Abstract
Mechanisms and signals that regulate transcriptional coactivators are still largely unknown. Here we provide genetic evidence for a repressor that interacts with and regulates the nuclear receptor coactivator PGC-1. Association with the repressor requires a PGC-1 protein interface that is similar to the one used by nuclear receptors. Removal of the repressor enhances PGC-1 coactivation of steroid hormone responses. We also provide evidence that interaction of the repressor with PGC-1 is regulated by mitogen-activated protein kinase (MAPK) signaling. Activation of the MAPK p38 enhances the activity of wild-type PGC-1 but not of a PGC-1 variant that no longer interacts with the repressor. Finally, p38 activation enhances steroid hormone response in a PGC-1-dependent manner. Our data suggest a model where the repressor and nuclear receptors compete for recruiting PGC-1 to an inactive and active state, respectively. Extracellular signals such as nuclear receptor ligands or activators of the MAPK p38 can shift the equilibrium between the two states.
Regulation of gene expression in response to signals is a dynamic and integrative process. Signaling information can in principle be transduced to three types of transcriptional regulators: transcription factors that bind specific DNA sequences in the vicinity of promoters; proteins of the basal transcriptional machinery, including the RNA polymerase, that recognize the promoters; and coactivators or corepressors that are recruited via protein–protein interactions. DNA-binding transcription factors have been extensively studied as the final targets of signal transduction pathways that regulate transcription. However, coactivators and corepressors are not necessarily passive partners that simply relay the state of the DNA-bound transcription factors to the basal machinery. Many coactivators are phosphoproteins, suggesting that they can integrate signals that do not directly affect their DNA-binding partner (1–4). The signals and mechanisms that regulate coactivator function are largely unknown.
The coactivator PGC-1 was isolated based on its ability to interact with the peroxisome proliferator activated receptor gamma (PPARγ) in a two-hybrid screen, and to enhance glucocorticoid responses in a functional genetic screen (5, 6). PPARγ and the glucocorticoid receptor (GR) belong to the family of nuclear receptors, which are ligand-regulated DNA-binding transcription factors (7). Ligands of these receptors include steroids, retinoids, thyroid hormone, and many other small lipophilic molecules. They regulate the activity of their cognate receptors by binding a conserved ligand binding domain (LBD) and inducing conformational changes that enable interactions with coactivators (reviewed in refs. 8 and 9). PGC-1 is only one of many coactivators that interact with nuclear receptors. The p160 coactivators SRC-1, TIF2, and pCIP (also referred to as GRIP-1, ACTR, AIB, or NcoA-3), the histone acetyl transferases CBP, p300, and pCAF, and subunits of the Mediator complex, all interact with a similar surface of the activated LBD of the receptors, and have been suggested to mediate their transcriptional activity (reviewed in refs. 8 and 9).
A major challenge is to understand the physiological role of the different receptor–coactivator complexes, and the mechanisms that regulate coactivator recruitment and activity in different cellular contexts. Studies on PGC-1 suggest that its coactivator function must be tightly regulated. Ectopic expression of PGC-1 in cultured muscle or heart cells induces mitochondrial biogenesis and respiration, indicating that PGC-1 activity is a limiting factor in this highly regulated process (10, 11). Transgenic mice with high levels of PGC-1 in heart develop cardiomyopathy (11), pointing to the undesirable effects that increased PGC-1 activity can have. One level at which regulation is exerted is the synthesis of PGC-1. PGC-1 mRNA is predominantly expressed in tissues with high energy demands or a role in adaptive thermogenesis, such as heart, skeletal muscle, kidney, liver, and brown fat (5, 6, 12). Moreover, PGC-1 levels are regulated by exposure to cold, fasting, exercise, and leptin (5, 11, 13, 14).
Less is known about regulation of PGC-1 at the posttranscriptional level. Studies of Puigserver et al. (15) have shown that one mechanism activating PGC-1 is the interaction with nuclear receptors. Binding of receptors induces a conformational change in PGC-1, and promotes interactions of PGC-1 with downstream effectors (15). In this report, we provide evidence that PGC-1 activity is regulated by an additional mechanism: via a putative repressor. The repressor and nuclear receptors recognize overlapping sites in PGC-1, suggesting a competition between the repressor and receptors for binding to PGC-1. In addition, we report that activation of the mitogen-activated protein kinase (MAPK) p38 pathway enhances PGC-1 function in a manner consistent with the p38 kinase regulating the PGC-1–repressor association.
Materials and Methods
Plasmids.
Expression plasmids for human PGC-1 have been described (6). The PGC-1 Leu-motif mutants were generated by fusion PCR (16) and subcloned into pcDNA3 (Invitrogen), pACT2 and pAS2-1 (CLONTECH), and pA4.7 (Gal4–PGC-1) (6). All PCR-derived constructs were verified by sequencing. More information about the plasmids is available on request.
Yeast Two-Hybrid Interaction and Transcriptional Activation Assays.
