Abstract
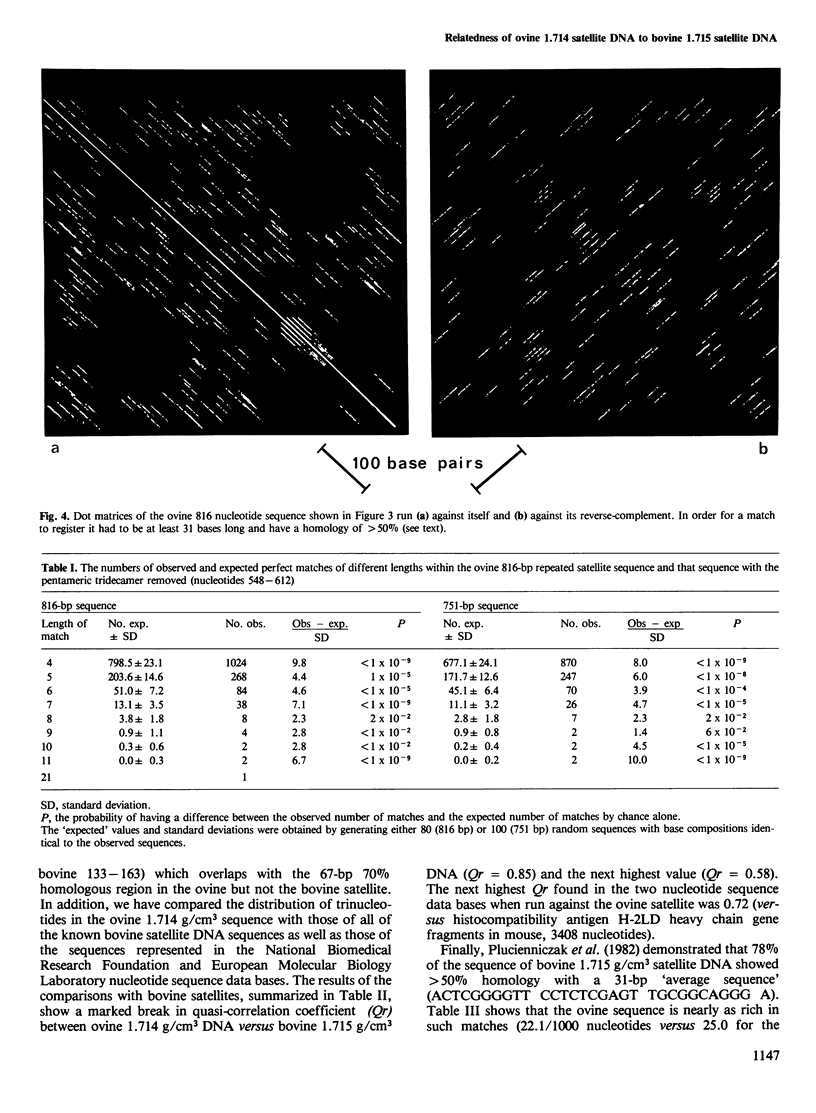
The nucleotide sequence of the principal component of ovine 1.714 g/cm3 satellite DNA was determined from a monomeric fragment inserted at the BamHI site of pBR322 and cloned in Escherichia coli strain RR1. The 816-bp tandemly repeated sequence contains a number of small repeated sequences dispersed within it, one group of which forms a pentameric tandem repeat of a 13-bp segment (positions 548-612). A 20-bp region (60-79) shows an 85% homology with the reverse-complement of the sequence from 455 through 474. There are two regions of 67 bp (75-141) and 59 bp (755-813) which show greater than 70% homology with regions of bovine 1.715 g/cm3 satellite DNA (1402 bp; positions 1218-1284 and 1079-1137, respectively) while a 31-bp region (ovine 62-92, bovine 133-163) shows 80% homology. Quasi-correlation coefficients (Qr) were determined using the triplet numbers of the sheep satellite versus all sequences in the National Biomedical Research Foundation and EMBL nucleotide sequence data bases. Qr equals 0.85 for ovine 1.714 g/cm3 satellite versus bovine 1.715 g/cm3 satellite. The next highest Qr for a bovine satellite segment was 0.58. Thus, the ovine 1.714 g/cm3 and bovine 1.715 g/cm3 satellite appear demonstrably related. Taking into account that sheep and cattle diverged 18-20 million years ago, this suggests that the material may be functional and that its function is related to its sequence.
