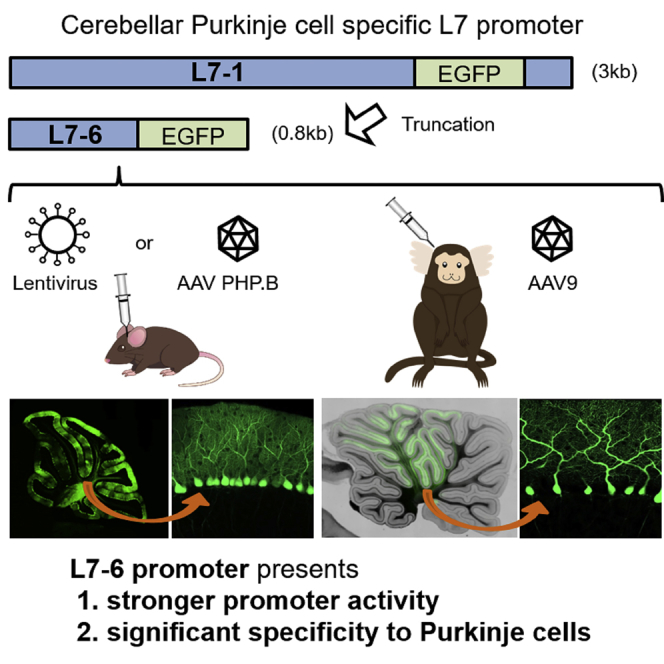
Abstract
Cell-type-specific promoters in combination with viral vectors and gene-editing technology permit efficient gene manipulation in specific cell populations. Cerebellar Purkinje cells play a pivotal role in cerebellar functions. Although the Purkinje cell-specific L7 promoter is widely used for the generation of transgenic mice, it remains unsuitable for viral vectors because of its large size (3 kb) and exceedingly weak promoter activity. Here, we found that the 0.8-kb region (named here as L7-6) upstream of the transcription initiation codon in the first exon was alone sufficient as a Purkinje cell-specific promoter, presenting a far stronger promoter activity over the original 3-kb L7 promoter with a sustained significant specificity to Purkinje cells. Intravenous injection of adeno-associated virus vectors that are highly permeable to the blood-brain barrier confirmed the Purkinje cell specificity of the L7-6 in the CNS. The features of the L7-6 were also preserved in the marmoset, a non-human primate. The high sequence homology of the L7-6 among mouse, marmoset, and human suggests the preservation of the promoter strength and Purkinje cell specificity features also in humans. These findings suggest that L7-6 will facilitate the cerebellar research targeting the pathophysiology and gene therapy of cerebellar disorders.
Keywords: Purkinje cell, L7, PCP2, cerebellum, lentivirus, adeno-associated virus, viral vector, cell type-specific promoter, marmoset, non-human primate
Graphical Abstract
Introduction
The cerebellum plays a critical role in motor coordination and motor learning.1, 2 Cerebellar impairments result in motor symptoms such as dysmetria, intention tremor, and dysarthria. In addition, recent studies have reported various cognitive deficits following damage to the cerebellum,3, 4, 5, 6, 7 suggesting its involvement in mental processes.8
Significant advances in the understanding of the molecular mechanisms underlying the cerebellar functions have been attained by the generation and analysis of gene-modified mice.9, 10, 11, 12, 13 In particular, mice with Purkinje cell-specific gene modifications played a pivotal role.14, 15, 16, 17, 18, 19 Purkinje cell-specific gene expression in most transgenic mice has used the Purkinje cell protein 2 (PCP2)/L7 promoter (henceforth L7), which drives the transcription of the L7 gene to produce the Purkinje cell-specific L7 protein.20, 21, 22 The L7 structural gene contains four exons that extend over about 2 kb in the genome.22 The rodent L7 promoter that was used in the Purkinje cell-specific transgenic mice consists of a 3-kb genomic region covering exons 1–4 with the upstream 0.9-kb promoter region.16, 23, 24, 25 The original start codon present in exon 2 and the potential downstream ATGs, in all reading frames, were removed from the exon regions.23 Therefore, the translation was initiated only from an ATG provided by an exogenous cDNA cloned into the unique BamHI site in exon 4.
Recently, the application of gene transfer using adeno-associated virus (AAV) vectors has markedly increased because of their efficient and broad transduction capacity in neuronal and glial cells. A major drawback of the AAV vectors, however, is their limited packaging capacity of a maximum of 5 kb, including the two inverted terminal repeats (ITRs), which is similar to the size of the parent AAV genome.26 Therefore, expressing a 2-kb transgene by the 3-kb L7 promoter is not guaranteed. Moreover, the L7 promoter has a considerably weaker promoter strength compared with constitutive virus promoters such as the cytomegalovirus (CMV) and murine stem cell virus (MSCV) promoters. Thus, identifying a minimal and strong L7 promoter sequence preserving the Purkinje cell specificity will broaden the range of the transgene size for the viral accommodation and will advance the utilization of AAV vectors in the cerebellar research. In a yet unpublished previous study using lentiviral vectors, we found that a 1-kb fragment of the L7 promoter exhibited strikingly greater promoter strength than the original 3-kb L7 promoter (US Patent 8912315 B2),27 although the experimental procedure to obtain the 1-kb fragment and the sequence have not yet been published. Subsequent utilization of the compact 1-kb promoter by several groups, including ours,28, 29, 30, 31 revealed overall maintenance of the Purkinje cell selectivity, but with some diffuse activity in off-target cells.
In the present study, we reported the identification of the 1-kb L7 fragment and its further truncation for a higher degree of Purkinje cell specificity. We showed that a 0.8-kb sequence upstream of the transcription initiation codon in the first exon of the L7 gene alone exhibited a great promoter strength that is almost comparable with the 1-kb fragment, but with a significantly higher specificity to Purkinje cells. Moreover, we demonstrated that this minimal 0.8-kb L7 promoter could work efficiently and specifically in Purkinje cells of a non-human primate.
Results
A 1 kb Genome Fragment Upstream of the Translational Start Codon in Exon 2 Holds Marked Promoter Strength with Purkinje Cell Specificity
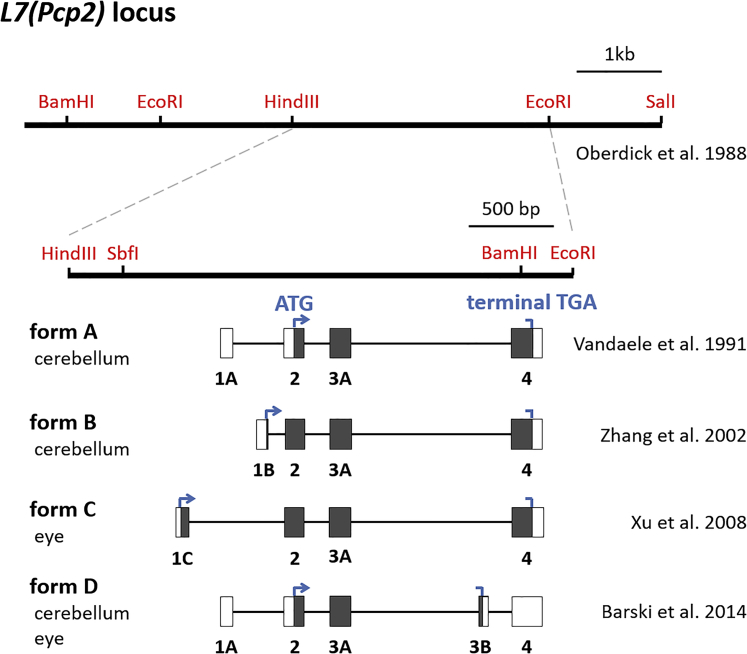
Since the initial report of the L7 protein in 1988,20, 21 four splice variants of the L7 gene consisting of four exons have been reported (Figure 1). Although those splice variants were designated by unique names in the original papers, they were re-named for clarity to form A,32 form B,33 form C,34 and form D.35 Oberdick et al.22 produced transgenic mice expressing LacZ (β-galactosidase) under the control of the 8-kb L7 genomic fragment comprising the transcribed region of the form A (1.9 kb), 4 kb upstream of the start codon present in exon 2, and 2 kb downstream of the polyadenylation signal. In accordance with the native L7 expression, the transgenic line showed β-galactosidase expression solely in Purkinje cells, indicating that the 8-kb genomic fragment involves regions critical for Purkinje cell-restricted expression. Subsequently, Smeyne et al.23 generated transgenic mice expressing an attenuated form of the diphtheria toxin under the control of the 3-kb L7 fragment in which the cDNA was inserted into a unique BamHI site present in exon 4, whereas the original start codon in exon 2 and other potential ATGs in exons were mutated (see L7-1 in Figure 2A). The transgenic mice expressing a transgene using the L7-1 promoter exhibited highly specific GFP expression in Purkinje cells,16, 23, 24, 25 which strongly indicates the presence of regions regulating the Purkinje cell-specific expression in the L7-1 promoter. Thus, we focused on the 3-kb L7-1 sequence for further investigation.
Figure 1.
Schematic Representation of the Former 8-kb L7 Promoter and Its Association with the Four Splice Forms
Four splice variants of L7 expressed in the cerebellum and/or eye were renamed chronologically as forms A–D. Boxes show the exons, in which the filled portions correspond to the coding region denoted with the start (ATG) and terminal (TGA) codons.