Yeast carrying Gal4-responsive β-gal reporters were transformed as described (6). For the two-hybrid assay, cells carried plasmid pAS2/GR.LBD [Gal4 DNA binding domain (DBD) fused to GR LBD (6)], and either pACT2 (empty vector) or pACT2/PGC-1 constructs [Gal4 activation domain (AD) fused to PGC-1 variants (6)]. For the transcriptional activation assay, yeast carried plasmids expressing the Gal4 DBD alone (pAS2-1) or Gal4 DBD fused to full-length PGC-1 variants (Gal4–PGC-1). Cells were grown and assayed for β-gal activity as described (6). Data represent the average ± standard deviation of eight independent yeast transformants.
Cell Culture.
COS7 and HeLa [HtTA-1 (17)] cells were cultured in DMEM supplemented with 9% FCS (charcoal-stripped when assaying hormone responses). The stable HeLa cell line (HtTA/HA-hPGC1-TAP clone 26) expresses human PGC-1 with a hemagglutinin (HA)-epitope tag at the N terminus and a TAP tag (18) at the C terminus, under the control of the tetracycline-regulatable promoter (Tet-Off system). Details are available on request (D.Kr. and A.K., unpublished data). The cells were cultured in media containing 100 μg/ml G418 and 100 μg/ml hygromycin. PGC-1 expression was repressed by the addition of 0.5 μg/ml doxycycline.
Transfection and Transcription Activation Assays.
Cells were transfected by calcium phosphate precipitation, as described (6). All transfections included 0.2 μg of p6RlacZ (19) for normalization of transfection efficiency. The amounts of reporter and expression plasmids per transfection were: 1 μg of the luciferase reporters pTAT3Luc (20) or pGK-1 (21); 0.5 or 1 μg of the GR expression plasmid p6RGR (22); 0.5 or 1 μg of the PGC-1 expression plasmid pcDNA3/HA-PGC-1 for coactivation assays; 50 ng of pA4.7-PGC-1/HA to measure the activity of Gal4–PGC-1 variants; and 1 μg of pcDNA3-MKK6bAA or pcDNA3-MKK6bE (23) to assay the effects of MKK6. Cell lysates were prepared 40 to 48 h after transfection and assayed for luciferase and β-gal activity (6). Luciferase values normalized to β-gal activity are referred to as relative luciferase units. Data shown represent the mean ± standard deviation of four to six values, from at least two independent experiments performed in duplicate, unless indicated otherwise.
Western Analysis.
Lysates from cells transfected as described above were subjected to Western analysis, using antibody HA.11 against the HA epitope (Babco, Richmond, CA) for HA-tagged PGC-1 or rabbit anti-protein A antibody (Sigma, 10 ng/ml) for the TAP-tagged PGC-1.
Results
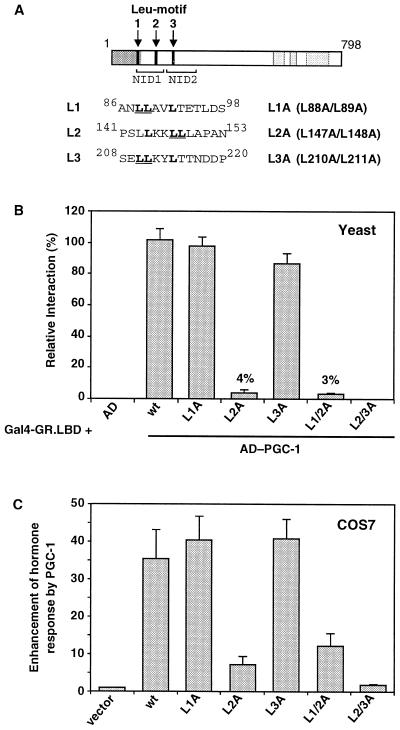
Hydrophobic sequences in amphipathic α-helices have been shown to mediate protein–protein interactions in several transcriptional regulatory complexes (24–29). The coactivator PGC-1 has three motifs that are rich in leucine residues (Leu-motifs) and predicted to be part of short α-helices (Fig. 1A). Leu-motif 1 (L1) resides in the N-terminal transcriptional AD. Leucine motifs 2 and 3 (L2, L3) are in two PGC-1 domains (NID1 and NID2, respectively) that interact with GR (6). The L2 motif fits a consensus LxxLL sequence (x being any amino acid) present in many proteins that interact with the LBD of nuclear receptors, and is important for the interaction of PGC-1 with PPARα and the estrogen receptor (30, 31). To investigate the role of these three Leu-motifs in PGC-1 function, we substituted the double leucine of each motif by a double alanine (Fig. 1A; L1A, L2A, and L3A), a mutation known to disrupt Leu-rich interaction surfaces (24, 29). We then determined the effects of these mutations on three aspects of PGC-1 function: physical interaction with GR, coactivation of glucocorticoid responses, and transcriptional activity of PGC-1.
Figure 1.
