentate gyrus in the hippocampus of rodent brain (Gage, 2000; Kriegstein and Alvarez-Buylla, 2009). The self-renewal and differentiation of NSCs are regulated by cell–cell and cell–matrix interactions and diffusible signals from other cells, such as endothelial cells (Shen et al., 2004) and microglia (Sierra et al., 2010). Microglia are the resident immune cells in the central nervous system. They defend against pathogens and foreign bodies and clear dead cells and debris (Kettenmann et al., 2011). Microglia also regulate neurogenesis in the early postnatal SVZ (Shigemoto-Mogami et al., 2014) and adult subgranular zone (Sierra et al., 2010). Soluble factors secreted by microglia at injury sites direct NSC migration to these sites and NSC differentiation (Aarum et al., 2003). Embryonic ventricular zone/SVZ basal progenitors recruit microglia for cerebral cortex development (Arnò et al., 2014). Postnatal NSCs secreting VEGF enhance the proliferation and function of resident microglia (Mosher et al., 2012). However, our understanding of the cross-regulation of NSCs and microglia is still very limited.
Autophagy is a highly conserved lysosomal-dependent degradation pathway that clears damaged organelles and protein aggregates (Klionsky et al., 2016). Autophagy plays important roles, balancing the effects of immunity and inflammation in cancer, infection, and autoimmune diseases (Levine et al., 2011; Shibutani et al., 2015). For example, carriers of the T300A mutation in Atg16L1, an autophagy-related gene, have a higher incidence of Crohn disease (Hampe et al., 2007) and show increased infiltration of inflammatory cells (Adolph et al., 2013). Our previous studies showed that ablation of FIP200, an essential component of the ULK1–Atg13–FIP200–Atg101 autophagy induction complex, resulted in increased infiltration of immune cells to the skin and tumor mass of mammary tumors (Wei et al., 2009, 2011). These findings suggest that autophagy may function non–cell autonomously during inflammation and oncogenic transformation.
Increasing data show crucial functions of autophagy in self-renewal and differentiation of stem cells, including postnatal NSCs (Guan et al., 2013; Wang et al., 2013), but the underlying mechanisms are not well understood. Whether autophagy plays a role in cross talk between NSCs and microglia in the SVZ has not been investigated. We used unique mouse models to explore the contributions and mechanisms of microglia as a factor in the regulation of NSCs by autophagy.
Results
Increased microglia infiltration in the SVZ upon FIP200 deletion in NSCs
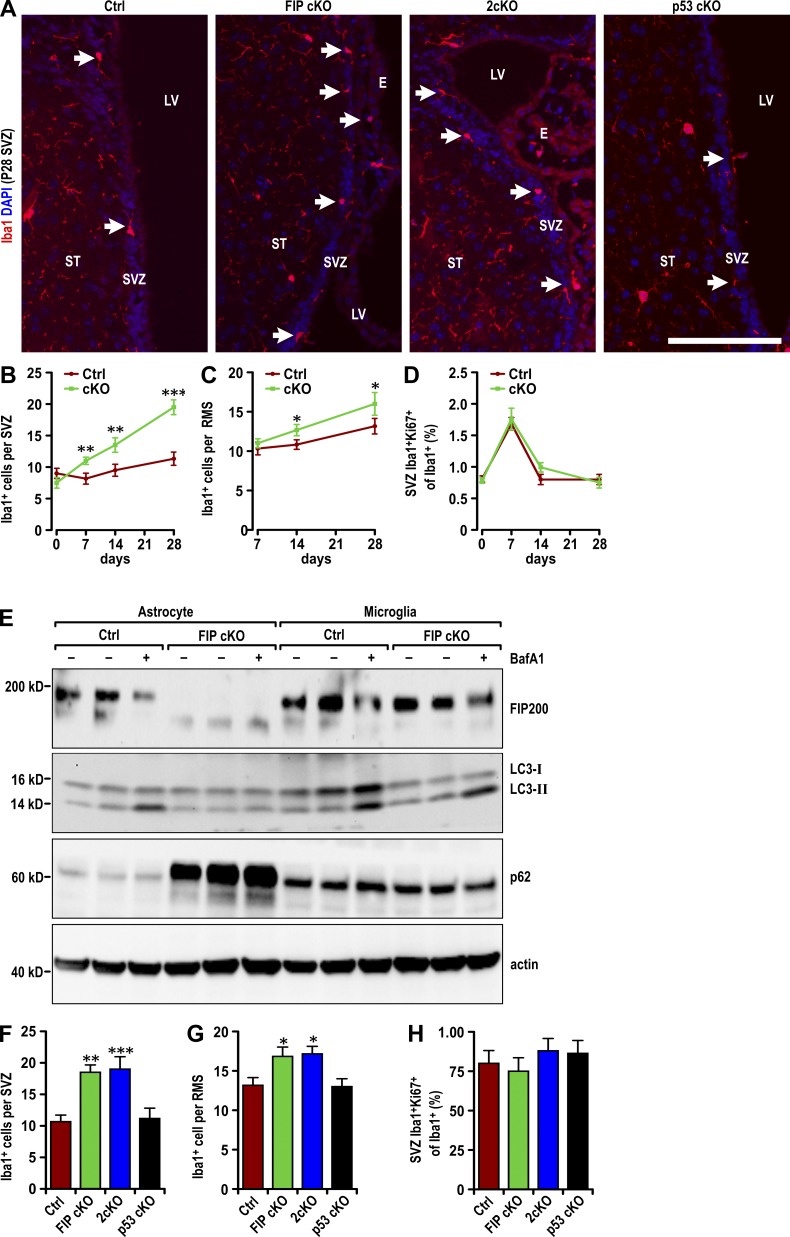
We first examined the number of microglia in different regions of the brain from FIP200hGFAP conditional knockout (FIP cKO) mice (Wang et al., 2013) using Iba1 as a marker, as described previously (Ito et al., 1998). The number of microglia significantly increased in the SVZ (Fig. 1, A and B) and rostral migratory stream (RMS; Fig. 1 C) of postnatal day 28 (P28) FIP cKO mice compared with control (Ctrl) mice. Time-course analysis showed that the number of microglia in the SVZ of FIP cKO and Ctrl mice was similar at P0 but gradually increased in FIP cKO mice compared with Ctrl mice at P7, P14, and P28 (Fig. 1, B and C; and Fig. S1 A). The number of microglia in the striatum and cerebral cortex (Fig. S1, B–D) was comparable between Ctrl and FIP cKO mice. Proliferation of microglia was low in the SVZ and comparable between groups (Figs. 1 D and S1 A), suggesting that the increased number of microglia was not caused by proliferation.
Figure 1.
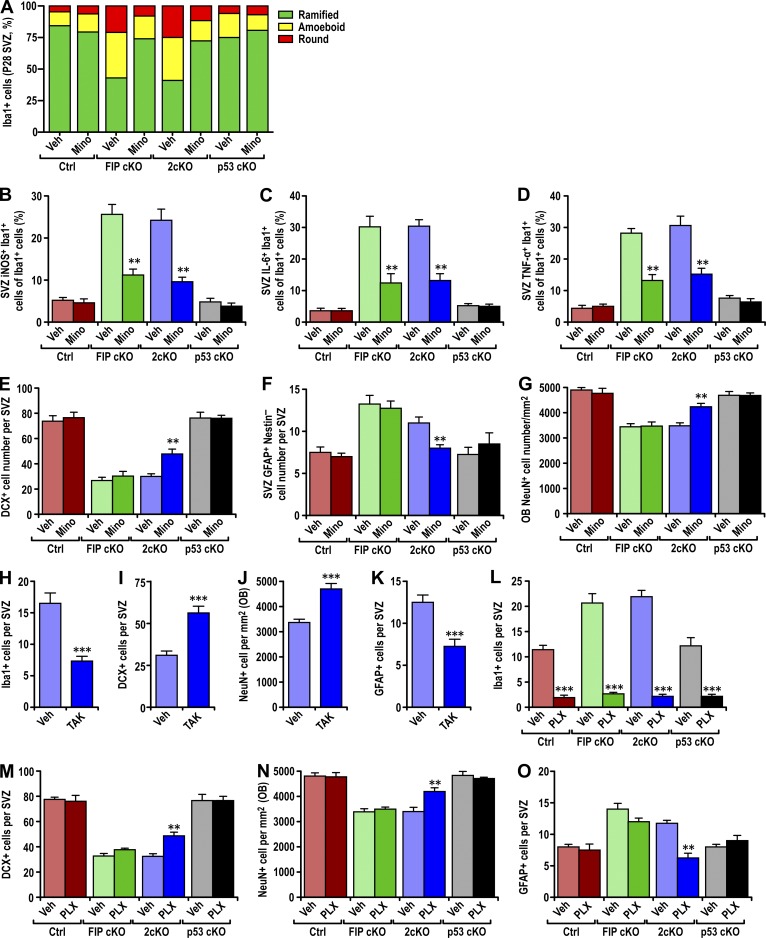
Increased infiltration of microglia in FIP200-deficient SVZ. (A) Immunofluorescence of Iba1 and DAPI in the SVZ (three mice each) in Ctrl, FIP cKO, 2cKO and p53 cKO mice at P28. Arrows indicate Iba1+ microglia. Bar, 40 µm. (B and C) Number of Iba1+ cells per SVZ (B) and RMS (C) section in Ctrl and FIP cKO mice at P0, P7, P14, and P28 (mean ± SEM; six mice per time point). (D) Percentage of Ki67+ microglia (Iba1+) per SVZ section in Ctrl and FIP cKO mice at P0, P7, P14, and P28 (mean ± SEM; >600 Iba1+ cells from six mice per time point). (E) Lysates were extracted from in vitro cultured astrocytes and microglia treated with DMSO or BafA1 for 1 h and analyzed by immunoblot to detect FIP200, LC3, p62, and actin. (F and G) Number of Iba1+ cells per SVZ (F) and RMS (G) section in Ctrl, FIP cKO, 2cKO, and p53 cKO mice at P28 (mean ± SEM; six mice each). (H) Percentage of Ki67+ microglia (Iba1+) per SVZ section of Ctrl, FIP cKO, 2cKO, and p53 cKO mice at P28 (mean ± SEM; >600 Iba1+ cells from six mice each). E, ependymal layer; LV, lateral ventricle; RMS, rostral migratory stream; ST, striatum; SVZ, subventricular zone. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIP200 was detected in astrocytes of Ctrl, but not FIP cKO, mice (Fig. 1 E, left six lanes), and a similar amount was found in microglia (Fig. 1 E, right six lanes), indicating no leaky expression of hGFAP-Cre in microglia (Ginhoux et al., 2010). FIP cKO mice showed significantly decreased levels of LC3-II in the presence of BafA1 and increased accumulation of p62 in astrocytes (Fig. 1 E, left six lanes) but comparable levels of LC3-II and p62 in microglia relative to Ctrl mice (Fig. 1 E, right six lanes). These results suggest that microglia infiltrate the SVZ via non–cell autonomous mechanisms in FIP cKO mice.
Our previous studies showed rescue of deficient self-renewal, but not defective differentiation, by p53 inactivation in FIP cKO (2cKO) mice (Wang et al., 2013). 2cKO mice, but not p53hGFAP cKO (p53 cKO) mice, showed an increase in microglia infiltration to the SVZ and RMS similar to that observed in FIP cKO mice at P28 (Fig. 1, A, F, and G). 2cKO mice and Ctrl mice showed a similar number of proliferative microglia (Fig. 1 H). Neither 2cKO nor p53 cKO mice showed increased microglia in other regions of the brain (Fig. 1, A, F, and G; and Fig. S1, E–G). These results suggest that the increase in the number of microglia in the SVZ upon FIP200 ablation in NSCs is not affected by p53 deletion and that microglia may contribute to their p53-independent defective differentiation.
