Abstract
The pregnane X receptor (PXR) and constitutive androstane receptor (CAR) are well-known xenobiotic-sensing nuclear receptors with overlapping functions. However, there lacks a quantitative characterization to distinguish between the PXR and CAR target genes and signaling pathways in liver. The present study performed a transcriptomic comparison of the PXR- and CAR-targets using RNA-Seq in livers of adult wild-type mice that were treated with the prototypical PXR ligand PCN (200 mg/kg, i.p. once daily for 4 days in corn oil) or the prototypical CAR ligand TCPOBOP (3 mg/kg, i.p., once daily for 4 days in corn oil). At the given doses, TCPOBOP differentially regulated many more genes (2125) than PCN (212), and 147 of the same genes were differentially regulated by both chemicals. As expected, the top pathways differentially regulated by both PCN and TCPOBOP were involved in xenobiotic metabolism, and they also up-regulated genes involved in retinoid metabolism, but down-regulated genes involved in inflammation and iron homeostasis. Regarding unique pathways, PXR activation appeared to overlap with the aryl hydrocarbon receptor signaling, whereas CAR activation appeared to overlap with the farnesoid X receptor signaling, acute-phase response, and mitochondrial dysfunction. The mRNAs of differentially regulated drug-processing genes (DPGs) partitioned into three patterns, namely TCPOBOP-induced, PCN-induced, as well as TCPOBOP-suppressed gene clusters. The cumulative mRNAs of the differentially regulated drug-processing genes (DPGs), phase-I and –II enzymes, as well as efflux transporters were all up-regulated by both PCN and TCPOBOPOP, whereas the cumulative mRNAs of the uptake transporters were down-regulated only by TCPOBOP. The absolute mRNA abundance in control and receptor-activated conditions was examined in each DPG category to predict the contribution of specific DPG genes in the PXR/CAR-mediated pharmacokinetic responses. The preferable differential regulation by TCPOBOP in the entire hepatic transcriptome correlated with a marked change in the expression of many DNA and histone epigenetic modifiers. In conclusion, the present study has revealed known and novel, as well as common and unique targets of PXR and CAR in mouse liver following pharmacological activation using their prototypical ligands. Results from this study will further support the role of these receptors in regulating the homeostasis of xenobiotic and intermediary metabolism in liver, and aid in distinguishing between PXR and CAR signaling at various physiological and pathophysiological conditions.
INTRODUCTION
Liver is a critical organ for drug metabolism, transport, as well as nutrient homeostasis. PXR and CAR are well known xenobiotic-sensing transcription factors that trans-activate a large battery of genes involved in xenobiotic metabolism and transport (together called “drug-processing genes” [DPGs]) (Kliewer et al., 1998; Wei et al., 2000; Kliewer et al., 2002; Hernandez et al., 2009). PXR was first characterized as a steroid-activating receptor (Kliewer et al., 1998). CAR was characterized the following year by Dr. David Moore’s group, as an important regulator for the response evoked by a class of xenobiotics known as “phenobarbital-like inducers” (Wei et al., 2000). Accumulating data in the literature have further characterized these two nuclear receptors regarding their tissue distribution, ligands, target genes, and physiological/pharmacological functions. Both PXR and CAR are preferably expressed in liver and gastrointestinal tract in humans and rodents (Nishimura et al., 2004; Bookout et al., 2006; Petrick and Klaassen, 2007).
PXR is a promiscuous transcription factor and it is activated by many xenobiotics, including various therapeutic drugs such as dexamethasone, paclitaxel, and rifampicin (Moore et al., 2003), environmental toxicants such as the flame retardants polybrominated diphenol ethers (Pacyniak et al., 2007), various pesticides (Lemaire et al., 2006), herbal supplements such as St. John’s wort (Moore and Kliewer, 2000; Moore et al., 2000), as well as intestinal microbial metabolites lithocholic acid and indole-3 propionic acid (Staudinger et al., 2001; Venkatesh et al., 2014). Pregnenolone-16α-carbonitrile (PCN) is a specific ligand for PXR in rodents, whereas rifampicin is a specific ligand for PXR in humans (Moore et al., 2003).
Regarding ligands for CAR, the endogenous CAR ligands androstenol and androstanol are repressors for the constitutive activity of CAR (Forman et al., 1998), whereas endogenous CAR activators are poorly characterized. Although phenobarbital and lithocholic acid are not direct CAR ligands, they can facilitate the nuclear localization of CAR and the subsequent up-regulation of the CAR target genes (Swales and Negishi, 2004; Beilke et al., 2009a; Beilke et al., 2009b). The 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-tetrachloro-1,4 bis(pyridyloxy)benzene (TCPOBOP) is a potent and selective CAR activator in mice, whereas 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) is a selective CAR activator in humans (Swales and Negishi, 2004). The flame retardant polybrominated diphenyl ether 47 is an activator of both mouse and human CAR (Sueyoshi et al., 2014).
Upon activation, both PXR and CAR are well known to up-regulate the transcription of numerous DPGs involved in phase-I and phase-II drug metabolism as well as transport (Cheng et al., 2005; Maher et al., 2005; Vyhlidal et al., 2006; Petrick and Klaassen, 2007; Buckley and Klaassen, 2009; Cui et al., 2009; Hernandez et al., 2009; Aleksunes and Klaassen, 2012; Cheng and Klaassen, 2012). Due to the critical roles of PXR and CAR in regulating the DPGs and the subsequent modifications on the metabolism and disposition of xenobiotics and endobiotics, these nuclear receptors are responsible for many adverse drug reactions (Willson and Kliewer, 2002).
Besides its critical role in drug metabolism and transport, liver also regulates intermediary metabolism such as lipogenesis, gluconeogenesis, protein synthesis, iron homeostasis, as well as cholesterol metabolism and bile acid synthesis (Nemeth and Ganz, 2006; Chiang, 2009a; Chiang, 2009b; Bechmann et al., 2012). Liver utilizes carbohydrates, free fatty acids, and amino acids to generate glucose and ketone bodies that can be utilized by other tissues to generate energy. In addition, liver produces very-low density proteins (VLDLs) to transport triglycerides to adipose tissues for storage (Zammit and Moir, 1994; Zammit, 1996; Casteels and Mathieu, 2003; Renaud et al., 2014).
Various, nuclear receptors coordinately regulate the interactions between xenobiotic and endobiotic metabolism, and are implicated in various liver diseases such as obesity and inflammation (Chiang, 2005; Arrese and Karpen, 2010; Wagner et al., 2011). Although PXR and CAR are well-known to enhance drug metabolism, recent studies in the literature have also unveiled their novel functions in intermediary metabolism. Activation of mouse PXR in liver leads to increased deposit of triglycerides in liver, and this is associated with increased expression of the free fatty acid transporter CD36, which is a direct PXR-target gene (Zhou et al., 2006; Zhou et al., 2008). Activation of human PXR in HepG2 cells promotes lipid synthesis, and this is associated with increased expression of stearoyl-CoA desaturase-1 (SCD1), which is a direct PXR-target gene (Zhang et al., 2013). Chronic exposure to rifaximin causes hepatic steatosis in PXR-humanized mice (Cheng et al., 2012); PXR-humanized mice fed a high-fat diet gain more body weight, exhibit hyperinsulinemia, and impaired glucose tolerance (Spruiell et al., 2014); whereas PXR depletion alleviates diet-induced and genetic obesity as well as insulin resistance in mice (He et al., 2013). Conversely, CAR has been found to be an anti-obesity sensor that regulates glucose and lipid metabolism, and improves insulin sensitivity in mice (Gao et al., 2009; Yan et al., 2015). In human hepatocytes, activation of CAR inhibits gluconeogenesis without suppressing fatty acid synthesis (Lynch et al., 2014). Therefore, it has been suggested that CAR represents an attractive therapeutic target to manage obesity and type-II diabetes (Nandchahal et al., 1990).
PXR and CAR share marked similarities regarding their transcriptional activities and target genes. Pharmacological activation of PXR and CAR lead to the up-regulation of many common DPGs in liver (Aleksunes and Klaassen, 2012). Both PXR and CAR prefer direct repeat-4 (DR-4) DNA binding motifs (Makinen et al., 2002; Frank et al., 2003; Cui et al., 2010), which likely explains the common regulatory patterns of their target genes. However, very little is known regarding the common and unique PXR- and CAR-target DPGs in the liver on a transcriptomic scale. Also, there lacks a systematic comparison of other target genes and signaling pathways in addition to the well-characterized DPGs in the liver. Therefore, the goal of the present study was to use a transcriptomic approach to unveil and compare the common and unique PXR- and CAR-target genes in mouse liver.
MATERIALS AND METHODS
Chemicals
The mouse PXR ligand pregnenolone-16α-carbonitrile (PCN) and the mouse CAR ligand 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-tetrachloro-1,4-bis(pyridyloxy)benzene (TCPOBOP) were purchased from Sigma Aldrich (St. Louis, MO).
Animals
Adult C57BL/6N wild-type male mice (approximately 12 weeks of age) were purchased from Charles River Laboratories (Wilmington, MA). All mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited facility at the University of Kansas Medical Center, with a 14-hour light/10-hour dark cycle, in a temperature- and humidity-controlled environment with ad libitum access to the Laboratory Rodent Chow 8604 (Harlan, Madison, WI) and drinking water. Prior to experiments, mice were acclimated for at least 1 week in the animal facilities. Mice were treated with PCN (200 mg/kg, i.p.), TCPOBOP (3 mg/kg, i.p.), or vehicle (corn oil, 5 ml/kg, i.p.) once daily for 4 days (n=3 per group). Tissues were collected 24-hours after the final dose. The dosing regimen is shown in Figure 1A. All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center.
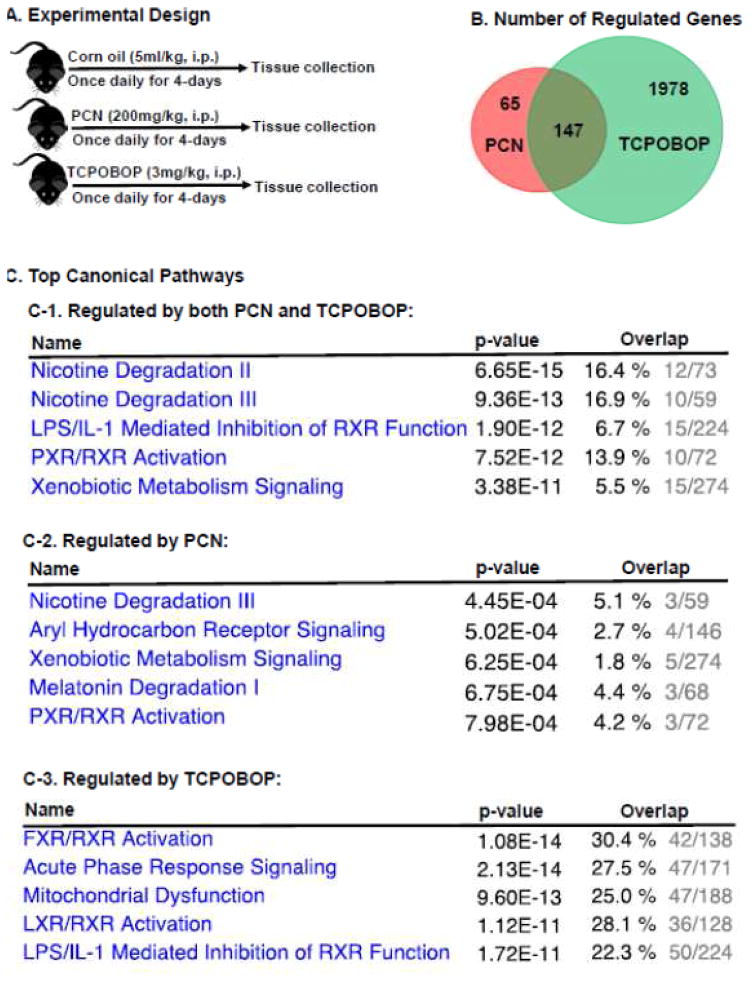
Figure 1.
A. A diagram illustrating the experimental design and dosing regimen of mice. B. A venn diagram demonstrating the number of differentially regulated genes in livers of wild-type mice treated with PCN or TCPOBOP as compared to vehicle-treated group. Differential expression was determined using Cuffdiff with FDR<0.05 as described in MATERIALS AND METHODS. C. Top canonical pathways of the liver genes that are differentially regulated by both PCN and TCPOBOP (C-1), by PCN only (C-2), and by TCPOBOP only (C-3). Data were analyzed using Ingenuity Pathway Analysis as described in MATERIALS AND METHODS.
RNA Isolation
Total RNA from livers of control and chemical-treated mice was extracted with RNA-Bee reagent (Tel-Test, Inc., Friendswood, TX) according to the manufacturer’s protocol. The concentration of total RNA was quantified spectrophotometrically at 260 nm using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE). RNA integrity was confirmed using a dual Agilent 2100 Bioanalyer (Agilent Technologies Inc., Santa Clara, CA).
RNA sequencing
The cDNA libraries and sequencing (2×100bp) of the mouse liver mRNA samples (n=3 per group) were performed using the Illumina standard operation pipeline using a similar method as we described previously (Selwyn et al., 2015).
Data Analysis
The quality control of all FASTQ files was performed using FastQC (Babraham Bioinformatics, Cambridge, UK), and data were trimmed to 90bp from each end to remove ambiguous reads. Data were mapped to the mouse reference genome NCBI GRCm38/mm10 using Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT) (Kim et al., 2015) (version 0.1.6 beta). The Sequence Alignment/Map (SAM) files were converted to binary format using Samtools (Li et al., 2009) (version 1.2). The transcript abundance was estimated using Cufflinks (Trapnell et al., 2010) (version 2.2.1). The mRNA abundance of genes was expressed in Fragments Per Kilobase of transcript per Million mapped Reads (FPKM). For the differential expression analysis, data were subjected to upper-quartile normalization, and a false discovery rate-adjusted p-value below 0.05 in at least one of the chemical-treated groups as compared to the vehicle-treated group was considered statistically significant (as shown by asterisks). The differentially regulated genes in 1) both PCN- and TCPOBOP-treated groups, 2) only in the PCN-treated group, as well as 3) only in the TCPOBOP-treated group were each analyzed using Ingenuity Pathway Analysis (IPA, QIAGEN, Vallencia, CA) to determine significantly altered gene networks. A two-way hierarchical clustering dendrogram was performed for all the differentially expressed drug-processing genes using the Ward Method and Standardized data using the individual FPKM values (JMP version 12, SAS, Cary, NC). The red color represents relatively higher expression, and the blue color represents relatively lower expression.
RESULTS
RNA-Seq generated approximately 47 to 68 million reads per sample, among which approximately 40 to 60 million reads were uniquely mapped to the mouse reference genome (NCBI GRCm38/mm10), and the percentage mapped was between 83.64% to 93.31% (Supplemental Table 1). Regarding PXR-signaling, PCN altered the mRNAs of 212 genes, among which 106 genes were up-regulated and 106 genes were down-regulated by PCN (Table S2–1) within the hepatic transcriptome. As shown in Table 1, the top 10 most up-regulated genes by PCN in the liver were almost exclusively involved in phase-I or –II drug metabolism, including Gstm3 (72.32-fold), Cyp2c55 (45.56-fold), the perinatal-specific Cyp3a isoform Cyp3a16 (32.95-fold), Akr1b7 (23.39-fold), Cyp3a59 (14.01-fold), Cyp3a44 (12.65-fold), Sult2a7 (10.28-fold), Cyp3a41b (10.03-fold), and Cyp2b10 (10.02-fold). Therefore, the drug metabolism pathway appeared to be the major inducible target following PXR activation. Ltf (lactotransferrin) was the only gene that was not a DPG among the top 10 most up-regulated genes by PCN (11.72-fold), and this gene encodes an iron glycoprotein that increases during inflammation, such as following lipopolysaccharide (LPS) exposure (Ahmad et al., 2011). The top 10 most down-regulated genes by PCN in the liver include metallothionein 1 (Mt1) and Mt2 (96% and 85%, respectively), which are important in protecting against cadmium toxicity (Klaassen et al., 1999); alpha-kinase 1 Alpk1 (12.81%), which is important in apical protein transport (Heine et al., 2005); monooxygenase DBH-like 1 (Moxd1) (15.98%), TRAF-interacting protein with forkhead-associated domain (Tifa) (17.85%), which is an inflammatory signaling adaptor and a tumor suppressive factor (Shen et al., 2015); the phase-I enzyme Cyp2c69 (18.80%); toll-like receptor 2 (Tlr2) (19.68%), which promotes inflammation (Ma et al., 2015a); 9430083A17Rik (21.15%); Gm7334 (21.33%); and major urinary protein 9 (Mup9) (21.46%).