PGC-1 interacts with GR via Leu-motifs L2 and L3. (A) PGC-1 has three Leu-motifs, L1 in the transcriptional activation domain (shaded dark gray), and L2 and L3 in the nuclear receptor interaction domains (NID1 and NID2) (6). The double leucines that were substituted by double alanines are underlined, and their residue numbers are indicated. (B) Interaction of PGC-1 variants with the LBD of GR, using a two-hybrid assay in yeast. Cells expressing Gal4-GR.LBD and either the Gal4 AD alone (AD) or the indicated PGC-1 variants fused to the AD were grown in the presence of 25 μM corticosterone and assayed for β-gal activity. Data are normalized to wild-type PGC-1 (wt) activity being equal to 100. (C) Enhancement of hormone response by PGC-1 variants. COS7 cells transfected with the GR expression plasmid p6RGR, the GR-responsive luciferase reporter pTAT3Luc, and either pcDNA3 vector or pcDNA3/HA-PGC-1 variants, were treated for 20 h with 50 nM corticosterone and assayed for luciferase activity. PGC-1 had no effect in the absence of hormone. Data are expressed as fold-enhancement of GR activity by PGC-1 in the presence of hormone.
Two Leu-Motifs, L2 and L3, Mediate PGC-1 Interactions with GR.
First, we assayed the PGC-1 mutants for their ability to interact physically with GR, using a two-hybrid assay in yeast. Wild-type PGC-1 fused to the Gal4 AD interacted with the LBD of GR in the presence of hormone (Fig. 1B), as reported (6). Mutations in either motif L1 or L3 alone had no effect on the interaction (L1A and L3A, Fig. 1B). In contrast, the disruption of motif L2 reduced drastically the ability of PGC-1 to interact with GR (L2A, Fig. 1B). The remaining activity of the L2A mutant (4% of wild-type PGC-1) depended on motif L3, because the double mutation of L2A and L3A abolished completely the interaction (L2/3A, Fig. 1B). Mutation of motif L1 in either the L2A or L3A context had no effect, indicating that L1 does not play a role in the PGC-1–GR interaction (Fig. 1B and data not shown). Taken together, our results suggest that PGC-1 interacts with GR via the two Leu-rich motifs L2 and L3. Motif L2 is the major site of interaction, whereas L3 has a secondary role that is apparent in the absence of a functional L2.
Next, we determined the ability of the mutants to enhance GR-mediated transcription, a PGC-1 function that requires both physical interaction with the receptor, and the N-terminal AD of PGC-1 (6). COS7 cells were transfected with expression vectors for GR and PGC-1, wild-type or mutant, and a glucocorticoid-responsive reporter. Wild-type PGC-1 enhanced strongly, by ≈35-fold, the transcriptional response to hormone (Fig. 1C). Mutations in L1 or L3 alone had no apparent effect. Mutant L2A displayed a dramatically reduced activity, enhancing the transcriptional response by only 6- to 7-fold. The residual activity of the L2A mutant depended on motif L3, because the double mutant L2/3A could no longer enhance response to hormone (Fig. 1C). Thus, the activities of the PGC-1 mutants in enhancing GR-mediated transcription correlate well with their abilities to interact physically with GR.
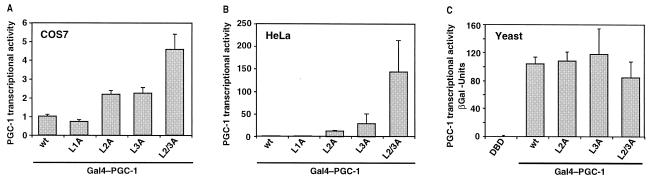
Disruption of Motifs L2 and L3 Increases PGC-1 Transcriptional Activity.
PGC-1 fused to a heterologous DBD is a strong activator of transcription (5, 6). Because the interaction with nuclear receptors has been shown to activate PGC-1 (15), we tested whether disruption of the Leu-motifs, in particular of L2 and L3, would affect PGC-1 transcriptional activity. For this, we determined the ability of the PGC-1 mutants fused to the Gal4 DBD (Gal4–PGC-1) to activate transcription from a Gal4 reporter. In COS7 cells, wild-type PGC-1 enhanced transcription by ≈200-fold, compared with the activity of Gal4 DBD alone. Disruption of the L1 motif resulted in a small decrease in transcriptional activity (Fig. 2A). Surprisingly, mutations in either of the two GR-interacting motifs, L2 or L3, caused modest increases in PGC-1 activity. The double mutant L2/3A, which is GR-binding deficient, was an even stronger activator (Fig. 2A). The increased activity was not due to altered expression or stability, because wild-type PGC-1 and the mutants were expressed at similar levels, as judged by Western analysis (data not shown). Hence, these results suggest that L2 and L3 confer an inhibitory regulation on PGC-1 activity, which is abolished by the L2/3A mutation.
Figure 2.
