Abstract
Glioblastoma (GBM) is the most common primary brain tumor and has a dismal prognosis. Amplification of chromosome 12q13–q15 [Cyclin-dependent kinase 4 (CDK4) amplicon] is frequently observed in numerous human cancers including GBM. Phosphoinositide 3-kinase enhancer (PIKE) is a group of GTP-binding proteins that belong to the subgroup of centaurin GTPase family, encoded by CENTG1 located in CDK4 amplicon. However, the pathological significance of CDK4 amplicon in GBM formation remains incompletely understood. In the current study, we show that co-expression of PIKE-A and CDK4 in TP53/PTEN double knockout GBM mouse model additively shortens the latency of glioma onset and survival compared to overexpression of these genes alone. Consequently, p-mTOR, p-Akt and p-ERK pathways are highly upregulated in the brain tumors, in alignment with their oncogenic activities by CDK4 and PIKE-A stably transfected in GBM cell lines. Hence, our findings support that PIKE amplification or overexpression coordinately acts with CDK4 to drive GBM tumorigenesis.
Keywords: glioblastoma, protein-protein interaction, PIKE-A, CDK4
Introduction
Glioblastoma (GBM; WHO grade IV), the most common type of primary brain tumors, are highly aggressive, infiltrative and destructive. The molecular pathogenesis of GBM studies concludes that GBM formation requires dysregulation in three core pathways: the receptor tyrosine kinase (RTK)/PI3K/Akt axis, p53 signaling, and Rb-mediated cell cycle progression (1). Recently, radiotherapy plus concomitant and adjuvant temozolomide (TMZ) was shown to significantly improve the survival of GBM patients without reducing their quality of life (2, 3). However, the aggressive nature of glioblastomas renders current treatments dismal, and the median survival after treatment is 14 months. Therefore, novel and effective therapeutic interventions for GBM are urgently needed. For this reason, tremendous effort has been spent to identify tumorigenic or tumor-promoting genomic events and molecular pathways that directly drive malignant transformation and tumor progression.
GBM comprehensive genomic profiles mapped by The Cancer Genome Atlas (TCGA) project reveal that one of the most common copy number alterations in GBM is amplification at chromosome 12q13.3–14.1. This amplification is also observed in many other human cancers including melanoma, breast cancers and lung cancers (4–7). Importantly, glioblastoma patients harboring this amplification had markedly decreased survival. The cyclin-dependent kinase 4 (CDK4) gene has been postulated as the target of this amplification. CDK4 promotes proliferation by inhibiting the Rb1 tumor suppressor and by sequestering p27Kip1 and p21Cip1, thereby promoting E2F- and Cdk2- dependent cell cycle progression (8). However, CDK4 overexpression alone does not induce spontaneous tumorigenesis in transgenic animal models, suggesting that CDK4 cooperates with other genetic changes to initiate tumorigenesis.
CENTG1 encodes a GTPase called PIKE, which directly binds and activates PI 3-kinase and Akt (9). PIKE-A associates with Akt and enhances Akt kinase activity, promoting GBM proliferation and invasion (10, 11). Our previous studies support that PIKE-A is frequently amplified in numerous human cancers and acts as a proto-oncogene, promoting cancer cell survival, invasion and migration (12). The expression of PKE-A, but not other PIKE isoforms, is selectively increased in primary gliomas as compared with non-neoplastic brain tissue. Indeed, from an automated network analysis on the core pathways of glioma formation, PIKE-A has recently been confirmed as a driver gene of glioblastoma (13). Moreover, CENTG1 is frequently co-amplified with Cdk4. Further, a recent report reveals that hsa-miR26a, CDK4, and PIKE-A comprise a functional integrated oncomir/oncogene DNA cluster, which promotes the aggressiveness in glioblastoma (14). Conceivably, PIKE amplification or overexpression coordinately acts with CDK4 to drive GBM tumorigenesis.
Since signaling transduction is mediated by dynamic molecular interactions, such as protein-protein interactions (PPIs). Oncogenic PPI offers opportunities to reveal their functional significance and potential therapeutic strategies. New class of drugs targeting PPIs has attracted much attention recently (15, 16). Previously, we also demonstrated that disrupting PIKA-A/Akt interaction suppressed GBM cell oncogenic activity (17). In the current study, we show that PIKE-A directly interacts with CDK4, and PIKE-A/CDK4 complex promotes cell proliferation and GBM tumorigenesis in vitro and in vivo. Our study provides foundation of PIKE-A/CDK4 being considered as a potential target for GBM treatment. The concomitant contribution of CDK4 and PIKEA/Akt signaling to tumor progression might also offer a selective target for future personalized anti-cancer drug therapy.
Results
Constructing PIKE-A/CDK4-inducible mouse model with GFAP-CreER; PtenloxP/loxP; Tp53loxP/loxP mouse and pTOMO lentivirus system
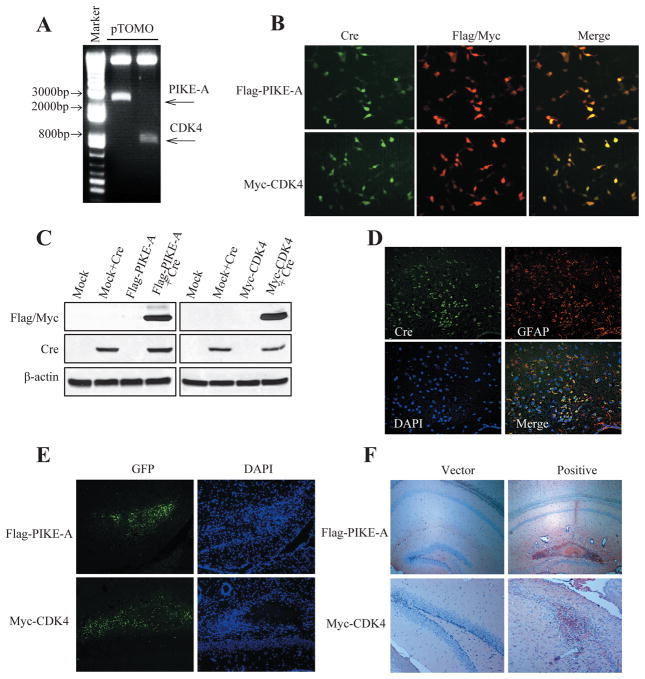
Dysregulation in three core pathways including RTK/PI3K, p53 and Rb signaling cascades are involved in human glioblastoma (GBM) formation and progression (1). Mutual exclusivity of alterations of components within each pathway or genes aberrations in all three pathways are required for glioblastoma pathogenesis. Accordingly, combined inactivation of Pten and Tp53 induce glioblastoma, which was achieved through GFAP-CreER; PtenloxP/loxP; Tp53loxP/loxP compound mice, hereafter termed Pten; p53 cKO mice (18). Cre activity was transiently induced in developmentally mature mice by tamoxifen administration after postnatal day P21 (range P20–P45) and 87% of Pten; p53 cKO brains developed tumors within the spectrum of HGA (High-grade gliomas), showing a range of histopathological features. Approximately 25% of tumors are classified as GBM. The survival median is 211 days and the onset day of the mice death is 90 day (18). To explore the pathological functions of over-expression of PIKE-A or CDK4 or both in mouse GBM model, we separately injected lentivirus expressing Flag-tagged PIKE-A or Myc-tagged Cdk4 or their combination into Pten; p53 cKO mouse (Supplemental Fig. S1) brain to determine their contribution to glioblastoma formation. In our preliminary study, we successfully packed Flag-PIKE-A or Myc-CDK4 virus using a Cre-loxP-controlled lentiviral vector (pTOMO) (19), named pTOMO-Flag-PIKE-A or pTOMO-Myc-CDK4 lentivirus. The viruses’ expression efficiency in HEK293 cells was validated in vitro by immunoblotting and immunofluorescent staining (Fig. 1A–C). To ensure the engineered genes can be expressed in the targeted tissue and cell types, we injected the lentivirus respectively into the hippocampus of mice, and conducted the immunofluorescent staining of Cre-recombinase and GFAP. As expected, the injected Flag-PIKE-A or Myc-Cdk4 was expressed in the GFAP positive cells in hippocampus (Fig. 1D–F).
