Abstract
To better understand the molecular mechanisms that underlie the tumorigenesis and progression of clear cell renal cell carcinoma (ccRCC), we studied the gene expression profiles of 29 ccRCC tumors obtained from patients with diverse clinical outcomes by using 21,632 cDNA microarrays. We identified gene expression alterations that were both common to most of the ccRCC studied and unique to clinical subsets. There was a significant distinction in gene expression profile between patients with a relatively nonaggressive form of the disease [100% survival after 5 years with the majority (15/17 or 88%) having no clinical evidence of metastasis] versus patients with a relatively aggressive form of the disease (average survival time 25.4 months with a 0% 5-year survival rate). Approximately 40 genes most accurately make this distinction, some of which have previously been implicated in tumorigenesis and metastasis. To test the robustness and potential clinical usefulness of this molecular distinction, we simulated its use as a prognostic tool in the clinical setting. In 96% of the ccRCC cases tested, the prediction was compatible with the clinical outcome, exceeding the accuracy of prediction by staging. These results suggest that two molecularly distinct forms of ccRCC exist and that the integration of expression profile data with clinical parameters could serve to enhance the diagnosis and prognosis of ccRCC. Moreover, the identified genes provide insight into the molecular mechanisms of aggressive ccRCC and suggest intervention strategies.
Renal cell carcinoma (RCC) is the most common malignancy arising in the adult kidney, representing 2% of all malignancies and 2% of cancer-related deaths. It is a clinicopathologically heterogeneous disease, subdivided into clear, papillary, granular, spindle, and mixed cell variants based on cytoplasmic features (1). The prognosis of RCC is based on tumor staging and histological grading (2), and patients with metastatic RCC (≈30% of all RCC cases) have a life expectancy averaging around 12 months (3, 4). Of the remaining patients with initially nonmetastatic disease, ≈30% relapse after surgery and usually succumb to the disease (5, 6). To date, there is little understanding of the underlying molecular mechanisms that may cause this variety in prognoses.
There is now strong evidence that global gene expression profiling can reveal subtypes of cancer based on underlying heterogeneity in transformation mechanisms, differentiation states, or cell types (7–12). For example, a recent study showed that two types of hereditary breast cancer (BRCA1 and -2) had distinguishable gene expression profiles (12), suggesting that the observed differences in gene expression resulted from differences in transforming genetic mutations. In another study, gene expression profiles of hepatocellular carcinoma differed according to whether patients were hepatitis B virus-positive or hepatitis C virus-positive (7), again suggesting that the mechanism of tumorigenesis affected the gene expression profile.
The clinical use of gene expression profiles could result in more accurate and objective diagnoses of cancers as well as prognoses of disease or response to treatment. Tumor samples with long-term followup information are required to assess the prognostic significance of certain gene expression profiles, such as in a recent study of diffuse large B-cell lymphoma tumors that showed significantly variant survival probabilities based on distinct gene expression profiles (9). Similar studies for other malignancies are anticipated but remain challenging because they require both proper storage of operated tissues and long-term followup information on patients.
The availability of 29 clear cell RCC (ccRCC) frozen tissue specimens with up to 12 years of followup information provided a tremendous opportunity to further our understanding of ccRCC classification and the role that tumor subclasses may play in the heterogeneity of ccRCC progression and aggressiveness. We obtained gene expression profiles of 29 ccRCC samples and studied the data to: (i) identify common alterations in ccRCC gene expression; (ii) identify expression signatures of ccRCC specific to particular clinical subsets of tumors; and (iii) assess the clinical usefulness of certain gene expression profiles by simulating clinical usage.
Materials and Methods
Patient Information and Tumor Samples.