Table 1.
Top differentially regulated genes
| Treatment | Up-regulated | Down-regulated | ||
|---|---|---|---|---|
| Genes | Fold-increase | Genes | % decrease | |
| PCN | Gstm3 | 72.32 | Mt1 | 94% |
| Cyp2c55 | 45.56 | Alpk1 | 87% | |
| Cyp3a16 | 32.95 | Mt2 | 84% | |
| Akr1b7 | 23.39 | Moxd1 | 84% | |
| Cyp3a59 | 14.01 | Tifa | 82% | |
| Cyp3a44 | 12.65 | Cyp2c69 | 81% | |
| Sult2a7 | 19.28 | Tlr2 | 80% | |
| Ltf | 11.72 | 9430083A17Rik | 79% | |
| Cyp3a41b | 10.03 | Gm7334 | 79% | |
| Cyp2b10 | 10.02 | Mup9 | 79% | |
| TCPOBOP | Gsta1 | 1115 | Dmbt1 | 99.8% |
| Gm3776 | 259.6 | Hsd3b5 | 99.3% | |
| Gstm3 | 249.7 | Serpina4-ps1 | 99% | |
| Cyp2b10 | 195.3 | Obp2a | 97% | |
| Akr1b7 | 192.3 | Mt2 | 97% | |
| Cyp2c55 | 170.1 | Slco1a1 | 97% | |
| Sult1e1 | 158.5 | Ces4a | 96% | |
| Ccnb1 | 98.83 | Susd4 | 95% | |
| Fndc5 | 95.43 | Dct | 95% | |
| Plk1 | 93.79 | Cyp2c40 | 94% | |
Regarding TCPOBOP-induced CAR-signaling within the hepatic transcriptome, TCPOBOP altered the mRNAs of 2125 genes, which was much more than the 212 genes of which the mRNAs were altered by PCN. Among the 2125 genes targeted by the CAR ligand, 1013 genes were up-regulated, and 1112 genes were down-regulated (Figure 1B, Table S2–2). As shown in Table 1, the top most up-regulated gene by TCPOBOP in liver is Gsta1 (1115-fold). Gm3776, which is a gene located adjacent to Gsta1, was the second most up-regulated (260-fold), suggesting that there is a “hot zone” for CAR-mediated gene trans-activation in this genomic neighborhood. There are many other DPGs that rank among the top 10 most up-regulated genes by TCPOBOP, including Gstm3 (250-fold), Cyp2b10 (prototypical CAR-target gene) (195-fold), Akr1b7 (192-fold), Cyp2c55 (170-fold), as well as Sult1e1 (159-fold). In addition, TCPOBOP markedly up-regulated cyclin B1 (Ccnb1) (99-fold), which is important for cell proliferation (Porter and Donoghue, 2003); fibronectin type III domain containing 5 (Fndc5) (94-fold), which is a precursor of the anti-obesity hormone irisin (Bostrom et al., 2012), as well as polo-like kinase 1 (Plk1) (95-fold), which controls several key steps during mitosis (Schoffski, 2009; Strebhardt, 2010). The top most down-regulated gene by TCPOBOP in liver is deleted in malignant brain tumors 1 (Dmbt1) (99.8%), which is important in tissue repair following liver injury (Bisgaard et al., 2002), followed by the hormone-metabolizing enzyme hydroxy-delta-5-steroid-dehydrogenase, 3 beta- and steroid delta-isomerase 5 (Hsd3b5) (99.3%); serine (or cysteine) peptidase inhibitor clade A member 4 pseudogene 1 (Serpina4-ps1) (98.7%), which is a pseudogene that transcribes but does not appear to form a protein; odorant binding protein 2A (Obp2a) (97.1%), which has a high affinity towards binding aldehydes and fatty acids (Tcatchoff et al., 2006); the stress-response metal-binding gene metallothionein Mt2 (97%) (Klaassen et al., 1999); dopachrome tautomerase (Dct) (95%), which is important for melanogenesis (Aroca et al., 1990); as well as sushi domain containing 4 (Susd4) (94.8%), which is a complement inhibitor (Holmquist et al., 2013). In addition, several DPGs such as the uptake transporter Slco1a1 (96.3%), phase-I Ces4a (96.3%), Cyp2c40 (94%), and Cyp4a10 (94%), also rank among the top 10 most down-regulated genes by TCPOBOP in liver.
Within the hepatic transcriptome, 65 genes were uniquely regulated by PCN but not by TCPOBOP, 1978 genes were uniquely regulated by TCPOBOP but not by PCN, whereas 147 genes were differentially regulated by both PCN and TCPOBOP (Figure 1B, Table S2–3). Among the 147 genes regulated by both chemicals, the majority (141 in total) were common targets of PCN and TCPOBOP (i.e. they were either both up-regulated [61 genes], or both down-regulated by these chemicals [80 genes]), suggesting that PXR and CAR share high similarity in their transcriptional activities. There were only 6 genes that were regulated by PCN and TCPOBOP in a completely opposite manner (Table S2–3), including up-regulation of hydroxysteroid (17-beta) dehydrogenase 6 (Hsd17b6), leucine rich repeat containing G protein coupled receptor 5 (Lgr5), and Ces1e by PCN but down-regulation of these genes by TCPOBOP, as well as a down-regulation of Cyp17a1, cyclin D1 (Ccnd1), and ribosomal protein L35 (Rpl35) by PCN but up-regulation of these genes by TCPOBOP in liver. These genes may serve as novel biomarkers for distinguishing a PXR-effect from a CAR-effect following a xenobiotic insult. The top canonical pathways of genes that were differentially regulated by both PCN and TCPOBOP significantly overlapped with nicotine degradation pathways, LPS/interleukin-1 (IL-1) mediated inhibition of retinoid X receptor (RXR) function, PXR/RXR activation, and xenobiotic signaling (Figure 1C–1). Unique PCN-target genes significantly overlapped with the aryl hydrocarbon receptor signaling and the melatonin degradation pathway (Figure 1C–2), whereas unique TCPOBOP-target genes significantly overlapped with the farnesoid X receptor (FXR)/RXR activation, acute phase response signaling, mitochondrial dysfunction, and liver X receptor (LXR)/RXR activation (Figure 1C–3).
To unveil the common and unique PXR- and CAR-targeted signaling pathways in liver following the chemical activation of these nuclear receptors, pathway analyses were performed in the common and unique PXR- and CAR-target genes. The top network that was commonly regulated by PCN and TCPOBOP is shown in Supplemental Figure 1, which includes the up-regulation of glutathione conjugation and retinoid metabolism, and suppression of inflammation and metal transport. Specifically, both PCN and TCPOBOP markedly up-regulated multiple glutathione S-transferases, which are important for glutathione conjugation of substrates and chemical detoxification (Parkinson et al., 2013). Regarding retinoid metabolism, both PCN and TCPOBOP markedly up-regulated Akr1b7, which is important in the metabolism of retinaldehyde (Ruiz et al., 2011) and the detoxification of lipid peroxidation byproducts (Liu et al., 2009); PCN and TCPOBOP also up-regulated Cyp26a1, which is important in the elimination of active retinoids (Topletz et al., 2015). In addition, Abcd2, which is an Abc transporter of which the promoter activity of is induced by 9-cis-retinoic acid (Pujol et al., 2000), was up-regulated by PCN and TCPOBOP. Upstream analysis demonstrated that both PCN and TCPOBOP appeared to activate the following regulators, PXR, CAR, estrogen receptor, interleukin (IL) receptor 10a, Cxcl12, insulin signaling, as well as the interaction between PXR/CAR with the nuclear receptor coactivator a (Ncoa) (Table S3–1).
Conversely, both PCN and TCPOBOP down-regulated NFκB-signaling, which is critically involved in the inflammatory response (Supplemental Figure 1). Specifically, PCN and TCPOBOP down-regulated chemokine (C-X-C motif) ligand 1 (Cxcl1, which is the mouse homolog of the human CXCL2), toll-like receptor 2 (Tlr2), STEAP family member 4, as well as serum amyloid A cluster (Saa [Saa1 and Saa2 in mice]), which are important for hepatic inflammation (Ramadoss et al., 2010; Luo et al., 2015; van der Heijden et al., 2015). The metal binding and transport pathway also appeared to be down-regulated by both PCN and TCPOBOP, evidenced by down-regulation of both metallothionein (Mt)1 and Mt2, divalent metal transporter Slc11a2 (also known as Dmt), as well as Steap4, which is a plasma membrane metalloreductase involved in the transport of iron and copper (Ramadoss et al., 2010). PCN and TCPOBOP also down-regulated a few genes important for intermediary metabolism, including the major urinary protein 1 (Mup1) that has been shown to be a key regulator of glucose and lipid metabolism in mice (Zhou et al., 2009), glucose-6-phosphatase (G6pc) that regulates gluconeogenesis and glycogenolysis, as well as fatty acid binding protein 5 (also known as bile acid ligase or Slc27a5), which regulates bile acid conjugation and promotes obesity (Hubbard et al., 2006). Upstream analysis demonstrated that both PCN and TCPOBOP appear to down-regulate IL1α and Ilβ, Iκβ kinase gamma (IKBKG), and interferon α.
The top network that was uniquely regulated by PCN is shown in Supplemental Figure 2A. Most notably, PCN down-regulated the inhibitor of cell proliferation, namely cyclin-dependent kinase inhibitor Cdkn1a (also known as p21), which is an important transcriptional target of several tumor suppressors and promotes cell cycle arrest in response to stimuli (Buitrago-Molina et al., 2013). This may contribute to PCN-mediated increase in cell proliferation. PCN also down-regulated multiple genes involved in intermediary metabolism, such as cysteine sulfinic acid decarboxylase (Csad), which is a rate-limiting enzyme in taurine de novo synthesis (Ma et al., 2015b); phospholipid scramblase 1 (Plscr1), which is involved in movement of phospholipids, immune-activation, and cell proliferation (Lu et al., 2007); peroxiredoxin 4 (Prdx4), which reduces oxidative-stress induced liver injury (Nabeshima et al., 2013), as well as the chemokine Cxcl9 and hemoglobin. Conversely, the uniquely up-regulated genes by PCN include the phase-I enzyme Aldh1b1; the antimicrobial peptide hepcidin 2 (hamp2), which is involved in inflammation, infection, and iron homeostasis (Lu et al., 2015); inositol polyphosphate multikinase (Ipmk), which regulates the central metabolic enzyme AMP-activated protein kinase (Ampk) for energy sensing (Dailey and Kim, 2012); the sodium/lithum/calcium exchanger Slc8b1; and immunoglobulin-like domain containing receptor 2 (Ildr2), which is an endoplasmic reticulum (ER) resident molecule mediating hepatic lipid homeostasis during ER stress (Watanabe et al., 2013). Upstream analysis demonstrated that PCN treatment appeared to activate Acox1, which is the first enzyme of fatty acid beta-oxidation, highlighting the important role of PXR in lipid metabolism (Table S3–2).
The top network that was uniquely regulated by TCPOBOP is shown in Supplemental Figure 2B. Most notably, TCPOBOP markedly increased the cell proliferation marker Mki67 (antigen identified by monoclonal antibody Ki-67), which has been shown to promote cell proliferation during liver regeneration (Gerlach et al., 1997), as well as the anti-obesity gene JAZF zinc finger 1 (Jazf1), which has been shown to reduce body weight gain and regulates lipid metabolism when consuming a high-fat diet treatment (Jang et al., 2014). The lipid-sensor PPARα as well as its target genes were down-regulated by TCPOBOP (Supplemental Figure 2B). Upstream analysis unveiled many regulators that were activated or suppressed by TCPOBOP treatment. For example, TCPOBOP appeared to activate T-box1 (Tbx1), which may contribute to tumorigenesis; Nrf2-signaling, which is important for reducing oxidative stress; vitamin D receptor; the orphan nuclear receptor nur77 (Nr4a1), etc. In contrast, TCPOBOP suppressed SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b (Smarcb1), which is a part of a complex that relieves repressive chromatin structures and allows the transcription machinery to access its targets more effectively. In addition, TCPOBOP appeared to suppress multiple interleukins, circadian rhythm related gene CLOCK, HNF4α-signaling, PPARα-signaling, as well as FXR (Nr1h4)-signaling (Table S3–3).
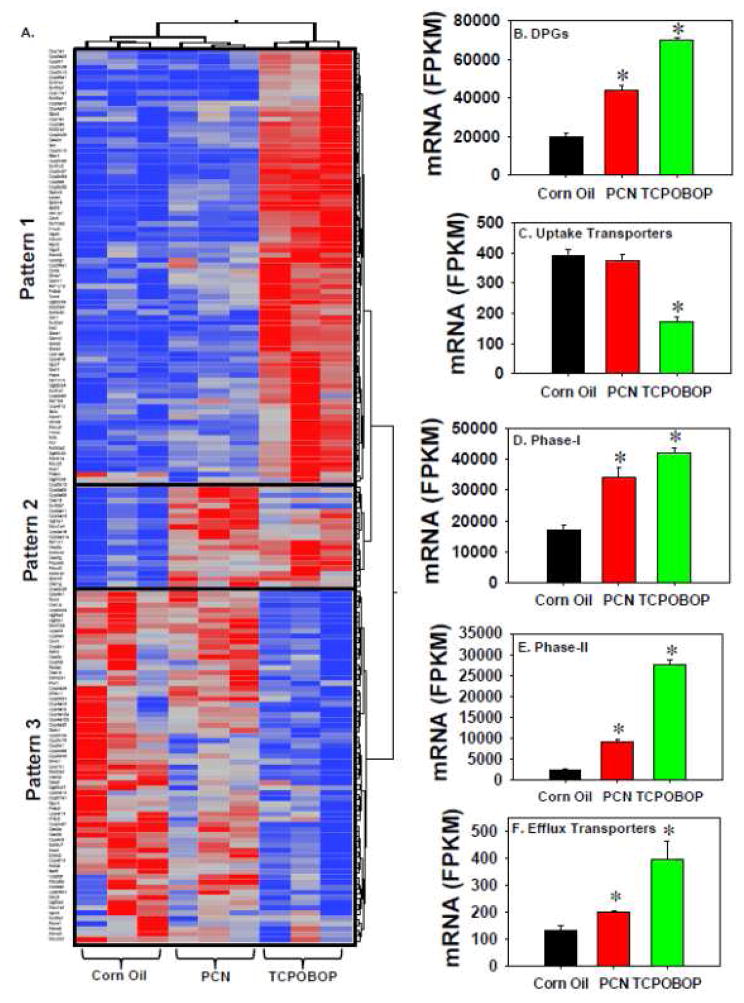
In summary, pharmacological activation of PXR and CAR leads to both common and unique changes in target genes. Both nuclear receptors appeared to up-regulate glutathione conjugation reactions but suppress inflammation and metal binding/transport pathways, whereas each receptor also uniquely regulates their own target genes involved in cell proliferation and intermediary metabolism. Because PXR and CAR are well known xenobiotic-sensing nuclear receptors, the mRNA expression of all DPGs (including 318 phase-I and 92 phase–II drug-metabolizing enzymes as well as 519 transporters) was examined in detail regarding their common and different transcriptional responses following PCN or TCPOBOP treatment. A detailed description of these DPGs is provided in Supplemental Material Section 4. A total of 1158 DPGs were examined based on a literature search. Among all DPGs, there were 172 genes that were differentially regulated by at least one of the two chemicals (PCN and/or TCPOBOP), including 119 phase-I enzymes, 35 phase-II enzymes, and 18 transporters. These 172 DPGs partitioned into 3 distinct expression patterns (Figure 2A). DPGs in Pattern 1, which was the largest cluster, were predominantly up-regulated by TCPOBOP, and to a much lesser extent by PCN. Therefore, these genes appeared to be primary CAR-targets. These genes include many Cyps, such as Cyp1a1, 1a2, 2a4, 2a5, 2a22, 2b10, 2b13, 2c29, 2c37, 2c39, 2c50, 2c54, 2c55, 2c65, 2g1, 2r1, 3a13, 4a31, 4f16, 4f15, 7a1 (bile acid synthetic enzyme), 17a1 (steroidogenic enzyme), 26a1 (retinoic acid-metabolizing enzyme), and 39a1 (oxysterol-metabolizng enzyme), as well as cytochrome b5 (Cyb5) and P450 (cytochrome) oxidoreductase (Por), which are important for the Cyp activities. Other DPGs in Pattern 1 include other phase-I enzymes, such as Akrs (Akr1b7, 1b8, 1c14, 1c19), Aldhs (Aldh1a7, 3a2, and 3b1), carbonyl reductase (Cbr) 1 and Cbr3, Dhrs7 and Dhrs9, Fmo4 and Fmo5, Gpx2 and Gpx7, Nqo1 and Nqo2, Prdx4 and Prdx6a, as well as Aox1, Ces2c, Ephx1, esterase D (Esd), Gclc, Gsr, peptidase D (Pepd), and Xdh. The phase-II enzymes in Pattern 1 include many Sults (Sult1e1, 1c2, 1d1, 1e1, 2a2, 3a1) and the enzymes that generate the glucuronidation co-substrates (Ugdh and Ugp2), many Gsts (Gsta1, a2, a4, m1, m2, m3, m4, t1, and t3), many Ugt2b family members (Ugt2b34, wb35, 2b36, and 2b38), as well as thiopurine methyltransferase (Tpmt). Transporters in Pattern 1 include the uptake transporters Slc10a2 (also known as Asbt) and Slc22a4 (also known as Octn1), as well as many Abc efflux transporters (Abcb1a/Mdr1a, Abcc2/Mrp2, Abcc4/Mrp4, and Abcd1-3).