Figure 1. Cre recombinase-dependent expression of Flag-PIKE-A/Myc-CDK4 by pTOMO PIKE-A/CDK4 lentiviral vector (LV) in vitro and in vivo.
A. Plasmids confirmation of pTOMO-Flag-PIKE-A and pTOMO-Myc-CDK4. pTOMO constructions were digested by BamHI an EcoRV. B. Immunocytochemistry showing Flag-PIKE-A/Myc-CDK4 expression induced by Cre recombinase. HEK293 cells were infected with pTOMO-Flag-PIKE-A/pTOMO-Myc-CDK4 LVs with Cre-expressing LVs. After fixation, the cells were stained with the indicated antibodies. C. Western blot showed Flag-PIKE-A/Myc-CDK4 expression induced by Cre recombinase. HEK293 cells were infected with pTOMO mock LVs or pTOMO PIKE-A/CDK4 LVs with or without Cre-expressing LVs, and the cell lysates were processed for western blotting with indicated antibodies. β-actin detection was used as a loading control. D. Confocal images of Cre recombinase specifically expressed in GFAP+ cells in GFAP-CreER mice. Brain sections were analyzed by immunofluorescence staining, Cre and GFAP expressed cells were stained as described in Experimental procedures. E. Tomo PIKE-A/CDK4 LVs were successfully injected into hippocampus (HP). Images were taken 10 days after injection of pTOMO PIKE-A/CDK4 LVs into HP. GFP signals (green) indicated the infected cells. F. Immunohistochemistry images showing Tomo PIKE-A/CDK4 were successfully expressed in mouse brains. Staining was taken 10 days after injection of pTOMO PIKE-A/CDK4 LVs into HPs. The brown cells showed the positive cells.
PIKE-A and CDK4 co-amplification accelerates glioblastoma formation
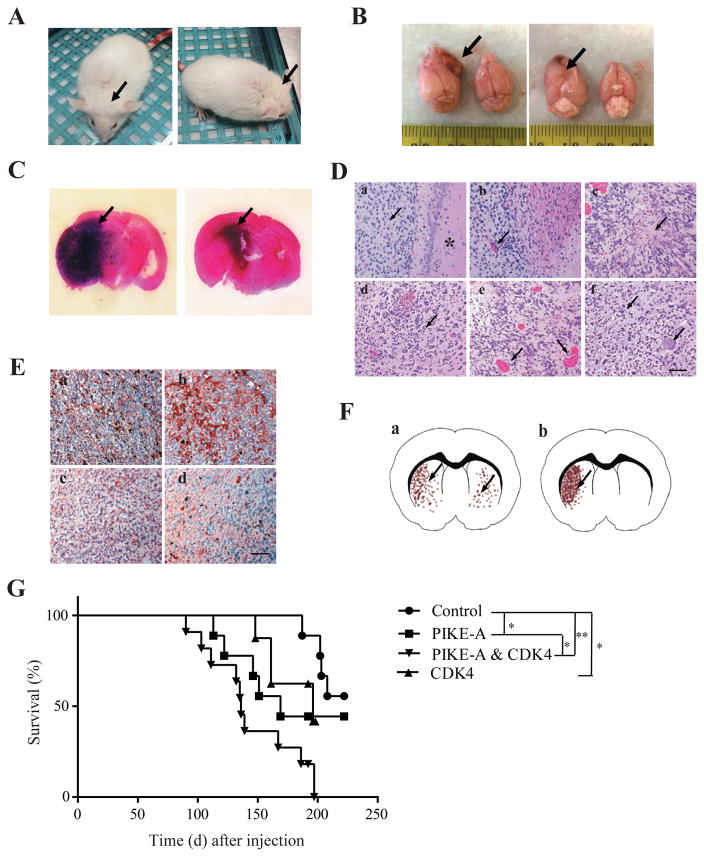
To test the pathological roles of PIKE-A and Cdk4 in the GBM formation, after virus injection into the GFAP-CreER; PtenloxP/loxP; Tp53loxP/loxP compound mice, we monitored GBM formation latency, mice survival rate and GBM pathologies. About 3 months after injection, mice with brain transduced with PIKE-A and CDK4 lentivirus showed an enlarged head and lethargy (Fig. 2A), indicating brain tumors formation, which was confirmed by the gross appearance of the brain (Fig. 2B) and H&E staining (Fig. 2C). On the H&E staining slide, we examined the characteristics of high grade gliomas (World Health Organization Grades III–IV), including increased cell density, pseudopalisading, and necrosis within the dense cellular region and perivascular invasion (Fig. 2D). In addition, IHC analysis (Fig. 2E) showed that the brain tumor contained cells positive for astrocyte marker GFAP, the oligodendrocyte marker myelin basic protein (MBP), the neuron-specific B III tubulin marker Tuj1, and a neural progenitor cell marker nestin, indicating the tumor contains various cell types, which is often found in human glioblastoma (18, 19). We also examined the survival rates of the various PIKE-A or CDK4 or their combination overexpressed groups using Kaplan-Meier survival analysis. PIKE-A or CDK4 overexpressed mice had a significantly reduced survival compared to Pten; p53 cKO mice. As expected, the shortest survival rates occurred to mice with both PIKE-A and CDK4 co-amplification (Fig. 2G) (19).
Figure 2. Glioblastoma induced by PIKE-A/CDK4 overexpressing in GFAP-CreER; Tp53 flox/flox; Pten flox/flox mice.
A. The representative images from GFAP-CreER; Tp53 flox/flox; Pten flox/flox mice showing tumor formation. GFAP-CreER; Tp53 flox/flox; Pten flox/flox mice injected with pTOMO PIKE-A/CDK4 into the right hippocampus showed an enlarged head (black arrows) after the injection. B. Photograph showing gross appearance of the brain, suggesting a large lesion in the cerebrum. Black arrows indicated the irregular surface of the cerebrum. C. Representative image of an H&E-stained section. A darker area denoted increased cellular density of the tumor. D. H&E staining shows the characteristics of glioblastoma: a, high cellular density, star indicating normal brain tissue; b, microvascular proliferation; c, necrosis within the dense cellular region; d, nuclear pleomorphism; e, intratumoral hemorrhage; f, multinucleated giant cell phenotype. Scar bar, 20 μm. E. The tumors induced by combined expression of PIKE-A and CDK4 in GFAP-CreER; Tp53 flox/flox; Pten flox/flox mice contain the cells expressing cell markers as glioblastoma. a, GFAP; b, nestin; c, MBP; d, Tuj1. Scar bar, 20 μm. F. Represent images of tumor growth patterns. a, tumor cells grow in the whole brain; b, tumor cell grow mainly in the injection side of brain. G. Effects of PIKE-A/CDK4 on tumor formation in GFAP-CreER; Tp53 flox/flox; Pten flox/flox mice. A Kaplan-Meier curve showing the survival of GFAP-CreER; Tp53 flox/flox; Pten flox/flox mice with vector/PIKE-A/CDK4/PIKE-A plus CDK4 LVs injection. GFAP-CreER; Tp53 flox/flox; Pten flox/flox mice were injected with vector, pTOMO- PIKE-A, pTOMO-CDK4, or pTOMO- PIKE-A and CDK4 into hippocampus. The median survivals are 240, 169, 196, and 136 days in vector, PIKE-A, CDK4, and PIKE-A plus CDK4 groups, respectively.