The Leu-motifs regulate the transcriptional activity of PGC-1. Disruption of motifs L2 and L3 increases PGC-1 activity in COS7 (A) and HeLa cells (B). Cells were transfected with expression plasmids for Gal4–PGC-1 variants and the Gal4-responsive luciferase reporter pGK-1, and assayed for luciferase activity. The activity of wild-type Gal4–PGC-1 (wt) was normalized to 1 within each experiment, and represents an average activation of 200-fold in COS7 and 50-fold in HeLa cells, compared with Gal4 DBD alone. (C) Disruption of motifs L2 and L3 does not affect PGC-1 activity in yeast. Yeast expressing the indicated Gal4–PGC-1 variants were assayed for activity from a Gal4-responsive β-gal reporter. Transcriptional activity in yeast expressing just the Gal4 DBD was set equal to 1.
One explanation for our findings is that the mutations in motifs L2 and L3 disrupt interactions with proteins that regulate negatively PGC-1. Alternatively, the L2/3A mutation causes a conformational change in PGC-1 that exposes the N-terminal AD. To determine whether the regulatory effect of L2/3 depends on the cellular context, we assayed the transcriptional activity of PGC-1 in two other cell types: HeLa and yeast cells. In HeLa cells, the L2A and L3A mutations also increased PGC-1 activity (Fig. 2B), albeit to a higher degree than in COS7 cells. The L2/3A mutation increased PGC-1 activity by more than 100-fold in HeLa cells, compared with the 4–5-fold increase in COS7 cells (L2/3A in Fig. 2 A and B). Interestingly, PGC-1 activity in yeast was unaffected by the L2/3A mutations, suggesting the absence of a regulatory mechanism via these sites in yeast (Fig. 2C). Our results show that the L2/3-mediated effect on PGC-1 activity depends on the cellular context, and is therefore unlikely to be due solely to intrinsic changes in PGC-1 conformation. For subsequent experiments we chose HeLa cells, as they show strong L2/3-dependent regulation of PGC-1 activity.
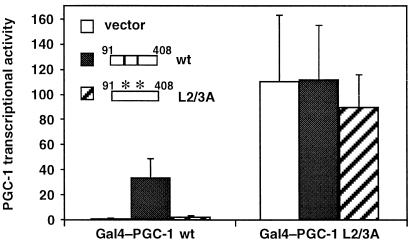
The Repression Mechanism Mediated by Motifs L2 and L3 Is Relieved by Competition.
Our data suggested that a protein could bind the L2/3 motifs and repress PGC-1 activity. To test this, we asked whether supplying the L2/3 sites in trans could compete the putative repressor away from PGC-1. We thus expressed together with Gal4–PGC-1 a PGC-1 fragment (amino acids 91–408) bearing the L2/3 sites. Coexpression of the 91–408 fragment increased strongly the activity of wild-type Gal4–PGC-1, supporting the hypothesis that this is a binding site for a repressor (Fig. 3). The increase in activity depended on the L2/3 sites of the competing fragment, because coexpression of the corresponding fragment with the L2/3A mutation had no significant effect. Moreover, coexpression was specifically affecting the regulatory mechanism mediated by the L2/3 sites, because neither the wild-type nor the L2/3A PGC-1 fragment affected the activity of the L2/3A PGC-1 mutant (Fig. 3). These findings suggest that the L2/3 motifs mediate interactions with a PGC-1 repressor.
Figure 3.
Coexpression of a fragment containing sites L2 and L3 enhances PGC-1 activity. HeLa cells were transfected with Gal4–PGC-1 [wild type (wt) or mutant (L2/3A)], together with 1 μg of vector alone or vector expressing amino acid 91–408 of PGC-1 bearing the L2/3 sites [wild type (wt) or mutant (L2/3A)], and the Gal4-responsive luciferase reporter pGK-1. Data are normalized to the luciferase activity of wild-type Gal4–PGC-1 in the presence of vector alone being equal to 1.
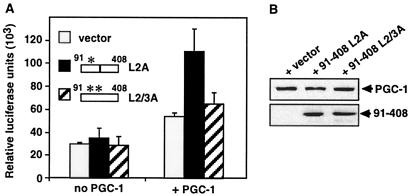
The Repressor Regulates Hormone Response.