Figure 2.
A. A two-way hierarchical clustering dendrogram of DPGs that were differentially regulated by PCN or TCPOBOP in livers of wild-type mice. B. Cumulative expression of all differentially regulated DPGs in livers of vehicle-, PCN- and TCPOBOP-treated mice. C. Cumulative expression of all differentially regulated Phase-I drug-metabolizing enzymes in livers of vehicle-, PCN- and TCPOBOP-treated mice. D. Cumulative expression of all differentially regulated Phase-II drug-metabolizing enzymes in livers of vehicle-, PCN- and TCPOBOP-treated mice. E. Cumulative expression of all differentially regulated uptake transporters in livers of vehicle-, PCN- and TCPOBOP-treated mice. F. Cumulative expression of all differentially regulated efflux transporters in livers of vehicle-, PCN- and TCPOBOP-treated mice. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
DPGs in Pattern 2 were predominantly up-regulated by PCN, and to a lesser extent by TCPOBOP, suggesting that they are primary PXR targets (Figure 2A). The phase-I enzymes in Pattern 2 include Cyps (Cyp2d12, 3a11, 3a16, 3a25, 3a41a, 3a44, and 3a59), Akr1d1, Aldh1a1 and 1b1, as well as Cess (Ces1d, 1g, 2a, and 2g). The phase-II enzymes in Pattern 2 include Sult2a7 and Papss2 (the co-substrate for sulfation reactions), and Ugt1a1. The uptake transporter Slco1a4 (also known as Oatp1a4) was the only transporter in Pattern 2.
DPGs in Pattern 3 were predominantly down-regulated by TCPOBOP, suggesting that CAR is either a trans-repressor or an indirect suppressor of these genes. The phase-I enzymes in Pattern 3 include many Cyps (Cyp2c6, 2c34, 2c38, 2c40, 2c44, 2c69, 2c70, 2d13, 2d26, 2d40, 2e1, 2f2, 2j5, 2j6, 2u1, 4a10, 4a12a, 4a12b, 4a14, 4a32, 4b1, 4f13, 4f13, 4v1, 7b1, 8b1, 26b1, and 27a1), Aldh2 and Aldh6a1, Cess (Ces1b, 1e, 2e, 3a, 3b, and 4a), Dhrs1 and Dhrs11, Gpx1 and Gpx3, Prdx2 and Prdx5, Sdr9c7 and Sdr42e1, as well as Aadac, Adh4, Aox3, Cela1, Dpyd, Ephx2, and Suox. The phase-II enzymes in Pattern 3 include Gstk1, Sult5a1, Ugts (2a3, 2b37, 3a1, and 3a2), Nat8, and Comt. Transporters in Pattern 3 include uptake transporters, such as Slc22a7 (also known as Oat2), Slc27a5 (also known as fatty acid binding protein 5 or bile acid ligase), Slco/Oatp1a1 and 2a1, as well as efflux transporters such as Abca1-3 and Abca8a.
To determine the overall contribution of PXR and CAR ligands on hepatic DPG expression, the cumulative FPKM of all differentially regulated DPGs was calculated as shown in Figure 2B–F. The cumulative hepatic mRNAs of all differentially regulated DPGs, phase-I and –II enzymes, and efflux transporters were all up-regulated by both PCN and TCPOBOP, and to a greater extent by TCPOBOP (Figure 2B–E). In contrast, the cumulative mRNAs of the uptake transporters were not influenced by PCN, but were down-regulated by TCPOBOP (Figure 2F).
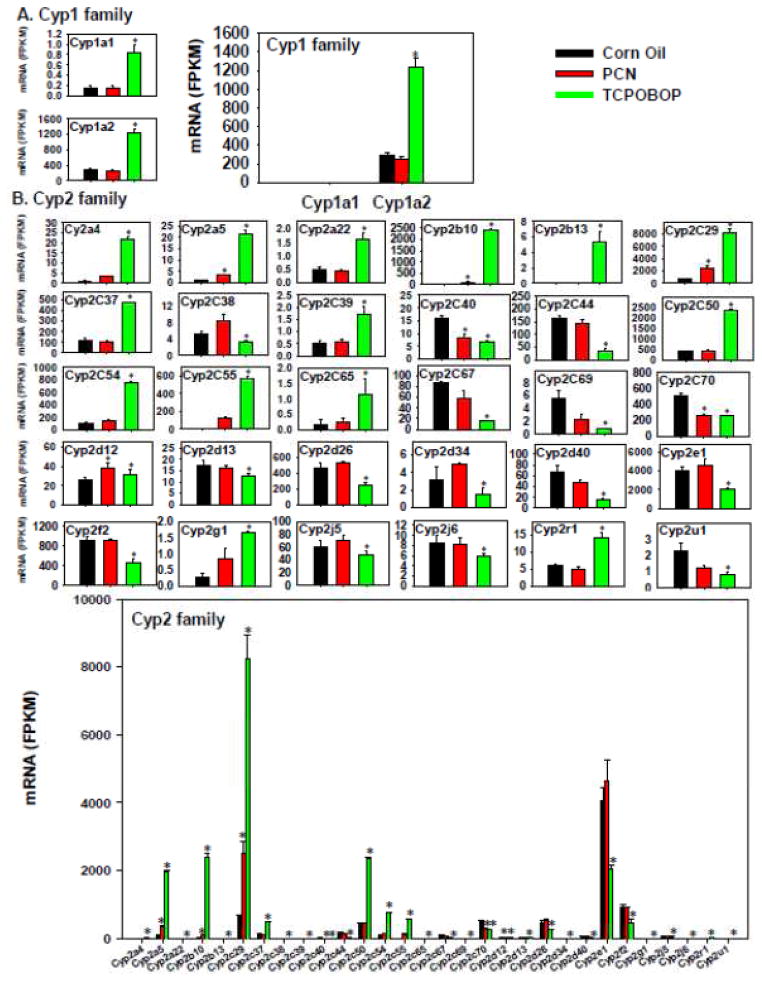
To compare the pharmacological activation of PXR and CAR on the regulation of DPGs, the expression of various phase-I and –II drug-metabolizing enzymes, transporters, as well as essential nuclear receptors and/or xenobiotic-sensing transcription factors was examined as shown in Figure 3 to Figure 12. Regarding the cytochrome P450s that are critically involved in drug metabolism, Cyp1a1 and 1a2 were the only Cyp1 isoforms that were differentially regulated, and they were both preferably up-regulated by TCPOBOP but not influenced by PCN (Figure 3A, left). Regarding the absolute mRNA abundance of these two Cyp1 genes, Cyp1a2 was the predominant Cyp1 isoform that was much more abundantly expressed in liver than Cyp1a1, and thus the potential TCPOBOP-mediated pharmacokinetic alteration of Cyp1a substrates is likely through Cyp1a2 in liver.
Figure 3.
Regulation of the Phase-I Cyp1 (A) and Cyp2 (B) drug-metabolizing enzymes by PCN and TCPOBOP in livers of wild-type mice. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
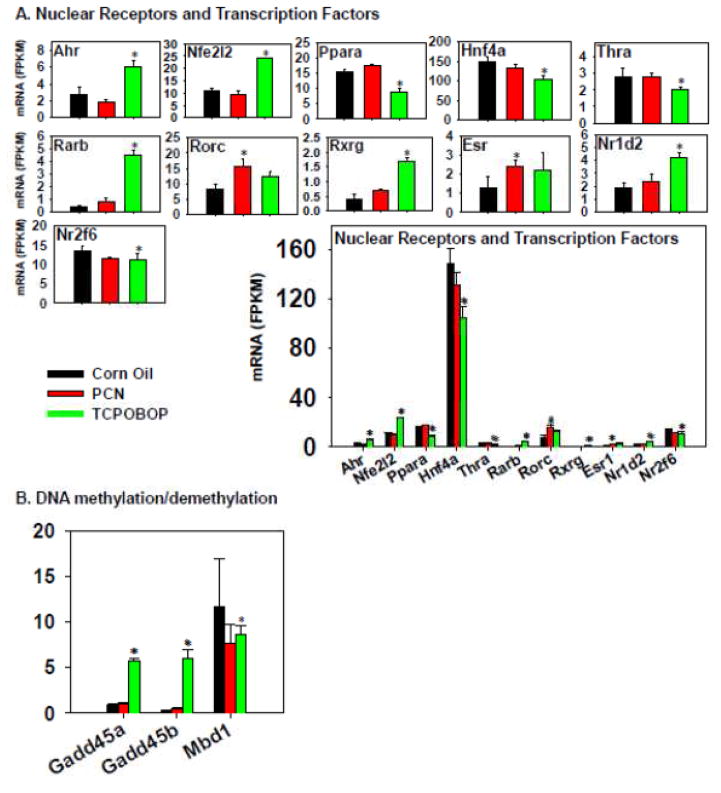
Figure 12.
Regulation of nuclear receptors and transcription factors by PCN and TCPOBOP in livers of wild-type mice. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
More genes in the Cyp2 family (Figure 3B) appeared to be differentially regulated by TCPOBOP than PCN. For example, TCPOBOP markedly increased the mRNAs of Cyp2a4, 2a5, 2a22, 2b10 (prototypical CAR-target gene), 2b13, 2c29, 2c37, 2c39, 2c50, 2c54, 2c55, 2c65, 2g1, and 2r1; but decreased the mRNAs of Cyp2c38, 2c40, 2c44, 2c67, 2c69, 2c70, 2d13, 2c26, 2d34, 2d40, 2e1, 2f2, 2j5, 2j6, and 2u1. PCN moderately increased the mRNAs of Cyp2b10, 2c29, and 2d12, but in general, the fold-increase was less than that produced by TCPOBOP, except the up-regulation of Cyp2d12 mRNA, which was similar by both chemicals. In addition, PCN down-regulated Cyp2c40 and 2c70 mRNAs. The Cyp2 genes that were co-regulated by both PCN and TCPOBOP include Cyp2a5, 2b10, 2c29, and 2d12 (up-regulated), as well as Cyp2c40 and 2c70 (down-regulated), whereas other genes were uniquely regulated only by TCPOBOP. Regarding the absolute mRNA abundance of these Cyp2 genes, Cyp2e1 mRNA was the highest in control livers; however, it was markedly decreased by TCPOBOP. Conversely, Cyp2c29 mRNA was lowly expressed in control livers, but was markedly up-regulated by TCPOBOP, and became the most abundantly expressed Cyp2. Cyp2a5, 2b10, and 2c50 were all lowly expressed in control livers, but were markedly increased by TCPOBOP to intermediate or high levels. Other Cyp2 genes were expressed at much lower levels under both basal and chemical-treated conditions. Considering the absolute mRNA abundance, the CAR ligand appears to have a more prominent influence on Cyp2 gene expression than the PXR ligand, and the majority of the abundantly expressed Cyp2a-c genes were up-regulated by the CAR ligand, whereas the highest expressed Cyp2 isoform Cyp2e1 was actually down-regulated by the CAR ligand.
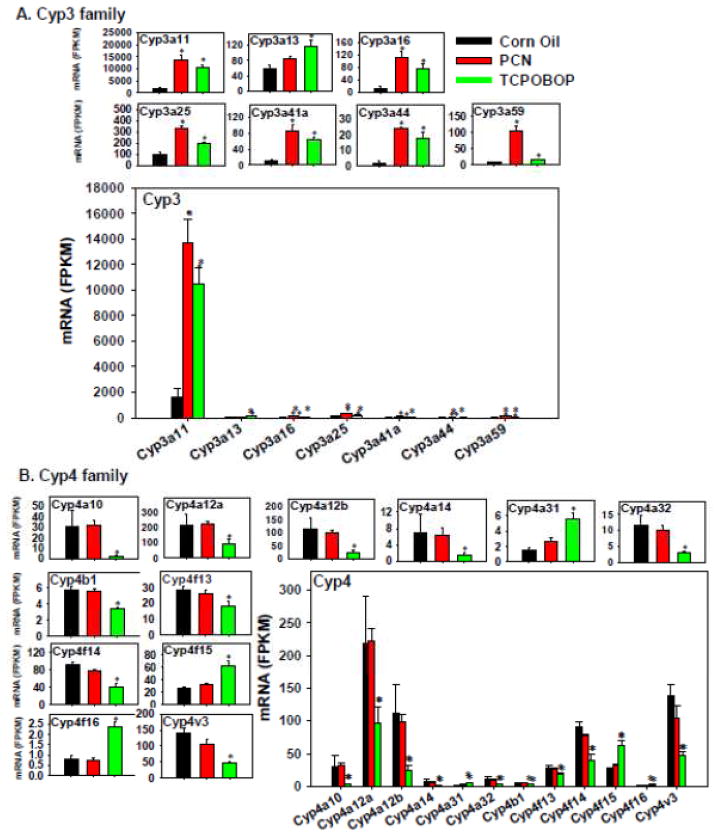
The effect of PXR and CAR agonists on the expression of the Cyp3a family members is shown in Figure 4A. Both PCN and TCPOBOP up-regulated Cyp3a11 (prototypical PXR-target gene), 3a16, 3a25, 3a44, 3a41a (which tended to increase although a statistical significance was not achieved), and 3a59. In general, PCN appeared to up-regulate the mRNAs of the Cyp3a genes more than did TCPOBOP (Figure 4A). In contrast, Cyp3a13 mRNA appeared to be preferably increased by TCPOBOP. The overwhelming highest expressed Cyp3a isoform was Cyp3a11 in all treatment conditions, whereas all other differentially expressed Cyp3a genes were expressed at much lower levels in all treatment conditions. The overall contribution of the PXR and CAR ligands appear to be an up-regulation of the Cyp3a genes, and especially through increasing the Cyp3a11 isoform.
Figure 4.
Regulation of the Phase-I Cyp3 (A) and Cyp4 (B) drug-metabolizing enzymes by PCN and TCPOBOP in livers of wild-type mice. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
The expression of the Cyp4 family members was not altered by PCN (Figure 4B); in contrast, TCPOBOP markedly decreased most Cyp4 genes, including Cyp4a10, 4a12a, 4a12b, 4a14, 4a32, 4b1, 4f13, 4f14, and 4v3. Conversely, TCPOBOP up-regulated Cyp4a31, 4f15, and 4f16. Therefore, Cyp4 genes appeared to be uniquely regulated by TCPOBOP. The absolute mRNA abundance of Cyp4a12a was the highest expressed Cyp4 isoform in control livers, followed by Cyp4v3, 4a12b, 4f14, 4f13 and 4a10, as well as 4f15; whereas other Cyp4 mRNAs were much lower expressed. The overall effect of the CAR ligand appears to be a down-regulation of the Cyp4 genes.