Based on the histopathological features of high grade gliomas (Figure 2D) and glioblastoma (Figure 2E) detected by H&E and IHC staining. Quantitative analysis indicated approximately 63.64% of tumor formation rate in control Pten; p53 cKO brain with 20% as glioblastomas. Overexpression of PIKE-A or CDK4, the ratios were elevated to 75% and 81.82%, respectively. Noticeably, glioblastoma rate was increased up to 42.85% by PIKE-A versus 28.57% by CDK4. Strikingly, 100% PIKE-A/CDK4 co-expressed Pten; p53 cKO brains harbored tumors within the spectrum of HGA. Approximately 66.67% of tumors were classified as glioblastomas (Table 1). Moreover, from the pathological analysis, we also noticed the difference of tumor formation patterns: tumor cells growing mainly in the half brain (Fig. 2F–b) with the injection site and tumor cells scattered in the whole brain (Fig. 2F–a). All the tumors were showing the latter growth pattern in the PIKE-A and CDK4 overexpressed brains. However, only 20.00% of brains showed the one site focused growth in control group. The focused growth was shown at 85.71% and 71.42% in PIKE-A and CDK4 groups, respectively. Therefore, co-expression of PIKE-A and CDK4 evidently decreased the survival period compared to these genes infected separately or the control mice, supporting that co-amplification of PIKE-A and CDK4 on the cdk4 amplicon elevates their oncogenic activity in GBM formation and progression.
Table 1.
Tumor formation data of groups of mice injected different kinds of virus.
| Parameters | Tum or cell distribution as Figure F -b (%) | Tumor formation rate (%) | Glioblastoma rate (%) |
|---|---|---|---|
| Groups | |||
| Control | 20.00 | 63.64 | 20.00 |
| PIKE-A | 85.71 | 75.00 | 42.85 |
| CDK4 | 71.42 | 81.82 | 28.57 |
| PIKE-A & CDK4 | 100.00 | 100.00 | 66.67 |
Co-amplification of PIKE-A and CDK4 enhances cell proliferation and suppresses apoptosis in GBM
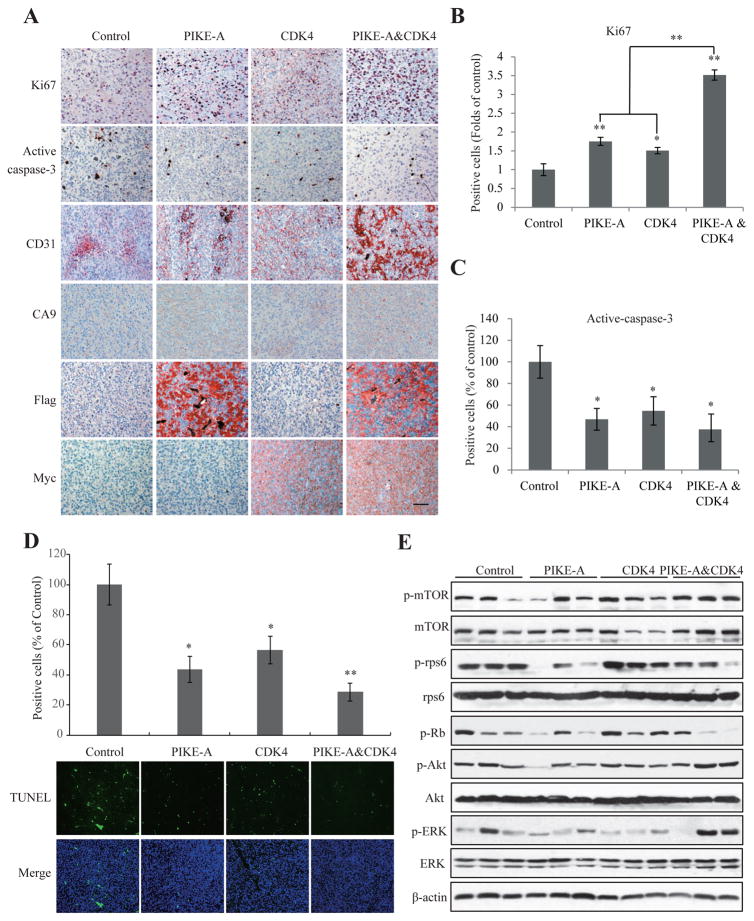
PIKEs directly interact with PI3K or Akt and enhance the kinase activities (10, 20). PIKE-A is frequently amplified in numerous human cancers including GBM and acts as a proto-oncogene, promoting cancer cell survival, invasion and migration (10, 12). CDK4 is the key regulator of the G1-S transition. In complex with cyclin D, CDK4 phosphorylates retinoblastoma protein (Rb) and drives the cell-cycle progression. Accordingly, overexpression of PIKE-A or CDK4 significantly stimulated cell proliferation, and the maximal effect occurred to the mixture of PIKE-A and CDK4, as shown by Ki67 immunohistochemistry (IHC) (Fig. 3A & B). Active Caspase-3, a marker of apoptosis, was also detectable in most tumors. Co-expression of PIKE-A and CDK4 evidently repressed the apoptosis than control (Fig. 3A & C), which was confirmed by TUNEL staining (Fig. 3D). CD31 is used primarily to demonstrate the presence of endothelial cells in histological tissue sections, which is a marker for evaluating the degree of tumor angiogenesis, which can imply rapidly growing tumors. Clearly, IHC analysis demonstrated that co-expression of PIKE-A and CDK4 elicited much more robust CD31 positive endothelial cell proliferation and malignant GBM growth than PIKE-A or CDK4 separately expressed alone (Fig. 3A, 3rd panels), which is confirmed by using immunochemical staining with antibody against Factor VIII (21), another marker of micro vessels (Supplemental Fig. S2). Since hypoxia is another a hallmark of GBM aggressiveness, we checked the expression of carbonic anhydrase (CA) 9 which is an indicator of hypoxia in glioblastoma (22). IHC staining showed that PIKE-A over-expression upregulated CA9 expression compared to control/CDK4 groups and PIKE-A & CDK4 group showed the strongest CA9 staining (Fig. 3A, 4th panel), indicating expression PIKE-A and CDK4 enhances the GBM aggressiveness in vivo. To further characterize the signaling cascades alteration by PIKE-A and Cdk4 co-amplification, we performed immunoblotting analysis with the brain tumors. Notably, p-ERK and p-Akt were highly activated in tumors with both PIKE-A/CDK4 co-amplification, so was p-mTOR. Interestingly, overexpression of PIKE-A decreased p-rps6 and p-Rb than control cells, though CDK4 overexpression elevated these two protein phosphorylation signals. However, both p-rps6 and p-Rb signals were reduced in the tumors from the combination of PIKE-A/CDK4 than the control (Fig. 3E).
Figure 3. PIKE-A and CDK4 additively promote tumor growth in vivo.
A. Immunohistochemistry analysis of cell proliferation, apoptosis and angiogenesis using Ki67, active-caspase-3, CD31, and CA9 in the brain tumors from indicated groups. Scar bar, 20 μm. B & C. Quantification of Ki67 (B) and active-caspase-3 (C) positive cells in the slides of brain tumors in different groups. D. Apoptosis detection/quantification using by TUNEL staining. Three fields were averaged in each tumor, and the averages for each animal yielded the final mean ± SEM (*P < 0.05, **P < 0.01, two-tailed Student’s t test, n = 3). E. Signaling characterization in the tumors formed in the brains infected by indicated lentivirus. Brain tumors were collected, lysed and equal amount of proteins in each sample were analyzed by western blotting with the indicated antibodies.