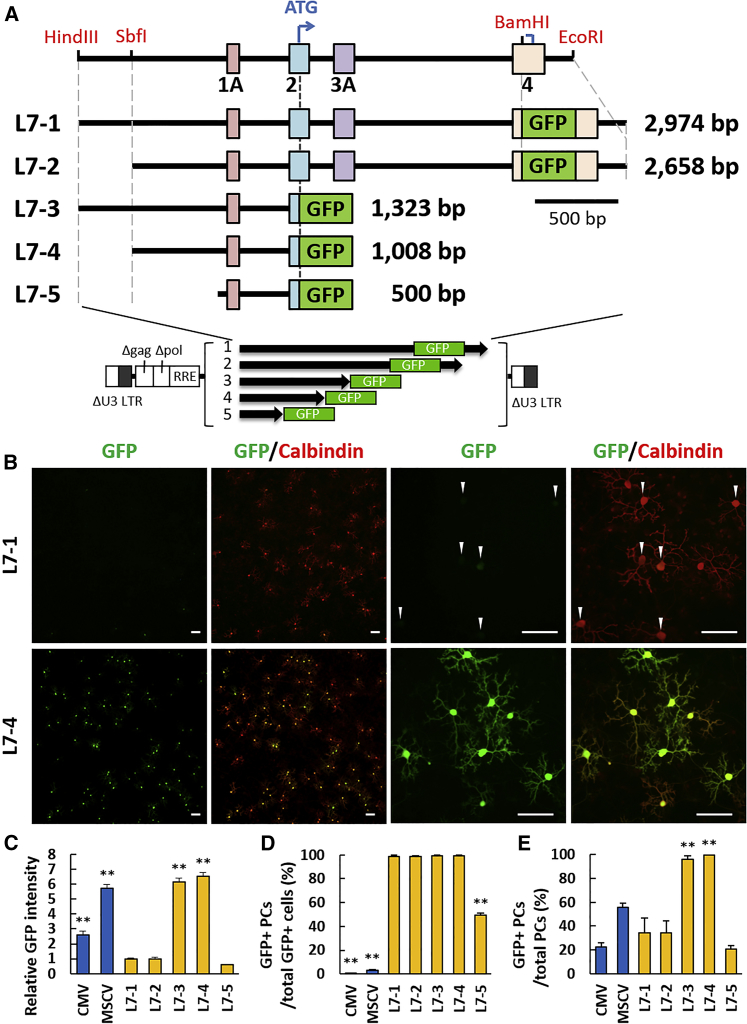
Figure 2.
Deletion of the L7 Distal Structural Gene Significantly Upregulates the Promoter Strength and Maintains the Specificity to Purkinje Cells
(A) Schema showing the original L7 promoter used for Purkinje cell-specific transgenic mouse (L7-1) and the deletion constructs (L7-2 to L7-5). The GFP gene was inserted at the BamHI site in exon 4 (L7-1 and L7-2) or after the original transcription initiation codon (ATG) present in exon 2 (L7-3 to L7-5). The size (base pair [bp]) of each construct without the GFP sequence is shown at the right side of each scheme. The L7 fragments were subcloned to the lentiviral plasmid as depicted. (B) Representative immunocytochemical images of the lentiviral GFP expression in cerebellar neurons under the control of L7-1 (upper) or L7-4 (lower). The neuronal culture was infected with lentiviral vectors at the day of plating and immunolabeled with GFP and calbindin D-28K at 14 days post-infection. Arrowheads indicate Purkinje cells. Scale bars, 100 μm. (C) Graph showing the GFP intensity relative to that induced by the L7-1 promoter. The GFP intensity in each promoter was calculated from 40 images randomly acquired from the culture. F(6,273) = 180.10; p < 0.001 by ANOVA. Error bars indicate SEM. (D) Graph showing the specificity of Purkinje cell transduction. Percent ratios of GFP-positive Purkinje cells to total GFP-positive cells are presented. F(6,33) = 4589.00; p < 0.001 by ANOVA. Error bars indicate SEM. (E) Graph showing the transduction efficiency of Purkinje cells. The ratios of GFP-positive Purkinje cells to total Purkinje cells are presented. F(6,33) = 55.50; p < 0.001 by ANOVA. Error bars indicate SEM. Data were obtained from eight (CMV, L7-3, and L7-4) and four (MSCV, L7-1, L7-2, and L7-5) independent cultures. **p < 0.01 by Bonferroni correction for post hoc test, as compared with the original L7-1 promoter.
We produced four truncated constructs (L7-2 to L7-5) by deleting the 5′ and/or 3′ sequence of L7-1 (Figure 2A). A major change was the site of GFP, which was shifted from exon 4 to exon 2; GFP in L7-3 to L7-5 was fused just downstream of the original start codon in exon 2. The five different lengths of the L7 gene constructs were subcloned into the lentiviral plasmid. Lentiviral vectors expressing GFP under the control of the constitutive CMV or MSCV promoter were used as controls. The primary culture of the rat cerebellum was infected with either of the lentiviral vectors (Figure S1). Compared with the L7-1 promoter, the GFP fluorescence intensity was more than six times higher when induced by the L7-3 promoter (6.13 ± 0.26; p < 0.001; Figures 2B and 2C; Figure S1), indicating a striking enhancement of the promoter strength by elimination of the structural gene downstream of the original start codon in exon 2. Notably, the Purkinje cell specificity, which was evaluated by the ratio of GFP-positive Purkinje cells relative to the total GFP-positive cells in the cell culture, was not compromised by the deletion of the distal half of the structural gene (98.88% ± 0.72% for L7-1 and 99.54 ± 0.30% for L7-3; p = 1.000; Figure 2D). Moreover, the transduction efficiency of the Purkinje cells, which was evaluated by the ratio of GFP-expressing Purkinje cells to total Purkinje cells in the culture, was significantly increased up to almost 100% in L7-3, in contrast with a low transduction efficiency in L7-1 (34.27% ± 12.45% for L7-1 and 96.13% ± 2.91% for L7-3; p < 0.001; Figure 2E). The L7-4, which lacked 0.3 kb from the 5′ end of the L7-3, showed similar Purkinje cell specificity to the L7-3 (99.54% ± 0.30% for L7-3 and 99.48% ± 0.52% for L7-4; p = 1.000). Similarly, the transduction efficiency of the Purkinje cells was 96.13% ± 2.91% for L7-3 and 99.61% ± 0.39% for L7-4; p = 1.000), but further deletion of 0.5 kb from the 5′ end of L7-4 (i.e., L7-5) significantly impaired the promoter strength (0.59 ± 0.04 times compared with the L7-1 promoter; p = 1.000), Purkinje cell specificity (49.20% ± 2.12%; p < 0.001 compared with L7-4), and transduction efficiency (20.83% ± 3.11%; p < 0.001 compared with L7-4; Figures 2C–2E). These results indicated that the 1-kb L7-4 fragment preserved the Purkinje cell specificity at least in the cerebellar culture and was superior to the 3-kb L7-1 in terms of the promoter strength.
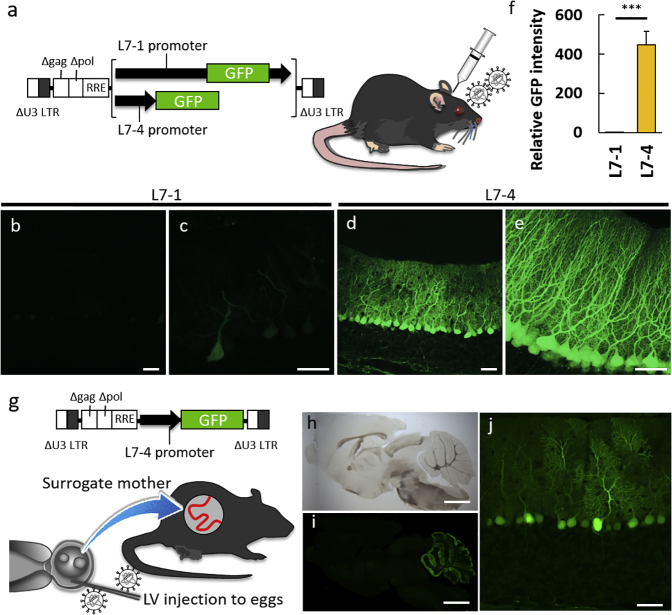
To examine whether these L7-4 properties were also maintained in vivo, we injected lentiviral vectors expressing GFP under the control of the L7-4 or L7-1 promoter into 4-week-old mouse cerebellum (Figure 3A). The cerebellum was examined 1 week after the injection using a confocal microscopy. Native GFP fluorescence from sagittal cerebellar sections revealed a Purkinje cell-specific GFP expression by both L7-1 and L7-4. However, consistent with the in vitro results, the GFP expression was significantly higher under the L7-4 control compared with L7-1, with a 449.04 ± 68.43 times higher relative intensity in the L7-4 compared with the L7-1 (p = 0.001; Figures 3B–3F). To confirm the Purkinje cell specificity of the L7-4 promoter, we generated a transgenic mouse by lentiviral injection into the perivitelline space of embryos (Figure 3G) as reported previously.36 Sagittal brain sections from the L7-4-GFP transgenic mouse revealed cerebellum-restricted (Figures 3H and 3I) and Purkinje cell-specific (Figure 3J) GFP expression. This is the first detailed description of the 1-kb L7-4 promoter and its selectivity to Purkinje cells since the patent application.27
Figure 3.