Tissue samples were obtained from 29 patients with ccRCC after radical nephrectomy at the University Hospital, School of Medicine, Tokushima University. Informed consent was obtained from patients to use their operated specimens and clinicopathological data for research purposes. The samples were anonymized before the study. Part of each tumor sample was frozen in liquid nitrogen immediately after the operation and stored at −80°C. Total RNA was isolated from the frozen tissue by using ISOGEN solution (Nippon Gene, Toyama, Japan), and poly(A)+ RNA was isolated from total RNA by using the Oligotex mRNA Mini Kit (Qiagen, Chatsworth, CA). The remaining parts of tumors were fixed with 10% buffered formalin, and the paraffin sections were stained with hematoxylin/eosin. The World Health Organization International Histological Classification of Tumors was used for histological evaluation of the specimens (1). Union Internationale Contre le Cancer TNM classification and stage groupings were used (13). Clinicopathological data are summarized in Table 1. Patients have been followed up for 3.2 to 137.2 months (median 83.7 months).
Table 1.
Patient clinical and followup data and corresponding prognosis classifications
| Patient | Grade | Stage | Outcome | Dur. | Outcome group | Prognosis group
|
|
|---|---|---|---|---|---|---|---|
| Staging | Gene expression | ||||||
| 46 | G1 | S1 | NED | 62.6 | L | L | L |
| 42 | G1 | S1 | NED | 77.3 | L | L | L |
| 41 | G1 | S1 | NED | 80.3 | L | L | L |
| 30 | G2 | S3 | NED | 87.1 | L | H* | H* |
| 7 | G1 | S1 | NED | 92.1 | L | L | L |
| 26 | G1 | S1 | NED | 96 | L | L | L |
| 24 | G1 | S1 | NED | 97.3 | L | L | L |
| 15 | G1 | S1 | OCD | 100.4 | L | L | L |
| 32 | G1 | S2 | OCD | 110.4 | L | L | L |
| 1 | G1 | S1 | NED | 111.6 | L | L | L |
| 21 | G1 | S1 | NED | 114.6 | L | L | L |
| 20 | G1 | S1 | NED | 115.8 | L | L | L |
| 35 | G1 | S3 | NED | 120.5 | L | H* | L |
| 9 | G1 | S3 | NED | 120.9 | L | H* | L |
| 3 | G1 | S1 | NED | 137.2 | L | L | L |
| 29 | G3 | S3 | AWC | 89.4 | L | H* | L |
| 54 | G1 | S4 | AWC | 105.6 | L | H* | L |
| 13 | G3 | S4 | Death | 3.2 | H | H | H |
| 48 | G2 | S4 | Death | 4.9 | H | H | H |
| 11 | G3 | S3 | Death | 18.8 | H | H | H |
| 60 | G3 | S4 | Death | 20.8 | H | H | H |
| 31 | G3 | S3 | Death | 22.6 | H | H | H |
| 53 | G3 | S4 | Death | 26.2 | H | H | H |
| 5 | G2 | S4 | Death | 31.7 | H | H | H |
| 12 | G2 | S4 | Death | 33.8 | H | H | H |
| 55 | G2 | S2 | Death | 55.8 | H | L* | H |
| 56 | G3 | S4 | AWC | 14.8 | U | H | L |
| 58 | G3 | S4 | AWC | 16.6 | U | H | H |
| 59 | G2 | S3 | NED | 41.1 | U | H | H |
Patient clinical data and corresponding prognostic classifications. Grade and stage information (columns 2 and 3) corresponds to the primary tumor. Outcomes (column 4) are: no evidence of disease (NED), alive with cancer (AWC), other cause of death (OCD), and cancer death. Duration (column 5) is months between nephrectomy and latest outcome assessment. Outcome group (column 6) is the risk group, based on actual patient outcome that was used for predictive gene set generation (L, low risk; H, high risk; U, unknown). Pathology prognosis group (column 7) is based on staging (L, stage I/II; H, stage III/IV). Gene expression prognosis group (column 8) is based on a gene expression prognosis test based on the selected genes.
, deviation in outcome from the predicted risk group.
Microarray Experiments.
cDNA microarray production and hybridizations were performed as described with slight modification (14, 15) (published as supplemental data on the PNAS web site, www.pnas.org).
Data Analysis.