PIKE-A interacts with CDK4 in human primary brain tumors
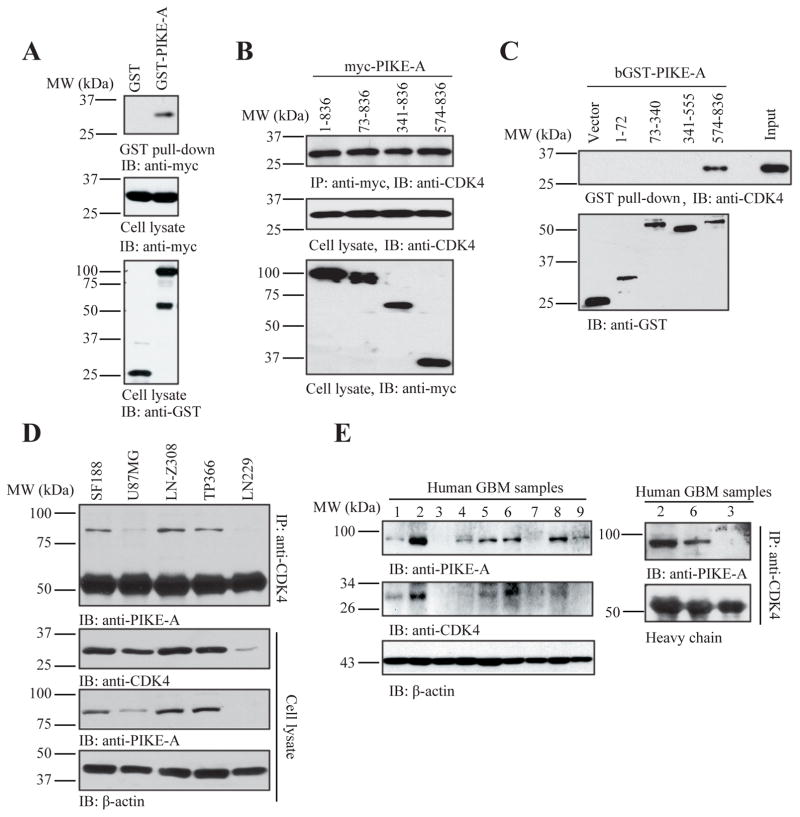
CDK4 and CENTG1 locate in Cdk4 amplicon and co-expression of both PIKE-A and CDK4 reveals the additive effect on cell proliferation and tumorigenesis, we wonder whether these two proteins may physically interact with each other. Indeed, protein-protein interaction studies with a protein fragment complementation assay in GBM revealed a physical interaction between PIKE-A and CDK4. To confirm this interaction, we conducted in vitro binding assay with GST pull-down assay. GST-PIKE-A but not GST control selectively interacted with Myc-CDK4, transfected in HEK293 cells (Fig. 4A). To explore which domain of PIKE-A binds to CDK4, we performed a truncation assay. Mapping study demonstrated that the C-terminal a.a. 574–836 on PIKE-A was sufficient to associate with CDK4, which was validated by both co-immunoprecipitation using transfected TP366 glioblastoma cell line and GST-pull down assay (Fig. 4B & C). We also extended the study into the endogenous proteins in various GBM cell lines. Co-immunoprecipitation assay revealed that CDK4 strongly associated with PIKE-A in SF188, LN-Z308 and TP366 cells, which contain the cdk 4 amplicon (10). Accordingly, PIKE-A and CDK4 were highly expressed in these cell lines than U87MG and LN229 GBM cells (Fig. 4D). Next, we explored the association between PIKE-A and CDK4 in human primary GBM samples. It is worth noting that both PIKE-A and CDK4 displayed the similar expression patterns in these tumor samples (Fig. 4E, left panels). Co-immunoprecipitation with Cdk4 antibody assay indicated that PIKE-A tightly associated with CDK4, when PIKE-A was demonstrable in the tumor samples (Fig. 4E, right panels). Hence, our findings support that PIKE-A directly associates with Cdk4 through its C-terminal Arf-GAP domain in primary gliomas.
Figure 4. PIKE-A interacts with CDK4 in vitro and in vivo.
A. GST-pull down assay for PIKE-A binding to CDK4. Cell lysates from HEK293 cells transfected with GST-tagged PIKE-A and Myc-tagged CDK4 were collected and analyzed by GST-pull down assay as described in Experimental procedures. Data indicated that PIKE-A interacted with CDK4 in cells compared to GST vector control. B & C. Truncation analysis indicates that the C-terminus of PIKE-A (a.a. 574–836) binds to CDK4. Cell lysates were collected from TP366 cells transfected with myc-tagged PIKE-A truncations and used for co-immunoprecipitation analysis with anti-myc antibody (B). Purified GST-tagged PIKE-A truncated recombinant proteins confirmed that C-terminus of PIKE-A (a.a. 574–836) interacted with CDK4 in GST pull-down assay (C). D. Interaction of PIKE-A and CDK in glioblastoma cells. Glioblastoma cells were collected and lysed for co-immunoprecipitation. Endogenous CDK4-associated proteins were collected by anti-CDK4 antibody and analyzed by indicated antibodies. E. Interaction of PIKE-A and CDK in human glioma tissues. Expression profiles of PIKE-A or CDK4 in human glioblastoma tissues (left panels). Co-immunoprecipitation assay was performed using the lysates form human glioma tissues with different PIKE-A expression levels. The CDK4-associated proteins were analyzed by western blotting showing that PIKE-A interacted with CDK4 (right panels).
Co-expression of PIKE-A and CDK4 displays additively oncogenic activity in tumorigenesis
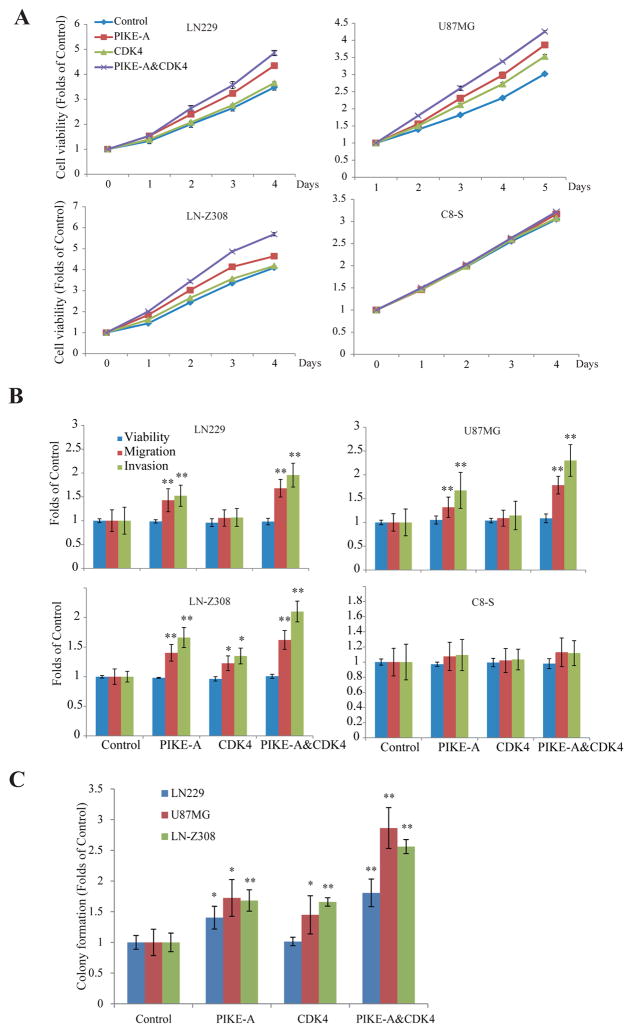
To explore the biological consequence of PIKE-A or CDK4 or their combination in tumorigenesis, we established several stable GBM cell lines with different genetic background. We stably transfected control vector, PIKE-A, CDK4 or both PIKE-A and CDK4 into LN229 (p53 mutant), U87MG (PTEN −/−) and LN-Z308 (TP53 −/− and PTEN −/−) glioblastoma cells as well as wild-type C8-S astrocyte cells. We monitored their effects on cell proliferation, migration, invasion, and colony formation. Interestingly, in C8-S cells, no difference on cell proliferation was observed regardless of overexpression of PIKE-A, CDK4 or their mixture. By contrast, PIKE-A overexpression revealed stronger activity than CDK4 in stimulating cell proliferation compared to vector control in GBM cell lines including LN229, U87MG and LN-Z308 cells. The maximal activity occurred to the mixture of PIKE-A and CDK4, indicating that they display the additive effect when co-expressed (Fig. 5A). Notably, migration and invasion assays basically exhibited the similar results in GBM cells lines, whereas PIKE-A or CDK4 had no effect in astrocyte C8-S cells (Fig. 5B). Again, PIKE-A exhibited stronger oncogenic activity than CDK4 in all of the GBM cell lines. Nonetheless, the colony formation assay revealed that PIKE-A and CDK4 revealed the comparable activity, and co-expression of these two proteins exhibited the additive activity as expected (Fig. 5C).