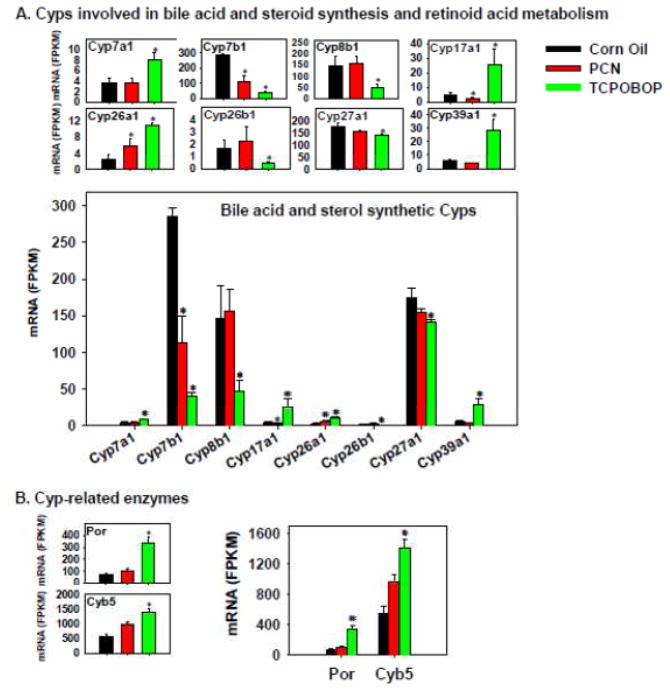
The Cyps involved in bile acid and steroid bio-synthesis as well as retinoic acid metabolism are shown in Figure 5A. The rate-limiting bile acid synthetic enzyme Cyp7a1 for the classic pathway of the de novo bile acid synthesis in liver was markedly up-regulated by TCPOBOP, but was not influenced by PCN. Conversely, the 12α-hydroxylase Cyp8b1, which is important for producing cholic acid, was markedly decreased by TCPOBOP, but was not influenced by PCN. The bile acid synthetic enzymes involved in the alternative pathway of bile acid synthesis, namely Cyp27a1 and 7b1, were both down-regulated by TCPOBOP; Cyp7b1 mRNA was also down-regulated by PCN. Cyp39a1, which has a minor contribution in the alternative pathway of bile acid synthesis in liver, was markedly up-regulated by TCPOBOP, but was not influenced by PCN. The steroidogenic enzyme Cyp17a1 was preferably up-regulated by TCPOBOP (and was only slightly decreased by PCN). The retinoic acid-metabolizing enzyme Cyp26a1 was up-regulated by both PCN and TCPOBOP, whereas Cyp26b1 was decreased by TCPOBOP. Regarding the absolute mRNA abundance, within the bile acid synthetic enzymes category, Cyp7b1 was the highest expressed gene in control livers, followed by Cyp27a1 and 8b1, whereas Cyp7a1 and 39a1 were the lowest expressed. The retinoic acid-metabolizing enzyme Cyp26a1 was more highly expressed than Cyp26b1 in all treatment conditions. Considering the absolute mRNA abundance among the bile acid synthetic genes, it appears that the CAR ligand has a more prominent influence on the bile acid synthesis pathway than the PXR ligand. Interestingly, the rate-limiting bile acid synthetic enzyme Cyp7a1 in the classic pathway of bile acid de novo synthesis was the lowest expressed gene (rating-limiting), and thus it is hypothesized that the TCPOBOP-mediated increase in Cyp7a1 gene expression will increase the 7α-hydroxylation in the bile acid synthesis pathway. In addition, TCPOBOP likely alters the bile acid composition by reducing the 12α-hydroxylated bile acids, such as cholic acid, as due to the marked down-regulation of the 12α-hydroxylase Cyp8b1. The expression of genes in the alternative pathway of bile acid synthesis appeared to be generally down-regulated by TCPOBOP but was not influenced by PCN. The steroid synthesis pathway and retinoic acid metabolism appeared to be increased by TCPOBOP, whereas PCN had less influence on these pathways.
Figure 5.
Regulation of the Cyps involved in bile acid and steroid synthesis and retinoic acid metabolism (A) and Cyp-related enzymes (B) by PCN and TCPOBOP in livers of wild-type mice. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
Both Por and Cyb5 are essential for Cyp activities, and their mRNAs were preferably up-regulated by TCPOBOP, but were not influenced by PCN (Figure 5B).
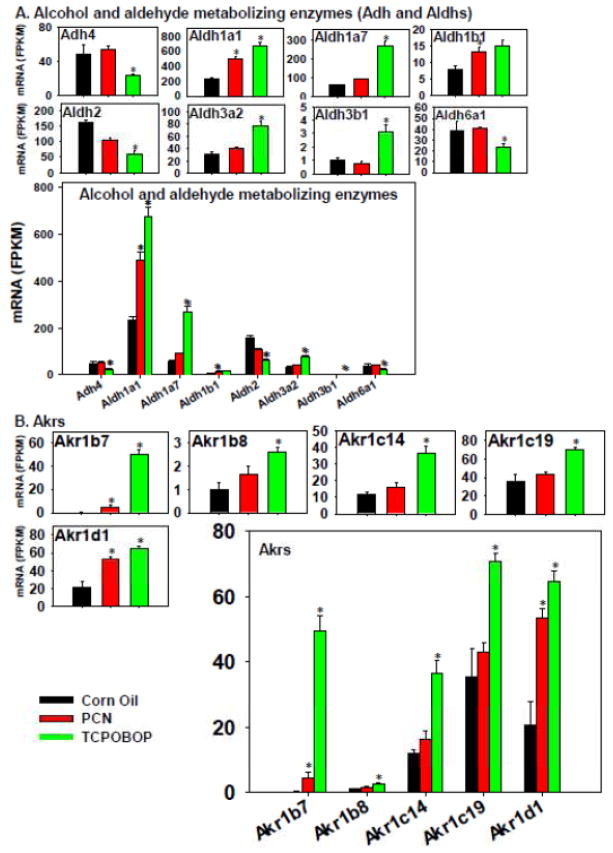
The effect of TCPOBOP and PCN on the expression of the phase-I enzymes responsible for the metabolism of alcohols (Adhs) aldehydes (Aldhs) is shown in Figure 6A. Adh4 was the only differentially regulated Adh, and its mRNA was preferably down-regulated by TCPOBOP but not altered by PCN. Among the Aldhs, Aldh1a1 and 1b1 mRNAs were up-regulated by PCN, but to a greater extent by TCPOBOP; the mRNAs of Aldh1a7, 3a2, and 2b1 were all markedly up-regulated uniquely by TCPOBOP. Conversely, TCPOBOP down-regulated the mRNAs of Aldh2 and 6a1. Aldh1a1 was the highest expressed Aldh in control livers, and it was even higher expressed after PCN and TCPOBOP treatment. Therefore, the overall contribution of PCN and TCPOBOP on aldehyde metabolism appears to be up-regulation.
Figure 6.
Regulation of other Phase-I drug-metabolizing enzymes by PCN and TCPOBOP in livers of wild-type mice: (A) alcohol and aldehyde metabolizing enzymes; (B) dehydrogenases and reductases. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
The phase-I aldo-keto reductases (Akrs) was in general induced more by TCPOBOP than PCN, evidenced by a marked increase in Akr1b7, 1b8, 1c14, 1c19, and 1d1 by TCPOBOP (Figure 6). Akr1b7 and 1d1 mRNAs were also up-regulated by PCN but to a lesser extent. Regarding the absolute mRNA abundance of these differentially regulated Akrs, Akr1c19 was the highest expressed Ahr in control livers, followed by Akr1d1 and 1c14, whereas Akr1b8 and Akr1b7 were very lowly expressed. However, following TCPOBOP treatment, Akr1b7 mRNA was substantially increased, and became one of the most abundantly expressed Akrs. In summary, the CAR ligand influences the Akr expression of the Akrs more than the PXR ligand, and the overall effect of CAR activation is up-regulation of these Akrs.
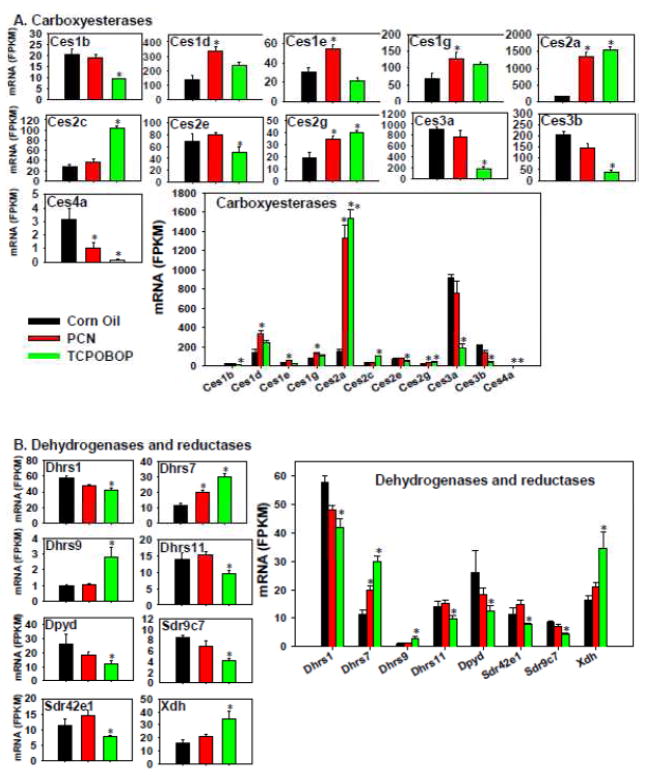
The following phase-I carboxyesterases (Cess) were up-regulated by PCN (Ces1d, 1e, 1g, 2a, and 2g), but PCN down-regulated Ces4a (Figure 7A). TCPOBOP up-regulated Ces2a, 2c, and 2g, but down-regulated Ces1b, 2e, 3a, 3b, and 4a. Ces2a, 2g, and 4a appeared to be co-regulated by both chemicals, whereas other differentially expressed Cess were uniquely regulated by one of the two chemicals. Regarding the absolute mRNA abundance, Ces3a was the highest expressed Ces in control livers, but it was markedly down-regulated by TCPOBOP. Ces2a was a relatively lowly expressed Ces in control livers, however PCN and TCPOBOP both substantially increased Ces2a mRNA to a level that exceeded Ces3a. Therefore, it is possible that Ces3a contributes more to the basal Ces-mediated hydrolysis, whereas Ces2a contributes more to the Ces-mediated hydrolysis following a xenobiotic insult that activates PXR or CAR.
Figure 7.
Regulation of other Phase-I drug-metabolizing enzymes by PCN and TCPOBOP in livers of wild-type mice: (A) carboxyesterases; (B) dehydrogenases and reductases. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05)
Only one of the eight phase-I dehydrogenases and reductases was increased by PCN, whereas three were increased and five were decreased by TCPOBOP (Figure 7B). TCPOBOP up-regulated Dhrs7, Dhrs9, and Xdh, but down-regulated in Dhrs1, Dhrs11, Dpyd, Sdr9c7, and Sdr42e1. PCN only altered the mRNA of Dhrs7, which was an up-regulation. Regarding the absolute mRNA abundance, Dhrs1 was the highest expressed in control livers, followed by Dpyd, Xdh, Dhrs11, Dhrs7, Sdr42e1, Sdr9c7, and Dhrs9. There were more genes that were down-regulated (5) than up-regulated by TCPOBOP (3). Therefore, the overall contribution of TCOPOBOP to genes in this category appeared to be down-regulation.
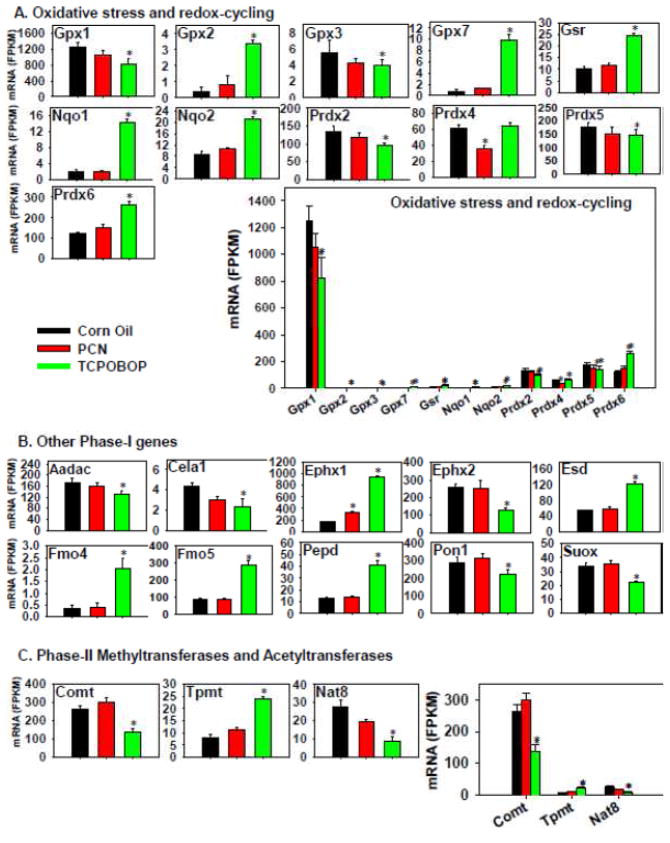
The genes related to oxidative stress and redox-cycling, namely Gpx2, Gpx7, Gsr, Nqo1-2, and Prdx6, were up-regulated more by TCPOBOP than PCN; whreas Gpx1, Gpx3, Prdx2, and Prdx5 were down-regulated more by TCPOBOP than PCN (Figure 8A). In comparison, PCN had minimal effect on these genes except for a decrease in Prdx4 mRNA. Gpx1 was the highest expressed among these genes in control livers, followed by Prdx5, Prdx2, Prdx6, and Prdx4 at intermediary levels, whereas other genes were lowly expressed.
Figure 8.
Regulation of genes involved in oxidative stress and redox-cycling (A), as well as other Phase-I genes (B) by PCN and TCPOBOP in livers of wild-type mice. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05)
The regulation of other phase-I genes by TCPOBOP and PCN are shown in Figure 8B. TCPOBOP increased the mRNAs of epoxide hydrolase 1 (Ephx1), esterase D (Esd), flavin containing monooxygenase (Fmo) 4–5, and peptidase D (Pepd), but down-regulated arylacetamide deacetylase (Aadac) (which was categorized into the Ces5 family but with a different structure from those of the other Ces proteins) (Satoh and Hosokawa, 2006; Hosokawa et al., 2008), chymotrypsin-like elastase 1 (Cela1), epoxide hydrolase 2 (Ephx2), paraoxonase 1 (Pon1), and sulfite oxidase (Suox). PCN had a minor effect on the expression of these genes, except for a moderate increase in Ephx1 mRNA.
The effect of PCN and TCPOBOP on the phase-II methyltransferases and acetyltransferases is shown in Figure 8C. PCN had minimal effect on the expression of these genes; whereas TCPOBOP up-regulated Tpmt but down-regulated Comt and Nat8. Comt was the highest expressed in control livers, whereas Nat8 and Tmpt were expressed at much lower levels.