Images were analyzed by using the software genepix pro 3.0 (Axon Instruments, Foster City, CA). Spots showing no signal or obvious defects were excluded from the analysis. The local background was subtracted from the remaining spots, and the ratios of net fluorescence from the Cy5-specific channel to the net fluorescence from the Cy3-specific channel were calculated for each spot, representing tumor mRNA expression relative to the corresponding normal kidney tissue. Ratios were log-transformed (base 2) and normalized so that the average log-transformed ratio equaled zero. Genes with good data present in 75% of the experiments and with expression ratios that varied at least 2-fold in at least two experiments were selected for the clustering analysis (3,184 genes). The gene ratios were “median polished” before hierarchical clustering by using cluster and visualized by using treeview (M. B. Eisen, http://rana.lbl.gov). The correlation distance calculated by cluster is equal to 1 − r, where r, the Pearson correlation coefficient (16), equals 1 for perfectly correlated series and −1 for perfectly opposite series.
We developed the program clusterfinder to identify and rank subclusters of genes that distinguish between two defined sample groups. Briefly, the red-to-green ratios of all of the genes within each subcluster, or node of a dendrogram, are averaged for each patient sample, and the averages are placed into two groups based on user-defined criteria. For each group of expression value averages, the mean (μ) and standard deviation (σ) is calculated. A discrimination score (DS) for each subcluster is calculated as DS = μ1 − μ2 /(σ1 + σ2), where the subscripts refer to the sample group. A large DS indicates that the genes in that cluster have great variation between the two groups but low variation within each group (8). A DS of 1.0 would approximate a significantly discriminating cluster (α = 0.05).
To calculate the ability of individual genes to distinguish between two sample groups by using the permutation t test (12), patients were randomly permuted into two groups 10,000 times, and for each gene, a t-statistic was calculated. The distribution of t-statistics defined a 99.9% significance threshold (α = 0.001). If a gene's t-statistic for the user-defined patient grouping passed the 99.9% significance threshold, the gene was considered to significantly distinguish the two groups.
Survival analyses based on stage, histological grade, and gene expression profile were performed by the Kaplan–Meier method and tested by the log-rank test. Correlation of histological grade or stage with the gene expression profile was analyzed as the Spearman correlation coefficient by the exact test with the sas/stat analysis package (Ver. 8.0, SAS Institute, Cary, NC). Three patients were excluded from the statistic analyses because of less than 5 years of followup.
Results
We compared the expression profiles of 29 ccRCC samples and patient-matched normal tissue samples by hybridization to 21,632 cDNA microarrays. We analyzed the data in two ways. First, we compared the gene expression of each tumor sample with its patient-matched normal tissue to identify gene expression alterations that occur in most ccRCC. Second, because all of the experiments shared a “common” normal tissue reference, the experiments could be compared with each other to identify gene expression patterns that correlated with differences in observed clinical features of the tumors.
Shared Gene Expression Alterations in ccRCC Tumors.
We first sought to identify gene expression alterations that were shared by all of the ccRCC tumors we studied. We selected genes that were at least 3-fold up- or down-regulated in at least 75% of the tumors. The 32 up- and 77 down-regulated genes that met the above criteria are summarized in Tables 2 and 3 (published as supplemental data on the PNAS web site, www.pnas.org). Both previously known and unknown genes were found to be significantly up- or down-regulated in many or all of the ccRCC. See Discussion for commentary on some of the individual genes.
Heterogeneity in ccRCC Gene Expression and Correlation with Clinical Phenotypes.
Having identified shared gene expression alterations in the ccRCC samples, we next studied differences in the gene expression profiles that could distinguish, and possibly give biological insight into, the clinical heterogeneity of the tumors.