Increased expression of chemokines Ccl5 and Cxcl10
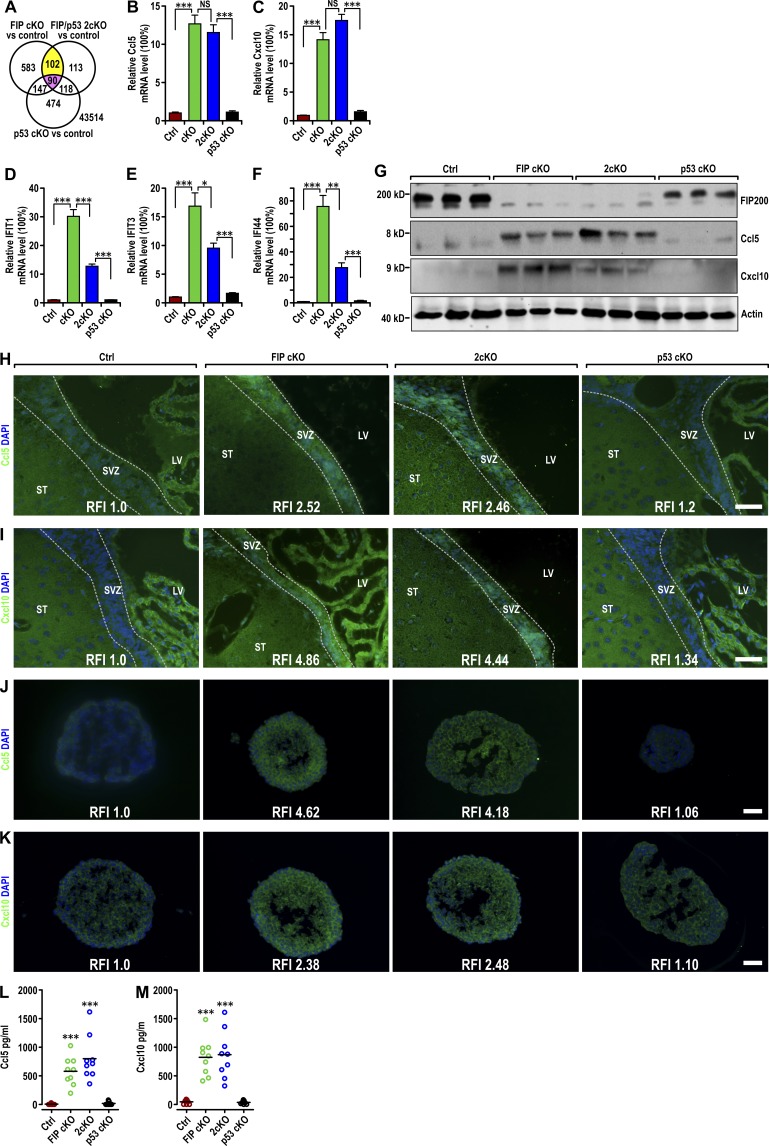
Gene expression profiles of neurospheres derived from FIP cKO, 2cKO, p53 cKO, and Ctrl mice were analyzed by microarray analyses. Of 922 genes with at least twofold changes in the FIP cKO sample compared with Ctrl samples, 192 were also changed in the 2cKO sample (potential targets not rescued by p53 inactivation; Fig. 2 A). Ninety were excluded as changed in the p53 cKO sample. Evaluation of the remaining 102 genes by gene ontology (Ashburner et al., 2000) indicated that the most prominent feature of the genome-wide and p53-independent alterations of FIP200-null NSCs was increased expression of genes involved in response to IFN stimulation, including Ccl5, Cxcl10, Ifi44, Ifit1, and Ifit3. Analysis of Ccl5, Cxcl10, and several other IFN-responsive genes by quantitative RT-PCR verified their up-regulation in FIP cKO neurospheres (Fig. 2, B–F). The elevated expression of Ccl5 and Cxcl10 was not rescued, but the increase in the expression of Ifit1, Ifit3, and Ifi44 was partially reversed in 2cKO neurospheres. Expression of these genes was not affected in p53 cKO neurospheres. These results suggest that the increased expression of chemokines and IFN-responsive genes may contribute to the p53-independent increase in microglia infiltration and defective differentiation of FIP200-null NSCs.
Figure 2.
p53-independent increase in Ccl5 and Cxcl10 expression in FIP200-deficient NSCs. (A) Venn diagram showing the overlap between genes with at least twofold increase or decrease compared with Ctrl mice in neurospheres derived from FIP cKO and 2cKO mice, but not those from p53 cKO mice (yellow) and those with similar changes in all samples (red). The number of genes is indicated. (B–F) Quantitative RT-PCR of Ccl5 (B), Cxcl10 (C), Ifit1 (D), Ifit3 (E), and Ifi44 (F) in neurospheres from Ctrl, FIP cKO, 2cKO, and p53 cKO mice (four mice each). (G) Lysates were extracted from SVZ of Ctrl, FIP cKO, 2cKO, and p53 cKO mice (three mice each) at P28 and analyzed by immunoblot using antibodies to detect FIP200, Ccl5, Cxcl10, and actin. (H–K) Immunofluorescence of Ccl5 (H and J) and Cxcl10 (I and K) with DAPI in the SVZ (H and I) and neurospheres (J and K; three mice each) of Ctrl, FIP cKO, 2cKO, and p53 cKO mice at P28. (L and M) Concentration of Ccl5 (L) and Cxcl10 (M) in conditioned neurosphere media (normalized to 1 mg total protein lysate of neurospheres) in Ctrl, FIP cKO, 2cKO, and p53 cKO mice (mean ± SEM; nine mice each). LV, lateral ventricle; ST:, striatum; SVZ, subventricular zone. Bars: (H and I) 50 µm; (J and K) 20 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001, NS, not significant; RFI, relative fluorescence intensity.
Ccl5 and Cxcl10 mediate the migration of immune cells, including microglia (Rappert et al., 2004; Lee et al., 2012), so we further examined the protein levels of these two chemokines in the SVZ of different mice and neurospheres derived from them. Immunoblot analysis of lysates from the SVZ and immunostaining of SVZ sections showed increased Ccl5 and Cxcl10 expression in FIP cKO and 2cKO mice compared with Ctrl and p53 cKO mice (Fig. 2, G–I). Immunostaining of neurospheres derived from SVZ cells of mice showed results consistent with these observations (Fig. 2, J and K). In conditioned media from neurospheres, we detected significantly elevated levels of Ccl5 and Cxcl10 from FIP cKO and 2cKO neurospheres and little to none in Ctrl and p53 cKO neurospheres by ELISA (Fig. 2, L and M). We also detected a moderate increase in the level of Ccl5 and Cxcl10 mRNA and secretion of these chemokines in FIP cKO astrocytes compared with Ctrl (Fig. S2, A–D), but much less than in NSCs (see Fig. 2, B, C, L, and M). These results further support that increased expression of Ccl5 and Cxcl10 by FIP200-null NSCs promotes microglia infiltration in FIP cKO and 2cKO mice via a non–cell autonomous and p53-independent mechanism.
Aberrant secretion of Ccl5 and Cxcl10 promotes microglia migration
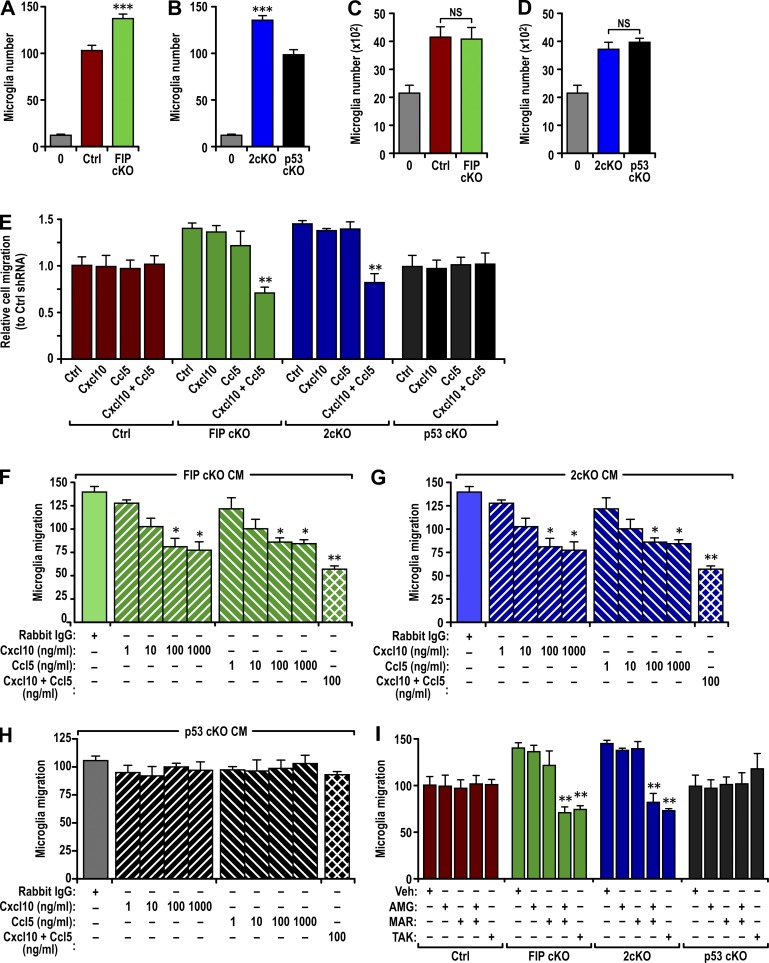
In a chemotaxis assay using conditioned media from various neurospheres, we found increased microglia migration in response to conditioned media from FIP cKO and 2cKO neurospheres compared with Ctrl and p53 cKO neurospheres (Fig. 3, A and B). Similar numbers of cells were found after plating in conditioned media from all samples (Fig. 3, C and D; and Fig. S3 A), suggesting that proliferation was not stimulated.
Figure 3.
Increased Ccl5 and Cxcl10 secretion by FIP200-deficient NSCs enhanced microglia migration. (A and B) Number of migrated microglia using naive media or conditioned media from neurospheres of Ctrl and FIP cKO mice (A) or 2cKO and p53 cKO mice (B) as attractant (mean ± SEM; six mice each). (C and D) Number of microglia cultured with naive media or conditioned media from neurospheres of Ctrl and FIP cKO mice (C) or 2cKO and p53 cKO mice (D; mean ± SEM; six mice each). (E) Normalized number of microglia migrating toward various conditioned media (mean ± SEM). Conditioned media are from neurospheres of Ctrl, FIP cKO, 2cKO, and p53 cKO mice that had been infected with recombinant lentiviruses encoding Ctrl shRNA, Ccl5 shRNA, Cxcl10 shRNA, or a combination of Ccl5 and Cxcl10 shRNAs, as indicated (three mice each). (F–H) Number of microglia migrating toward conditioned media from neurospheres of FIP cKO (F), 2cKO (G), and p53 cKO (H) mice (mean ± SEM). Conditioned media was treated with an unrelated antibody, Ccl5 antibody, Cxcl10 antibody, or a combination of Ccl5 and Cxcl10 antibodies at different concentrations (three mice each). (I) Number of microglia migrating toward conditioned media from neurospheres of Ctrl, FIP cKO, 2cKO, and p53 cKO mice (mean ± SEM). Conditioned media was treated with DMSO vehicle (Veh), 1 µM AMG-487 (AMG), 1 µM maraviroc (MAR), or 10 µM TAK-779 (TAK; three mice each). CM, conditioned media. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We then prepared recombinant lentiviruses encoding shRNAs for Ccl5 and Cxcl10 (Fig. S2, E and F). Knockdown of either Ccl5 or Cxcl10 in neurospheres did not affect the ability of the conditioned media to induce microglia migration (Fig. 3 E). However, knockdown of both in FIP cKO and 2cKO neurospheres abolished the ability of their conditioned media to increase microglia migration but did not affect basal microglia migration by conditioned media of Ctrl and p53 cKO neurospheres (Fig. 3 E). Knockdown of Ccl5 and Cxcl10 individually or in combination had no effect on microglia proliferation (Fig. S3 B) or neurosphere formation (Fig. S3, C and D).