Figure 5. PIKE-A and CDK4 complex additively promotes GBM tumorigenesis.
A, Additive effect of PIKE-A and CDK4 on cell proliferation in glioblastoma cells. PIKE-A, CDK4, and PIKE-A/CDK4 stably transfected human glioblastoma LN229 (A), U87MG (B), LN-Z308 (C), and C8-S astrocyte (D) cells were seeded in 96-well plates and cell proliferation were examined every day for total 4 days using MTT assay. B & C, Additive effect of PIKE-A and CDK4 on cell migration and invasion in glioblastoma cells. PIKE-A, CDK4, and PIKE-A/CDK4 stably transfected human glioblastoma LN229, U87MG, LN-Z308, and C8-S astrocyte cells were examined by cell migration and invasion and colony formation assays.
Since PIKE-A and CDK4 overexpression blocked apoptosis in primary brain tumors, we further examined cell apoptosis in the LN-Z308 isogenic cell lines. Following Annexin V/PI staining, flow cytometry analysis indicated that under normal culture condition (medium with 10% FBS), ratios of the apoptosis cells among the stable transfected cell lines are almost the same. However, following starvation, cells with overexpression of PIKE-A and CDK4 decreased the apoptosis compared to others (Supplemental Fig. S3), which is consistent with the in vivo data (Fig. 3). To assess whether these effectors mediate the cell cycle progression or not, we conducted the flow cytometry analysis following PI staining. As shown in Supplemental Fig. S4, compared to control cell lines, PIKE-A/CDK4 co-expression did not change the cell cycle profiles significantly in either C8-S or LN229 isogenic cell line, indicating that the difference of the effect of PIKE-A/CDK4 on cell proliferation in C8-S and GBM cell lines might not be due to regulation of cell cycle progression.
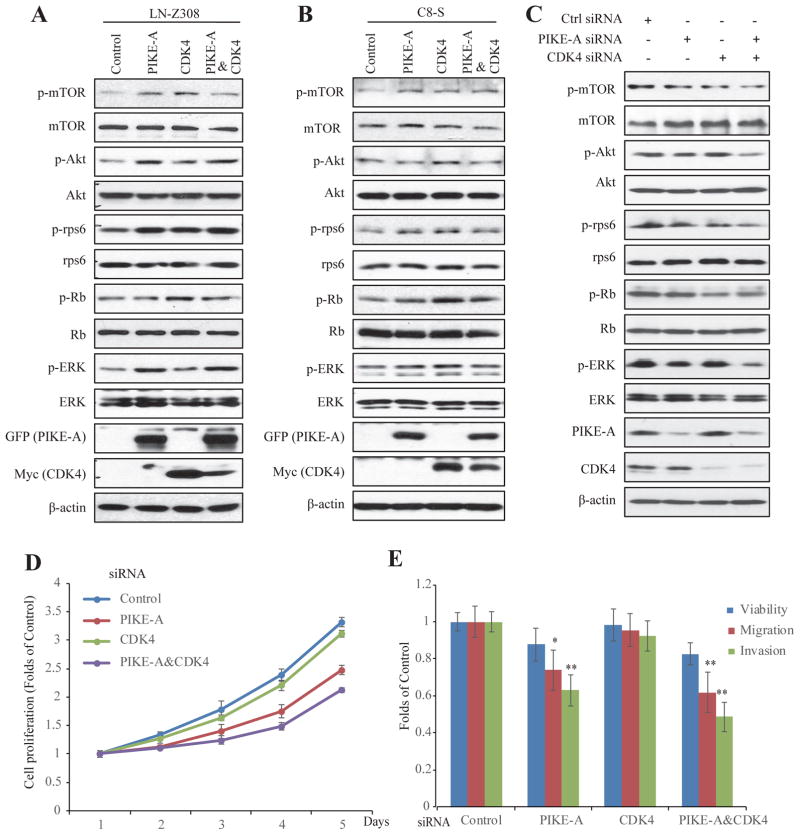
To explore the effects of PIKE-A, CDK4 and their mixture’s effect on different signaling cascades, we performed immunoblotting analysis with these stable cell lines. Overexpression of PIKE-A or CDK4 elevated mTOR phosphorylation, and its downstream effector p-rps6 signals were escalated by PIKE-A or CDK4 compared to control samples (Fig. 6A, Supplemental Fig. S5). In alignment with what observed in primary brain tumors, overexpression of PIKE-A strongly enhanced p-Akt, so was p-ERK (Fig. 6A, Supplemental Fig. S5). Since Rb is one of the major downstream targets of CDK4, p-Rb signals were prominently elevated, when Cdk4 was overexpressed in U87MG (Supplemental Fig. S5A), LN-Z308 and C8-S cells (Fig. 6A & 6B). Since p-Rb baseline was very high in control samples in LN229, the effect by CDK4 on p-Rb was not as clear as in other cell lines (Supplemental Fig. S5B). Furthermore, to define the biological roles of PIKE-A and CDK4 on the endogenous levels, we knocked down PIKE-A and CDK4 in LN-Z308 cells. In alignment with the upregulation of p-mTOR, p-Akt, and p-ERK in over-expression system, PIKE-A and CDK4 knocking down decreased signalings described above (Fig. 6C), validating the effects of PIKE-A and CDK4 on oncogenic pathways. Functional studies showed that cells with PIKE-A deficiency displayed decreased proliferation, migration/invasion compared to controls and together with CDK4 deficiency, the oncogenic activities were further suppressed (Fig. 6D&E). In addition, rapamycin, an mTOR signaling pathway inhibitor, completely blocked PIKE-A/CDK4 induced cell proliferation (Supplemental Fig. S6). Collectively, these data support that co-expression of PIKE-A and CDK4 additively exerts the oncogenic activity, which is involved in up-regulation of mTOR, Akt, and ERK signaling pathways.
Figure 6. Effects of PIKE-A, CDK4, and PIKE-&CDK4 on oncogenic signaling in LN-Z308 GBM cells and C8-S astrocyte.
PIKE-A, CDK4, and PIKE-A/CDK4 stable transfected LN-Z308 (A) and C8-S astrocyte (B) cells were collected and cell lysates were used for signaling examination by western blot with the indicated antibodies. C. In LN-Z308 cells, PIKE-A or/and CDK4 knocking down by siRNA, cell lysates were used for signaling examination by western blot with the indicated antibodies. D&E. Effect of PIKE-A/CDK4 knocking down on cell proliferation, migration and invasion. Following siRNA treatment, LN-Z308 cells were analyzed by cell proliferation, migration, and invasion assays described in Materials and Methods.
Discussion
In the current study, we demonstrate that PIKE-A directly interacts with CDK4 in vitro and in vivo, and this protein complex promotes GBM tumorigenesis, in addition to the genetic co-localization on chromosomal 12q13.1–14 (Cdk4 amplicon) by these two genes. Interestingly, in vitro CDK4 kinase assay or p-Rb signals in PIKE-A overexpressed GBM cells was not altered, suggesting that PIKE-A may not regulate CDK4 kinase activity. On the other hand, CDK4 overexpression does not affect Akt activation by PIKE-A either. However, when both PIKE-A and Cdk4 are co-amplified, p-Akt and p-ERK signals were highly upregulated in both brain tumors and GBM cell lines (Fig. 3E, 6A–C, and Supplemental Fig. S5), which might be due to the mTOR signaling activation. It is worth noting that the downstream effector of mTORC1, rps6, was robustly phosphorylated in PIKE-A or CDK4 stably cell lines, and the maximal effect occurred in the co-amplified cell lines (Supplemental Fig. S5, Fig. 6). However, p-rps6 was inhibited, when PIKE-A was overexpressed in the brain tumors (Fig. 3E, lane 4–6; 10–12). The molecular mechanism accounting for this discrepancy remains unclear. Presumably, the heterogenous cell types in primary brain tumors may partially contribute to this effect.