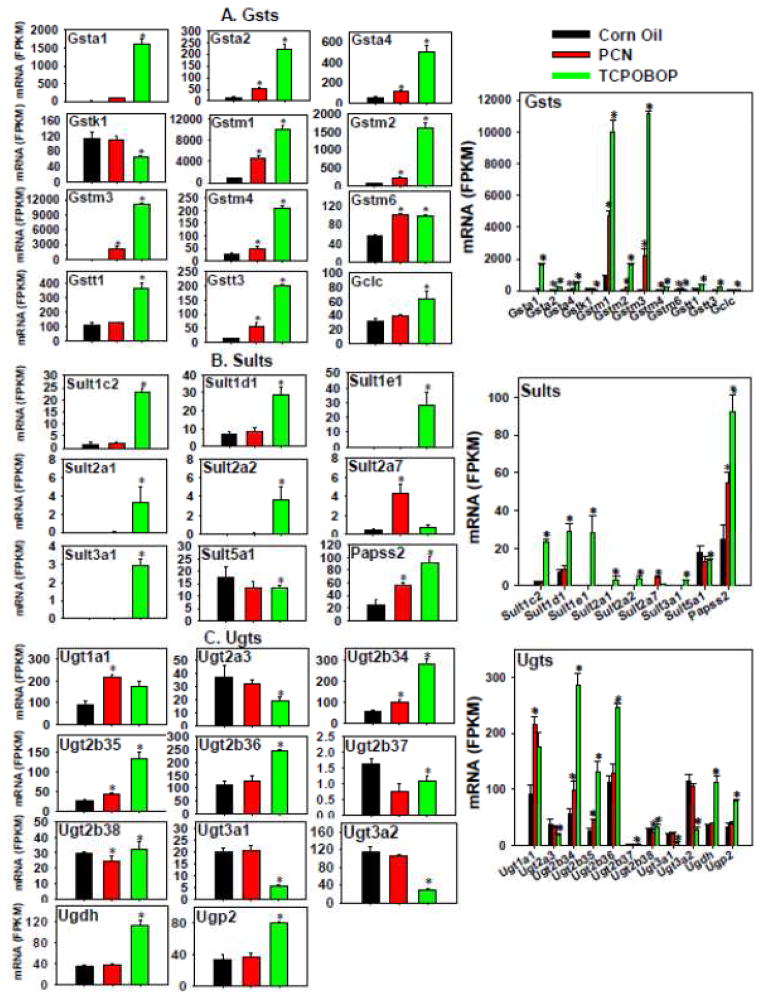
Most of the differentially regulated Gsts (Figure 9A) were up-regulated preferably by TCPOBOP and to a much lesser extent by PCN, including Gsta1 (PCN tended to increase Gsta1 although a statistical significance was not achieved), Gsta2, Gstm1-4, and Gstt3. Gstm6 mRNA was up-regulated to a similar extent by PCN and TCPOBOP. Gstk1 mRNA was not altered by PCN but was moderately decreased by TCPOBOP. Glutamate-cysteine ligase catalytic subunit (Gclc), which is involved in the synthesis of GSH and the cosubstrate of Gsts, was up-regulated uniquely by TCPOBOP. Among all these genes, Gstm1 was the highest expressed in control livers; however, following TCPOBOP or PCN treatment, both Gstm3 and Gstm1 were the highest expressed. Other genes were relatively lowly expressed, and a more notable expression was generally observed after TCPOBOP treatment. Therefore, TCPOBOP, and PCN to a lesser extent, appeared to generally up-regulate the phase-II glutathione conjugation reactions.
Figure 9.
Regulation of Phase-II glutathione S-transferases (A), sulfotransferases (B), and UDP glucuronosyltransferases (C) by PCN and TCPOBOP in livers of wild-type mice. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
The mRNAs of Sult1c2, 1d1, 1e1, 2a1, 2a2, 3a1, as well as Papss2, which is the enzyme that synthesizes the cosubstrate for sulfation, were markedly increased by TCPOBOP (Figure 9B). PCN had less of an effect except for an increase in Sult2a7 mRNA, but a moderate decrease in Sult5a1 mRNA. Papss2 was the highest expressed in control livers, followed by Sult5a1, 1d1, and 1c2; whereas other Sults were minimally expressed. However, after TCPOBOP treatment, not only the abundantly expressed Sults and Papss2 were further up-regulated, but also the lowly expressed Sults were induced, except for Sult5a1 which was slightly decreased. Therefore, TCPOBOP appeared to contribute more to the increase in sulfation reactions than did PCN.
The effect of TCPOBOP and PCN on the expression of the phase-II Ugts is shown in Figure 9C. TCPOBOP increased the mRNAs of Ugt2b34, 2b35, 2b36, 2b38, as well as Ugdh and Ugp2, which are enzymes that synthesize the co-substrate; whereas it decreased the mRNAs of Ugt2a3, 2b37, 3a1, and 3a2. PCN had less of an effect, except for an increase in the mRNAs of Ugt1a1, Ugt2b34, Ugt2b35, but a slight decrease in Ugt2b38. Ugt2b36 and 3a2 were the highest expressed Ugts in control livers, however, after TCPOBOP treatment, Ugt2b34 and 2b36 became the highest expressed Ugts; whereas after PCN treatment, Ugt1a1 became the highest expressed Ugt. Other Ugts were expressed at intermediary levels, whereas Ugt2b37 was minimally expressed under all treatment conditions. Therefore, TCPOBOP appeared to have more of an influence than PCN on glucuronidation than PCN, and the major effect appeared to be an up-regulation.
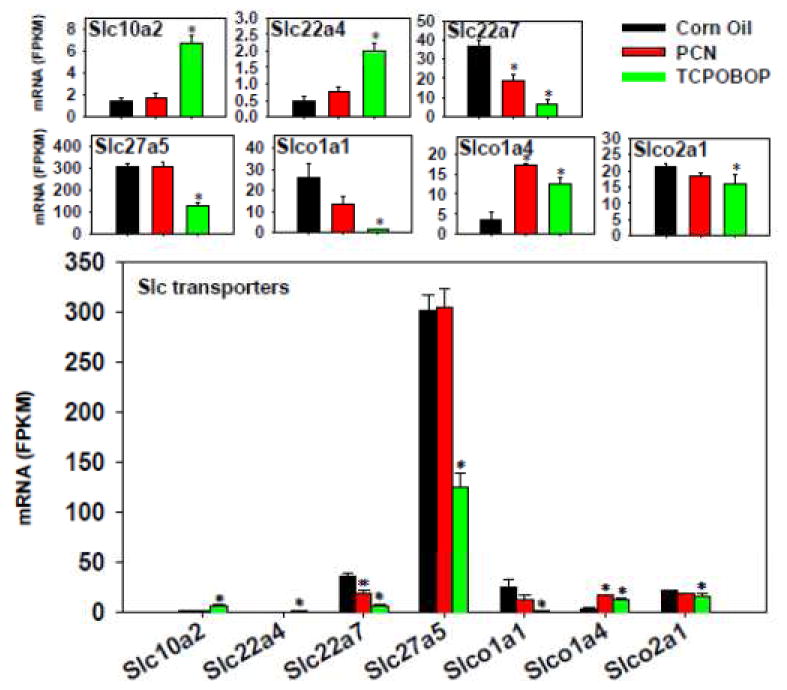
The effect of TCPOBOP and PCN on the hepatic uptake solute carrier (Slc) transporters is shown in Figure 10. TCPOBOP had a more prominent effect than PCN, as noted by a TCPOBOP-mediated up-regulation of Slc10a2/Asbt, Slc22a4/Octn1, and Slco1a4/Oatp1a4, but a down-regulation of Slc22a7/Oat2, Slc27a5/Fatp5, Slco1a1/Oatp1a1, and Slco2a1/Oatp2a1. PCN only altered the expression of two Slc genes, namely a down-regulation of Slc22a7/Octn1 but a marked up-regulation of Slco1a4/Oatp1a4. Regarding the absolute mRNA abundance, Slc27a5/Fatp5 was the highest expressed in control livers, followed by Slc22a7/Octn1, Slco1a1/Oatp1a1, and Slco2a1/Oatp2a1, whereas other Slcs were lowly expressed. Slco1b2/Oatp1b2 mRNA was not altered by TCPOBOP or PCN.
Figure 10.
Regulation of Slc transporters by PCN and TCPOBOP in livers of wild-type mice. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
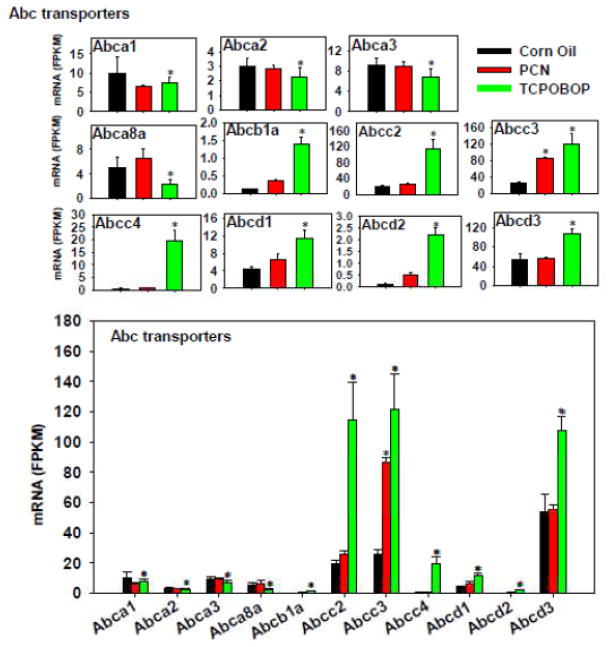
The regulation of the hepatic efflux transporters by TCPOBOP and PCN is shown in Figure 11. TCPOBOP had a major effect in up-regulating Abcb1a/Mdr1a and many Abcc/Mrp and Abcd transporters, including Abcc2-3/Mrp2-3 and Abcd1-3; however, TCPOBOP down-regulated Abca1-3, as well as Abca8a. PCN had relatively minor contributions to the regulation of these Abc transporters, except for a moderate increase in Abcc3 mRNA. Regarding the absolute mRNA abundance, Abcd2 was the highest expressed in control livers, followed by Abcc3/Mrp3 and Abcc2/Mrp2, whereas other transporters were expressed at much lower levels. Following TCPOBOP treatment, all three most abundantly expressed Abcs were further increased. Therefore, the overall contribution of the CAR ligand is likely to be an increase in Abc-mediated efflux, whereas the PXR ligand appears to have less effect.
Figure 11.
Regulation of Abc transporters by PCN and TCPOBOP in livers of wild-type mice. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
The effect of TCPOBOP and PCN on the expression of genes encoding nuclear receptors and other important xenobiotic-sensing transcription factors is shown in Figure 12. TCPOBOP markedly increased the mRNAs of Ahr/AhR, Nfe2l2/Nrf2, Rarb/RARβ), Rxrg/RXRγ, and Nr1d2/Rev-erb, but decreased the mRNAs of Ppara/PPARα, Hnf4a/HNF4α, Thra/TRα, and Nr2f6/COUP-TF3. PCN had relatively less influence on the expression of these genes, except for an increase in Esr/ERα mRNA. The absolute mRNA abundance of Hnf4a/HNF4α was the highest expressed in all treatment conditions, whereas other transcription factors were expressed at intermediate to low levels. The expression of PXR and CAR was not altered by either PCN or TCPOBOP (data not shown).
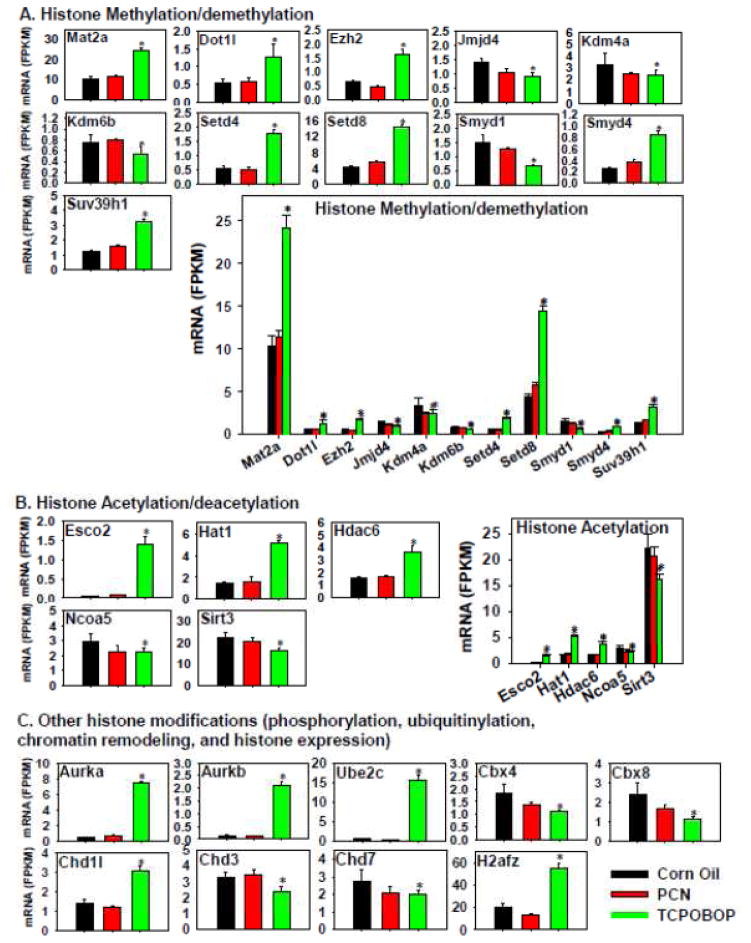
In summary, regarding the regulation of the hepatic transcription as well as the DPGs as a specific category, TCPOBOP in general had a more prominent effect on the differential regulation of these genes in comparison to PCN, suggesting that there may be global chromatin epigenetic changes as a consequence of pharmacological activation of CAR. Therefore, critical genes involved in DNA and histone modifications were examined in detail (Figure 13 and supplemental Figure 3). Regarding DNA methylation, interestingly, the expression of important genes involved in DNA methylation and gene silencing, such as DNA methyltransferases and methyl-CpG binding domain proteins, was in general not influenced by PCN or TCPOBOP treatment (except for a moderate decrease in Mbp1 mRNA by TCPOBOP). Conversely, there was a marked TCPOBOP-mediated increase in the growth arrest and DNA damage-inducible protein 45α and β (Gadd45α and β), which are involved in DNA demethylation (Ma et al., 2009) (Figure 12B). This indicates that TCPOBOP and CAR-signaling may promote DNA methylation to re-activate gene expression in liver. Regarding histone methylation and demethylation pathways, as shown in Figure 13A, TCPOBOP, but not PCN, markedly increased the hepatic mRNAs of methionine adenosyltransferase 2a (Mat2a), which synthesizes S-adenosylmethionine and promotes histone H3 lysine 9 tri-methylation (H3K9Me3) (Kera et al., 2013); DOT1- like histone H3 methyltransferase (Dot1l), which is involved in H3K79 methylation (Feng et al., 2010); enhancer of zeste 2 polycomb repressive complex 2 (Ezh2), which is involved in histone H3 lysine-27 tri-methylation (H3K27Me3) (Kondo et al., 2008); SET domain containing 4 (Setd4) and Setd8, which are histone lysine methyltransferases (Kouzarides, 2002; Zhang et al., 2003); SET and MYND domain containing 4 (Smyd4), which contains histone methyltransferase activity; as well as Suv39h1, which is involved in maintaining H3K9 methylation at pericentric heterochromatin (Rea et al., 2000). TCPOBOP also decreased the mRNAs of lysine (K)-specific demethylase 4A (Kdm4a), which is involved in the demethylation of H3K9Me2/3, H3K36Me2/3, as well as H1.4K26 (Berry and Janknecht, 2013; Cascante et al., 2014); Kdm6b, which is involved in H3K27Me3; as well as Smyd1, which has transcription corepressor activity (Burchfield et al., 2015). Considering the absolute mRNA abundance, TCPOBOP in general appeared to promote methylation of histones, whereas PCN had little effect on histone methylation/demethylation.
Figure 13.
Regulation of genes involved in histone methylation and demethylation (A), histone acetylation (B), and other histone modifications (phosphorylation, ubiquitination, chromatin remodeling, and histone expression) (C), by PCN and TCPOBOP in livers of wild-type mice. Only the differentially expressed genes in either PCN- or TCPOBOP-treated groups are presented. Genes in the same family are graphed together to quantitatively compare the mRNA abundance, and are graphed individually to better visualize the mRNA fold-changes by PCN or TCPOBOP of all genes. Asterisks represent statistically significant differences as compared to corn oil-treated group (FDR-adjusted p value <0.05).