L7-4 Works as a Purkinje Cell-Specific Promoter in the Mouse Cerebellum In Vivo
(A) Schema depicting the lentiviral vectors expressing GFP by the L7-1 or L7-4 promoter and the viral injection to the mouse cerebellum. (B–E) Representative images of native GFP fluorescence on sagittal cerebellar sections at 1 week after viral injection. (B–E) The sections expressing GFP by the L7-1 (B and C) or L7-4 (D and E) promoter are presented. (F) Graph showing the relative GFP intensity of cerebellar sections lentivirally expressing GFP by the L7-4 promoter as compared with those induced by the original L7-1 promoter (n = 4 mice; t(6) = −6.55; ***p = 0.001 by Student’s t test). Error bars indicate SEM. (G) Schema describing the method of lentivirus-based generation of a transgenic mouse expressing GFP by the L7-4 promoter. (H–J) Bright-field (H) and native GFP fluorescent (I and J) images of a sagittal section of the whole brain (H and I) and the enlarged GFP image of the cerebellar cortex (J). Scale bars, 50 μm (A–D and J) and 2 mm (H and I).
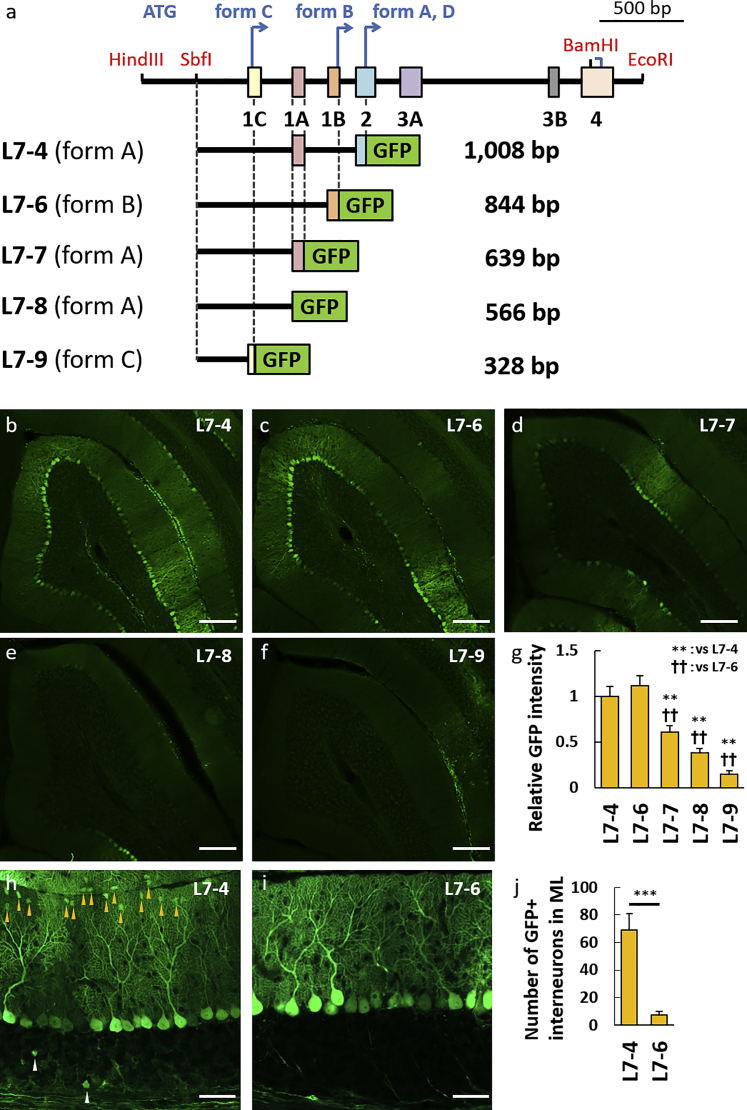
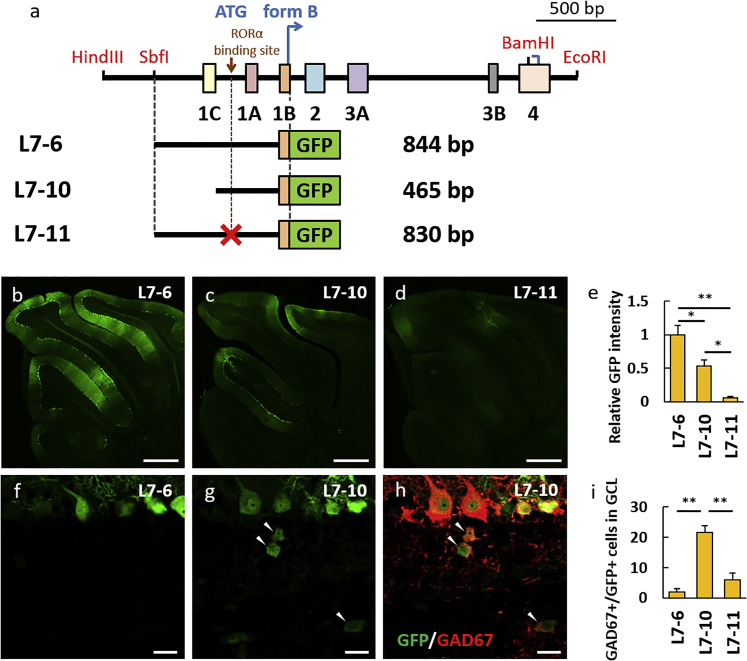
The 844 bp Fragment, L7-6, Upstream of the Start Codon in Exon 1B Serves as a Highly Purkinje Cell-Specific Promoter
Since the identification of the L7-4 sequence as an effective Purkinje cell-specific promoter suitable for viral vectors, the promoter has been widely used to date.28, 29, 30, 31 Nevertheless, we and others have noted a diffuse transduction in non-Purkinje cells in the molecular and granule cell layers. In addition, the L7 splice variants that use different ATG in exon 1 as a translation initiation codon (forms B and C) prompted us to narrow down the L7 further to a shorter promoter with a more restricted leakage to other cell types. As a result, four more constructs were produced by the 3′ deletion of the L7-4 sequence, in which GFP was fused following ATG in exon 1B (L7-6), following the 3′ end of exon 1A (L7-7), following the 5′ end of exon 1A (L7-8), and following ATG in exon 1C (L7-9; Figure 4A). After subcloning of these constructs in pCL20c, lentiviral vectors were produced and injected into 4-week-old mouse cerebellum to assess the GFP expression patterns in terms of the promoter strength and Purkinje cell specificity. The L7-6 promoter strength was comparable with that of L7-4, with a relative GFP intensity of 1.12 ± 0.11 times compared with that of L7-4 (p = 1.000). Compared with L7-4, the relative GFP intensity of the other deleted promoters, namely L7-7, L7-8, and L7-9, indicated a significantly lower promoter strength, with a relative GFP intensity of 0.61 ± 0.07 times (p = 0.006), 0.38 ± 0.05 times (p < 0.001), and 0.15 ± 0.04 times (p < 0.001), respectively (Figures 4B–4G). Notably, the leakage of the promoter activity into non-Purkinje cells in the molecular and granule cell layers was almost eliminated in L7-6, where the number of GFP-positive interneurons in the molecular layer of the lobule III was 68.83 ± 12.33 for L7-4 compared with 7.17 ± 2.97 for L7-6 (p = 0.001; Figures 4H–4J; Figure S2).
Figure 4.
The 0.8-kb Genome Region Upstream of the Transcription Initiation Codon in Exon 1B, L7-6, Shows a Similar Promoter Strength as L7-4 and Much Less Leaky Activity in Cortical Cells Other Than Purkinje Cells
(A) Schema depicting the L7-4 promoter and the 5′ side deletion constructs. Whereas the L7-4 promoter uses the transcription initiation codon in exon 2 as the form A and form D, the L7-6 and L7-9 promoters use different transcription initiation codons in exon 1B (L7-6) and exon 1C (L7-9) as the form B and form C, respectively. The L7-7 and L7-8 promoters are present or absent from exon 1A in the form A, respectively. The size (base pairs [bp]) of each construct without the GFP sequence is shown at the right side of each scheme. (B–F) Representative native GFP fluorescent images on sagittal sections of the cerebellum virally expressing GFP by the deleted promoters as depicted. Robust GFP expression was observed by L7-4 (B) and L7-6 (C), whereas the GFP expression was significantly decreased by deletion of the 3′ side (D–F). (G) Summary graph showing the GFP intensity relative to that by the L7-4 promoter. F(4,39) = 19.96; p < 0.001, ANOVA; **p < 0.01; ††p < 0.01 by Bonferroni correction for post hoc test, as compared with L7-4 or L7-6 promoter, respectively. Error bars indicate SEM. (H and I) Enlarged GFP fluorescent images of the cerebellar cortex virally expressing GFP by the L7-4 (H) or L7-6 (I) promoter. Arrowheads indicate GFP-labeled non-Purkinje cells. (J) Graph showing the number of GFP-positive non-Purkinje cells in the molecular layer. t(10) = 4.86; ***p = 0.001 by Student’s t test. Error bars indicate SEM. Statistical data were obtained using 11 (L7-4), 12 (L7-6), 8 (L7-7), 8 (L7-8), and 5 (L7-9) mice. Scale bars, 200 μm (B–F) and 50 μm (H and I).