Antibody depletion of Ccl5 and Cxcl10 also inhibited microglia migration in response to the conditioned media from FIP cKO and 2cKO neurospheres at 100 ng/ml (Fig. 3, F and G). Increasing the antibody concentration to 1 µg/ml did not increase inhibition, suggesting that 100 ng/ml was sufficient to deplete the respective chemokines. The combination of both antibodies at 100 ng/ml inhibited microglia migration to a greater extent, suggesting redundant roles of Ccl5 and Cxcl10 on chemoattraction. The use of similar antibody amounts individually or in combination did not affect the basal migration in response to Ctrl (not depicted) and p53 cKO (Fig. 3 H) conditioned media. Antibody depletion did not affect microglia proliferation (Fig. S3 E).
Lastly, we examined the effect of inhibitors for Ccl5 and Cxcl10 receptors (i.e., Ccr5 and Cxcr3) in microglia (Albright et al., 1999; Biber et al., 2002). Addition of either the Ccr5 inhibitor (±) AMG-487 or the Cxcr3 inhibitor maraviroc alone had no effect on microglia migration in response to the conditioned media from FIP cKO and 2cKO neurospheres (Fig. 3 I). However, the combination of these two inhibitors or the use of TAK-779 (TAK), a Ccr5 inhibitor later found to also inhibit Cxcr3 (Suzaki et al., 2008), significantly reduced microglia migration under the same conditions. None of the inhibitor treatments affected microglia proliferation (Fig. S3 F). These results indicate that increased production of Ccl5 and Cxcl10 with FIP200 deletion in NSCs plays a major role in the increased microglia migration and infiltration into the SVZ of FIP cKO and 2cKO mice through a p53-independent mechanism.
Activated infiltrated microglia contribute to defective NSC differentiation
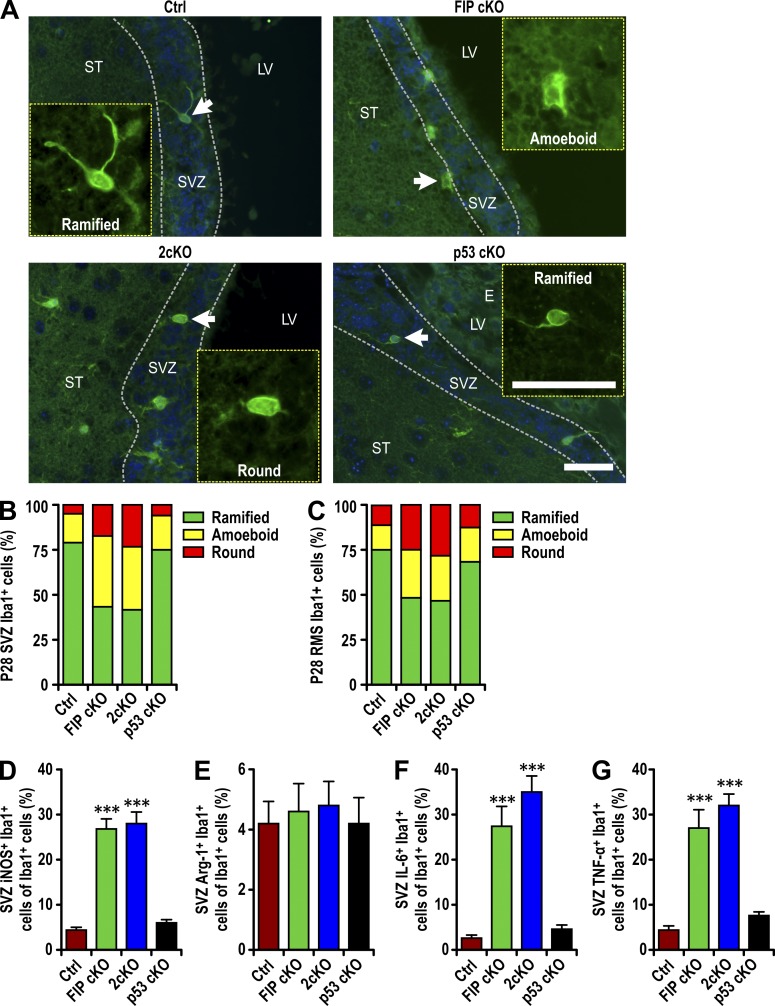
Microglia have been shown to influence NSC differentiation both positively and negatively, depending on activation status (Sato, 2015). In the SVZ of FIP cKO and Ctrl mice at P0, microglia showed similar morphology, with large somata, a few thick processes, and large lamellipodia extending to neighboring cells (see Fig. S1 A, arrows). At P28, however, >50% of microglia in the SVZ of FIP cKO mice exhibited activated morphology (amoeboid and round shapes), whereas >75% of microglia in Ctrl had resting morphology with multiple ramified processes (Fig. 4, A and B). Similar increases in activated microglia were also found in the SVZ of 2cKO, but not p53 cKO, mice. The increased fraction of activated microglia was also apparent in the RMS of FIP cKO and 2cKO mice compared with Ctrl and p53 cKO mice (Fig. 4 C). No differences in the fraction of activated microglia were found in other regions of the brain (Fig. S4, A and B).
Figure 4.
Infiltrated microglia are activated in FIP200-deficient SVZ. (A) Immunofluorescence of Iba1 and DAPI in SVZ (three mice each) of Ctrl, FIP cKO, 2cKO, and p53 cKO mice at P28. Arrows indicate cells shown in more detail for Iba1 staining in insets. The dotted lines indicate the boundaries of the SVZ. Bars, 30 µm. (B and C) The percentage of ramified, round, and amoeboid microglia per SVZ (B) and RMS (C) section in Ctrl, FIP cKO, 2cKO, and p53 cKO mice at P28 (>300 cells from three mice each). (D–G) Percentage of iNOS+ microglia (Iba1+ cells; D), Arginase 1+ microglia (E), IL-6+ microglia (F), and TNF+ microglia per SVZ section (mean ± SEM; five mice each) in Ctrl, FIP cKO, 2cKO, and p53 cKO mice at P28. LV, lateral ventricle; ST, striatum; SVZ, subventricular zone. ***, P < 0.001.
We also stained SVZ sections using markers that functionally classify microglia that can inhibit (M1) or promote (M2) NSC differentiation (Ekdahl et al., 2003; Monje et al., 2003; Walter et al., 2011). We found a significant increase in the fraction of M1 microglia in the SVZ of P28 FIP cKO and 2cKO mice compared with Ctrl and p53 cKO mice, measured by the M1 marker iNOS (Fig. 4 D). Similar fractions of M2 (Arginase1) microglia were found in the SVZ, suggesting that the increased infiltrated microglia showed preferential M1 activation, which could contribute to the defective NSC differentiation.
M1 macrophages can secrete proinflammatory cytokines, including IL-6 and TNF, which are known to inhibit postnatal neurogenesis (Vallières et al., 2002; Iosif et al., 2006). To examine this, SVZ sections were analyzed for microglia-associated expression of IL-6 and TNF. Significantly increased fractions of IL-6+ and TNF+ activated microglia were detected in FIP cKO and 2cKO SVZs compared with Ctrl (Fig. 4, F and G; and Fig. S4, C and D). These results confirm that activated infiltrated microglia with M1 phenotype inhibited NSC neurogenesis in FIP cKO and 2cKO mice in a p53-independent manner.
Rescue of defective neurogenesis by inhibition of microglia activation and migration
We used minocycline, an inhibitor of microglia activation (Fan et al., 2007; Kobayashi et al., 2013), to treat mice every other day from P7 to P28. Minocycline treatment did not alter the number of infiltrated microglia, consistent with activation after infiltration. Minocycline treatment reversed the increased round and amoeboid microglia in FIP cKO and 2cKO mice (Fig. 5 A) compared with Ctrl and p53 cKO, suggesting inhibition of their activation. We also found significant decreases in the percentage of iNOS+, IL-6+, and TNF+ microglia in FIP cKO and 2cKO with minocycline treatment (Fig. 5, B–D). These results indicate inhibition of activated M1 microglia expressing proinflammatory cytokines.
Figure 5.
Suppression of microglia activation and infiltration rescued the defective neurogenesis in 2cKO mice. (A) The percentage of ramified, round, and amoeboid microglia per SVZ section of Ctrl, FIP cKO, 2cKO, and p53 cKO mice treated with vehicle (Veh) or minocycline (Mino; three mice each). (B–D) Number of iNOS+ microglia (B), IL-6+ microglia (C), or TNF+ microglia (D) per SVZ section in Ctrl, FIP cKO, 2cKO, and p53 cKO mice treated with vehicle (Veh) or minocycline (Mino; mean ± SEM; three mice each). (E–G) Number of DCX+ neuroblasts (E) and GFAP+ astrocytes (F) per SVZ section or the density of NeuN+ neurons per olfactory bulb section (G) in Ctrl, FIP cKO, 2cKO, and p53 cKO mice treated with vehicle or minocycline are shown (mean ± SEM; three mice each). (H) Number of Iba1+ microglia per SVZ section of 2cKO mice treated with vehicle (Veh) or TAK-779 (TAK; mean ± SEM; three mice each). (I–K) Number of DCX+ neuroblasts (I) and GFAP+ astrocytes (K) per SVZ section or the density of NeuN+ neurons per olfactory bulb section (J) in 2cKO mice treated with vehicle (Veh) or TAK-779 (TAK; mean ± SEM; three mice each). (L) Number of Iba1+ microglia per SVZ section in Ctrl, FIP cKO, 2cKO, and p53 cKO mice treated with vehicle (Veh) or PLX3397 (PLX; mean ± SEM; four mice each). (M–O) Number of DCX+ neuroblasts (M) and GFAP+ astrocytes (O) per SVZ section or the density of NeuN+ neurons per olfactory bulb section (N) in Ctrl, FIP cKO, 2cKO, and p53 cKO mice treated with vehicle (Veh) or PLX3397 (PLX) are shown (mean ± SEM; four mice each). **, P < 0.01; ***, P < 0.001.
We then examined the effects of inhibition of microglia activation on differentiation using various lineage markers, as described previously (Wang et al., 2013). Consistent with lack of effect on basal activation, minocycline did not affect normal differentiation of NSCs measured by DCX (neuroblasts), GFAP (astrocytes), and NeuN (olfactory bulb neurons; Fig. 5, E–G). To our surprise, however, minocycline did not rescue the decrease in neurogenesis and the increase in the number of astrocytes in FIP cKO mice, despite its significant inhibition of microglia activation. These results suggested that the increased number of microglia alone (Fig. 5, A–D) was sufficient to cause aberrant differentiation of FIP200-null NSCs. Alternatively, inhibition of microglia activation alone may not be sufficient to rescue FIP200-null NSC defects when their self-renewal is also blocked (Wang et al., 2013). In support of the latter, we found that minocycline significantly increased the number of SVZ DCX+ neuroblasts and olfactory bulb NeuN+ neurons, whereas it reduced the number of increased GFAP+ astrocytes in the SVZ of 2cKO mice (Fig. 5, E–G), in which defective self-renewal of NSC was rescued by p53 inactivation (Wang et al., 2013). Minocycline did not affect the maintenance of NSCs of FIP cKO or 2cKO mice (Fig. S4, D and E).
We also treated 2cKO mice every day from P7 to P28 with TAK, which is capable of penetrating the brain (Takami et al., 2002). TAK treatment blocked microglia migration in response to conditioned media from FIP200-null NSCs (Fig. 3 I). TAK treatment also significantly decreased the number of microglia in the SVZ of 2cKO mice (Figs. 5 H), suggesting that infiltration of microglia depends on Ccr5 and Cxcr3 activation by their ligands. TAK treatment significantly increased the number of DCX+ neuroblasts in the SVZ (Figs. 5 I) and olfactory bulb NeuN+ neurons (Fig. 5 J), whereas it reduced the GFAP+ astrocytes (Fig. 5 K) in the SVZ of 2cKO mice (Wang et al., 2013). TAK treatment did not affect NSC maintenance in these mice (Fig. S4 F). Lastly, we used PLX3397, a specific CSF1R antagonist used to deplete microglia (Elmore et al., 2014), and we examined its effect on neurogenesis. PLX3397 treatment significantly decreased the number of microglia in the SVZ (Figs. 5 L and S4 I) and other regions of all mice (not depicted). PLX3397 treatment also significantly increased the number of SVZ DCX+ neuroblasts and olfactory bulb NeuN+ neurons but reversed the increase in SVZ GFAP+ astrocytes in 2cKO mice, but not Ctrl, FIP cKO, and p53 cKO mice (Fig. 5, M–O). NSC maintenance was not affected (Fig. S4, J and K). These results demonstrate that the increased infiltration and activation of microglia in response to increased Ccl5 and Cxcl10 secretion by FIP200-null NSCs plays a direct role in their p53-independent, aberrant differentiation when their self-renewal defects are corrected by p53 ablation.