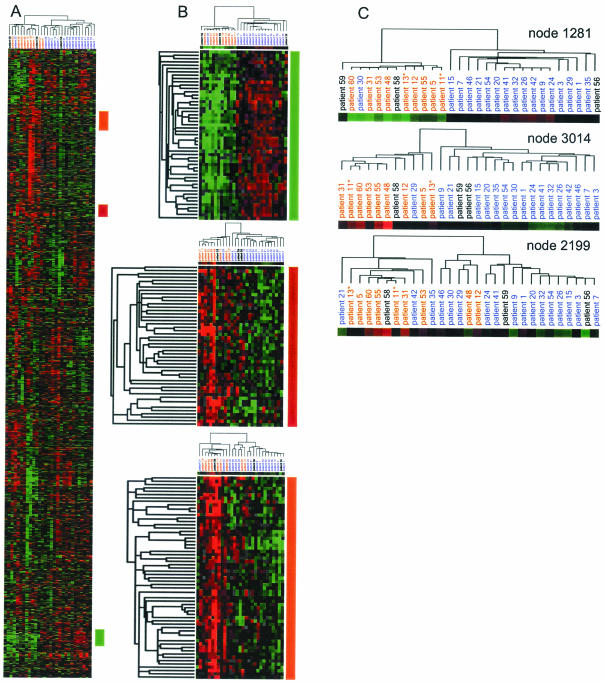
We used hierarchical clustering (16) to look at the variation in gene expression among the tumors. The clustering algorithm groups both genes and tumors by similarity in expression pattern. The pattern of groupings depends on the set of genes and tumors used. For example, a set of genes with highly correlated expression patterns may dominate and define a cluster pattern, but clustering based on another less correlated set of genes may result in a very different pattern of clustering. Clustering based on the total gene expression profile, approximated by a selected 3,184 gene set, revealed great variation in up- and down-regulated genes among the tumors (Fig. 1A). The tumors clustered into two main groups, as shown by the dendrogram at the top of the cluster pattern. The grouping largely correlated with cause-specific survival at 5 years, with only two tumors that did not cluster by that parameter.
Figure 1.
Clustering of subsets of genes in 29 ccRCC tumors. Rows represent individual cDNAs, and columns represent individual patient tumor samples. The color of each square represents the median-polished normalized ratio of gene expression in a tumor relative to patient-matched normal kidney tissue. Red indicates gene expression above the median; green, below the median; black, equal to the median; and gray, inadequate or missing data. The color saturation indicates the degree of divergence from the median. (A) Clustering of all 3,184 genes and 29 tumors. The colored bars on the right of the diagram indicate clusters with high discrimination scores. (B) Reclustering of genes and tumors by using three “predictive” clusters. The color coding of the patient list indicates cause-specific survival at 5 years: red indicates cancer death, blue indicates alive, and black indicates short followup. The color bars beneath the dendrograms represent the average expression values for the subsets of genes. (C) Expanded view of the patient dendrograms from B, showing the structure of similarity relationships between the gene expression profiles.
We next sought to identify particular subsets of genes that most strongly defined the division of patients by cause-specific survival at 5 years and to ascertain whether other gene sets could distinguish other clinical parameters. We used the program clusterfinder to identify and rank the subclusters of genes by their ability to differentiate two user-defined groups of tumors. When we applied the program to 10 random groupings of tumors, no gene clusters differentiated the groups above a DS of 1.0, our previously determined threshold for significance. Because tumor staging is often used to determine the prognosis of ccRCC (2), we tested whether any gene sets defined this parameter and could possibly have value as indicators of cancer progression. Surprisingly, no gene clusters significantly differentiated between stage I + II and stage III + IV tumors (data not shown).
When we applied clusterfinder to a tumor grouping based on cause-specific survival at 5 years, multiple clusters with a high DS were found. Cluster 687, containing 24 genes, and its parent, cluster 1281, containing 51 genes, had the highest DS (1.70), whereas cluster 3014, with 48 genes, and cluster 2199, with 61 genes, also had passing scores (1.46 and 1.011, respectively). The tumors were reclustered on the basis of these gene sets (Fig. 1B). Clustering based on the genes of cluster 1281 grouped the tumors by outcome, except for patient 30. This patient showed similar expression with the poor outcome group but has no evidence of disease after 5 years. The dendrogram for cluster 1281 shows the strict segregation of patients by outcome (Fig. 1C), reflected in a high correlation score for cluster 1281 (0.839). Clustering by the genes of clusters 3014 and 2199 resulted in one and five misgroupings by patient outcome, respectively, with lower correlation scores (0.516 and 0.692, respectively).