The repressor and GR interact with the same PGC-1 domain, suggesting that they may compete with each other for binding to the coactivator. If so, the repressor could limit PGC-1 recruitment by GR and reduce the transcriptional response to hormone. To test this, we asked whether removing the repressor by supplying its binding site in trans altered the ability of PGC-1 to enhance GR-mediated transcription. As a binding site, we could not use the wild-type PGC-1 91–408 fragment, because this fragment can also bind GR (6) and interfere with the GR–PGC-1 interaction. To circumvent this problem, we made use of the apparent inverse preference of GR and the repressor for motifs L2 and L3. Whereas mutation of L2 affects drastically the interaction with GR, mutation of L3 affects preferentially the interaction with the repressor (see Figs. 1B and 2B). We thus measured the ability of PGC-1 to enhance GR activity in the absence or presence of the 91–408 L2A PGC-1 fragment, which carries a mutated L2 (so that it does not interact with GR) and a wild-type L3 (so that it binds the repressor). To favor competition by an excess of the binding site, we used HeLa cells that express low levels of PGC-1 from a stably integrated tetracycline-regulated locus. Coexpression of the 91–408 L2A fragment with GR in these cells increased the transcriptional response (Fig. 4A). The increase depended on the L3 site of the competing fragment, because expression of a similar fragment with both L2 and L3 mutated had no significant effect (L2/3A, Fig. 4A). Furthermore, the L2A fragment had no effect on GR activity in HeLa cells that do not express PGC-1 (Fig. 4A). The effects were due to changes in PGC-1 activity, because expression of the fragments did not alter PGC-1 expression levels (Fig. 4B). Thus, our results demonstrate that the putative PGC-1 repressor can determine the efficiency with which the coactivator enhances GR activity.
Figure 4.
Competition of the repressor increases the ability of PGC-1 to enhance the hormone response. (A) HeLa cells expressing PGC-1 stably, under the control of a tetracycline-regulatable promoter, were transfected with 1 μg of vector alone or vector expressing either the L2A or the L2/3A mutant 91–408 PGC-1 fragment, together with the GR expression plasmid p6RGR and the GR-responsive luciferase reporter pTAT3Luc. Cells were cultured in the presence of doxycycline (0.5 μg/ml) to repress PGC-1 expression (no PGC-1) or absence of doxycycline (+ PGC-1), treated for 24 h with 5 nM corticosterone, and assayed for luciferase activity. (B) Protein levels of the stably expressed full-length PGC-1 and the transiently expressed 91–408 PGC-1 fragments. Extracts were from cells grown in the absence of doxycycline (+ PGC-1); no PGC-1 was detected in cells grown in the presence of doxycycline. Proteins were detected with an anti-protein A antibody for full-length PGC-1 (Upper) and an anti-HA antibody for the 91–408 fragments (Lower).
The MAPK p38 Regulates PGC-1–Repressor Interaction and PGC-1 Coactivation Function.
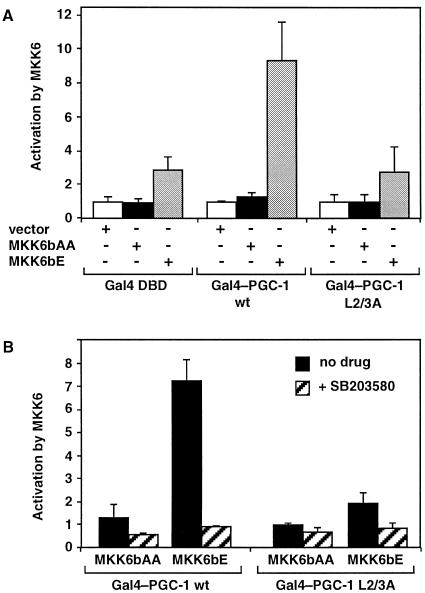
Interactions between macromolecules are often regulated by signal transduction pathways, via phosphorylation of the interacting partners. PGC-1 is likely to be a phosphoprotein. First, its protein sequence includes putative phosphorylation targets for protein kinase A, protein kinase C, casein kinase II, and MAPKs. Second, treatment of cells with okadaic acid, which inhibits protein phosphatase 2A, causes a strong shift in PGC-1 mobility in SDS/PAGE gels (D.K. and A.K., unpublished data). To determine whether phosphorylation could modulate the interaction of PGC-1 with the repressor, we tested whether activated kinases could alter the activity of wild-type PGC-1, without affecting that of the repressor-free L2/3A mutant. As seen in Fig. 5A, coexpression of a constitutively active variant of the MAPK kinase MKK6 (MKK6bE) with Gal4–PGC-1 enhanced strongly and preferentially the activity of wild-type PGC-1. The modest increase seen with the L2/3A mutant was not specific to PGC-1, but was also observed with the Gal4 DBD alone (Fig. 5A). The inability of MKK6bE to activate the repressor-free L2/3A PGC-1 was not due to the already high activity of this mutant. Even when we lowered the expression levels of the mutant, so that the transcriptional output was similar to that in cells with high levels of wild-type PGC-1, MKK6bE did not affect its activity (data not shown). Coexpression of the kinase-deficient MKK6 variant (MKK6bAA) had no effect on PGC-1, demonstrating that the kinase activity of MKK6 was required for the enhancement.
Figure 5.