Aggregated p62 promotes chemokine expression through NF-κB
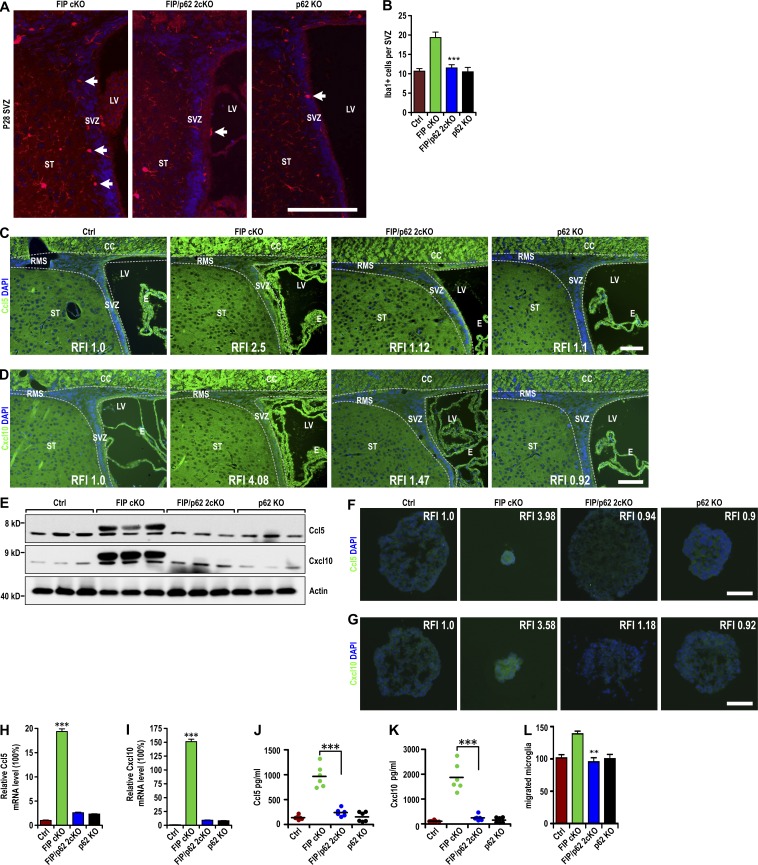
Our previous data showed that abnormal p62 aggregation plays an important role in defective NSC maintenance and differentiation (Wang et al., 2013, 2016) and might be responsible for the p53-independent Ccl5 and Cxcl10 up-regulation in FIP200-null NSCs. We analyzed FIP200f/f;p62−/−;hGFAP-Cre mice (FIP/p62 2cKO; Wang et al., 2016) to determine whether p62 ablation could rescue the increase in Ccl5 and Cxcl10 expression and microglia infiltration in FIP cKO mice. P28 FIP/p62 2cKO mice showed a reduced number of microglia in the SVZ that was comparable to Ctrl and p62 KO mice (Fig. 6, A and B). Most microglia (>75%) in the SVZ of FIP/p62 2cKO mice had multiple ramified processes, in contrast to >50% amoeboid or round microglia in the FIP cKO SVZ. The increased levels of Ccl5 and Cxcl10 in the SVZ of FIP cKO mice were also abrogated (Fig. 6, C–E). Expression of Ccl5 and Cxcl10 in neurospheres (Fig. 6, F–I) and conditioned media (Fig. 6, J and K) was reverted to control levels. Conditioned media failed to increase microglia migration (Fig. 6 L). These results suggest an essential role of p62 in mediating elevated Ccl5 and Cxcl10 expression in FIP200-deificient NSCs.
Figure 6.
Accumulated p62-mediated increase in Ccl5 and Cxcl10 expression and microglia infiltration in the FIP200-deficient SVZ. (A) Immunofluorescence of Iba1 and DAPI in the SVZ (three mice each) of FIP cKO, FIP/p62 2cKO and p62 KO mice at P28. Arrows indicate Iba1+ microglia. (B) Number of Iba1+ microglia per SVZ section in Ctrl, FIP cKO, FIP/p62 2cKO, and p62 KO mice at P28 (mean ± SEM; six mice each). (C and D) Immunofluorescence of Ccl5 (C) and Cxcl10 (D) with DAPI in the SVZ (three mice each) of Ctrl, FIP cKO, FIP/p62 2cKO, and p62 KO mice at P28. Dotted lines indicate the boundaries of SVZ. (E) Lysates were extracted from the SVZ of Ctrl, FIP cKO, FIP/p62 2cKO, and p62 KO mice at P28 and analyzed by immunoblot using antibodies to detect Ccl5, Cxcl10, and actin (three mice each). (F and G) Immunofluorescence of Ccl5 (F) and Cxcl10 (G) with DAPI in neurospheres from Ctrl, FIP cKO, FIP/p62 2cKO, and p62 KO mice at P28 (three mice each). (H and I) mRNA levels of Ccl5 (H) and Cxcl10 (I) in neurospheres from Ctrl, FIP cKO, FIP/p62 2cKO, and p62 KO mice (mean ± SEM; four mice each). (J and K) Concentration of Ccl5 (J) and Cxcl10 (K) in conditioned medium of neurospheres (normalized to 1 mg of total protein lysate of neurospheres) from Ctrl, FIP cKO, FIP/p62 2cKO, and p62 KO mice are shown (mean ± SEM; six mice each). (L) Number of migrated microglia in conditioned media from neurospheres of Ctrl, FIP cKO, FIP/p62 2cKO, and p62 KO mice as attractant (mean ± SEM; four mice each). CC, corpus callosum; E, ependymal layer; LV, lateral ventricle; RFI, relative fluorescence intensity; RMS, rostral migratory stream; ST, striatum; SVZ, subventricular zone. Bars: (A) 40 µm; (C and D) 30 µm; (F and G) 20 µm. **, P < 0.01; ***, P < 0.001.
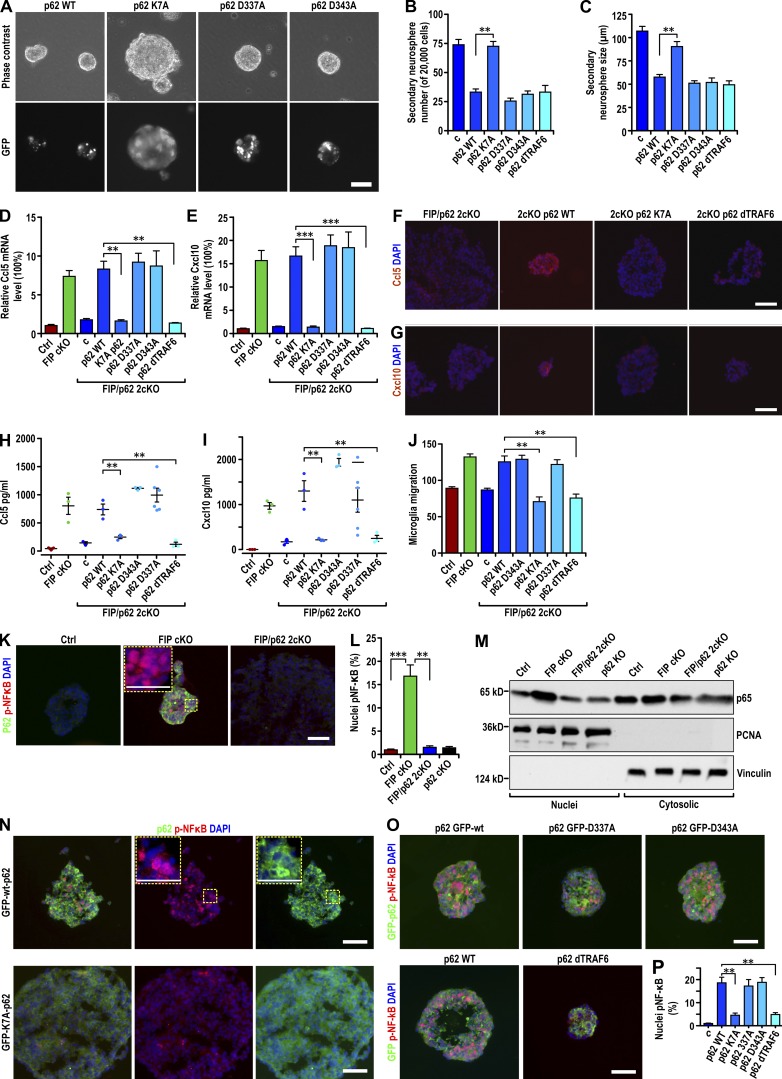
To study the mechanisms involved, recombinant retroviruses encoding p62 mutants D337A (GFP-p62-D337A) and D343A (GFP-p62-D343A) disrupting binding to LC3 (Ichimura et al., 2008), K7A/D69A mutant (GFP-p62-K7A) lacking self-aggregation (Lamark et al., 2003; Itakura and Mizushima, 2011), and wild-type p62 (GFP-p62-wt) were analyzed in neurospheres from FIP/p62 2cKO mice. Reexpression of GFP-p62-wt led to the inhibition of neurosphere self-renewal (Fig. 7, A–C) and p62 aggregation (Fig. 7 A, bottom). GFP-p62-D337A and GFP-p62-D343A showed similar phenotypes to wild-type p62, suggesting that LC3 binding is not important for the inhibition of NSC self-renewal and is disposable for p62 aggregation, as shown previously (Ichimura et al., 2008; Itakura and Mizushima, 2011). GFP-p62-K7A did not affect the self-renewal of FIP/p62 2cKO neurospheres and, as reported previously (Lamark et al., 2003; Itakura and Mizushima, 2011), did not form aggregates, suggesting that self-aggregation of p62 is necessary to inhibit NSC self-renewal. Reexpression of wild-type p62 or mutants had no effect on the formation and growth of neurospheres from p62 KO mice (Fig. S5, A and B), indicating a specific role of p62 in FIP200-deficient NSCs. Increased expression of Ccl5 and Cxcl10 mRNA upon reexpression of GFP-p62-wt, GFP-p62-D337A, or GFP-p62-D343A, but not GFP-p62-K7A (Fig. 7, D and E), was seen. Examination of neurosphere sections by immunofluorescent staining confirmed these results at the protein level (Fig. 7, F and G; and not depicted). Increased secretion of Ccl5 and Cxcl10 in conditioned media and microglia migration was also seen (Fig. 7, H–J). These results demonstrate that the self-aggregation activity of p62, but not its LC3-binding function, is essential for promoting Ccl5 and Cxcl10 expression in FIP200-deficient NSCs.
Figure 7.