The effect of PXR and CAR activation on the expression of genes related to histone acetylation and deacetylation is shown in Figure 13B. TCPOBOP, but not PCN, markedly increased the mRNAs of establishment of cohesion 1 homolog 2 (Esco2), which is an acetyltransferase that promotes cohesin acetylation to speed up the replication fork (Terret et al., 2009); Hat1, which catalyzes the histone and non-histone targets (Lu et al., 2012); and Hdac6, which is involved in histone deacetylation (de Ruijter et al., 2003). Conversely, TCPOBOP slightly decreased the mRNAs of nuclear receptor co-activator 5 (Ncoa5), which serves as a co-activator to recruit histone acetyltransferases or methyltransferases (Lu et al., 2012); as well as the histone deacetylase sirtuin 3 (Sirt3) (de Ruijter et al., 2003). Considering absolute mRNA abundance, TCPOBOP in general appeared to promote histone acetylation, and this could favor active gene transcription and cell replication, whereas PCN had minimal effect on histone acetylation/deacetylation.
In addition to histone methylation and acetylation, as shown in Figure 13C, TCPOBOP also increased the mRNAs of aurora kinases a and b (Aurka and b), which are involved in histone phosphorylation (Pascreau et al., 2003); ubiquitin-conjugating enzyme E2C (Ube2c), which is involved in histone ubiquitylation (Komander and Rape, 2012); chromatin remodeler chromodomain helicase DNA binding protein 1-like (Chd1l) (Ahel et al., 2009); as well as the histone H2A family member Z (H2afz). TCPOBOP moderately decreased the chromobox (Cbx) proteins 4 and 8, as well as the chromatin remodelers Chd3 and 7.
In summary, TCPOBOP appeared to be a potent epigenetic remodeler that transcriptionally modulates multiple epigenetic regulatory pathways involved in DNA demethylation, as well as histone methylation, acetylation, phosphorylation, as well as ubiquitylation and chromatin remodeling. Alterations in the expression of these epigenetic marks may at least partially explain the preferable differentiation of the hepatic transcription by TCPOBOP as compared to PCN.
DISCUSSION
The present study has systematically compared the in vivo PXR and CAR target genes in the mouse transcriptome, with a primary focus on critical genes involved in xenobiotic biotransformation and epigenetics. Results have demonstrated that PXR and CAR share many common targets, among which xenobiotic metabolism is the top category, but genes involved in inflammation, iron homeostasis, and retinoic acid metabolism are also co-regulated. Similar to CAR, PXR also binds to the direct-repeat 4 (DR-4) like motifs, which are the most prevalent PXR-DNA binding response elements in the mouse liver (Cui et al., 2010). This may at least in part explain the co-regulated gene profiles by PXR and CAR. However, PXR and CAR also have unique target gene profiles, most notably, CAR up-regulates many genes involved in cell proliferation, bile acid synthesis and composition, as well as epigenetic remodeling, whereas PXR has less effect on these pathways. In regard to the DPGs, the cumulative mRNAs of phase-I and –II drug-metabolizing enzymes as well as efflux transporters are up-regulated by PCN and to a greater extent by TCPOBOP, however, the cumulative mRNAs of the efflux transporters are down-regulated only by TCPOBOP. The absolute mRNA abundance of each DPG category has unveiled the major responder genes following exposure to PCN or TCPOBOP, and this aids in the prediction of the most predominant pharmacokinetic modifications in liver following PXR or CAR activation.
The regulation of many hepatic DPGs by PXR and CAR ligands has been characterized in mice using branched DNA amplification (bDNA) assays (Cheng et al., 2005; Maher et al., 2005; Alnouti and Klaassen, 2008a; Alnouti and Klaassen, 2008b; Knight et al., 2008; Buckley and Klaassen, 2009; Cui et al., 2009; Aleksunes and Klaassen, 2012; Cheng and Klaassen, 2012; Zhang et al., 2012; Pratt-Hyatt et al., 2013). The present study using RNA-Seq is consistent with these previous studies regarding the up-regulation of Cyp1a2, Cyp2b10, Aldh1a7, Gsta1, Gsta4, Gstm1-3, Gstt1, Sult1e1, Papss2, Ugt2b34-36, and Mrp2-4, most notably by the chemical activation of CAR by TCPOBOP but also by PCN to a lesser extent; whereas PCN also up-regulates Gstm1-3 and Mrp3 in both studies to a much lesser extent than does TCPOBOP (Figure 3, 8, 11, 12, and 13) (Maher et al., 2005; Alnouti and Klaassen, 2008a; Knight et al., 2008; Buckley and Klaassen, 2009). The present study is also consistent with these previous studies regarding the co-up-regulated genes Cyp3a11 and Oatp1a4 by both PCN and TCPOBOP (Figure 10) (Aleksunes and Klaassen, 2012). Although the up-regulation of the prototypical CAR-target gene Cyp2b10 by the PXR ligand PCN was observed in the present study (Figure 3) and in another study (Pacyniak et al., 2007), it only tended to be induced by PCN without statistical significance in male mice, but was significant in female mice (Aleksunes and Klaassen 2012). Similarly, the PCN-mediated up-regulation of the mRNAs of Aldh1a1, Aldh1b1, Gsta4, Gstm4, and Papss2, the TCPOBOP-mediated up-regulation of Aldh3a2 mRNA; as well as the TCPOBOP-mediated down-regulation of Cyp4a14 and Aldh6a2, were not observed previously in male mice (Aleksunes and Klaassen 2012). The PCN-mediated increase of Aldh1a1 and 1b1 mRNAs was consistent with another study (Alnouti and Klaassen, 2008a). The discrepancies are likely due to small fold-change in gene expression, sample size, statistics, or different platforms of mRNA quantification (such as probe design strategies in bDNA assay and mapping strategies in RNA-Seq). In female mice, for certain DPGs discussed above, they were differentially regulated by PCN or TCPOBOP (Aleksunes and Klaassen, 2012), suggesting that gender also plays a role in PXR and CAR signaling in liver.
The data on aldo-ketoreductase in the present study are consistent with a previous study regarding the PCN- and TCPOBOP-mediated up-regulation in the Akr1b7 mRNA, as well as the TCPOBOP-mediated up-regulation in the mRNAs of Akr1c19 and 1d1 (Pratt-Hyatt et al., 2013). The present study has also determined the regulation of all other Akrs in the mouse genome, and demonstrated that TCPOBOP up-regulates Akr1b8 mRNA, which was not included in the previous study. The TCPOBOP-mediated increase in Akr1c14 mRNA and PCN-mediated increase in Akr1d1 mRNA observed in the present study were not observed in the previous study. Regarding the carboxyesterases, the present study is consistent with a previous study on the PCN- and TCPOBOP-mediated increase in Ces2a mRNA, TCPOBOP-mediated increase in Ces2c mRNA, but decrease in the mRNAs of Ces3a and Aadac (Zhang et al., 2012). In addition, the present study has provided new information regarding the regulation of all other Ces genes that was not included in the previous study, and has demonstrated that TCPOBOP uniquely down-regulates the mRNAs of Ces1b and 3a, whereas both PCN and TCPOBOP up-regulate Ces2g mRNA but down-regulate Ces4a mRNA. There are certain discrepancies between the present study and the study by Zhang and Klaassen (2012), for example, the PCN-mediated increase in the mRNAs of Ces1d, 1e, and 1g was observed in the present study but not in the previous study. Ces2e mRNA was down-regulated slightly by TCPOBOP in the present study but not in the previous study; whereas PCN increased Ces2c mRNA in the previous study but not in the present study. As mentioned before, the discrepancies are likely due to small fold-change in gene expression, sample size, and different methodology.
The present study has provided new information regarding the regulation of many other categories of DPGs by PCN and TCPOBOP, which was not included in previous studies; these gene categories include the alcohol dehydrogenases (Figure 6A), dehydrogenases and reductases (Figure 7B), glutathione peroxidases and peroxiredoxins (Figure 8A), flavin containing monooxygenases, methyltransferases and acetyltransferases (Figure 8B–C), as well as the regulation of xenobiotic-sensing transcription factors (Figure 12) and epigenetic modifiers (Figure 13 and supplemental Figure 1). In addition, regarding the gene categories that have been characterized before, due to technical challenges in designing gene-specific primers or specific target-quantification on a multiplexing platform, previous studies only included a limited number of DPGs in that particular gene category; whereas the present has included all the DPG isoforms using RNA-Seq. Thus the present study has further expanded our knowledge regarding the PXR- and CAR-mediated regulation of all DPGs in liver.
The present study has demonstrated that at the given doses, the CAR ligand TCPOBOP differentially regulates many more genes that the PXR ligand PCN in mouse liver (Figure 1), and this appears to correlate with a marked change in the expression of many epigenetic regulators by TCPOBOP (Figure 13). Indeed, CAR has been shown in previous studies to be a potent epigenetic reprogramming factor. For example, neonatal activation of CAR results in epigenetic memory, evidenced by a permanent increase of H3K4Me1, H3K4Me2, and H3K4Me3, but a decrease of H3K9Me3 around the Cyp2b10 gene locus (Chen et al., 2012). The present study has demonstrated that TCPOBOP increases the mRNAs of multiple histone methyltransferases, but a decrease in H3K9Me3 demethylase Kdma, which are consistent with the previous findings (Chen et al., 2012). TCPOBOP increased the mRNA of Gadd45β, which is involved in cell proliferation and DNA demethylation, and is consistent with the previous studies that demonstrated that Gadd45β is induced in a CAR-dependent pathway to promote liver hyperplasia (Columbano et al., 2005). In fact, loss of Gadd45β in hepatocytes has been shown to attenuate TCPOBOP-mediated up-regulation of Cyp2b10 mRNA (Columbano et al., 2005; Tian et al., 2011), and the CAR-activator phenobarbital-mediated epigenetic switch in DNA methylation has been observed around the Cyp2b10 gene promoter (Lempiainen et al., 2011). Conversely, although many epigenetic modifiers have been shown to be involved in PXR-signaling (Xie et al., 2009; Habano et al., 2011; Tian, 2013; Ma et al., 2015c), pharmacological activation of PXR in liver does not appear to alter the expression of these genes at the transcriptional level.
Although PXR and CAR are well-known xenobiotic-sensing receptors, the present study demonstrates that many other pathways appear to be regulated by PXR and CAR ligands. For example, TCPOBOP appears to regulate bile acid synthesis and composition, because it markedly increased the mRNA of cholesterol 7α hydroxylase (Cyp7a1), which is the rate-limiting enzyme for bile acid synthesis in the classic pathway, but decreased the mRNA of the 12α-hydroxylase (Cyp8b1) that synthesizes cholic acid, as well as Cyp27a1 (note: is up-regulated by PCN also) and Cyp7b1 that is involved in the alternative pathway of bile acid synthesis (Figure 5). Decreased Cyp8b1 expression by TCPOBOP was also observed in previous studies (Stedman et al., 2005; Miao et al., 2006); however, Cyp7a1 mRNA was found to be decreased in other studies following PCN or TCPOBOP treatment (Staudinger et al., 2001; Stedman et al., 2005; Miao et al., 2006), likely because the doses used in these studies lead to a cholestatic-like condition that activates other receptors, such as FXR and intestinal Fgf15 to down-regulate Cyp7a1. Deletion of CAR has been shown to result in a less severe repression of Cyp7b1 and Cyp8b1 expression after bile duct ligation, indicating that CAR is at least partly responsible for the down-regulation of these genes (Stedman et al., 2005). The present study is also consistent with the literature regarding the PXR- and CAR-mediated up-regulation in the efflux transporters Mrp2-4, as well as the uptake transporter Oatp1a4 in liver (Tabbot and Robson, 2006).
The present study has shown that both PXR and CAR ligands appear to down-regulate multiple genes involved in the pro-inflammatory response. PXR has been shown to inhibit inflammation, evidenced by decreased Tumor necrosis factor α and interleukin 1-α mRNA expression, and attenuated NF-κB-signaling both in mouse liver (Wallace et al., 2010) and in primary culture of hepatocytes (Sun et al., 2015). Relatively less is known regarding the contribution of CAR in hepatic inflammation, but the RNA-Seq data in the present study also suggests the potential anti-inflammatory function of CAR in liver.
Regarding the cross-talk between nuclear receptors, the present study is consistent with the previous literature regarding CAR-mediated suppression of HNF4α (Gallina et al., 1989) (Figure 12). The CAR-mediated suppression of HNF4α may lead to a decrease in glucose and lipid metabolism pathways. There are apparent interactions between CAR and other receptors such as AhR, PXR, and PPARα, because the activators for these other receptors can also activate CAR-signaling (Oshida et al., 2015). The present study has shown that activation of CAR by TCPOBOP up-regulates the AhR-target genes Cyp1a as well as many PXR-target genes, however, it actually down-regulates the PPARα-target genes in the Cyp4a family. A comparison between PXR and CAR activation by lithocholic acid in wild-type and PXR/CAR-null mice has shown that CAR predominantly mediates the induction of Cyp3a11 and Mrp3, whereas PXR is the major regulator of Oatp1a4 (Zhang et al., 2004). However, the present study has shown that pharmacological activation of PXR and CAR can both lead to the up-regulation of these genes.
There are species-differences in the pharmacodynamic functions of PXR and CAR between rodents and humans, thus caution needs to be made when extrapolating the data from mice. Similar to mice, the human (h) PXR and hCAR also have overlapping but also distinct biological functions in human hepatocytes (Maglich et al., 2003). In contrast to mice, hPXR and hCAR only support the hypertrophic but not the hyperplastic response to the mouse non-genotoxic hepatocarcinogens phenobarbital and chlordane (which are both CAR and PXR activators) in livers of hPXR/hCAR-humanized mice (Ross et al., 2010). In human HepaRG cells, many genes promoting cell proliferation and tumorigenesis were up-regulated in hCAR-knockout cells, suggesting that hCAR plays an important role in cell growth that differs from mouse CAR (Li et al., 2015). Indeed, the mode of action for phenobarbital-induced rodent liver tumor formation is considered to be qualitatively not plausible in humans, supported by data from various epidemiological studies conducted in human populations that were chronically exposed to phenobarbital, in which there was no observed evidence for increased risk for liver tumors (Elcombe et al., 2014). Regarding drug metabolism pathways, similar to the mouse receptors, in HepG2 cells hPXR and hCAR activated by herbal supplements also up-regulate the expression and activities of CYP2B6, which is the human homolog of mouse Cyp2b10 (Xu et al., 2015). Regarding hPXR signaling, the hPXR ligand rifampicin treated hPXR/hCAR-double-humanized mice had increased hepatic microsomal protein, total Cyp contents, Cyp reductase activity, and UGT activity (Lee et al., 2015). Regarding hCAR signaling, genes encoding drug-metabolizing enzymes are among the main clusters altered by both the hCAR ligand CITCO, and the species-unspecific CAR activator phenobarbital (which can also activate PXR) (Li et al., 2015), whereas similar results were observed in livers of mice treated with TCPOBOP (Figure 2). At the given concentrations, CITCO differentially regulated the expression of 135 genes in a hCAR-dependent manner, whereas phenobarbital altered the expression of 227 genes, among which 94 genes were up-regulated in a hCAR-independent manner (likely due to hPXR effect) (Li et al., 2015).
In conclusion, the present study has revealed known and novel, as well as common and unique targets of the PXR and CAR receptors in mouse liver following chemical activation using their prototypical ligands. Results from this study further support the role of these important nuclear receptors in regulating the homeostasis of xenobiotic and intermediary metabolism in liver, and aid in distinguishing between PXR and CAR signaling at various physiological and pathophysiological conditions.
Supplementary Material
Acknowledgments
The authors would like to thank members in Dr. Klaassen’s laboratory for help in tissue collection, Mr. Clark Bloomer for the technical assistance in library construction and sequencing, as well as members in Dr. Cui’s laboratory for help in bioinformatics.