Activation of the MAPK p38 increases Gal4–PGC-1 activity in a repressor-dependent manner. (A) Expression of the constitutively active MKK6 (MKK6bE) activates the wild-type PGC-1 (Gal4–PGC-1 wt), but not the L2/3A mutant (Gal4–PGC-1 L2/3A). Data are expressed as activation by coexpression of the indicated MKK6 variant. (B) The p38 inhibitor SB203580 inhibits the enhancement by MKK6bE and reduces Gal4–PGC-1 activity. SB203580 (30 μM) or vehicle DMSO were added to cells 12 h after transfection. Data are expressed as activation by coexpression of the indicated MKK6 variant, in the absence or presence of SB203580 (A and B). HeLa cells were transfected with the Gal4-reporter pGK-1 and plasmids expressing either Gal4 DBD, Gal4–PGC-1 wild-type (wt), or Gal4–PGC-1 mutant (L2/3A). Empty vector, pcDNA3-MKK6bAA (inactive kinase mutant), or pcDNA3-MKK6bE (constitutively active variant) was cotransfected as indicated. Activity in the presence of vector alone and the absence of SB203580 drug was set equal to 1 for all Gal4 constructs.
MKK6 phosphorylates and activates the MAPK p38, which can then target several transcriptional regulators (32). To determine whether the p38 kinase mediates the effect of MKK6 on PGC-1, we used a specific inhibitor of p38, SB203580 (33). As expected for a p38-dependent event, SB203580 prevented the activation of wild-type PGC-1 by MKK6bE (Fig. 5B). Inhibition of p38 had no significant effect on Gal4–PGC-1 L2/3A, suggesting again that p38 activity does not affect the PGC-1 mutant. In conclusion, our findings show that the MKK6/p38 pathway regulates the activity of PGC-1 only when the latter is competent to associate with the repressor. This is consistent with the notion that phosphorylation by the p38 kinase causes a dissociation or destabilization of the PGC-1–repressor interaction.
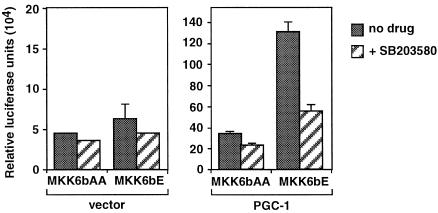
If the MKK6/p38 pathway activates PGC-1 by removing the repressor, then it should also enhance the ability of PGC-1 to coactivate GR-mediated transcription. Thus, we tested whether activation or inhibition of the p38 kinase affects the transcriptional response to corticosterone, in the absence or presence of PGC-1. MKK6bE, MKK6bAA, and the p38 inhibitor SB203580 had no significant effects on GR activity in the absence of PGC-1 expression, suggesting that the MKK6/p38 pathway does not act directly on either the GR or the endogenous coactivators of GR in HeLa cells (Fig. 6). In contrast, when PGC-1 was present, coexpression of the active MKK6bE variant enhanced the transcriptional response by more than 3-fold. The enhancement was prevented by treatment with the inhibitor SB203580, indicating that it is mediated by the p38 kinase. Expression of the kinase-inactive MKK6bAA had no effect (Fig. 6). In conclusion, activation of the MKK6/p38 pathway enhanced the glucocorticoid response in a PGC-1-dependent manner, suggesting that either PGC-1 or a PGC-1 regulator is the p38 target.
Figure 6.
Activation of the MKK6/p38 pathway enhances PGC-1-mediated activation of glucocorticoid response. Expression vectors for GR (p6RGR), constitutively active or inactive MKK6 (MKK6bE or MKK6bAA), and the GR reporter pTAT3Luc were transfected into HeLa cells together with either just vector (Left) or PGC-1 expression plasmid pcDNA3/HA-PGC-1 (Right). SB203580 (30 μM) or DMSO vehicle were added 15 h after transfection, and corticosterone (5 nM) 5 h later. Luciferase activity was assayed 16 h after hormone addition. Data are from one experiment performed in duplicate and are representative of four independent experiments.
Discussion
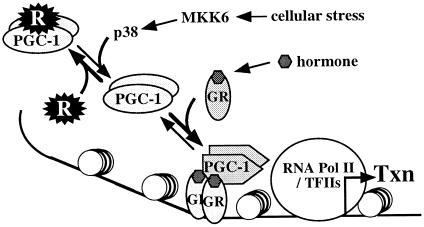
In this study we provide evidence for a model in which negative and positive regulators use overlapping sites to recruit the coactivator PGC-1 to inactive and active states, respectively (Fig. 7). Interaction with the putative repressor decreases PGC-1 activity in the context of Gal4–PGC-1 acting on Gal4-responsive promoters, as well as native PGC-1 acting on GR-responsive reporters. Nuclear receptors such as GR constitute positive regulators, because they may displace the repressor and recruit PGC-1 to active sites at promoters. Consistent with our model, Puigserver et al. (15) have shown that nuclear receptor binding induces a PGC-1 conformational change, which we propose may “lock” PGC-1 to the active state. Signaling via the MAPK p38 pathway can also regulate PGC-1, by affecting the repressor–PGC-1 interaction. Consequently, p38 can regulate GR activity in a PGC-1-dependent manner (Fig. 7).
Figure 7.