Aggregated p62 promoted chemokine expression by activating NF-κB in FIP200-deficient NSCs. (A) Phase contrast (top) and GFP fluorescent (bottom) images of FIP/p62 2cKO retrovirus-infected neurospheres encoding GFP-p62-wt and its mutants as indicated. (B and C) Number (B) and the size (C) of secondary FIP/p62 2cKO retrovirus-infected neurospheres encoding GFP-p62-wt and various p62 mutations as indicated (mean ± SEM; six mice each in B and five mice each in C). (D and E) mRNA levels of Ccl5 (D) and Cxcl10 (E) in Ctrl, FIP cKO, and FIP/p62 2cKO neurospheres and FIP/p62 2cKO retrovirus-infected neurospheres encoding GFP-p62-wt and various p62 mutations as indicated (mean ± SEM; six mice each). (F and G) Immunofluorescence of Ccl5 (F) and Cxcl10 (G) with DAPI in FIP/p62 2cKO neurospheres and FIP/p62 2cKO retrovirus-infected neurospheres encoding GFP-p62-wt and its mutants as indicated (three mice each). (H and I) Concentration of Ccl5 (H) and Cxcl10 (I) in conditioned media of neurospheres (normalized to 1 mg total protein lysate of neurospheres) from Ctrl, FIP cKO, FIP/p62, and 2cKO neurospheres and FIP/p62 2cKO retrovirus-infected neurospheres encoding GFP-p62-wt and various p62 mutations as indicated (mean ± SEM; three mice each). (J) Number of migrated microglia in conditioned media from Ctrl, FIP cKO, and FIP/p62 2cKO neurospheres and FIP/p62 2cKO retrovirus-infected neurospheres encoding GFP-p62-wt and various p62 mutations as indicated (mean ± SEM; six mice each). (K) Immunofluorescence of p62 and phosphorylated p65 with DAPI in neurospheres (3threemice each) of Ctrl, FIP cKO, and FIP/p62 2cKO mice at P28. (L) Percentage of nuclei localized phosphorylated p65 from neurospheres of Ctrl, FIP cKO, FIP/p62 2cKO, and p62 KO mice are shown (five mice each). (M) Nuclear and cytoplasmic lysates extracted from neurospheres of Ctrl, FIP cKO, FIP/p62 2cKO, and p62 KO mice were analyzed by immunoblot using antibodies to detect p65, PCNA, and vinculin. (N and O) Immunofluorescence of GFP and phosphorylated p65 with DAPI in neurospheres (three mice each) of FIP/p62 2cKO retrovirus-infected mice encoding GFP-p62-wt and various p62 mutations as indicated. (P) Percentage of nuclei localized phosphorylated p65 from FIP/p62 2cKO neurospheres and FIP/p62 2cKO retrovirus-infected neurospheres encoding GFP-p62-wt and various p62 mutations as indicated (mean ± SEM; five mice each). Bars: (A) 40 µm; (F, G, K, M, and N) 20 µm; (K and N, insets) 10 µm. **, P < 0.01; ***, P < 0.001.
As a multifunctional adaptor protein, p62 participates in other intracellular signaling events besides autophagy (Bjørkøy et al., 2005; Pankiv et al., 2007). One function is as an adaptor for TRAF6 (TNF receptor–associated factor 6) to activate NF-κB (Duran et al., 2008). We tested the effect of a p62 mutant lacking binding to TRAF6 (p62-dTRAF6; Fang et al., 2014) on Ccl5 and Cxcl10 expression in FIP200-null NSCs. When reexpressed in FIP/p62 2cKO neurospheres, p62-dTRAF6 formed aggregates (Fig. S5 C) and led to decreases in the number and size of neurospheres similar to wild-type p62 (Fig. 7, B and C), suggesting that TRAF6 binding is not required for p62 to mediate inhibition of NSC self-renewal. However, reexpression of p62-dTRAF6 did not result in the increased expression and secretion of Ccl5 and Cxcl10 in FIP/p62 2cKO neurospheres (Fig. 7, D–I) or the ability of the conditioned medium to increase microglia migration (Fig. 7 J). These results suggested that p62 binding to TRAF6 is important for mediating expression of Ccl5 and Cxcl10 in FIP200-null NSCs. p62 binding to TRAF6 is required for NF-κB activation, which controls expression of many cytokines and chemokines (Tak and Firestein, 2001; Wooten et al., 2005; Hayden et al., 2006). We found a significantly increased level of phosphorylated p65 in the nucleus of FIP cKO neurospheres compared with Ctrl neurospheres, in contrast to FIP/p62 2cKO neurospheres (Fig. 7, K and L). An increased level of p65 was detected in the nuclear fraction of FIP cKO, but not FIP/p62 2cKO or p62 KO, neurospheres when compared with Ctrl neurospheres (Fig. 7 M). Comparable levels of p65 were detected in the cytoplasmic fractions of all samples. Reexpression of GFP-p62-wt in FIP/p62 2cKO neurospheres led to increased nuclear phosphorylated p65 (Fig. 7, N–P). Reexpression of GFP-p62-D337A or GFP-p62-D343A, but not GFP-p62-K7A, also led to increased phosphorylated nuclear p65 (Fig. 7, O and P). Reexpression of p62-dTRAF6 did not increase p65 levels, consistent with its inability to promote Ccl5 and Cxcl10 expression. These results suggest an essential role for aggregated p62 in the activation of NF-κB to elevate Ccl5 and Cxcl10 expression in FIP200-deificient NSCs.
Discussion
Our recent studies have shown that the self-renewal and differentiation of postnatal NSCs are regulated by the essential autophagy gene FIP200 through both p53-dependent and p53-independent mechanisms (Wang et al., 2013). Increased p53 expression induced by superoxide upon deletion of FIP200 (Wang et al., 2016) is responsible for the deficient self-renewal of FIP200-null NSCs, but ablation of p53 did not rescue the defective differentiation of these cells (Wang et al., 2013). In this study, we reveal a novel non–cell autonomous mechanism by which FIP200 deletion in NSCs leads to increased infiltration of activated M1 microglia to inhibit their differentiation in a p53-independent manner.
The migration, proliferation, and activation of microglia and consequent microgliosis in neurodegenerative diseases and ageing brain are well documented in recent studies (Cunningham, 2013; Keren-Shaul et al., 2017). However, our observed gradual increase of microglia in postnatal FIP200-deficient SVZ was independent of proliferation (see Fig. 1), indicating a difference from the classic mechanism in injury models and neurodegenerative diseases (Dheen et al., 2007). Microglia from FIP cKO mice did not exhibit FIP200 deletion or any abnormality of autophagy, supporting that increased microglia infiltration is caused by FIP200-null NSCs through non–cell autonomous mechanisms. Other studies suggested that prenatal and postnatal NSCs play active roles in regulating surrounding cells. For example, it was reported that NSCs promote the formation of endothelial cell tubes in coculture (Ford et al., 2006) and increase the proliferation and phagocytotic activities of microglia (Mosher et al., 2012). NSCs have also been shown to influence local immune activity in neurodegenerative disease models (Kokaia et al., 2012). Our findings suggest that normal NSC differentiation requires the correct environment or “niche,” orchestrated by NSCs, and highlight postnatal NSCs as crucial to homeostasis.
Several previous studies suggested potential mechanisms of regulation of microglia by NSCs, including stimulation of proliferation of microglia (Mosher et al., 2012), but no increased proliferation of microglia was observed in our study. Increased numbers of microglia in FIP200-null SVZ was not caused by SVZ degeneration or increased apoptosis. Both of these defects were rescued by p53 ablation in 2cKO mice (Wang et al., 2013), yet increased numbers of microglia were still found. On the other hand, our systematic transcriptome analysis revealed increased expression of the chemokines Ccl5 and Cxcl10 in the SVZ of both FIP cKO and 2cKO mice. A study showed that microglia respond to Ccl5 from bone marrow–derived mesenchymal stem cells with migration in a mouse model (Lee et al., 2012). Cxcl10 plays a crucial role in the recruitment of microglia to damaged brain sites (Rappert et al., 2004). Increased Ccl5 and Cxcl10 were found in FIP200-null NSC-conditioned media that induced migration of microglia in vitro. Concomitant blockade of both Ccl5 and Cxcl10 signaling, but not either one alone, significantly decreased microglia migration, indicating a redundant role for the two chemokines. A dual Ccr5 and Cxcr3 inhibitor, TAK-779 (Suzaki et al., 2008), effectively reduced the infiltration of microglia into FIP200-deficient SVZ in vivo, providing compelling support.
Autophagy’s impact on inflammation and immunity has been increasingly recognized (Shibutani et al., 2015; Zhong et al., 2016). For example, ULK1, a component of the ULK1–Atg13–FIP200 autophagy induction complex (Hara et al., 2008; Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009), has been shown to phosphorylate STING (stimulator of interferon genes) to prevent sustained interferon response (Konno et al., 2013). Loss of Atg9L1 causes hyperaggregation of STING. Our findings are similar to previous findings of elevated levels of Ccl5 and Cxcl10 when FIP200 was ablated in mammary tumor cells, triggering increased cytotoxic T cell infiltration (Wei et al., 2011). Unlike FIP200-null tumor cells or mouse embryonic fibroblasts, FIP200-null NSCs showed increased numbers of chemokines with or without other stimulation, suggesting different mechanisms. Unlike FIP200 deletion, ablation of Atg5 reduced the levels of another inflammatory cytokine, IL-1β, in bone marrow–derived macrophages (Dupont et al., 2011) and IL-6 in Ras-transformed MCF10A cells (Lock et al., 2014). The mechanism of the opposing effects of the loss of FIP200 versus other autophagy genes like Atg5 warrants further investigation. We have previously observed differences between FIP200 deletion versus Atg5, Atg16L1, or Atg7 on the maintenance and function of NSCs (Wang et al., 2016).
Our previous studies showed accumulation of p62 aggregates upon FIP200 deletion in different cells (Wei et al., 2011; Wang et al., 2013, 2016). In FIP200-null NSCs, unlike p53 deletion, which restored self-renewal, but not differentiation (Wang et al., 2013), ablation of p62 restored both defects (Wang et al., 2016). This suggested a potential role for aggregated p62 in increased Ccl5 and Cxcl10 expression. This was demonstrated directly by showing increased Ccl5 and Cxcl10 expression with reexpressed p62 in FIP200/p62-null NSCs. These results were similar to the previously reported function of p62 to promote an inflammatory response (Lee et al., 2011). p62 is a multifunctional scaffold protein that regulates signaling pathways by binding to various proteins (Sanz et al., 2000; Wooten et al., 2005; Komatsu et al., 2010; Duran et al., 2011). Analysis of p62 mutants indicated that the ability to self-aggregate, but not the ability to bind to LC3, is required for stimulation of Ccl5 and Cxcl10. Because LC3 binding is important for p62 as a cargo receptor (Pankiv et al., 2007), these data suggest that increased expression of Ccl5 and Cxcl10 is unlikely a consequence of blockade of particular cargos for autophagic degradation. On the other hand, because p62 was often observed in aggregates in neurodegenerative diseases with neuroinflammation, our findings raise the possibility that p62-containing aggregates may also trigger chemokines that contribute to pathogenesis. The interaction between p62 and Traf6 was also required for increased Ccl5 and Cxcl10 expression, implicating a role for NF-κB signaling, as suggested previously (Sanz et al., 2000; Duran et al., 2008). NF-κB activation was observed upon reexpression of wild-type, but not mutant, p62 lacking self-aggregation or binding to Traf6 in FIP200/p62-null NSCs. Our results suggest a working model in which p62 aggregate formation upon FIP200 deletion activates NF-κB through its interaction with Traf6 to promote Ccl5 and Cxcl10 expression in FIP200-null NSCs.
Recent studies suggested beneficial roles of activated microglia on neurogenesis (Aarum et al., 2003; Ekdahl, 2012; Vukovic et al., 2012; Shigemoto-Mogami et al., 2014). However, activated microglia in inflammatory backgrounds decrease neurogenesis in adult mice (Ekdahl et al., 2003; Monje et al., 2003). Our findings of an increase in the number of proinflammatory activated microglia, including elevated expression of TNF and IL-6, are consistent with the latter. Our data clearly demonstrate a role for Ccl5 and Cxcl10 in the induction of microglia migration to FIP200-deficient SVZ, but it is unclear whether they also play a role in microglia activation. Ccl5 has been reported to induce a proinflammatory profile of microglia in vitro, although it required the presence of lipopolysaccharide (Skuljec et al., 2011). Cxcl10 signaling in hippocampus from N-methyl-d-aspartate did not activate microglia (van Weering et al., 2011). We did not observe the activation of microglia by addition of Ccl5 and Cxcl10 to the culture media (unpublished data), suggesting that microglia activation occurred through other mechanisms. Consistent with this, inhibition of microglia activation by minocycline did not affect the increased infiltration, supporting the idea that increased microglia migration and activation are separate events. These results suggest that microglia activation is not required for migration and likely follows the infiltration of microglia into the FIP200-decifient SVZ.