L7-6 Serves as a Minimal Purkinje Cell-Specific Promoter That Is Activated upon the Binding of RORα
We further attempted to delete the 5′ side of the L7-6 without compromising the promoter characteristics. The L7-10 was produced by the deletion of 0.4 kb from the 5′ side of the L7-6 (Figure 5A). Subsequently, lentiviral vectors expressing GFP by the L7-10 promoter were injected into the mouse cerebellum. One week after the viral injection, confocal microscopic analysis revealed a significantly lower GFP expression evidenced by the GFP intensity (0.53 ± 0.10; p = 0.015) and lower specificity to Purkinje cells compared with L7-6-induced GFP expression (Figures 5B, 5C, and 5E). Numerous GFP/GAD67 double-positive cells were detected in the granule cell layer, which were significantly higher in the L7-10 (21.60 ± 2.25) compared with the L7-6 (2.00 ± 1.08; p < 0.001; Figures 5G–5I), indicating a significantly higher transduction of Golgi cells by the L7-10 compared with the L7-6 promoter. To examine whether the retinoic-acid-receptor-related orphan receptor α (RORα) binding to the L7-6 promoter was critical for the promoter strength, as reported in a previous study,37 we disrupted the RORα binding to the L7-6 by eliminating the binding site (L7-11 in Figure 5A). Subsequently, lentiviral vectors expressing GFP by the L7-11 promoter were injected into the mouse cerebellum. As evidenced by the GFP intensity, the GFP expression by the L7-11 (0.06 ± 0.03) was drastically decreased to about 5% of that induced by the L7-6 promoter (p < 0.001; Figures 5B, 5D, and 5E). These results confirm the critical role of RORα binding to the L7-6 sequence in the promoter activity.
Figure 5.
Regions Critical for Promoter Strength and Purkinje Cell Specificity in the L7-6
(A) Schema depicting the deletion constructs of the L7-6. L7-10 lacks approximately 380-bp deletion upstream of the L7-6. L7-11 is almost identical to L7-6 but lacks the RORα binding sequence. (B–D) Representative native GFP fluorescent images on sagittal sections of the cerebellum virally expressing GFP by the promoters as depicted. The bright GFP fluorescence was observed in the slice transduced by the L7-6 (B), which was significantly decreased in the slice transduced by L7-10 (C) and L7-11 (D). (E) Summary graph showing the GFP intensity relative to that by the L7-6 promoter. F(2,11) = 24.78; p < 0.001 by ANOVA. Error bars indicate SEM. (F–H) Enlarged images of the cerebellar cortex virally expressing GFP by the L7-6 (F) or L7-10 (G and H) promoter. Sections were double-labeled with GFP and GAD67. Arrowheads indicate examples of non-Purkinje cells double-labeled for GFP and GAD67 in the granule cell layer. (I) Graph showing the number of cells double-labeled for GFP and GAD67 in the granule cell layer. F(2,11) = 25.72; p < 0.001 by ANOVA. Error bars indicate SEM. Data were obtained using four (L7-6), five (L7-10), and five (L7-11) mice. *p < 0.05; **p < 0.01 by Bonferroni correction for post hoc test. Scale bars, 500 μm (B–D) and 20 μm (F–H).
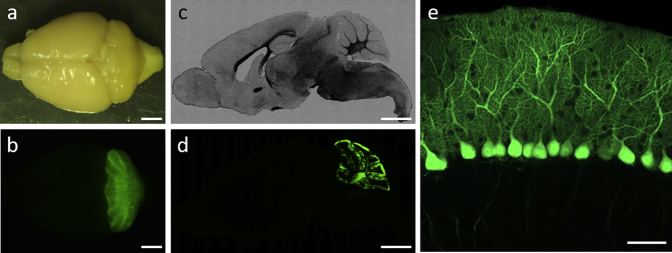
To verify the specificity of the L7-6 promoter in the CNS, we used AAV-PHP.B, which efficiently crosses the blood-brain barrier leading to diffuse transgene expression in almost the entire CNS regions when a ubiquitous promoter such as the CMV-β-actin-intron-β-globin hybrid (CAG) promoter is used.38 We administered AAV-PHP.B that expressed GFP under the control of the L7-6 promoter via the orbital venous plexus. Three weeks after the AAV-PHP.B injection, stereoscopic and fluorescent microscopic observations of the whole brain and the sagittal brain sections revealed marked GFP expression only in the cerebellum (Figures 6A–6D), which was specifically noted in Purkinje cells (Figure 6E). These findings confirm that the L7-6 promoter functions exclusively in the CNS Purkinje cells.
Figure 6.
Validation of the Cellular Specificity of the L7-6 by Intravenous Administration of AAV PHP.B Vectors That Are Highly Permeable to the Blood-Brain Barrier
(A–D) Bright-field (A and C) and native GFP fluorescent (B and D) images of the whole brain and the sagittal section. The mouse received intravenous injection of 100 μL of AAV PHP.B (3.0 × 1013 viral genomes/mL) and was sacrificed 3 weeks post-injection. (E) Magnification of the GFP fluorescent image at the cerebellar cortex. Scale bars, 2 mm (AD) and 50 μm (E).
In a transgenic mouse line that expressed high levels of humanized GFP under the control of the original L7-1 promoter, aggressive behavior reflected by bitten females was observed.39 Therefore, we intravenously injected high-titer AAV-PHP.B expressing GFP under the control of the L7-6 to adult male (n = 4) and female (n = 4) mice, and observed their behavior over a period of 2 months. Based on the results, none of the treated mice displayed aberrant behavioral phenotypes during the observation period, suggesting little influence of intravenously administered AAV-PHP.B and L7-6-driven GFP expression on the behavior of adult mice.
L7-6 Also Serves as a Purkinje Cell-Specific Promoter in Non-human Primates
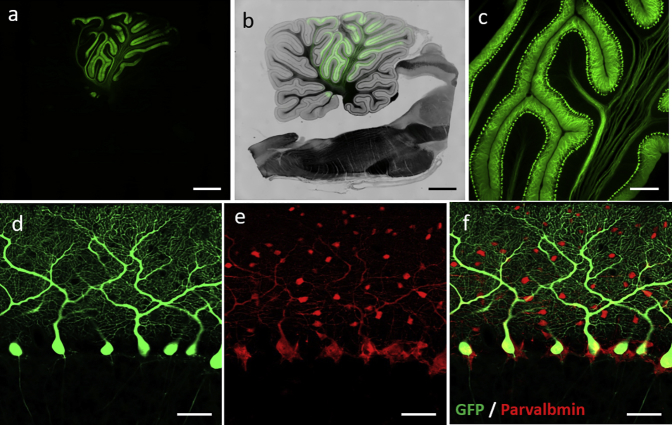
Because our observations were so far observed solely using rodents, we attempted to verify whether the Purkinje cell specificity of the L7-6 promoter is valid across species. For the in vivo transduction of the marmoset cerebellum, AAV9 vectors, instead of lentiviral vectors, were used because they are significantly smaller (∼20 nm) than lentiviral vectors (∼100 nm), diffuse more efficiently in the brain parenchyma, and are suitable for the transduction of a broader area in large animals. Thus, we injected AAV9 vectors expressing GFP under the control of the L7-6 promoter into the cerebellum of a common marmoset, which is a New World monkey widely used in neuroscience research.40, 41, 42, 43 Five weeks after the viral injection, the sagittal brain sections revealed a highly specific GFP expression in Purkinje cells (Figures 7A–7C), which was confirmed by immunohistochemical analysis against Parvalbumin (Figures 7D–7F). These results suggest that the L7-6 promoter is also Purkinje cell specific in non-human primates.
Figure 7.
The L7-6 Promoter Behaves Identically as a Purkinje Cell-Specific Promoter in Marmoset Brain
(A–C) Native GFP fluorescent image of the sagittal section of the marmoset cerebellum 5 weeks after the viral injection. A 1.9-year-old marmoset received cerebellar injection of 100 μL of AAV9 vectors expressing GFP by the mouse L7-6 promoter (1.0 × 1013 viral genomes/mL). Low-magnified images of GFP fluorescence alone (A) and GFP fluorescence superimposed on the bright field (B) and the enlarged GFP fluorescent image (C) were presented. (D–F) Immunohistochemical analysis of the sagittal section of the cerebellum expressing GFP under the control of the L7-6 promoter. The section was double-labeled with GFP and parvalbumin. Fluorescent images of GFP (D) and parvalbumin (E) and the merged image (F) are shown. Scale bars, 2 mm (A and B), 400 μm (C), and 50 μm (D–F).
Discussion
Truncation of promoters and their evaluation in vivo by viral vector-mediated expression is a useful strategy to engineer better promoters. Indeed, we previously succeeded in identifying minimal and critical promoter regions required for promoter strength and cell-type specificity in both the astrocyte-specific glial fibrillary acidic protein promoter44 and the neuron-specific enolase promoter.45 In the present study, using a similar approach, we demonstrated that the L7-6 sequence comprising the 844-bp genome region upstream of the transcription start codon in exon 1B was sufficient for a robust promoter strength and Purkinje cell-specific gene expression in the cerebellum of mouse and common marmoset. Moreover, the use of AAV-PHP.B, which easily crossed the blood-brain barrier and attained global neuronal and glial transduction in the CNS when administered intravenously,38 confirmed the absence of the L7-6 promoter activity in extra-cerebellar regions, at least in the viral vector-mediated expression system.
In the first part of this study, we used the proximal 0.9-kb promoter sequence and the L7 (form A) structural gene sequence containing four exons (L7-1 in Figure 2A) to identify the minimal sequence required for a Purkinje cell-specific and robust transgene expression. This region was selected because the 3-kb L7-1 sequence has been widely used for the generation of transgenic mice with Purkinje cell-specific transgene expression.14, 16, 17, 22, 24, 25 In the L7-1 composition used for those transgenic mice, the original transcription start codon in exon 2 and other possible start codons in every exon were disrupted, and the transgene containing a start codon was inserted into the BamHI site of exon 4. Obviously, this composition was unnatural. Therefore, we returned the transcription start codon to the original position in exon 2: we deleted the distal half of the structural gene and placed the transgene in exon 2 following the original ATG codon with a reading frame adjusted (L7-3). The lentivirus-mediated GFP expression by the L7-3 or L7-4 promoter, which had a 0.3-kb deletion at the 5′ side of the L7-3 promoter, exhibited a greater promoter strength compared with L7-1, without substantially compromising the specificity to Purkinje cells (Figure 2). This superior feature of the L7-4 promoter over the L7-1 promoter was confirmed also in the mouse cerebellum in vivo (Figures 3A–3F).