Model for the regulation of the coactivator PGC-1. PGC-1 associates with a repressor (R), which maintains the coactivator in an inactive state. Hormone-activated GR interacts with PGC-1, displaces the repressor and recruits the coactivator to sites of transcription. A conformational change in PGC-1 upon receptor binding, as seen by Puigserver et al. (15), is symbolized by the different shape of PGC-1 and may stabilize the active state. Phosphorylation by the MAPK p38 favors the release of the repressor, thereby enhancing PGC-1 activity and glucocorticoid responses.
Several lines of evidence suggest that the mutations in motifs L2/3 disrupt an interaction with a repressor, rather than relieve an autoinhibitory intramolecular mechanism. First, the effect of the L2/3A mutation is cell-type-dependent, suggesting that another cellular factor is required. Second, overexpression of the L2/3 sites in trans titrates an inhibitory factor and increases PGC-1 activity. The L2/3-containing fragment used in this experiment does not interact with PGC-1 (unpublished data), indicating that it does not act by preventing an intramolecular PGC-1 interaction. Third, the ability of the L2/3 motifs to mediate interactions with GR points to their capacity as protein interaction surfaces. The deletion of amino acids 170–350 of PGC-1, which include motif L3, has been shown previously to increase PGC-1 activity (15). Our model is consistent with these earlier observations and suggests that nuclear receptors may activate PGC-1 in two steps: first, by displacing the repressor, and second, by inducing a PGC-1 conformational change that enables interactions with downstream effectors (15).
The requirements for PGC-1 interaction with the repressor and nuclear receptors are similar, but not identical. GR association depends primarily on motif L2, whereas repressor association depends on both L2 and L3. Motif L2 may be the major interaction site for several nuclear receptors, because L2 mutations also disrupt PGC-1 association with PPARα and the estrogen receptor (30, 31). Despite the difference in motif dependence, we considered whether nuclear receptors could themselves act as repressors of Gal4–PGC-1. Experiments by Puigserver et al. suggest that this is unlikely, because they find that coexpression of nuclear receptors activates rather than represses Gal4–PGC-1 (15). We also find that coexpression of the thyroid hormone receptor and retinoid X receptor, in the absence or presence of their respective hormones, can activate Gal4–PGC-1 (see Fig. 8, which is published as supplemental data on the PNAS web site, www.pnas.org), supporting the notion that nuclear receptors displace the repressor. Overexpression of GR had no effect on Gal4–PGC-1 activity (data not shown). This may reflect the inability of GR, when not bound to DNA, to efficiently dimerize and interact with PGC-1. In no case did we observe that nuclear receptor overexpression could repress Gal4–PGC-1.
The putative PGC-1 repressor could play more than one role. First, it may enable the maintenance of a “silent” pool of this potent coactivator that could be readily activated by appropriate signals. The requirement to keep coactivators inactive when not recruited by DNA-bound transcription factors may be general. The transcriptional coactivator TAZ is negatively regulated by 14-3-3 proteins, which bind the phosphorylated TAZ and keep it in the cytoplasm (34). Nucleocytoplasmic partitioning does not play a role in the L2/3-mediated repression of PGC-1, because both wild-type PGC-1 and the L2/3A mutant show similar nuclear localization in HeLa cells (see Fig. 9, which is published as supplemental data). In addition, the repressor may set distinct thresholds for PGC-1 recruitment by nuclear receptors. The levels of the repressor, as well as the affinities of different nuclear receptors relative to the affinity of the repressor for PGC-1, could determine utilization of PGC-1.
The association of PGC-1 with the repressor presents an entry point for other signal transduction pathways to crosstalk with steroid hormone signaling. The mechanism by which the MAPK p38 activates PGC-1 is not yet clear. Our experiments suggest that p38-mediated phosphorylation counteracts the repressor effect, possibly by encouraging the release of the repressor from PGC-1. Whether p38 phosphorylates directly PGC-1, which harbors putative MAPK sites, or the yet unidentified repressor remains to be determined. An interesting prediction of our model is that p38 signaling will only have an impact on PGC-1 function in cells expressing the repressor.
Nuclear receptors interact with a multitude of coactivators, among which they have to choose a partner (8, 9). Our findings bear on two important aspects of coactivator specificity. First, they suggest that third factors, such as the repressor, influence the choice for a specific coactivator. Second, they illustrate how the presence of a coactivator can confer new properties to nuclear receptor signaling, such as regulation by other pathways. The role of crosstalk between p38 and GR is not immediately clear. It is however interesting that the two molecules are components of pathways that mediate responses to different types of stress, such as systemic stress for GR and cellular stress for p38. The fact that other coactivators like CBP, AIB1, and SRC-1 are activated by distinct MAPK kinases (p42/p44) suggests that coactivators may present a versatile target via which signaling pathways impinge on nuclear receptor activity (1, 35–37).
The biological consequences of PGC-1 overexpression suggest that PGC-1 activity is limiting in muscle and heart cells, and that mechanisms regulating PGC-1 are important for energy homeostasis (10, 11). Our studies reveal an unexpected level of regulation by a PGC-1-associating repressor and, when this repressor is present, by cellular stress signaling pathways. Identification of the repressor will be crucial for understanding its role in PGC-1 function.