We also used a permutation t test to assess each gene's individual ability to distinguish between two groups of patients (12). Ninety-four percent of the genes from cluster 1281 significantly (α < 0.001) differentiated the patient groups, but only 19 and 7% of the genes from clusters 3014 and 2199, respectively, significantly differentiated the patient groups.
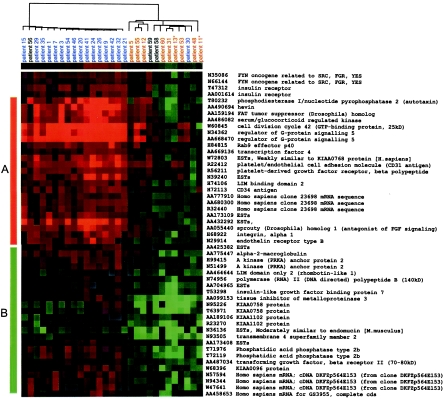
The tumors were reclustered on the basis of genes of node 1281 by using nonmedian centered expression values, so that the colors represented the actual gene expression level of the tumor relative to the normal (Fig. 2). The resulting cluster pattern was different from that by using median-centered values (Fig. 1 B and C), but the grouping by cause-specific survival remains distinct. This cluster is comprised of genes up-regulated in the tumors with the good outcome and genes down-regulated in tumors with the poor outcome.
Figure 2.
Clustering of the 51 genes of cluster 1281 by using nonmedian centered values. In this case, the color of each square corresponds to actual normalized gene expression level relative to normal kidney tissue, by using the same scale as in Fig. 1. (A) Genes mostly up-regulated in tumors with the good outcome. (B) Genes mostly down-regulated in tumors with the poor outcome.
Simulation of Clinical Diagnosis and Prognosis.
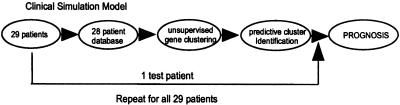
The groups of genes that differentiate the tumors by patient outcome may provide insight into classifications and molecular mechanisms of ccRCC and also may have value as potential clinical diagnostic and prognostic markers. As such, we simulated the clinical diagnostic and prognostic use of particular gene sets for each tumor sample (Fig. 3). A “test” tumor was removed from the group, a new set of predictive genes was generated from the remaining 28 tumors by using clusterfinder, and the test tumor was clustered with the other samples by using the predictive gene set. The test tumor was classified as high or low risk, depending on whether it clustered with the poor-outcome or good-outcome tumors, respectively. This process was repeated for each tumor. The prognostic classification of each tumor was considered “correct” if it corresponded to the actual outcome. The predictive gene set was slightly different at each iteration, but on average 95% of the genes were conserved.
Figure 3.
Clinical simulation model (see Results for description).
Table 1 presents the results of the simulation. The “outcome group” column indicates the grouping by using 5-year cause-specific survival as the cutoff, and the two “prognosis group” columns show the risk group classification by staging (stage I + II = low risk, stage III + IV = high risk) or by expression profiling. Prognostic classification by expression profiling had a better prediction than staging in five patients (patients 35, 9, 29, 54, and 55). Patient 29, who had a grade 3 tumor invading into the renal vein at operation (high risk by staging), had a low-risk gene expression profile and has survived the operation by 7.5 years. Patient 55, who had a stage II, grade 2 tumor (low risk by staging), had a high-risk gene expression profile and died of ccRCC 4.6 years after the operation. Patient 54, classified as low-risk by gene expression profiling, had bone metastasis at the initial diagnosis but is still alive with stable bone metastasis 8.8 years after surgery. One patient, who was misgrouped by both methods (patient 30), presented with stage III cancer and had a high-risk gene expression profile, but 7 years later has no evidence of disease.