Supported by National Institute of Health R-01 grants ES019487, ES025708, and GM11138, as well as start-up funds from University of Washington Center for Ecogenetics and Environmental Health (P30ES007033).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad G, Sial GZ, Ramadori P, Dudas J, Batusic DS, Ramadori G. Changes of hepatic lactoferrin gene expression in two mouse models of the acute phase reaction. Int J Biochem Cell Biol. 2011;43:1822–1832. doi: 10.1016/j.biocel.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Aleksunes LM, Klaassen CD. Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARalpha-, and Nrf2-null mice. Drug Metab Dispos. 2012;40:1366–1379. doi: 10.1124/dmd.112.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. J Pharmacol Exp Ther. 2008a;324:612–621. doi: 10.1124/jpet.107.129650. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci. 2008b;101:51–64. doi: 10.1093/toxsci/kfm280. [DOI] [PubMed] [Google Scholar]
- Aroca P, Garcia-Borron JC, Solano F, Lozano JA. Regulation of mammalian melanogenesis. I: Partial purification and characterization of a dopachrome converting factor: dopachrome tautomerase. Biochim Biophys Acta. 1990;1035:266–275. doi: 10.1016/0304-4165(90)90088-e. [DOI] [PubMed] [Google Scholar]
- Arrese M, Karpen SJ. Nuclear receptors, inflammation, and liver disease: insights for cholestatic and fatty liver diseases. Clin Pharmacol Ther. 2010;87:473–478. doi: 10.1038/clpt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J Hepatol. 2012;56:952–964. doi: 10.1016/j.jhep.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Beilke LD, Aleksunes LM, Holland RD, Besselsen DG, Beger RD, Klaassen CD, Cherrington NJ. Constitutive androstane receptor-mediated changes in bile acid composition contributes to hepatoprotection from lithocholic acid-induced liver injury in mice. Drug Metab Dispos. 2009a;37:1035–1045. doi: 10.1124/dmd.108.023317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilke LD, Aleksunes LM, Olson ER, Besselsen DG, Klaassen CD, Dvorak K, Cherrington NJ. Decreased apoptosis during CAR-mediated hepatoprotection against lithocholic acid-induced liver injury in mice. Toxicol Lett. 2009b;188:38–44. doi: 10.1016/j.toxlet.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard HC, Holmskov U, Santoni-Rugiu E, Nagy P, Nielsen O, Ott P, Hage E, Dalhoff K, Rasmussen LJ, Tygstrup N. Heterogeneity of ductular reactions in adult rat and human liver revealed by novel expression of deleted in malignant brain tumor 1. Am J Pathol. 2002;161:1187–1198. doi: 10.1016/S0002-9440(10)64395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosron WF, Yin SJ, Li TK. Purification and characterization of human liver beta 1 beta 1, beta 2 beta 2 and beta Ind beta Ind alcohol dehydrogenase isoenzymes. Prog Clin Biol Res. 1985;174:193–206. [PubMed] [Google Scholar]
- Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, Kajimura S, Zingaretti MC, Vind BF, Tu H, Cinti S, Hojlund K, Gygi SP, Spiegelman BM. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DB, Klaassen CD. Induction of mouse UDP-glucuronosyltransferase mRNA expression in liver and intestine by activators of aryl-hydrocarbon receptor, constitutive androstane receptor, pregnane X receptor, peroxisome proliferator-activated receptor alpha, and nuclear factor erythroid 2-related factor 2. Drug Metab Dispos. 2009;37:847–856. doi: 10.1124/dmd.108.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitrago-Molina LE, Marhenke S, Longerich T, Sharma AD, Boukouris AE, Geffers R, Guigas B, Manns MP, Vogel A. The degree of liver injury determines the role of p21 in liver regeneration and hepatocarcinogenesis in mice. Hepatology. 2013;58:1143–1152. doi: 10.1002/hep.26412. [DOI] [PubMed] [Google Scholar]
- Burchfield JS, Li Q, Wang HY, Wang RF. JMJD3 as an epigenetic regulator in development and disease. Int J Biochem Cell Biol. 2015;67:148–157. doi: 10.1016/j.biocel.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascante A, Klum S, Biswas M, Antolin-Fontes B, Barnabe-Heider F, Hermanson O. Gene-specific methylation control of H3K9 and H3K36 on neurotrophic BDNF versus astroglial GFAP genes by KDM4A/C regulates neural stem cell differentiation. J Mol Biol. 2014;426:3467–3477. doi: 10.1016/j.jmb.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Casteels K, Mathieu C. Diabetic ketoacidosis. Rev Endocr Metab Disord. 2003;4:159–166. doi: 10.1023/a:1022942120000. [DOI] [PubMed] [Google Scholar]
- Chen WD, Fu X, Dong B, Wang YD, Shiah S, Moore DD, Huang W. Neonatal activation of the nuclear receptor CAR results in epigenetic memory and permanent change of drug metabolism in mouse liver. Hepatology. 2012;56:1499–1509. doi: 10.1002/hep.25766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Krausz KW, Tanaka N, Gonzalez FJ. Chronic exposure to rifaximin causes hepatic steatosis in pregnane X receptor-humanized mice. Toxicol Sci. 2012;129:456–468. doi: 10.1093/toxsci/kfs211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Klaassen CD. Hormonal and chemical regulation of paraoxonases in mice. J Pharmacol Exp Ther. 2012;342:688–695. doi: 10.1124/jpet.112.194803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Maher J, Dieter MZ, Klaassen CD. Regulation of mouse organic anion-transporting polypeptides (Oatps) in liver by prototypical microsomal enzyme inducers that activate distinct transcription factor pathways. Drug Metab Dispos. 2005;33:1276–1282. doi: 10.1124/dmd.105.003988. [DOI] [PubMed] [Google Scholar]
- Chiang JY. Nuclear receptor regulation of lipid metabolism: potential therapeutics for dyslipidemia, diabetes, and chronic heart and liver diseases. Curr Opin Investig Drugs. 2005;6:994–1001. [PubMed] [Google Scholar]
- Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009a;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY. Hepatocyte nuclear factor 4alpha regulation of bile acid and drug metabolism. Expert Opin Drug Metab Toxicol. 2009b;5:137–147. doi: 10.1517/17425250802707342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbano A, Ledda-Columbano GM, Pibiri M, Cossu C, Menegazzi M, Moore DD, Huang W, Tian J, Locker J. Gadd45beta is induced through a CAR-dependent, TNF-independent pathway in murine liver hyperplasia. Hepatology. 2005;42:1118–1126. doi: 10.1002/hep.20883. [DOI] [PubMed] [Google Scholar]
- Cui JY, Gunewardena SS, Rockwell CE, Klaassen CD. ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Res. 2010;38:7943–7963. doi: 10.1093/nar/gkq654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YJ, Cheng X, Weaver YM, Klaassen CD. Tissue distribution, gender-divergent expression, ontogeny, and chemical induction of multidrug resistance transporter genes (Mdr1a, Mdr1b, Mdr2) in mice. Drug Metab Dispos. 2009;37:203–210. doi: 10.1124/dmd.108.023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey MJ, Kim S. Inositol polyphosphate multikinase: an emerging player for the central action of AMP-activated protein kinase. Biochem Biophys Res Commun. 2012;421:1–3. doi: 10.1016/j.bbrc.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson PB. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3:561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcombe CR, Peffer RC, Wolf DC, Bailey J, Bars R, Bell D, Cattley RC, Ferguson SS, Geter D, Goetz A, Goodman JI, Hester S, Jacobs A, Omiecinski CJ, Schoeny R, Xie W, Lake BG. Mode of action and human relevance analysis for nuclear receptor-mediated liver toxicity: A case study with phenobarbital as a model constitutive androstane receptor (CAR) activator. Crit Rev Toxicol. 2014;44:64–82. doi: 10.3109/10408444.2013.835786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yang Y, Ortega MM, Copeland JN, Zhang M, Jacob JB, Fields TA, Vivian JL, Fields PE. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood. 2010;116:4483–4491. doi: 10.1182/blood-2010-03-276501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando L. Multiple personality disorder. Br J Psychiatry. 1990;157:150. doi: 10.1192/bjp.157.1.150b. [DOI] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Frank C, Gonzalez MM, Oinonen C, Dunlop TW, Carlberg C. Characterization of DNA complexes formed by the nuclear receptor constitutive androstane receptor. J Biol Chem. 2003;278:43299–43310. doi: 10.1074/jbc.M305186200. [DOI] [PubMed] [Google Scholar]
- Gallina G, Cumbo V, Messina P, Caprera V, Lio D, Caruso C. Lack of correlation between HLA-B35 resistance against herpes labialis and antibody titers to HSV-1. Oral Surg Oral Med Oral Pathol. 1989;68:167–170. doi: 10.1016/0030-4220(89)90187-4. [DOI] [PubMed] [Google Scholar]
- Gao J, He J, Zhai Y, Wada T, Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J Biol Chem. 2009;284:25984–25992. doi: 10.1074/jbc.M109.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C, Sakkab DY, Scholzen T, Dassler R, Alison MR, Gerdes J. Ki-67 expression during rat liver regeneration after partial hepatectomy. Hepatology. 1997;26:573–578. doi: 10.1002/hep.510260307. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Cytochrome p450 and chemical toxicology. Chem Res Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- Habano W, Gamo T, Terashima J, Sugai T, Otsuka K, Wakabayashi G, Ozawa S. Involvement of promoter methylation in the regulation of Pregnane X receptor in colon cancer cells. BMC Cancer. 2011;11:81. doi: 10.1186/1471-2407-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Gao J, Xu M, Ren S, Stefanovic-Racic M, O'Doherty RM, Xie W. PXR ablation alleviates diet-induced and genetic obesity and insulin resistance in mice. Diabetes. 2013;62:1876–1887. doi: 10.2337/db12-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine M, Cramm-Behrens CI, Ansari A, Chu HP, Ryazanov AG, Naim HY, Jacob R. Alpha-kinase 1, a new component in apical protein transport. J Biol Chem. 2005;280:25637–25643. doi: 10.1074/jbc.M502265200. [DOI] [PubMed] [Google Scholar]
- Hernandez JP, Mota LC, Baldwin WS. Activation of CAR and PXR by Dietary, Environmental and Occupational Chemicals Alters Drug Metabolism, Intermediary Metabolism, and Cell Proliferation. Curr Pharmacogenomics Person Med. 2009;7:81–105. doi: 10.2174/187569209788654005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist E, Okroj M, Nodin B, Jirstrom K, Blom AM. Sushi domain-containing protein 4 (SUSD4) inhibits complement by disrupting the formation of the classical C3 convertase. Faseb J. 2013;27:2355–2366. doi: 10.1096/fj.12-222042. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Furihata T, Yaginuma Y, Yamamoto N, Watanabe N, Tsukada E, Ohhata Y, Kobayashi K, Satoh T, Chiba K. Structural organization and characterization of the regulatory element of the human carboxylesterase (CES1A1 and CES1A2) genes. Drug Metab Pharmacokinet. 2008;23:73–84. doi: 10.2133/dmpk.23.73. [DOI] [PubMed] [Google Scholar]
- Hubbard B, Doege H, Punreddy S, Wu H, Huang X, Kaushik VK, Mozell RL, Byrnes JJ, Stricker-Krongrad A, Chou CJ, Tartaglia LA, Lodish HF, Stahl A, Gimeno RE. Mice deleted for fatty acid transport protein 5 have defective bile acid conjugation and are protected from obesity. Gastroenterology. 2006;130:1259–1269. doi: 10.1053/j.gastro.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Jang WY, Bae KB, Kim SH, Yu DH, Kim HJ, Ji YR, Park SJ, Kang MC, Jeong JI, Lee SG, Lee I, Kim MO, Yoon D, Ryoo ZY. Overexpression of Jazf1 reduces body weight gain and regulates lipid metabolism in high fat diet. Biochem Biophys Res Commun. 2014;444:296–301. doi: 10.1016/j.bbrc.2013.12.094. [DOI] [PubMed] [Google Scholar]
- Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- Kakhnover NB, Khmelevskii Iu V. Glutathione-S-transferases as detoxication enzymes. Ukr Biokhim Zh (1978) 1983;55:86–92. [PubMed] [Google Scholar]
- Kera Y, Katoh Y, Ohta M, Matsumoto M, Takano-Yamamoto T, Igarashi K. Methionine adenosyltransferase II-dependent histone H3K9 methylation at the COX-2 gene locus. J Biol Chem. 2013;288:13592–13601. doi: 10.1074/jbc.M112.429738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev. 2010;62:1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Liu J, Choudhuri S. Metallothionein: an intracellular protein to protect against cadmium toxicity. Annu Rev Pharmacol Toxicol. 1999;39:267–94. doi: 10.1146/annurev.pharmtox.39.1.267. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev. 2002;23:687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Knight TR, Choudhuri S, Klaassen CD. Induction of hepatic glutathione S-transferases in male mice by prototypes of various classes of microsomal enzyme inducers. Toxicol Sci. 2008;106:329–338. doi: 10.1093/toxsci/kfn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Lee SY, Lee JY, Kim YM, Kim SK, Oh SJ. Expression of hepatic cytochrome P450s and UDP-glucuronosyltransferases in PXR and CAR double humanized mice treated with rifampicin. Toxicol Lett. 2015;235:107–115. doi: 10.1016/j.toxlet.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Lemaire G, Mnif W, Pascussi JM, Pillon A, Rabenoelina F, Fenet H, Gomez E, Casellas C, Nicolas JC, Cavailles V, Duchesne MJ, Balaguer P. Identification of new human pregnane X receptor ligands among pesticides using a stable reporter cell system. Toxicol Sci. 2006;91:501–509. doi: 10.1093/toxsci/kfj173. [DOI] [PubMed] [Google Scholar]
- Lempiainen H, Muller A, Brasa S, Teo SS, Roloff TC, Morawiec L, Zamurovic N, Vicart A, Funhoff E, Couttet P, Schubeler D, Grenet O, Marlowe J, Moggs J, Terranova R. Phenobarbital mediates an epigenetic switch at the constitutive androstane receptor (CAR) target gene Cyp2b10 in the liver of B6C3F1 mice. PLoS One. 2011;6:e18216. doi: 10.1371/journal.pone.0018216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Mackowiak B, Brayman TG, Mitchell M, Zhang L, Huang SM, Wang H. Genome-wide analysis of human constitutive androstane receptor (CAR) transcriptome in wild-type and CAR-knockout HepaRG cells. Biochem Pharmacol. 2015;98:190–202. doi: 10.1016/j.bcp.2015.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Takahashi Y, Wada T, He J, Gao J, Tian Y, Li S, Xie W. The aldo-keto reductase Akr1b7 gene is a common transcriptional target of xenobiotic receptors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol. 2009;76:604–611. doi: 10.1124/mol.109.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, Kuo I, Perozich J, Lindahl R, Hempel J, Wang BC. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4:317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- Lu B, Sims PJ, Wiedmer T, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. Expression of the phospholipid scramblase (PLSCR) gene family during the acute phase response. Biochim Biophys Acta. 2007;1771:1177–1185. doi: 10.1016/j.bbalip.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Lu H, Cui JY, Gunewardena S, Yoo B, Zhong XB, Klaassen CD. Hepatic ontogeny and tissue distribution of mRNAs of epigenetic modifiers in mice using RNA-sequencing. Epigenetics. 2012;7:914–929. doi: 10.4161/epi.21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Gunewardena S, Cui JY, Yoo B, Zhong XB, Klaassen CD. RNA-sequencing quantification of hepatic ontogeny and tissue distribution of mRNAs of phase II enzymes in mice. Drug Metab Dispos. 2013;41:844–857. doi: 10.1124/dmd.112.050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Seravalli J, Harrison-Findik D. Inductively coupled mass spectrometry analysis of biometals in conditional Hamp1 and Hamp1 and Hamp2 transgenic mouse models. Transgenic Res. 2015;24:765–773. doi: 10.1007/s11248-015-9879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MX, Wong SH, Chan MT, Yu L, Yu SS, Wu F, Xiao Z, Wang X, Zhang L, Cheng AS, Ng SS, Chan FK, Cho CH, Yu J, Sung JJ, Wu WK. Autophagy Mediates HBx-Induced Nuclear Factor-kappaB Activation and Release of IL-6, IL-8, and CXCL2 in Hepatocytes. J Cell Physiol. 2015;230:2382–2389. doi: 10.1002/jcp.24967. [DOI] [PubMed] [Google Scholar]