Tp53 gene is frequently mutated in human glioblastoma and is associated with malignant transformation astrocytoma (23). And mutation of p53 is usually used as a tool to study other genes function in glioblastoma formation and progression, such as EGFR (24), Akt (19), Ras, and PTEN. In the current study, we used GFAP-CreER; PTEN loxP/loxP; Tp53loxP/loxP compound mice (18). Here we show that overexpression of PIKE-A increased GBM formation from 20% in PTEN; p53 cKO mice up to 42.85%, and co-expression of both PIKE-A and Cdk4 enhanced the effect to 66.67%. Accordingly, the median survivals were 240, 169, 196, and 136 days in vector, PIKE-A, Cdk4, and PIKE-A plus Cdk4 groups, respectively, underscoring that PIKE-A plays a critical role in GBM transformation. It is reported that p53 cKO mice has a long latency (6–14 months) to tumor formation, and overexpression of EGFRvIII in p53 knockout mice can shorten the latency to 26 days with a 100% glioblastoma formation ratio. As a part of downstream effectors of EGFR, PIKE-A amplification displays a fraction of amplified RTK (e.g. EGFRvIII) oncogenic effect fits with the observed in vivo activities (Fig. 2 & 3). There are several other genetically engineered mouse GBM models, such as EGFR WT/WT; lnkΔ2/3−/−;PTEN2lox and EGFRVIII/VIII; lnkΔ2/3−/−(p16Ink4a/p19Arf knockout); PTEN2lox models, the median survival are 15 and 5 weeks, respectively(24). Since glioblastoma formation needs mutual exclusivity of alterations of components within each pathway (RTK/PI3K, p53 and Rb) or genes aberrations in all three pathways (1, 25). If EGFR/EGFRvIII overexpression model was employed, CDK4 overexpression should be required. EGFR/EGFRVIII have multiple downstream signaling pathways and PI3K/Akt pathway is one of them, which is not suitable for characterizing PIKE-A functions.
PIKE-A belongs to PIKE family which has other two members, PIKE-L and PIKE-S. All of the three can be expressed in brains. However, only PIKE-A is highly amplified in glioblastoma, which has been considered as one key pro-oncogene responsible glioblastoma formation (10, 17). Both PIKE-S and PIKE-L bind to PI3K and enhance its activity. However, PIKE-A does not interplay with PI3K. Instead, it interacts with the downstream effector Akt and promotes its activity (10, 12). Our previous works have demonstrated that PIKE-A enhances Akt activity in vitro and in vivo (10). However, p-Akt levels in tumors that overexpress PIKE-A are not upregulated. This discrepancy may be caused by the GBM mouse model which is tamoxifen-induced tp53 and pten double deleting in GFAP expressed cells in brain based on the floxP/Cre system. The model can form glioma/glioblastoma by itself following tamoxifen treatment. The lentivirus harboring PIKE-A/CDK4 also acted based on the floxP/Cre system. Conceivably, the brain tumor formed in the mouse model is mediated by at least two gene manipulation. It remains unclear why overexpression of PIKE-A alone did not elevate p-Akt or p-mTOR pathways; nevertheless, these signals cascades were highly augmented when co-expressed with Cdk4, indicating that CDK4 may somehow contribute to PIKE-A’s stimulatory effect on p-Akt and p-mTOR signaling. In addition, the primary tumor tissues contain a mix of cell types, which may be also the reason that PIKE is not expressed across all GBM specimens (Fig. 4E); yet it is rather well expressed across in GBM cell lines. In addition, although over-expression PIKE-A or CDK4 slightly increase the levels of p-mTOR, p-rps6, p-Akt, p-Rb, and p-ERK in C8-S cells, functional analysis does not reflect the induction of oncogenic activity (proliferation, migration, and invasion) by PIKE-A or CDK4, which is different from GBM cell systems (Fig. 5). This may be caused by different degrees of the signaling activation among various cell lines (Fig. 6A&B, Supplemental Fig. S5). Another reason may depend on the genomic background. As mentioned before, LN229 (p53 mutant), U87MG (PTEN −/−) and LN-Z308 (TP53 −/− and PTEN −/−) are GBM cell lines with oncogenic mutations and there should be many other gene mutations contributing cell malignancy, which is different from normal astrocytes cells. Among the three cell lines, LN229 and U87MG cells have low endogenous PIKE-A and CDK4, whereas LN-Z308 cells highly express PIKE-A and CDK4, which is irrelevant of the oncogenic effects induced by co-amplification of PIKE-A and CDK4.
Our data demonstrate that co-amplified PIKE-A and CDK4 substantially increased GBM cell migration and invasion, however, their effect on cell proliferation is not so dramatic (Fig. 5). Cell proliferation integrates varies biological processes, such as cell survival, cell cycle progression and apoptosis etc.. GBM cells possess numerous mutations implicated in controlling these processes. The tested GBM cell lines (U87MG, LN229, LN-Z308) are dividing very fast, conceivably, co-expression of extra PIKE-A and CDK4 may not greatly further enhance their proliferation. It is worth noting that co-amplification of PIKE-A and CDK4 decreased apoptosis in primary brain tumors (Fig. 3). Furthermore, flow cytometry analysis indicated that under normal conditions, cells among the groups showed subtle differences in apoptosis. However, following starvation, cells with overexpression of PIKE-A and CDK4 decreased the apoptosis compared to others (Supplemental Fig. S3). Moreover, co-expression both of them elicited prominent tumorigenesis activities demonstrated by colony formation assay (Fig. 5C). All of these effects are in alignment with the robust activities on p-Akt/p-mTOR and p-ERK by PIKE-A and Cdk4 co-amplification in GBM. Except for ERK, the MAPK family in mammals includes c-Jun NH2-terminal kinase (JNK), p38 MAPK. It is demonstrated that ERK signaling closely integrates with mTOR signalings (26, 27). Recently studies show that inhibition of p38 MAPK decreases sensitivity to PI3K/mTOR inhibition (28) and JNK contributes tumorigenic activity through mTOR pathway in cholangiocarcinoma (29). Therefore, it is possible that the p38 MAPK and JNK signaling involved in GBM formation and progression.
Previous study has shown that phosphorylation of PIKE-A by Cdk5 mediates growth factor-induced migration and invasion of human glioblastoma cells (30). Mechanisms by which interactions of PIKE-A and CDK4 stimulates oncogenic signaling as well as the mutual regulation between PIKE-A and CDK4 need to be further defined. Here, we provide evidence that PIKE-A interact CDK4 and that PIKE-A/CDK4 promotes tumorigenesis in vitro and in vivo. Hence, PIKE amplification or overexpression coordinately acts with CDK4 to drive GBM tumorigenesis. Conceivably, selectively disrupting this oncogenic protein complex may provide an innovative therapeutic target for GBM treatment.