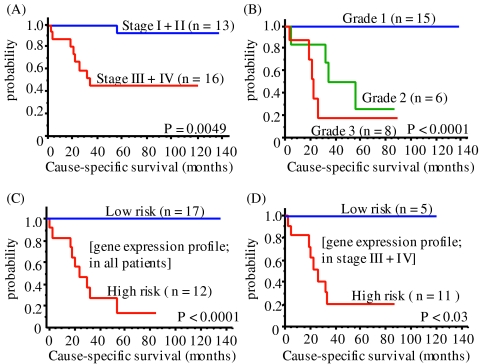
We used Kaplan–Meier survival analysis to further compare the significance of the prognostic classifications made by stage, grade, and gene expression profile (Fig. 4). Classification by grade (Fig. 4B, P < 0.0001) was better than that by stage (Fig. 4A, P = 0.0049). Classification by expression profile (Fig. 4C) was similar in significance to that by grade. In addition, histological grade and gene expression classification were highly correlated (correlation coefficient = 0.7703, P < 0.0001), suggesting that histological grades are affected by gene expression profiles. Within the high-risk group defined by staging (stage III + IV), gene expression profiling significantly distinguished two groups of patients with different outcomes (Fig. 4D). Multivariate analysis of these parameters was also attempted but prohibited by the sample size of our cohort. A larger cohort of patients would enable further investigation of the correlation of expression profile with other covariates, such as grade, gender and age, through multivariate analysis.
Figure 4.
Cause-specific survival curve with staging (A), histological grading (B), gene expression profiling in all patients (C), and patients with stage III/IV (D), by the Kaplan–Meier method.
Discussion
Common Alterations in ccRCC Gene Expression.
The above analysis revealed that there are both similarities and differences in the gene expression profiles of ccRCCs. Genes with altered expression in most ccRCCs may serve as molecular markers for the diseased state or may play a causal role in renal transformation, therefore serving as candidate therapeutic targets. For example, ceruloplasmin, a protein involved in iron and copper homeostasis, had the highest increase in expression in the ccRCC relative to corresponding normal kidney tissues (Table 2, www.pnas.org). Interestingly, only a handful of reports have associated ceruloplasmin with RCC. One reported its secretion by RCC (17) and another its elevation in the serum of RCC patients (18). Another copper-related protein, lysyl oxidase (an extracellular enzyme involved in the connective tissue maturation pathway), was also up-regulated in ccRCC. It has been reported to be highly expressed in invasive breast cancer cell lines (19) but has never been studied in RCC. In 96% of the tumors, high expression of vascular endothelial growth factor (VEGF), a well-known protein highly expressed in ccRCC (20, 21) and found elevated in the serum of ccRCC patients (22–24), was found.
In addition, many identified down-regulated genes (Table 3, www.pnas.org) may be involved in the tumorigenesis of ccRCC. Most strikingly, kininogen was found to be more than 27-fold down-regulated in the tumors. Kininogen, a molecule involved in the activation of the contact system, recently was shown to be an inhibitor of angiogenesis (25). Its down-regulation may concur with up-regulation of VEGF, resulting in hypervascularization, a characteristic of ccRCC. We also found the metallothionein (MT) family to be coordinately down-regulated. Differential expression of this class of genes has been reported in many cancers (26), and several subtypes (MT-1A, -1G, and -1H) were reported down-regulated in RCC (27, 28). Our study supported these reports and additionally found MT-1L and -1E to be down-regulated.
Expression Signatures of RCC Specific to Particular Clinical Subsets of Tumors.
The diversity in the gene expression profiles largely defined two patient groups that were distinguishable by cause-specific survival at 5 years. These findings may reflect the existence of distinct subclasses of ccRCC that differ in clinical behavior. We showed that, whereas no statistically significant groups of genes correlated with random groupings of tumors or with staging of tumors, multiple clusters of dozens of genes with high statistical significance correlated with cause-specific survival at 5 years. This result showed that only certain groupings of patients have distinguishing gene expression signatures, likely only the groupings that have an underlying biological basis. Therefore, the two groups of ccRCC identified by gene expression profiling may represent two classes of ccRCC, an aggressive and a nonaggressive class, that have distinct molecular bases and distinct mechanisms of progression.