- Lynch C, Pan Y, Li L, Heyward S, Moeller T, Swaan PW, Wang H. Activation of the constitutive androstane receptor inhibits gluconeogenesis without affecting lipogenesis or fatty acid synthesis in human hepatocytes. Toxicol Appl Pharmacol. 2014;279:33–42. doi: 10.1016/j.taap.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JQ, Li Z, Xie WR, Liu CM, Liu SS. Quercetin protects mouse liver against CCl(4)-induced inflammation by the TLR2/4 and MAPK/NF-kappaB pathway. Int Immunopharmacol. 2015a;28:531–539. doi: 10.1016/j.intimp.2015.06.036. [DOI] [PubMed] [Google Scholar]
- Ma Q, Zhao J, Cao W, Liu J, Cui S. Estradiol decreases taurine level by reducing cysteine sulfinic acid decarboxylase via the estrogen receptor-alpha in female mice liver. Am J Physiol Gastrointest Liver Physiol. 2015b;308:G277–286. doi: 10.1152/ajpgi.00107.2014. [DOI] [PubMed] [Google Scholar]
- Ma X, Chen J, Tian Y. Pregnane X receptor as the “sensor and effector” in regulating epigenome. J Cell Physiol. 2015c;230:752–757. doi: 10.1002/jcp.24838. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, Moore JT. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- Maher JM, Cheng X, Slitt AL, Dieter MZ, Klaassen CD. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos. 2005;33:956–962. doi: 10.1124/dmd.105.003798. [DOI] [PubMed] [Google Scholar]
- Makinen J, Frank C, Jyrkkarinne J, Gynther J, Carlberg C, Honkakoski P. Modulation of mouse and human phenobarbital-responsive enhancer module by nuclear receptors. Mol Pharmacol. 2002;62:366–378. doi: 10.1124/mol.62.2.366. [DOI] [PubMed] [Google Scholar]
- McCarver DG, Hines RN. The ontogeny of human drug-metabolizing enzymes: phase II conjugation enzymes and regulatory mechanisms. J Pharmacol Exp Ther. 2002;300:361–366. doi: 10.1124/jpet.300.2.361. [DOI] [PubMed] [Google Scholar]
- Miao J, Fang S, Bae Y, Kemper JK. Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J Biol Chem. 2006;281:14537–14546. doi: 10.1074/jbc.M510713200. [DOI] [PubMed] [Google Scholar]
- Moore JT, Kliewer SA. Use of the nuclear receptor PXR to predict drug interactions. Toxicology. 2000;153:1–10. doi: 10.1016/s0300-483x(00)00300-0. [DOI] [PubMed] [Google Scholar]
- Moore JT, Moore LB, Maglich JM, Kliewer SA. Functional and structural comparison of PXR and CAR. Biochim Biophys Acta. 2003;1619:235–238. doi: 10.1016/s0304-4165(02)00481-6. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, Collins JL, Kliewer SA. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- Nabeshima A, Yamada S, Guo X, Tanimoto A, Wang KY, Shimajiri S, Kimura S, Tasaki T, Noguchi H, Kitada S, Watanabe T, Fujii J, Kohno K, Sasaguri Y. Peroxiredoxin 4 protects against nonalcoholic steatohepatitis and type 2 diabetes in a nongenetic mouse model. Antioxid Redox Signal. 2013;19:1983–1998. doi: 10.1089/ars.2012.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandchahal KK, Routh J, Mathur S, Bhartiya HC. Radiation response of plasma protein and albumin of peripheral blood and its modification by MPG (2-Mercaptopropionylglycine) in mice. Strahlenther Onkol. 1990;166:306–309. [PubMed] [Google Scholar]
- Nebert DW, Gonzalez FJ. P450 genes: structure, evolution, and regulation. Annu Rev Biochem. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Wikvall K, Miller WL. Human cytochromes P450 in health and disease. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120431. doi: 10.1098/rstb.2012.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr. 2006;26:323–342. doi: 10.1146/annurev.nutr.26.061505.111303. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Naito S, Yokoi T. Tissue-specific mRNA expression profiles of human nuclear receptor subfamilies. Drug Metab Pharmacokinet. 2004;19:135–149. doi: 10.2133/dmpk.19.135. [DOI] [PubMed] [Google Scholar]
- Oshida K, Vasani N, Jones C, Moore T, Hester S, Nesnow S, Auerbach S, Geter DR, Aleksunes LM, Thmas RS, Applegate D, Klaassen CD, Corton JC. Identification of chemical modulator of the constitutive activated receptor (CAR) in a gene expression compendium. Nucl Recept Signal. 2015;13:e002. doi: 10.1621/nrs.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacyniak EK, Cheng X, Cunningham ML, Crofton K, Klaassen CD, Guo GL. The flame retardants, polybrominated diphenyl ethers, are pregnane X receptor activators. Toxicol Sci. 2007;97:94–102. doi: 10.1093/toxsci/kfm025. [DOI] [PubMed] [Google Scholar]
- Parkinson A, Ogilvie BW, Buckley DB, Kazmi F, Czerwinski M, Parkinson O. Biotransformation of Xenobiotics. In: Klaassen CD, editor. 2013 Casarett & Doull’s Toxicology, The Basic Science of Poisons. 8 [Google Scholar]
- Pascreau G, Arlot-Bonnemains Y, Prigent C. Phosphorylation of histone and histone-like proteins by aurora kinases during mitosis. Prog Cell Cycle Res. 2003;5:369–374. [PubMed] [Google Scholar]
- Peng L, Cui JY, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong XB. RNA-sequencing quantification of hepatic ontogeny of phase-I enzymes in mice. Drug Metab Dispos. 2013;41:2175–2186. doi: 10.1124/dmd.113.054635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong XB. RNA sequencing reveals dynamic changes of mRNA abundance of cytochromes P450 and their alternative transcripts during mouse liver development. Drug Metab Dispos. 2012;40:1198–1209. doi: 10.1124/dmd.112.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrick JS, Klaassen CD. Importance of hepatic induction of constitutive androstane receptor and other transcription factors that regulate xenobiotic metabolism and transport. Drug Metab Dispos. 2007;35:1806–1815. doi: 10.1124/dmd.107.015974. [DOI] [PubMed] [Google Scholar]
- Porter LA, Donoghue DJ. Cyclin B1 and CDK1: nuclear localization and upstream regulators. Prog Cell Cycle Res. 2003;5:335–347. [PubMed] [Google Scholar]
- Pratt-Hyatt M, Lickteig AJ, Klaassen CD. Tissue distribution, ontogeny, and chemical induction of aldo-keto reductases in mice. Drug Metab Dispos. 2013;41:1480–1487. doi: 10.1124/dmd.113.051904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol A, Troffer-Charlier N, Metzger E, Chimini G, Mandel JL. Characterization of the adrenoleukodystrophy-related (ALDR, ABCD2) gene promoter: inductibility by retinoic acid and forskolin. Genomics. 2000;70:131–139. doi: 10.1006/geno.2000.6367. [DOI] [PubMed] [Google Scholar]
- Ramadoss P, Chiappini F, Bilban M, Hollenberg AN. Regulation of hepatic six transmembrane epithelial antigen of prostate 4 (STEAP4) expression by STAT3 and CCAAT/enhancer-binding protein alpha. J Biol Chem. 2010;285:16453–16466. doi: 10.1074/jbc.M109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Renaud HJ, Cui YJ, Lu H, Zhong XB, Klaassen CD. Ontogeny of hepatic energy metabolism genes in mice as revealed by RNA-sequencing. PLoS One. 2014;9:e104560. doi: 10.1371/journal.pone.0104560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Plummer SM, Rode A, Scheer N, Bower CC, Vogel O, Henderson CJ, Wolf CR, Elcombe CR. Human constitutive androstane receptor (CAR) and pregnane X receptor (PXR) support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogens phenobarbital and chlordane in vivo. Toxicol Sci. 2010;116:452–466. doi: 10.1093/toxsci/kfq118. [DOI] [PubMed] [Google Scholar]
- Ruiz FX, Moro A, Gallego O, Ardevol A, Rovira C, Petrash JM, Pares X, Farres J. Human and rodent aldo-keto reductases from the AKR1B subfamily and their specificity with retinaldehyde. Chem Biol Interact. 2011;191:199–205. doi: 10.1016/j.cbi.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Hosokawa M. Structure, function and regulation of carboxylesterases. Chem Biol Interact. 2006;162:195–211. doi: 10.1016/j.cbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Schoffski P. Polo-like kinase (PLK) inhibitors in preclinical and early clinical development in oncology. Oncologist. 2009;14:559–570. doi: 10.1634/theoncologist.2009-0010. [DOI] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Bammler TK, Prasad B, Vrana M, Klaassen C, Cui JY. Developmental Regulation of Drug-Processing Genes in Livers of Germ-Free Mice. Toxicol Sci. 2015;147:84–103. doi: 10.1093/toxsci/kfv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Chang A, Wang J, Zhou W, Gao R, Li J, Xu Y, Luo X, Xiang R, Luo N, Stupack DG. TIFA, an inflammatory signaling adaptor, is tumor suppressive for liver cancer. Oncogenesis. 2015;4:e173. doi: 10.1038/oncsis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruiell K, Jones DZ, Cullen JM, Awumey EM, Gonzalez FJ, Gyamfi MA. Role of human pregnane X receptor in high fat diet-induced obesity in pre-menopausal female mice. Biochem Pharmacol. 2014;89:399–412. doi: 10.1016/j.bcp.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman CA, Liddle C, Coulter SA, Sonoda J, Alvarez JG, Moore DD, Evans RM, Downes M. Nuclear receptors constitutive androstane receptor and pregnane X receptor ameliorate cholestatic liver injury. Proc Natl Acad Sci U S A. 2005;102:2063–2068. doi: 10.1073/pnas.0409794102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9:643–660. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Li L, Wang H, Moore R, Kodavanti PR, Lehmler HJ, Negishi M, Birnbaum LS. Flame retardant BDE-47 effectively activates nuclear receptor CAR in human primary hepatocytes. Toxicol Sci. 2014;137:292–302. doi: 10.1093/toxsci/kft243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Cui W, Woody SK, Staudinger JL. Pregnane X receptor modulates the inflammatory response in primary cultures of hepatocytes. Drug Metab Dispos. 2015;43:335–343. doi: 10.1124/dmd.114.062307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swales K, Negishi M. CAR, driving into the future. Mol Endocrinol. 2004;18:1589–1598. doi: 10.1210/me.2003-0397. [DOI] [PubMed] [Google Scholar]
- Tabbot PN, Robson MG. The New Jersey Residential Well-Testing Program--a case study: Randolph Township. J Environ Health. 2006;69:15–19. [PubMed] [Google Scholar]
- Tcatchoff L, Nespoulous C, Pernollet JC, Briand L. A single lysyl residue defines the binding specificity of a human odorant-binding protein for aldehydes. FEBS Lett. 2006;580:2102–2108. doi: 10.1016/j.febslet.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–234. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Huang H, Hoffman B, Liebermann DA, Ledda-Columbano GM, Columbano A, Locker J. Gadd45beta is an inducible coactivator of transcription that facilitates rapid liver growth in mice. J Clin Invest. 2011;121:4491–4502. doi: 10.1172/JCI38760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. Epigenetic regulation of pregnane X receptor activity. Drug Metab Rev. 2013;45:166–172. doi: 10.3109/03602532.2012.756012. [DOI] [PubMed] [Google Scholar]
- Topletz AR, Tripathy S, Foti RS, Shimshoni JA, Nelson WL, Isoherranen N. Induction of CYP26A1 by metabolites of retinoic acid: evidence that CYP26A1 is an important enzyme in the elimination of active retinoids. Mol Pharmacol. 2015;87:430–441. doi: 10.1124/mol.114.096784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden RA, Bijzet J, Meijers WC, Yakala GK, Kleemann R, Nguyen TQ, de Boer RA, Schalkwijk CG, Hazenberg BP, Tietge UJ, Heeringa P. Obesity-induced chronic inflammation in high fat diet challenged C57BL/6J mice is associated with acceleration of age-dependent renal amyloidosis. Sci Rep. 2015;5:16474. doi: 10.1038/srep16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, Fleet JC, Kortagere S, Mukherjee P, Fasano A, Le Ven J, Nicholson JK, Dumas ME, Khanna KM, Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyhlidal CA, Gaedigk R, Leeder JS. Nuclear receptor expression in fetal and pediatric liver: correlation with CYP3A expression. Drug Metab Dispos. 2006;34:131–137. doi: 10.1124/dmd.105.005967. [DOI] [PubMed] [Google Scholar]
- Wagner M, Zollner G, Trauner M. Nuclear receptors in liver disease. Hepatology. 2011;53:1023–1034. doi: 10.1002/hep.24148. [DOI] [PubMed] [Google Scholar]
- Wallace K, Cowie DE, Konstantinou DK, Hill SJ, Tjelle TE, Axon A, Koruth M, White SA, Carlsen H, Mann DA, Wright MC. The PXR is a drug target for chronic inflammatory liver disease. J Steroid Biochem Mol Biol. 2010;120:137–148. doi: 10.1016/j.jsbmb.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Watson E, Cremona ML, Millings EJ, Lefkowitch JH, Fischer SG, LeDuc CA, Leibel RL. ILDR2: an endoplasmic reticulum resident molecule mediating hepatic lipid homeostasis. PLoS One. 2013;8:e67234. doi: 10.1371/journal.pone.0067234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Wermuth B, Platts KL, Seidel A, Oesch F. Carbonyl reductase provides the enzymatic basis of quinone detoxication in man. Biochem Pharmacol. 1986;35:1277–1282. doi: 10.1016/0006-2952(86)90271-6. [DOI] [PubMed] [Google Scholar]
- Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1:259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- Xie Y, Ke S, Ouyang N, He J, Xie W, Bedford MT, Tian Y. Epigenetic regulation of transcriptional activity of pregnane X receptor by protein arginine methyltransferase 1. J Biol Chem. 2009;284:9199–9205. doi: 10.1074/jbc.M806193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Luo M, Jiang H, Yu L, Zeng S. Involvement of CAR and PXR in the transcriptional regulation of CYP2B6 gene expression by ingredients from herbal medicines. Xenobiotica. 2015;45:773–781. doi: 10.3109/00498254.2015.1020076. [DOI] [PubMed] [Google Scholar]
- Yan J, Chen B, Lu J, Xie W. Deciphering the roles of the constitutive androstane receptor in energy metabolism. Acta Pharmacol Sin. 2015;36:62–70. doi: 10.1038/aps.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin SJ, Bosron WF, Magnes LJ, Li TK. Human liver alcohol dehydrogenase: purification and kinetic characterization of the beta 2 beta 2, beta 2 beta 1, alpha beta 2, and beta 2 gamma 1 “Oriental” isoenzymes. Biochemistry. 1984;23:5847–5853. doi: 10.1021/bi00319a026. [DOI] [PubMed] [Google Scholar]
- Zammit VA. Role of insulin in hepatic fatty acid partitioning: emerging concepts. Biochem J. 1996;314(Pt 1):1–14. doi: 10.1042/bj3140001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit VA, Moir AM. Monitoring the partitioning of hepatic fatty acids in vivo: keeping track of control. Trends Biochem Sci. 1994;19:313–317. doi: 10.1016/0968-0004(94)90068-x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wei Y, Hu B, Huang M, Xie W, Zhai Y. Activation of human stearoyl-coenzyme A desaturase 1 contributes to the lipogenic effect of PXR in HepG2 cells. PLoS One. 2013;8:e67959. doi: 10.1371/journal.pone.0067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang W, Qatanani M, Evans RM, Moore dd. The constitutive androstane receptor and pregane X receptor function cordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem. 2004;279:49517–22. doi: 10.1074/jbc.M409041200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cheng X, Aleksunes L, Klaassen CD. Transcription factor-mediated regulation of carboxylesterase enzymes in livers of mice. Drug Metab Dispos. 2012;40:1191–1197. doi: 10.1124/dmd.111.043877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Febbraio M, Wada T, Zhai Y, Kuruba R, He J, Lee JH, Khadem S, Ren S, Li S, Silverstein RL, Xie W. Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology. 2008;134:556–567. doi: 10.1053/j.gastro.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, Xie W. A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem. 2006;281:15013–15020. doi: 10.1074/jbc.M511116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Jiang L, Rui L. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem. 2009;284:11152–11159. doi: 10.1074/jbc.M900754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.