The prior 8-kb22 and 3-kb L7 promoters contain a 47-bp UTR at the 3′ end (3′ UTR) from the L7 gene, between the termination codon and the polyadenylation signal sequence.32 The microRNA (miRNA) sequence database, miRBase,46 predicted four miRNA binding elements in this region. Because miRNAs potentially silence mRNAs, the predicted miRNAs could contribute to the Purkinje cell-specific expression if the Purkinje cells selectively lacked the miRNA expression. However, the present results demonstrated that the distal half of the structural gene containing the 3′ UTR was overall dispensable for the Purkinje cell specificity of the L7 promoter. Alternatively, these miRNAs could silence the mRNA molecule in Purkinje cells, which could explain the weaker promoter strength of the L7-1 and presumably 8-kb promoters, compared with the L7-3, L7-4, and L7-6 promoters that lack the 3′ UTR from the L7 gene.
Previously, we and others have reported a subtle, but obvious, leakage of the L7-4 promoter activity into interneurons in the molecular layer (Figures 4H and 4J; Figure S2). Further deletion of 164 bp from the 3′ end of the L7-4 promoter (L7-6) drastically decreased the leakage rate (Figures 4I and 4J), suggesting that the leakage observed in the L7-4 is not due to the absence of the downstream structural gene (E3–E4). The 164-bp region deleted from the L7-4 promoter to obtain L7-6 contains a unique consensus sequence (5′-CACGTG-3′) for binding the upstream stimulatory transcription factor (USF)-1, a member of the eukaryotic evolutionary conserved basic-Helix-Loop-Helix-Leucine Zipper transcription factor family. Because the USF-1 participates in the activation of numerous genes implicated in a variety of functions and networks,47 the loss of its binding element may be responsible for the enhanced Purkinje cell specificity of the L7-6 promoter over the L7-4 promoter.
Using Patch, a pattern-based program for predicting transcription factor binding sites, we searched the L7-6 sequence for transcription factor binding elements other than RORα that were potentially critical for directing the transcription specificity in Purkinje cells. We identified a binding element for REV-ERBα, a nuclear receptor48 that competes with RORα for the RORE (retinoic-acid-related orphan receptor response element) site and represses transcription.49, 50, 51 Because RORα is abundant in Purkinje cells in addition to thalamic neurons,52, 53 the transcription upregulation by RORα likely dominates the repression by REV-ERBα. However, the L7 promoter activity is thought to be suppressed in other cell populations, in which the expression of REV-ERBα surpasses that of RORα.
On the other hand, the deletion of 379 bp from the 5′ side of the L7-6 promoter, corresponding to L7-10 (Figure 5A), increased significantly the leakage of the promoter activity into GAD67-positive cells in the granule cell layer (Figures 5A–5I). These results suggested that the 379 bp from the 5′ side of the L7-6 promoter play a role in suppressing the L7 promoter activity in off-target cells. Additionally, using Patch, we searched possible binding elements for transcription repressors in the 379-bp sequence and identified an element for the orphan nuclear receptor chicken ovalbumin upstream promoter-transcription factor (COUP-TF). COUP-TF binds the thyroid hormone response element (TRE) in the 5′ upstream region (A1TRE)54, 55 located within the 379-bp sequence and represses the triiodothyronine (T3)-dependent transcriptional activation of the L7 in immature Purkinje cells. COUP-TF is expressed in the fetal and early neonatal Purkinje cell (at around post-natal day [P] 6) and is diminished onward, leading to T3-dependent activation of the L7 promoter.56 COUP-TF was detected until P12 in other brain regions including the granule cell layer even after its disappearance from the Purkinje cells, although its expression in the adult brain has not yet been validated. Deletion of the 379-bp sequence containing the COUP-TF element may explain the emergence of the L7-10 activity in off-target cells such as GAD67-positive cells in the granule cell layer (Figures 5G and 5H).
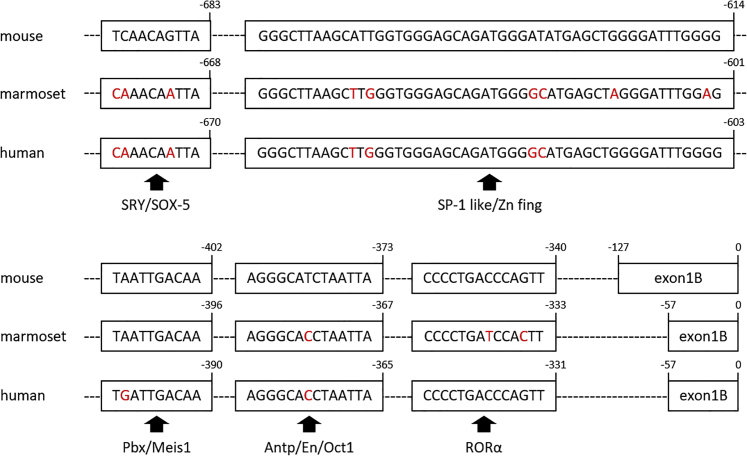
Common marmosets are gaining increasing attention as a potential experimental animal linking between rodents and humans.57 Furthermore, they are increasingly used in biomedical studies because of their many advantages over macaques, including: (1) the small size and easy handling, (2) the high reproductive efficiency, and (3) the lack of fatal zoonotic diseases such as the herpes B virus.41 Thus, viral vector-mediated expression of a transgene specifically in Purkinje cells of marmosets could advance the understanding of higher cerebellar functions such as fine finger movements and its involvement in mental processes. The structure and sequence of the L7 gene is comparable between humans and rodents. Indeed, similarly to rodents, the human L7 also has two alternative splice variants (forms A and B) and the human exon 1B, which contains the transcriptional start site for form B, lies within intron 1 of form A.33 This suggests a shared mechanism regulating the L7 gene expression between rodents and primates. Moreover, possible transcription factor binding sites including RORα are highly preserved among mice, marmosets, and humans (Figure 8). This was supported by our results that confirmed the mouse-derived L7-6 sequence to be also a Purkinje cell-specific promoter in marmosets (Figure 7). Thus, the mechanism underlying the robust and Purkinje cell-restricted expression is shared across those species, and the mouse L7-6 or the homologous human genome sequence may become available also for humans as a tool for gene therapy targeting Purkinje cells.
Figure 8.
Sequences for Validated RORα and Putative Transcription Factor Binding Sites in L7-6 Promoter Are Highly Conserved among Mouse, Marmoset, and Human
Boxed regions are proposed transcription factor binding sites of the L7-6 promoter.37 Numbers above the sequence are relative positions when the transcription initiation codon begins from +1. Notably, the retinoic-acid-receptor-related orphan receptor α (RORα) binding sites completely match between mouse and human, suggesting that the mouse L7-6 promoter effectively works in the human brain as proven in the marmoset brain.
Materials and Methods
Animals
All animal procedures were performed according to the institutional and national guidelines and were approved by the Institutional Committee of the Gunma University (12-013). We obtained Wistar rats, C57BL/6J (BL6), and ICR mice from Japan SLC. The mice were maintained on a 12-hr light/dark cycle with light turning on at 9 a.m. Food and water were freely accessible. One adult male common marmoset (Callithrix jacchus) born and raised at the Gunma University Bioresource Center was used for the AAV9 injection. The common marmoset was kept in a large cage (375 mm × 550 mm × 762 mm) equipped with wood branches and iron perches, and was housed under a 12-hr light/dark cycle with light turning on at 7 a.m. Temperature (26°C–28°C) and humidity (20%–70%) were also controlled. The marmoset had free access to water and a daily portion of food consisting of 40–50 g of soaked monkey chow (CMS-1; CLEA Japan) with vitamin supplements, fresh fruit, vegetables, boiled chicken, or milk powder. The handling of all animals was based on the Guide for the Care and Use of Laboratory Animals (8th edition). All efforts were made to minimize suffering and reduce the number of animals used.
Lentiviral Vector
The L7 promoter sequences were amplified using the primer sets summarized in Table S1 and were subcloned into pCL20c. To delete the RORα binding site, we amplified the DNA fragments upstream and downstream of the RORα binding site by using PCR with primer pairs of F1 and R1 or F2 and R2, respectively. Subsequently, two DNA fragments were ligated to generate L7-11 sequence using the In-Fusion HD Cloning Kit (Takara Bio). The lentiviral vectors were produced as previously described.58 Four plasmids (pCAGkGP1R, pCAG4RTR2, pCAG-VSV-G, and pCL20c/L7-GFP) were transfected into the HEK293T cells using the calcium phosphate precipitation method. The supernatant-containing virus particles were harvested 48 hr after the transfection, followed subsequently by ultracentrifugation. The precipitated viral particles were re-suspended in 70 μL of PBS. To measure the viral genomic titer, we obtained the genomic RNA from 1 μL of the viral solution using the RNeasy Mini Kit (QIAGEN). Subsequently, the genomic RNA was reverse transcribed by the ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo). Finally, real-time qPCR was performed using the Takara thermal cycler Dice TP800 system (Takara Bio) and SYBR Premix Ex TaqII (Takara Bio). The following primer pairs were used: EGFP-F, 5′-CGACCACTACCAGCAGAACAC-3′ and EGFP-R, 5′-TGTGATCGCGCTTCTCGTTGG-3′.