Many of the 51 cDNAs (40 genes) in cluster 1281 that most effectively discriminate between patients with the good and poor outcome gave insight into the biology of the two groups of ccRCC. For example, sprouty, the mammalian homolog of the Drosophila melanogaster angiogenesis inhibitor, was up-regulated exclusively in the good outcome group, suggesting that failure to properly inhibit angiogenesis may contribute to the aggressive form of ccRCC. The regulator of G-protein signaling 5 was exclusively up-regulated in the good outcome tumors and may be important for the proper control of cancer progression. Transforming growth factor (TGF)β, TGFβ receptor II (TGFβRII) and its down-stream effector, tissue inhibitor of metalloproteinase 3 (TIMP3), were exclusively down-regulated in the poor outcome group. Loss of the TGFβII signaling pathway previously was shown to be important for the development of aggressive cancers (29), and loss of TIMP3 expression by promoter methylation was shown to increase tumorigenicity because of unregulated matrix metalloproteinases (30). The identification of this pathway as down-regulated in aggressive ccRCC suggests numerous targets for intervention to supplement the still low response rate of current adjuvant therapies, such as IFN-α and IL-2 injection. For example, a recent study demonstrated the inhibition of invasion in melanoma cell lines by overexpressing TIMP3 by adenovirus-mediated gene delivery (31).
Clinical Usefulness of the Gene Expression Profiles.
The long-term followup information on each tumor allowed us to test the usefulness of gene expression profiling for prognosis. Gene expression profiling very accurately grouped the patients by risk group, demonstrated by a clinical simulation test (Fig. 3 and Table 1) and by Kaplan–Meier survival analysis (Fig. 4 C and D). Gene expression profiling more accurately predicted risk group than staging alone (Fig. 4A) and identified two risk groups within the stage III + IV tumors. This result is encouraging for the eventual development of more accurate clinical diagnostic and prognostic methods for ccRCC patients.
In conclusion, we have identified gene expression alterations that are both common to most ccRCC cases and unique to clinical subsets. The identified genes may give insight into ccRCC tumorigenesis and progression and may also suggest intervention strategies. The segregation of patients by 5-year survival based on expression profiles and histological grading may indicate that there are at least two subclasses of ccRCC that have distinct behaviors and underlying mechanisms of progression. If so, the treatment of ccRCC patients should be based in part on the knowledge of this subclassification. For example, physicians may be advised to provide careful observation and adjuvant therapies to patients presenting with low-stage but high-risk expression profile cancers. On the contrary, for patients with the low-risk expression profile and high-stage cancer, we may be able to avoid aggressive treatments under careful observation.
Supplementary Material
Acknowledgments
We thank Jeremy Miller, John Marcus, Tom Wilson, Weng Onn Lui, and Xiang Guo for technical assistance, Eric Kort for statistical help, and George Vande Woude for support and constructive discussion. This research was funded by the Van Andel Research Institute.
Abbreviations
- RCC
renal cell carcinoma
- ccRCC
clear cell RCC
- DS
discrimination score
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mostfi F K, Davis C J. WHO International Histological Classification of Tumours. Berlin: Springer; 1998. [Google Scholar]
- 2.Tsui K H, Shvarts O, Smith R B, Figlin R A, deKernion J B, Belldegrun A. J Urol. 2000;163:1090–1095. doi: 10.1016/s0022-5347(05)67699-9. [DOI] [PubMed] [Google Scholar]
- 3.Minasian L M, Motzer R J, Gluck L, Mazumdar M, Vlamis V, Krown S E. J Clin Oncol. 1993;11:1368–1375. doi: 10.1200/JCO.1993.11.7.1368. [DOI] [PubMed] [Google Scholar]