Full text
PDF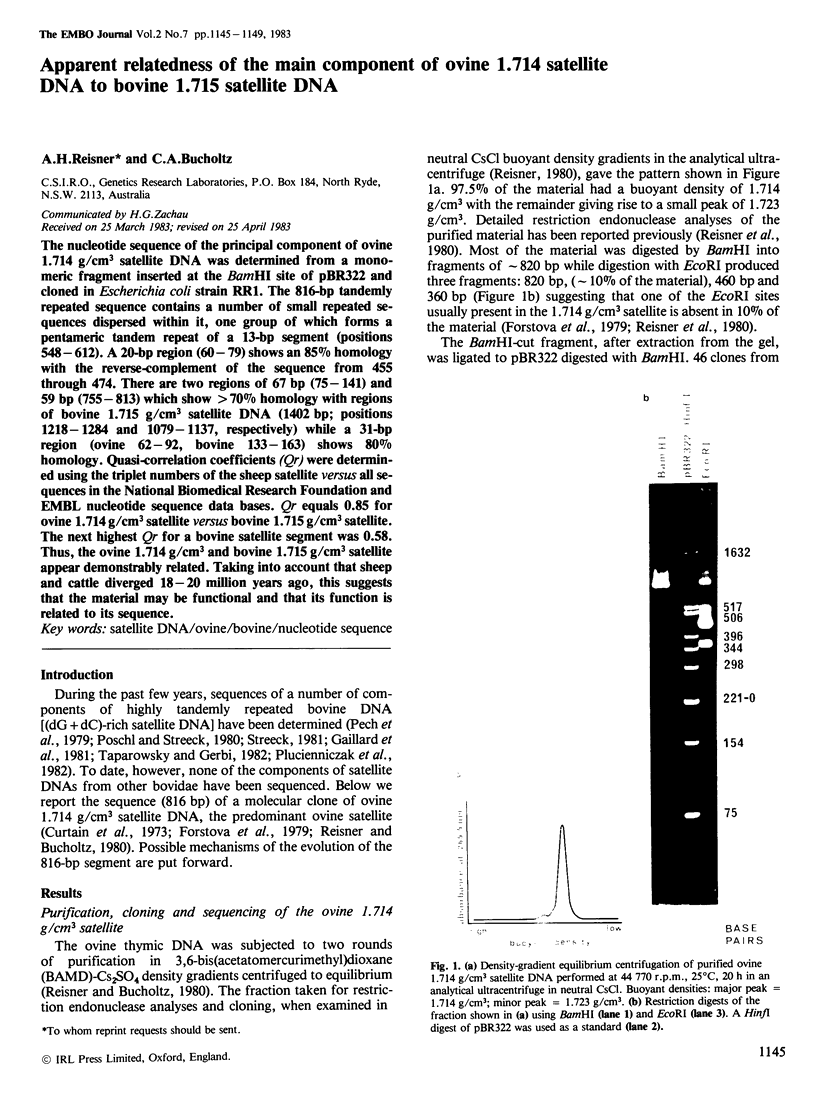



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Both G. W., Sleigh M. J. Complete nucleotide sequence of the haemagglutinin gene from a human influenza virus of the Hong Kong subtype. Nucleic Acids Res. 1980 Jun 25;8(12):2561–2575. doi: 10.1093/nar/8.12.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschi S., Wells J. R. Vertebrate histone gene transcription occurs from both DNA strands. Nucleic Acids Res. 1981 Apr 10;9(7):1591–1597. doi: 10.1093/nar/9.7.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünemann H., Dattagupta N. On the binding and specificity of 3,6-bis-(acetatomercurimethyl)-dioxane to DNAs of different base composition. Biochim Biophys Acta. 1973 Dec 21;331(3):341–348. doi: 10.1016/0005-2787(73)90020-8. [DOI] [PubMed] [Google Scholar]
- Cortadas J., Macaya G., Bernardi G. An analysis of the bovine genome by density gradient centrifugation: fractionation in Cs2SO4/3,6-bis(acetatomercurimethyl)dioxane density gradient. Eur J Biochem. 1977 Jun 1;76(1):13–19. doi: 10.1111/j.1432-1033.1977.tb11565.x. [DOI] [PubMed] [Google Scholar]
- Curtain C. C., Pascoe G., Hayman R. Satellite DNA in the sheep and goat. Biochem Genet. 1973 Nov;10(3):253–262. doi: 10.1007/BF00485703. [DOI] [PubMed] [Google Scholar]
- Forstová J., Votavová H., Guttmann T., Pivec L., Doskocil J. A comparative study of G+C-rich satellite components of sheep and goat DNA. Nucleic Acids Res. 1979 Jan;6(1):57–70. doi: 10.1093/nar/6.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard C., Doly J., Cortadas J., Bernardi G. The primary structure of bovine satellite 1.715. Nucleic Acids Res. 1981 Nov 25;9(22):6069–6082. doi: 10.1093/nar/9.22.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A. J., McIntyre G. A. The diagram, a method for comparing sequences. Its use with amino acid and nucleotide sequences. Eur J Biochem. 1970 Sep;16(1):1–11. doi: 10.1111/j.1432-1033.1970.tb01046.x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pech M., Streeck R. E., Zachau H. G. Patchwork structure of a bovine satellite DNA. Cell. 1979 Nov;18(3):883–893. doi: 10.1016/0092-8674(79)90140-5. [DOI] [PubMed] [Google Scholar]