Cerebellar Neuronal Culture and Immunocytochemistry
Dissociated cerebellar neuronal cultures were prepared using 20- to 21-day-old Wistar rat fetuses according to a published protocol for Purkinje cell culture.59 In brief, rat cerebellar neurons were plated on a 12-well plate covered with plastic coverslips (200,000 cells per well), at a Purkinje-to-granule cells ratio of 1:20. After 3 hr, 1 mL of serum-free culture medium was added to each well. Half of the medium was replaced at 9 days in vitro (DIV). At 14 DIV, the cerebellar cultures were fixed in 4% paraformaldehyde in PBS for 2 hr at 4°C and then blocked with 2% bovine serum albumin and 0.4% Triton X-100 in PBS for 30 min. The cultures were incubated with the following primary antibodies for 2 hr at 24°C: rat monoclonal anti-GFP (1:1,000; 04404-84; Nacalai) and mouse monoclonal anti-calbindin D-28K (1:1,000; AB1778; Merck Millipore). Subsequently, the cultures were incubated with the following secondary antibodies for 1 hr at 24°C: Alexa Fluor 488 goat anti-rat IgG (1:1,000; A11006; Thermo Fisher Scientific) and Alexa Fluor 568 goat anti-mouse IgG (1:1,000; A11036; Thermo Fisher Scientific). Eight coverslips were used for the assay of the CMV, L7-3, and L7-4 promoters, whereas four coverslips were used for the MSCV, L7-1, L7-2, and L7-5 promoters. After fixation at 14 DIV, we counted the number of GFP-expressing Purkinje cells, the total number of Purkinje cells, and the number of GFP-expressing cells in each coverslip.
For the comparison of the relative GFP intensity, we randomly selected 40 visual fields per promoter and acquired the GFP fluorescent images using a fluorescent microscope (DMI6000 B; Leica Microsystems). Subsequently, the GFP fluorescence intensity was measured using ImageJ; the intensity obtained after subtraction of the background intensity was used for the comparison.
AAV9 and AAV-PHP.B Vectors
The packaging plasmid for the AAV-PHP.B (pAAV-PHP.B) was constructed by replacing the wild-type fragment between the BsiWI and PmeI sites of pAAV2/9 (kindly provided by Dr. J. Wilson) with the mutant capsid gene fragment. The mutant capsid gene between the BsiWI and PmeI sites was synthesized according to the PHP.B VP1 sequence (GenBank: KU056473 [3]; Eurofins Genomics). Subsequently, the mutant capsid gene fragment was subcloned into the pAAV2/9. Recombinant single-strand AAV9 or AAV-PHP.B vectors were generated by co-transfection of HEK293T cells with three plasmids: pAAV/L7-6-GFP-WPRE (woodchuck hepatitis post-transcriptional regulatory element [WPRE]), pHelper (Stratagene), and pAAV2/9 or pAAV-PHP.B. The viral particles were harvested from the media at day 6 post-transfection and were concentrated by precipitation with 8% polyethylene glycol 8000 (Sigma-Aldrich) with 500 mM sodium chloride. The precipitated viral particles were re-suspended in PBS and purified with iodixanol (OptiPrep; Axis-Shield Diagnostics) continuous gradient centrifugation. The viral solution was further concentrated and formulated in PBS using Vivaspin 20 (100,000 molecular weight cutoff [MWCO] polyethersulfone [PES]; Sartorius). The genomic titers of the purified AAV9 and AAV-PHP.B vectors were determined by real-time qPCR using THUNDERBIRD SYBR qPCR Mix (Toyobo) with the primers 5′-CTGTTGGGCACTGACAATTC-3′ and 5′-GAAGGGACGTAGCAGAAGGA-3′, targeting the WPRE sequence. The expression plasmid vectors were used as standards. The viral titers were 1.1 × 1014 vector genomes (vg)/mL (AAV9) and 3.7 × 1013 vg/mL (AAV-PHP.B).
Generation of the L7-4-GFP Transgenic Mouse
The L7-4-GFP transgenic mouse was generated as previously described.36, 58 Briefly, C57BL/6 mice were treated with serotropin (5 IU; ASKA Pharmaceutical) and gonadotropin (5 IU; ASKA Pharmaceutical) to induce superovulation. Subsequently, two-cell embryos were harvested by oviduct perfusion. Lentiviral vectors expressing GFP under the control of the L7-4 promoter were injected into the perivitelline space of these two-cell embryos using a Femto Jet microinjector (Eppendorf AG) through the Femto Tip (Eppendorf AG). The lentivirus-injected embryos were subsequently transplanted into the oviduct of pseudopregnant ICR mice.
Injection of the Lentiviral Vectors into the Mouse Cerebellum
After deep anesthesia by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg), a C57BL/6 mouse at P22–P27 was fixed on a stereotactic frame. The scalp was cut and a hole through the occipital bone was made 2 mm caudal from the lambda. The tip of a Hamilton syringe (33G) with an attached micro pump (Ultra Micro Pump II; World Precision Instrument [WPI]) was stereotactically inserted into the cerebellar vermis at a depth of 1.75 mm from the cranial bone surface. A total of 10 μL of the viral solution was injected using a microprocessor-based controller (Micro4; WPI) at a rate of 450 nL/min.
Injection of the AAV-PHP.B Vectors into the Mouse Orbital Venous Plexus
C57BL/6 mice at P36–P46 were anesthetized with the same procedure described above. The AAV-PHP.B vectors were loaded into a 29G syringe (SS-10M2913A; TERUMO). The mice were placed on their left side, and the facial skin was stretched to immobilize the face during the injection. Along the medial angle of the right eye, the needle was inserted into the right orbit until the tip reached the orbital bone, where the orbital venous plexus is located. An amount of 100 μL of the viral solution was injected during 1 min.
Mouse Histological Analysis
Mice were histologically analyzed at P28 (transgenic mouse) or 1 (lentiviral vectors) or 3 (AAV-PHP.B) weeks after the viral injection. After deep anesthesia, the mice were transcardially perfused through the left ventricle with PBS followed by 4% paraformaldehyde in 0.1 M phosphate buffer. Subsequently, 50-μm-thick sagittal sections of the whole brain or cerebellum were made using a microslicer (VT1000 S; Leica Microsystems). The intensity of GFP fluorescence of the cerebellar sections from lentivirus-treated mice was compared as described previously.44 Briefly, three sections per mouse were randomly selected, and a GFP fluorescence image of each slice was captured using a fluorescence microscope (BZ-X700; Keyence). The measurement of the fluorescence intensity was performed using ImageJ. The background intensity was subtracted for the comparisons.
For immunohistochemical analysis, free-floating brain sections were permeabilized with 0.1% Triton X-100 for 30 min at 24°C. Subsequently, sections were incubated either in a blocking solution (5% normal donkey serum, 0.5% Triton X-100, and 0.05% NaN3 in phosphate buffer) for 1 day at 4°C with the primary antibodies rat monoclonal anti-GFP (1:1000; 04404-84; Nacalai Tesque) and rabbit polyclonal anti-Parvalbumin (1:200; PV-Rb-Af750; Frontier Institute), or in a blocking solution (10% normal donkey serum, 3% bovine serum albumin, and 0.05% NaN3 in phosphate buffer) for 2 days at 24°C with mouse monoclonal anti-GAD67 antibody (1:200; MAB5406; Merck Millipore). To visualize the bound primary antibodies, we subsequently incubated sections with Alexa Fluor 488 donkey anti-rat IgG (1:1,000; Life Technologies), Alexa Fluor 568 donkey anti-rabbit IgG (1:1,000; Life Technologies), and Alexa Fluor 568 donkey anti-mouse IgG (1:1,000; Life Technologies) in blocking solution for 3 hr at 24°C. After the secondary antibody reaction, Nissl bodies were stained using the NeuroTrace 640/660 (1:200; Thermo Fisher Scientific) in PBS for 1 hr (or as required) at 24°C.
Injection of the AAV9 Vectors into the Marmoset Cerebellum
A 1.9-year-old male marmoset with a body weight (BW) of 339 g received an intramuscular injection of ketamine (25 mg/kg) and xylazine (2.0 mg/kg) for anesthesia. The anesthesia was further maintained through the constant inhalation of 2.0% isoflurane. After placing the marmoset into the stereotactic frame, the muscles on the occipital bone were separated bilaterally to make a burr hole at 3 mm posterior to the cross point of the sagittal and lambdoid sutures. The hole was made using a hand drill (SD-101; Narishige) with a drill head (BS1201; MINITOR). An AAV9 vector suspension (100 μL, 1.0 × 1013 vg/mL) was injected into the cerebellar cortex 6 mm deep from the occipital bone surface at a rate of 3.3 μL/min. The syringe was kept in place for 2 min after the injection to avoid the viral reversion. Vital signs of the marmoset were monitored throughout the procedure.
Marmoset Histological Analysis
Five weeks after the viral injection, the marmoset was anesthetized as described above and perfused intracardially with PBS followed by 4% paraformaldehyde in 0.1 M phosphate buffer. The cerebellum was subsequently dissected, and 100-μm-thick sections were cut and immunostained using the same protocol as in mice. Primary antibodies used were rat monoclonal anti-GFP (1:1,000; 04404-84; Nacalai Tesque) and rabbit polyclonal anti-Parvalbumin (1:200: PV-Rb-Af750; Frontier Institute), whereas the secondary antibodies were Alexa Fluor 488 donkey anti-rat IgG (1:1,000; Life Technologies) and Alexa Fluor 568 donkey anti-rabbit IgG (1:1,000; Life Technologies).