Microglia migration and activation are both important for the defective differentiation of FIP200-null NSCs. Blocking microglia activation, abolishing microglia migration, or depleting microglia all rescued deficient NSC differentiation. However, no treatment restored the defective NSC differentiation of FIP cKO mice, which also have defective NSC self-renewal. These results suggest that self-renewal and differentiation of NSCs may be coupled and that the defective self-renewal of FIP200-null NSCs prevented their normal differentiation. Another explanation may be that sufficient numbers of NSCs in 2cKO mice, but not FIP cKO mice, are necessary to observe the restoration of NSC differentiation after drug treatment.
In summary, our study clarified a novel FIP200–p62–Ccl5 and Cxcl10 cascade involved in the cross talk between postnatal NSCs and microglia. FIP200 ablation and consequent autophagy inhibition in NSCs leads to their defective differentiation by increased infiltration and activation of microglia through a p53-independent and non–cell autonomous mechanism, which is triggered by the increased Ccl5 and Cxcl10 expression in response to accumulated p62 aggregates in FIP200-null NSCs. These findings provide new insight into the understanding of how autophagy genes impact immunity and inflammation to regulate stem cell differentiation.
Materials and methods
Mice
FIP200f/f, p53f/f, hGFAP-Cre transgenic, and p62 total knockout mice were described previously (Wang et al., 2013, 2016). Male and female mice with 50% FVB and 50% C57/B6 backgrounds were used in all experiments. Age- and littermate-matched control and mutant mice were randomly collected based on their genotypes. Mice were housed and handled according to local, state, and federal regulations. All experimental procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee at the University of Cincinnati. Mouse genotyping for FIP200, p53, p62, and Cre alleles were performed by PCR analysis of tail DNA, essentially as described previously (Wang et al., 2013, 2016).
Cell cultures
Neurosphere formation was performed as previously described (Wang et al., 2013, 2016). In brief, SVZ tissue was isolated under a dissection microscope and cut into ∼1 mm3 cubes. Tissue was digested in 0.2% trypsin/EDTA to obtain single-cell suspensions. Cells were cultured in neural basal media supplemented with B27 (Invitrogen), 10 ng/ml basic FGF, and 20 ng/ml EGF (Invitrogen) in Ultra-Low Attachment Plates (Corning). Neurospheres with diameters larger than 50 µm were counted 10 d after culturing.
Mixed cortical cultures were prepared essentially as described previously (Gao et al., 2014). In brief, newborn pups (P0–P2) were decapitated and the cortices were removed from meninges, hippocampi, basal ganglia, and midbrain and kept in cold DMEM. The cortical tissue was cut into ∼1 mm3 cubes, digested in 0.2% trypsin/EDTA at 37°C for 20 min, and mechanically triturated. After filtration with 70-µm mesh, the cell suspension was plated in mixed cortical medium (DMEM with 10% FBS) onto tissue culture dishes and cultured at 37°C and 95% humidity. Media was changed 3 d after plating. Microglia were harvested 10 d after plating by adding 15 mM lidocaine (Sigma-Aldrich) into the medium for 15 min at room temperature. The medium containing the floating microglia was collected and centrifuged at 400 g for 5 min. The cell pellet was then resuspended, and the cell number was counted. Dispersed microglia were seeded in DMEM with 1% FBS, and the purity of the microglial cultures was >98% as examined by Iba-1 staining. The microglia was used for experiments after 24 to 48 h of culture.
Primary astrocytes were cultured as described previously (Gao et al., 2013). Newborn pups were decapitated, and the cortices were removed from meninges, hippocampi, basal ganglia, and midbrain and kept in cold DMEM. The cortical tissue was cut into ∼1 mm3 cubes and vortexed at the highest speed for 90 s. After filtration with 70-µm and 30-µm mesh, the cell suspension was plated in DMEM with 10% FBS onto tissue culture dishes (37°C and 95% humidity). Medium was changed 3 d after plating, and FBS concentration was decreased to 7% 2 wk after seeding. Astrocytes and astrocyte conditioned media was collected 4 wk after seeding for biochemistry and molecular assays.
Minocycline, TAK-779, and PLX3397 treatments
Minocycline (Sigma-Aldrich) and vehicle (PBS) were administrated i.p. at 10 mg/kg every the other day into mice at P7, and the treated mice were euthanized at P28. TAK-779 (Sigma-Aldrich) was administrated i.p. at 20 mg/kg every day into P7 mice, and the treated mice were euthanized at P28. PLX3397 (ApexBio) was dissolved in DMSO and then added immediately before using to a solution of 0.5% methyl cellulose (M0512; Sigma-Aldrich) and 1.0% Tween 80 (P1754; Sigma-Aldrich). PLX3397 was gavaged into P7 mice at 75 mg/kg every day with a 24G 1-inch stainless steel gavage needle (Braintree Scientific) as described previously (Butchbach et al., 2007). PLX3397-treated mice were euthanized at P28.
Cell migration assay
Cell migration was measured based on the 48-well Boyden chamber assays system (Micro Chemotaxis Chamber AP48, Neuro Probe) to measure the migration of microglia using 8-µm pore polycarbonate membranes as described previously (Wang et al., 2010). In brief, 2 × 104 primary microglia were added to the top compartment of the chamber and either microglia media or diluted conditioned media to the bottom compartment of the chamber. After 4-h incubation, a cotton swab was used to remove the cells on the top of the membrane, and microglia on the bottom part of the membrane were stained with Giemsa (1:10 dilution in H2O) for 60 min at room temperature. Cells were counted under a light microscope as described previously. Data were obtained from three to six independent experiments.
Proliferation assay
To assess microglia proliferation, we seeded 2 × 103 primary microglia per well in a 48-well plate with microglia media (DMEM with 1% FBS), or a one to three dilution of microglia media to neurospheric conditioned media. FBS, neural basal media, and B27 were used for all dilutions. After 48 h, the cells were trypsinized and collected to count the number of cells with an automatic cell counter (Countess II; Thermo Fisher Scientific). Data were obtained from three to five independent experiments.
Preparation of Ccl5 and Cxcl10 lentiviral shRNA and p62 retroviruses and infection of NSCs
The psPAX2 and pMD2G vectors and the pGIPZ lentiviral vectors (GE Healthcare) encoding shRNA targeting mouse Ccl5 and Cxcl10 were purchased through the University of Michigan shRNA Core Facility. HEK293 cells were transfected with 10 µg pGIPZ lentiviral vector encoding each shRNA, 10 µg psPAX2, and 5 µg pMD2G by the calcium phosphate method according to the instructions recommended by the manufacturer. 12 h after transfection, the media was replaced with DMEM containing 5% FBS. The conditioned media was then collected twice at 1-d intervals and combined. After centrifugation and filtration, the supernatant was used to infect neural cells isolated from the SVZ. The infected neurospheres were selected with 1 µg/ml puromycin in neurosphere media to obtain pools that stably expressed shRNA. The number and size of neurospheres were analyzed essentially as described previously (Wang et al., 2013).
Mouse pMXs-puro-GFP-p62 WT and pMXs-puro-GFP-p62 LRS mutant 1 (L343A), pMXs-puro-GFP-p62 LRS mutant 2 (D337A, D338A, and D339A), and pMXs-puro-GFP-p62 K7A and D69A mutants were purchased from Addgene (deposited by N. Mizushima, University of Tokyo, Tokyo, Japan; Itakura and Mizushima, 2011). Full-length MSCV-IRES-GFP-p62 and MSCV-IRES-GFP-p62ΔTB were obtained from D.T. Starczynowski (Cincinnati Children’s Hospital, Cincinnati, OH; Fang et al., 2014). Retroviral plasmids were used to transfect HEK293 cells to generate recombinant retroviruses. NSCs infected with recombinant lentivirus or retroviruses were selected in spherical medium containing 1 µg/ml puromycin.
Antibody-mediated immunodepletion
For Ccl5 (R&D Systems) and Cxcl10 (PeproTech) immunodepletion from conditioned media of NSCs, we incubated antibodies against Ccl5 and Cxcl10 at 4°C for 60 min with continuous shaking. After incubation, Protein G PLUS-agarose beads were added to each sample and then incubated at 4°C for 3 h. The agarose beads together with the immunoprecipitation were then cleared by centrifugation and supernatants collected. As a control, conditional media was processed in an identical way with the addition of isotype-matched irrelevant antibodies.
ELISA
Mouse Ccl5 (900-M124) and Cxcl10 (900-M153) ELISA (PeproTech) was performed according to the manufacturer’s instructions. In brief, a 96-well plate was coated overnight with capturing antibody. The antibody was then blocked with assay diluent for 1 h at room temperature. 100 µl sample or cytokine standard diluted in assay diluent was added and incubated for 2 h at room temperature. Biotin-conjugated detection antibody, avidin-HRP reagent, substrate solution, and stop solution were then added into the wells sequentially and incubated for specific time periods as per the protocol. Extensive washes were performed after each step. Color development was monitored with an ELISA plate reader at 405 nm with wavelength correction set at 650 nm. Cytokine release was calculated and the values normalized to neuronal conditioned medium–treated samples.
Antibodies and reagents
Primary antibodies used were mouse anti-BrdU (1:200, G3G4; Developmental Studies Hybridoma Bank), anti–β-actin (1:5,000, A5316; Sigma-Aldrich), antivinculin (1:5,000, V9131; Sigma-Aldrich), anti-GFAP (1:100, 3670T; Cell Signaling Technology), anti-NOS2 (1:100, SC-7271; Santa Cruz Biotechnology), antinestin (1:50, Rat-401; Developmental Studies Hybridoma Bank), anti-NeuN (1:500, MAB377; EMD Millipore), anti-TNF (1:100, SC-52746; Santa Cruz Biotechnology), rat anti-Ki67 (1:100, 652401; BioLegend), rabbit anti-Ccl5 (1:100, 500-P118; PeproTech), anti-Cxcl10 (1:100, 500-P129; PeproTech; and 1:100; Biorbyt), anti-FIP200 (1:1,000, 12436S; Cell Signaling Technology), anti-Iba1 (1:1000, 019–19741; Wako Pure Chemical Industries), anti-Ki67 (1:100, M3060; Spring Bioscience), anti-GFAP (1:400, Z033429-2; Dako), anti-LC3 (1:1,000, 2775; Cell Signaling Technology), anti-p65 (1:1,000, 8242; Cell Signaling Technology), anti-Sox2 (1:500, AB5603; EMD Millipore); goat anti–Ccl-5 (1:100, AF478; R&D Systems), anti–IL-6 (1:100, SC-1265; Santa Cruz Biotechnology), anti-PCNA (1:1,000, SC-9857; Santa Cruz Biotechnology), anti–Arginase 1 (1:100, SC-18351; Santa Cruz Biotechnology), and guinea pig anti-DCX (AB2253, 1:1,000, EMD Millipore). Secondary antibodies were goat anti–rabbit IgG-FITC (1:400), goat anti–rabbit IgG-Texas red (1:400), goat anti–mouse IgG-FITC (1:200), goat anti–mouse IgG-Texas red (1:400), donkey anti–guinea pig IgG–Texas red (1:400), rabbit anti goat IgG-FITC (1:200), rabbit anti–rat IgG-FITC (1:200), goat anti–mouse IgG-HRP (1:5000), goat anti–rabbit IgG-HRP (1:5000; all from Jackson ImmunoResearch Laboratories).
Cxcr3 inhibitor (±) AMG-487 was from Tocris Bioscience, and Ccr5 inhibitor maraviroc was from Sigma-Aldrich. Both compounds were dissolved in DMSO.