Materials and Methods
Cell lines, reagents, and mice
The human embryonic kidney 293 (HEK293) cells, human glioblastoma LN-Z308, U87MG, LN229, and C8-S astrocyte cells were cultured in DMEM with 10% FBS. All the cell lines were from Dr. Keqing Ye’s lab and reported recently (17, 31, 32). They were used before being tested for mycoplasma contamination. U87MG, LN299, LN-Z308 human glioblastoma cells and C8-S astrocyte cells were stably transfected with vector control, PIKE-A (GFP-tagged), CDK4 (Myc-tagged), PIKE-A/CDK4 were maintained in DMEM with 10% FBS and 1×Pen/Strep/glutamine supplemented with various selection antibiotics (PIKE-A, 400 μg/ml G418; CDK4, 150 μg/ml hygromycin). Anti-β-actin (A5441), anti-Flag (F1804), anti-Myc (MAB8864), anti-BMP (M3821), and anti-Tuj1 (MAB1637) antibodies were purchased from Sigma (St. Louis, MO). Antibody against GFAP (sc-71143) and Protein A/G agarose (sc-2003) beads were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against Nestin (ab6142), Ki67 (ab15580) and CA9 (ab184006) were purchased from Abcam (Cambridge, MA). The Horseradish peroxidase-linked IgG secondary antibodies and Glutathione Sepharose 4B were from GE healthcare. Antibodies of Cre (15036S), Active-caspase-3 (9664S), and CD31 (3508S were obtained from Cell signaling (Beverly, MA). Antibody against Factor VIII (RB281A1), Aneexin V/PI staining kit (V13241), and the Histostain-SP AEC kit was obtained from Invitrogen, Inc. (USA). All of the chemicals not included above were from Sigma. We obtained GFAP-CreER; Tp53 flox/flox; Pten flox/flox compound mice from St. Jude Children’s Research Hospital. Animal care was in accordance with guidelines of Division of Animal Resources in Emory University.
Cell proliferation, migration, invasion, and colony formation assay
Cells were seeded in 96-well plates at a density of 3,000 cells per well. Cell were incubated at 37 °C for indicated times. The cell proliferation was monitored by MTT incorporation (Cell signaling, Beverly, MA) assays according to the manufacturer’s protocols. Cell migration assay was performed using BD Falcon cell culture inserts for 24-well plates with 8.0 μm pore filter according to manufacturer’s instructions. Invasion of cells through Matrigel was determined using a transwell system (10 mm diameter, 8 μm pore size with polycarbonate membrane; Corning Costar). Cells (equal number of viable cells counted by Trypan blue staining) were seeded into the upper chamber of the insert in serum-free media, and lower chamber was filled with media containing 5% FBS. After 16 hours (migration) or 24 hours (invasion), cells were fixed using 100% methanol and stained using 0.05% crystal violet. Cells in upper chamber were carefully removed, and cells migrated through the filter were determined by counting the cells attached on the filters. For colony formation assay, a base 0.6% agar gel with 10% FBS in DMEM was prepared and added to the wells of a six-well culture dish. Cells were plated at a density of 5,000 cells per well on top of the base agar for anchorage-independent growth analysis in 0.4% agar gel with 10% FBS in DMEM supplemented specific antibiotics. The cells were maintained at 37 °C and allowed to grow for 3 weeks. The colonies were scored with staining with using MTT dye.
Flow cytometric analysis
For cell cycle analysis, cells were washed twice with ice-cold PBS, and fixed in 70 % ethanol. Tubes containing the cell pellets were stored at −20 °C for at least 24 hours. After centrifugation for 10 minutes, the supernatant was discarded. Cells were then washed with 5 ml of PBS and incubated with propidium iodide (20 μg/ml)/R Nase A (20 μg/ml) in PBS for 45 minutes. For cell apoptosis analysis, cells were collected and stained with AnnexinV/PI according to the data sheet of the product. The samples were analyzed on a Coulter Elite flow cytometer;
Western blotting, co-immunoprecipitation, and GST-pull down assay
Western blotting was performed using standard protocol. Cell lysate was quantified using Brandford assay. Equally amount of protein (20–50 μg) was loading for blotting with specific antibodies indicated. For immunoprecipitation, after centrifuge, the supernatants were collected, quantified and antibody was mixed with the supernatant for 1 hour, and protein A/G agarose beads were added in for overnight. Immunoprecipitates were washed four times with lysis buffer, and proteins were eluted with SDS-PAGE sample buffer. For GST-pull down assay, transfected cells were collected and lysed in the lysis buffer described above, and then centrifuged for 10 min at 13,000 × g at 4 °C. The supernatant was collected and 20 μl glutathione epharose 4B was added into the supernatant, incubated with slow rotation for 3 hours, and washed five times with lysis buffer. The beads were then eluted and boiled with sample buffer. Eluted proteins associated with the A/D agarose beads or glutathione beads were analyzed by western blotting assay.
Mice treatment and virus injection
To generate pTOMO-PIKE-A/pTOMO-CDK4 lentiviral vectors, we inserted a Flag-tagged PIKE-A or Myc-tagged CDK4 fragment between EcoRV and BamH1 sites of the pTOMO vector (Addgene). Other lentiviral vectors and packaged virus were provided by the Viral Vector Core at Emory University. Based on the literatures (18, 19), mice (P25–P49, n=12/group) were injected with virus using stereotaxic apparatus. Virus (2 μl, 2 × 108 international units) was injected stereotactically at coordinates posterior 1.8 mm, lateral 1.6 mm, ventral 1.8 mm relative to bregma followed by treatment with tamoxifen (0.225 g/kg body weight, i.p.) for constituent 5 days. No significant difference associated with the age of induction in the time from tamoxifen administration to tumor onset, or in the histology of the resulting tumors. To inject a mixture of pTOMO-PIKE-A and pTOMO-CDK4 lentivral vectors, we mixed the two viral preparations (1:1) and injected 2 μl. Animals were treated blindly during the experiments and the tumor tissue collection was also done blindly. The animal experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Emory University.
Immunohistochemistry and immunofluorescence staining
Samples were fixed in 4% paraformaldehyde overnight followed by paraffin embedding. Sections (8 μm) were deparaffinized in xylene and rehydrated in graded alcohols. Endogenous peroxidase activity was blocked by 3% hydrogen peroxide for 5 minutes and all slides were boiled in 10 mM citrate buffer (pH 6.0) for 10 minutes. Aim proteins were detected using specific primary antibodies and Zymed Histostain-SP AEC kit. Slides were then counterstained with hematoxylin. For immunofluorescent staining, the tissue sections were deparaffinized in xylene, rehydrated in graded alcohols and were boiled in 10 mM sodium citrate buffer (pH 6.0) for 10 minutes. The sections were blocked with 1% BSA in PBS at 37 °C for 30 min followed primary antibodies incubation at 4 °C for overnight. On the second day, the sections were washed with PBS and incubated with Alexa Fluor 488-labeled goat anti-rabbit IgG antibody or Alexa Fluor 555-labeled goat anti-mouse IgG antibody (1: 800) at room temperature for 60 minutes followed rinsing with PBS for 10 minutes and staining with DAPI for another 10 minutes at room temperature. Cells were seeded on coverslips and fixed with 4% paraformaldehyde for (prepared in PBS) for 30 min and permeabilized with 0.1% Triton X-100 for 30 minutes. Following blocking in 3% FBS in PBS for 1 hour, cells were incubated with antibody at 4 °C for overnight. After washing with PBS for three times, cells were incubated with secondary antibodies (1:1000) for 2 hours at room temperature. Then the cells were counterstained with 4′ 6-diamidino-2-phenlindole (DAPI) and examined under a fluorescence microscope. After mounting, sections were examined under a fluorescence microscope.
Statistics analysis
Data are presented as mean ± S. E. M. from three independent experiments. Statistical evaluation was carried out by Student’s t-test or one-way ANOVE. Data were considered statistically significant when P< 0.05 *, P< 0.01 **. All statistical analysis was performed by program Prism (GraphPad Software, La Jolla, CA, USA).