Experimental Design and Statistical Analysis
We used the Student’s t test to compare between the two experimental groups, whereas ANOVA and Bonferroni correction for post hoc test were used to compare among more than three groups. Data were expressed as the mean ± SEM. All statistical analyses were performed using SPSS v22 (IBM Japan). The sample size in each experiment was estimated by the power analysis using R v3.3.1 (R Foundation for Statistical Computing). Briefly, we used a power of 80% and a significance level of 0.05. Subsequently, a set of parameters including delta and SD were selected when using the Student’s t test, whereas a set of parameters including between variance and within variance were selected when using ANOVA. In the rat primary cerebellar neuronal cultures, more than four samples for GFP intensity (Figure 2C) or four coverslips for the transduction ratios (Figures 2D and 2E) were required per promoter based on calculations using a power of 80%, significance level of 0.05, and a between and within variance of 1 each. Data were obtained from two to three independent cultures, and quantification was performed by at least two persons who were blinded to the experimental conditions.
In a similar manner, to compare the GFP intensity among mouse groups virally expressing GFP by different L7 promoter constructs, we required four (Figure 3F), five (Figure 4G), or four (Figure 5E) mice per promoter. To compare the number of GFP-positive interneurons in the molecular layer (Figure 4J) or the number of GAD67/GFP double-positive cells (Figure 5I), six or four mice per promoter were necessary, respectively. We used three tissue sections per mouse and utilized the average value for the mouse data.
Four mice received an intravenous injection of AAV-PHP.B to validate the presence or absence of the L7-6 promoter activity in extra-cerebellar regions. To examine the Purkinje cell specificity of the L7-6 promoter in the non-human primate cerebellum, we used only one marmoset to minimize the number of animals used.
Author Contributions
Conceptualization, K.N., Y.M., H.H.; Methodology, K.N., Y.M., H.H.; Investigation, K.N., Y.M., A.K.; Writing – Original Draft, K.N., Y.M., A.K., H.H.; Writing – Review & Editing, K.N., Y.M., A.K., H.H.; Supervision, H.H.
Conflicts of Interest
The Purkinje cell-tropic viral vector has been patented.27
Acknowledgments
The authors thank Asako Ohnishi, Junko Sugiyama, Motoko Uchiyama, and Minako Noguchi for the AAV9 vector production, mice genotyping, and raising the marmoset. This study was partially supported by the program for Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from the Ministry of Education, Culture, and Sports Science (MEXT), Japan Agency for Medical Research and Development (AMED), and Gunma University Initiative for Advanced Research (GIAR) (to H.H.).
Footnotes
Supplemental Information includes two figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2017.07.006.
Supplemental Information
References
- 1.Bellebaum C., Daum I. Cerebellar involvement in executive control. Cerebellum. 2007;6:184–192. doi: 10.1080/14734220601169707. [DOI] [PubMed] [Google Scholar]
- 2.Peterburs J., Desmond J.E. The role of the human cerebellum in performance monitoring. Curr. Opin. Neurobiol. 2016;40:38–44. doi: 10.1016/j.conb.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collinson S.L., Anthonisz B., Courtenay D., Winter C. Frontal executive impairment associated with paraneoplastic cerebellar degeneration: a case study. Neurocase. 2006;12:350–354. doi: 10.1080/13554790601076697. [DOI] [PubMed] [Google Scholar]
- 4.Fiez J.A., Petersen S.E., Cheney M.K., Raichle M.E. Impaired non-motor learning and error detection associated with cerebellar damage. A single case study. Brain. 1992;115:155–178. doi: 10.1093/brain/115.1.155. [DOI] [PubMed] [Google Scholar]
- 5.Gebhart A.L., Petersen S.E., Thach W.T. Role of the posterolateral cerebellum in language. Ann. N Y Acad. Sci. 2002;978:318–333. doi: 10.1111/j.1749-6632.2002.tb07577.x. [DOI] [PubMed] [Google Scholar]
- 6.Janssen G., Messing-Jünger A.M., Engelbrecht V., Göbel U., Bock W.J., Lenard H.G. Cerebellar mutism syndrome. Klin. Padiatr. 1998;210:243–247. doi: 10.1055/s-2008-1043886. [DOI] [PubMed] [Google Scholar]
- 7.Leggio M.G., Silveri M.C., Petrosini L., Molinari M. Phonological grouping is specifically affected in cerebellar patients: a verbal fluency study. J. Neurol. Neurosurg. Psychiatry. 2000;69:102–106. doi: 10.1136/jnnp.69.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito M. Control of mental activities by internal models in the cerebellum. Nat. Rev. Neurosci. 2008;9:304–313. doi: 10.1038/nrn2332. [DOI] [PubMed] [Google Scholar]
- 9.Aiba A., Kano M., Chen C., Stanton M.E., Fox G.D., Herrup K., Zwingman T.A., Tonegawa S. Deficient cerebellar long-term depression and impaired motor learning in mGluR1 mutant mice. Cell. 1994;79:377–388. [PubMed] [Google Scholar]
- 10.Kakizawa S., Kishimoto Y., Hashimoto K., Miyazaki T., Furutani K., Shimizu H., Fukaya M., Nishi M., Sakagami H., Ikeda A. Junctophilin-mediated channel crosstalk essential for cerebellar synaptic plasticity. EMBO J. 2007;26:1924–1933. doi: 10.1038/sj.emboj.7601639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kina S., Tezuka T., Kusakawa S., Kishimoto Y., Kakizawa S., Hashimoto K., Ohsugi M., Kiyama Y., Horai R., Sudo K. Involvement of protein-tyrosine phosphatase PTPMEG in motor learning and cerebellar long-term depression. Eur. J. Neurosci. 2007;26:2269–2278. doi: 10.1111/j.1460-9568.2007.05829.x. [DOI] [PubMed] [Google Scholar]
- 12.Maejima T., Hashimoto K., Yoshida T., Aiba A., Kano M. Presynaptic inhibition caused by retrograde signal from metabotropic glutamate to cannabinoid receptors. Neuron. 2001;31:463–475. doi: 10.1016/s0896-6273(01)00375-0. [DOI] [PubMed] [Google Scholar]
- 13.Offermanns S., Hashimoto K., Watanabe M., Sun W., Kurihara H., Thompson R.F., Inoue Y., Kano M., Simon M.I. Impaired motor coordination and persistent multiple climbing fiber innervation of cerebellar Purkinje cells in mice lacking Galphaq. Proc. Natl. Acad. Sci. USA. 1997;94:14089–14094. doi: 10.1073/pnas.94.25.14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Zeeuw C.I., Hansel C., Bian F., Koekkoek S.K., van Alphen A.M., Linden D.J., Oberdick J. Expression of a protein kinase C inhibitor in Purkinje cells blocks cerebellar LTD and adaptation of the vestibulo-ocular reflex. Neuron. 1998;20:495–508. doi: 10.1016/s0896-6273(00)80990-3. [DOI] [PubMed] [Google Scholar]
- 15.Goossens J., Daniel H., Rancillac A., van der Steen J., Oberdick J., Crépel F., De Zeeuw C.I., Frens M.A. Expression of protein kinase C inhibitor blocks cerebellar long-term depression without affecting Purkinje cell excitability in alert mice. J. Neurosci. 2001;21:5813–5823. doi: 10.1523/JNEUROSCI.21-15-05813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirai H., Miyazaki T., Kakegawa W., Matsuda S., Mishina M., Watanabe M., Yuzaki M. Rescue of abnormal phenotypes of the delta2 glutamate receptor-null mice by mutant delta2 transgenes. EMBO Rep. 2005;6:90–95. doi: 10.1038/sj.embor.7400312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichise T., Kano M., Hashimoto K., Yanagihara D., Nakao K., Shigemoto R., Katsuki M., Aiba A. mGluR1 in cerebellar Purkinje cells essential for long-term depression, synapse elimination, and motor coordination. Science. 2000;288:1832–1835. doi: 10.1126/science.288.5472.1832. [DOI] [PubMed] [Google Scholar]
- 18.Korhonen L., Hansson I., Maugras C., Wehrle R., Kairisalo M., Borgkvist A., Jokitalo E., Sotelo C., Fisone G., Dusart I., Lindholm D. Expression of X-chromosome linked inhibitor of apoptosis protein in mature Purkinje cells and in retinal bipolar cells in transgenic mice induces neurodegeneration. Neuroscience. 2008;156:515–526. doi: 10.1016/j.neuroscience.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka S., Kawaguchi S.Y., Shioi G., Hirano T. Long-term potentiation of inhibitory synaptic transmission onto cerebellar Purkinje neurons contributes to adaptation of vestibulo-ocular reflex. J. Neurosci. 2013;33:17209–17220. doi: 10.1523/JNEUROSCI.0793-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordquist D.T., Kozak C.A., Orr H.T. cDNA cloning and characterization of three genes uniquely expressed in cerebellum by Purkinje neurons. J. Neurosci. 1988;8:4780–4789. doi: 10.1523/JNEUROSCI.08-12-04780.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberdick J., Levinthal F., Levinthal C. A Purkinje cell differentiation marker shows a partial DNA sequence homology to the cellular sis/PDGF2 gene. Neuron. 1988;1:367–376. doi: 10.1016/0896-6273(88)90186-9. [DOI] [PubMed] [Google Scholar]
- 22.Oberdick J., Smeyne R.J., Mann J.R., Zackson S., Morgan J.I. A promoter that drives transgene expression in cerebellar Purkinje and retinal bipolar neurons. Science. 1990;248:223–226. doi: 10.1126/science.2109351. [DOI] [PubMed] [Google Scholar]
- 23.Smeyne R.J., Chu T., Lewin A., Bian F., Sanlioglu S., Kunsch C., Lira S.A., Oberdick J. Local control of granule cell generation by cerebellar Purkinje cells. Mol. Cell. Neurosci. 1995;6:230–251. doi: 10.1006/mcne.1995.1019. [DOI] [PubMed] [Google Scholar]
- 24.Tomomura M., Rice D.S., Morgan J.I., Yuzaki M. Purification of Purkinje cells by fluorescence-activated cell sorting from transgenic mice that express green fluorescent protein. Eur. J. Neurosci. 2001;14:57–63. doi: 10.1046/j.0953-816x.2001.01624.x. [DOI] [PubMed] [Google Scholar]
- 25.Torashima T., Koyama C., Iizuka A., Mitsumura K., Takayama K., Yanagi S., Oue M., Yamaguchi H., Hirai H. Lentivector-mediated rescue from cerebellar ataxia in a mouse model of spinocerebellar ataxia. EMBO Rep. 2008;9:393–399. doi: 10.1038/embor.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirai, H., and Torashima, T. December 2007. Purkinje cell-tropic viral vector. U.S. Patent 8912315 B2.