Histology and immunofluorescence
Mice were euthanized using CO2, and a complete tissue set was harvested during necropsy. Fixation was performed for 16 h at 4°C using 4% (wt/vol) freshly made, prechilled PBS-buffered paraformaldehyde. Brain tissue was embedded in paraffin and sagittal sectioned at 5 µm. Slides from histologically comparable positions (triangular lateral ventricle with intact RMS) were stained with hematoxylin and eosin for routine histological examination or left unstained for immunofluorescence. Hematoxylin and eosin–stained sections were examined under a BX41 light microscope (Olympus), and images were captured with an Olympus digital camera (model DP70) using DP Controller software (version 1.2.1.10 8). For immunofluorescence, unstained tissues were first deparaffinized in three washes of xylene (3 min each) and then rehydrated in graded ethanol solutions (100%, 95%, 70%, 50%, and 30%). After heat-activated antigen retrieval (Retriever 2000; PickCell Laboratories B.V.) according to the manufacturer’s specifications, sections were treated with Protein Block Serum Free (Dako) at room temperature for 10 min. Slices were then incubated with the primary antibodies at 4°C for 16 h in a humidified chamber, washed in PBS three times (5 min each), and incubated with 1:200 secondary antibodies for 1 h at room temperature. After three washes in PBS (5 min each), nuclei were stained with DAPI and mounted with Vectashield mounting medium (Vector Laboratories). Digital photography was performed as described previously (Wei et al., 2011). In some experiments (e.g., iNOS, TNF, and IL-6 staining), fixed brains were consecutively transferred to 20% and 30% sucrose to settle overnight. They were embedded in optiomal cutting temparature compound (Sakura) and cryosectioned at 10 µm (3050 Cryostat; Leica Biosystems), and the slices were kept at −80°C before being used for immunofluorescence.
The fluorescence intensity of Ccl5 and Cxcl10 was quantified with ImageJ. The fluorescence was quantified from three to four fields of view for each sample. In brief, the images files are open with ImageJ (http://imagej.nih.gov/ij/). The green channel pictures are selected. The SVZ or neurospheres regions are selected using the drawing/selection tools. Set Measurements is selected from the “analyze” menu. Area, integrated density, and mean gray value are selected from the popup menu. The menu “measure” is selected from the analyze menu. The same process is repeated to measure the other samples.
Morphometric analyses of microglial phenotypes
The morphometric analyses of microglia were based on the assessment of Iba1-immunoreactivity according to previous publications with minor modifications (Lehrmann et al., 1997; Torres-Platas et al., 2014). Two to three consecutive sections containing the SVZ area were used to identify the cell body and processes of microglia. For each sample, >100 cells were quantified. Ramified microglia displayed several highly branched processes (at least three or four per cell) longer than the cell body. Amoeboid microglia presented amoeboid-shaped cell bodies. The processes extended by amoeboid microglia were fewer (usually one per cell) and shorter than the cell body diameter. Round microglia had smaller and round cell body. They were either devoid of processes or with few unbranched processes (usually one per cell) within the length of the cell body diameter.
Microarray
Neurospheres from SVZ cells of Ctrl, FIP cKO, 2cKO, and p53 cKO mice were collected at P0. Neurospheres from four mice were pooled for mRNA preparation and reverse transcription to generate probes for the Affymetrix gene chip mouse genome 430 2.0 array. Microarray and data analyses were performed by the University of Michigan Comprehensive Cancer Center Affymetrix and Microarray Core Facility according to standard protocols. Genes were selected if their signals were up-regulated or down-regulated at least twofold in FIP cKO, 2cKO, and p53 cKO samples compared with controls.
RNA extraction, reverse transcription, and quantitative real-time PCR
Total RNA was extracted from neurospheres and freshly isolated microglia using TRIzol reagent (Ambion) and RNeasy Mini kit (QIAGEN) according to the manufacturer’s directions. Revere transcription and Q-PCR using the SYBR Green PCR Core reagents system (QIAGEN) were performed as described previously (Wei et al., 2011). The primers for mouse Ccl5, Cxcl10, IFIT1, IFIT3, and IFI44 sequences will be sent upon request, and an internal control (sequence as described previously; Wei et al., 2011) was obtained from Thermo Fischer Scientific.
Protein extraction, SDS-PAGE, and immunoblotting
SVZ tissue was microdissected from 300-µm-thick coronal sections of P28 brains and used for protein extraction by homogenization in modified radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 1% Triton X-100, 0.2% sodium deoxycholate, 0.2% SDS, and 1 mm sodium EDTA) supplemented with protease inhibitors (5 µg/ml leupeptin, 5 µg/ml aprotinin, and 1 mm PMSF). In other experiments, astrocytes, microglia, and neurospheres were dissolved with the same lysis buffer. After removing tissue and cell debris by centrifugation at 13,000 rpm for 10 min at 4°C, protein concentration was determined using Bio-Rad Laboratories protein assay reagent. The lysates were boiled for 5 min in 1×SDS sample buffer (50 mm Tris-HCl, pH 6.8, 12.5% glycerol, 1% SDS, and 0.01% bromphenol blue) containing 5% β-mercaptoethanol and then analyzed by SDS-PAGE, followed by immunoblotting using various antibodies, as described previously (Wang et al., 2016).
Nuclear and cytoplasmic fractionation
Fractionation was performed essentially as described previously (Wu et al., 2004). In brief, cells were lysed in a lysis buffer (20 mM Hepes, pH 7.2, 10 mM KCl, 2 mM MgCl2, 0.5% Nonidet P40, 1 mM Na3VO4, 1 mM PMSF, and proteasome inhibitors) and homogenized. The homogenate was centrifuged at 1,500 g for 5 min to sediment nuclei. The supernatant was sedimented at 15,000 g for 10 min and the resulting supernatant formed the cytoplasmic fraction. The nuclear pellet was washed three times with lysis buffer and resuspended in the same lysis buffer with 0.5 M NaCl. The extracted material was sedimented at 15,000 g for 10 min, and the resulting supernatant was harvested as the nuclear fraction. The harvested samples were examined under Western blot.
Statistical analysis
Lengths, areas, and the number of cells from comparable sections were quantified using ImageJ. Image analysis was not double blinded. No data points were excluded in the analysis. The distribution of the variables in each experimental group was approximately normal. Statistical significance was evaluated by unpaired two-tailed Student’s t test, with P < 0.05 indicating statistical significance, using GraphPad Prism (Version 5.0). Power analysis was used to justify the sample size. The number of animals used for quantification is indicated in the figure legends.
Online supplemental material
Fig. S1 shows infiltration of microglia into P0 SVZ and other brain regions of P28 mice. Fig. S2 shows increased Ccl5 and Cxcl10 levels in FIP200-null astrocytes and shRNA knockdown efficiency for Ccl5 and Cxcl10. Fig. S3 shows that Ccl5 and Cxcl10 depleting antibodies, shRNAs, and their receptor antagonists had no effect on microglia proliferation and neurosphere formation in vitro. Fig. S4 indicates that reactive microglia express high levels of IL-6 and TNF-α. It also shows that minocycline and PLX3397 did not affect NSCs maintenance in FIP200-deficient SVZ. Fig. S5 shows the maintenance of p62 KO neurospheres infected with WT p62 and mutant p62 retroviruses.
Supplementary Material
Acknowledgments
We thank Dr. Dan Starczynowski for providing retroviral vectors encoding p62 and p62-dTRAF6 mutant and Dr. Song Chen for the preparation of retroviral vectors encoding GFP-p62-wt, GFP-p62-D337A, GFP-p62-D343A, and K7A/D69A. We also thank Dr. Stella Tsirka of Stony Brook University and members of the Guan laboratory for critical reading and helpful comments on the manuscript, Dr. Belinda Peace for professional editing, and Glenn Doerman for figure preparation.
This research was supported by National Institutes of Health grants R03NS097887 (to C. Wang) and R01NS094144 and R01CA163493 to (J.-L. Guan).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- cKO
- conditional knockout
- Ctrl
- control
- NSC
- neural stem cell
- P
- postnatal day
- RMS
- rostral migratory stream
- SVC
- subventricular zone
References
- Aarum J., Sandberg K., Haeberlein S.L., and Persson M.A.. 2003. Migration and differentiation of neural precursor cells can be directed by microglia. Proc. Natl. Acad. Sci. USA. 100:15983–15988. 10.1073/pnas.2237050100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolph T.E., Tomczak M.F., Niederreiter L., Ko H.J., Böck J., Martinez-Naves E., Glickman J.N., Tschurtschenthaler M., Hartwig J., Hosomi S., et al. . 2013. Paneth cells as a site of origin for intestinal inflammation. Nature. 503:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albright A.V., Shieh J.T., Itoh T., Lee B., Pleasure D., O’Connor M.J., Doms R.W., and González-Scarano F.. 1999. Microglia express CCR5, CXCR4, and CCR3, but of these, CCR5 is the principal coreceptor for human immunodeficiency virus type 1 dementia isolates. J. Virol. 73:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnò B., Grassivaro F., Rossi C., Bergamaschi A., Castiglioni V., Furlan R., Greter M., Favaro R., Comi G., Becher B., et al. . 2014. Neural progenitor cells orchestrate microglia migration and positioning into the developing cortex. Nat. Commun. 5:5611 10.1038/ncomms6611 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. The Gene Ontology Consortium . 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biber K., Dijkstra I., Trebst C., De Groot C.J., Ransohoff R.M., and Boddeke H.W.. 2002. Functional expression of CXCR3 in cultured mouse and human astrocytes and microglia. Neuroscience. 112:487–497. 10.1016/S0306-4522(02)00114-8 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., and Johansen T.. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614. 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchbach M.E., Edwards J.D., Schussler K.R., and Burghes A.H.. 2007. A novel method for oral delivery of drug compounds to the neonatal SMNDelta7 mouse model of spinal muscular atrophy. J. Neurosci. Methods. 161:285–290. 10.1016/j.jneumeth.2006.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C. 2013. Microglia and neurodegeneration: The role of systemic inflammation. Glia. 61:71–90. 10.1002/glia.22350 [DOI] [PubMed] [Google Scholar]
- Dheen S.T., Kaur C., and Ling E.A.. 2007. Microglial activation and its implications in the brain diseases. Curr. Med. Chem. 14:1189–1197. 10.2174/092986707780597961 [DOI] [PubMed] [Google Scholar]
- Dupont N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., and Deretic V.. 2011. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 30:4701–4711. 10.1038/emboj.2011.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A., Linares J.F., Galvez A.S., Wikenheiser K., Flores J.M., Diaz-Meco M.T., and Moscat J.. 2008. The signaling adaptor p62 is an important NF-κB mediator in tumorigenesis. Cancer Cell. 13:343–354. 10.1016/j.ccr.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Duran A., Amanchy R., Linares J.F., Joshi J., Abu-Baker S., Porollo A., Hansen M., Moscat J., and Diaz-Meco M.T.. 2011. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell. 44:134–146. 10.1016/j.molcel.2011.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdahl C.T. 2012. Microglial activation: Tuning and pruning adult neurogenesis. Front. Pharmacol. 3:41 10.3389/fphar.2012.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdahl C.T., Claasen J.H., Bonde S., Kokaia Z., and Lindvall O.. 2003. Inflammation is detrimental for neurogenesis in adult brain. Proc. Natl. Acad. Sci. USA. 100:13632–13637. 10.1073/pnas.2234031100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore M.R., Najafi A.R., Koike M.A., Dagher N.N., Spangenberg E.E., Rice R.A., Kitazawa M., Matusow B., Nguyen H., West B.L., and Green K.N.. 2014. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 82:380–397. 10.1016/j.neuron.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R., Xu F., Previti M.L., Davis J., Grande A.M., Robinson J.K., and Van Nostrand W.E.. 2007. Minocycline reduces microglial activation and improves behavioral deficits in a transgenic model of cerebral microvascular amyloid. J. Neurosci. 27:3057–3063. 10.1523/JNEUROSCI.4371-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Barker B., Bolanos L., Liu X., Jerez A., Makishima H., Christie S., Chen X., Rao D.S., Grimes H.L., et al. . 2014. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-κB gene network. Cell Reports. 8:1328–1338. 10.1016/j.celrep.2014.07.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.C., Bertram J.P., Hynes S.R., Michaud M., Li Q., Young M., Segal S.S., Madri J.A., and Lavik E.B.. 2006. A macroporous hydrogel for the coculture of neural progenitor and endothelial cells to form functional vascular networks in vivo. Proc. Natl. Acad. Sci. USA. 103:2512–2517. 10.1073/pnas.0506020102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage F.H. 2000. Mammalian neural stem cells. Science. 287:1433–1438. 10.1126/science.287.5457.1433 [DOI] [PubMed] [Google Scholar]