- 4.Hermanek P, Schrott K M. J Urol. 1990;144:238–241. doi: 10.1016/s0022-5347(17)39420-x. [DOI] [PubMed] [Google Scholar]
- 5.Ljungberg B, Alamdari F I, Rasmuson T, Roos G. BJU Int. 1999;84:405–411. doi: 10.1046/j.1464-410x.1999.00202.x. [DOI] [PubMed] [Google Scholar]
- 6.Levy D A, Slaton J W, Swanson D A, Dinney C P. J Urol. 1998;159:1163–1167. [PubMed] [Google Scholar]
- 7.Okabe H, Satoh S, Kato T, Kitahara O, Yanagawa R, Yamaoka Y, Tsunoda T, Furukawa Y, Nakamura Y. Cancer Res. 2001;61:2129–2137. [PubMed] [Google Scholar]
- 8.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 9.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 10.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature (London) 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 11.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Nature (London) 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 12.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi O P, et al. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 13.Sobin L H, Wittekind C, editors. International Union Against Cancer. TNM Classification of Malignant Tumours. 5th Ed. New York: Wiley; 1997. [Google Scholar]
- 14.Eisen M B, Brown P O. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 15.Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes J E, Snesrud E, Lee N, Quackenbush J. BioTechniques. 2000;29:548–550. doi: 10.2144/00293bi01. , 552–554, 556. [DOI] [PubMed] [Google Scholar]
- 16.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito K, Saito T, Draganac P S, Andrews R B, Lange R D, Etkin L D, Farkas W R. Biochem Med. 1985;33:45–52. doi: 10.1016/0006-2944(85)90125-5. [DOI] [PubMed] [Google Scholar]
- 18.Pejovic M, Djordjevic V, Ignjatovic I, Stamenic T, Stefanovic V. Int Urol Nephrol. 1997;29:427–432. doi: 10.1007/BF02551108. [DOI] [PubMed] [Google Scholar]
- 19.Kirschmann D A, Seftor E A, Nieva D R, Mariano E A, Hendrix M J. Breast Cancer Res Treat. 1999;55:127–136. doi: 10.1023/a:1006188129423. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi A, Sasaki H, Kim S J, Tobisu K, Kakizoe T, Tsukamoto T, Kumamoto Y, Sugimura T, Terada M. Cancer Res. 1994;54:4233–4237. [PubMed] [Google Scholar]
- 21.Thelen P, Hemmerlein B, Kugler A, Seiler T, Ozisik R, Kallerhoff M, Ringert R H. Anticancer Res. 1999;19:1563–1565. [PubMed] [Google Scholar]
- 22.Sato K, Tsuchiya N, Sasaki R, Shimoda N, Satoh S, Ogawa O, Kato T. Jpn J Cancer Res. 1999;90:874–879. doi: 10.1111/j.1349-7006.1999.tb00829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wechsel H W, Bichler K H, Feil G, Loeser W, Lahme S, Petri E. Anticancer Res. 1999;19:1537–1540. [PubMed] [Google Scholar]
- 24.Edgren M, Lennernas B, Larsson A, Nilsson S. Anticancer Res. 1999;19:869–873. [PubMed] [Google Scholar]
- 25.Zhang J C, Claffey K, Sakthivel R, Darzynkiewicz Z, Shaw D E, Leal J, Wang Y C, Lu F M, McCrae K R. FASEB J. 2000;14:2589–2600. doi: 10.1096/fj.99-1025com. [DOI] [PubMed] [Google Scholar]
- 26.Janssen A M, van Duijn W, Oostendorp-Van De Ruit M M, Kruidenier L, Bosman C B, Griffioen G, Lamers C B, van Krieken J H, van De Velde C J, Verspaget H W. J Pathol. 2000;192:293–300. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH712>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen A, Jing Z, Mahoney P S, Davis R, Sikka S C, Agrawal K C, Abdel-Mageed A B. Cancer Lett. 2000;160:133–140. doi: 10.1016/s0304-3835(00)00534-6. [DOI] [PubMed] [Google Scholar]
- 28.Izawa J I, Moussa M, Cherian M G, Doig G, Chin J L. Urology. 1998;52:767–772. doi: 10.1016/s0090-4295(98)00323-9. [DOI] [PubMed] [Google Scholar]
- 29.Engel J D, Kundu S D, Yang T, Lang S, Goodwin S, Janulis L, Cho J S, Chang J, Kim S J, Lee C. Urology. 1999;54:164–170. doi: 10.1016/s0090-4295(99)00093-x. [DOI] [PubMed] [Google Scholar]
- 30.Bachman K E, Herman J G, Corn P G, Merlo A, Costello J F, Cavenee W K, Baylin S B, Graff J R. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 31.Ahonen M, Baker A H, Kahari V M. Cancer Res. 1998;58:2310–2315. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.