- 28.Miki T., Hirai H., Takahashi T. Activity-dependent neurotrophin signaling underlies developmental switch of Ca2+ channel subtypes mediating neurotransmitter release. J. Neurosci. 2013;33:18755–18763. doi: 10.1523/JNEUROSCI.3161-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawada Y., Kajiwara G., Iizuka A., Takayama K., Shuvaev A.N., Koyama C., Hirai H. High transgene expression by lentiviral vectors causes maldevelopment of Purkinje cells in vivo. Cerebellum. 2010;9:291–302. doi: 10.1007/s12311-010-0161-1. [DOI] [PubMed] [Google Scholar]
- 30.Sawada Y., Konno A., Nagaoka J., Hirai H. Inflammation-induced reversible switch of the neuron-specific enolase promoter from Purkinje neurons to Bergmann glia. Sci. Rep. 2016;6:27758. doi: 10.1038/srep27758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uesaka N., Uchigashima M., Mikuni T., Nakazawa T., Nakao H., Hirai H., Aiba A., Watanabe M., Kano M. Retrograde semaphorin signaling regulates synapse elimination in the developing mouse brain. Science. 2014;344:1020–1023. doi: 10.1126/science.1252514. [DOI] [PubMed] [Google Scholar]
- 32.Vandaele S., Nordquist D.T., Feddersen R.M., Tretjakoff I., Peterson A.C., Orr H.T. Purkinje cell protein-2 regulatory regions and transgene expression in cerebellar compartments. Genes Dev. 1991;5:1136–1148. doi: 10.1101/gad.5.7.1136. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X., Zhang H., Oberdick J. Conservation of the developmentally regulated dendritic localization of a Purkinje cell-specific mRNA that encodes a G-protein modulator: comparison of rodent and human Pcp2(L7) gene structure and expression. Brain Res. Mol. Brain Res. 2002;105:1–10. doi: 10.1016/s0169-328x(02)00379-0. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y., Sulaiman P., Feddersen R.M., Liu J., Smith R.G., Vardi N. Retinal ON bipolar cells express a new PCP2 splice variant that accelerates the light response. J. Neurosci. 2008;28:8873–8884. doi: 10.1523/JNEUROSCI.0812-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barski J.J., Denker B.M., Guan J., Lauth M., Spreafico F., Fertala A., Meyer M. Developmental upregulation of an alternative form of pcp2 with reduced GDI activity. Cerebellum. 2014;13:207–214. doi: 10.1007/s12311-013-0529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oue M., Handa H., Matsuzaki Y., Suzue K., Murakami H., Hirai H. The murine stem cell virus promoter drives correlated transgene expression in the leukocytes and cerebellar Purkinje cells of transgenic mice. PLoS ONE. 2012;7:e51015. doi: 10.1371/journal.pone.0051015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serinagaoglu Y., Zhang R., Zhang Y., Zhang L., Hartt G., Young A.P., Oberdick J. A promoter element with enhancer properties, and the orphan nuclear receptor RORalpha, are required for Purkinje cell-specific expression of a Gi/o modulator. Mol. Cell. Neurosci. 2007;34:324–342. doi: 10.1016/j.mcn.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X., Baader S.L., Bian F., Müller W., Oberdick J. High level Purkinje cell specific expression of green fluorescent protein in transgenic mice. Histochem. Cell Biol. 2001;115:455–464. doi: 10.1007/s004180100283. [DOI] [PubMed] [Google Scholar]
- 40.Burkart J.M., Finkenwirth C. Marmosets as model species in neuroscience and evolutionary anthropology. Neurosci. Res. 2015;93:8–19. doi: 10.1016/j.neures.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Kishi N., Sato K., Sasaki E., Okano H. Common marmoset as a new model animal for neuroscience research and genome editing technology. Dev. Growth Differ. 2014;56:53–62. doi: 10.1111/dgd.12109. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell J.F., Leopold D.A. The marmoset monkey as a model for visual neuroscience. Neurosci. Res. 2015;93:20–46. doi: 10.1016/j.neures.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okano H., Mitra P. Brain-mapping projects using the common marmoset. Neurosci. Res. 2015;93:3–7. doi: 10.1016/j.neures.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 44.Shinohara Y., Konno A., Takahashi N., Matsuzaki Y., Kishi S., Hirai H. Viral vector-based dissection of marmoset GFAP promoter in mouse and marmoset brains. PLoS ONE. 2016;11:e0162023. doi: 10.1371/journal.pone.0162023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinohara Y., Ohtani T., Konno A., Hirai H. Viral vector-based evaluation of regulatory regions in the neuron-specific enolase (NSE) promoter in mouse cerebellum in vivo. Cerebellum. 2017 doi: 10.1007/s12311-017-0866-5. Published online May 15, 2017. [DOI] [PubMed] [Google Scholar]
- 46.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 47.Corre S., Galibert M.D. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 48.Ueda H.R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 49.Guillaumond F., Dardente H., Giguère V., Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 50.Kojetin D.J., Burris T.P. REV-ERB and ROR nuclear receptors as drug targets. Nat. Rev. Drug Discov. 2014;13:197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sierk M.L., Zhao Q., Rastinejad F. DNA deformability as a recognition feature in the reverb response element. Biochemistry. 2001;40:12833–12843. doi: 10.1021/bi011086r. [DOI] [PubMed] [Google Scholar]
- 52.Jetten A.M., Joo J.H. Retinoid-related orphan receptors (RORs): roles in cellular differentiation and development. Adv. Dev. Biol. 2006;16:313–355. doi: 10.1016/S1574-3349(06)16010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konno A., Shuvaev A.N., Miyake N., Miyake K., Iizuka A., Matsuura S., Huda F., Nakamura K., Yanagi S., Shimada T., Hirai H. Mutant ataxin-3 with an abnormally expanded polyglutamine chain disrupts dendritic development and metabotropic glutamate receptor signaling in mouse cerebellar Purkinje cells. Cerebellum. 2014;13:29–41. doi: 10.1007/s12311-013-0516-5. [DOI] [PubMed] [Google Scholar]
- 54.Hagen S.G., Larson R.J., Strait K.A., Oppenheimer J.H. A Purkinje cell protein-2 intronic thyroid hormone response element binds developmentally regulated thyroid hormone receptor-nuclear protein complexes. J. Mol. Neurosci. 1996;7:245–255. doi: 10.1007/BF02737062. [DOI] [PubMed] [Google Scholar]
- 55.Zou L., Hagen S.G., Strait K.A., Oppenheimer J.H. Identification of thyroid hormone response elements in rodent Pcp-2, a developmentally regulated gene of cerebellar Purkinje cells. J. Biol. Chem. 1994;269:13346–13352. [PubMed] [Google Scholar]
- 56.Anderson G.W., Larson R.J., Oas D.R., Sandhofer C.R., Schwartz H.L., Mariash C.N., Oppenheimer J.H. Chicken ovalbumin upstream promoter-transcription factor (COUP-TF) modulates expression of the Purkinje cell protein-2 gene. A potential role for COUP-TF in repressing premature thyroid hormone action in the developing brain. J. Biol. Chem. 1998;273:16391–16399. doi: 10.1074/jbc.273.26.16391. [DOI] [PubMed] [Google Scholar]
- 57.Izpisua Belmonte J.C., Callaway E.M., Caddick S.J., Churchland P., Feng G., Homanics G.E., Lee K.F., Leopold D.A., Miller C.T., Mitchell J.F. Brains, genes, and primates. Neuron. 2015;86:617–631. doi: 10.1016/j.neuron.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuzaki Y., Oue M., Hirai H. Generation of a neurodegenerative disease mouse model using lentiviral vectors carrying an enhanced synapsin I promoter. J. Neurosci. Methods. 2014;223:133–143. doi: 10.1016/j.jneumeth.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 59.Hirai H., Launey T. The regulatory connection between the activity of granule cell NMDA receptors and dendritic differentiation of cerebellar Purkinje cells. J. Neurosci. 2000;20:5217–5224. doi: 10.1523/JNEUROSCI.20-14-05217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.