Supplementary Material
Acknowledgments
We thank G. Werlen for the MKK6 expression plasmids; T. Grange, M. Hall, U. Müller, and Kralli lab members for discussions and comments on the manuscript. This work was supported by the Basel Chemical Industry (D.Kn.), the Swiss National Science Foundation, the University of Basel, and the Max Cloëtta foundation (A.K.).
Abbreviations
- GR
glucocorticoid receptor
- MAPK
mitogen activated protein kinase
- LBD
ligand binding domain
- DBD
DNA binding domain
- AD
activation domain
- Ln
Leu-motif n
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Font de Mora J, Brown M. Mol Cell Biol. 2000;20:5041–5047. doi: 10.1128/mcb.20.14.5041-5047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 3.Hu S C, Chrivia J, Ghosh A. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- 4.Rowan B G, Weigel N L, O'Malley B W. J Biol Chem. 2000;275:4475–4483. doi: 10.1074/jbc.275.6.4475. [DOI] [PubMed] [Google Scholar]
- 5.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 6.Knutti D, Kaul A, Kralli A. Mol Cell Biol. 2000;20:2411–2422. doi: 10.1128/mcb.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glass C K, Rosenfeld M G. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 9.Robyr D, Wolffe A P, Wahli W. Mol Endocrinol. 2000;14:329–347. doi: 10.1210/mend.14.3.0411. [DOI] [PubMed] [Google Scholar]
- 10.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla R C, Spiegelman B M. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 11.Lehman J J, Barger P M, Kovacs A, Saffitz J E, Medeiros D M, Kelly D P. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esterbauer H, Oberkofler H, Krempler F, Patsch W. Genomics. 1999;62:98–102. doi: 10.1006/geno.1999.5977. [DOI] [PubMed] [Google Scholar]
- 13.Goto M, Terada S, Kato M, Katoh M, Yokozeki T, Tabata I, Shimokawa T. Biochem Biophys Res Commun. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- 14.Kakuma T, Wang Z W, Pan W, Unger R H, Zhou Y T. Endocrinology. 2000;141:4576–4582. doi: 10.1210/endo.141.12.7804. [DOI] [PubMed] [Google Scholar]
- 15.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman B M. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 16.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 19.Pearce D, Yamamoto K R. Science. 1993;259:1161–1165. doi: 10.1126/science.8382376. [DOI] [PubMed] [Google Scholar]
- 20.Iniguez-Lluhi J A, Lou D Y, Yamamoto K R. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- 21.Webb P, Lopez G N, Uht R M, Kushner P J. Mol Endocrinol. 1995;9:443–456. doi: 10.1210/mend.9.4.7659088. [DOI] [PubMed] [Google Scholar]
- 22.Godowski P J, Rusconi S, Miesfeld R, Yamamoto K R. Nature (London) 1987;325:365–368. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Lee J D, Jiang Y, Li Z, Feng L, Ulevitch R J. J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 24.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 25.Nagy L, Kao H Y, Love J D, Li C, Banayo E, Gooch J T, Krishna V, Chatterjee K, Evans R M, Schwabe J W. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perissi V, Staszewski L M, McInerney E M, Kurokawa R, Krones A, Rose D W, Lambert M H, Milburn M V, Glass C K, Rosenfeld M G. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu X, Lazar M A. Nature (London) 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 28.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto K R, Darimont B D, Wagner R L, Iniguez-Lluhi J A. Cold Spring Harbor Symp Quant Biol. 1998;63:587–598. doi: 10.1101/sqb.1998.63.587. [DOI] [PubMed] [Google Scholar]
- 30.Vega R B, Huss J M, Kelly D P. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tcherepanova I, Puigserver P, Norris J D, Spiegelman B M, McDonnell D P. J Biol Chem. 2000;275:16302–16308. doi: 10.1074/jbc.M001364200. [DOI] [PubMed] [Google Scholar]
- 32.Widmann C, Gibson S, Jarpe M B, Johnson G L. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 33.Cuenda A, Rouse J, Doza Y N, Meier R, Cohen P, Gallagher T F, Young P R, Lee J C. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 34.Kanai F, Marignani P A, Sarbassova D, Yagi R, Hall R A, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley L C, Yaffe M B. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y Z, Chrivia J C, Latchman D S. J Biol Chem. 1998;273:32400–32407. doi: 10.1074/jbc.273.49.32400. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y Z, Thomas N S, Latchman D S. NeuroReport. 1999;10:1239–1243. doi: 10.1097/00001756-199904260-00016. [DOI] [PubMed] [Google Scholar]
- 37.Rowan B G, Garrison N, Weigel N L, O'Malley B W. Mol Cell Biol. 2000;20:8720–8730. doi: 10.1128/mcb.20.23.8720-8730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.