- Ganley I.G., Lam H., Wang J., Ding X., Chen S., and Jiang X.. 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284:12297–12305. 10.1074/jbc.M900573200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K., Wang C.R., Jiang F., Wong A.Y., Su N., Jiang J.H., Chai R.C., Vatcher G., Teng J., Chen J., et al. . 2013. Traumatic scratch injury in astrocytes triggers calcium influx to activate the JNK/c-Jun/AP-1 pathway and switch on GFAP expression. Glia. 61:2063–2077. 10.1002/glia.22577 [DOI] [PubMed] [Google Scholar]
- Gao Z., Nissen J.C., Ji K., and Tsirka S.E.. 2014. The experimental autoimmune encephalomyelitis disease course is modulated by nicotine and other cigarette smoke components. PLoS One. 9:e107979 10.1371/journal.pone.0107979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., et al. . 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J.L., Simon A.K., Prescott M., Menendez J.A., Liu F., Wang F., Wang C., Wolvetang E., Vazquez-Martin A., and Zhang J.. 2013. Autophagy in stem cells. Autophagy. 9:830–849. 10.4161/auto.24132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F.M., Briggs J., et al. . 2007. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39:207–211. 10.1038/ng1954 [DOI] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., and Mizushima N.. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., West A.P., and Ghosh S.. 2006. NF-κB and the immune response. Oncogene. 25:6758–6780. 10.1038/sj.onc.1209943 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. . 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 20:1981–1991. 10.1091/mbc.E08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., and Komatsu M.. 2008. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283:22847–22857. 10.1074/jbc.M802182200 [DOI] [PubMed] [Google Scholar]
- Iosif R.E., Ekdahl C.T., Ahlenius H., Pronk C.J., Bonde S., Kokaia Z., Jacobsen S.E., and Lindvall O.. 2006. Tumor necrosis factor receptor 1 is a negative regulator of progenitor proliferation in adult hippocampal neurogenesis. J. Neurosci. 26:9703–9712. 10.1523/JNEUROSCI.2723-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., and Mizushima N.. 2011. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 192:17–27. 10.1083/jcb.201009067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D., Imai Y., Ohsawa K., Nakajima K., Fukuuchi Y., and Kohsaka S.. 1998. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res. 57:1–9. 10.1016/S0169-328X(98)00040-0 [DOI] [PubMed] [Google Scholar]
- Jung C.H., Jun C.B., Ro S.H., Kim Y.M., Otto N.M., Cao J., Kundu M., and Kim D.H.. 2009. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 20:1992–2003. 10.1091/mbc.E08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. . 2017. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 169:1276–1290. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kettenmann H., Hanisch U.K., Noda M., and Verkhratsky A.. 2011. Physiology of microglia. Physiol. Rev. 91:461–553. 10.1152/physrev.00011.2010 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C.M., Adams P.D., Adeli K., et al. . 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 12:1-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Imagama S., Ohgomori T., Hirano K., Uchimura K., Sakamoto K., Hirakawa A., Takeuchi H., Suzumura A., Ishiguro N., and Kadomatsu K.. 2013. Minocycline selectively inhibits M1 polarization of microglia. Cell Death Dis. 4:e525 10.1038/cddis.2013.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokaia Z., Martino G., Schwartz M., and Lindvall O.. 2012. Cross-talk between neural stem cells and immune cells: the key to better brain repair? Nat. Neurosci. 15:1078–1087. 10.1038/nn.3163 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. . 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12:213–223. [DOI] [PubMed] [Google Scholar]
- Konno H., Konno K., and Barber G.N.. 2013. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 155:688–698. 10.1016/j.cell.2013.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A., and Alvarez-Buylla A.. 2009. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32:149–184. 10.1146/annurev.neuro.051508.135600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., and Johansen T.. 2003. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278:34568–34581. 10.1074/jbc.M303221200 [DOI] [PubMed] [Google Scholar]
- Lee H.M., Shin D.M., Yuk J.M., Shi G., Choi D.K., Lee S.H., Huang S.M., Kim J.M., Kim C.D., Lee J.H., and Jo E.K.. 2011. Autophagy negatively regulates keratinocyte inflammatory responses via scaffolding protein p62/SQSTM1. J. Immunol. 186:1248–1258. 10.4049/jimmunol.1001954 [DOI] [PubMed] [Google Scholar]
- Lee J.K., Schuchman E.H., Jin H.K., and Bae J.S.. 2012. Soluble CCL5 derived from bone marrow-derived mesenchymal stem cells and activated by amyloid β ameliorates Alzheimer’s disease in mice by recruiting bone marrow-induced microglia immune responses. Stem Cells. 30:1544–1555. 10.1002/stem.1125 [DOI] [PubMed] [Google Scholar]
- Lehrmann E., Christensen T., Zimmer J., Diemer N.H., and Finsen B.. 1997. Microglial and macrophage reactions mark progressive changes and define the penumbra in the rat neocortex and striatum after transient middle cerebral artery occlusion. J. Comp. Neurol. 386:461–476. [DOI] [PubMed] [Google Scholar]
- Levine B., Mizushima N., and Virgin H.W.. 2011. Autophagy in immunity and inflammation. Nature. 469:323–335. 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R., Kenific C.M., Leidal A.M., Salas E., and Debnath J.. 2014. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 4:466–479. 10.1158/2159-8290.CD-13-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje M.L., Toda H., and Palmer T.D.. 2003. Inflammatory blockade restores adult hippocampal neurogenesis. Science. 302:1760–1765. 10.1126/science.1088417 [DOI] [PubMed] [Google Scholar]
- Mosher K.I., Andres R.H., Fukuhara T., Bieri G., Hasegawa-Moriyama M., He Y., Guzman R., and Wyss-Coray T.. 2012. Neural progenitor cells regulate microglia functions and activity. Nat. Neurosci. 15:1485–1487. 10.1038/nn.3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Rappert A., Bechmann I., Pivneva T., Mahlo J., Biber K., Nolte C., Kovac A.D., Gerard C., Boddeke H.W., Nitsch R., and Kettenmann H.. 2004. CXCR3-dependent microglial recruitment is essential for dendrite loss after brain lesion. J. Neurosci. 24:8500–8509. 10.1523/JNEUROSCI.2451-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Diaz-Meco M.T., Nakano H., and Moscat J.. 2000. The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1-TRAF6 pathway. EMBO J. 19:1576–1586. 10.1093/emboj/19.7.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K. 2015. Effects of microglia on neurogenesis. Glia. 63:1394–1405. 10.1002/glia.22858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Goderie S.K., Jin L., Karanth N., Sun Y., Abramova N., Vincent P., Pumiglia K., and Temple S.. 2004. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 304:1338–1340. 10.1126/science.1095505 [DOI] [PubMed] [Google Scholar]
- Shibutani S.T., Saitoh T., Nowag H., Münz C., and Yoshimori T.. 2015. Autophagy and autophagy-related proteins in the immune system. Nat. Immunol. 16:1014–1024. 10.1038/ni.3273 [DOI] [PubMed] [Google Scholar]
- Shigemoto-Mogami Y., Hoshikawa K., Goldman J.E., Sekino Y., and Sato K.. 2014. Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J. Neurosci. 34:2231–2243. 10.1523/JNEUROSCI.1619-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A., Encinas J.M., Deudero J.J., Chancey J.H., Enikolopov G., Overstreet-Wadiche L.S., Tsirka S.E., and Maletic-Savatic M.. 2010. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell. 7:483–495. 10.1016/j.stem.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuljec J., Sun H., Pul R., Bénardais K., Ragancokova D., Moharregh-Khiabani D., Kotsiari A., Trebst C., and Stangel M.. 2011. CCL5 induces a pro-inflammatory profile in microglia in vitro. Cell. Immunol. 270:164–171. 10.1016/j.cellimm.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Suzaki Y., Hamada K., Nomi T., Ito T., Sho M., Kai Y., Nakajima Y., and Kimura H.. 2008. A small-molecule compound targeting CCR5 and CXCR3 prevents airway hyperresponsiveness and inflammation. Eur. Respir. J. 31:783–789. 10.1183/09031936.00111507 [DOI] [PubMed] [Google Scholar]
- Tak P.P., and Firestein G.S.. 2001. NF-κB: A key role in inflammatory diseases. J. Clin. Invest. 107:7–11. 10.1172/JCI11830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami S., Minami M., Katayama T., Nagata I., Namura S., and Satoh M.. 2002. TAK-779, a nonpeptide CC chemokine receptor antagonist, protects the brain against focal cerebral ischemia in mice. J. Cereb. Blood Flow Metab. 22:780–784. 10.1097/00004647-200207000-00003 [DOI] [PubMed] [Google Scholar]
- Torres-Platas S.G., Comeau S., Rachalski A., Bo G.D., Cruceanu C., Turecki G., Giros B., and Mechawar N.. 2014. Morphometric characterization of microglial phenotypes in human cerebral cortex. J. Neuroinflammation. 11:12 10.1186/1742-2094-11-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallières L., Campbell I.L., Gage F.H., and Sawchenko P.E.. 2002. Reduced hippocampal neurogenesis in adult transgenic mice with chronic astrocytic production of interleukin-6. J. Neurosci. 22:486–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weering H.R., Boddeke H.W., Vinet J., Brouwer N., de Haas A.H., van Rooijen N., Thomsen A.R., and Biber K.P.. 2011. CXCL10/CXCR3 signaling in glia cells differentially affects NMDA-induced cell death in CA and DG neurons of the mouse hippocampus. Hippocampus. 21:220–232. 10.1002/hipo.20742 [DOI] [PubMed] [Google Scholar]
- Vukovic J., Colditz M.J., Blackmore D.G., Ruitenberg M.J., and Bartlett P.F.. 2012. Microglia modulate hippocampal neural precursor activity in response to exercise and aging. J. Neurosci. 32:6435–6443. 10.1523/JNEUROSCI.5925-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Honsek S.D., Illes S., Wellen J.M., Hartung H.P., Rose C.R., and Dihné M.. 2011. A new role for interferon gamma in neural stem/precursor cell dysregulation. Mol. Neurodegener. 6:18 10.1186/1750-1326-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yoo Y., Fan H., Kim E., Guan K.L., and Guan J.L.. 2010. Regulation of Integrin β 1 recycling to lipid rafts by Rab1a to promote cell migration. J. Biol. Chem. 285:29398–29405. 10.1074/jbc.M110.141440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liang C.C., Bian Z.C., Zhu Y., and Guan J.L.. 2013. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat. Neurosci. 16:532–542. 10.1038/nn.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chen S., Yeo S., Karsli-Uzunbas G., White E., Mizushima N., Virgin H.W., and Guan J.L.. 2016. Elevated p62/SQSTM1 determines the fate of autophagy-deficient neural stem cells by increasing superoxide. J. Cell Biol. 212:545–560. (published erratum appears in J. Cell Biol. 2016. 212:879) 10.1083/jcb.201507023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Gan B., Wu X., and Guan J.L.. 2009. Inactivation of FIP200 leads to inflammatory skin disorder, but not tumorigenesis, in conditional knock-out mouse models. J. Biol. Chem. 284:6004–6013. 10.1074/jbc.M806375200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Wei S., Gan B., Peng X., Zou W., and Guan J.L.. 2011. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 25:1510–1527. 10.1101/gad.2051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten M.W., Geetha T., Seibenhener M.L., Babu J.R., Diaz-Meco M.T., and Moscat J.. 2005. The p62 scaffold regulates nerve growth factor-induced NF-κB activation by influencing TRAF6 polyubiquitination. J. Biol. Chem. 280:35625–35629. 10.1074/jbc.C500237200 [DOI] [PubMed] [Google Scholar]
- Wu X., Suetsugu S., Cooper L.A., Takenawa T., and Guan J.L.. 2004. Focal adhesion kinase regulation of N-WASP subcellular localization and function. J. Biol. Chem. 279:9565–9576. 10.1074/jbc.M310739200 [DOI] [PubMed] [Google Scholar]
- Zhong Z., Sanchez-Lopez E., and Karin M.. 2016. Autophagy, inflammation, and immunity: A troika governing cancer and its treatment. Cell. 166:288–298. 10.1016/j.cell.2016.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.