Supplementary Material
Acknowledgments
This work has been funded in part with Federal funds from National Cancer Institute (NCI), National Institutes of Health (NIH), under the NCI Chemical Biology Consortium Contract HHSN261200800001E (H.F.; K.Y.), NIH grant RO1 CA186918 (K.Y.), U01 CA168449 (H.F.), SBTF Foundation (K.Y), National Natural Science Foundation of China (No.81672781). The authors are thankful to Dr. Suzanne J. Baker for the PTEN/P53 loxP/loxP mice and to the Winship Cancer Institute of Emory University Cancer Center Support Grant P30-CA138292.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005 Mar 10;352(10):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Grzmil M, Hemmings BA. Deregulated signalling networks in human brain tumours. Biochimica et biophysica acta. 2010 Mar;1804(3):476–83. doi: 10.1016/j.bbapap.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Khatib ZA, Matsushime H, Valentine M, Shapiro DN, Sherr CJ, Look AT. Coamplification of the CDK4 gene with MDM2 and GLI in human sarcomas. Cancer research. 1993 Nov 15;53(22):5535–41. [PubMed] [Google Scholar]
- 5.Reifenberger G, Ichimura K, Reifenberger J, Elkahloun AG, Meltzer PS, Collins VP. Refined mapping of 12q13–q15 amplicons in human malignant gliomas suggests CDK4/SAS and MDM2 as independent amplification targets. Cancer research. 1996 Nov 15;56(22):5141–5. [PubMed] [Google Scholar]
- 6.Wikman H, Nymark P, Vayrynen A, Jarmalaite S, Kallioniemi A, Salmenkivi K, et al. CDK4 is a probable target gene in a novel amplicon at 12q13.3-q14.1 in lung cancer. Genes, chromosomes & cancer. 2005 Feb;42(2):193–9. doi: 10.1002/gcc.20122. [DOI] [PubMed] [Google Scholar]
- 7.Muthusamy V, Hobbs C, Nogueira C, Cordon-Cardo C, McKee PH, Chin L, et al. Amplification of CDK4 and MDM2 in malignant melanoma. Genes, chromosomes & cancer. 2006 May;45(5):447–54. doi: 10.1002/gcc.20310. [DOI] [PubMed] [Google Scholar]
- 8.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes & development. 2004 Nov 15;18(22):2699–711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 9.Ye K, Snyder SH. PIKE GTPase: a novel mediator of phosphoinositide signaling. Journal of cell science. 2004 Jan 15;117(Pt 2):155–61. doi: 10.1242/jcs.00924. [DOI] [PubMed] [Google Scholar]
- 10.Ahn JY, Hu Y, Kroll TG, Allard P, Ye K. PIKE-A is amplified in human cancers and prevents apoptosis by up-regulating Akt. Proceedings of the National Academy of Sciences of the United States of America. 2004 May 4;101(18):6993–8. doi: 10.1073/pnas.0400921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn JY, Rong R, Kroll TG, Van Meir EG, Snyder SH, Ye K. PIKE (phosphatidylinositol 3-kinase enhancer)-A GTPase stimulates Akt activity and mediates cellular invasion. The Journal of biological chemistry. 2004 Apr 16;279(16):16441–51. doi: 10.1074/jbc.M312175200. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Hu Y, Hao C, Rempel SA, Ye K. PIKE-A is a proto-oncogene promoting cell growth, transformation and invasion. Oncogene. 2007 Jul 26;26(34):4918–27. doi: 10.1038/sj.onc.1210290. [DOI] [PubMed] [Google Scholar]
- 13.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PloS one. 2010 Feb 12;5(2):e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, Huang W, Jiang X, Pennicooke B, Park PJ, Johnson MD. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proceedings of the National Academy of Sciences of the United States of America. 2010 Feb 02;107(5):2183–8. doi: 10.1073/pnas.0909896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007 Dec 13;450(7172):1001–9. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov AA, Khuri FR, Fu H. Targeting protein-protein interactions as an anticancer strategy. Trends in pharmacological sciences. 2013 Jul;34(7):393–400. doi: 10.1016/j.tips.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi Q, He K, Liu X, Pham C, Meyerkord C, Fu H, et al. Disrupting the PIKE-A/Akt interaction inhibits glioblastoma cell survival, migration, invasion and colony formation. Oncogene. 2013 Feb 21;32(8):1030–40. doi: 10.1038/onc.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow LM, Endersby R, Zhu X, Rankin S, Qu C, Zhang J, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer cell. 2011 Mar 8;19(3):305–16. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marumoto T, Tashiro A, Friedmann-Morvinski D, Scadeng M, Soda Y, Gage FH, et al. Development of a novel mouse glioma model using lentiviral vectors. Nature medicine. 2009 Jan;15(1):110–6. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye K, Hurt KJ, Wu FY, Fang M, Luo HR, Hong JJ, et al. Pike. A nuclear gtpase that enhances PI3kinase activity and is regulated by protein 4.1N. Cell. 2000 Dec 08;103(6):919–30. doi: 10.1016/s0092-8674(00)00195-1. [DOI] [PubMed] [Google Scholar]
- 21.Uras N, Oguz SS, Zergeroglu S, Akdag A, Polat B, Dizdar EA, et al. CD31 and Factor VIII in angiogenesis of normal and pre-eclamptic human placentas. Journal of obstetrics and gynaecology : the journal of the Institute of Obstetrics and Gynaecology. 2012 Aug;32(6):533–6. doi: 10.3109/01443615.2012.677875. [DOI] [PubMed] [Google Scholar]
- 22.Proescholdt MA, Merrill MJ, Stoerr EM, Lohmeier A, Pohl F, Brawanski A. Function of carbonic anhydrase IX in glioblastoma multiforme. Neuro-oncology. 2012 Nov;14(11):1357–66. doi: 10.1093/neuonc/nos216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang FF, Miller DC, Pisharody S, Koslow M, Newcomb EW. High frequency of p53 protein accumulation without p53 gene mutation in human juvenile pilocytic, low grade and anaplastic astrocytomas. Oncogene. 1994 Mar;9(3):949–54. [PubMed] [Google Scholar]
- 24.Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009 Feb 24;106(8):2712–6. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Network TC. Corrigendum: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2013 Feb 06;494(7438):506. doi: 10.1038/nature11903. [DOI] [PubMed] [Google Scholar]
- 26.Ma Z, Liu X, Li F, Wang Y, Xu Y, Zhang M, et al. Perfluorooctanoic acid induces human Ishikawa endometrial cancer cell migration and invasion through activation of ERK/mTOR signaling. Oncotarget. 2016 Aug 29; doi: 10.18632/oncotarget.11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathak P, Kumar A, Jha P, Purkait S, Faruq M, Suri A, et al. Genetic alterations related to BRAF-FGFR genes and dysregulated MAPK/ERK/mTOR signaling in adult pilocytic astrocytoma. Brain pathology. 2016 Sep 08; doi: 10.1111/bpa.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muranen T, Selfors LM, Hwang J, Gallegos LL, Coloff JL, Thoreen CC, et al. ERK and p38 MAPK Activities Determine Sensitivity to PI3K/mTOR Inhibition via Regulation of MYC and YAP. Cancer research. 2016 Dec 15;76(24):7168–80. doi: 10.1158/0008-5472.CAN-16-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng C, He K, Zhang C, Su S, Li B, Li Y, et al. JNK contributes to the tumorigenic potential of human cholangiocarcinoma cells through the mTOR pathway regulated GRP78 induction. PloS one. 2014;9(2):e90388. doi: 10.1371/journal.pone.0090388. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Liu R, Tian B, Gearing M, Hunter S, Ye K, Mao Z. Cdk5-mediated regulation of the PIKE-A-Akt pathway and glioblastoma cell invasion. Proceedings of the National Academy of Sciences of the United States of America. 2008 May 27;105(21):7570–5. doi: 10.1073/pnas.0712306105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He K, Qi Q, Chan CB, Xiao G, Liu X, Tucker-Burden C, et al. Blockade of glioma proliferation through allosteric inhibition of JAK2. Science signaling. 2013 Jul 09;6(283):ra55. doi: 10.1126/scisignal.2003900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Qi Q, Chan CB, Zhou W, Chen J, Luo HR, et al. Fyn-phosphorylated PIKE-A binds and inhibits AMPK signaling, blocking its tumor suppressive activity. Cell death and differentiation. 2016 Jan;23(1):52–63. doi: 10.1038/cdd.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.