- Pöschl E., Streeck R. E. Prototype sequence of bovine 1.720 satellite DNA. J Mol Biol. 1980 Oct 15;143(1):147–153. doi: 10.1016/0022-2836(80)90128-x. [DOI] [PubMed] [Google Scholar]
- Płucienniczak A., Skowroński J., Jaworski J. Nucleotide sequence of bovine 1.715 satellite DNA and its relation to other bovine satellite sequences. J Mol Biol. 1982 Jun 25;158(2):293–304. doi: 10.1016/0022-2836(82)90434-x. [DOI] [PubMed] [Google Scholar]
- Reisner A. H. Analyses of DNA by automated logging and processing of data from the analytical ultracentrifuge. Anal Biochem. 1980 Jun;105(1):24–31. doi: 10.1016/0003-2697(80)90417-0. [DOI] [PubMed] [Google Scholar]
- Reisner A. H., Bucholtz C. A. A physical study of the ovine genome. Biochim Biophys Acta. 1980 Aug 26;609(1):97–106. doi: 10.1016/0005-2787(80)90204-x. [DOI] [PubMed] [Google Scholar]
- Reisner A. H., Colless V. M., Bucholtz C. A. Physical analysis of a (dG + dC)-rich fraction of DNA obtained from the ovine genome. Aust J Biol Sci. 1980 Dec;33(6):623–632. doi: 10.1071/bi9800623. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Sleigh M. J., Both G. W., Brownlee G. G. A new method for the size estimation of the RNA genome segments of influenza virus. Nucleic Acids Res. 1979 Apr;6(4):1309–1321. doi: 10.1093/nar/6.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh M. J., Both G. W., Brownlee G. G. The influenza virus haemagglutinin gene: cloning and characterisation of a double-stranded DNA copy. Nucleic Acids Res. 1979 Oct 25;7(4):879–893. doi: 10.1093/nar/7.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck R. E. A multicopy insertion sequence in the bovine genome with structural homology to the long terminal repeats of retroviruses. Nature. 1982 Aug 19;298(5876):767–769. doi: 10.1038/298767a0. [DOI] [PubMed] [Google Scholar]
- Streeck R. E. Inserted sequences in bovine satellite DNA's. Science. 1981 Jul 24;213(4506):443–445. doi: 10.1126/science.6264600. [DOI] [PubMed] [Google Scholar]
- Taparowsky E. J., Gerbi S. A. Sequence analysis of bovine satellite I DNA (1.715 gm/cm3). Nucleic Acids Res. 1982 Feb 25;10(4):1271–1281. doi: 10.1093/nar/10.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]



